Abstract
The importance of cell migration for both normal physiological functions and disease processes has been clear for the past 50 years. Although investigations of two-dimensional (2D) migration in regular tissue culture have elucidated many important molecular mechanisms, recent evidence suggests that cell migration depends profoundly on the dimensionality of the extracellular matrix (ECM). Here we review a number of evolving concepts revealed when cell migration is examined in different dimensions.
Introduction
Cell migration is crucial for numerous physiological and developmental processes, including gastrulation, organ formation, immune function, and wound healing. In addition, aberrant cell motility contributes to diseases such as cancer metastasis [1,2]. Initial characterizations of fibroblast motility in tissue culture helped to establish key concepts about cell migration based on adhesion and interactions with a 2D planar surface. These observations continue to guide current research on the intracellular regulation of signaling pathways involved in migration. However, the physical characteristics of an ECM can also strongly modulate cell migration by outside-in signaling from the microenvironment. Over the past decade, modeling of cell motility in three-dimensional (3D) ECM models that mimic more-physiological in vivo conditions has revealed substantial differences between 2D and 3D cellular migration. Besides these 3D models, simplified reductionist model systems have allowed analysis of matrix regulation of migration under more controllable experimental conditions [3–7]. In this review, we will explore recent conceptual advances in cell migration from investigations of cell migration in different dimensions using a variety of model systems. We will focus on how the unique dimensional aspects of 2D planar substrates, 3D scaffolds, and simplified one-dimensional (1D) fibers can help regulate migration rate, the mode of migration, cellular mechanotransduction, and cell signaling of mesenchymal-derived fibroblasts, but allude to other cell types when appropriate.
Overview of dimensional concepts in cell migration
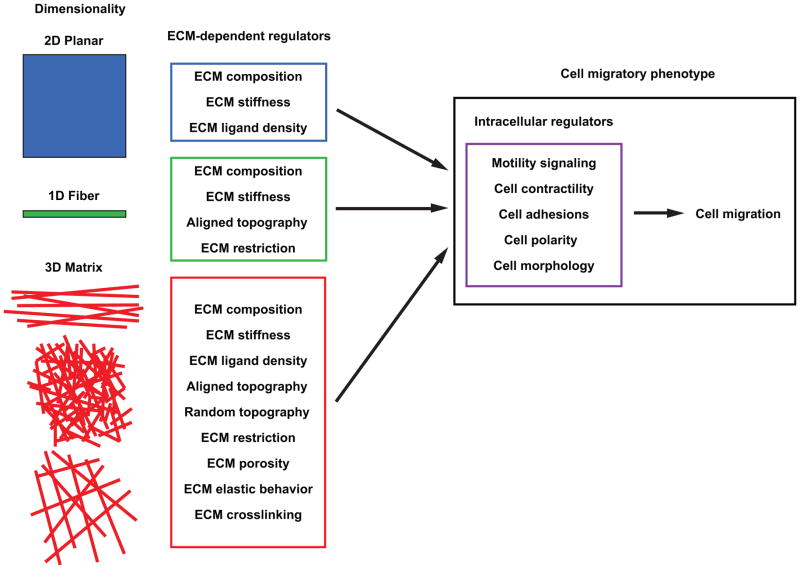
As illustrated in Figure 1 (right panel), multiple intracellular regulatory mechanisms are known to govern adhesion-dependent fibroblast migration. Compounding this internal regulation, it is now clear that a host of ECM microenvironmental properties can directly influence these intracellular regulatory mechanisms to control the mode and rates of cell migration (Figure 1, left panels). The three primary classes of dimensionality involve 2D planar substrates classically used in cell culture, 1D fibers, and 3D matrix; the latter can exist as parallel fibers, dense random networks, or more porous matrices. Specific ECM properties can become important regulators of migration (central boxes) depending on the type of ECM dimensionality. For example, even though ECM composition and ECM stiffness can regulate migration rates regardless of dimensionality, in 3D matrices, many other physical properties of the ECM including porosity and elastic behavior become important regulators of migration.
Figure 1. Dimensional regulation of cell migration.
Illustration of the numerous unique ECM-dependent regulators (center column) associated with 2D, 1D, and 3D migration. These microenvironmental regulators in turn influence intracellular regulatory pathways that govern the migratory phenotype (right panel) and determine how cell migration proceeds.
In some cases, reductionist approaches in model migration systems can provide a clearer understanding of the roles in migration of a specific feature or property of the ECM, such as by using a single ECM fiber, a micropatterned line [3–5], a derivatized 3D biomaterial [8,9] or a range of ECM pore sizes using 3D microtracks or microchannels [6,7,10]. Although the concept of cell behavioral plasticity controlled by the microenvironment is well-established for 3D migration (e.g., see ref. [11]), recent investigations have expanded this concept of the importance of matrix-dependent regulation to all dimensional conditions. Our review will show that matrix regulation of cell motility is highly context-dependent – it depends on both dimensionality and each set of specific physical and biochemical conditions in a given ECM microenvironment. Table 1 summarizes the differences in cell migration depending on dimensional conditions discussed in this review.
Table 1.
Key migration differences associated with 2D, 3D, and 1D ECMs*.
| 2D surface | 3D matrix | 1D fiber | |
|---|---|---|---|
|
| |||
| ECM structure or topography | Planar adsorbed globular ECM molecules[12,52,53] | Fibrillar matrices (3D CDM, COL I, FB)[6,7,12,13,15,16,21,23,24,48,51,54–57] Non-fibrillar matrices (Matrigel, PEG hydrogels)[8,9] |
Single fibronectin-based fibers [55] ECM adsorbed to micropatterns [3–5] |
|
| |||
| Centrosome position** (assay) | Front [5] (wound assay) | Rear (3D-CDM) [5] (single cells) | Rear [5] (single cells) |
|
| |||
| Ligand density effect on migration rate | Biphasic [20,58] | ND [24] | Increases to a plateau [5] |
|
| |||
| Restriction/confinement | None | Matrix pore size dependence [24,43] | Reduction in lateral spreading[3–5] |
|
| |||
| Migration mode | LA, FIL | LA, FIL, LOB, Bleb | LA, FIL, LOB, Bleb |
|
| |||
| Elastic behavior (ECM) | Linear (PAA gels) [59] | Linear (3D-CDM) [33] Non-linear (FB) [35] Non-linear (COL I) [33,35] |
Linear (PAA gels) [3] |
|
| |||
| Fibroblast migration rate compared to 2D | Not applicable | 3D-CDM: 1.5x faster [5,12,16] Collagen: elevated [12] Fibrin: elevated [12] BME: slower [12] |
1.5x faster (FN)[4,5] |
|
| |||
| Loss of contractility | Increased migration [5,53,60] | Decreased migration [5,24] | Decreased migration [4,5] |
|
| |||
| Signaling Polarity | |||
| RhoA | Yes (front) [33,45] | Yes (Collagen), No (3D-CDM) [33] | ND |
| Rac | Yes (front) [33,45] | Yes (Collagen), No (3D-CDM) [33] | ND |
| Cdc42 | Yes (front) [33,45] | Yes (Collagen), No (3D-CDM) [33] | ND |
| PIP2 | Yes (front) [33] | Yes (Collagen), No (3D-CDM) [33] | ND |
|
| |||
| Protein requirement for efficient migration | |||
| Vinculin | No [48] | Yes (COL I) [48] | ND |
| NEDD9 | No [51] | Yes (COL I)[51] | ND |
| Myosin IIA | No [4] | ND | Yes (FN) [4] |
| Myosin IIB | No [4] | Yes (COL I) [63] | No (FN) [4] |
| N-WASP | No [47] | Yes (COL I) [47] | ND |
| Scar/WAVE | Yes [47] | No (COL I) [47] | ND |
| Cdc42 | Yes [61] | Yes (COL I) [61] | ND |
| Rac1 | Yes (directional) [62] | No (3D-CDM) [5,33] | ND |
| RhoA | Yes [62] | Yes (COL I)[13] No (3D-CDM) [33] | ND |
For 3D models, the specific type of ECM is listed.
With respect to the nucleus. ND: Not determined, LA: lamellipodia, FIL: filopodia, LOB: Lobopodia, COL I: Collagen I gels, FB: Fibrin gels, BME: basement membrane extract, FN: fibronectin.
Control of cell migration through ECM topography
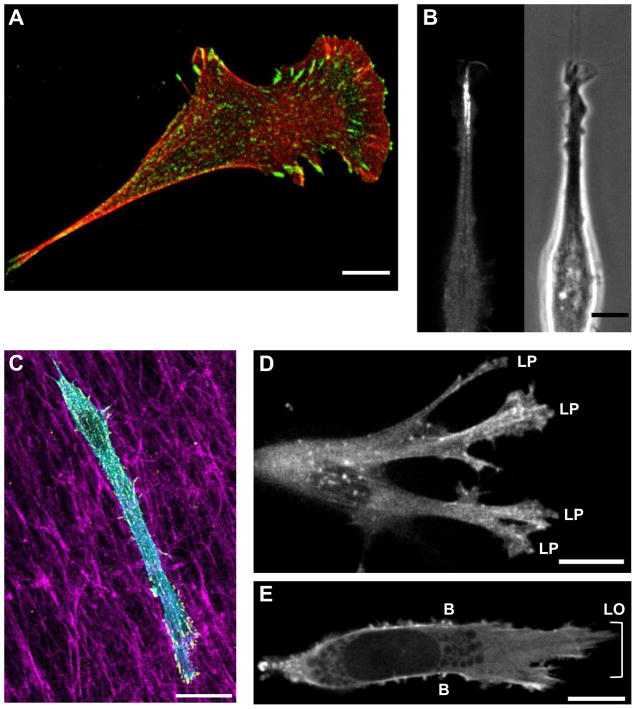
When comparing migration in different dimension, a key ECM-dependent regulator involves differences in ECM topography. In a classic 2D migration model, ECM molecules are presented to cells as a flat sheet of globular molecules without appreciable fibrillar structure. This planar ECM topography promotes a spread cell morphology, and fibroblasts acquire a “hand-mirror” appearance (Figure 2A) with apical/basal polarity in cell adhesions and most of the contractile apparatus associated with the 2D surface. This contrasts with 3D ECMs (Figure 2C), in which fibrillar topography and ECM pore size decrease lateral spreading [12] and do not impose apical/basal polarity on either adhesions or the actin cytoskeleton.
Figure 2. Mode of cellular protrusion as determined by ECM dimensionality.
A) NIH/3T3 fibroblast demonstrating a classic hand-mirror morphology on a 2D substrate; red is phalloidin staining and green shows activated β1 integrin adhesions. B) eGFP-VASP (left) and phase contrast (right) image of a NIH/3T3 fibroblast migrating along a 1D micropatterned line. C) NIH/3T3 fibroblast within a 3D-CDM showing staining for F-actin (phalloidin, cyan), paxillin (yellow), and fibronectin (magenta). D). eGFP-actin expressed in a human foreskin fibroblast illustrating lamellipodia in non-linear 3D collagen. E) Lobopodia (LO) and lateral blebs (B) shown by eGFP-actin as a human foreskin fibroblast migrates through a linear-elastic 3D-CDM. Scale bars: 10 μm.
In fibrillar 3D ECMs, the polymeric fibrils of fibronectin, collagen I, or fibrin can have a random or aligned topography; the latter increases migration velocity and directionality in vitro [13] and is often found in tumor-associated stroma and cell-derived ECMs [14,15]. 1D migration models mimic single fibers in aligned native 3D ECMs and have clarified the important role of topography and contact guidance during migration. 1D ECM restricts cell shape by preventing lateral cell spreading and promoting a uniaxial phenotype (Figure 2B); it stimulates migration to rates matching those in aligned 3D cell-derived matrices (3D-CDMs;1.5-fold over 2D substrates), [5,16]. Although 1D fiber models are engineered, similar aligned ECM structures exist in vivo and are thought to promote tumor metastasis [13], as well as helping direct stem cell migration after transplantation into the rat auditory nerve and after spinal cord injuries [17,18].
Dimensional control of cell migration through ECM-ligand interactions
Interactions between integrins and the ECM can profoundly affect migration rate and cell phenotype. 2D fibroblast migration rates demonstrate a biphasic dependence on ECM ligand density: cells on too little ECM fail to generate adhesions to the underlying substrate, whereas too much ECM inhibits cell tail retraction, reducing leading edge protrusion, and slowing migration rate [19]. Optimal 2D migration rate occurs at intermediate levels, but the optimum can be shifted towards higher or lower ECM concentrations depending on integrin availability [20]. In contrast, cells on increasing levels of a 1D fibrillar ECM substrate show increasing migration rates that plateau with no decrease at even high ECM concentrations [5]. 3D systems add more complexity, because changing the protein concentration of 3D collagen and other polymer-based 3D ECMs also alters: a) ECM stiffness, b) matrix pore size, c) ECM confinement (restriction) of cells, and d) ECM crosslinking. In addition, 3D collagen gel polymerization techniques can vary greatly between research groups, which affects all of these matrix physical properties. Small pore sizes reduce cell migration rate, in part by restricting ability of the nucleus to squeeze between matrix fibrils, which can often also requires proteolytic degradation of the matrix [21]. While it is difficult to determine which ECM-dependent regulator is predominant, initial investigations in 3D collagen indicate that migration rate is particularly dependent on pore size [22,23]. Recently, Wolf et al.[24] carefully evaluated many ECM-dependent regulators of 3D migration, including ECM porosity, ECM compliance, collagen fiber size, and collagen concentration. They concluded that deformation of the nucleus of a cell migrating through a 3D lattice is the main rate limiting factor and is linearly related to cell migration rate for HT-1080 fibrosarcoma cancer cells and polymorphonuclear neutrophils [24]. In addition, matrix degradation through matrix metalloproteinases (MMPs) and mechanical coupling to the surrounding ECM to provide support for actinomyosin contractile force generation were required to maintain migration rates in matrices with pore sizes below 30 μm2 [24].
We emphasize that direct comparisons of cell migration rates between 2D globular and 3D native fibrillar ECMs are not normally performed at the same ECM concentrations. Such concentrations often differ immensely, and combined with the fibrillar structure of many ECM proteins, will probably differentially affect integrin clustering and cell-matrix adhesions. Furthermore, variations in 2D/3D migration rates will likely vary for different cell types. The closest comparisons between 2D to 3D indicate higher migration rates in 3D-CDMs, 3D collagen I, and fibrin gels compared to corresponding flattened or 2D-adsorbed ECMs [12], while directionality is not affected. The exception is single-cell migration in 3D basement membrane extract (BME) which has extremely fine porosity, lacks fibrillar architecture, and does not support fibroblast spreading or polarization.
Regulation of cell migration by ECM rheological readout
The rheological or elastic properties of an ECM can be “sensed” by a cell and can directly regulate intracellular functions. At the heart of this physical sensing mechanism is the mechanical link between the ECM and the actin-myosin contractile apparatus through cell adhesion sites [25–27]. Cells demonstrate a relatively proportional contractile response to the rigidity of the local microenvironment: adhesion size and number of stress fibers increase with ECM rigidity [28,29]. Because of this cellular response, alterations to ECM rigidity and/or cellular contractility affect the efficiency of cell migration, with dimensionality playing an important role. In general, cells preferentially associate with rigid matrices, which reduces 2D migration rate at a given ECM concentration [30]. However, an inverse relationship exists in 2D between ECM rigidity and concentration, allowing optimal rates of migration at any given ECM stiffness if its concentration is adjusted [31]. As is the case for other matrix-dependent regulators of 3D migration, ECM rigidity can vary substantially, for example, in collagen I gels because of the interdependence between rigidity and the other physical aspects of the 3D environment (e.g., concentration, porosity, gel thickness, crosslinking[32]. Recent evidence using artificial poly(ethylene glycol)-based ECMs where matrix pore size is not altered shows that lower ECM rigidity enhances cell migration [8].
Another unique rheological aspect of 3D ECMs is their elastic behavior, which differs depending on molecular composition [33]. The elastic behavior of a material is characterized by how its rigidity changes in response to repeated and increasing force. Non-linear elastic materials undergo strain-stiffening – that is, their rigidity increases with the application of increasing force, while the rigidity of linear elastic materials does not [34]. This elastic behavior originates at the fiber level within the matrix. Non-linear elastic ECMs, e.g., collagen and fibrin 3D gels, have a meshwork of entangled fibers that can move independently of one another in response to an applied force, whereas in linear elastic 3D ECMs, such as fibronectin-containing 3D-CDMs, individual fibers are covalently linked to each other, leading to restricted matrix movement [35]. Elastic behavior is most apparent in 3D materials and has recently been shown to govern the mechanism or mode of 3D cell migration [33].
The dependence of migration mode on ECM elastic behavior
Cells migrating on 2D surfaces predominantly use lamellipodia-based motility, in which actin polymerization against the plasma membrane over a broad area pushes the leading edge forward, followed by adhesion to the underlying substrate via integrin-dependent adhesions [36–38]. However, lamellipodia (Figure 2D) are only one of several types of protrusive elements generated by motile cells (recently reviewed in [39]), and in 3D environments the elastic behavior of the ECM in part governs the choice between lamellipodial and lobopodial migration. Lobopodia (Figure 2E) are blunt, cylindrical protrusions that extend the cell’s leading edge independently of lamellipodia formation in a process combining elevated actomyosin contractility with adhesion to the surrounding matrix [33]. Lobopodia-based migration appears to be restricted to linear elastic 3D environments, such as 3D cell-derived matrix (CDM) and skin dermis [33]. Covalently crosslinking 3D collagen converts it into a linear elastic material which supports lobopodial migration; conversely, trypsinizing CDM removes the covalent crosslinks holding the fibers together and switches it to a non-linear elastic material which triggers lamellipodia-based motility. 3D collagen gels can also be converted to linear elastic material by including relatively low amounts of agarose to restrict degrees of freedom of the collagen fibers [40]. Similar to covalently crosslinked collagen and fibroblasts, agarose-containing collagen can switch cancer cells from a mesenchymal to more amoeboid form of migration. In the future, it will be important to investigate whether cellular mechanisms involved in rigidity sensing are also important for detecting differences in ECM elastic behavior.
Dimensionality and signaling in cell migration
Traditional 2D cell migration models have facilitated the identification of cell-matrix receptors, cytoskeletal machinery, and other intracellular regulators required for migration. However, the relative importance and roles of many of these molecules differ in recent analyses comparing migration in 3D ECM. As discussed earlier, cells in 3D and 1D fibrillar matrix models require actomyosin contractility for efficient migration; inhibition of myosin II activity decreases migration rates in 1D and 3D environments while increasing 2D migration [5]. Further analysis by genetic ablation of contractile myosin II isoforms reveal that myosin IIA is essential for efficient migration in 1D environments [5]. This actomyosin contractility depends on RhoA and its downstream effector Rho-associated protein kinase (ROCK) [33]. In contrast to 2D migration, this pathway is a key determinant of both mode and efficiency of migration in 3D. Modulating RhoA-ROCK signaling switches 3D modes of motility in primary fibroblasts between lamellipodial and lobopodial-driven migration in 3D cell-derived matrix [33]. Additionally, the mode of cancer cell migration in 3D collagen matrices depends on both traditional and non-canonical RhoA signaling pathways: ameboid (RhoA-ROCK) and mesenchymal migration (Cdc42-MRCK) [41], as well as migratory efficiency (RhoA-ROCK1/ROCK2, RhoC) [42]. Furthermore, activation of Rho/ROCK-regulated contractility is necessary for remodeling and alignment of matrix fibers to provide contact guidance for 3D malignant epithelial cell migration [13]. Additionally, a novel role for caveolin-1 has been described in 3D cultures for maintaining the activity and localization of RhoA through p190 RhoGAP signaling to drive the local matrix remodeling required for fibroblast elongation and migration [15]. Moreover, cell migration in 2D and 1D models does not require matrix-degrading proteases, whereas cells in 3D can adopt either proteolytic or non-proteolytic modes of movement, depending on the molecular composition, porosity, and rigidity of the 3D substrate [11,24,43,44].
Understanding the signaling mechanisms driving migration in 3D is in its infancy, with most observed differences attributed to changes in dimensionality rather than to a specific aspect of the ECM. During traditional 2D migration, Rac1, Cdc42, and RhoA are spatiotemporally polarized and activated at the leading edge of cells [45]. However during migration in 3D, polarization of Rac1 and Cdc42 can be absent during migration [33], or the cell can exhibit variable dependence on the activity of a particular GTPase depending on matrix rigidity and composition [46]. Requirements for the regulators of actin assembly and lamellipodial protrusions N-WASP and Scar/WAVE differ for 2D and 3D migration [47]. In addition, genetic ablation or knockdown of key cell adhesion components such as vinculin [48], zyxin [49], and NEDD9 [50,51] demonstrate that while these proteins hinder 2D migration on rigid surfaces, they are essential for normal migration within 3D environments. Therefore, to understand migratory signaling in different dimensions, reductionist approaches will be needed to correlate specific cellular responses with changes in individual chemical and physical 3D substrate regulators to progress beyond current studies testing only effects of altered dimensionality.
Conclusions
Our review has touched upon selected recent investigations that illustrate important differences associated with cell migration in different dimensions, as well as the high context-dependence of migration due to specific ECM regulators of migration in each dimension. ECM composition, ECM stiffness, topography, elastic behavior, and other key biochemical and physical properties of the microenvironment initiate cellular adaptations that alter the overall mode of cell migration and the signaling that controls cell motility. In future research on migration in 3D environments, it will be important to isolate experimentally and conceptually the specific roles of each of the many ECM regulators in cell migration, which we have seen influence each other to define the ECM microenvironmental context, e.g., the various physical characteristics of a particular 3D matrix. These insights should ultimately help to determine the important extracellular regulatory cues that are altered in developmental defects and diseases.
Acknowledgments
We thank Emily Joo and Duy Tran for critically reviewing this manuscript and Tim Lammermann for helpful suggestions. Support was provided by the intramural research program of the National Institute of Dental and Craniofacial Research at the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1•.Nguyen-Ngoc KV, Cheung KJ, Brenot A, Shamir ER, Gray RS, Hines WC, Yaswen P, Werb Z, Ewald AJ. ECM microenvironment regulates collective migration and local dissemination in normal and malignant mammary epithelium. Proc Natl Acad Sci U S A. 2012;109:E2595–2604. doi: 10.1073/pnas.1212834109. Malignant breast cancer cells did not invade into 3D Matrigel, but a change in local microenvironment to 3D collagen allowed invasive migration; this study thus identifies ECM-specific migration programs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 3.Chang SS, Guo WH, Kim Y, Wang YL. Guidance of cell migration by substrate dimension. Biophys J. 2013;104:313–321. doi: 10.1016/j.bpj.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doyle AD, Kutys ML, Conti MA, Matsumoto K, Adelstein RS, Yamada KM. Micro-environmental control of cell migration--myosin IIA is required for efficient migration in fibrillar environments through control of cell adhesion dynamics. J Cell Sci. 2012;125:2244–2256. doi: 10.1242/jcs.098806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doyle AD, Wang FW, Matsumoto K, Yamada KM. One-dimensional topography underlies three-dimensional fibrillar cell migration. J Cell Biol. 2009;184:481–490. doi: 10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ilina O, Bakker G-J, Vasaturo A, Hoffman RM, Friedl P. Two-photon laser-generated microtracks in 3D collagen lattices: principles of MMP-dependent and -independent collective cancer cell invasion. Physical Biology. 2011;8:029501–029501. doi: 10.1088/1478-3975/8/1/015010. [DOI] [PubMed] [Google Scholar]
- 7.Kraning-Rush CM, Carey SP, Lampi MC, Reinhart-King CA. Microfabricated collagen tracks facilitate single cell metastatic invasion in 3D. Integr Biol (Camb) 2013;5:606–616. doi: 10.1039/c3ib20196a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehrbar M, Sala A, Lienemann P, Ranga A, Mosiewicz K, Bittermann A, Rizzi SC, Weber FE, Lutolf MP. Elucidating the role of matrix stiffness in 3D cell migration and remodeling. Biophys J. 2011;100:284–293. doi: 10.1016/j.bpj.2010.11.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Legant WR, Miller JS, Blakely BL, Cohen DM, Genin GM, Chen CS. Measurement of mechanical tractions exerted by cells in three-dimensional matrices. Nat Methods. 2010;7:969–971. doi: 10.1038/nmeth.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hawkins R, Piel M, Faure-Andre G, Lennon-Dumenil A, Joanny J, Prost J, Voituriez R. Pushing off the Walls: A Mechanism of Cell Motility in Confinement. Physical Review Letters. 2009:102. doi: 10.1103/PhysRevLett.102.058103. [DOI] [PubMed] [Google Scholar]
- 11.Friedl P, Wolf K. Plasticity of cell migration: a multiscale tuning model. J Cell Biol. 2010;188:11–19. doi: 10.1083/jcb.200909003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hakkinen KM, Harunaga JS, Doyle AD, Yamada KM. Direct comparisons of the morphology, migration, cell adhesions, and actin cytoskeleton of fibroblasts in four different three-dimensional extracellular matrices. Tissue Eng Part A. 2011;17:713–724. doi: 10.1089/ten.tea.2010.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13••.Provenzano PP, Inman DR, Eliceiri KW, Trier SM, Keely PJ. Contact guidance mediated three-dimensional cell migration is regulated by Rho/ROCK-dependent matrix reorganization. Biophys J. 2008;95:5374–5384. doi: 10.1529/biophysj.108.133116. Illustrates the requirement of contractile force for regulating collagen reorganization by tumor cell but it is dispensible for migration in aligned matrices. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amatangelo MD, Bassi DE, Klein-Szanto AJ, Cukierman E. Stroma-derived three-dimensional matrices are necessary and sufficient to promote desmoplastic differentiation of normal fibroblasts. Am J Pathol. 2005;167:475–488. doi: 10.1016/S0002-9440(10)62991-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15••.Goetz JG, Minguet S, Navarro-Lerida I, Lazcano JJ, Samaniego R, Calvo E, Tello M, Osteso-Ibanez T, Pellinen T, Echarri A, et al. Biomechanical remodeling of the microenvironment by stromal caveolin-1 favors tumor invasion and metastasis. Cell. 2011;146:148–163. doi: 10.1016/j.cell.2011.05.040. Defines a novel role for caveloin-1 in regulating extracellular matrix remodeling in both in vitro and in vivo 3D model systems, and in both normal and cancerous cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 17.Palmgren B, Jiao Y, Novozhilova E, Stupp SI, Olivius P. Survival, migration and differentiation of mouse tau-GFP embryonic stem cells transplanted into the rat auditory nerve. Exp Neurol. 2012;235:599–609. doi: 10.1016/j.expneurol.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 18.Tysseling VM, Sahni V, Pashuck ET, Birch D, Hebert A, Czeisler C, Stupp SI, Kessler JA. Self-assembling peptide amphiphile promotes plasticity of serotonergic fibers following spinal cord injury. J Neurosci Res. 2010;88:3161–3170. doi: 10.1002/jnr.22472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palecek SP, Huttenlocher A, Horwitz AF, Lauffenburger DA. Physical and biochemical regulation of integrin release during rear detachment of migrating cells. J Cell Sci. 1998;111 (Pt 7):929–940. doi: 10.1242/jcs.111.7.929. [DOI] [PubMed] [Google Scholar]
- 20••.Palecek SP, Loftus JC, Ginsberg MH, Lauffenburger DA, Horwitz AF. Integrin-ligand binding properties govern cell migration speed through cell-substratum adhesiveness. Nature. 1997;385:537–540. doi: 10.1038/385537a0. A definitive study examining the regulation of 2D cell migration by cell/ECM interactions. [DOI] [PubMed] [Google Scholar]
- 21.Wolf K, Wu YI, Liu Y, Geiger J, Tam E, Overall C, Stack MS, Friedl P. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat Cell Biol. 2007;9:893–904. doi: 10.1038/ncb1616. [DOI] [PubMed] [Google Scholar]
- 22.Carey SP, Kraning-Rush CM, Williams RM, Reinhart-King CA. Biophysical control of invasive tumor cell behavior by extracellular matrix microarchitecture. Biomaterials. 2012;33:4157–4165. doi: 10.1016/j.biomaterials.2012.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miron-Mendoza M, Seemann J, Grinnell F. The differential regulation of cell motile activity through matrix stiffness and porosity in three dimensional collagen matrices. Biomaterials. 2010;31:6425–6435. doi: 10.1016/j.biomaterials.2010.04.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24••.Wolf K, te Lindert M, Krause M, Alexander S, te Riet J, Willis AL, Hoffman RM, Figdor CG, Weiss SJ, Friedl P. Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J Cell Biol. 2013:201. doi: 10.1083/jcb.201210152. in press. A well-controlled study that takes into account the effect of ECM regulators upon each other and demonstrates that matrix porosity is the limiting factor during 3D cell migration. It further illustrates that cancer cells require both cellular mechanotransduction and matrix proteolysis to adapt to ECM confinement. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedland JC, Lee MH, Boettiger D. Mechanically activated integrin switch controls alpha5beta1 function. Science. 2009;323:642–644. doi: 10.1126/science.1168441. [DOI] [PubMed] [Google Scholar]
- 26.Plotnikov SV, Pasapera AM, Sabass B, Waterman CM. Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration. Cell. 2012;151:1513–1527. doi: 10.1016/j.cell.2012.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwarz US, Gardel ML. United we stand: integrating the actin cytoskeleton and cell-matrix adhesions in cellular mechanotransduction. J Cell Sci. 2012;125:3051–3060. doi: 10.1242/jcs.093716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prager-Khoutorsky M, Lichtenstein A, Krishnan R, Rajendran K, Mayo A, Kam Z, Geiger B, Bershadsky AD. Fibroblast polarization is a matrix-rigidity-dependent process controlled by focal adhesion mechanosensing. Nat Cell Biol. 2011;13:1457–1465. doi: 10.1038/ncb2370. [DOI] [PubMed] [Google Scholar]
- 29.Pasapera AM, Schneider IC, Rericha E, Schlaepfer DD, Waterman CM. Myosin II activity regulates vinculin recruitment to focal adhesions through FAK-mediated paxillin phosphorylation. J Cell Biol. 2009;188:877–890. doi: 10.1083/jcb.200906012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lo CM, Wang HB, Dembo M, Wang YL. Cell movement is guided by the rigidity of the substrate. Biophys J. 2000;79:144–152. doi: 10.1016/S0006-3495(00)76279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peyton SR, Putnam AJ. Extracellular matrix rigidity governs smooth muscle cell motility in a biphasic fashion. J Cell Physiol. 2005;204:198–209. doi: 10.1002/jcp.20274. [DOI] [PubMed] [Google Scholar]
- 32.Arevalo RC, Urbach JS, Blair DL. Size-dependent rheology of type-I collagen networks. Biophys J. 2010;99:L65–67. doi: 10.1016/j.bpj.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33••.Petrie RJ, Gavara N, Chadwick RS, Yamada KM. Nonpolarized signaling reveals two distinct modes of 3D cell migration. J Cell Biol. 2012;197:439–455. doi: 10.1083/jcb.201201124. This paper is the first to show that RhoA, ROCK, and myosin II activity responds to the elastic behavior of 3D ECM to regulate signaling polarity and mode of migration in normal fibroblasts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Storm C, Pastore JJ, MacKintosh FC, Lubensky TC, Janmey PA. Nonlinear elasticity in biological gels. Nature. 2005;435:191–194. doi: 10.1038/nature03521. [DOI] [PubMed] [Google Scholar]
- 35.Pedersen JA, Swartz MA. Mechanobiology in the third dimension. Ann Biomed Eng. 2005;33:1469–1490. doi: 10.1007/s10439-005-8159-4. [DOI] [PubMed] [Google Scholar]
- 36.Giannone G, Dubin-Thaler BJ, Döbereiner H-G, Kieffer N, Bresnick AR, Sheetz MP. Periodic Lamellipodial Contractions Correlate with Rearward Actin Waves. Cell. 2004;116:431–443. doi: 10.1016/s0092-8674(04)00058-3. [DOI] [PubMed] [Google Scholar]
- 37.Wu C, Asokan Sreeja B, Berginski Matthew E, Haynes Elizabeth M, Sharpless Norman E, Griffith Jack D, Gomez Shawn M, Bear James E. Arp2/3 Is Critical for Lamellipodia and Response to Extracellular Matrix Cues but Is Dispensable for Chemotaxis. Cell. 2012;148:973–987. doi: 10.1016/j.cell.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giannone G, Dubin-Thaler BJ, Rossier O, Cai Y, Chaga O, Jiang G, Beaver W, Dobereiner HG, Freund Y, Borisy G, et al. Lamellipodial actin mechanically links myosin activity with adhesion-site formation. Cell. 2007;128:561–575. doi: 10.1016/j.cell.2006.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petrie RJ, Yamada KM. At the leading edge of three-dimensional cell migration. Journal of Cell Science. 2012;125:5917–5926. doi: 10.1242/jcs.093732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40•.Ulrich TA, Jain A, Tanner K, MacKay JL, Kumar S. Probing cellular mechanobiology in three-dimensional culture with collagen-agarose matrices. Biomaterials. 2010;31:1875–1884. doi: 10.1016/j.biomaterials.2009.10.047. Demonstrates that glioma cells can respond to linear elasticity in 3D collagen gels by changing their morphology. [DOI] [PubMed] [Google Scholar]
- 41.Sahai E, Marshall CJ. Differing modes of tumour cell invasion have distinct requirements for Rho/ROCK signalling and extracellular proteolysis. Nat Cell Biol. 2003;5:711–719. doi: 10.1038/ncb1019. [DOI] [PubMed] [Google Scholar]
- 42.Vega FM, Fruhwirth G, Ng T, Ridley AJ. RhoA and RhoC have distinct roles in migration and invasion by acting through different targets. J Cell Biol. 2011;193:655–665. doi: 10.1083/jcb.201011038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolf K, Friedl P. Extracellular matrix determinants of proteolytic and non-proteolytic cell migration. Trends Cell Biol. 2011;21:736–744. doi: 10.1016/j.tcb.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 44••.Sabeh F, Shimizu-Hirota R, Weiss SJ. Protease-dependent versus -independent cancer cell invasion programs: three-dimensional amoeboid movement revisited. J Cell Biol. 2009;185:11–19. doi: 10.1083/jcb.200807195. This paper provides a comprehensive overview of the unique extracellular matrix features that dictate the requirement for proteases in 3D migration. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Machacek M, Hodgson L, Welch C, Elliott H, Pertz O, Nalbant P, Abell A, Johnson GL, Hahn KM, Danuser G. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461:99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deakin NO, Turner CE. Distinct roles for paxillin and Hic-5 in regulating breast cancer cell morphology, invasion, and metastasis. Mol Biol Cell. 2011;22:327–341. doi: 10.1091/mbc.e10-09-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang H, Li A, Bi J, Veltman DM, Zech T, Spence HJ, Yu X, Timpson P, Insall RH, Frame MC, et al. Loss of Scar/WAVE complex promotes N-WASP- and FAK-dependent invasion. Curr Biol. 2013;23:107–117. doi: 10.1016/j.cub.2012.11.059. [DOI] [PubMed] [Google Scholar]
- 48.Mierke CT, Kollmannsberger P, Zitterbart DP, Diez G, Koch TM, Marg S, Ziegler WH, Goldmann WH, Fabry B. Vinculin facilitates cell invasion into three-dimensional collagen matrices. J Biol Chem. 2010;285:13121–13130. doi: 10.1074/jbc.M109.087171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fraley SI, Feng Y, Giri A, Longmore GD, Wirtz D. Dimensional and temporal controls of three-dimensional cell migration by zyxin and binding partners. Nat Commun. 2012;3:719. doi: 10.1038/ncomms1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ahn J, Sanz-Moreno V, Marshall CJ. The metastasis gene NEDD9 product acts through integrin beta3 and Src to promote mesenchymal motility and inhibit amoeboid motility. J Cell Sci. 2012;125:1814–1826. doi: 10.1242/jcs.101444. [DOI] [PubMed] [Google Scholar]
- 51.Zhong J, Baquiran JB, Bonakdar N, Lees J, Ching YW, Pugacheva E, Fabry B, O’Neill GM. NEDD9 stabilizes focal adhesions, increases binding to the extra-cellular matrix and differentially effects 2D versus 3D cell migration. PLoS One. 2012;7:e35058. doi: 10.1371/journal.pone.0035058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen L, Vicente-Manzanares M, Potvin-Trottier L, Wiseman PW, Horwitz AR. The integrin-ligand interaction regulates adhesion and migration through a molecular clutch. PLoS One. 2012;7:e40202. doi: 10.1371/journal.pone.0040202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gupton SL, Waterman-Storer CM. Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration. Cell. 2006;125:1361–1374. doi: 10.1016/j.cell.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 54.Grinnell F, Ho CH, Tamariz E, Lee DJ, Skuta G. Dendritic fibroblasts in three-dimensional collagen matrices. Mol Biol Cell. 2003;14:384–395. doi: 10.1091/mbc.E02-08-0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kubow KE, Klotzsch E, Smith ML, Gourdon D, Little WC, Vogel V. Crosslinking of cell-derived 3D scaffolds up-regulates the stretching and unfolding of new extracellular matrix assembled by reseeded cells. Integr Biol (Camb) 2009;1:635–648. doi: 10.1039/b914996a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Provenzano PP, Keely PJ. The role of focal adhesion kinase in tumor initiation and progression. Cell Adh Migr. 2009;3:347–350. doi: 10.4161/cam.3.4.9458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wolf K, Mazo I, Leung H, Engelke K, von Andrian UH, Deryugina EI, Strongin AY, Brocker EB, Friedl P. Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J Cell Biol. 2003;160:267–277. doi: 10.1083/jcb.200209006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DiMilla PA, Stone JA, Quinn JA, Albelda SM, Lauffenburger DA. Maximal migration of human smooth muscle cells on fibronectin and type IV collagen occurs at an intermediate attachment strength. J Cell Biol. 1993;122:729–737. doi: 10.1083/jcb.122.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pelham RJ, Jr, Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci U S A. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Even-Ram S, Doyle AD, Conti MA, Matsumoto K, Adelstein RS, Yamada KM. Myosin IIA regulates cell motility and actomyosin-microtubule crosstalk. Nat Cell Biol. 2007;9:299–309. doi: 10.1038/ncb1540. [DOI] [PubMed] [Google Scholar]
- 61.Sipes NS, Feng Y, Guo F, Lee HO, Chou FS, Cheng J, Mulloy J, Zheng Y. Cdc42 regulates extracellular matrix remodeling in three dimensions. J Biol Chem. 2011;286:36469–36477. doi: 10.1074/jbc.M111.283176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pankov R, Endo Y, Even-Ram S, Araki M, Clark K, Cukierman E, Matsumoto K, Yamada KM. A Rac switch regulates random versus directionally persistent cell migration. J Cell Biol. 2005;170:793–802. doi: 10.1083/jcb.200503152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meshel AS, Wei Q, Adelstein RS, Sheetz MP. Basic mechanism of three-dimensional collagen fibre transport by fibroblasts. Nat Cell Biol. 2005;7:157–164. doi: 10.1038/ncb1216. [DOI] [PubMed] [Google Scholar]