Abstract
The suprachiasmatic nucleus (SCN) is the primary circadian pacemaker in mammals. Individual SCN neurons in dispersed culture can generate independent circadian oscillations of clock gene expression and neuronal firing. However, SCN rhythmicity depends on sufficient membrane depolarization and levels of intracellular calcium and cAMP. In the intact SCN, cellular oscillations are synchronized and reinforced by rhythmic synaptic input from other cells, resulting in a reproducible topographic pattern of distinct phases and amplitudes specified by SCN circuit organization. The SCN network synchronizes its component cellular oscillators, reinforces their oscillations, responds to light input by altering their phase distribution, increases their robustness to genetic perturbations, and enhances their precision. Thus, even though individual SCN neurons can be cell-autonomous circadian oscillators, neuronal network properties are integral to normal function of the SCN.
Keywords: circadian, clock, coupling
Introduction
Mammalian physiology and behavior is organized in a daily program that allows coordinated anticipation of the 24 hr environmental light/dark cycle. To serve this purpose, mammalian cells contain “circadian clocks” composed of genes that interact in oscillatory transcriptional networks within cells (1, 2) and regulate the expression of many other genes critical for cell physiology and metabolism. Circadian regulation is remarkably pervasive: nearly half of all mammalian genes are expressed rhythmically in one or more tissues (3). In recent years, there has been a growing recognition of the importance of clock genes and circadian regulation for health; circadian clock genes have been directly implicated not only in sleep disorders but also in diabetes, cancer, and bipolar disorder (4).
For proper functioning of the circadian timing system, all the circadian clocks in the body must be kept synchronized with one another and to the 24 hr day; this is the function of the master circadian pacemaker, the suprachiasmatic nucleus (SCN) (5). Like other cells (6), SCN neurons can generate autonomous circadian rhythms (7). But SCN neurons are special in several important respects. First, they receive direct photic input from the retina, which allows them to synchronize to the day/night cycle (8). Second, they have distinct, topographically organized coupling mechanisms which allow them to remain synchronized to one another even in constant darkness (9). Third, they generate a pronounced circadian rhythm of neuronal firing frequency which allows them, through a variety of direct and indirect output pathways, to synchronize other cells throughout the body (10). Thus, the SCN master pacemaker synchronizes (“entrains”) to the light/dark cycle, and in turn synchronizes other subsidiary cellular oscillators. Further, as a result of internal coupling, the SCN also generates a coherent output signal even in the absence of a light/dark cycle, accounting for the “free-running” circadian (ca. 24 hr) rhythms of physiology and behavior that persist under constant conditions.
But the SCN is more than just a set of coupled oscillator cells receiving photic input and providing coherent rhythmic output. Even though SCN neurons can oscillate independently, they are designed to be “team players” within a tissue, depending more on input from other cells to generate rhythms than fibroblasts do (11). Through phase diversity among its constituent cells, the SCN pacemaker can provide differently phased output signals to different tissues (12). Furthermore, changes in phase relationships among SCN neurons can alter the strength or waveform of output rhythms at the whole animal level, accounting for effects of constant light (13), seasonal changes in day length (14), and certain mutations affecting SCN coupling (15). As a tissue, the SCN provides a much more precise rhythmic output signal than is generated by single cells (16). SCN rhythmic output is also more robust to genetic perturbations than that of single cells (17). In this review, we will discuss the function of the SCN as a master circadian pacemaker tissue, focusing on cell autonomy and network properties.
Basic Structure and Function
Intracellular mechanisms
The circadian clock that operates in mammalian cells is based on delayed negative feedback (18) in a core transcriptional feedback loop (4, 19). CLOCK/BMAL1 dimers act at E-box elements to promote transcription of a family of Period (Per1, Per2, Per3) and Cryptochrome (Cry1, Cry2) genes, leading to increases in PER and CRY levels. After delays associated with transcription, translation, dimerization, and nuclear entry, PER/CRY dimers inhibit transcription of their own genes. This, in concert with degradation regulated by the ubiquitin ligase complexes beta-TrCP1 and FBXL3, leads to declines in PER and CRY levels, thus relieving the inhibition and permitting a new cycle to begin. In addition to this core loop, another negative feedback loop, in which REV-ERBα acts at ROR elements to inhibit Bmal1 transcription, also contributes to clock precision and robustness. Recent work reveals yet another important negative feedback loop linking the clock to cell metabolism (20, 21). Thus, the intracellular circadian timekeeper can be conceptualized as a genetic network, a web of interconnected negative feedback loops regulating transcription of core clock genes (2) and output genes.
On the other hand, investigators have recently questioned the view of a mammalian clock based exclusively on transcriptional mechanisms (22), especially in light of the dramatic demonstration that cyanobacterial circadian clock function can be reconstituted in a cell-free system with only three proteins and ATP (23). In particular, membrane depolarization, intracellular calcium, and cAMP appear to be important regulators of the mammalian transcriptional clock. Neuronal firing rhythms in SCN slices are calcium-dependent (24, 25). Also, rhythmic expression of clock genes in SCN neurons requires sufficient membrane depolarization, cytoplasmic calcium, and/or cAMP (26–28). These effects may be mediated through calcium/cAMP response element binding protein (CREB), which when activated by phosphorylation binds to calcium/cAMP regulatory elements (CREs) on DNA. CRE sequences are found in the promoters of several core clock genes (29). Notably, CREs in Per1 and Per2 promoters bind CREB in SCN nuclear extracts (30). Furthermore, as membrane potential (31), calcium (25), and cAMP (28) are also rhythmic themselves in SCN, i.e. they are outputs of as well as inputs to the transcriptional clock, they may constitute positive feedback loops that contribute to rhythm generation, reinforcing the transcriptional cycle of the intracellular clock (11, 32, 33). In support of this view, holding cAMP constant even at high levels (forskolin + IBMX) damps rhythms of single neurons in SCN slices, suggesting that cAMP is not merely permissive for clock gene transcription, but that rhythmic drive from cAMP signaling makes a substantial contribution to the amplitude of clock gene rhythms, at least in SCN slices (28). Whether the same is true in dispersed SCN neurons, which lack rhythmic synaptic input from other cells, is not yet clear.
Structure
The SCN is a paired neuronal structure located in the anteroventral hypothalamus, on either side of the third ventricle, just above the optic chiasm (5). In the mouse, each unilateral SCN contains ~10,000 neurons in two anatomic subdivisions: a ventral “core” region which abuts the optic chiasm and receives retinal input, and a dorsal “shell” region which partially envelops and receives input from the core (34) (Figure 1). The core projects densely to the shell, which projects only sparsely back to the core (35). Neuronal cell bodies in the SCN are fairly small (~10 μm), have simple dendritic arbors, and may be closely apposed. Neurons in core and shell subregions are distinguished by neurochemical content. Within each unilateral core are ~1100 neurons containing vasoactive intestinal polypeptide (VIP; 10% of all SCN cells), and smaller numbers containing calretinin (900), neurotensin (NT; 700), and gastrin releasing peptide (GRP; 500). Within the shell are ~2100 neurons containing arginine vasopressin (AVP, ~20% of all SCN cells), ~1100 containing angiotensin II, and ~500 containing met-enkephalin. In most SCN neurons, neuropeptides are colocalized with GABA (36), though some cells may be glutamatergic (37). Most synapses among SCN neurons are GABAergic (38). SCN intrinsic anatomy and neuropeptides vary considerably across species (39), and are more complex than might be suggested by the simple core/shell terminology (40).
Figure 1.
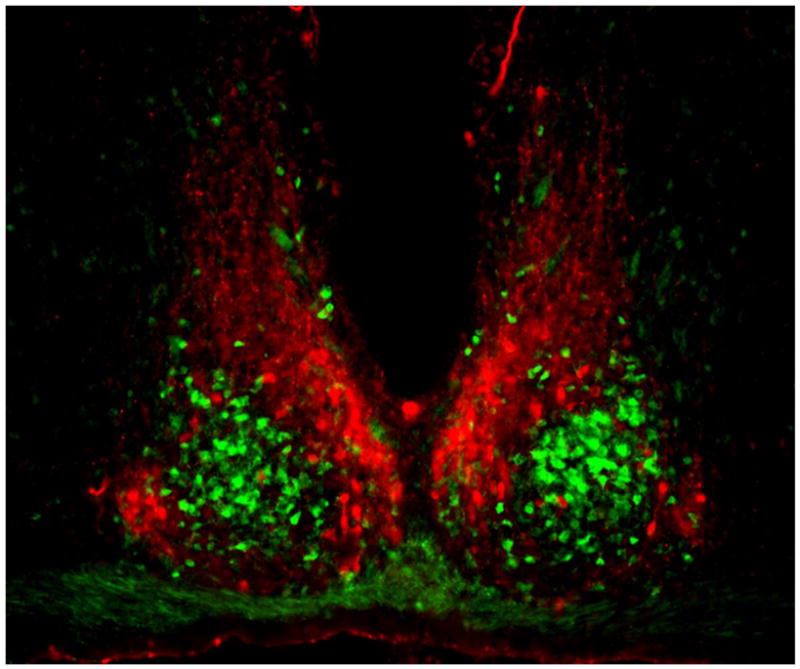
Coronal section of mouse SCN, showing the ventral core region delineated by green fluorescent protein (GFP) expressed in GRP neurons (green) and the dorsal shell region delineated by immunofluorescent labeling for AVP (red). Between left and right SCN is the third ventricle, and below is the optic chiasm. Reprinted from Karatsoreos et al. (185), with permission pending from the Society for Neuroscience.
Pacemaker function
Lesion and transplant studies have established that the SCN is the dominant circadian pacemaker in mammals. In rodents, complete SCN lesions abolish circadian rhythmicity in behavioral and endocrine variables (41, 42). Circadian rhythms can be partially restored to animals bearing SCN lesions by implantation of fetal SCN tissue (43), even in a genetically non-rhythmic host (44). In experiments using mutant animals with abnormally short or long circadian periods, the period of the restored rhythm is determined by the genotype of the SCN donor, not the SCN-lesioned host (44, 45). The SCN also generates pronounced circadian rhythms in frequency of spontaneous neuronal firing when physically isolated, either in vivo (46) or in vitro (47–49), although recent data indicate this may not be a unique property (50). The firing rhythm in SCN neurons is mediated partly by circadian regulation of membrane potassium channels (51).
Glia
The SCN is composed of glial cells as well as neurons, and glia could potentially contribute to pacemaker function. Glial astrocytes exhibit circadian rhythms of clock gene expression (52), and there are remarkable circadian variations in glial morphology in the SCN, as revealed by immunolabeling for glial fibrillary acidic protein (GFAP) (53). GFAP mutant mice have altered locomotor activity rhythms in constant light (54). Also, fluorocitrate, a drug that inhibits glial metabolism, disrupts neuronal firing rhythms in SCN slices (55). Moreover, glial signaling can affect neuronal function in other brain areas (56–58).
Input
In the retinohypothalamic tract, axons of special photosensitive retinal ganglion cells (59) project to the SCN, where they release glutamate and pituitary adenylate cyclase-activating polypeptide (PACAP) at synaptic contacts with SCN neurons (8). The neuropeptide PACAP has a modulatory role, enhancing the effects of glutamate (60). Glutamate acts at NMDA and AMPA receptors to depolarize the membrane, which leads both directly (through NMDA receptors) and indirectly (through voltage-sensitive channels) to calcium influx (61). Calcium activates a complex web of kinases (CamK, MAPK, PKA), leading to phosphorylation of CREB (62). Activated CREB then induces transcription by binding to calcium/cAMP response elements (CREs) of genes, in cooperation with accessory proteins such as CREB binding protein (CBP). In the SCN, CREB phosphorylation at Ser133 and Ser142 occurs within minutes of a light pulse, but only during the subjective night, when light pulses are effective in shifting the phase of the clock (63, 64). This circadian “gating” of light input is achieved at least partly through circadian changes in sensitivity to glutamate at the receptor level (65, 66). CREB targets (as reported by CRE:βGal) are induced in the SCN by light, again only during subjective night (32). Per1 and Per2 promoters contain CREs that bind CREB (30) and are induced by light in SCN (67–69). Behavioral phase shifts are attenuated in mutant S142A mice in which Ser142 phosphorylation cannot occur (64), and by CREB-decoy (70), Per1 antisense (70, 71), or Per2 antisense oligodeoxynucleotides (72) in SCN. As Per1 is induced within 5–15 min of a light pulse and is involved directly in the core clock feedback loop, its induction could be largely responsible for clock resetting. However, other CREB targets may also play a role: c-fos and other immediate early genes also have CREs and are induced by light in SCN (though not as rapidly as Per1), and c-fos + junB antisense can block phase shifts (73). In any event, CREB-mediated transcription appears to be essential for photic resetting of the SCN clock. Photic input is modulated by various non-photic inputs, notably from the thalamic intergeniculate leaflet (IGL) and the midbrain raphé (8).
Output
SCN output signals are largely mediated by circadian variation of neuronal firing and transmitter release at SCN axon terminals, but some data also indicate a role for humoral output. Modest circadian periodicity in locomotor activity can be restored after SCN lesions by implantation of fetal SCN tissue, even when encapsulated in a membrane which allows passage of small molecules but does not permit axonal fiber outgrowth into host tissue (74). SCN grafts do not, however, restore other rhythms (75). Furthermore, disruptions of SCN efferent axon function by knife cuts (46) or tetrodotoxin (TTX) (76) generally abolish circadian output rhythms, supporting a predominant role for specific neuronal projections in SCN output. The SCN core and shell regions send distinct efferent projections to fairly local primary targets, primarily in the medial areas of hypothalamus and thalamus (77) (Figure 1). Factors secreted from SCN which may regulate behavioral rhythms include: TGF-alpha (78), prokineticin-2 (79), and cardiotrophin-like cytokine (CLC) (80).
As a master pacemaker, the SCN synchronizes other oscillators throughout the brain (50) and peripheral tissues (10). This is accomplished through diverse pathways, including autonomic neural connections (81) and hormones (82–84), as well as less directly through circadian modulation of body temperature (85) and feeding behavior (86, 87). Rhythms in most tissues gradually damp out in the absence of the SCN (88). In the case of cultured fibroblasts, the damping is due to desynchrony among cells (6, 89). However, very sensitive reporters can still detect low amplitude rhythms in some tissues after many days in vitro (90), so local coupling mechanisms could still be present in some non-SCN tissues, particularly retina (91) and olfactory bulb (92), which can oscillate persistently without the SCN. Thus, SCN signals are required to coordinate rhythmicity among tissues, and also contribute to synchronizing cells within most tissues.
Some circadian rhythms in local tissues are driven by local circadian oscillators, but others are driven by the SCN. The olfactory bulb drives odor sensitivity rhythms in SCN-lesioned mice (93). Genetic manipulations selectively disabling local oscillators have revealed that local clocks drive physiological rhythms in the retina (94) and liver (95), and that circadian regulation of gene expression in liver is driven partly by the liver clock and partly by signals emanating from the SCN (96) that drive feeding rhythms. Thus, the SCN not only synchronizes other oscillators, but can also drive rhythms in other tissues.
SCN Cell Autonomy
Dispersed cells
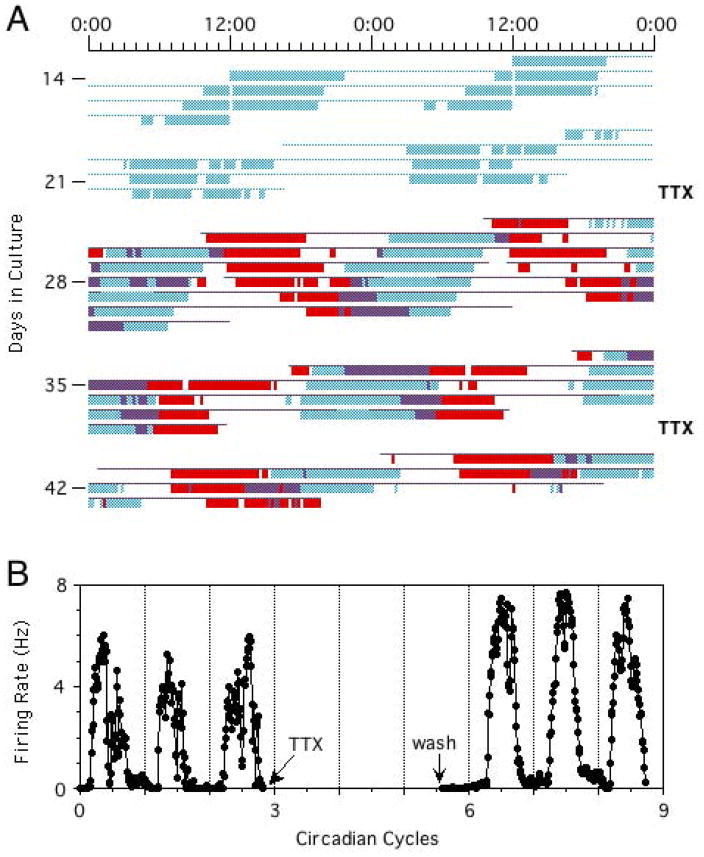
Dispersed SCN neurons generate independent circadian oscillations in rate of spontaneous neuronal firing, as revealed by multielectrode recordings (7, 97–99). Cells in the same culture dish show rhythms with a wide range of periods, and therefore progressively varying phase relationships (Figure 2). Dissociating SCN cells apparently releases them from the coupling forces that normally keep them synchronized in vivo and allows the cells to oscillate independently, despite the development of abundant functional synaptic connections in the cultures. Phases of individual cells are preserved even after blocking neuronal firing with TTX (7). Only at very high cell densities can some limited coupling be detected (100). At low density, phases of the cellular rhythms are randomly distributed. Thus, individual SCN neurons cultured at low density are autonomous circadian oscillators, in the sense that they do not require rhythmic input from other cells to generate circadian rhythms of firing rate.
Figure 2.
(A) Circadian rhythms of neuronal firing recorded from two SCN neurons in dispersed culture over a period of four weeks, showing that the cells oscillate independently with distinct circadian periods. Colored bars represent firing rates above the daily mean for the two cells (blue and red), with time of day plotted left to right and successive days top to bottom. The oscillations continue with unaltered phases after blockade of neuronal firing for 2.5 days by TTX (marked at right). (B) Circadian firing rhythm of one cell before and after TTX. Reprinted from Welsh et al. (7), with permission from Elsevier.
Period determination
The circadian period imposed by the SCN master pacemaker (44, 45) is determined at the level of a single cell, as revealed by studies of mutant SCN neurons. Behavioral circadian rhythms of tau mutant hamsters have short periods, and SCN neurons from those hamsters also have short periods (97). Behavioral circadian rhythms of Cry2−/− mice, on the other hand, have long periods, and SCN neurons from those mice also have long periods (17). Similar results apply for mice and SCN neurons heterozygous for the Clock d19 mutation, which exhibit long periods (99). Thus, the circadian period measured at the whole animal level reflects the intrinsic properties of circadian oscillators within individual SCN neurons; in this sense, circadian period is a cell-autonomous property.
PER2::LUC imaging
One limitation of multielectrode studies is that only a small minority of cells within a local area can be recorded, leaving open the possibility that small clusters of SCN neurons may form local coupled networks, and that these networks may be necessary for rhythmicity. This possibility was addressed by bioluminescence imaging of PER2 expression in dispersed SCN neurons (17). Cells were cultured from PER2::LUC knockin mice, expressing a reporter protein containing PER2 fused to firefly luciferase, to reflect both transcriptional and post-transcriptional regulation (90). In this study, phases of cells were randomly distributed by 6–7 days after a medium change (Movie S1). Even among cells less than 500 μm apart, cells that were closer together did not tend to have more similar phases or periods (17)(Welsh & Kay, unpublished results). This rules out any measurable local coupling, and confirms that SCN neurons are indeed autonomous oscillators, in the sense that rhythmic input is unnecessary for them to oscillate.
Isolated cells
Cells were not completely isolated in these experiments, however, so it was still possible that SCN neuron rhythms might require tonic input from other cells. This possibility was addressed in a recent study from Erik Herzog’s group, in which SCN neurons were cultured at various densities (101). In multielectrode array experiments, reducing cell density to ~500 cells/sq mm reduced the proportion of rhythmic cells from 61% to 33%. In addition, SCN neurons were cultured from PER2::LUC mice at very low density (100 cells/sq mm). In these cultures, only 27% of cells expressed PER2 at detectable levels for 3 days, but of these cells 65% (252/386) were rhythmic. In a few cases, a single rhythmic neuron was selected, and all other neurons in the same dish were mechanically ablated; 3 out of 5 of these isolated neurons remained rhythmic. Thus, tonic input from other cells is not absolutely required for SCN neurons to oscillate, but it does appear to increase levels of PER2 expression and the proportion of rhythmic cells.
Subtypes?
Not all SCN neurons are rhythmic, and rhythmic cells have different periods (7, 98); this raises the question of whether there might be stable functional subclasses of SCN neurons, identifiable on the basis of neuropeptide expression or other molecular markers. Early studies, however, found too high a proportion of rhythmic cells (50–60%) to be readily accounted for by any single neuropeptide population (7). Erik Herzog’s group recently addressed this issue by combining PER2::LUC imaging with immunolabeling for neuropeptides (101). They found no association between circadian phenotype (rhythmic/not, period, or amplitude) and neuropeptide expression (AVP or VIP). Moreover, mutant SCN neurons sometimes exhibit rare, intermittent PER2 oscillations (17), suggesting that rhythmicity could be a stochastic phenomenon even in wild type SCN neurons. Indeed, wild type SCN neurons can switch from rhythmic to non-rhythmic (or vice versa), either spontaneously or upon adding forskolin (101). Furthermore, PER2::LUC rhythms persist in a minority of SCN neurons during application of tetrodotoxin (TTX), but this is not a stable subpopulation (101). Thus, there is no evidence that cell-autonomous circadian phenotypes define stable, identifiable subtypes of SCN neurons.
Non-SCN cells
Cell-autonomous circadian oscillation is not exclusive to SCN neurons. Circadian rhythms have long been observed in unicellular organisms, including Gonyaulax (102, 103) and cyanobacteria (104, 105). In the latter, imaging of luciferase reporters confirms cell-autonomous oscillations (106). In isolated basal retinal neurons of the sea snail Bulla gouldiana, circadian rhythms of membrane conductance have been measured (by cross-sectional sampling) (107). Zebrafish cells exhibit cell-autonomous rhythms of clock gene expression, monitored by bioluminescence imaging (108). Chick pinealocytes in dispersed culture release melatonin in a circadian rhythm (109). In mammals, clock gene expression is rhythmic in many non-SCN tissues (88) and cell populations (110). Although non-SCN rhythms do tend to damp out gradually over time, low amplitude rhythms persist for many days (90). PER2::LUC imaging of cultured fibroblasts reveal that individual fibroblast oscillations persist without damping, and that the damping observed at the population level is due to desynchrony among cells (6). Cells that are closer together do not have more similar phases, but rather oscillate independently. Similar results were obtained in NIH3T3 cells using a fluorescent reporter of Rev-Erbα (89). Thus, like SCN neurons, mammalian fibroblasts (and probably most other cell types) are autonomous circadian oscillators, leading to the suggestion that perhaps “the rat-1 fibroblast should replace the SCN as the in vitro model of choice” (111).
Limits of autonomy
While the unassuming fibroblast may be a good substitute for the SCN neuron from the point-of-view of a molecular biologist studying molecular mechanisms of the intracellular clock (112), there is clearly much more to the SCN than its cell-autonomous circadian oscillation (113–115). Unlike a population of fibroblasts, the SCN is a neuronal network of coupled cellular oscillators, with a specific pattern of interconnections. As a tissue, the SCN shows specific modes of coupling mediated by specialized mechanisms, provides more robust and more precise time-keeping relative to single cells, responds to light input in complex and flexible ways, and produces variously phased output signals. In addition, unlike in other cells, the amplitude of the SCN intracellular clock is highly responsive to and dependent on intercellular signaling, which effectively reinforces the collective rhythmicity of the network and may silence errant cells. SCN neurons are not designed to function alone.
SCN Coupling
The most prominent network property of the SCN is the remarkable coupling of its constituent cellular oscillators (Figure 3) to produce a coherent circadian oscillation at the tissue level. In striking contrast to the independent oscillations of dissociated cells, neurons within SCN tissue adopt identical circadian periods and similar phases. Coordinated rhythms of clock gene expression can be measured by two complementary approaches: at one point in time for the entire SCN in vivo by in situ hybridization (mRNA) or immunohistochemistry (protein), or longitudinally in an SCN slice in vitro by bioluminescence imaging using Per1-luc or PER2::LUC reporters (116, 117) (Movie S2). PER cycling is much more pronounced in shell than in core, where it is reliably detected only by the more sensitive in vitro methods. In each circadian cycle, activation of PER expression begins in dorsomedial SCN, and proceeds ventrally and laterally, followed by marked activation of an “inner shell” or “cap” just dorsal to the core. Deactivation tends to proceed in reverse order, like a tide. This general pattern of dorsal AVP-rich shell peaking earlier than ventral VIP-rich core is consistent with patterns of neuropeptide release from SCN slices (118). A minority of cells, however, may be ~12 hrs out of phase with the population average (118–120). The precise relative phasing of individual cells is preserved from cycle to cycle, as well as after manipulations that delay or stop the clock (by inhibiting protein synthesis with cycloheximide) or severely damp gene expression rhythms (by inhibiting neuronal firing with TTX), suggesting that it is a function of SCN circuitry (116). One potential advantage of the dispersal of phase among different cell populations within the SCN is that it provides an opportunity for multiple, variously phased output signals for more flexible regulation of peripheral rhythms (12). Thus, when SCN tissue organization is preserved in vivo or in slice preparations in vitro, SCN neuron oscillations are generally coherent, but follow a complex, consistent pattern of distinct phases and amplitudes. This argues strongly that SCN coupling is mediated by specific neural circuits, as opposed to homogeneous coupling schemes involving uniform direct neuronal interactions (97, 121) or a diffusible coupling factor (122).
Figure 3.
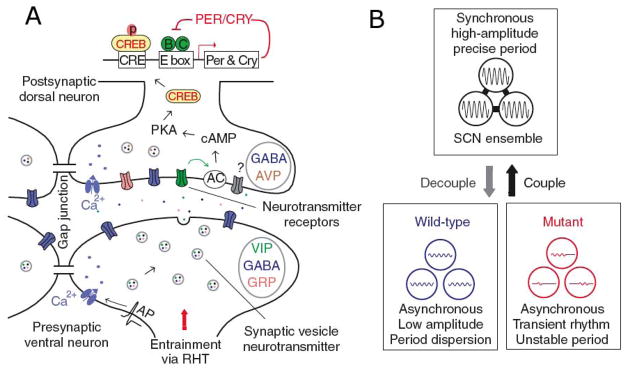
(A) Schematic diagram of intercellular signaling mechanisms within SCN, showing that action potentials arriving at synaptic terminals of core neurons trigger release of GABA and colocalized neuropeptides such as VIP or GRP. VIP, for example, binds to postsynaptic VPAC2 receptors of shell neurons, leading to membrane depolarization, elevated calcium and cAMP levels, phosphorylation of CREB, and induction of Per and Cry transcription, thus altering the phase and amplitude of the intracellular oscillator. Adjacent SCN neurons are also coupled by gap junctions. (B) The SCN network synchronizes and reinforces cellular oscillations, increases precision, and rescues rhythmicity of mutant oscillators. Reprinted from Liu et al. (209), with permission from McMillan Publishers Ltd.
Synapses
Consistent with the involvement of specific neuronal connections, there is evidence that neuronal firing and chemical synapses are required for circadian coupling within SCN. Blocking action potentials with TTX for several days allows SCN neurons in slice culture or high density dispersed culture to desynchronize (116, 123). In multielectrode studies of dispersed cells, excitatory or inhibitory synaptic interactions between cells can be detected by the presence of positive or negative cross-correlations of neuronal firing with short synaptic delays (7). In a study by Shirakawa et al. (124), circadian firing rhythms of 81 out of 126 SCN neuron pairs were not synchronized; short-term firing was not correlated within these pairs. Circadian firing rhythms of 42 pairs were synchronized with a small phase angle; for these pairs, cross-correlations indicated an excitatory synaptic interaction. Circadian firing rhythms of 3 pairs were synchronized with a larger phase angle; for these pairs, cross-correlations indicated an inhibitory interaction. Thus, there was a perfect correlation between circadian synchrony and presence of either excitatory or inhibitory synaptic interaction. These data strongly implicate neuronal firing and chemical synaptic interactions in SCN coupling.
Gap junctions
Electrical synapses (gap junctions) might also be involved in SCN coupling, though the evidence is less compelling. An early indication that non-synaptic mechanisms may be important was that circadian rhythms of metabolic activity (measured by 2-deoxyglucose uptake) appear uniform throughout E19 embryonic rat SCN (125), even though chemical synapses are rare in the SCN at this stage (<1/cell) (126). Also, short-term firing synchrony can be observed in SCN slices even when synaptic transmission is blocked by use of calcium-free medium (127). Subsequent studies have demonstrated dye coupling in SCN slices, i.e. dyes introduced into one neuron diffuse into others, presumably through gap junctions (128). The coupling is inhibited by hyperpolarization, is lower in subjective night phase, and occurs selectively within dorsal or ventral SCN but not between these subregions (129). There is also direct evidence of electrical coupling between SCN neurons, which is eliminated in knockout mice lacking the neuronal gap junction protein connexin-36 (Cx36) (130). Functionally, these knockout mice have weak locomotor activity rhythms, which would be consistent with compromised SCN coupling, but this has not been directly tested, and the relatively modest effect on behavioral rhythms has not yet been replicated using mice on a uniform genetic background. In SCN slices, the gap junction blockers octanol or halothane disrupt or damp circadian rhythms of neuronal firing or neuropeptide release (55, 131), but these agents are relatively non-specific. A recent study found modest numbers of very small Cx36-containing gap junctions between SCN neurons by freeze-fracture electron microscopy (132). These studies suggest that electrical synapses via gap junctions may contribute to SCN coupling, but conventional chemical synapses are probably more important.
Role of the core
The core region is crucial for maintaining coupling within the SCN. Selective lesions of the core abolish circadian rhythms of locomotor activity, body temperature, heart rate, melatonin, and cortisol, whereas similar lesions sparing the core do not (133, 134). When SCN slices are bisected into dorsal 2/3 and ventral 1/3 pieces, Per1-luc imaging shows that cells in the ventral piece remain synchronized, whereas cells in the dorsal piece are still rhythmic but lose synchrony (116). Similarly, in more rostral SCN slices lacking a core, PER2::LUC imaging shows low amplitude rhythms, without the precisely ordered spatiotemporal organization seen in slices containing both core and shell (117). Furthermore, studies of neuropeptide release from SCN slices show that the isolated dorsal (shell) SCN runs with a shorter circadian period than either the isolated ventral (core) SCN or the intact SCN slice containing both core and shell, suggesting that the core normally synchronizes the shell, imposing its longer period (135). This is consistent with anatomical data showing dense projections from core to shell but sparse projections from shell to core (35). This asymmetry could also be related to the finding that GABA is excitatory in the shell but inhibitory in the core (136). Thus, the core is important for coupling within the SCN; without this coupling, there is no coherent output signal from the SCN, and circadian rhythms at the whole animal level are abolished.
VIP
Within the core, VIP is the most prevalent neuropeptide transmitter, and recent studies strongly implicate VIP in SCN coupling. VIP is released rhythmically from the core (131) and acts through VPAC2 receptors in both core and shell (137). VIP depolarizes most SCN neurons by closing potassium channels (138). It also induces Per1 and Per2 expression, at least partly via protein kinase A (PKA) and CREB (139). In mice lacking VIP or its receptor, SCN cells are hyperpolarized (138) and have low levels of Per1 and Per2 expression (140). These mice also have weak behavioral rhythms (140, 141), often with multiple period components (100). In SCN slices from mutant mice, neuronal firing (142, 143) and clock gene rhythms (reported by Per1-GFP, Per1-luc, or PER2::LUC) (15, 144, 145) are suppressed, largely due to desynchrony among cells (15, 143, 144). The extent of desynchrony correlates (across genotype) with the degree of behavioral disruption in mice from which the slices are taken (145). Similar but less dramatic results are observed in high density dispersed SCN neurons: the limited coupling present in wild type cells is absent in mutant cells (100). Daily application of a VIP agonist to mutant SCN cultures restores synchrony (100), even in the presence of GABA antagonists (146). Finally, VIP application to (synchronized) wild type SCN in vivo (147) or in vitro (148, 149) produces phase-specific phase shifts similar to those of light. Thus, the synchronizing function of the SCN core requires VIP release from synaptic terminals at target neurons throughout the SCN.
GRP, NT, GABA
Other transmitters located in SCN core neurons may also play a role in SCN coupling. GRP, which is found in ~4% of SCN neurons, acts through BB2 receptors to induce Per1, Per2, and c-fos expression in the dorsal SCN (150). Like VIP, GRP produces a light-like phase-dependent pattern of phase shifts when applied to the SCN, either in vivo (150) or in vitro (151). In SCN slices from mice lacking receptors for VIP, acutely applied GRP induces Per1 expression and synchronizes cells (15). Neurotensin, which is present in ~7% of SCN neurons, also has phase-shifting effects on SCN slices (152). The most common transmitter in the SCN is GABA, which is present in most or perhaps all SCN neurons, including those of the core (36). Although GABA is classically inhibitory, it may be excitatory under some conditions in SCN (136, 153, 154). GABA, through GABA-A receptors, causes phase shifts when applied to dissociated SCN neurons, and daily application synchronizes them (155). On the other hand, neurons in SCN slices remain synchronized in the presence of GABA-A and GABA-B antagonists (146), indicating that GABA is sufficient but not necessary for SCN coupling. Finally, inhibiting Gi/o G-protein function with pertussis toxin (PTX) causes desynchronization of neurons in SCN slices, suggesting that another unidentified ligand may be important for SCN coupling (146). In summary, VIP is necessary for SCN coupling, but other transmitters including GRP, neurotensin, and GABA likely also contribute or play a modulatory role.
Effects of Light on SCN Coupling
SCN coupling is altered by light input. For example, the phase distribution of SCN neurons is different in slices from mice kept in a light/dark cycle vs. constant darkness (156). Also, shifts in the light/dark cycle (jet lag), seasonal changes in day length (photoperiod), or certain artificial lighting schedules perturb coupling in characteristic ways.
Jet lag
After a shift in the light/dark cycle, the retinorecipient SCN core shifts more quickly than the dorsal shell, which must rely on coupling signals from the shifted core. This lag of the shell, which accounts for part of the “lag” in jet lag, can be detected in vivo by in situ hybridization for clock genes (157) as well as in vitro by electrophysiology (136) or Per1-luc imaging (158). Single cell PER2::LUC imaging reveals that after a 6 hr advance of the light/dark cycle, SCN cells are initially more widely distributed in phase, that more dorsal cells shift more slowly, and that full resynchronization of SCN cells requires at least 8 days (159). Consistent with a role for VIP in this process, VIP application to the SCN produces light-like phase-dependent phase shifts in vivo (147) or in vitro (148, 149), and behavioral phase shifts after a shift of the light/dark cycle are accelerated in mice overexpressing the VPAC2 receptor for VIP (160). Mice lacking this receptor have reduced or absent behavioral phase shifts (141), although this could be due to aberrant effects of light signals in the core (161, 162). GABA may also contribute to this process, as the GABA-A antagonist bicuculline blocks transmission of excitation from core to shell during the re-synchronization after a delay of the light/dark cycle (136).
Photoperiod
Seasonal changes in day length (photoperiod) in rodents result in changes in waveform of circadian rhythms, notably locomotor activity and melatonin release, the latter being an important signal for seasonal changes in reproductive function and other physiological processes (163). These changes in waveform of output rhythms reflect changes within the SCN, including alterations in clock gene expression and neuronal firing rhythms (164). Based on analysis of activity rhythms in hamsters, Pittendrigh and Daan (165) proposed decades ago that photoperiod might be encoded in the phase relationship between separate evening (E) and morning (M) oscillators. Consistent with this idea, neuronal firing rhythms are broader (166) or even bimodal (167) in long photoperiod. Furthermore, single cell firing peaks are relatively narrow (168), and do not change with photoperiod (169), leading to the conclusion that photoperiod is encoded in the phase distribution of single cell oscillators with the SCN.
Regional differences in how SCN cell rhythms respond to photoperiod complicate this picture somewhat (170). For instance, under long photoperiod, single cell firing rhythms are indeed more widely distributed in ventral SCN, but the single unit profiles themselves are wider in dorsal SCN (171). Also, rostral and caudal SCN cells respond differently to changing photoperiod (172). Single cell Per1-luc imaging reveals that caudal SCN cells track activity offset (dawn), whereas rostral SCN cells track activity onset (dusk) but also adopt a bimodal phase distribution in long photoperiod (14). While the precise encoding of photoperiod is not yet fully understood, it is clear that the SCN responds to seasonal changes in day length by changing the phase distribution of its single cell oscillators.
By this mechanism, seasonal changes in day length alter the waveform of the SCN output signal, modulating behavioral rhythms and inducing melatonin-mediated changes in reproductive function (163). The seasonal change in single cell phasing also explains why the SCN is more sensitive to phase-shifting effects of light in winter: in short days, single cell rhythms of light sensitivity (“phase response curves”) are more tightly aligned in phase, leading to enhanced aggregate phase-shifting responses (173).
Partial decoupling
Exotic lighting schedules can induce various modes of partial SCN decoupling, revealing “fracture planes” of weakness in coupling among SCN subregions. Constant light can induce cellular desynchrony throughout the SCN (13, 174), and in hamsters sometimes induces “splitting”, in which two separate components of behavioral activity oscillate independently and then adopt a stable anti-phase relationship ~12 hrs apart. Remarkably, in situ hybridization reveals corresponding anti-phase rhythms of clock gene expression in left and right SCNs of split hamsters, indicating decoupling between left and right SCN (13, 175). Some cells within the core, however, seem to oscillate strongly in phase with the contralateral SCN in these animals (176). Short light/dark cycles (e.g., 22 hrs) can induce a different type of decoupling in rats, in which SCN core and shell oscillate independently (177). This has been used as an experimental tool to show that the circadian rhythm of REM sleep is predominantly under the control of the shell subregion (178). A similar phenomenon induced by even shorter light/dark cycles (e.g., 12 hrs) (179) may also reflect decoupling of core and shell (180). These data indicate that coupling is stronger within left and right SCN, and within core and shell, than across the boundaries of these subcompartments.
SCN Network Reinforces Cellular Rhythmicity
Stimuli activating CREB can affect amplitude as well as phase of cellular rhythms (181). Another critical feature of the SCN network is that it reinforces cellular rhythmicity in individual SCN neurons, through rhythmic input from other cells in the network. As noted above, rhythmic expression of clock genes in SCN neurons requires sufficient levels of membrane depolarization, cytoplasmic calcium, and cAMP (26, 28). Furthermore, at least in the case of cAMP, rhythmic drive (not just some minimal permissive level) is required for full amplitude clock gene rhythms in SCN slices (28). Therefore, either tonic or rhythmic synaptic input that depolarizes cells or increases calcium or cAMP levels, should reinforce cellular rhythmicity (11, 33). VIP, for example, which has an important role in SCN synchrony, increases cAMP levels and depolarizes SCN neurons (138), and would therefore be predicted to reinforce SCN cell rhythms. Conversely, manipulations that disrupt such intercellular signaling should impair rhythmicity in SCN neurons.
Effects of disrupted VIP signaling have been studied extensively in knockout mice lacking VIP or its VPAC2 receptor, and there is substantial evidence for impaired rhythms. Knockout mice have weak activity rhythms in constant darkness (100, 140, 141). Neuronal firing rhythms in SCN slices from these mice are suppressed (142), even at a single cell level (100, 143, 182). Neuronal firing rhythms are also weaker and less prevalent in dissociated SCN neurons (100). Clock gene rhythms of SCN neurons in slice cultures are also suppressed (15, 144), though not always (145). In dispersed cultures, rhythms can be restored by daily application of a VPAC2 receptor agonist (100). In slice cultures, they can be restored by application of GRP (15, 182) or depolarization with 40 mM potassium (15). One study found that firing rhythms were weaker in mice lacking the VPAC2 receptor than in mice lacking VIP until adding a receptor antagonist (143); this suggests that PACAP, a transmitter which acts at the same receptor and is released from retinal afferent terminals in the SCN core, may reinforce rhythms in the core, just as VIP reinforces rhythms in both core and shell.
Disrupting SCN intercellular communication by mechanical dissociation, TTX, or PTX also impairs rhythmicity. SCN neurons in low density dispersed culture exhibit a lower prevalence of rhythmic clock gene expression (~65%) (17, 101) than in SCN slice cultures (>95%) (15, 116). Blocking neuronal firing with TTX dramatically and reversibly damps clock gene rhythms in both SCN slice (116, 162) and dispersed cultures (Welsh & Kay, unpublished). Disrupting signaling via PTX-sensitive G proteins also damps rhythms of SCN cells in slices (146); the corresponding ligand is unknown, but to reinforce rhythms by reducing CREB activation it would have to act rhythmically and peak during subjective night.
The relative contributions of rhythmic vs. tonic input of the SCN network in reinforcing cellular rhythms is not clear, but decoupling is not invariably accompanied by suppression of rhythms. Constant light desynchronizes Per1-GFP rhythms of cells in SCN slices without substantially suppressing their amplitude (13, 174). Similar results were found in a recent study of knockout mice lacking VIP (145). Also, some completely isolated SCN neurons can generate strong PER2::LUC rhythms (101). It appears that if an SCN neuron is sufficiently depolarized, either autonomously or through synaptic input, then it will be rhythmic, but that rhythmic input can further reinforce rhythms.
Reinforcement of cellular rhythmicity by the SCN network is not uniform throughout the SCN; it is greater in the SCN shell than in the core. Clock gene rhythms are more prominent in the shell than in the core, whether measured in vivo (183–185) or in SCN slices (15, 117). Neuronal firing rhythms are also more prevalent in the dorsal shell than in the ventral core of SCN slices (118). However, when dissociated, AVP shell neurons and VIP core neurons do not differ in prevalence, amplitude, or period of PER2::LUC rhythms (101). This suggests that the position of these cells in the SCN circuit, and the associated balance of synaptic input, may alter membrane potential, calcium, cAMP, or other signals, thereby specifying the region-specific variations in rhythmic phenotypes seen in vivo or in SCN slices.
Robustness of SCN Network
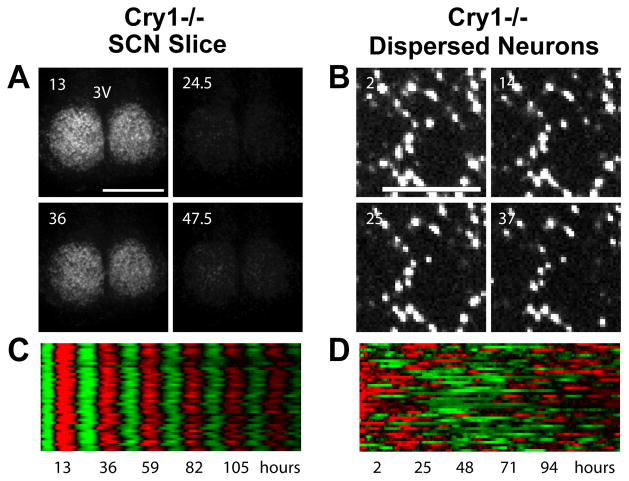
The coupled SCN network is more resistant to genetic perturbations than single SCN neurons. The first evidence for this principle came from a multielectrode study of SCN neurons from Clock d19 mutant mice (186), which express a dominant negative form of CLOCK (187). In this study, SCN neurons homozygous for this mutation were compared to wild type cells in both slice and dispersed cultures. As expected, cellular rhythms were less prevalent in dispersed cultures than in slices, due to loss of reinforcement by the SCN network. However, only 20% of mutant neurons were rhythmic in dispersed culture (vs. 46% of wild type cells), whereas 77% were rhythmic in slice cultures (vs. 95% of wild type cells). Thus, effects of the Clock d19 mutation are more pronounced in the absence of the SCN network. More dramatic results were found in a recent PER2::LUC imaging study of SCN neurons and fibroblasts from knockout mice lacking functional Cry and Per genes (17). In this study, dispersed Cry2−/− neurons (and fibroblasts) exhibited strong PER2::LUC rhythms with long period, similar to Cry2−/− neurons in SCN slices. In contrast, even though neurons in Per1−/− and Cry1−/− SCN slices also exhibited strong rhythms, dispersed Per1−/− and Cry1−/− neurons (and fibroblasts) had very weak rhythms (Figure 4). These mutant cells were able to generate only rare, intermittent rhythmicity, such that when sampled for 6–7 days only 5–10% had detectable rhythmicity. This indicates that the SCN network is substantially more robust to certain genetic perturbations, and probably other kinds of perturbations as well, compared to SCN neurons that are oscillating independently in dispersed culture.
Figure 4.
Patterns of PER2::LUC bioluminescence recorded from mutant Cry1−/− neurons in a cultured slice (A, C) or in dispersed culture (B, D) over a period of six days. Bioluminescence images are shown above (A, B), from indicated times in hours after start of the experiment. Raster plots are shown below (C, D), with each horizontal line representing one cell, and values above and below the mean shown in red and green, respectively. Cry1−/− neurons are rhythmic and synchronized in SCN slices, but are not rhythmic in dispersed culture. Figure from Liu et al. (17), with permission from Elsevier.
This is consistent with theoretical results showing that a multicellular, coupled SCN network should provide redundancy and increased robustness to noise (121, 188) and to variance of single cell periods (189), as well as to genetic perturbations that impair single cell rhythmicity (17).
Determination of a Precise Period
SCN neurons interact within the SCN to determine a compromise period in the intact SCN network. This is consistent with several studies showing that dispersed mutant cells exhibit periods similar to the behavioral period of corresponding mutant animals in vivo (17, 97, 99, 186). It has also been demonstrated directly in a genetic chimera experiment (190). In this study, SCNs of chimeric mice were composed of a mixture of wild type cells and Clock d19 mutant cells, which exhibit a long period in vitro. Behavioral rhythms of these mice often showed intermediate circadian periods. Thus, neurons of different periods can interact within an SCN network to determine a single compromise circadian period of the whole SCN pacemaker.
The period of the intact SCN pacemaker is more precise compared to the periods of independently oscillating SCN neurons. The idea that a coupled population of circadian oscillators can be more precise than its isolated components originated with Enright (191). When circadian rhythms of dispersed SCN neurons were first observed in multielectrode studies, it was noted that the cells exhibited a remarkably wide range of circadian periods, e.g. range 21–26 hrs, SD 1.2 hrs (7, 97–99, 192), substantially greater than cells in SCN slices, or mice on running wheels. Herzog et al. (16) examined this issue in detail in a study comparing neuronal firing rhythms of dispersed SCN neurons, Per1-luc rhythms of SCN slices, and running wheel activity rhythms of mice. The SDs of circadian period across samples were 1.28 hrs for cells, 0.32 hrs for slices, and 0.13 hrs for mice. Mice and slices with longer periods (near 24 hrs) were more precise. The median cycle-to-cycle SDs within samples were 2.07 hrs for cells, 0.66 hrs for slices, and 0.42 hrs for mice. Expressed as percentages of mean circadian period, these were 8.8% for cells, 2.7% for slices, and 1.8% for mice. Thus, mice were remarkably precise (cycle-to-cycle SD <2% of period), comparable to previous results (193), and mice or slices were significantly more precise than single cells.
Cycle-to-cycle variability of circadian period can be partitioned into pacemaker variance and variance due to other processes downstream from the pacemaker, using a procedure developed by Pittendrigh and Daan (193). This is based on the concept that a negative serial correlation (i.e., shorter cycles tend to follow longer cycles, and vice versa) reflects variability in lag time between pacemaker and output. Using this procedure, Herzog et al. (16) found that SDs for cycle-to-cycle variability due to the pacemaker alone were only 0.76 hrs for cells, 0.52 hrs for slices, and 0.32 hrs for mice. Thus, much of the extra variance seen in cells relative to slices or mice may be due to variance in processes downstream from the pacemaker, so it would be interesting to repeat this experiment using a more direct measure of clock function in single cells, such as PER2::LUC imaging.
Quantitative Models
Several models of the coupled SCN network have been developed. Even a model using very simple phase-only relaxation oscillators can capture a remarkable range of circadian behavior (194). However, most recent models use more realistic, heterogeneous cellular oscillators (17, 188, 189, 195, 196). In one model focusing on the special role of the core in SCN coupling, there are “gate cells” in the core and “oscillator cells” in the shell (197, 198). Gate cells integrate the output from oscillator cells; when this exceeds a threshold, it triggers a signal to synchronize oscillator cells. This model accounts for SCN synchrony in constant darkness and entrainment to a limited range of light/dark cycles.
One of the most realistic models so far is that of Bernard et al. (188), comprising a heterogeneous set of damped cellular oscillators, and a coupling agent that induces Per/Cry transcription through cAMP and CREB. Connectivity patterns are adjustable to simulate the three-dimensional (3D) SCN in vivo or the two-dimensional (2D) SCN slice, and there are separate core and shell compartments. In this model, synchronization is tighter for 3D than 2D configurations, and more robust with more than ~40 cells, or higher connectivity. Noise can either promote or impair synchrony, depending on whether it is uniform or not. After a shift of the light/dark cycle, there is a 2 day lag of the core and a 10 day lag of the shell. The period of the coupled system is approximately equal to the mean of the uncoupled single cells, but slower oscillators have higher amplitude, and thus a disproportionate effect on the period of the coupled system. Single cells, even though they are defined as damped oscillators, can oscillate autonomously with autocrine feedback, even without input from other cells. Reducing sensitivity to the coupling agent damps network oscillations, simulating TTX or VIP knockout results. The model predicts that cells will lose rhythmicity in the presence of constant high levels of the coupling agent. Another interesting prediction is that coupled cells either synchronize or damp, but do not run out of phase, so they do not interfere with the network.
A recent model by Locke et al. (189), a refinement of an earlier model by Gonze et al. (122), also makes some interesting predictions: 1) synchronization may be more efficient for damped oscillators than for sustained oscillators, 2) realistically fast resetting after a shift of the light/dark cycle is possible when VIP reinforces cellular rhythms by positive feedback, even if only some cells are directly retinorecipient, and 3) a system of coupled damped oscillators is robust to large variance of cellular oscillator periods.
Parallels in Drosophila
The function and organization of the SCN network has striking similarities to the Drosophila circadian system. At the cellular level, the mammalian and Drosophila clocks are both composed of a web of interlocked transcriptional feedback loops, and many of the molecular components of the Drosophila clock have one or more homologs in the mammalian clock (199). Depolarization, calcium, and cAMP are important for intracellular clock function in both SCN and Drosophila clocks, and the mammalian work in this area has been inspired by Drosophila studies (200); Harrisingh, 2007 #7473; Levine, 1994 #3239]. Just as remarkable are similarities at the network level (201, 202). In the SCN, ventral cells containing the neuropeptide VIP transmit photic information to dorsal cells, and photoperiod is encoded in phase distribution of component cellular oscillators. Similarly, in the Drosophila clock, ventral cells containing the neuropeptide pigment dispersing factor (PDF) transmit photic information to dorsal cells, and photoperiod is again encoded in phase distribution of component cellular oscillators (203, 204). Mice lacking VIP have disrupted behavioral rhythms; SCN neurons from these mice are uncoupled and show impaired rhythmicity. Similarly, flies lacking PDF have disrupted behavioral rhythms (205); clock neurons from these flies are uncoupled and show impaired rhythmicity (206–208). Thus, the mammalian SCN is remarkably similar to the Drosophila circadian clock; in the coming years, advances in one species will continue to inspire investigators studying the other, as we continue to explore the mysteries of the circadian clock.
Conclusion
The suprachiasmatic nucleus (SCN) is the primary circadian pacemaker in mammals. Individual SCN neurons are capable of generating independent circadian oscillations of neuronal firing rate and clock gene expression in dispersed culture. On this basis, it is commonly agreed that single SCN neurons are autonomous circadian oscillators. However, SCN neurons deficient in certain molecular clock components (Cry1−/− or Per1−/−) can generate only rare, intermittent oscillations in dispersed culture, whereas rhythmicity is intact in Cry1−/− or Per1−/− SCN slice preparations or in vivo. This suggests that SCN network properties can compensate for certain genetic defects, conferring on the SCN a robustness to perturbations that is lacking in single cells. This same principle may also apply to genetically intact SCN cells, as clock gene rhythms are substantially more prevalent among wild type SCN neurons in slice preparations (>95%) than in dispersed culture (~65%). Furthermore, the circadian periods of dispersed SCN neurons show substantial variability, both among cells and from cycle to cycle in a single cell, whereas circadian periods are synchronized and more precise when SCN tissue organization is preserved. Thus, it is clear that SCN network properties are important for synchronizing SCN cellular oscillators to one another to produce a precise, coherent output signal, as well as for synchronizing to the environmental light/dark cycle. Genetic, pharmacological, and selective dissection studies indicate that the ventral core region of the SCN plays an important role in this synchronization process. The complexity of the SCN circuits required for such synchronization is illustrated by the specific, topographically ordered sequence of clock gene activation across the SCN, which depends on an intact SCN core. Thus, even though individual SCN neurons can be cell-autonomous oscillators, neuronal network properties are integral to normal function of the SCN.
Supplementary Material
Circadian rhythms of PER2::LUC bioluminescence recorded from mouse SCN neurons in dispersed culture over a period of two weeks, showing that the cells oscillate independently. Bioluminescence intensity of one cell highlighted near the center of the image is plotted below.
Circadian rhythms of PER2::LUC bioluminescence recorded from mouse SCN neurons in a cultured slice over a period of seven days, showing that the cells oscillate with the same periods and similar phases, in a complex spatiotemporal pattern. This is a coronal slice of the ventral hypothalamus, including the bright bioluminescent left and right SCN, the third ventricle between them, and the optic chiasm below.
Summary Points.
SCN neurons in dispersed culture can generate independent circadian oscillations of clock gene expression and neuronal firing.
However, SCN neurons are not always rhythmic: sufficient depolarization, cytoplasmic calcium, and cAMP levels are required to sustain oscillations.
In the SCN network, synaptic interactions synchronize and reinforce cellular oscillations, producing a specific topographic pattern of oscillations of distinct phases and amplitudes.
The retinorecipient ventral core of the SCN and the VIPergic neurons it contains play a particularly important role in synchronizing SCN cells to one another and to the light/dark cycle.
Light input to the SCN alters the coupling and phase distribution of its component cellular oscillators in characteristic ways that reveal the network’s structure and function.
The SCN network increases the robustness of cellular oscillators to genetic perturbations and enhances their precision.
Many features of SCN function and organization have parallels in Drosophila circadian pacemaker neurons.
Acknowledgments
Supported by NIH grants R01 MH082945 (DKW), P50 MH074924 (JST), R01 MH078024 (JST), and R01 MH51573 (SAK), and a V.A. Career Development Award to DKW. JST is an Investigator in the Howard Hughes Medical Institute.
Glossary
- cAMP
3′-5′-cyclic adenosine monophosphate
- ATP
adenosine triphosphate
- AVP
arginine vasopressin
- CBP
CREB binding protein
- CRE
calcium/cAMP response element
- CREB
calcium/cAMP response element binding protein
- GABA
gamma-aminobutyric acid
- GFP
green fluorescent protein
- GRP
gastrin releasing peptide
- IGL
intergeniculate leaflet of the lateral geniculate
- NT
neurotensin
- PACAP
pituitary adenylate cyclase-activating peptide
pigment dispersing factor
- PTX
pertussis toxin
- REM sleep
rapid eye movement sleep
- SCN
suprachiasmatic nucleus
- TTX
tetrodotoxin
- VIP
vasoactive intestinal polypeptide
References
- 1.Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Gen. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ueda HR, et al. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005;37:187–92. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 3.Yan J, Wang H, Liu Y, Shao C. Analysis of gene regulatory networks in the mammalian circadian rhythm. PLoS Comput Biol. 2008;4:e1000193. doi: 10.1371/journal.pcbi.1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–75. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein DC, Moore RY, Reppert SM, editors. Suprachiasmatic Nucleus: The Mind’s Clock. New York: Oxford University Press; 1991. [Google Scholar]
- 6.Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol. 2004;14:2289–95. doi: 10.1016/j.cub.2004.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Welsh DK, Logothetis DE, Meister M, Reppert SM. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron. 1995;14:697–706. doi: 10.1016/0896-6273(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 8.Morin LP, Allen CN. The circadian visual system, 2005. Brain Res Rev. 2006;51:1–60. doi: 10.1016/j.brainresrev.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Aton SJ, Herzog ED. Come together, right... now: synchronization of rhythms in a mammalian circadian clock. Neuron. 2005;48:531–4. doi: 10.1016/j.neuron.2005.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gachon F, Nagoshi E, Brown SA, Ripperger J, Schibler U. The mammalian circadian timing system: from gene expression to physiology. Chromosoma. 2004;113:103–12. doi: 10.1007/s00412-004-0296-2. [DOI] [PubMed] [Google Scholar]
- 11.Hastings MH, Maywood ES, O’Neill JS. Cellular circadian pacemaking and the role of cytosolic rhythms. Curr Biol. 2008;18:R805–R815. doi: 10.1016/j.cub.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 12.Kalsbeek A, et al. SCN outputs and the hypothalamic balance of life. J Biol Rhythms. 2006;21:458–69. doi: 10.1177/0748730406293854. [DOI] [PubMed] [Google Scholar]
- 13.Ohta H, Yamazaki S, McMahon DG. Constant light desynchronizes mammalian clock neurons. Nature Neurosci. 2005;30:30. doi: 10.1038/nn1395. [DOI] [PubMed] [Google Scholar]
- 14.Inagaki N, Honma S, Ono D, Tanahashi Y, Honma K. Separate oscillating cell groups in mouse suprachiasmatic nucleus couple photoperiodically to the onset and end of daily activity. Proc Natl Acad Sci U S A. 2007;104:7664–9. doi: 10.1073/pnas.0607713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maywood ES, et al. Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Curr Biol. 2006;16:599–605. doi: 10.1016/j.cub.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 16.Herzog ED, Aton SJ, Numano R, Sakaki Y, Tei H. Temporal precision in the mammalian circadian system: a reliable clock from less reliable neurons. J Biol Rhythms. 2004;19:35–46. doi: 10.1177/0748730403260776. [DOI] [PubMed] [Google Scholar]
- 17.Liu AC, et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–16. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sato TK, et al. Feedback repression is required for mammalian circadian clock function. Nat Genet. 2006;38:312–9. doi: 10.1038/ng1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siepka SM, Yoo SH, Park J, Lee C, Takahashi JS. Genetics and neurobiology of circadian clocks in mammals. Cold Spring Harb Symp Quant Biol. 2007;72:251–9. doi: 10.1101/sqb.2007.72.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramsey KM, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324:651–4. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–7. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lakin-Thomas PL. Transcriptional feedback oscillators: maybe, maybe not. J Biol Rhythms. 2006;21:83–92. doi: 10.1177/0748730405286102. [DOI] [PubMed] [Google Scholar]
- 23.Nakajima M, et al. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–5. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 24.Shibata S, Newman GC, Moore RY. Effects of calcium ions on glucose utilization in the rat suprachiasmatic nucleus in vitro. Brain Res. 1987;426:332–338. doi: 10.1016/0006-8993(87)90886-9. [DOI] [PubMed] [Google Scholar]
- 25.Ikeda M, et al. Circadian dynamics of cytosolic and nuclear Ca2+ in single suprachiasmatic nucleus neurons. Neuron. 2003;38:253–63. doi: 10.1016/s0896-6273(03)00164-8. [DOI] [PubMed] [Google Scholar]
- 26.Lundkvist GB, Kwak Y, Davis EK, Tei H, Block GD. A calcium flux is required for circadian rhythm generation in mammalian pacemaker neurons. J Neurosci. 2005;25:7682–6. doi: 10.1523/JNEUROSCI.2211-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nahm SS, Farnell YZ, Griffith W, Earnest DJ. Circadian regulation and function of voltage-dependent calcium channels in the suprachiasmatic nucleus. J Neurosci. 2005;25:9304–8. doi: 10.1523/JNEUROSCI.2733-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Neill JS, Maywood ES, Chesham JE, Takahashi JS, Hastings MH. cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science. 2008;320:949–53. doi: 10.1126/science.1152506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X, et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci U S A. 2005;102:4459–64. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Travnickova-Bendova Z, Cermakian N, Reppert SM, Sassone-Corsi P. Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc Natl Acad Sci U S A. 2002;99:7728–33. doi: 10.1073/pnas.102075599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Jeu M, Hermes N, Pennartz C. Circadian modulation of membrane properties in slices of rat suprachiasmatic nucleus. Neuroreport. 1998;9:3725–3729. doi: 10.1097/00001756-199811160-00028. [DOI] [PubMed] [Google Scholar]
- 32.Obrietan K, Impey S, Smith D, Athos J, Storm DR. Circadian regulation of cAMP response element-mediated gene expression in the suprachiasmatic nuclei. Journal of Biological Chemistry. 1999;274:17748–56. doi: 10.1074/jbc.274.25.17748. [DOI] [PubMed] [Google Scholar]
- 33.Nitabach MN, Holmes TC, Blau J. Membranes, ions, and clocks: testing the Njus-Sulzman-Hastings model of the circadian oscillator. Methods Enzymol. 2005;393:682–93. doi: 10.1016/S0076-6879(05)93036-X. [DOI] [PubMed] [Google Scholar]
- 34.Abrahamson EE, Moore RY. Suprachiasmatic nucleus in the mouse: retinal innervation, intrinsic organization and efferent projections. Brain Res. 2001;916:172–91. doi: 10.1016/s0006-8993(01)02890-6. [DOI] [PubMed] [Google Scholar]
- 35.Leak RK, Card JP, Moore RY. Suprachiasmatic pacemaker organization analyzed by viral transynaptic transport. Brain Research. 1999;819:23–32. doi: 10.1016/s0006-8993(98)01317-1. [DOI] [PubMed] [Google Scholar]
- 36.Moore RY, Speh JC. GABA is the principal neurotransmitter of the circadian system. Neurosci Lett. 1993;150:112–116. doi: 10.1016/0304-3940(93)90120-a. [DOI] [PubMed] [Google Scholar]
- 37.Cui LN, Coderre E, Renaud LP. Glutamate and GABA mediate suprachiasmatic nucleus inputs to spinal-projecting paraventricular neurons. Am J Physiol. 2001;281:R1283–R1289. doi: 10.1152/ajpregu.2001.281.4.R1283. [DOI] [PubMed] [Google Scholar]
- 38.Strecker GJ, Wuarin JP, Dudek FE. GABA(A)-mediated local synaptic pathways connect neurons in the rat suprachiasmatic nucleus. J Neurophysiol. 1997;78:2217–2220. doi: 10.1152/jn.1997.78.4.2217. [DOI] [PubMed] [Google Scholar]
- 39.Morin LP, Shivers KY, Blanchard JH, Muscat L. Complex organization of mouse and rat suprachiasmatic nucleus. Neuroscience. 2006;137:1285–97. doi: 10.1016/j.neuroscience.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 40.Morin LP. SCN organization reconsidered. J Biol Rhythms. 2007;22:3–13. doi: 10.1177/0748730406296749. [DOI] [PubMed] [Google Scholar]
- 41.Moore RY, Eichler VB. Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic nucleus lesions in the rat. Brain Res. 1972;42:201–206. doi: 10.1016/0006-8993(72)90054-6. [DOI] [PubMed] [Google Scholar]
- 42.Stephan FK, Zucker I. Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc Natl Acad Sci USA. 1972;69:1583–1586. doi: 10.1073/pnas.69.6.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lehman MN, et al. Circadian rhythmicity restored by neural transplant. Immunocytochemical characterization of the graft and its integration with the host brain. J Neurosci. 1987;7:1626–1638. doi: 10.1523/JNEUROSCI.07-06-01626.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sujino M, et al. Suprachiasmatic nucleus grafts restore circadian behavioral rhythms of genetically arrhythmic mice. Curr Biol. 2003;13:664–8. doi: 10.1016/s0960-9822(03)00222-7. [DOI] [PubMed] [Google Scholar]
- 45.Ralph MR, Foster RG, Davis FC, Menaker M. Transplanted suprachiasmatic nucleus determines circadian period. Science. 1990;247:975–978. doi: 10.1126/science.2305266. [DOI] [PubMed] [Google Scholar]
- 46.Inouye ST, Kawamura H. Persistence of circadian rhythmicity in a mammalian hypothalamic “island” containing the suprachiasmatic nucleus. Proc Natl Acad Sci USA. 1979;76:5962–5966. doi: 10.1073/pnas.76.11.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Green DJ, Gillette R. Circadian rhythm of firing rate recorded from single cells in the rat suprachiasmatic brain slice. Brain Res. 1982;245:198–200. doi: 10.1016/0006-8993(82)90361-4. [DOI] [PubMed] [Google Scholar]
- 48.Groos G, Hendriks J. Circadian rhythms in electrical discharge of rat suprachiasmatic neurones recorded in vitro. Neurosci Lett. 1982;34:283–288. doi: 10.1016/0304-3940(82)90189-6. [DOI] [PubMed] [Google Scholar]
- 49.Shibata S, Oomura Y, Kita H, Hattori K. Circadian rhythmic changes of neuronal activity in the suprachiasmatic nucleus of the rat hypothalamic slice. Brain Res. 1982;247:154–158. doi: 10.1016/0006-8993(82)91041-1. [DOI] [PubMed] [Google Scholar]
- 50.Guilding C, Piggins HD. Challenging the omnipotence of the suprachiasmatic timekeeper: are circadian oscillators present throughout the mammalian brain? Eur J Neurosci. 2007;25:3195–216. doi: 10.1111/j.1460-9568.2007.05581.x. [DOI] [PubMed] [Google Scholar]
- 51.Kuhlman SJ, McMahon DG. Encoding the ins and outs of circadian pacemaking. J Biol Rhythms. 2006;21:470–81. doi: 10.1177/0748730406294316. [DOI] [PubMed] [Google Scholar]
- 52.Prolo LM, Takahashi JS, Herzog ED. Circadian rhythm generation and entrainment in astrocytes. J Neurosci. 2005;25:404–8. doi: 10.1523/JNEUROSCI.4133-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lavialle M, Servière J. Circadian fluctuations in GFAP distribution in the Syrian hamster suprachiasmatic nucleus. NeuroReport. 1993;4:1243–1246. doi: 10.1097/00001756-199309000-00008. [DOI] [PubMed] [Google Scholar]
- 54.Moriya T, et al. Involvement of glial fibrillary acidic protein (GFAP) expressed in astroglial cells in circadian rhythm under constant lighting conditions in mice. J Neurosci Res. 2000;60:212–8. doi: 10.1002/(SICI)1097-4547(20000415)60:2<212::AID-JNR10>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 55.Prosser RA, Edgar DM, Heller HC, Miller JD. A possible glial role in the mammalian circadian clock. Brain Res. 1994;643:296–301. doi: 10.1016/0006-8993(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 56.Nedergaard M. Direct signaling from astrocytes to neurons in cultures of mammalian brain cells. Science. 1994;263:1768–1771. doi: 10.1126/science.8134839. [DOI] [PubMed] [Google Scholar]
- 57.Fellin T, Carmignoto G. Neurone-to-astrocyte signalling in the brain represents a distinct multifunctional unit. J Physiol. 2004;559:3–15. doi: 10.1113/jphysiol.2004.063214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pascual O, et al. Astrocytic purinergic signaling coordinates synaptic networks. Science. 2005;310:113–6. doi: 10.1126/science.1116916. [DOI] [PubMed] [Google Scholar]
- 59.Berson DM. Strange vision: ganglion cells as circadian photoreceptors. Trends Neurosci. 2003;26:314–20. doi: 10.1016/S0166-2236(03)00130-9. [DOI] [PubMed] [Google Scholar]
- 60.Michel S, Itri J, Han JH, Gniotczynski K, Colwell CS. Regulation of glutamatergic signalling by PACAP in the mammalian suprachiasmatic nucleus. BMC Neurosci. 2006;7:15. doi: 10.1186/1471-2202-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meijer JH, Schwartz WJ. In search of the pathways for light-induced pacemaker resetting in the suprachiasmatic nucleus. J Biol Rhythms. 2003;18:235–49. doi: 10.1177/0748730403018003006. [DOI] [PubMed] [Google Scholar]
- 62.Carlezon WA, Jr, Duman RS, Nestler EJ. The many faces of CREB. Trends Neurosci. 2005;28:436–45. doi: 10.1016/j.tins.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 63.Ginty DD, et al. Regulation of CREB phosphorylation in the suprachiasmatic nucleus by light and a circadian clock. Science. 1993;260:238–241. doi: 10.1126/science.8097062. [DOI] [PubMed] [Google Scholar]
- 64.Gau D, et al. Phosphorylation of CREB Ser142 regulates light-induced phase shifts of the circadian clock. Neuron. 2002;34:245–53. doi: 10.1016/s0896-6273(02)00656-6. [DOI] [PubMed] [Google Scholar]
- 65.Colwell CS. NMDA-evoked calcium transients and currents in the suprachiasmatic nucleus: gating by the circadian system. Eur J Neurosci. 2001;13:1420–1428. doi: 10.1046/j.0953-816x.2001.01517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pennartz CM, Hamstra R, Geurtsen AM. Enhanced NMDA receptor activity in retinal inputs to the rat suprachiasmatic nucleus during the subjective night. J Physiol. 2001;532:181–94. doi: 10.1111/j.1469-7793.2001.0181g.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shearman LP, Zylka MJ, Weaver DR, Kolakowski LF, Jr, Reppert SM. Two period homologs: circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron. 1997;19:1261–1269. doi: 10.1016/s0896-6273(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 68.Shigeyoshi Y, et al. Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mper1 transcript. Cell. 1997;91:1043–1053. doi: 10.1016/s0092-8674(00)80494-8. [DOI] [PubMed] [Google Scholar]
- 69.Albrecht U, Sun ZS, Eichele G, Lee CC. A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell. 1997;91:1055–1064. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 70.Tischkau SA, Mitchell JW, Tyan SH, Buchanan GF, Gillette MU. Ca2+/cAMP response element-binding protein (CREB)-dependent activation of Per1 is required for light-induced signaling in the suprachiasmatic nucleus circadian clock. J Biol Chem. 2003;278:718–23. doi: 10.1074/jbc.M209241200. [DOI] [PubMed] [Google Scholar]
- 71.Akiyama M, et al. Inhibition of light- or glutamate-induced mPer1 expression represses the phase shifts into the mouse circadian locomotor and suprachiasmatic firing rhythms. J Neurosci. 1999;19:1115–21. doi: 10.1523/JNEUROSCI.19-03-01115.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wakamatsu H, et al. Additive effect of mPer1 and mPer2 antisense oligonucleotides on light-induced phase shift. Neuroreport. 2001;12:127–31. doi: 10.1097/00001756-200101220-00033. [DOI] [PubMed] [Google Scholar]
- 73.Wollnik F, et al. Block of c-fos and junB expression by antisense oligonucleotides inhibits light-induced phase shifts of the mammalian circadian clock. Eur J Neurosci. 1995;7:388–393. doi: 10.1111/j.1460-9568.1995.tb00334.x. [DOI] [PubMed] [Google Scholar]
- 74.Silver R, LeSauter J, Tresco PA, Lehman MN. A diffusible coupling signal from the transplanted suprachiasmatic nucleus controlling circadian locomotor rhythms. Nature. 1996;382:810–813. doi: 10.1038/382810a0. [DOI] [PubMed] [Google Scholar]
- 75.Meyer-Bernstein EL, et al. Effects of suprachiasmatic transplants on circadian rhythms of neuroendocrine function in golden hamsters. Endocrinology. 1999;140:207–18. doi: 10.1210/endo.140.1.6428. [DOI] [PubMed] [Google Scholar]
- 76.Schwartz WJ, Gross RA, Morton MT. The suprachiasmatic nuclei contain a tetrodotoxin-resistant circadian pacemaker. Proc Natl Acad Sci USA. 1987;84:1694–1698. doi: 10.1073/pnas.84.6.1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Leak RK, Moore RY. Topographic organization of suprachiasmatic nucleus projection neurons. J Comp Neurol. 2001;433:312–334. doi: 10.1002/cne.1142. [DOI] [PubMed] [Google Scholar]
- 78.Kramer A, et al. Regulation of daily locomotor activity and sleep by hypothalamic EGF receptor signaling. Science. 2001;294:2511–5. doi: 10.1126/science.1067716. [DOI] [PubMed] [Google Scholar]
- 79.Cheng MY, et al. Prokineticin 2 transmits the behavioural circadian rhythm of the suprachiasmatic nucleus. Nature. 2002;417:405–10. doi: 10.1038/417405a. [DOI] [PubMed] [Google Scholar]
- 80.Kraves S, Weitz CJ. A role for cardiotrophin-like cytokine in the circadian control of mammalian locomotor activity. Nat Neurosci. 2006;9:212–9. doi: 10.1038/nn1633. [DOI] [PubMed] [Google Scholar]
- 81.Vujovic N, Davidson AJ, Menaker M. Sympathetic input modulates, but does not determine, phase of peripheral circadian oscillators. Am J Physiol Regul Integr Comp Physiol. 2008;295:R355–60. doi: 10.1152/ajpregu.00498.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Balsalobre A, et al. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–7. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- 83.Le Minh N, Damiola F, Tronche F, Schutz G, Schibler U. Glucocorticoid hormones inhibit food-induced phase-shifting of peripheral circadian oscillators. EMBO J. 2001;20:7128–36. doi: 10.1093/emboj/20.24.7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nakamura TJ, Sellix MT, Menaker M, Block GD. Estrogen directly modulates circadian rhythms of PER2 expression in the uterus. Am J Physiol Endocrinol Metab. 2008;295:E1025–31. doi: 10.1152/ajpendo.90392.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brown SA, Zumbrunn G, Fleury-Olela F, Preitner N, Schibler U. Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Curr Biol. 2002;12:1574–83. doi: 10.1016/s0960-9822(02)01145-4. [DOI] [PubMed] [Google Scholar]
- 86.Damiola F, et al. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–61. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- 88.Yamazaki S, et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–5. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- 89.Nagoshi E, et al. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 90.Yoo SH, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tosini G, Menaker M. Circadian rhythms in cultured mammalian retina. Science. 1996;272:419–421. doi: 10.1126/science.272.5260.419. [DOI] [PubMed] [Google Scholar]
- 92.Abraham U, Prior JL, Granados-Fuentes D, Piwnica-Worms DR, Herzog ED. Independent circadian oscillations of Period1 in specific brain areas in vivo and in vitro. J Neurosci. 2005;25:8620–6. doi: 10.1523/JNEUROSCI.2225-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Granados-Fuentes D, Tseng A, Herzog ED. A circadian clock in the olfactory bulb controls olfactory responsivity. J Neurosci. 2006;26:12219–25. doi: 10.1523/JNEUROSCI.3445-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Storch KF, et al. Intrinsic circadian clock of the mammalian retina: importance for retinal processing of visual information. Cell. 2007;130:730–41. doi: 10.1016/j.cell.2007.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci U S A. 2008 Sep 8; doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu C, Weaver DR, Strogatz SH, Reppert SM. Cellular construction of a circadian clock: period determination in the suprachiasmatic nuclei. Cell. 1997;91:855–860. doi: 10.1016/s0092-8674(00)80473-0. [DOI] [PubMed] [Google Scholar]
- 98.Honma S, Shirakawa T, Katsuno Y, Namihira M, Honma K. Circadian periods of single suprachiasmatic neurons in rats. Neurosci Lett. 1998;250:157–160. doi: 10.1016/s0304-3940(98)00464-9. [DOI] [PubMed] [Google Scholar]
- 99.Herzog ED, Takahashi JS, Block GD. Clock controls circadian period in isolated suprachiasmatic nucleus neurons. Nature Neurosci. 1998;1:708–713. doi: 10.1038/3708. [DOI] [PubMed] [Google Scholar]
- 100.Aton SJ, Colwell CS, Harmar AJ, Waschek J, Herzog ED. Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat Neurosci. 2005;8:476–83. doi: 10.1038/nn1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Webb AB, Angelo N, Herzog ED. Individual SCN neurons are weak oscillators with changeable circadian phenotypes (P105) Soc Res Biol Rhythms Abstr. 2008;20:147. [Google Scholar]
- 102.Hastings JW, Sweeney BM. On the mechanisms of temperature independence in a biological clock. Proc Natl Acad Sci USA. 1957;43:804–811. doi: 10.1073/pnas.43.9.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hastings JW. The Gonyaulax clock at 50: translational control of circadian expression. Cold Spring Harb Symp Quant Biol. 2007;72:141–4. doi: 10.1101/sqb.2007.72.026. [DOI] [PubMed] [Google Scholar]
- 104.Kondo T, et al. Circadian rhythms in prokaryotes: luciferase as a reporter of circadian gene expression in cyanobacteria. Proc Natl Acad Sci USA. 1993;90:5672–5676. doi: 10.1073/pnas.90.12.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kondo T. A cyanobacterial circadian clock based on the Kai oscillator. Cold Spring Harb Symp Quant Biol. 2007;72:47–55. doi: 10.1101/sqb.2007.72.029. [DOI] [PubMed] [Google Scholar]
- 106.Mihalcescu I, Hsing W, Leibler S. Resilient circadian oscillator revealed in individual cyanobacteria. Nature. 2004;430:81–5. doi: 10.1038/nature02533. [DOI] [PubMed] [Google Scholar]
- 107.Michel S, Geusz ME, Zaritsky JJ, Block GD. Circadian rhythm in membrane conductance expressed in isolated neurons. Science. 1993;259:239–241. doi: 10.1126/science.8421785. [DOI] [PubMed] [Google Scholar]
- 108.Carr AJ, Whitmore D. Imaging of single light-responsive clock cells reveals fluctuating free-running periods. Nat Cell Biol. 2005;7:319–21. doi: 10.1038/ncb1232. [DOI] [PubMed] [Google Scholar]
- 109.Robertson LM, Takahashi JS. Circadian clock in cell culture: I. Oscillation of melatonin release from dissociated chick pineal cells in flow-through microcarrier culture. J Neurosci. 1988;8:12–21. doi: 10.1523/JNEUROSCI.08-01-00012.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 111.Rosbash M. Why the rat-1 fibroblast should replace the SCN as the in vitro model of choice. Cell. 1998;93:917–919. doi: 10.1016/s0092-8674(00)81197-6. [DOI] [PubMed] [Google Scholar]
- 112.Yagita K, Tamanini F, van Der Horst GT, Okamura H. Molecular mechanisms of the biological clock in cultured fibroblasts. Science. 2001;292:278–281. doi: 10.1126/science.1059542. [DOI] [PubMed] [Google Scholar]
- 113.Hastings MH, Herzog ED. Clock genes, oscillators, and cellular networks in the suprachiasmatic nuclei. J Biol Rhythms. 2004;19:400–13. doi: 10.1177/0748730404268786. [DOI] [PubMed] [Google Scholar]
- 114.Antle MC, Silver R. Orchestrating time: arrangements of the brain circadian clock. Trends Neurosci. 2005;28:145–51. doi: 10.1016/j.tins.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 115.Herzog ED. Neurons and networks in daily rhythms. Nat Rev Neurosci. 2007;8:790–802. doi: 10.1038/nrn2215. [DOI] [PubMed] [Google Scholar]
- 116.Yamaguchi S, et al. Synchronization of cellular clocks in the suprachiasmatic nucleus. Science. 2003;302:1408–1412. doi: 10.1126/science.1089287. [DOI] [PubMed] [Google Scholar]
- 117.Yan L, et al. Exploring spatiotemporal organization of SCN circuits. Cold Spring Harb Symp Quant Biol. 2007;72:527–41. doi: 10.1101/sqb.2007.72.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nakamura W, Honma S, Shirakawa T, Honma K. Regional pacemakers composed of multiple oscillator neurons in the rat suprachiasmatic nucleus. Eur J Neurosci. 2001;14:666–674. doi: 10.1046/j.0953-816x.2001.01684.x. [DOI] [PubMed] [Google Scholar]
- 119.Herzog ED, Geusz ME, Khalsa SBS, Straume M, Block GD. Circadian rhythms in mouse suprachiasmatic nucleus explants on multimicroelectrode plates. Brain Res. 1997;757:285–290. doi: 10.1016/s0006-8993(97)00337-5. [DOI] [PubMed] [Google Scholar]
- 120.King VM, et al. A hVIPR transgene as a novel tool for the analysis of circadian function in the mouse suprachiasmatic nucleus. Eur J Neurosci. 2003;17:822–32. [PubMed] [Google Scholar]
- 121.Kunz H, Achermann P. Simulation of circadian rhythm generation in the suprachiasmatic nucleus with locally coupled self-sustained oscillators. J Theor Biol. 2003;224:63–78. doi: 10.1016/s0022-5193(03)00141-3. [DOI] [PubMed] [Google Scholar]
- 122.Gonze D, Bernard S, Waltermann C, Kramer A, Herzel H. Spontaneous synchronization of coupled circadian oscillators. Biophys J. 2005;89:120–9. doi: 10.1529/biophysj.104.058388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Honma S, Shirakawa T, Nakamura W, Honma K. Synaptic communication of cellular oscillations in the rat suprachiasmatic neurons. Neurosci Lett. 2000;294:113–6. doi: 10.1016/s0304-3940(00)01558-5. [DOI] [PubMed] [Google Scholar]
- 124.Shirakawa T, Honma S, Katsuno Y, Oguchi H, Honma KI. Synchronization of circadian firing rhythms in cultured rat suprachiasmatic neurons. Eur J Neurosci. 2000;12:2833–8. doi: 10.1046/j.1460-9568.2000.00170.x. [DOI] [PubMed] [Google Scholar]
- 125.Reppert SM, Schwartz WJ. The suprachiasmatic nuclei of the fetal rat: characterization of a functional circadian clock using 14C-labeled deoxyglucose. J Neurosci. 1984;4:1677–1682. doi: 10.1523/JNEUROSCI.04-07-01677.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Moore RY, Bernstein ME. Synaptogenesis in the rat suprachiasmatic nucleus demonstrated by electron microscopy and synapsin I immunoreactivity. J Neurosci. 1989;9:2151–2162. doi: 10.1523/JNEUROSCI.09-06-02151.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bouskila Y, Dudek FE. Neuronal synchronization without calcium-dependent synaptic transmission in the hypothalamus. Proc Natl Acad Sci USA. 1993;90:3207–3210. doi: 10.1073/pnas.90.8.3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jiang ZG, Yang YQ, Allen CN. Tracer and electrical coupling of rat suprachiasmatic nucleus neurons. Neuroscience. 1997;77:1059–1066. doi: 10.1016/s0306-4522(96)00539-8. [DOI] [PubMed] [Google Scholar]
- 129.Colwell CS. Rhythmic coupling among cells in the suprachiasmatic nucleus. J Neurobiol. 2000;43:379–388. doi: 10.1002/1097-4695(20000615)43:4<379::aid-neu6>3.0.co;2-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Long MA, Jutras MJ, Connors BW, Burwell RD. Electrical synapses coordinate activity in the suprachiasmatic nucleus. Nat Neurosci. 2005;8:61–66. doi: 10.1038/nn1361. [DOI] [PubMed] [Google Scholar]
- 131.Shinohara K, Funabashi T, Mitushima D, Kimura F. Effects of gap junction blocker on vasopressin and vasoactive intestinal polypeptide rhythms in the rat suprachiasmatic nucleus in vitro. Neurosci Res. 2000;38:43–7. doi: 10.1016/s0168-0102(00)00141-3. [DOI] [PubMed] [Google Scholar]
- 132.Rash JE, et al. Connexin36 vs connexin32, “miniature” neuronal gap junctions, and limited electrotonic coupling in rodent suprachiasmatic nucleus. Neuroscience. 2007;149:350–71. doi: 10.1016/j.neuroscience.2007.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.LeSauter J, Silver R. Localization of a suprachiasmatic nucleus subregion regulating locomotor rhythmicity. J Neurosci. 1999;19:5574–85. doi: 10.1523/JNEUROSCI.19-13-05574.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kriegsfeld LJ, LeSauter J, Silver R. Targeted microlesions reveal novel organization of the hamster suprachiasmatic nucleus. J Neurosci. 2004;24:2449–57. doi: 10.1523/JNEUROSCI.5323-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Noguchi T, Watanabe K, Ogura A, Yamaoka S. The clock in the dorsal suprachiasmatic nucleus runs faster than that in the ventral. Eur J Neurosci. 2004;20:3199–202. doi: 10.1111/j.1460-9568.2004.03784.x. [DOI] [PubMed] [Google Scholar]
- 136.Albus H, Vansteensel MJ, Michel S, Block GD, Meijer JH. A GABAergic mechanism is necessary for coupling dissociable ventral and dorsal regional oscillators within the circadian clock. Curr Biol. 2005;15:886–93. doi: 10.1016/j.cub.2005.03.051. [DOI] [PubMed] [Google Scholar]
- 137.Kallo I, et al. Transgenic approach reveals expression of the VPAC2 receptor in phenotypically defined neurons in the mouse suprachiasmatic nucleus and in its efferent target sites. Eur J Neurosci. 2004;19:2201–11. doi: 10.1111/j.0953-816X.2004.03335.x. [DOI] [PubMed] [Google Scholar]
- 138.Pakhotin P, Harmar AJ, Verkhratsky A, Piggins H. VIP receptors control excitability of suprachiasmatic nuclei neurones. Pflugers Arch. 2006;452:7–15. doi: 10.1007/s00424-005-0003-z. [DOI] [PubMed] [Google Scholar]
- 139.Nielsen HS, Hannibal J, Fahrenkrug J. Vasoactive intestinal polypeptide induces per1 and per2 gene expression in the rat suprachiasmatic nucleus late at night. Eur J Neurosci. 2002;15:570–4. doi: 10.1046/j.0953-816x.2001.01882.x. [DOI] [PubMed] [Google Scholar]
- 140.Harmar AJ, et al. The VPAC(2) receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell. 2002;109:497–508. doi: 10.1016/s0092-8674(02)00736-5. [DOI] [PubMed] [Google Scholar]
- 141.Colwell CS, et al. Disrupted circadian rhythms in VIP- and PHI-deficient mice. Am J Physiol Regul Integr Comp Physiol. 2003;285:R939–49. doi: 10.1152/ajpregu.00200.2003. [DOI] [PubMed] [Google Scholar]
- 142.Cutler DJ, et al. The mouse VPAC2 receptor confers suprachiasmatic nuclei cellular rhythmicity and responsiveness to vasoactive intestinal polypeptide in vitro. Eur J Neurosci. 2003;17:197–204. doi: 10.1046/j.1460-9568.2003.02425.x. [DOI] [PubMed] [Google Scholar]
- 143.Brown TM, Colwell CS, Waschek J, Piggins HD. Disrupted neuronal activity rhythms in the suprachiasmatic nuclei of vasoactive intestinal polypeptide-deficient mice. J Neurophysiol. 2007;97:2553–8. doi: 10.1152/jn.01206.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hughes AT, et al. Live imaging of altered period1 expression in the suprachiasmatic nuclei of Vipr2−/− mice. J Neurochem. 2008;106:1646–57. doi: 10.1111/j.1471-4159.2008.05520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ciarleglio CM, et al. Population encoding by circadian clock neurons organizes circadian behavior. J Neurosci. 2009;29:1670–6. doi: 10.1523/JNEUROSCI.3801-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Aton SJ, Huettner JE, Straume M, Herzog ED. GABA and Gi/o differentially control circadian rhythms and synchrony in clock neurons. Proc Natl Acad Sci U S A. 2006;103:19188–93. doi: 10.1073/pnas.0607466103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Piggins HD, Antle MC, Rusak B. Neuropeptides phase shift the mammalian circadian pacemaker. J Neurosci. 1995;15:5612–5622. doi: 10.1523/JNEUROSCI.15-08-05612.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Watanabe K, Vanecek J, Yamaoka S. In vitro entrainment of the circadian rhythm of vasopressin-releasing cells in suprachiasmatic nucleus by vasoactive intestinal polypeptide. Brain Res. 2000;877:361–366. doi: 10.1016/s0006-8993(00)02724-4. [DOI] [PubMed] [Google Scholar]
- 149.Reed HE, Meyer-Spasche A, Cutler DJ, Coen CW, Piggins HD. Vasoactive intestinal polypeptide (VIP) phase-shifts the rat suprachiasmatic nucleus clock in vitro. Eur J Neurosci. 2001;13:839–43. doi: 10.1046/j.0953-816x.2000.01437.x. [DOI] [PubMed] [Google Scholar]
- 150.Aida R, et al. Gastrin-releasing peptide mediates photic entrainable signals to dorsal subsets of suprachiasmatic nucleus via induction of Period gene in mice. Mol Pharmacol. 2002;61:26–34. doi: 10.1124/mol.61.1.26. [DOI] [PubMed] [Google Scholar]
- 151.McArthur AJ, et al. Gastrin-releasing peptide phase-shifts suprachiasmatic nuclei neuronal rhythms in vitro. J Neurosci. 2000;20:5496–502. doi: 10.1523/JNEUROSCI.20-14-05496.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Meyer-Spasche A, Reed HE, Piggins HD. Neurotensin phase-shifts the firing rate rhythm of neurons in the rat suprachiasmatic nuclei in vitro. Eur J Neurosci. 2002;16:339–44. doi: 10.1046/j.1460-9568.2002.02067.x. [DOI] [PubMed] [Google Scholar]
- 153.De Jeu M, Pennartz C. Circadian modulation of GABA function in the rat suprachiasmatic nucleus: excitatory effects during the night phase. J Neurophysiol. 2002;87:834–44. doi: 10.1152/jn.00241.2001. [DOI] [PubMed] [Google Scholar]
- 154.Choi HJ, et al. Excitatory actions of GABA in the suprachiasmatic nucleus. J Neurosci. 2008;28:5450–9. doi: 10.1523/JNEUROSCI.5750-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Liu C, Reppert SM. GABA synchronizes clock cells within the suprachiasmatic circadian clock. Neuron. 2000;25:123–8. doi: 10.1016/s0896-6273(00)80876-4. [DOI] [PubMed] [Google Scholar]
- 156.Quintero JE, Kuhlman SJ, McMahon DG. The biological clock nucleus: a multiphasic oscillator network regulated by light. J Neurosci. 2003;23:8070–6. doi: 10.1523/JNEUROSCI.23-22-08070.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Nagano M, et al. An abrupt shift in the day/night cycle causes desynchrony in the mammalian circadian center. J Neurosci. 2003;23:6141–51. doi: 10.1523/JNEUROSCI.23-14-06141.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Nakamura W, Yamazaki S, Takasu NN, Mishima K, Block GD. Differential response of Period 1 expression within the suprachiasmatic nucleus. J Neurosci. 2005;25:5481–7. doi: 10.1523/JNEUROSCI.0889-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Davidson AJ, Castanon-Cervantes O, Leise TL, Molyneux PC, Harrington ME. Visualizing jet lag in the mouse suprachiasmatic nucleus and peripheral circadian timing system. Eur J Neurosci. 2008;21:21. doi: 10.1111/j.1460-9568.2008.06534.x. [DOI] [PubMed] [Google Scholar]
- 160.Shen S, et al. Overexpression of the human VPAC2 receptor in the suprachiasmatic nucleus alters the circadian phenotype of mice. Proc Natl Acad Sci U S A. 2000;97:11575–80. doi: 10.1073/pnas.97.21.11575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hughes AT, Fahey B, Cutler DJ, Coogan AN, Piggins HD. Aberrant gating of photic input to the suprachiasmatic circadian pacemaker of mice lacking the VPAC2 receptor. J Neurosci. 2004;24:3522–6. doi: 10.1523/JNEUROSCI.5345-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Maywood ES, O’Neill JS, Chesham JE, Hastings MH. Minireview: The circadian clockwork of the suprachiasmatic nuclei--analysis of a cellular oscillator that drives endocrine rhythms. Endocrinology. 2007;148:5624–34. doi: 10.1210/en.2007-0660. [DOI] [PubMed] [Google Scholar]
- 163.Goldman BD. Mammalian photoperiodic system: formal properties and neuroendocrine mechanisms of photoperiodic time measurement. J Biol Rhythms. 2001;16:283–301. doi: 10.1177/074873001129001980. [DOI] [PubMed] [Google Scholar]
- 164.Johnston JD. Measuring seasonal time within the circadian system: regulation of the suprachiasmatic nuclei by photoperiod. J Neuroendocrinol. 2005;17:459–65. doi: 10.1111/j.1365-2826.2005.01326.x. [DOI] [PubMed] [Google Scholar]
- 165.Pittendrigh CS, Daan S. A functional analysis of circadian pacemakers in nocturnal rodents: V. Pacemaker structure: a clock for all seasons. J Comp Physiol [A] 1976;106:333–355. [Google Scholar]
- 166.Mrugala M, Zlomanczuk P, Jagota A, Schwartz WJ. Rhythmic multiunit neural activity in slices of hamster suprachiasmatic nucleus reflect prior photoperiod. Am J Physiol. 2000;278:R987–R994. doi: 10.1152/ajpregu.2000.278.4.R987. [DOI] [PubMed] [Google Scholar]
- 167.Jagota A, de La Iglesia HO, Schwartz WJ. Morning and evening circadian oscillations in the suprachiasmatic nucleus in vitro. Nat Neurosci. 2000;3:372–376. doi: 10.1038/73943. [DOI] [PubMed] [Google Scholar]
- 168.Schaap J, et al. Heterogeneity of rhythmic suprachiasmatic nucleus neurons: Implications for circadian waveform and photoperiodic encoding. Proc Natl Acad Sci U S A. 2003;100:15994–9. doi: 10.1073/pnas.2436298100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.VanderLeest HT, et al. Seasonal encoding by the circadian pacemaker of the SCN. Curr Biol. 2007;17:468–73. doi: 10.1016/j.cub.2007.01.048. [DOI] [PubMed] [Google Scholar]
- 170.Naito E, Watanabe T, Tei H, Yoshimura T, Ebihara S. Reorganization of the suprachiasmatic nucleus coding for day length. J Biol Rhythms. 2008;23:140–9. doi: 10.1177/0748730408314572. [DOI] [PubMed] [Google Scholar]
- 171.Brown TM, Piggins HD. Spatiotemporal heterogeneity in the electrical activity of suprachiasmatic nuclei neurons and their response to photoperiod. J Biol Rhythms. 2009;24:44–54. doi: 10.1177/0748730408327918. [DOI] [PubMed] [Google Scholar]
- 172.Hazlerigg DG, Ebling FJ, Johnston JD. Photoperiod differentially regulates gene expression rhythms in the rostral and caudal SCN. Curr Biol. 2005;15:R449–50. doi: 10.1016/j.cub.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 173.vanderLeest HT, Rohling JH, Michel S, Meijer JH. Phase shifting capacity of the circadian pacemaker determined by the SCN neuronal network organization. PLoS ONE. 2009;4:e4976. doi: 10.1371/journal.pone.0004976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ohta H, Mitchell AC, McMahon DG. Constant light disrupts the developing mouse biological clock. Pediatr Res. 2006;60:304–8. doi: 10.1203/01.pdr.0000233114.18403.66. [DOI] [PubMed] [Google Scholar]
- 175.de la Iglesia HO, Meyer J, Carpino A, Jr, Schwartz WJ. Antiphase oscillation of the left and right suprachiasmatic nuclei. Science. 2000;290:799–801. doi: 10.1126/science.290.5492.799. [DOI] [PubMed] [Google Scholar]
- 176.Yan L, Foley NC, Bobula JM, Kriegsfeld LJ, Silver R. Two antiphase oscillations occur in each suprachiasmatic nucleus of behaviorally split hamsters. J Neurosci. 2005;25:9017–26. doi: 10.1523/JNEUROSCI.2538-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.de la Iglesia HO, Cambras T, Schwartz WJ, Diez-Noguera A. Forced desynchronization of dual circadian oscillators within the rat suprachiasmatic nucleus. Curr Biol. 2004;14:796–800. doi: 10.1016/j.cub.2004.04.034. [DOI] [PubMed] [Google Scholar]
- 178.Lee ML, Swanson BE, de la Iglesia HO. Circadian Timing of REM Sleep Is Coupled to an Oscillator within the Dorsomedial Suprachiasmatic Nucleus. Curr Biol. 2009;15:15. doi: 10.1016/j.cub.2009.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Gorman MR, Elliott JA. Entrainment of 2 subjective nights by daily light:dark:light:dark cycles in 3 rodent species. J Biol Rhythms. 2003;18:502–12. doi: 10.1177/0748730403260219. [DOI] [PubMed] [Google Scholar]
- 180.Watanabe T, et al. Bimodal clock gene expression in mouse suprachiasmatic nucleus and peripheral tissues under a 7-hour light and 5-hour dark schedule. J Biol Rhythms. 2007;22:58–68. doi: 10.1177/0748730406295435. [DOI] [PubMed] [Google Scholar]
- 181.Pulivarthy SR, et al. Reciprocity between phase shifts and amplitude changes in the mammalian circadian clock. Proc Natl Acad Sci U S A. 2007;104:20356–61. doi: 10.1073/pnas.0708877104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Brown TM, Hughes AT, Piggins HD. Gastrin-releasing peptide promotes suprachiasmatic nuclei cellular rhythmicity in the absence of vasoactive intestinal polypeptide-VPAC2 receptor signaling. J Neurosci. 2005;25:11155–64. doi: 10.1523/JNEUROSCI.3821-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hamada T, LeSauter J, Venuti JM, Silver R. Expression of Period genes: rhythmic and nonrhythmic compartments of the suprachiasmatic nucleus pacemaker. J Neurosci. 2001;21:7742–7750. doi: 10.1523/JNEUROSCI.21-19-07742.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Yan L, Okamura H. Gradients in the circadian expression of Per1 and Per2 genes in the rat suprachiasmatic nucleus. Eur J Neurosci. 2002;15:1153–62. doi: 10.1046/j.1460-9568.2002.01955.x. [DOI] [PubMed] [Google Scholar]
- 185.Karatsoreos IN, Yan L, LeSauter J, Silver R. Phenotype matters: identification of light-responsive cells in the mouse suprachiasmatic nucleus. J Neurosci. 2004;24:68–75. doi: 10.1523/JNEUROSCI.1666-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Nakamura W, Honma S, Shirakawa T, Honma KI. Clock mutation lengthens the circadian period without damping rhythms in individual SCN neurons. Nat Neurosci. 2002;5:399–400. doi: 10.1038/nn843. [DOI] [PubMed] [Google Scholar]
- 187.King DP, et al. The mouse clock mutation behaves as an antimorph and maps within the w-19h deletion, distal of kit. Genetics. 1997;146:1049–1060. doi: 10.1093/genetics/146.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Bernard S, Gonze D, Cajavec B, Herzel H, Kramer A. Synchronization-induced rhythmicity of circadian oscillators in the suprachiasmatic nucleus. PLoS Comput Biol. 2007;3:e68. doi: 10.1371/journal.pcbi.0030068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Locke JC, Westermark PO, Kramer A, Herzel H. Global parameter search reveals design principles of the mammalian circadian clock. BMC Syst Biol. 2008;2:22. doi: 10.1186/1752-0509-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Low-Zeddies SS, Takahashi JS. Chimera analysis of the Clock mutation in mice shows that complex cellular integration determines circadian behavior. Cell. 2001;105:25–42. doi: 10.1016/s0092-8674(01)00294-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Enright JT. Temporal precision in circadian systems: a reliable neuronal clock from unreliable components? Science. 1980;209:1542–5. doi: 10.1126/science.7433976. [DOI] [PubMed] [Google Scholar]
- 192.Honma S, Nakamura W, Shirakawa T, Honma K. Diversity in the circadian periods of single neurons of the rat suprachiasmatic nucleus depends on nuclear structure and intrinsic period. Neurosci Lett. 2004;358:173–6. doi: 10.1016/j.neulet.2004.01.022. [DOI] [PubMed] [Google Scholar]
- 193.Pittendrigh CS, Daan S. A functional analysis of circadian pacemakers in nocturnal rodents: I. The stability and lability of spontaneous frequency. J Comp Physiol [A] 1976;106:223–252. [Google Scholar]
- 194.Beersma DG, van Bunnik BA, Hut RA, Daan S. Emergence of circadian and photoperiodic system level properties from interactions among pacemaker cells. J Biol Rhythms. 2008;23:362–73. doi: 10.1177/0748730408317992. [DOI] [PubMed] [Google Scholar]
- 195.To T-L, Henson MA, Herzog ED, Doyle FJ. A computational model for intercellular synchronization in the mammalian circadian clock. Proc Natl Acad Sci USA. 2006 doi: 10.1529/biophysj.106.094086. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Li Y, Liu Z, Zhang J, Wang R, Chen L. Synchronisation mechanisms of circadian rhythms in the suprachiasmatic nucleus. IET Syst Biol. 2009;3:100. doi: 10.1049/iet-syb.2007.0057. [DOI] [PubMed] [Google Scholar]
- 197.Antle MC, Foley DK, Foley NC, Silver R. Gates and oscillators: a network model of the brain clock. J Biol Rhythms. 2003;18:339–50. doi: 10.1177/0748730403253840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Antle MC, Foley NC, Foley DK, Silver R. Gates and oscillators II: zeitgebers and the network model of the brain clock. J Biol Rhythms. 2007;22:14–25. doi: 10.1177/0748730406296319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Yu W, Hardin PE. Circadian oscillators of Drosophila and mammals. J Cell Sci. 2006;119:4793–5. doi: 10.1242/jcs.03174. [DOI] [PubMed] [Google Scholar]
- 200.Nitabach MN, Blau J, Holmes TC. Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell. 2002;109:485–95. doi: 10.1016/s0092-8674(02)00737-7. [DOI] [PubMed] [Google Scholar]
- 201.Vansteensel MJ, Michel S, Meijer JH. Organization of cell and tissue circadian pacemakers: a comparison among species. Brain Res Rev. 2008;58:18–47. doi: 10.1016/j.brainresrev.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 202.Nitabach MN, Taghert PH. Organization of the Drosophila circadian control circuit. Curr Biol. 2008;18:R84–93. doi: 10.1016/j.cub.2007.11.061. [DOI] [PubMed] [Google Scholar]
- 203.Stoleru D, Peng Y, Agosto J, Rosbash M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature. 2004;431:862–8. doi: 10.1038/nature02926. [DOI] [PubMed] [Google Scholar]
- 204.Stoleru D, Peng Y, Nawathean P, Rosbash M. A resetting signal between Drosophila pacemakers synchronizes morning and evening activity. Nature. 2005;438:238–42. doi: 10.1038/nature04192. [DOI] [PubMed] [Google Scholar]
- 205.Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- 206.Peng Y, Stoleru D, Levine JD, Hall JC, Rosbash M. Drosophila free-running rhythms require intercellular communication. PLoS Biol. 2003;1:E13. doi: 10.1371/journal.pbio.0000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Lin Y, Stormo GD, Taghert PH. The neuropeptide pigment-dispersing factor coordinates pacemaker interactions in the Drosophila circadian system. J Neurosci. 2004;24:7951–7. doi: 10.1523/JNEUROSCI.2370-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Yoshii T, et al. The neuropeptide pigment-dispersing factor adjusts period and phase of Drosophila’s clock. J Neurosci. 2009;29:2597–610. doi: 10.1523/JNEUROSCI.5439-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Liu AC, Lewis WG, Kay SA. Mammalian circadian signaling networks and therapeutic targets. Nat Chem Biol. 2007;3:630–639. doi: 10.1038/nchembio.2007.37. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Circadian rhythms of PER2::LUC bioluminescence recorded from mouse SCN neurons in dispersed culture over a period of two weeks, showing that the cells oscillate independently. Bioluminescence intensity of one cell highlighted near the center of the image is plotted below.
Circadian rhythms of PER2::LUC bioluminescence recorded from mouse SCN neurons in a cultured slice over a period of seven days, showing that the cells oscillate with the same periods and similar phases, in a complex spatiotemporal pattern. This is a coronal slice of the ventral hypothalamus, including the bright bioluminescent left and right SCN, the third ventricle between them, and the optic chiasm below.