Abstract
The dynamics of adaptation determines which mutations fix in a population, and hence how reproducible evolution will be. This is central to understanding the spectra of mutations recovered in evolution of antibiotic resistance1, the response of pathogens to immune selection2,3, and the dynamics of cancer progression4,5. In laboratory evolution experiments, demonstrably beneficial mutations are found repeatedly6–8, but are often accompanied by other mutations with no obvious benefit. Here we use whole-genome whole-population sequencing to examine the dynamics of genome sequence evolution at high temporal resolution in 40 replicate Saccharomyces cerevisiae populations growing in rich medium for 1,000 generations. We find pervasive genetic hitchhiking: multiple mutations arise and move synchronously through the population as mutational “cohorts.” Multiple clonal cohorts are often present simultaneously, competing with each other in the same population. Our results show that patterns of sequence evolution are driven by a balance between these chance effects of hitchhiking and interference, which increase stochastic variation in evolutionary outcomes, and the deterministic action of selection on individual mutations, which favors parallel evolutionary solutions in replicate populations.
Evolutionary adaptation is driven by the accumulation of beneficial mutations. The traditional view is that these dynamics are dominated by rare beneficial “driver” mutations that occasionally survive drift and increase in frequency until they fix (a “selective sweep”)9,10. This implicitly assumes that at most a single beneficial mutation is present in the population at once. Recent experiments, however, have shown that even for modestly sized populations of microbes and viruses, beneficial mutation rates are large enough11,12 that multiple driver mutations spread simultaneously, an effect known as “clonal interference.” This means that the fate of each mutation depends not only on its own effect on fitness, but also on the rest of the variation in the population: neutral or deleterious mutations can fix if they occur in very fit genetic backgrounds, and beneficial mutations occurring in unfit lineages cannot succeed13–17.
Recent work has uncovered important consequences of these clonal interference effects. For example, interference alters the rate of adaptation18,19, the fate of marked lineages20,21, and the distribution of fitness effects of fixed mutations12,16,17. The underlying basis of these effects at the genomic sequence level, however, has not been directly observed. What is the fate of those mutations that occur? How does the frequency of each mutation change over time? How do these sequence-level dynamics determine the rate and repeatability of adaptation? Recent studies have sequenced clones or whole-population samples from microbial evolution experiments6,22–25, but apart from studies in viral systems26–28, this work has been limited to individual clones or populations or to widely separated timepoints that lack the temporal resolution to address these questions.
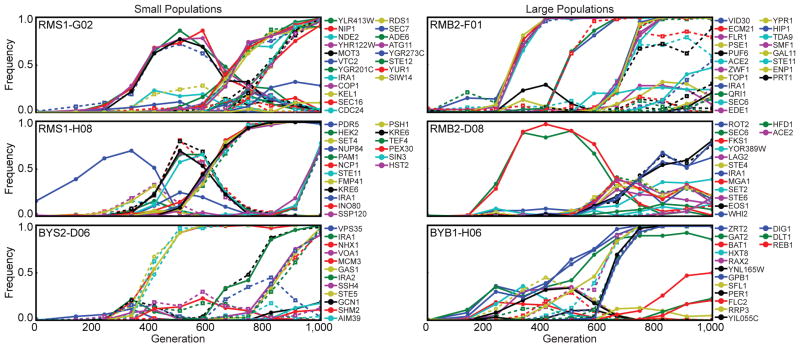
Here we describe the first direct and detailed view of the dynamics of genomic sequence evolution across many replicate microbial populations. In previous work21, we adapted ~600 replicate haploid yeast populations to growth in rich medium for 1,000 generations, half at “large” (106) and half at “small” (105) population sizes. Here we report the sequencing of whole-population samples from 40 of these populations (14 large and 26 small), chosen because we previously followed a single marker above a frequency of 0.121. Each population was sequenced to 100-fold depth at 12 timepoints (approximately every 80 generations) for a total of 480 sequenced timepoints. Distinguishing mutations from sequencing errors in this whole-population sequence data is challenging. The high temporal resolution of our data, however, permits the identification of mutations even at relatively low frequency by leveraging multiple timepoints. We developed two independent pipelines for this purpose, which rely on the fact that real mutations (but not sequencing/alignment errors) have frequencies that are correlated through time (Methods). This strategy allowed us to identify mutations that rose to a frequency of at least ~0.1 and to track these mutations through the rest of the timecourse. Across the 40 populations, we identified a total of 1,020 mutations, 253 of which fix; we annotated each to a gene or intergenic region and classified coding mutations as synonymous or nonsynonymous (Supplementary Table 1). Fig. 1 shows six representative populations; the remaining populations exhibit similar patterns (Supplementary Fig. 1).
Figure 1. The fates of individual spontaneously arising mutations.
We show the frequency of all identified mutations through 1,000 generations in 6 of the 40 sequenced populations. Nonsynonymous mutations are solid lines with solid circles, while synonymous and intergenic mutations are dotted lines with open circles and squares respectively. Populations in the left and right columns were evolved at small (105) and large (106) population sizes, respectively. We observe qualitatively similar patterns in the other populations (Supplementary Fig. 1).
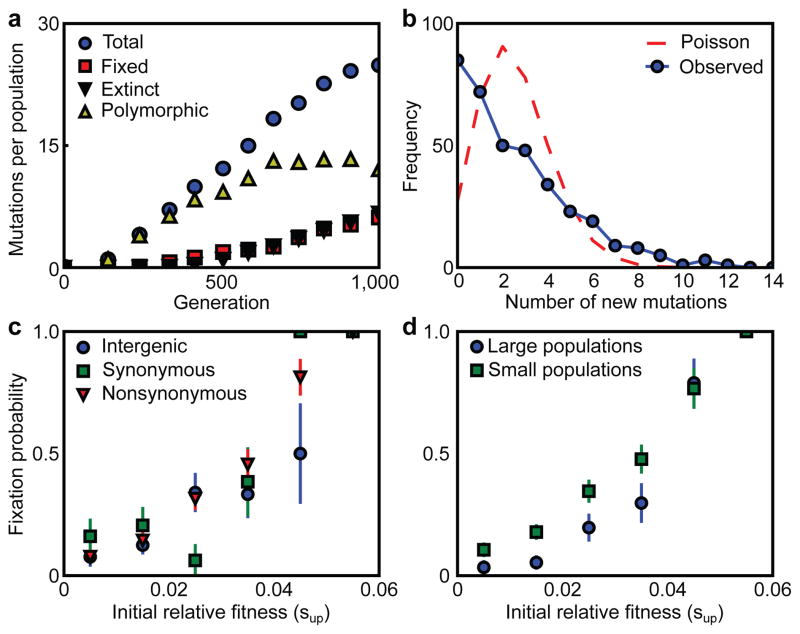
Averaged across all 40 populations, the rate at which mutations appeared and subsequently went extinct or fixed was constant through 1,000 generations (Fig. 2a). The average within-population polymorphism increased steadily through the first 600 generations, before saturating thereafter (Fig. 2a). In individual populations, however, the appearance of mutations is highly punctuated. This leads to the most striking feature of our results: selective sweeps are rarely single mutation/single phase events. Instead, mutations often move through the populations as temporal clusters (“cohorts”) of functionally unrelated mutations, synchronously escaping drift and tracking tightly with one another through time. We quantify this temporal clustering of mutations in Fig. 2b, showing that it leads to a significant overrepresentation of timepoints at which either many or no mutations appeared, compared to the null expectation of mutations reaching detectable frequency at a constant rate (p<10−6).
Figure 2. Statistical analysis across 40 replicate populations.
a, The per-population number of total mutations, fixed mutations, extinct mutations, and mutations that are currently polymorphic over the course of the 1,000 generations. b, The distribution of the number of new mutations detected at each timepoint (solid blue line; see Methods for details) and a Poisson distribution with the same mean (dashed red line). c–d, Mutation fixation probability as a function of initial relative fitness. Data are mean±s.e.m.
As is apparent in Fig. 1, multiple mutations or cohorts of mutations are often present simultaneously, and selective sweeps are often “nested” – that is, one sweep initiates before the preceding sweep has completed. Cohorts and nesting of mutations are forms of genetic hitchhiking, where individual mutations are helped (or hindered) by the genetic background in which they happen to arise. This includes both hitchhiking of likely neutral synonymous mutations, as well as “quasi-hitchhiking” of multiple beneficial mutations that act together as co-drivers. In addition, frequent interference between competing cohorts often leads to the extinction of beneficial mutations even after they reach substantial frequency. Drawing from the full aggregate data set as well as individual “case study” populations, we now show how this pervasive hitchhiking and interference strikes a balance between chance and determinism in governing evolutionary outcomes.
To investigate the repeatability of adaptation and identify those mutations that are driving adaptation, we looked for genes in which we observed mutations more often than expected by chance. Of the 995 nuclear mutations we identified, 723 fall within coding regions. If these mutations were distributed randomly over the 5,799 yeast genes, we expect only two genes with three or more mutations. Instead, we find 24 genes hit three or more times (Table 1, Supplementary Table 2, Supplementary Fig. 2). This parallelism is at the gene level; mutations in different populations are different at the nucleotide level, with four exceptions (Methods). These 24 putative drivers represent ~0.6% of the yeast genome by size but account for 14% of the observed mutations, and are more likely to fix in the population (52/140, 37%) compared to all other nonsynonymous mutations (110/476, 23%, p < 0.005). Only 1 of the 141 mutations in these putative drivers is synonymous (<1%), compared to 19% for the 472 mutations that fall in genes that are hit only once (Supplementary Table 3). Putative drivers are similarly depleted for missense mutations, and are enriched for nonsense and frameshift mutations (Supplementary Table 3). This mutational spectrum differs between functional categories of putative driver mutations. For genes in the mating pathway and negative regulators of Ras, we observe 14 missense, 8 nonsense, and 10 frameshift mutations, suggesting that selection at these loci is for loss of function (Supplementary Fig. 2). In contrast, all 13 mutations observed in cell wall assembly genes are missense, suggesting that at these loci selection is for alteration or attenuation, not loss, of function (Supplementary Fig. 2).
Table 1.
Repeatedly hit genes are putative drivers of adaptation
| Gene | Hits | Fixed | Biological process* |
|---|---|---|---|
| IRA1 | 21 | 10 | Negative regulator of Ras |
| ROT2 | 11 | 2 | Cell wall biogenesis |
| YUR1 | 11 | 5 | Cell wall biogenesis |
| ACE2 | 9 | 4 | Cytokinesis |
| STE11 | 9 | 1 | Mating |
| STE12 | 9 | 2 | Mating |
| PDR5 | 8 | 5 | Multidrug transport |
| WHI2 | 7 | 2 | General stress response |
| STE4 | 6 | 1 | Mating |
| IRA2 | 5 | 3 | Negative regulator of Ras |
| KRE6 | 4 | 1 | Cell wall assembly |
| SFL1 | 4 | 1 | Regulation of flocculation genes |
| STE5 | 4 | 3 | Mating |
| ANP1 | 3 | 1 | Protein glysosylation |
| CNE1 | 3 | 2 | Protein folding |
| GAS1 | 3 | 3 | Cell wall assembly |
| GCN1 | 3 | 1 | Regulation of translation |
| GPB1 | 3 | 1 | Negative regulator of Ras |
| GPB2 | 3 | 1 | Negative regulator of Ras |
| KEG1 | 3 | 0 | Cell wall assembly |
| KRE5 | 3 | 1 | Cell wall assembly |
| RPO31 | 3 | 0 | RNA ploymerase III transcription |
| SET4 | 3 | 2 | unknown |
| YJL171C | 3 | 0 | unknown |
Biological process was manually curated from the Saccharomyces Genome Database (www.yeastgenome.org).
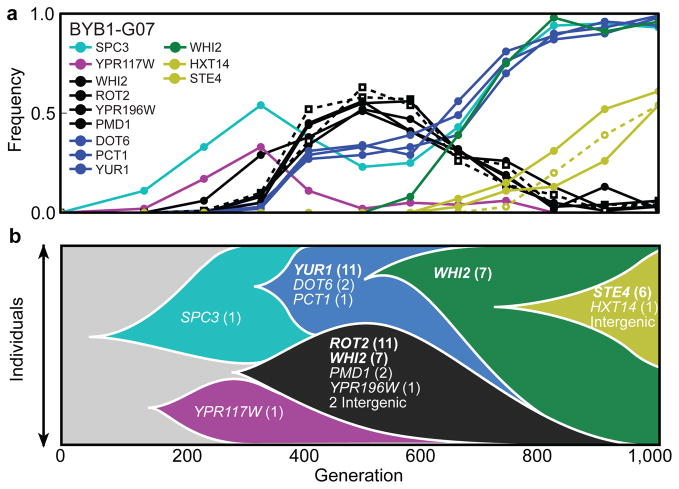
This evidence argues that mutations in multi-hit genes provided strong fitness advantages that made them parallel adaptive solutions in multiple replicate populations (related arguments have been made in bacterial6,7 and viral29 systems). The fate of each mutation, however, also depends on random hitchhiking and interference effects, which increase variation in evolutionary outcomes. Even beneficial driver mutations must often quasi-hitchhike as co-drivers with others in a larger cohort if they are to succeed. For example, in population BYB1-G07, a mutation in SPC3 began to sweep within the first 300 generations (Fig. 3), before a competing cohort appeared containing mutations in the multi-hit genes WHI2 and ROT2. The WHI2/ROT2 cohort rose in frequency at the expense of SPC3, until the SPC3 genotype was partially rescued by a mutation in the multi-hit gene YUR1. Finally, a second and distinct mutation in WHI2 appeared in the SCP3/YUR1 background. This genotype fixed, forcing the WHI2/ROT2 cohort to extinction. These dynamics illustrate how a balance between the fitness advantages of individual driver mutations and random hitchhiking and interference effects determines evolutionary outcomes.
Figure 3. The dynamics of sequence evolution in BYB1-G07.
a, The trajectories of the 15 mutations that attain a frequency of at least 30%, hierarchically clustered into several distinct mutation “cohorts,” each of which is represented by a different color (Methods). b, Muller diagram showing the dynamics of the six main cohorts in the population. The number of times a mutation was observed in a given gene across all 40 populations is indicated in parentheses. Mutations in genes observed in more than three replicate populations (Table 1) are indicated in bold.
While the dynamics of any individual population are highly stochastic, a statistical analysis across replicate populations sheds light on the factors that determine the fate of each mutation. To this end, we measured the initial rate of increase in frequency of each mutation (Methods). We have previously21 referred to this “initial relative fitness” as sup. It measures the combined fitness effect of a mutation together with the genetic background in which it arose, relative to the average of all other genetic backgrounds currently in the population. The probability that a mutation fixes increases with sup (Fig. 2c). Nonsynonymous mutations tended to have higher sup than synonymous mutations (p<.05) and nonsynonymous mutations in multi-hit genes tended to have higher sup than those in single-hit genes (p<.02), as we would expect if the former classes tend to confer a larger fitness advantage. However, given a particular value of sup, all types of mutations were equally likely to succeed. In other words, a weak or neutral mutation on a good background is just as likely to fix as a strongly beneficial mutation on a poor background; all that matters is the initial relative fitness of the mutation combined with the background in which it occurred.
In theory, population size could be predicted to either increase or decrease the patterns of reproducibility between replicate populations. Larger populations will sample more possible mutations, and thus favor the best genotypes in replicate populations13,16. But larger populations also maintain more genetic variation, making each mutation more likely to be influenced by chance associations16. Our data make it possible to determine experimentally the influence of population size on the reproducibility of evolutionary outcomes. Of the 40 sequenced populations, 14 were evolved at a large (106) and 26 at a small (105) population size. We find that putative driver mutations are more commonly observed in large populations (Table 2, p<0.025). However, given a particular value of sup, a mutation is less likely to fix in a large population—that is, subsequent chance associations are more likely to interfere (Fig. 2d, p<10−5). Together, these results show that beneficial mutations occur more consistently in larger populations, but that each mutation has a more random fate once it has occurred.
Table 2.
Summary of the fates of nuclear mutations observed throughout the experiment
| Class of mutation | All populations (40) | Small populations (26) | Large populations (14) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | Fixed | % Fixed | Total | Fixed | % Fixed | Total | Fixed | % Fixed | |
|
|
|
|
|||||||
| All | 995 | 246 | 25% | 703 | 191 | 27% | 292 | 55 | 19% |
| Intergenic | 272 | 58 | 21% | 207 | 48 | 23% | 65 | 10 | 15% |
| Synonymous | 107 | 26 | 24% | 77 | 21 | 27% | 30 | 5 | 17% |
| Nonsynonymous | 616 | 162 | 26% | 419 | 122 | 29% | 197 | 40 | 20% |
| Hit 1x | 381 | 86 | 23% | 273 | 65 | 24% | 108 | 21 | 19% |
| Hit 2x | 95 | 24 | 25% | 63 | 17 | 27% | 32 | 7 | 22% |
| Hit ≥3x | 140 | 52 | 37% | 83 | 40 | 48% | 57 | 12 | 21% |
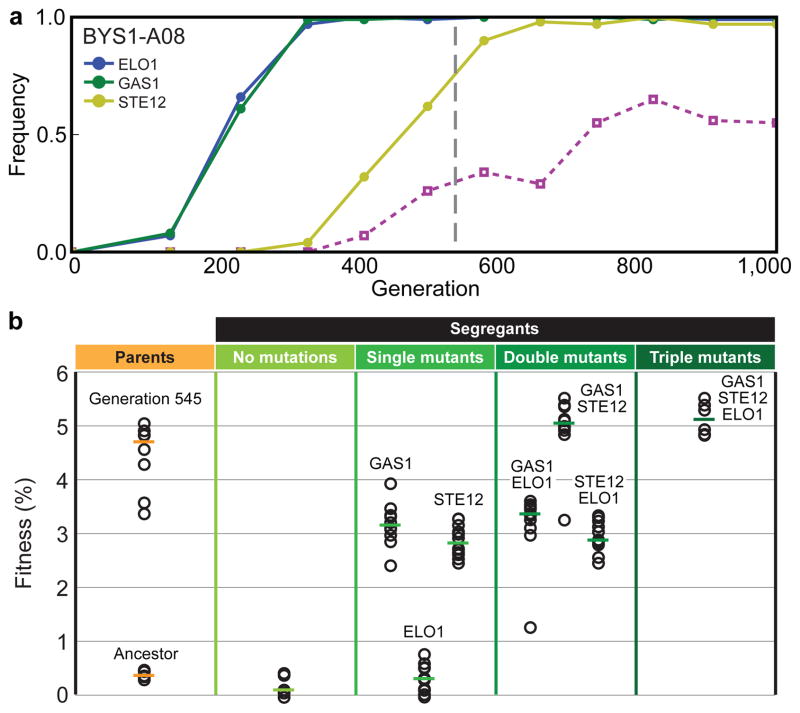
To demonstrate how our system can be used to dissect the fitness effects of the individual mutations that underlie these dynamics, we chose for genetic dissection a population that displayed simple sequence-level dynamics. In BYS1-A08, two mutations (ELO1 and GAS1) have fixed and a third (STE12) is on its way to fixation by generation 545 (Fig. 4a). Clones from this time point had gained, on average, a 4.3% fitness advantage relative to the ancestor. To determine how these three mutations contribute to fitness, we crossed three clones from generation 545 to the ancestor and isolated 80 haploid progeny. Each haploid was genotyped at these three loci and assayed for fitness, allowing us to quantify the fitness effect of each mutation individually and in combination. We find that mutations in both GAS1 and STE12 provide a selective advantage, while the mutation in ELO1 is a neutral hitchhiker (Fig. 4b). Consistent with this, mutations in GAS1 and STE12 are observed in three and nine replicate populations, respectively (Table 1), but ELO1 only once.
Figure 4. Genetic dissection of BYS1-A08.
a, The trajectories of observed mutations. b, We crossed evolved clones from generation 545 to the ancestor; shown here are the fitnesses and genotypes of parental clones and 80 haploid progeny.
Our analysis has shown that the combination of experimental evolution and whole-genome whole-population sequencing over a dense timecourse is a powerful tool. Our data demonstrates the importance of pervasive hitchhiking and clonal interference among cohorts of mutations in determining the molecular dynamics of adaptation. Further work is needed to determine the mechanism underlying the formation of these cohorts. Interestingly, cohorts and genetic hitchhiking have been described in other systems, such as influenza evolution2 and the somatic evolution of cancers30, suggesting that these dynamics represent a general mode of adaptation. Our data also highlight the relatively small subset of genes that repeatedly provide driver mutations, suggesting a limited number of open pathways to substantially increased fitness. This work is a first step towards a complete understanding of the dynamics of adaptation under conditions where multiple beneficial mutations spread simultaneously, and illustrates the importance of both chance and selection in determining evolutionary outcomes.
METHODS
DNA Sequencing
Cells were grown by inoculating 8 μl of each frozen population from our earlier experiment21 into 20 ml YPD (yeast extract, peptone, dextrose) + ampicillin (100 μg/ml) and tetracycline (25 μg/ml) and grown overnight to saturation. Cells were pelleted and washed once with water. Genomic DNA was prepared using a modified glass bead lysis method. Cells were resuspended in 400 μl of DNA Extraction Buffer (2% Triton X-100, 1% SDS, 100 mM NaCl, 10 mM Tris pH 8.0, and 1 mM EDTA). To the resuspended cells, 600 μL of acid washed glass beads (425–600 μm, acid-washed; Sigma) and 400 μL of phenol:chloroform:isoamyl alcohol (25:24:1, Tris saturated) was added and the cells were mechanically lysed for 2.5 minutes using a bead beater. After centrifugation, the supernatant was removed and incubated at 37°C with RNaseA for 1 hour, followed by a second phenol:chloroform:isoamyl alcohol extraction. The aqueous supernatant was removed and genomic DNA was precipitated with ethanol and resuspended in water. Paired-end Illumina sequencing libraries of 500 bp fragments were prepared at The Genome Institute, Washington University School of Medicine, St. Louis, and the libraries were run on the Illumina HiSeq with average of 100-fold coverage.
Identifying mutations from raw sequencing data
We developed two independent methods for identifying mutations from the raw sequencing data and for distinguishing bona fide mutations from spurious calls that resulted from either sequencing or alignment errors by leveraging time course information. Both pipelines identified base-pair substitutions (BPS), small insertion/deletion mutations (InDel) and complex mutations involving both BPS and InDels. We note, however, that neither pipeline is well suited to identify certain types of mutations, such as copy number variation, inversions, or large insertions or deletions. Both pipelines produced similar results. The data presented in the paper were produced using Pipeline 1. Supplementary Table 1 reports the results of both pipelines.
In Pipeline 1, we used the software package Breseq (barricklab.org/breseq) to align Illumina reads and make initial polymorphism calls. We ran Breseq on each timepoint of each population independently and constructed a list of all mutations called in any timepoint. For each mutation, we used SAMTOOLS31 to calculate the frequency of reads supporting the mutation in all timepoints of the population where the mutation was called. We then applied a series of filters based on the frequency trajectories to eliminate false positives. Mutations that did not change frequency over the course of the entire experiment are likely to be sequencing or alignment errors. Therefore, we required the maximum frequency to be at least 0.1 greater than the minimum frequency. We also required the absolute difference between the maximum or minimum frequency and the frequency at generation zero to be at least 0.1. The frequency trajectories of real mutations are expected to be autocorrelated, whereas those of false positives should be uncorrelated from timepoint to timepoint. We rejected any mutation with an autocorrelation coefficient less than 0.2. Generation zero was not expected to contain any mutations. Therefore, we rejected any mutation detected by Breseq in generation zero of more than five populations. Also, for any mutation detected by Breseq in generation zero of more than two populations, we required the autocorrelation coefficient to be at least 0.5. Finally, for any mutation with a frequency greater than 0.01 in generation zero, we required the autocorrelation coefficient to be at least 0.35.
In Pipeline 2, for each population and for each time point, we aligned the raw reads to a SNP/Indel corrected W303 reference genome (reference available upon request) using BWA for Illumina version 1.2.232 using default parameters (except “Disallow insertion/deletion within [value] bp towards the end” set to 0 and “Gap open penalty” set to 5). Mutations were called relative to the SNP/Indel corrected W303 reference genome using Freebayes version 0.8.9.a, (Marth Laboratory, Boston College) using default parameters (except “Pooled” set to “True,” and “Base alignment quality (BAQ) adjustment” set to “True”). For each population we merged the 12 resulting .vcf files (one for each time point) using the “vcf-merge” included in the VCFtools package (http://vcftools.sourceforge.net/perl_module.html). We wrote two perl scripts to analyze the resulting merged .vcf file (programs available upon request). The script “allele_counts.pl” calculated the frequencies of mutant alleles for each time point in the series and “composite_scores.pl” scored the trajectories of each mutation across the twelve time points based on six attributes: autocorrelation, area under the curve relative to time zero, minimum frequency, maximum frequency, max step (the largest difference in frequency in adjacent time points), and the number of called alternate alleles. We developed a heuristic composite score with which to rank the trajectories by their likelihood of being a bona fide mutation.
Any mutation called in either Pipeline was validated manually using the Integrative Genome Viewer 33,34.
Annotating mutations
For each mutation, we aligned the surrounding 2kb region to the annotated s288c genome using NCBI-BLAST35. We then used NCBI-BLAST’s CDS feature option to identify the gene or intergenic region containing the mutation and the identity of any amino-acid changes.
All of the observed nuclear mutations represent unique alterations to the yeast genome with four exceptions: two cases of recurrent mutation at the same position and two instances of pre-existing mutations in the seed culture that reached detectable frequency during the evolution experiment. In ROT2 and STE12, recurrent frameshift mutations were observed within homopolymeric runs of seven T’s and eight G’s, respectively. For ROT2 all four occurrences of mutations in this homopolymeric run were T insertions. For STE12, two mutations were G insertions and two were G deletions. In addition to recurrent mutations, we observed two pre-existing mutations. In the initial evolution experiment, two nearly isogenic haploid ancestral strains (B and R) were used to seed ~300 populations each. Of the sequenced populations reported here, 30 are derived from the B progenitor and 10 from the R progenitor. We observed several occurrences were the same mutation was observed in multiple populations. The same single base-pair deletion in IRA1 was observed in four populations derived from the B ancestor. In each case this allele was observed early and prior to the first selective sweep suggesting that this mutation was present at low frequency in the starting B population. In all 10 R populations, the same T to C substitution in PDR5 was initially at 15% at Generation 0. This mutation quickly fixed in two populations, slowly fixed in another, rose to above 50% before going extinct in two and quickly went extinct in the other five (Supplementary Fig. 3).
Analysis of trajectories
In order to assess the relationship between fitness and fixation probability, we estimated sup: the fitness of clones containing a given mutation relative to the mean fitness of the population when we first detected the mutation. For each mutation, we identified t1 and t2, the first consecutive timepoints such that the frequency of the mutation at t1 was greater than zero and the frequency at t2 was greater than 0.1. We then calculated
where f(t) is the frequency of the mutation at time t. This quantity estimates the combined effects of the focal mutation and the background it occurred on. For instance, a mutation conferring a 3% fitness advantage on a neutral background will have the same value of sup as a neutral mutation occurring on a background that is 3% fitter than the population average.
Identifying mutation cohorts
The most striking feature of our results is that mutations often move through populations as temporal clusters of functionally unrelated mutations, tracking tightly with one another through time. We have termed these “cohorts.” In order to empirically assign mutations to cohorts, we treated each frequency trajectory as a vector in twelve dimensions. We used the hierarchical clustering package in SciPy (www.scipy.org) to cluster the mutations in each population based on the Euclidean distance between frequency vectors. Because low-frequency mutations contain too little information for reliable clustering, we excluded mutations with maximum frequencies less than 0.3. We then flattened the hierarchies using a cutoff distance of 0.275.
Fitness Assays and Genetic Dissection
Fitness assays were performed as described previously21. To measure the fitness of evolved clones from frozen stock, we struck to singles from population BYS1-A08 from generation 545. We selected seven single colonies at random and measured their fitness relative to an mCherry-expressing reference strain. The experimental and reference strains were grown separately in 96-well plates, then mixed 50:50 and propagated by diluting 1:1,024 every 24 hours. At generations 10, 20, and 30, and 40, we transferred 4 μl of saturated culture into 100 μl of cold PBST and the ratio of nonfluorescent (experimental) and mCherry-positive (reference) cells was determined by flow cytometry using an LSRII flow cytometer (BD Biosciences, San Jose, CA) counting 50,000 total cells for each sample. The fitness difference between the experimental and reference strain was calculated as the rate of the change in the ln ratio of experimental to reference versus generations36. To determine fitness effects of the three evolved mutations in BYS1-A08 (GAS1, ELO1, STE12), we chose three of the seven clones and backcrossed them to a MATα version of the ancestral strain. From these three diploids we sporulated and selected 80 haploid MATa segregants. Each segregant was genotyped by SNP-specific PCR using the following primers: GAS1_Forward (5′ TTTTC GTGCC GCAAA CGTGG 3′), GAS1_WT_Reverse (5′ ATTGG AAGAG TAGCC AACTG 3′), GAS1_Mutant_Reverse (5′ ATTGG AAGAG TAGCC AACTA 3′), ELO1_Forward (5′ AACAC AACAA ATCGC AAGCC 3′), ELO1_WT_Reverse (5′ TAACC AACCA ATTGA TTATA 3′), ELO1_Mutant_Reverse (5′ TAACC AACCA ATTGA TTATG 3′), STE12_Reverse (5′ TGAGC AGAAT CTTCG TCACC 3′), STE12_WT_Forward (5′ AATCT CACAA CTCTG GCCAG 3′), and STE12_Mutant_Forward (5′ AAATC TCACA ACTCT GCCAA 3′). The fitness of each of the haploid segregants was measured relative to the mCherry-expressing reference strain as described above.
Supplementary Material
Acknowledgments
We thank the production team led by Lucinda Fulton and Robert Fulton at the Genome Institute at Washington University for sample management and data production, and Elizabeth Lobos for coordinating the project. We thank Lance Parsons and John Wiggins for assistance with data management, Pat Gibney for assistance with sample preparation, and Tina DeCoste for assistance with flow cytometry. We thank Katya Kosheleva for discussions, and Andrew Murray, Chris Marx, Michael McDonald, Gavin Sherlock, and Dan Kvitek for comments on the manuscript. D.P.R. acknowledges support from an NSF Graduate Research Fellowship. D.B. acknowledges support from NIGMS Centers of Excellence grant GM071508 and NIH grant GM046406. M.M.D. acknowledges support from the James S. McDonnell Foundation, the Alfred P. Sloan Foundation, and the Harvard Milton Fund.
Footnotes
AUTHOR CONTRIBUTIONS
G.I.L., D.B., and M.M.D. designed the project; E.S. and G.M.W. generated the sequencing data; G.I.L., D.P.R., M.J.H., and M.M.D. analyzed the sequencing data; G.I.L. performed the experiments; G.I.L., D.P.R., D.B., and M.M.D. wrote the paper.
Genome sequence data have been deposited to Genbank under the BioProject identifier PRJNA205542.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
REFERENCES CITED
- 1.Weinreich DM, Delaney NF, DePristo MA, Hartl DL. Darwinian Evolution Can Follow Only Very Few Mutational Paths to Fitter Proteins. Science. 2006;312:111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- 2.Strelkowa N, Lässig M. Clonal Interference in the Evolution of Influenza. Genetics. 2012;192:671–682. doi: 10.1534/genetics.112.143396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levin BR, Bull JJ. Short-sighted evolution and the virulence of pathogenic microorganisms. Trends in Microbiology. 1994;2:76–81. doi: 10.1016/0966-842x(94)90538-x. [DOI] [PubMed] [Google Scholar]
- 4.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sprouffske K, Merlo Lauren MF, Gerrish Philip J, Maley Carlo C, Sniegowski Paul D. Cancer in Light of Experimental Evolution. Current biology : CB. 2012;22:R762–R771. doi: 10.1016/j.cub.2012.06.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tenaillon O, et al. The Molecular Diversity of Adaptive Convergence. Science. 2012;335:457–461. doi: 10.1126/science.1212986. [DOI] [PubMed] [Google Scholar]
- 7.Woods R, Schneider D, Winkworth CL, Riley MA, Lenski RE. Tests of parallel molecular evolution in a long-term experiment with Escherichia coli. Proceedings of the National Academy of Sciences. 2006;103:9107–9112. doi: 10.1073/pnas.0602917103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saxer G, Doebeli M, Travisano M. The Repeatability of Adaptive Radiation During Long-Term Experimental Evolution of Escherichia coli in a Multiple Nutrient Environment. PLoS One. 2010;5:e14184. doi: 10.1371/journal.pone.0014184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Atwood KC, Schneider LK, Ryan FJ. Periodic selection in Escherichia coli. Proceedings of the National Academy of Sciences. 1951;37:146–155. doi: 10.1073/pnas.37.3.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pacquin C, Adams J. Frequency of fixation of adaptive mutations is higher in evolving diploid than haploid yeast populations. Nature. 1983;302:495–500. doi: 10.1038/302495a0. [DOI] [PubMed] [Google Scholar]
- 11.Joseph SB, Hall DW. Spontaneous Mutations in Diploid Saccharomyces cerevisiae: More Beneficial Than Expected. Genetics. 2004;168:1817–1825. doi: 10.1534/genetics.104.033761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perfeito L, Fernandes L, Mota C, Gordo I. Adaptive mutations in bacteria: High rate and small effects. Science. 2007;317:813–815. doi: 10.1126/science.1142284. [DOI] [PubMed] [Google Scholar]
- 13.Gerrish P, Lenski R. The Fate of Competing Beneficial Mutations in an Asexual Population. Genetica. 1998;102/103:127–144. [PubMed] [Google Scholar]
- 14.Desai MM, Fisher DS. Beneficial Mutation-Selection Balance and the Effect of Linkage on Positive Selection. Genetics. 2007;176:1759–1798. doi: 10.1534/genetics.106.067678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rouzine I, Wakeley J, Coffin J. The Solitary Wave of Asexual Evolution. PNAS. 2003;100:587–592. doi: 10.1073/pnas.242719299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Good BH, Rouzine IM, Balick DJ, Hallatschek O, Desai MM. The rate of adaptation and the distribution of fixed beneficial mutations in asexual populations. Proceedings of the National Academy of Sciences. 2012;109:4950–4955. doi: 10.1073/pnas.1119910109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schiffels S, Szöllősi GJ, Mustonen V, Lässig M. Emergent Neutrality in Adaptive Asexual Evolution. Genetics. 2011;189:1361–1375. doi: 10.1534/genetics.111.132027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desai MM, Fisher DS, Murray AW. The Speed of Evolution and Maintenance of Variation in Asexual Populations. Current Biology. 2007;17:385–394. doi: 10.1016/j.cub.2007.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Visser J, Zeyl CW, Gerrish PJ, Blanchard JL, Lenski RE. Diminishing returns from mutation supply rate in asexual populations. Science. 1999;283:404–406. doi: 10.1126/science.283.5400.404. [DOI] [PubMed] [Google Scholar]
- 20.Kao KC, Sherlock G. Molecular Characterization of Clonal Interference During Adaptive Evolution in Asexual Populations of Saccharomyces cerevisiae. Nature Genetics. 2008;40:1499–1504. doi: 10.1038/ng.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lang GI, Botstein D, Desai MM. Genetic variation and the fate of beneficial mutations in asexual populations. Genetics. 2011;188:647–661. doi: 10.1534/genetics.111.128942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barrick JE, et al. Genome Evolution and Adaptation in a Long-Term Experiment with Escherichia Coli. Nature. 2009;461:1243–1247. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- 23.Barrick JE, Lenski RE. Genome-wide mutational diversity in an evolving population of Escherichia coli. Cold Spring Harb Symp Quant Biol. 2009;74:119–129. doi: 10.1101/sqb.2009.74.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dettman JR, et al. Evolutionary insight from whole-genome sequencing of experimentally evolved microbes. Molecular Ecology. 2012;21:2058–2077. doi: 10.1111/j.1365-294X.2012.05484.x. [DOI] [PubMed] [Google Scholar]
- 25.Gresham D, et al. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS genetics. 2008;4:e1000303. doi: 10.1371/journal.pgen.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bollback JP, Huelsenbeck JP. Clonal Interference Is Alleviated by High Mutation Rates in Large Populations. Mol Biol Evol. 2007;24:1397–1406. doi: 10.1093/molbev/msm056. [DOI] [PubMed] [Google Scholar]
- 27.Betancourt AJ. Genomewide Patterns of Substitution in Adaptively Evolving Populations of the RNA Bacteriophage MS2. Genetics. 2009;181:1535–1544. doi: 10.1534/genetics.107.085837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller CR, Joyce P, Wichman HA. Mutational Effects and Population Dynamics During Viral Adaptation Challenge Current Models. Genetics. 2011;187:185–202. doi: 10.1534/genetics.110.121400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wichman HA, Badgett MR, Scott LA, Boulianne CM, Bull JJ. Different Trajectories of Parallel Evolution During Viral Adaptation. Science. 1999;285:422–424. doi: 10.1126/science.285.5426.422. [DOI] [PubMed] [Google Scholar]
- 30.Nik-Zainal S, et al. The Life History of 21 Breast Cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2012 doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 36.Hartl D. A primer of population genetics. Sinauer Associates; 2000. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.