Abstract
How does the brain compute? Answering this question necessitates neuronal connectomes, annotated graphs of all synaptic connections within defined brain areas. Further, understanding the energetics of the brain's computations requires vascular graphs. The assembly of a connectome requires sensitive hardware tools to measure neuronal and neurovascular features in all three dimensions, as well as software and machine learning for data analysis and visualization. We present the state of the art on the reconstruction of circuits and vasculature that link brain anatomy and function. Analysis at the scale of tens of nanometers yields connections between identified neurons, while analysis at the micrometer scale yields probabilistic rules of connection between neurons and exact vascular connectivity.
Introduction
What are the algorithms that animals use to sample their environment, compute strategies for survival, and control the motion and rhythms of their body? Answers to these questions are essential in neuroscience, the study of how brains work (Bullock, 1993). The nature of these algorithms and the concomitant dynamics that underlie neuronal computations are constrained by the architecture of the particular nervous system. Likewise, the delivery of nutriments to sustain nervous systems and the interactions between neurons and vasculature are constrained by the architecture of the underlying vasculature network. Recent and ongoing technical advances in histology and all facets of imaging suggest that we are entering a renaissance in terms of our ability to decipher these networks. The description of brain networks can be cast as an annotated graph, commonly called a connectome, in which edges correspond to weighted connections between nodes.
An especially simple connectome is the description of blood vessels that supply nutrients to the brain. Each vessel defines an undirected edge, as blood is permitted to flow in either direction. The junction where different vessel segments meet, which occurs for triplets in the mammalian brain, defines a node. Additional information may be added by annotating individual edges and nodes. For example, each edge in this graph can be labeled with the length and radius of the vessel. A node may be labeled by its association with a control element, such as a pericyte. The connectivity matrix and annotation associated with the vascular connectome, or angiome, is sufficient to calculate flow patterns in cortex.
Neuronal circuits can also be described in terms of a graph. Here, each synaptic connection defines an edge. Presynaptic and postsynaptic cells form the nodes. In contrast to the case of brain vasculature, a node typically connects to thousands of inputs and outputs, which define the edges. An edge is directed for chemical synapses and undirected for electrical connections. Annotation defines the chemical composition and strength of the synapse along with a multitude of other attributes. Annotation may also be used to record details about the somata. It is important to be mindful that the graphical description of nervous systems has limitations. When the electrical space constant of a neuron is small compared to the extent of the dendrites, different parts of the neuron may function independently. While this is ordinarily not an issue with cortical interneurons, it is an issue for cells in which electrical activity in the dendrites is decoupled from that in the soma. This occurs, for example, in starburst amacrine cells in the retina (Euler et al., 2002; Hausselt et al., 2007). In these cases, each neuron may be represented as a superset of nodes and the value of a connectome is retained.
The recent advent of high-throughput anatomical techniques offers the realistic possibility of completing neuronal “wiring” diagrams and vascular “plumbing” diagrams of large brains. What are the precedents to compare this work against? With respect to nervous systems, the pioneering serial section electron microscopy study of White et al. (1986) largely completed the connectome for Caenorhabditis elegans, which contains both electrical and chemical synapses. After a hiatus of 25 years, a near-finished wiring diagram based on the original data and new micrographs have been obtained (Varshney et al., 2011). An analysis of the data yields statistical insights into the network along with a hypothesized mapping of circuit motifs for egg-laying behavior (Zhang et al., 2008) and pheromone sensing (Macosko et al., 2009). Fractions of underlying neuronal circuitry have been worked out for “simpler” nervous systems and include circuits for rhythmic neuronal activity, such as those that drive the heart in Hirudo medicinalis (Kristan et al., 2005), swimming in H. medicinalis (Stent et al., 1978), swimming in Clione limacina (Satterlie, 1985), and the escape swim response in Tritonia diomedea (Getting, 1983; Kleinfeld and Sompolinsky, 1988), among other stereotypic behaviors (Delcomyn, 1980; Shepherd and Grillner, 2010). Yet the dream of a complete connectome in systems more complex than C. elegans remains unfulfilled. With respect to brain vascular systems, the statistics of the connections among blood vessels have been addressed in mice (Tsai et al., 2009b), monkeys (Weber et al., 2008), and man (Lauwers et al., 2008), and flow in small pieces of the subsurface vasculature has been examined (Fang et al., 2008). Yet a full pattern of connectivity exists only for the two-dimensional network of pial vessels (Blinder et al., 2010).
In light of past efforts described above, and cognizant of the growing interest in obtaining connectomes across a variety of animals (Sporns et al., 2005; Lichtman et al., 2008; Bohland et al., 2009; Lu et al., 2009; Seung, 2009; DeFelipe, 2010; Swanson and Bota, 2010), we summarize the status of six ongoing projects that use high-throughput automated histology in the pursuit of brain connectomes.
Connectomes derived from nanostructural (10–100 nm resolution) data
We consider a set of projects in which electron microscopy is used to define the connectome that underlies three computations from vision. In each case, the connections between pairs of neurons are mapped to a connectome with a low degree of false-positive errors, as erroneous connections are judged to be worse than missing connections. Electron microscopy is required since axons and spine necks can be 50 nm or less in diameter and the structures of interest are densely packed (Sorra and Harris, 2000). Recently developed methods based on scanning electron microscopy offer exceptionally high axial resolution and reliability of throughput, using block face imaging of the tissue (Denk and Horstmann, 2004; Knott et al., 2008; Briggman et al., 2011), while methods based on transmission electron microscopy currently maintain a significant advantage in terms of the speed of imaging (Bock et al., 2011).
As a first example, circuits in the optic lobe medulla of the fly are reconstructed using serial section transmission electron microscopy (ssTEM) and related to motion detection (Chklovskii et al., 2010). A connectome is formed among all cells in the reconstructed volume. Second, the receptive fields of direction-selective ganglion cells in the mammalian retina are measured and, using serial block-face scanning electron microscopy (SBEM) to delineate synaptic connectivity, modeled by the pattern of input from starburst amacrine cells (Briggman et al., 2011). The connectome is restricted to synapses onto neurons that were functionally probed. Last, the receptive fields of a set of cortical neurons are assessed physiologically. Then, using ssTEM, a connectome of their local synaptic interconnections is formed. These data reveal promiscuous connectivity from excitatory pyramidal cells onto inhibitory interneurons (Bock et al., 2011). The latter two studies emphasize that the combination of physiological recordings with subsequent anatomical reconstructions of the recorded neurons is critical to correlate the neuronal function with network structure.
In search of the mechanism of motion detection: serial electron microscopy of the fruit fly visual system (Chklovskii et al., 2010)
Visual systems excel at the detection of moving objects. Yet, the circuit that underlies motion detection is not fully understood. More than 50 years ago, Hassenstein and Reichardt (1956) proposed an elementary motion detector that computes correlations between visual signals offset in time and space (Fig. 1A). Their seminal work was followed by theoretical (Poggio and Reichardt, 1976; Adelson and Bergen, 1985; Hildreth and Koch, 1987; Potters and Bialek, 1994) and experimental (Buchner, 1976, 1984; Borst and Egelhaaf, 1987; Egelhaaf and Reichardt, 1987; Egelhaaf and Borst, 1989; Egelhaaf et al., 1989; Schuling et al., 1989; Zanker et al., 1999; Borst et al., 2005, 2010; Clark et al., 2011; Eichner et al., 2011) investigations whose conclusions were largely consistent with the original proposal. However, there has never been a “smoking gun” that identifies particular neurons with the specific computations within the elementary motion detector.
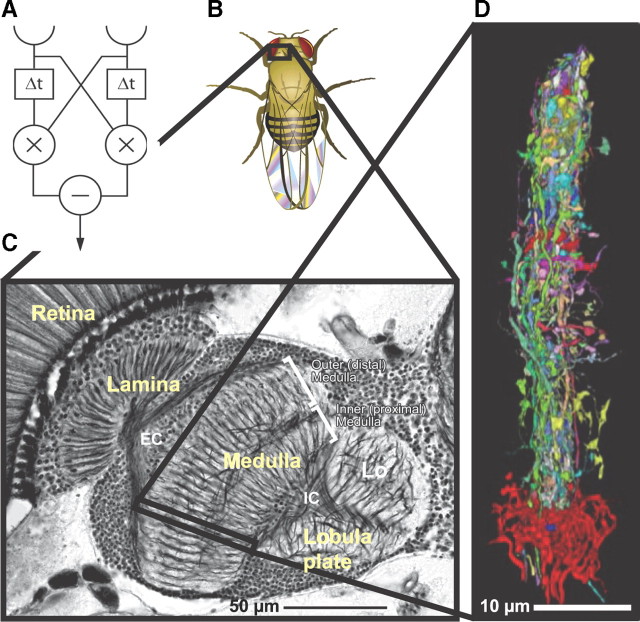
Figure 1.
Motion detection circuit in fruit fly. A, Hassenstein–Reichardt elementary motion detection compares correlations in light intensity signals that are offset in space and time. B, Fruit fly. C, A light microscopic section through fruit fly optic lobe showing the first three stages of visual processing: lamina, medulla, and lobula plate. D, Electron microscopy reconstruction of a medulla column, different colors correspond to different neurons. C is adapted from Takemura et al. (2008).
The combination of two lines of experimental evidence suggests that elementary motion detection is performed in the optic lobe medulla in the fly (Fig. 1B,C). First, the upstream lamina does not contain directionally selective neurons (Laughlin and Hardie, 1978; Laughlin, 1994; Zheng et al., 2006, 2009). Second, the immediate downstream processing stage, the lobula plate, integrates elementary motion signals over the visual field (Brotz and Borst, 1996; Haag and Borst, 2003; Schnell et al., 2010; Seelig et al., 2010). These suggest that the neuronal mechanism of motion detection may be revealed through a reconstruction of the medulla of the fruit fly. The realization of such a reconstruction is motivated by the small size of the fly, stereotypic brain geometry from fly to fly, and the repeating structure of the medulla column, so that the reconstructed volume needs to include only one repeating column. It is also aided by the availability of genetic tools for physiological manipulations of individual classes of neurons (Pfeiffer et al., 2008), which provides a means to combine structural information with functional studies.
Serial section transmission electron microscopy is used to reconstruct the neural circuits of the optic lobe medulla (Clements et al., 2008). The tissue is cut into a series of 40-nm-thick sections through the entire depth of the medulla, and each section is imaged at 3 × 3 nm2 per pixel across a field of 90 × 90 μm2 as a set of 4000 × 4000 pixel images that form a mosaic. This resolution reveals individual presynaptic and postsynaptic sites and permits thin neuronal processes to be traced through the series. The resultant dataset across each fly consists of 2 × 105 micrographs, or 3 teravoxels. A semiautomated pipeline with five distinct tasks was developed for high-throughput circuit reconstruction from these datasets (Chklovskii et al., 2010). Registration places the separately acquired images of different sections and different parts from the same section into alignment with each other. Segmentation partitions the stack of images into sets of voxels that define profiles for distinct neurons. Linkages connect adjacent voxels in consecutive sections that belong to the same cell. Proofreading is a manual step to correct errors in the automated reconstruction. Last, annotation identifies presynaptic and postsynaptic terminals and neuronal classes.
The results of reconstructing ssTEM sections of the medulla column show that it contains several dozen columnar neurons and tens of thousands of synaptic terminals. The connectome was assembled by attributing synapses to specific neurons that were identified by comparing their shape with the light microscopy images of Golgi impregnations (Fischbach and Dittrich, 1989). Finally, most neurons reconstructed with ssTEM could be matched to light microscopy images of transgenic GAL4-UAS lines that expressed green fluorescent protein as an extension of past methods (Pfeiffer et al., 2008). These lines will allow genetic access to neurons of specific classes to determine their repertoire of neurotransmitters and synaptic receptors, manipulate their physiological properties, and monitor their activity. The emerging connectome, along with future physiological investigations, should be sufficient to conclusively resolve the nature of elementary motion detection mechanism in fruit fly, as well as provide insight into the other computations of the visual system.
The specificity of synaptic connections that underlie direction selectivity in the mammalian retina (Denk and Horstmann, 2004; Briggman and Denk, 2006; Briggman et al., 2011)
An automated method to acquire volumetric electron microscopy data, SBEM (Denk and Horstmann, 2004), may be used to reconstruct all neuronal structures at resolutions of tens of nanometers. This high resolution is crucial for the reliable tracing of fine neuronal processes in all three dimensions and to ensure that the connections between neurons are inferred with relatively few false-positive errors. In this technique, scanning electron microscopy is used to image backscattered electrons from the surface of a plastic embedded tissue block. The top tens of nanometers of a tissue block are first imaged (Fig. 2A); the block is then advanced upward, and a thin section is cut from the block face in situ with a custom-made microtome that is mounted within the vacuum chamber of the scanning electron microscope (Fig. 2B). This provides a fresh surface for imaging. Repeated imaging and sectioning of the block face are completely automated.
Figure 2.
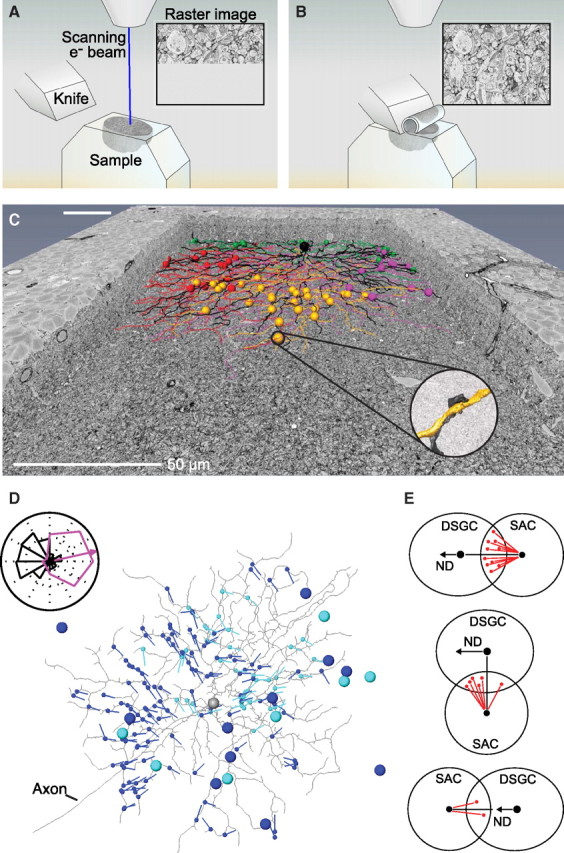
A schematic view of serial block-face scanning electron microscopy technology. A, The electron beam first raster scans the surface of a tissue sample that is embedded in a plastic block. B, After the acquisition of an image, a diamond knife cuts a thin section from the surface of the block. C, A cutaway of the volume of mouse retina acquired for the study of the direction-selectivity circuit in retina. The image shows a reconstructed starburst amacrine cell in black and the locations of its synapses (see inset) onto direction-selective retinal ganglion cells. The synapse locations are color coded by the functionally recorded preferred directions of the direction-selective retinal ganglion cells. The scale bar is shorter near the top as a consequence of perspective. D, Starburst amacrine cell somata (large blue and cyan dots) and the direction of their dendrites (blue and cyan vectors) that form synapses (small blue and cyan dots) onto a direction-selective ganglion cell (gray). Inset, A polar histogram of the vector directions (black) indicate an asymmetric distribution opposite to the functionally recorded preferred direction of the ganglion cell (purple tuning curve). E, Schematic wiring diagrams of starburst amacrine cell (SAC) input to direction-selective ganglion cells (DSGC) for three different locations of the starburst cell soma. The starburst cell dendrite directions are biased toward the null direction (ND). A and B are courtesy of Julia Kuhl. C–E are adapted from Briggman et al. (2011).
The specificity of connections from starburst amacrine to direction-selective retinal ganglion cells in the mammalian retina is used to demonstrate the capabilities of SBEM. Both of these cell types are involved in computing the direction of movement. Starburst amacrine cells make inhibitory synapses onto direction-selective retinal ganglion cells. Each dendrite of an amacrine cell is preferentially activated by visual stimuli that move in a particular direction (Euler et al., 2002), while different retinal ganglion cells have different preferred directions. It has been hypothesized, based on the results of physiological studies (Fried et al., 2002; Wei et al., 2011), that On–Off direction-selective retinal ganglion cells gain their direction selectivity in part by an asymmetric pattern of inputs from directionally sensitive starburst amacrine cells.
Two experimental steps are required to test this hypothesis. The first is to record the functional response properties of several directionally sensitive ganglion cells. Two-photon laser scanning microscopy (Denk et al., 1990) (TPLSM) was used to image calcium transients in a population of several hundred ganglion cell somata (Denk and Detwiler, 1999; Briggman and Euler, 2011) in response to stimulation with a bar of light that moved in one of eight possible directions. The retina was then fixed and the tissue prepared for SBEM. The second step is to reconstruct the connections in a volume that contained the nearly complete dendritic trees of the recorded cells and their presynaptic starburst amacrine cells. The acquired volume, 60 × 350 × 300 μm3 at a resolution of 16.5 × 16.5 × 23 nm3 for a dataset size of 1 teravoxel, contained all of the imaged somata and encompassed the typical diameter of the dendritic tree of retinal ganglion cells. In the example of Figure 2C, the dendritic trees for six direction-selective ganglion cells and 24 starburst amacrine cells were manually traced and the synaptic contacts between pairs of cells were tabulated.
The analysis of the reconstructed synaptic contacts revealed that the dendrites of starburst amacrine cells that are oriented along the direction for which the postsynaptic ganglion cell has a minimal calcium repose to motion, i.e., the null direction, are much more likely to form inhibitory synapses than the dendrites oriented along the direction for the maximum response of the ganglion cell (Fig. 2C,D). This result confirms the hypothesis that asymmetric wiring of this circuit is essential for the computation of the direction of motion in the retina. In addition, these studies uncovered evidence that information about the direction of individual starburst amacrine cell dendrites is likely used during development to create the wiring specificity (Fig. 2E). While manual methods were used to reconstruct only a small fraction of the neurons contained in the tissue collected for this study, further analysis of this and future volumes of tissue will greatly benefit from the use of high-throughput tracing efforts (Helmstaedter et al., 2011) and automated methods (Jain et al., 2010).
Anatomy and in vivo physiology from a neuronal network in visual cortex (Bock et al., 2011)
In work similar to the above case of circuits in the retina, an understanding of the relationship between function and structure in neuronal circuits of the cerebral cortex is achieved by relating the simultaneous activity of many neurons to the anatomical connections among these cells. To achieve this goal, in vivo TPLSM for intracellular calcium measurements (Svoboda et al., 1997; Stosiek et al., 2003; Kerr et al., 2005; Ohki et al., 2005) was combined with large-scale ssTEM to examine the local microcircuit formed by these cells (Bock et al., 2011). The use of TPLSM to record calcium signals, as opposed to microelectrode recordings of cell voltages, permits functional data as well as anatomical landmarks to be obtained. All measurements involved mouse primary visual cortex.
The first step in this process is to characterize the sensory physiology of neurons in visual cortex while the animal watched drifting black and white bars of varying orientations. The animal was then perfused and the brain was removed and processed for ssTEM (Harris et al., 2006). A custom-built camera array was used to acquire data (Fig. 3A). For the example of Figure 3, a 450 × 350 × 50 μm3 volume was sampled at 4 × 4 × 45 nm3 per voxel; the final set of stitched and aligned data is 10 teravoxels. Brain vasculature served as a fiducial to achieve a cell-level correspondence between in vivo imaging and electron microscopy image stacks. In this same example, the proximal dendritic and axonal arbors of 13 physiologically characterized cells were manually traced. For each synapse along the axonal arbor, the postsynaptic dendrite was traced either to the edge of the ssTEM-imaged volume or to the postsynaptic cell body (Fig. 3B). The postsynaptic processes were reliably categorized as either inhibitory or excitatory based on a variety of morphological criteria. This led to a connectome that was annotated with in vivo physiology with local network anatomy in cortex (Fig. 3C).
Figure 3.
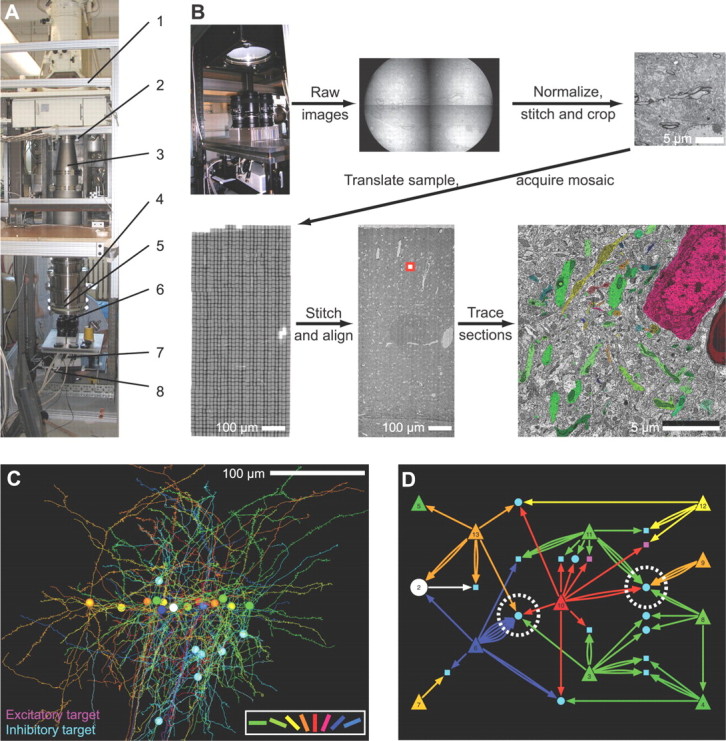
The process of realizing a connectome from the network anatomy for mouse visual cortex. A, Custom camera array for transmission electron microscopy. Labeled components: 1, motorized positioner knobs; 2, lead shielding; 3, vacuum chamber extension; 4, scintillator; 5, leaded glass; 6, 2 × 2 array of f/2.0 optical lenses coupled to 50 MHz readout of 11 megapixel CCD cameras; 7, camera trigger breakout box; 8, cables to four acquisition computers. B, Acquisition and reconstruction overview. Top row, Left to right, Four cameras with overlapping fields of view of the scintillator; raw camera images; normalized, stitched, and cropped camera images into a single “tile.” Bottom row, Left to right, Mosaic of tiles resulting from sample motion; stitched mosaic, with red square delineating zoom area in the next image; zoom of a single tile showing area tracing. Not shown: imaging of multiple sections, alignment of serial section mosaics, tracing through multiple sections, and validation of tracing. C, Three-dimensional tracing of the 14 physiologically characterized cells and their postsynaptic targets; only those targets receiving convergent synaptic input from two or more physiologically characterized cells are shown. Physiologically characterized orientation preference for the cell given in the key, bottom right. D, Connectivity graph of the anatomical objects shown in C. Magenta targets are excitatory; cyan targets are inhibitory. Circles are postsynaptic dendrites that could be traced back to the cell body; squares are dendrites that left the imaged volume before the cell body was reached. C and D are adapted from Bock et al. (2011).
The first realization of this process showed that inhibitory cells received convergent anatomical input from diversely tuned neighboring excitatory cells, without apparent regard to orientation selectivity (Bock et al., 2011). This finding is in agreement with intracellular measurements of pairwise synaptic connectivity (Hofer et al., 2011) and is further consistent with the observation that most inhibitory interneurons show weak or no orientation selectivity (Sohya et al., 2007; Liu et al., 2009; Kerlin et al., 2010; Niell and Stryker, 2010; Kuhlman et al., 2011; Zariwala et al., 2011), although some subtypes may be more selective than others (Ma et al., 2010; Runyan et al., 2010). Finally, when orientation-selective inhibitory neurons are found, their preferred orientation matches the bias shown by neighboring pyramidal cells (Kerlin et al., 2010). Together, these anatomical and physiological results suggest a model in which inhibitory interneurons usually pool the activity of nearby pyramidal cells. Continued increases in the number of physiologically characterized cells and the size of the reconstructed volumes should further clarify this issue and are anticipated to uncover additional motifs that link circuit function and anatomical connectivity.
Connectomes derived from microstructural (0.1–1 μm resolution) data
In studies complementary to the connectomes derived above, the complete inventory of structures in murine cortex is assessed with high-throughput optical tools and protein-specific markers. The molecular signature and structural characteristics of synapses across the depth of cortex, at single-synapse resolution, are revealed by array tomography (Micheva et al., 2010). This serves as essential information to annotate edges in the connectome. Reconstruction of the connections among all blood vessels relative to their surrounding brain cells suggests strategies for synaptic control of blood flow by contractile elements (Blinder et al., 2010). Last, the locations of all neuronal somata within a single column, their identification in terms of specific cell types, the assignment of canonical morphologies, and the estimate of synaptic contacts between different cell types form a connectome based on estimated connectivity, and allow the assessment of putative pathways for self-motion versus touch (Oberlaender et al., 2011a).
The diversity of cortical synaptic connections as revealed by array tomography (Micheva and Smith, 2007; Micheva et al., 2010)
The connectome involves annotation of synaptic components in addition to a graph that tabulates connections. In the case of neuronal circuitry, as opposed to electrical circuits, the components are extremely diverse and schemes to classify synaptic components are only emerging (Bug et al., 2008). Neuronal cell diversity has been studied intensively since the pioneering morphological investigations of the 19th century, and ongoing electrophysiological and molecular studies make it clear that such cellular diversity is enormous; e.g., there are at least 24 neuronal types in the CA1 region of the hippocampus alone (Klausberger and Somogyi, 2008). Synaptic diversity is an even greater enigma. This occurs because synapses are orders of magnitude more numerous than neurons and because synapses have proved far more difficult than neurons to study at the unitary level. Nonetheless, the limited morphological, gene expression, and proteomic data presently available strongly imply that synapse diversity must be enormous.
Array tomography is a high-resolution proteomic imaging method that provides new opportunities to explore and map synapse molecular diversity (Fig. 4). First, array tomography allows individual synapses within brain tissue to be resolved with wide-field fluorescent microscopy at the theoretical maximum for lateral resolution, i.e., ∼200 nm, and subdiffraction resolution in the axial direction, i.e., ∼70 nm, by physically sectioning the samples. Second, array tomography can detect the distribution of dozens of antigens within the same synapse by eluting antibodies and restaining the sample multiple times. The ability to multiplex the binding capability of structural proteins and organelles provides a broader context to decipher molecular architecture than that gleamed through conventional immunofluorescence. Third, array tomography can acquire large volumes, which contain millions of synapses, since both immunological labeling and imaging are not limited by depth within the tissue. The planar images from each section are stitched. While there is no fundamental limit to the final size, current datasets typically encompass 1500 × 500 × 12 μm3 regions with ∼20 different molecular markers. As a result of these three features, array tomography data can reveal a previously unseen level of synaptic diversity.
Figure 4.
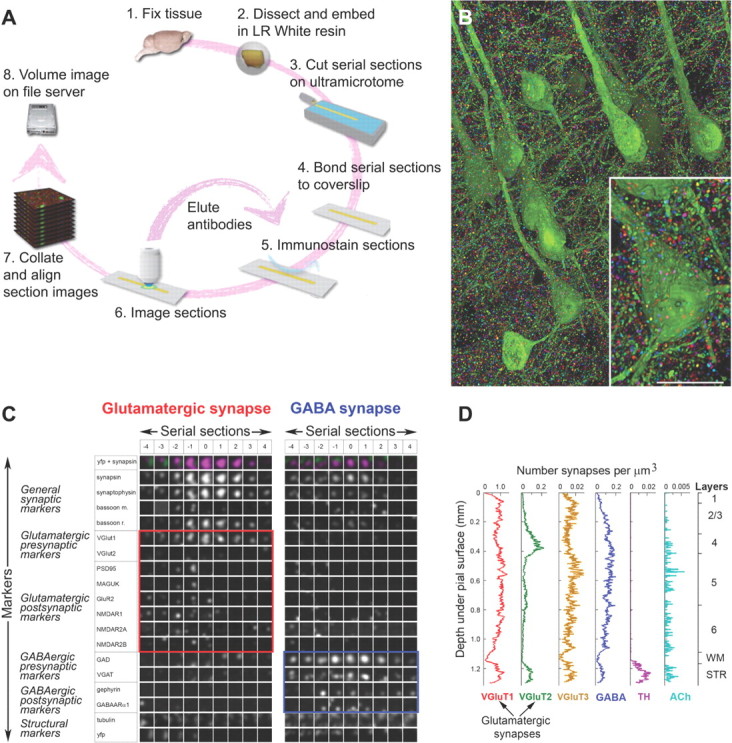
Array tomography allows the resolution, classification and quantification of individual synapses. A, The array tomography proteomic imaging cycle involves multiple rounds of immunostaining, imaging, and antibody elution and allows collection of immunofluorescence data of multiple antibody channels; 24 demonstrated to date. B, Volume rendering of layer 5 of the somatosensory cortex of a Thy-1 YFP-H mouse (Feng et al., 2000), immunolabeled for the general synaptic marker synapsin. Individual synapsin puncta have been segmented digitally and rendered in random color to emphasize that each synapsin punctum is fully resolvable in all three dimensions. Scale bar, 10 μm. C, High dimensional proteomic data are viewed as “synaptograms” with columns representing individual serial sections through a synapse and rows representing each marker. The two synaptograms show examples of a glutamatergic synapse with glutamatergic markers boxed in red and a GABAergic synapse with the respective markers boxed in blue. Each square represents an area of 1 × 1 μm2. D, The density of six synapse subtypes in the somatosensory cortex of a C57BL/6J mouse plotted as a function of depth through the cortical layers, white matter (WM), and part of the striatum (STR).
Data acquired by array tomography are represented by “synaptograms” (Fig. 4C), which allow the simultaneous display of both the spatial and high proteomic content of each synapse. The columns in a synaptogram represent serial sections through a synapse and the rows represent individual markers. Synapses are classified into subtypes based on the coexpression and spatial relation of a number of relevant markers. For the example of Figure 4C, the synaptogram on the left represents a glutamatergic VGluT1 synapse, based on the colocalization of a number of presynaptic and postsynaptic glutamatergic markers, while the synaptogram on the right contains GABAergic presynaptic and postsynaptic markers and represents a GABAergic synapse. At least a dozen of different synapse subtypes in mouse cerebral cortex have been identified in this way so far, using neurotransmitter, vesicular transporter, calcium-binding protein, and receptor markers. Manually classified synapse samples are used to train machine-learning algorithms for automatic classification and quantification of all synapses within the sample (Fig. 4D). Last, synapse subtypes can be related to their parent neurons through the use of transgenic animals that express fluorescent proteins in specific neuronal subsets (Feng et al., 2000) or in individual neurons (Li et al., 2010).
The details of synaptic diversity as observed with array tomography represent only a snapshot of the state of brain synapses at a fixed moment of time. However, it may be possible to address temporal changes in synaptic proteomics that relate to activity levels, plasticity, circadian rhythms, and development by a combination of live imaging followed by array tomographic analysis of the same tissue. In this way, potentiated synapses at dendritic spines (Murakoshi et al., 2011) or newly formed spines during learning (Xu et al., 2009; Yang et al., 2009) could be analyzed to find the molecular signatures defining a potentiated or newly formed synapse. In the simplest form, one can analyze synapses that emanate from neurons that were previously filled with dye. In this way, the scope of array tomography can offer a dynamic view of synapse composition and diversity that is integrated into a neuronal circuit schematic.
Dynamics and control of cortical blood flow in relation to the angiome (Tsai et al., 2003, 2009b; Blinder et al., 2010)
The cerebral vascular system services the constant demand for energy during neuronal activity in the brain. The determination of the geometry of this network is mandatory to understand how neurovascular control elements modulate the distribution of blood flow in the brain and, by extension, how to interpret the signals from functional magnetic resonance imaging (fMRI) techniques that infer the activation of neurons based on changes in blood flow and oxygenation. It has been suggested that the vasculature in mammalian cortex is organized as modules around neuronal units and thus has a columnar organization (Duvernoy et al., 1981; Duong et al., 2001; Zhao et al., 2005; Lauwers et al., 2008). However, fMRI-based studies do not achieve sufficient spatial resolution to analyze the underlying microvascular structure. For example, Bolan et al. (2006) attained a resolution of 24 × 24 × 78 μm3/voxel across the brain, which was sufficient to resolve major arteries and veins but insufficient to resolve microvessels (Tsai et al., 2009b). In addition, high-resolution anatomical studies of the human (Cassot et al., 2006; Lauwers et al., 2008) and primate (Weber et al., 2008) brain vasculature were not colocalized with cortical columns. To achieve the necessary combination of resolution and spatial extent, as well as colocalization of vessels and neurons, all optical histology (Tsai et al., 2003) was used to simultaneously acquire datasets of the vasculature and the cytoarchitecture of the murine primary somatosensory cortex from a fixed and labeled brain (Fig. 5A).
Figure 5.
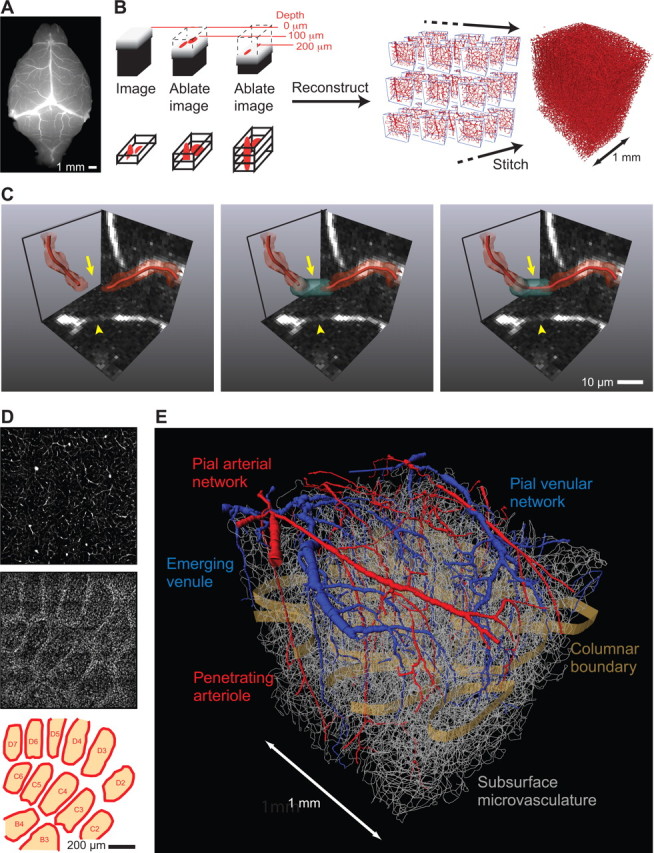
Cortical angiome and its relation with neuronal columns revealed with all optical histology. A, Gelatin-based fluorescent vascular cast of the entire vasculature of the mouse brain. B, Schematics of all optical histology block-face imaging. A sample that contains one or more fluorescently labeled structures is imaged by TPLSM to collect optical sections through the ablated surface. Sections are collected until scattering of the incident light reduces the signal-to-noise ratio below a useful value. The top of the now-imaged region of the tissue is cut away with amplified ultrashort laser pulses to expose a new surface for imaging. The sample is again imaged down to a maximal depth, the new optical sections are added to the previously stored stack, and the process of ablation and imaging is repeated. This process generates thousands of overlapping blocks that are stitched together to generate a contiguous volumes. The position of each block on the global coordinate system is determined by the least-square solution of a minimization problem defined by all cross-correlations between overlapping blocks. C, The raw images are vectorized by fitting the data to tubes. However, sections of low signal-to-noise along the vasculature can result in “gaps,” several to tens of micrometers in length, in the reconstructed microvasculature. These are corrected with an automated technique. Left, A region of low signal-to-noise ratio (arrowhead) and the resulting “gap” (arrow) seen in the max-projections of the grayscale data and its volumetric representation (red isosurface) as well as in the vectorization (red tubes). Center, Automated local threshold relaxation finds a bridging mask (cyan, arrow). Right, A corrected vectorization is computed through the gap (red tube, arrow). D, Localization of cortical columns (“barrels”) using data on all fluorescently labeled neurons. Top, Axially oriented maximal projections across 50 μm of the microvasculature in layer IV. Middle, Cell somata using fluorescent channels for a fluorophore linked to the pan-neuronal antibody αNeuN in the same dataset as the vasculature; columnar boundaries are distinguished in somata channel but not in the vasculature. Bottom, Columns outlines from the cell densities in the middle panel and labeled per convention. E, Vectorized reconstruction across a 1.5 mm3 volume of somatosensory cortex; same dataset as in D.
All optical histology combines TPLSM and plasma-mediated ablation (Oraevsky et al., 1996; Tsai et al., 2009a) to sequentially image and remove portions of the tissue (Fig. 5B). This process of imaging and ablation is repeated iteratively to acquire volumes of several cubic millimeters, with separate channels for different fluorescent indicators, e.g., a fluorescein-conjugated gel that fills the lumen of all vasculature, DAPI that stains all nuclei, and Alexa 594-conjugated to the pan-neuronal marker αNeuN to identify neuronal nuclei. This method was used to reconstruct the entire vasculature around several cortical columns in primary vibrissa cortex of mouse, i.e., “barrel” cortex (Woolsey and Van der Loos, 1970; Fox, 2008; Lefort et al., 2009), in which the input from a single facial macrovibrissa is conveyed by a set of highly organized thalamic afferents into a respective cortical column (Van Der Loos, 1976; Meyer et al., 2010b; Wimmer et al., 2010). Images were obtained at 1 × 1 × 1 μm3 sampling to yield datasets of 1 gigavoxel per channel for a cubic millimeter. The raw grayscale data for each channel is processed to obtain a vectorized representation. The vasculature is transformed into an angiome with the edges, i.e., vessel segments, annotated by their length and radius. All cell nuclei are represented by their centroid coordinates and are classified as neurons or non-neurons (Tsai et al., 2009b). One critical aspect of this process is the stitching of data blocks in all three dimensions (Fig. 5B). A second is the correction of gaps in the vascular data, for which a threshold-relaxation algorithm connects nearby ends when the gel in the lumen contains a discontinuity (Kaufhold et al., 2008) (Fig. 5C). Finally, the data on neuronal positions is used to determine the location of columnar boundaries (Fig. 5D).
The pial vasculature of cortex forms a fully connected two-dimensional graph that robustly transports blood across the cortical mantle (Schaffer et al., 2006) (Fig. 5E). Here, as with all aspects of the cortical vasculature, the coordination number for each node is three. Blood is actively redistributed from regions with low need to those with high need (Devor et al., 2007; Blinder et al., 2010). The neurons are fed by a subsurface network of tortuous microvessels that form a fully connected graph. The surface vasculature network connects to the subsurface microvasculature through a series of penetrating vessels (Fig. 5E). The issue of structural domains in the microvasculature was addressed by an exhaustive search for a statistically meaningful partition of this network (Frey and Dueck, 2007). By definition, structural modules will have higher interconnectivity between vertices within a module than between vertices of different modules. Overall, no significant topological clusters were identified, neither in the entire microvasculature nor in layer IV separately. In particular, there is no evidence for a microvascular unit that overlays a column. These findings suggest that neuronal units are embedded in a continuum of microvessels. Thus, local regulation of perfusion, and not the vasculature structure itself, governs the flow patterns observed by fMRI.
Cell type-specific reconstruction of the rodent brain at cellular resolution (Oberlaender et al., 2009b, 2011b; Meyer et al., 2010a; Lang et al., 2011)
Reverse engineering of the cortical column provides a means to link an anatomically realistic network model, which includes the structure of neurons and their short- and long-range synaptic connections, to the flow of electrical signals in the column (Helmstaedter et al., 2007; Lang et al., 2011). This reconstruction program involves four essential steps (Oberlaender et al., 2011b): (1) reconstruction of the spatial locations of neuronal somata in large brain structures, independent of the type of cell; (2) assignment of a type to each neuronal somata; (3) reconstruction of the dendritic and axonal projection patterns for each type of neuron; and (4) estimation of the synaptic connections based on the probability of overlap between dendrites and axons. The connections between cells are thus known only probabilistically. The focus is on the cortical columns in vibrissa cortex of rodent, as was the case for the vascular connectome (Fig. 5E).
The first step, reconstruction of the location of all neuronal somata, proceeds by labeling sections with αNeuN to specifically visualize all neuronal nuclei. Manually detected somata in high-resolution confocal image stacks led to a measurement of the density of neurons within entire columns in rat (Fig. 6); the resulting neuron numbers per layer in the average column were 60, 2000, 3700, 4400, 1700, 2200, 3800, and 1100 for layers 1, 2, 3, 4, 5A, 5B, 6A, and 6B respectively, for a total of 19,000 neurons per cortical column (Meyer et al., 2010a).
Figure 6.
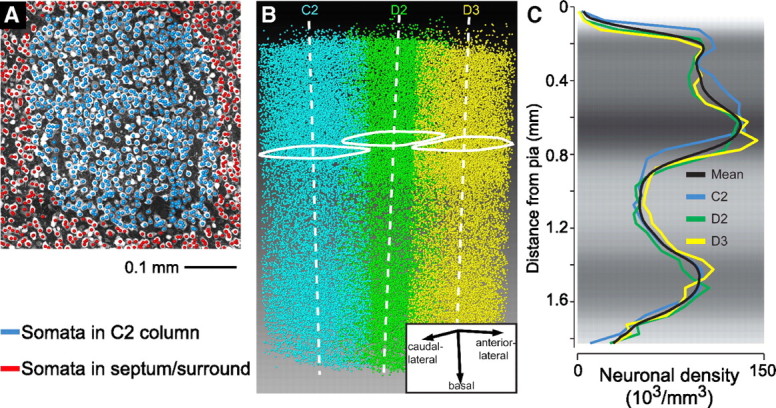
Manual detection of neuronal somata in cortical columns from vibrissa cortex of rat. A, Overlay of a maximum-intensity α-NeuN fluorescence image with markers that were manually placed at the midpoints of α-NeuN-positive somata. B, All NeuN-positive somata in the C2, D2, and D3 columns, defined by extrapolation of the barrel outline in L4 along the vertical axis. C, One-dimensional profiles of α-NeuN density along the respective vertical column axis for the C2, D2, D3, and the mean column. Adapted from Meyer et al. (2010a).
An automated image-processing pipeline facilitated a combined analysis of the first two steps, reconstruction of the location of neuronal somata and the assignment of a cell type to each neuron, within an entire rodent brain (Oberlaender et al., 2009a). In the example of Figure 7 for mouse cortex, sections were labeled with antibodies against GAD67, to specifically visualize GABAergic interneurons, in addition to α-NeuN (Meyer et al., 2011). Reconstruction of the distribution of excitatory and inhibitory neurons is achievable from 210 serial sections that were 50 μm thick. High-speed confocal microscopy at 0.36 × 0.36 × 0.6 μm3 resolution was used to form a mosaic of images across an entire section (Fig. 7B), resulting in files of ∼60 gigavoxels per section per channel or ∼25 teravoxels per mouse brain. The end result is the set of position landmarks of all excitatory and inhibitory neuron somata (Fig. 7C,D). Semiautomated postprocessing of the image stacks can be used to compensate for tissue preparation artifacts, such as tilt or local distortions of individual brain sections (Fig. 7E) and the alignment of consecutive serial sections with cellular precision (Fig. 7F). The resultant high-resolution image stack comprises the entire mouse brain and allows its reconstruction at cellular resolution (Fig. 7G).
Figure 7.
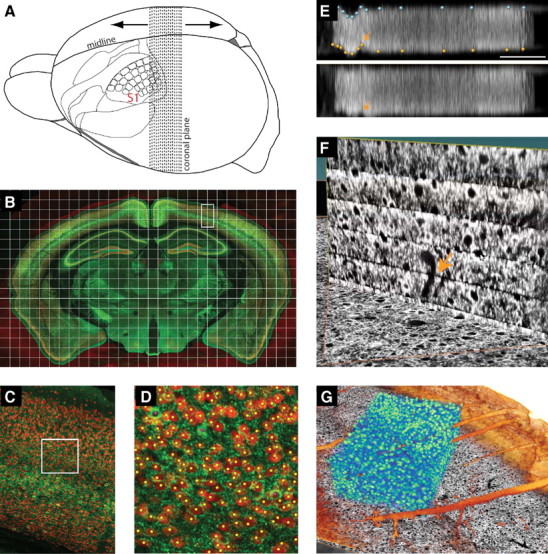
Automated methods to reconstruct an entire mouse brain at cellular resolution. A, The brain is cut along the coronal plane into ∼210 consecutive sections. B, Sections are labeled within α-NeuN (red) and α-GAD67 (green) and imaged as a mosaic with high-speed confocal microscopy. C, Enlargement of one box in B. D, Enlargement of one box in C. Excitatory (yellow) and inhibitory (pink) somata are detected and labeled using an automated image-processing pipeline. E, Tilt and/or distortions of individual brain sections are corrected by semiautomated postprocessing. F, The quality of each image stack after postprocessing is sufficient to align consecutive sections with cellular precision. G, The resultant image stack comprises the entire mouse brain and allows segmentation of anatomical structures, e.g., blood vessels or pia (brown), and quantification of soma densities (green).
For the third step, the spatial distribution of cell types within a brain region may be determined by post hoc anatomical reconstructions of dendrite and axon morphologies (Oberlaender et al., 2007, 2009a, 2011a,b) and by colocalization of neuron somata in transgenic mouse lines where specific cell types express a fluorescent protein (Groh et al., 2010). The final, fourth step is to assemble an average circuit model that combines the locations of all neurons in the reconstructed large brain areas with tracings of cell type-specific dendritic and axonal projections. The synaptic connectivity between neurons within the column and with intercolumnar projections are then estimated from the density of the overlap between axons and dendrites to form an anatomically realistic circuit (Oberlaender et al., 2011b).
In time, the serial section and block face techniques described earlier (Figs. 2, 3) may advance to the stage where entire cortical columns may be exactly reconstructed. This should reveal the nature and magnitude of possible deviations from the statistical view of connectivity. One possibility, implied by findings from studies on neuronal connectivity measured in brain slices (Thomson et al., 2002; Song et al., 2005; Otsuka and Kawaguchi, 2009; Ko et al., 2011; Perin et al., 2011), is that cortical neurons have biases in their connectivity and that some connections may be particularly strong. However, findings with slice preparations can be biased as a consequence of lesioned connections. For now, the circuits derived by this statistical approach provide the state-of-the-art basis for large-scale numerical simulations of sensory-evoked signal flow in a cortical column (www.neurodune.org) (Lang et al., 2011).
Epilogue: computation and connectomes
The computational capabilities of a neuronal circuit, and the patterns of electrical activity generated by that circuit, are constrained by the connectome. At the very minimum, the wiring diagram specified by the connectome can be used to distinguish among algorithms that support a particular computation (Seung, 2009). Lessons from past theoretical work and the analysis of simpler nervous systems provide some guidance on this issue. Tree- or bus-like architectures are suggestive of perceptrons (Minsky and Papert, 1969), in which the output of a single cell depends on the summation of weighted inputs. This is a canonical model for receptive fields based on summation across a set of receptors (Zipser and Andersen, 1988), of which the Reichardt motion detector is a particular example (Hassenstein and Reichardt, 1956). Sparse feedforward networks can function as synfire chains (Abeles, 1982), in which sparse spike activity hops along clusters of connected neurons. This is a potential model for temporal sequence generation in bird song (Long et al., 2010). In contrast, feedback loops formed by recurrent neuronal connections can support multiple patterns of persistent activity and are conjectured to be used as memories (Hopfield, 1982; Seung, 2009) and as control structures (Seung, 1996). In the simplest case, triplets of neurons with unidirectional wiring can function as a three-phase clock (Kling and Székely, 1968); this arrangement forms the aforementioned pattern generator for swimming in leech (Stent et al., 1978; Kristan et al., 2005). Studies on larger recurrent networks, such as the stomatogastric rhythm generator in crustaceans, provide qualitative support for the notion that a network can support multiple patterns of persistent activity (Miller and Selverston, 1982; Marder and Bucher, 2007). Contrariwise, different connectomes that realize the same recurrent network topology can support identical mean patterns of activity (Sompolinsky, 1988; Amit, 1989).
Considerable effort was spent toward mapping specific neuronal computations, primarily in vision, onto network architectures (Figs. 1–3). All of these involve large nervous systems that enjoy the advantage of genetic manipulation. Yet one must consider the potential value in revisiting the analysis of a simple system, such as the leech. Further, while much current emphasis in high-throughput anatomy is placed on connectomes, it would be foolish to dismiss anatomical tracing efforts that take a purely mesoscopic approach (Bohland et al., 2009; Swanson and Bota, 2010). In support of the latter possibility, recent work considered the computational capabilities of a model of the primate visual system based on only feedforward connections between brain regions (Serre et al., 2007). This model could be used to successfully detect a range of salient features in the visual field, such as a person walking into the street. All told, given the results reported here and the potential value of extensive datasets on all scales, cautious optimism is called for in the realization of connectomes across different species and brain areas.
Footnotes
The concept for the symposium that is summarized in this preview originated at the 2010 program “Emerging Techniques in Neuroscience” at the Kavli Center for Theoretical Physics at the University of California, Santa Barbara. This program was supported by the Gatsby Charitable Foundation and the National Science Foundation (Grant PHY05-51164). The collected research in this preview was supported by the Center for Brain Science at Harvard, the Gatsby Charitable Trust, the Howard Hughes Medical Institute, the Max Planck Society, Microsoft Research, the National Institutes of Health (Grants EB003832, EY010115, EY018742, MH072570, MH085499, NS059832, NS063210, NS075252, and RR006009), and the Stanford BioX Program. We thank Harvey J. Karten, Thomas D. Mrsic-Flogel, and H. Sebastian Seung for discussions and Beth Friedman, William B. Kristan, Jr, David W. Matthews, and the anonymous reviewer for critical comments on this manuscript.
References
- Abeles M. Local cortical circuits: an electrophysiological study. Berlin: Springer; 1982. [Google Scholar]
- Adelson EH, Bergen JR. Spatiotemporal energy models for the perception of motion. J Opt Soc Am A. 1985;2:284–299. doi: 10.1364/josaa.2.000284. [DOI] [PubMed] [Google Scholar]
- Amit DJ. Modeling brain function: the world of attractor neural networks. Cambridge: Cambridge UP; 1989. [Google Scholar]
- Blinder P, Shih AY, Rafie C, Kleinfeld D. Topological basis for the robust distribution of blood to rodent neocortex. Proc Natl Acad Sci U S A. 2010;107:12670–12675. doi: 10.1073/pnas.1007239107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock DD, Lee WC, Kerlin AM, Andermann ML, Hood G, Wetzel AW, Yurgenson S, Soucy ER, Kim HS, Reid RC. Network anatomy and in vivo physiology of visual cortical neurons. Nature. 2011;471:177–182. doi: 10.1038/nature09802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohland JW, Wu C, Barbas H, Bokil H, Bota M, Breiter HC, Cline HT, Doyle JC, Freed PJ, Greenspan RJ, Haber SN, Hawrylycz M, Herrera DG, Hilgetag CC, Huang ZJ, Jones A, Jones EG, Karten HJ, Kleinfeld D, Kötter R, et al. A proposal for a coordinated effort for the determination of brainwide neuroanatomical connectivity in model organisms at a mesoscopic scale. PLoS Comput Biol. 2009;5:e1000334. doi: 10.1371/journal.pcbi.1000334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolan PJ, Yacoub E, Garwood M, Ugurbil K, Harel N. In vivo micro-MRI of intracortical neurovasculature. Neuroimage. 2006;32:62–69. doi: 10.1016/j.neuroimage.2006.03.027. [DOI] [PubMed] [Google Scholar]
- Borst A, Egelhaaf M. Temporal modulation of luminance adapts time constant of fly movement detectors. Biol Cybern. 1987;56:209–215. [Google Scholar]
- Borst A, Flanagin VL, Sompolinsky H. Adaptation without parameter change: dynamic gain control in motion detection. Proc Natl Acad Sci U S A. 2005;102:6172–6176. doi: 10.1073/pnas.0500491102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst A, Haag J, Reiff DF. Fly motion vision. Annu Rev Neurosci. 2010;33:49–70. doi: 10.1146/annurev-neuro-060909-153155. [DOI] [PubMed] [Google Scholar]
- Briggman KL, Denk W. Towards neural circuit reconstruction with volume electron microscopy techniques. Curr Opin Neurobiol. 2006;16:562–570. doi: 10.1016/j.conb.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Briggman KL, Euler T. Bulk electroporation and population calcium imaging in the adult mammalian retina. J Neurophysiol. 2011;105:2601–2609. doi: 10.1152/jn.00722.2010. [DOI] [PubMed] [Google Scholar]
- Briggman KL, Helmstaedter M, Denk W. Wiring specificity in the direction-selectivity circuit of the retina. Nature. 2011;471:183–188. doi: 10.1038/nature09818. [DOI] [PubMed] [Google Scholar]
- Brotz TM, Borst A. Cholinergic and GABAergic receptors on fly tangential cells and their role in visual motion detection. J Neurophysiol. 1996;76:1786–1799. doi: 10.1152/jn.1996.76.3.1786. [DOI] [PubMed] [Google Scholar]
- Buchner E. Elementary movement detectors in an insect visual-system. Biol Cybern. 1976;24:85–101. [Google Scholar]
- Buchner E. Behavioural analysis of spatial vision in insects. In: Ali MA, editor. Photoreception and vision in invertebrates. New York: Plenum; 1984. pp. 561–621. [Google Scholar]
- Bug WJ, Ascoli GA, Grethe JS, Gupta A, Fennema-Notestine C, Laird AR, Larson SD, Rubin D, Shepherd GM, Turner JA, Martone ME. The NIFSTD and BIRNLex vocabularies: building comprehensive ontologies for neuroscience. Neuroinformatics. 2008;6:175–194. doi: 10.1007/s12021-008-9032-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock TH. How do brains work? Boston: Birkhäuser; 1993. [Google Scholar]
- Cassot F, Lauwers F, Fouard C, Prohaska S, Lauwers-Cances V. A novel three-dimensional computer-assisted method for a quantitative study of microvascular networks of the human cerebral cortex. Microcirculation. 2006;13:1–18. doi: 10.1080/10739680500383407. [DOI] [PubMed] [Google Scholar]
- Chklovskii DB, Vitaladevuni S, Scheffer LK. Semi-automated reconstruction of neural circuits using electron microscopy. Curr Opin Neurobiol. 2010;20:667–675. doi: 10.1016/j.conb.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Clark DA, Bursztyn L, Horowitz MA, Schnitzer MJ, Clandinin TR. Defining the computational structure of the motion detector in Drosophila. Neuron. 2011;70:1165–1177. doi: 10.1016/j.neuron.2011.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements J, Lu Z, Gehring WJ, Meinertzhagen IA, Callaerts P. Central projections of photoreceptor axons originating from ectopic eyes in Drosophila. Proc Natl Acad Sci U S A. 2008;105:8968–8973. doi: 10.1073/pnas.0803254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFelipe J. From the connectome to the synaptome: an epic love story. Science. 2010;330:1198–1201. doi: 10.1126/science.1193378. [DOI] [PubMed] [Google Scholar]
- Delcomyn F. Neural basis of rhythmic behavior in animals. Science. 1980;210:492–498. doi: 10.1126/science.7423199. [DOI] [PubMed] [Google Scholar]
- Denk W, Detwiler PB. Optical recording of light-evoked calcium signals in the functionally intact retina. Proc Natl Acad Sci U S A. 1999;96:7035–7040. doi: 10.1073/pnas.96.12.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denk W, Horstmann H. Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol. 2004;2:e329. doi: 10.1371/journal.pbio.0020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- Devor A, Tian P, Nishimura N, Teng IC, Hillman EM, Narayanan SN, Ulbert I, Boas DA, Kleinfeld D, Dale AM. Suppressed neuronal activity and concurrent arteriolar vasoconstriction may explain negative blood oxygenation level-dependent signaling. J Neurosci. 2007;27:4452–4459. doi: 10.1523/JNEUROSCI.0134-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong TQ, Kim DS, Uğurbil K, Kim SG. Localized cerebral blood flow response at submillimeter columnar resolution. Proc Natl Acad Sci U S A. 2001;98:10904–10909. doi: 10.1073/pnas.191101098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvernoy HM, Delon S, Vannson JL. Cortical blood vessels of the human brain. Brain Res Bull. 1981;7:519–579. doi: 10.1016/0361-9230(81)90007-1. [DOI] [PubMed] [Google Scholar]
- Egelhaaf M, Borst A. Transient and steady-state response properties of movement detectors. J Opt Soc Am A. 1989;6:116–127. doi: 10.1364/josaa.6.000116. [DOI] [PubMed] [Google Scholar]
- Egelhaaf M, Reichardt W. Dynamic-response properties of movement detectors: theoretical-analysis and electrophysiological investigation in the visual-system of the fly. Biol Cybern. 1987;56:69–87. [Google Scholar]
- Egelhaaf M, Borst A, Reichardt W. Computational structure of a biological motion-detection system as revealed by local detector analysis in the fly's nervous system. J Opt Soc Am A. 1989;6:1070–1087. doi: 10.1364/josaa.6.001070. [DOI] [PubMed] [Google Scholar]
- Eichner H, Joesch M, Schnell B, Reiff DF, Borst A. Internal structure of the fly elementary motion detector. Neuron. 2011;70:1155–1164. doi: 10.1016/j.neuron.2011.03.028. [DOI] [PubMed] [Google Scholar]
- Euler T, Detwiler PB, Denk W. Directionally selective calcium signals in dendrites of starburst amacrine cells. Nature. 2002;418:845–852. doi: 10.1038/nature00931. [DOI] [PubMed] [Google Scholar]
- Fang Q, Sakadzić S, Ruvinskaya L, Devor A, Dale AM, Boas DA. Oxygen advection and diffusion in a three-dimensional vascular anatomical network. Optics Express. 2008;16:17530–17541. doi: 10.1364/oe.16.17530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Fischbach KF, Dittrich APM. The optic lobe of Drosophila melanogaster. 1. A Golgi analysis of wild-type structure. Cell Tissue Res. 1989;258:441–475. [Google Scholar]
- Fox K. Barrel cortex. Cambridge, UK: Cambridge UP; 2008. [Google Scholar]
- Frey BJ, Dueck D. Clustering by passing messages between data points. Science. 2007;315:972–976. doi: 10.1126/science.1136800. [DOI] [PubMed] [Google Scholar]
- Fried SI, Münch TA, Werblin FS. Mechanisms and circuitry underlying directional selectivity in the retina. Nature. 2002;420:411–414. doi: 10.1038/nature01179. [DOI] [PubMed] [Google Scholar]
- Getting PA. Mechanisms of pattern generation underlying swimming in Tritonia. II. Network reconstruction. J Neurophysiol. 1983;49:1017–1035. doi: 10.1152/jn.1983.49.4.1017. [DOI] [PubMed] [Google Scholar]
- Groh A, Meyer HS, Schmidt EF, Heintz N, Sakmann B, Krieger P. Cell-type specific properties of pyramidal neurons in neocortex underlying a layout that is modifiable depending on the cortical area. Cereb Cortex. 2010;20:826–836. doi: 10.1093/cercor/bhp152. [DOI] [PubMed] [Google Scholar]
- Haag J, Borst A. Orientation tuning of motion-sensitive neurons shaped by vertical-horizontal network interactions. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2003;189:363–370. doi: 10.1007/s00359-003-0410-6. [DOI] [PubMed] [Google Scholar]
- Harris KM, Perry E, Bourne J, Feinberg M, Ostroff L, Hurlburt J. Uniform serial sectioning for transmission electron microscopy. J Neurosci. 2006;26:12101–12103. doi: 10.1523/JNEUROSCI.3994-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassenstein B, Reichardt W. Courellis SH, Marmarelis VZ, editors. Structure of a mechanism of perception of optical movement. First international conference on cybernetics. 1956:797–801. [Google Scholar]
- Hausselt SE, Euler T, Detwiler PB, Denk W. Dendrite-autonomous mechanism for direction selectivity in retinal starburst amacrine cells. PLoS Biol. 2007;5:e185. doi: 10.1371/journal.pbio.0050185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmstaedter M, de Kock CP, Feldmeyer D, Bruno RM, Sakmann B. Reconstruction of an average cortical column in silico. Brain Res Rev. 2007;55:193–203. doi: 10.1016/j.brainresrev.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Helmstaedter M, Briggman KL, Denk W. High-accuracy neurite reconstruction for high-throughput neuroanatomy. Nat Neurosci. 2011;14:1081–1088. doi: 10.1038/nn.2868. [DOI] [PubMed] [Google Scholar]
- Hildreth EC, Koch C. The analysis of visual motion: from computational theory to neuronal mechanisms. Annu Rev Neurosci. 1987;10:477–533. doi: 10.1146/annurev.ne.10.030187.002401. [DOI] [PubMed] [Google Scholar]
- Hofer SB, Ko H, Pichler B, Vogelstein J, Ros H, Zeng H, Lein E, Lesica NA, Mrsic-Flogel TD. Differential connectivity and response dynamics of excitatory and inhibitory neurons in visual cortex. Nat Neurosci. 2011;14:1045–1052. doi: 10.1038/nn.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfield JJ. Neural networks and physical systems with emergent collective computational abilities. Proc Natl Acad Sci U S A. 1982;79:2554–2558. doi: 10.1073/pnas.79.8.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain V, Seung HS, Turaga SC. Machines that learn to segment images: a crucial technology for connectomics. Curr Opin Neurobiol. 2010;20:653–666. doi: 10.1016/j.conb.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufhold J, Tsai PS, Blinder P, Kleinfeld D. Third workshop on microscopic image analysis with applications in biology. New York: Medical Image Computing and Computer Assisted Intervention Society; 2008. Threshold relaxation is an effective means to connect gaps in 3D images of complex microvascular networks. [Google Scholar]
- Kerlin AM, Andermann ML, Berezovskii VK, Reid RC. Broadly tuned response properties of diverse inhibitory neuron subtypes in mouse visual cortex. Neuron. 2010;67:858–871. doi: 10.1016/j.neuron.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr JN, Greenberg D, Helmchen F. Imaging input and output of neocortical networks in vivo. Proc Natl Acad Sci U S A. 2005;102:14063–14068. doi: 10.1073/pnas.0506029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausberger T, Somogyi P. Neuronal diversity and temporal dynamics: the unity of hippocampal circuit operations. Science. 2008;321:53–57. doi: 10.1126/science.1149381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinfeld D, Sompolinsky H. Associative neural network model for the generation of temporal patterns: theory and application to central pattern generators. Biophys J. 1988;54:1039–1051. doi: 10.1016/S0006-3495(88)83041-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kling U, Székely G. Simulation of rhythmic activities I. Function of networks with cyclic inhibitions. Kybernetik. 1968;5:89–103. doi: 10.1007/BF00288899. [DOI] [PubMed] [Google Scholar]
- Knott G, Marchman H, Wall D, Lich B. Serial section scanning electron microscopy of adult brain tissue using focused ion beam milling. J Neurosci. 2008;28:2959–2964. doi: 10.1523/JNEUROSCI.3189-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko H, Hofer SB, Pichler B, Buchanan KA, Sjöström PJ, Mrsic-Flogel TD. Functional specificity of local synaptic connections in neocortical networks. Nature. 2011;473:87–91. doi: 10.1038/nature09880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristan WB, Jr, Calabrese RL, Friesen WO. Neuronal basis of leech behaviors. Prog Neurobiol. 2005;76:279–327. doi: 10.1016/j.pneurobio.2005.09.004. [DOI] [PubMed] [Google Scholar]
- Kuhlman SJ, Tring E, Trachtenberg JT. Fast-spiking interneurons have an initial orientation bias that is lost with vision. Nat Neurosci. 2011;14:1121–1123. doi: 10.1038/nn.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang S, Dercksen VJ, Sakmann B, Oberlaender M. Simulation of signal flow in 3D reconstructions of an anatomically realistic neural network in rat vibrissal cortex. Neural Netw. 2011;24:998–1011. doi: 10.1016/j.neunet.2011.06.013. [DOI] [PubMed] [Google Scholar]
- Laughlin SB. Progress in retinal and eye research. Oxford: Pergamon; 1994. Matching coding, circuits, cells, and molecules to signals: general principles of retinal design in the fly's eye; pp. 167–196. [Google Scholar]
- Laughlin SB, Hardie RC. Common strategies for light adaptation in the peripheral visual systems of fly and dragonfly. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1978;128:319–340. [Google Scholar]
- Lauwers F, Cassot F, Lauwers-Cances V, Puwanarajah P, Duvernoy H. Morphometry of the human cerebral cortex microcirculation: general characteristics and space-related profiles. Neuroimage. 2008;39:936–948. doi: 10.1016/j.neuroimage.2007.09.024. [DOI] [PubMed] [Google Scholar]
- Lefort S, Tomm C, Floyd Sarria J-C, Petersen CCH. The excitatory neuronal network of the C2 barrel column in mouse primary somatosensory cortex. Neuron. 2009;61:301–316. doi: 10.1016/j.neuron.2008.12.020. [DOI] [PubMed] [Google Scholar]
- Li L, Tasic B, Micheva KD, Ivanov VM, Spletter ML, Smith SJ, Luo L. Visualizing the distribution of synapses from individual neurons in the mouse brain. PLoS One. 2010;5:e11503. doi: 10.1371/journal.pone.0011503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtman JW, Livet J, Sanes JR. A technicolour approach to the connectome. Nat Rev Neurosci. 2008;9:417–422. doi: 10.1038/nrn2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu BH, Li P, Li YT, Sun YJ, Yanagawa Y, Obata K, Zhang LI, Tao HW. Visual receptive field structure of cortical inhibitory neurons revealed by two-photon imaging guided recording. J Neurosci. 2009;29:10520–10532. doi: 10.1523/JNEUROSCI.1915-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long MA, Jin DZ, Fee MS. Support for a synaptic chain model of neuronal sequence generation. Nature. 2010;468:394–399. doi: 10.1038/nature09514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Tapia JC, White OL, Lichtman JW. The interscutularis muscle connectome. PLoS Biol. 2009;7:e32. doi: 10.1371/journal.pbio.1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma WP, Liu BH, Li YT, Huang ZJ, Zhang LI, Tao HW. Visual representations by cortical somatostatin inhibitory neurons: selective but with weak and delayed responses. J Neurosci. 2010;30:14371–14379. doi: 10.1523/JNEUROSCI.3248-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Pokala N, Feinberg EH, Chalasani SH, Butcher RA, Clardy J, Bargmann CI. A hub-and-spoke circuit drives pheromone attraction and social behaviour in C. elegans. Nature. 2009;458:1171–1175. doi: 10.1038/nature07886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marder E, Bucher D. Understanding circuit dynamics using the stomatogastric nervous system of lobsters and crabs. Annu Rev Physiol. 2007;69:291–316. doi: 10.1146/annurev.physiol.69.031905.161516. [DOI] [PubMed] [Google Scholar]
- Meyer HS, Wimmer VC, Oberlaender M, de Kock CP, Sakmann B, Helmstaedter M. Number and laminar distribution of neurons in a thalamocortical projection column of rat vibrissal cortex. Cereb Cortex. 2010a;20:2277–2286. doi: 10.1093/cercor/bhq067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HS, Wimmer VC, Hemberger M, Bruno RM, de Kock CP, Frick A, Sakmann B, Helmstaedter M. Cell type-specific thalamic innervation in a column of rat vibrissal cortex. Cereb Cortex. 2010b;20:2287–2303. doi: 10.1093/cercor/bhq069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HS, Schwarz D, Wimmer VC, Schmitt AC, Kerr JND, Sakmann B, Helmstaedter M. Inhibitory interneurons in a cortical column form hot zones of inhibition in layers 2 and 5A. Proc Natl Acad Sci U S A. 2011;108:16807–16812. doi: 10.1073/pnas.1113648108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheva KD, Smith SJ. Array tomography: a new tool for imaging the molecular architecture and ultrastructure of neural circuits. Neuron. 2007;55:25–36. doi: 10.1016/j.neuron.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheva KD, Busse B, Weiler NC, O'Rourke N, Smith SJ. Single-synapse analysis of a diverse synapse population: proteomic imaging methods and markers. Neuron. 2010;68:639–653. doi: 10.1016/j.neuron.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JP, Selverston AI. Mechanisms underlying pattern generation in lobster stomatogastric ganglion as determined by selective inactivation of identified neurons. IV. Network properties of pyloric system. J Neurophysiol. 1982;48:1416–1432. doi: 10.1152/jn.1982.48.6.1416. [DOI] [PubMed] [Google Scholar]
- Minsky ML, Papert SA. Perceptrons. Cambridge: MIT Press; 1969. [Google Scholar]
- Murakoshi H, Wang H, Yasuda R. Local, persistent activation of Rho GTPases during plasticity of single dendritic spines. Nature. 2011;472:100–104. doi: 10.1038/nature09823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niell CM, Stryker MP. Modulation of visual responses by behavioral state in mouse visual cortex. Neuron. 2010;65:472–479. doi: 10.1016/j.neuron.2010.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberlaender M, Bruno RM, Sakmann B, Broser PJ. Transmitted light brightfield mosaic microscopy for three-dimensional tracing of single neuron morphology. J Biomed Opt. 2007;12 doi: 10.1117/1.2815693. 064029. [DOI] [PubMed] [Google Scholar]
- Oberlaender M, Broser PJ, Sakmann B, Hippler S. Shack-Hartmann wave front measurements in cortical tissue for deconvolution of large three-dimensional mosaic transmitted light brightfield micrographs. J Microsc. 2009a;233:275–289. doi: 10.1111/j.1365-2818.2009.03118.x. [DOI] [PubMed] [Google Scholar]
- Oberlaender M, Dercksen VJ, Egger R, Gensel M, Sakmann B, Hege HC. Automated three-dimensional detection and counting of neuron somata. J Neurosci Methods. 2009b;180:147–160. doi: 10.1016/j.jneumeth.2009.03.008. [DOI] [PubMed] [Google Scholar]
- Oberlaender M, Boudewijns ZS, Kleele T, Mansvelder HD, Sakmann B, de Kock CP. Three-dimensional axon morphologies of individual layer 5 neurons indicate cell type-specific intracortical pathways for whisker motion and touch. Proc Natl Acad Sci U S A. 2011a;108:4188–4193. doi: 10.1073/pnas.1100647108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberlaender M, de Kock CPJ, Bruno RM, Ramirez A, Meyer HS, Dercksen VJ, Helmstaedter M, Sakmann B. Cell type-specific three-dimensional structure of thalamocortical networks in a barrel column in rat vibrissal cortex. Cereb Cortex. 2011b doi: 10.1093/cercor/bhr317. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohki K, Chung S, Ch'ng YH, Kara P, Reid RC. Functional imaging with cellular resolution reveals precise microarchitecture in visual cortex. Nature. 2005;433:597–603. doi: 10.1038/nature03274. [DOI] [PubMed] [Google Scholar]
- Oraevsky A, Da Silva L, Rubenchik A, Feit M, Glinsky M, Perry M, Mammini B, Small W, Stuart B. Plasma mediated ablation of biological tissues with nanosecond-to-femtosecond laser pulses: relative role of linear and nonlinear absorption. IEEE J Sel Top Quantum Electron. 1996;2:801–809. [Google Scholar]
- Otsuka T, Kawaguchi Y. Cortical inhibitory cell types differentially form intralaminar and interlaminar subnetworks with excitatory neurons. J Neurosci. 2009;29:10533–10540. doi: 10.1523/JNEUROSCI.2219-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perin R, Berger TK, Markram H. A synaptic organizing principle for cortical neuronal groups. Proc Natl Acad Sci U S A. 2011;108:5419–5424. doi: 10.1073/pnas.1016051108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, Mungall C, Svirskas R, Kadonaga JT, Doe CQ, Eisen MB, Celniker SE, Rubin GM. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci U S A. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poggio T, Reichardt W. Visual control of orientation behaviour in the fly. Part II. Towards the underlying neural interactions. Q Rev Biophys. 1976;9:377–438. doi: 10.1017/s0033583500002535. [DOI] [PubMed] [Google Scholar]
- Potters M, Bialek W. Statistical mechanics and visual signal processing. J Phys I (France) 1994;4:1755–1775. [Google Scholar]
- Runyan CA, Schummers J, Van Wart A, Kuhlman SJ, Wilson NR, Huang ZJ, Sur M. Response features of parvalbumin-expressing interneurons suggest precise roles for subtypes of inhibition in visual cortex. Neuron. 2010;67:847–857. doi: 10.1016/j.neuron.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterlie RA. Reciprocal inhibition and postinhibitory rebound produces reverberation in a locomotor pattern generator. Science. 1985;229:402–404. doi: 10.1126/science.229.4711.402. [DOI] [PubMed] [Google Scholar]
- Schaffer CB, Friedman B, Nishimura N, Schroeder LF, Tsai PS, Ebner FF, Lyden PD, Kleinfeld D. Two-photon imaging of cortical surface microvessels reveals a robust redistribution in blood flow after vascular occlusion. PLoS Biol. 2006;4:e22. doi: 10.1371/journal.pbio.0040022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell B, Joesch M, Forstner F, Raghu SV, Otsuna H, Ito K, Borst A, Reiff DF. Processing of horizontal optic flow in three visual interneurons of the Drosophila brain. J Neurophysiol. 2010;103:1646–1657. doi: 10.1152/jn.00950.2009. [DOI] [PubMed] [Google Scholar]
- Schuling FH, Mastebroek HAK, Bult R, Lenting BPM. Properties of elementary movement detectors in the fly Calliphora erythrocephala. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1989;165:179–192. [Google Scholar]
- Seelig JD, Chiappe ME, Lott GK, Dutta A, Osborne JE, Reiser MB, Jayaraman V. Two-photon calcium imaging from head-fixed Drosophila during optomotor walking behavior. Nat Methods. 2010;7:535–540. doi: 10.1038/nmeth.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serre T, Oliva A, Poggio T. A feedforward architecture accounts for rapid categorization. Proc Natl Acad Sci U S A. 2007;104:6424–6429. doi: 10.1073/pnas.0700622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seung HS. How the brain keeps the eyes still. Proc Natl Acad Sci U S A. 1996;93:13339–13344. doi: 10.1073/pnas.93.23.13339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seung HS. Reading the book of memory: sparse sampling versus dense mapping of connectomes. Neuron. 2009;62:17–29. doi: 10.1016/j.neuron.2009.03.020. [DOI] [PubMed] [Google Scholar]
- Shepherd GM, Grillner S. Handbook of brain microcircuits. Oxford: Oxford UP; 2010. [Google Scholar]
- Sohya K, Kameyama K, Yanagawa Y, Obata K, Tsumoto T. GABAergic neurons are less selective to stimulus orientation than excitatory neurons in layer II/III of visual cortex, as revealed by in vivo functional Ca2+ imaging in transgenic mice. J Neurosci. 2007;27:2145–2149. doi: 10.1523/JNEUROSCI.4641-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sompolinsky H. Statistical mechanics of neural networks. Phys Today. 1988;41:70–80. [Google Scholar]
- Song S, Sjöström PJ, Reigl M, Nelson S, Chklovskii DB. Highly nonrandom features of synaptic connectivity in local cortical circuits. PLoS Biol. 2005;3:e350. doi: 10.1371/journal.pbio.0030068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorra KE, Harris KM. Overview on the structure, composition, function, development, and plasticity of hippocampal dendritic spines. Hippocampus. 2000;10:501–511. doi: 10.1002/1098-1063(2000)10:5<501::AID-HIPO1>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Sporns O, Tononi G, Kötter R. The human connectome: a structural description of the human brain. PLoS Comput Biol. 2005;1:e42. doi: 10.1371/journal.pcbi.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stent GS, Kristan WB, Jr, Friesen WO, Ort CA, Poon M, Calabrese RL. Neuronal generation of the leech swimming movement. Science. 1978;200:1348–1357. doi: 10.1126/science.663615. [DOI] [PubMed] [Google Scholar]
- Stosiek C, Garaschuk O, Holthoff K, Konnerth A. In vivo two-photon calcium imaging of neuronal networks. Proc Natl Acad Sci U S A. 2003;100:7319–7324. doi: 10.1073/pnas.1232232100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda K, Denk W, Kleinfeld D, Tank DW. In vivo dendritic calcium dynamics in neocortical pyramidal neurons. Nature. 1997;385:161–165. doi: 10.1038/385161a0. [DOI] [PubMed] [Google Scholar]
- Swanson LW, Bota M. Foundational model of structural connectivity in the nervous system with a schema for wiring diagrams, connectome, and basic plan architecture. Proc Natl Acad Sci U S A. 2010;107:20610–20617. doi: 10.1073/pnas.1015128107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura SY, Lu Z, Meinertzhagen IA. Synaptic circuits of the Drosophila optic lobe: the input terminals to the medulla. J Comp Neurol. 2008;509:493–513. doi: 10.1002/cne.21757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson AM, West DC, Wang Y, Bannister AP. Synaptic connections and small circuits involving excitatory and inhibitory neurons in layers 2–5 of adult rat and cat neocortex: triple intracellular recordings and biocytin labelling in vitro. Cereb Cortex. 2002;12:936–953. doi: 10.1093/cercor/12.9.936. [DOI] [PubMed] [Google Scholar]
- Tsai PS, Friedman B, Ifarraguerri AI, Thompson BD, Lev-Ram V, Schaffer CB, Xiong Q, Tsien RY, Squier JA, Kleinfeld D. All-optical histology using ultrashort laser pulses. Neuron. 2003;39:27–41. doi: 10.1016/s0896-6273(03)00370-2. [DOI] [PubMed] [Google Scholar]
- Tsai PS, Blinder P, Migliori BJ, Neev J, Jin Y, Squier JA, Kleinfeld D. Plasma-mediated ablation: an optical tool for submicrometer surgery on neuronal and vascular systems. Curr Opin Biotechnol. 2009a;20:90–99. doi: 10.1016/j.copbio.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai PS, Kaufhold JP, Blinder P, Friedman B, Drew PJ, Karten HJ, Lyden PD, Kleinfeld D. Correlations of neuronal and microvascular densities in murine cortex revealed by direct counting and colocalization of cell nuclei and microvessels. J Neurosci. 2009b;29:14553–14570. doi: 10.1523/JNEUROSCI.3287-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Loos H. Barreloids on the mouse somatosensory thalamus. Neurosci Lett. 1976;2:1–6. doi: 10.1016/0304-3940(76)90036-7. [DOI] [PubMed] [Google Scholar]
- Varshney LR, Chen BL, Paniagua E, Hall DH, Chklovskii DB. Structural properties of the Caenorhabditis elegans neuronal network. PLoS Comput Biol. 2011;7:e1001066. doi: 10.1371/journal.pcbi.1001066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber B, Keller AL, Reichold J, Logothetis NK. The microvascular system of the striate and extrastriate visual cortex of the macaque. Cereb Cortex. 2008;18:2318–2330. doi: 10.1093/cercor/bhm259. [DOI] [PubMed] [Google Scholar]
- Wei W, Hamby AM, Zhou K, Feller MB. Development of asymmetric inhibition underlying direction selectivity in the retina. Nature. 2011;469:402–406. doi: 10.1038/nature09600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Wimmer VC, Bruno RM, de Kock CP, Kuner T, Sakmann B. Dimensions of a projection column and architecture of VPM and POm axons in rat vibrissal cortex. Cereb Cortex. 2010;20:2265–2276. doi: 10.1093/cercor/bhq068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolsey TA, Van der Loos H. The structural organization of layer IV in the somatosensory region (SI) of mouse cerebral cortex. Brain Res. 1970;17:205–242. doi: 10.1016/0006-8993(70)90079-x. [DOI] [PubMed] [Google Scholar]
- Xu T, Yu X, Perlik AJ, Tobin WF, Zweig JA, Tennant K, Jones T, Zuo Y. Rapid formation and selective stabilization of synapses for enduring motor memories. Nature. 2009;462:915–919. doi: 10.1038/nature08389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Pan F, Gan WB. Stably maintained dendritic spines are associated with lifelong memories. Nature. 2009;462:920–924. doi: 10.1038/nature08577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanker JM, Srinivasan MV, Egelhaaf M. Speed tuning in elementary motion detectors of the correlation type. Biol Cybern. 1999;80:109–116. doi: 10.1007/s004220050509. [DOI] [PubMed] [Google Scholar]
- Zariwala HA, Madisen L, Ahrens KF, Bernard A, Lein ES, Jones AR, Zeng H. Visual tuning properties of genetically identified layer 2/3 neuronal types in the primary visual cortex of cre-transgenic mice. Front Syst Neurosci. 2011;4:162. doi: 10.3389/fnsys.2010.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Chung SH, Fang-Yen C, Craig C, Kerr RA, Suzuki H, Samuel AD, Mazur E, Schafer WR. A self-regulating feed-forward circuit controlling C. elegans egg-laying behavior. Curr Biol. 2008;18:1445–1455. doi: 10.1016/j.cub.2008.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F, Wang P, Hendrich K, Kim SG. Spatial specificity of cerebral blood volume-weighted fMRI responses at columnar resolution. Neuroimage. 2005;27:416–424. doi: 10.1016/j.neuroimage.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Zheng L, de Polavieja GG, Wolfram V, Asyali MH, Hardie RC, Juusola M. Feedback network controls photoreceptor output at the layer of first visual synapses in Drosophila. J Gen Physiol. 2006;127:495–510. doi: 10.1085/jgp.200509470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Nikolaev A, Wardill TJ, O'Kane CJ, de Polavieja GG, Juusola M. Network adaptation improves temporal representation of naturalistic stimuli in Drosophila eye: I. Dynamics. PLoS One. 2009;4:e4307. doi: 10.1371/journal.pone.0004307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipser D, Andersen RA. A back-propagation programmed network that simulates response properties of a subset of posterior parietal neurons. Nature. 1988;331:679–684. doi: 10.1038/331679a0. [DOI] [PubMed] [Google Scholar]