Abstract
BACKGROUND AND OBJECTIVES:
Methionine synthase reductase (MTRR) is a vital enzyme of homocysteine/methionine metabolic pathway and is required for the conversion of inactive form of methionine synthase (MTR) to its active form. A clinically important allelic variant of MTRR A66G, with less enzymatic activity is reported with worldwide prevalence rate of ~ 30%. The present study was designed to determine the frequency of MTRR A66G polymorphism in rural Sunni Muslim population of Eastern Uttar Pradesh.
MATERIALS AND METHODS:
Total 56 subjects were analyzed for MTRR A66G polymorphism. A66G mutation analysis was carried out according to the polymerase chain reaction-restriction fragment length polymorphism method of Wilson et al. [1] amplification with MTRR specific primers followed by amplicon digestion with NdeI enzyme was used for the identification of different MTRR genotypes in subjects.
RESULTS AND DISCUSSION:
The AA genotype was found in 5 subjects, AG in 23 subjects, and GG genotype in 28 subjects. Genotype frequencies of AA, AG, and GG were 0.089, 0.41, and 0.5 respectively. The allele frequency of A allele was found to be 0.298 and G allele was 0.705.
CONCLUSION:
It is evident from the present study that the percentage of homozygous genotype GG and frequency of G allele is high in the target Muslim population.
Keywords: A66G, allele, genotype, methionine synthase reductase, polymorphism
Introduction
Methionine synthase reductase (MTRR) enzyme is a member of electron transferase family and plays a vital role in folate dependent homocysteine/methionine metabolism. MTRR enzyme catalyzes the conversion of inactive form of methionine synthase (MTR) (another enzyme of the homocysteine metabolic pathway) into its active form, by regeneration of cofactor methyl (III) cobalamin of MTR. The enzyme has three characteristic sites which bind FMN, FAD and NADH. In 1999, working on homocysteinuria, Wilson et al. and colleagues[1] identified a variant of MTRR (A66G) enzyme in which methionine replaces isoleucine at amino acid 22 (I22M). The frequency of the I22M variant enzyme is reported ~30% worldwide.[1–4] The I22M substitution is located in the FMN-binding domain of the MTRR enzyme, which is interacted with methionine synthase.[5]
MTRR gene is a house keeping gene and located at chromosome 5p15.2-15.3 and its expression is low and almost the same in all examined tissues. The size of MTRR gene is ~ 34kb and is made of 15 exons (10% of the whole gene), its RNA can be alternatively spliced.[5,6] The most common polymorphism is A66G substitution, in which adenine is replaced by guanine and this substitution reduces the efficiency of binding of MTRR to MTR-Cob (I) alamine complex, thereby decreasing the rate of activation of MTR enzyme. Gaughan et al.[2] have reported that MTRR A66G polymorphism is associated with elevated plasma homocysteine concentration. Kluijtmans et al.[7] also detected a trend toward higher homocysteine concentration with the presence of the G allele. Other studies however, could not find such an effect of MTRR A66G polymorphism on homocysteine concentration.[1,8–11] Olteanu et al.[12,13] have reported that the I22M variant (A66G) MTRR enzyme exhibits four-fold lower activity than the wild-type protein in the reactivation of MTR in vivo. Hence, the level of active MTR is reduced and leads to DNA hypomethylation and it was pointed out by several studies that the DNA hypomethylation is the main causative factor in the chromosome missegregation, micronucleus formation, and defective gene expression etc.[14] Several epidemiological and case control studies have reported that the GG genotype may be a risk factor for several disease/disorders like Neural tube defects,[1,6,15–18] Down syndrome,[1,19–21] Coronory artery disease,[4,22,23] male infertility,[24,25] Cancer[26] etc., Hence, the aim of the present study is to determine the frequency of clinically important A66G polymorphism in rural Sunni Muslim population of Eastern Uttar Pradesh. Muslims of India make up more than 12% of the population.[27] They belong to two major sects; Sunnis and Shias, while each sect has different Biradaris grouped under Ashraf and Ajlafs,[28] these groups are based on traditional, social and occupational divisions. Ashraf comprises higher rank Muslims and Ajlaf includes lower rank Muslims like – Qureshi, Saifi, Ansari, and other lower occupation.[29] In the present study, MTRR A66G polymorphism analysis was carried out in 56 subjects belonging to a backward lower rank community of Sunni Muslims.
Materials and Methods
Samples
Ethical Clearance Certificate for the present study was obtained from Institutional Ethics Committee of VBS Purvanchal University, Jaunpur. Total 56 healthy unrelated subjects belonging to Sunni Muslim religion, between the age group of 18 years to70 years were randomly selected for the present study. Out of 56 subjects, 28 were males and 28 were females. After taking prior informed written consent, 3 ml blood samples were collected from each subject. The inclusion criteria of subjects for present study are that they should be domicile of Uttar Pradesh, and healthy without any individual/family history of genetic and metabolic disorders. Study was conducted in the Human Molecular Genetics Laboratory, Department of Biotechnology, VBS Purvanchal University, Jaunpur, India during the period 2009-2010.
Genomic DNA extraction
Genomic DNA was extracted according to the method of Bartlett and White[30] and extracted genomic DNA was kept at –20°C until the genotype analysis.
MTRR genotype analysis
Analysis of the MTRR A66G mutation was based on the method of Wilson et al.[1] Polymerase chain reaction (PCR) was performed using genomic DNA and the primers 5’-GCAAAGGCCATCGCAGAAGACAT-3’ and 5’- GTGAAGATCTGCAGAAAATCCATGTA-3’ to generate 66-bp amplicon. PCR was performed in MJ Mini thermocycler (Bio-Rad, USA). The amplified product was digested with NdeI restriction enzyme (Genei, India) and digested products were analyzed in 4% agarose (Fermentas) gel electrophoresis for allele/genotype identification. Allele frequencies were calculated using the gene counting method and χ2test was performed to test Hardy-Weinberg. The frequencies of three genotypes and alleles were represented with 95% confidence interval (CI). All statistical analysis was done by online calculator tool quick calcs (program for scientists from Graph-pad Software).[31]
Results and Discussion
Genotype number and allele frequencies observed in the present study are presented in Table 1. The GG genotype was found in 28 subjects (50%), followed by AG genotype in 23 subjects (41%) and AA genotype in 5 subjects (8.9%). Genotype frequencies of AA, AG, and GG were 0.089 (95% CI = 0.033 to 0.187), 0.41 (95% CI = 0.288 to 0.542) and 0.50 (95% CI = 0.371 to 0.629) respectively. The frequency of A allele was found to be 0.295 (95% CI = 0.216 to 0.384) and G allele was 0.705 (95% CI = 0.626 to 0.784). Amplification with MTRR gene specific primer generated 66-bp amplicon, and after NdeI digestion homozygous AA genotype produced two bands of 44 bp and 22 bp, heterozygous AG genotype produced three bands 66 bp, 44 bp and 22 bp and homozygous mutated GG genotype remained uncut [Figure 1].
Table 1.
Distribution of MTRR genotypes and allele frequencies among Sunni Muslims
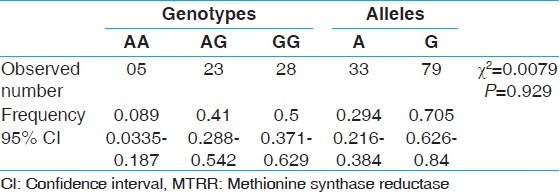
Figure 1.
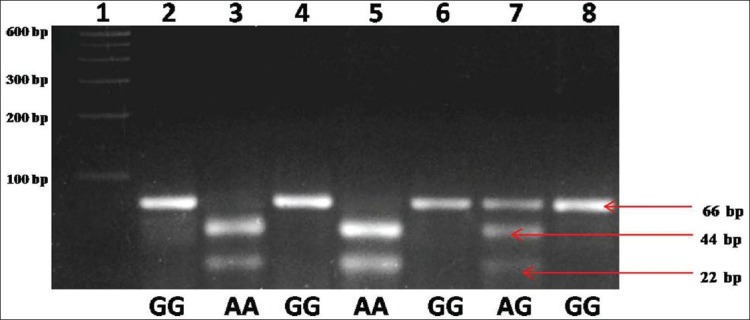
Agarose gel picture showing NdeI digested different methionine synthase reductase genotypes
During the homocysteine/methionine cycle, methionine is synthesized from homocysteine by vitamin B12-dependent methionine synthase (MTR). The methyl groups released upon the transformation of methionine to homocysteine (S-adenosylmethionine [SAM] being the most important methyl-group donor) facilitate the methylation of DNA, lipids and proteins. MTRR is required for the conversion of inactive MTR into active MTR. Olteanu et al.[13] demonstrated that the required molar ratio of MTRR to MTR was four fold higher for maximal activation for the M22 variant compared to the I22 variant. This suggests that the I22M variant in the MTRR enzyme disrupts the binding of MTRR and MTR. When MTRR function is impaired, it is likely that less methionine will be formed, leading to decreased SAM formation. Decreased formation of SAM will subsequently lead to reduced transfer of methyl groups by SAM, thereby disturbing the methylation, and thus, expression and function, of genes and proteins involved in fetal development. An additional effect of an impaired MTRR enzyme is the reduced conversion of 5-methylTHF to tetrahydrofolate, which may subsequently lead to a decreased availability of folate for purine and thymidine synthesis. During the fetal development, especially at neurulation, cell proliferation is extremely fast and DNA requirement is subsequently high. Decreased availability of DNA nucleotide could therefore lead to a disturbed fetal development[18] and resulting in congenital malformation/birth defects.
Different reports showed that the prevalence of the A66G polymorphism of the MTRR gene differ dramatically among human populations. Numerous case-control studies reported 66G allele frequency in different countries viz. - America,[3,19] Brazil,[32] Canada,[1] China,[21] France,[22,33,34] Ireland,[9] Italy,[20] Korea,[24] Netherland,[18] United Kingdom,[17] and also from India.[35,36,37] Evidence of this dynamism can be observed in many reports: frequency variations between populations that are geographically very close, even in the same country; changes found in the same race or ethnic group such as Africans and African-Americans. The very high frequency of 66G allele (greater than normal allele) was reported from Italy (0.51),[20] France (0.65),[34] Ireland (0.55),[9] Netherland (0.613),[18] and also from India (0.711).[36] Rady et al.[3] reported G allele frequency in different ethnic populations and they observed lowest G allele frequency in Hispanic population (0.28), compared to 0.34 among African-American, 0.431 among Ashkenazi Jews and 0.545 among Caucasians. Whereas, according to Centre for Disease Control (CDC, USA), the MTRR 66G allele frequency in Hispanic black, non-Hispanics White and Mexican American is 0.287,0.528 and 0.261, respectively.[38] There is numerous interpretation of this gene diversity, and most tend to be related to adaptation to external conditions such as climate or nutritional status. Dependence on folate deficiency/degradation, vitamin B12 deficiency, nutritional habits or human intervention during peri-conceptional periods could explain this genetic variation.
Frequency of G allele in Hapmap populations viz. - European, Nigerian, Japanese, and Chinese was reported to be 0.448, 0.233, 0.302, and 0.250 respectively.[39] The G allele frequency observed in present study (0.705) was quite high in comparison to Hapmap Asian populations, but was well fell in the range reported in other studies, where frequencies were ranging from 0.43 (Brazilian population)[28] to 0.733 (British population)[17] suggesting that this population might be susceptible to some type of factor contributing to the modification of the MTRR A66G allele frequencies. In this population, the mutant allele with lower enzymatic activity has higher fitness than the wild type. Wilson et al.[1] found equal frequency of both the alleles (A and G allele) in Canadian population and it was difficult for them to designate the wild and mutated alleles, so on the basis of sequence homology with other flavin binding proteins of other species they consider A allele as wild allele. Except one report,[37] population frequency of this clinically important polymorphism is not well reported from Indian population, only few reports are available which are based on the case-control studies.[35,36] So it is urgently required to screen the Indian ethnic and geographical populations for this clinically important polymorphism.
Acknowledgment
We are grateful to the subjects who participated in the present study without their cooperation; this study could not be completed. The financial support from Department of Biotechnology (No BT/PR98887/SPD/11/1028/2007) as major research project to Vandana Rai is gratefully acknowledged.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
References
- 1.Wilson A, Platt R, Wu Q, Leclerc D, Christensen B, Yang H, et al. A common variant in methionine synthase reductase combined with low cobalamin (vitamin B12) increases risk for spina bifida. Mol Genet Metab. 1999;67:317–23. doi: 10.1006/mgme.1999.2879. [DOI] [PubMed] [Google Scholar]
- 2.Gaughan DJ, Kluijtmans LA, Barbaux S, McMaster D, Young IS, Yarnell JW, et al. The methionine synthase reductase (MTRR) A66G polymorphism is a novel genetic determinant of plasma homocysteine concentrations. Atherosclerosis. 2001;157:451–6. doi: 10.1016/s0021-9150(00)00739-5. [DOI] [PubMed] [Google Scholar]
- 3.Rady PL, Szucs S, Grady J, Hudnall SD, Kellner LH, Nitowsky H, et al. Genetic polymorphisms of methylenetetrahydrofolate reductase (MTHFR) and methionine synthase reductase (MTRR) in ethnic populations in Texas; a report of a novel MTHFR polymorphic site, G1793A. Am J Med Genet. 2002;107:162–8. doi: 10.1002/ajmg.10122. [DOI] [PubMed] [Google Scholar]
- 4.Brilakis ES, Berger PB, Ballman KV, Rozen R. Methylenetetrahydrofolate reductase (MTHFR) 677C>T and methionine synthase reductase (MTRR) 66A>G polymorphisms: Association with serum homocysteine and angiographic coronary artery disease in the era of flour products fortified with folic acid. Atherosclerosis. 2003;168:315–22. doi: 10.1016/s0021-9150(03)00098-4. [DOI] [PubMed] [Google Scholar]
- 5.Leclerc D, Wilson A, Dumas R, Gafuik C, Song D, Watkins D, et al. Cloning and mapping of a cDNA for methionine synthase reductase, a flavoprotein defective in patients with homocystinuria. Proc Natl Acad Sci U S A. 1998;95:3059–64. doi: 10.1073/pnas.95.6.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gos M, Jr, Szpecht-Potocka A. Genetic basis of neural tube defects. II. Genes correlated with folate and methionine metabolism. J Appl Genet. 2002;43:511–24. [PubMed] [Google Scholar]
- 7.Kluijtmans LA, Young IS, Boreham CA, Murray L, McMaster D, McNulty H, et al. Genetic and nutritional factors contributing to hyperhomocysteinemia in young adults. Blood. 2003;101:2483–8. doi: 10.1182/blood.V101.7.2483. [DOI] [PubMed] [Google Scholar]
- 8.Brown CA, McKinney KQ, Kaufman JS, Gravel RA, Rozen R. A common polymorphism in methionine synthase reductase increases risk of premature coronary artery disease. J Cardiovasc Risk. 2000;7:197–200. doi: 10.1177/204748730000700306. [DOI] [PubMed] [Google Scholar]
- 9.O’Leary VB, Parle-McDermott A, Molloy AM, Kirke PN, Johnson Z, Conley M, et al. MTRR and MTHFR polymorphism: Link to Down syndrome. Am J Med Genet. 2002;107:151–5. doi: 10.1002/ajmg.10121. [DOI] [PubMed] [Google Scholar]
- 10.Jacques PF, Bostom AG, Selhub J, Rich S, Ellison RC, Eckfeldt JH, et al. Effects of polymorphisms of methionine synthase and methionine synthase reductase on total plasma homocysteine in the NHLBI Family Heart Study. Atherosclerosis. 2003;166:49–55. doi: 10.1016/s0021-9150(02)00204-6. [DOI] [PubMed] [Google Scholar]
- 11.Feix A, Winkelmayer WC, Eberle C, Sunder-Plassmann G, Födinger M. Methionine synthase reductase MTRR 66A>G has no effect on total homocysteine, folate, and Vitamin B12 concentrations in renal transplant patients. Atherosclerosis. 2004;174:43–8. doi: 10.1016/j.atherosclerosis.2003.12.036. [DOI] [PubMed] [Google Scholar]
- 12.Olteanu H, Banerjee R. Human methionine synthase reductase, a soluble P-450 reductase-like dual flavoprotein, is sufficient for NADPH-dependent methionine synthase activation. J Biol Chem. 2001;276:35558–63. doi: 10.1074/jbc.M103707200. [DOI] [PubMed] [Google Scholar]
- 13.Olteanu H, Munson T, Banerjee R. Differences in the efficiency of reductive activation of methionine synthase and exogenous electron acceptors between the common polymorphic variants of human methionine synthase reductase. Biochemistry. 2002;41:13378–85. doi: 10.1021/bi020536s. [DOI] [PubMed] [Google Scholar]
- 14.Zijno A, Andreoli C, Leopardi P, Marcon F, Rossi S, Caiola S, et al. Folate status, metabolic genotype, and biomarkers of genotoxicity in healthy subjects. Carcinogenesis. 2003;24:1097–103. doi: 10.1093/carcin/bgg064. [DOI] [PubMed] [Google Scholar]
- 15.Bailey LB, Moyers S, Gregory JF. Folate. In: Bowman BA, Russell RM, editors. Present Knowledge in Nutrition. Washington DC: International Life Sciences Institute; 2001. pp. 214–29. [Google Scholar]
- 16.Pietrzyk JJ, Bik-Multanowski M, Sanak M, Twardowska M. Polymorphisms of the 5,10-methylenetetrahydrofolate and the methionine synthase reductase genes as independent risk factors for spina bifida. J Appl Genet. 2003;44:111–3. [PubMed] [Google Scholar]
- 17.Relton CL, Wilding CS, Pearce MS, Laffling AJ, Jonas PA, Lynch SA, et al. Gene-gene interaction in folate-related genes and risk of neural tube defects in a UK population. J Med Genet. 2004;41:256–60. doi: 10.1136/jmg.2003.010694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Linden IJ, Afman LA, Heil SG, Blom HJ. Genetic variation in genes of folate metabolism and neural-tube defect risk. Proc Nutr Soc. 2006;65:204–15. doi: 10.1079/pns2006495. [DOI] [PubMed] [Google Scholar]
- 19.Hobbs CA, Sherman SL, Yi P, Hopkins SE, Torfs CP, Hine RJ, et al. Polymorphisms in genes involved in folate metabolism as maternal risk factors for Down syndrome. Am J Hum Genet. 2000;67:623–30. doi: 10.1086/303055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scala I, Granese B, Sellitto M, Salomè S, Sammartino A, Pepe A, et al. Analysis of seven maternal polymorphisms of genes involved in homocysteine/folate metabolism and risk of Down syndrome offspring. Genet Med. 2006;8:409–16. doi: 10.1097/01.gim.0000228206.21793.82. [DOI] [PubMed] [Google Scholar]
- 21.Wang SS, Qiao FY, Feng L, Lv JJ. Polymorphisms in genes involved in folate metabolism as maternal risk factors for Down syndrome in China. J Zhejiang Univ Sci B. 2008;9:93–9. doi: 10.1631/jzus.B0710599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guéant-Rodriguez RM, Juilliére Y, Candito M, Adjalla CE, Gibelin P, Herbeth B, et al. Association of MTRRA66G polymorphism (but not of MTHFR C677T and A1298C, MTRA2756G, TCN C776G) with homocysteine and coronary artery disease in the French population. Thromb Haemost. 2005;94:510–5. doi: 10.1160/TH05-04-0262. [DOI] [PubMed] [Google Scholar]
- 23.Laraqui A, Allami A, Carrié A, Coiffard AS, Benkouka F, Benjouad A, et al. Influence of methionine synthase (A2756G) and methionine synthase reductase (A66G) polymorphisms on plasma homocysteine levels and relation to risk of coronary artery disease. Acta Cardiol. 2006;61:51–61. doi: 10.2143/AC.61.1.2005140. [DOI] [PubMed] [Google Scholar]
- 24.Lee HC, Jeong YM, Lee SH, Cha KY, Song SH, Kim NK, et al. Association study of four polymorphisms in three folate-related enzyme genes with non-obstructive male infertility. Hum Reprod. 2006;21:3162–70. doi: 10.1093/humrep/del280. [DOI] [PubMed] [Google Scholar]
- 25.Ravel C, Chantot-Bastaraud S, Chalmey C, Barreiro L, Aknin-Seifer I, Pfeffer J, et al. Lack of association between genetic polymorphisms in enzymes associated with folate metabolism and unexplained reduced sperm counts. PLoS One. 2009;4:e6540. doi: 10.1371/journal.pone.0006540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang FF, Terry MB, Hou L, Chen J, Lissowska J, Yeager M, et al. Genetic polymorphisms in folate metabolism and the risk of stomach cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:115–21. doi: 10.1158/1055-9965.EPI-06-0513. [DOI] [PubMed] [Google Scholar]
- 27.Shariff A. Relative economic and social deprivation of Indian Muslims. J Obj Stud. 1998;10:2–18. [Google Scholar]
- 28.Ansari G. Ethnographic and Folk Culture Society. 1959. Muslims caste in UP Lucknow. [Google Scholar]
- 29.Ahmad I. Endogamy and status mobility among the Siddiqui Sheikhs of Allahabad, UP. In: Ahmad I, editor. Caste and Stratification among the Muslims of India. New Delhi: Manohar Publication; 1978. [Google Scholar]
- 30.Bartlett JM, White A. Extraction of DNA from whole blood. In: Bartlett JM, Stirling D, editors. Methods in Molecular Biology, PCR Protocls. 2nd ed. Totowa, NJ: Humana Press Inc; 2003. [Google Scholar]
- 31. Available from: http://www.graphpad.com/wuickcalcs/
- 32.de Silva LR, Vergani N, Galdieri Lde C, Ribeiro Porto MP, Longhitano SB, Brunoni D, et al. Relationship between polymorphisms in genes involved in homocysteine metabolism and maternal risk for Down syndrome in Brazil. Am J Med Genet A. 2005;135:263–7. doi: 10.1002/ajmg.a.30591. [DOI] [PubMed] [Google Scholar]
- 33.Bosco P, Guéant-Rodriguez RM, Anello G, Barone C, Namour F, Caraci F, et al. Methionine synthase (MTR) 2756 (A ->G) polymorphism, double heterozygosity methionine synthase 2756 AG/methionine synthase reductase (MTRR) 66 AG, and elevated homocysteinemia are three risk factors for having a child with Down syndrome. Am J Med Genet A. 2003;121A:219–24. doi: 10.1002/ajmg.a.20234. [DOI] [PubMed] [Google Scholar]
- 34.Chango A, Fillon-Emery N, Mircher C, Bléhaut H, Lambert D, Herbeth B, et al. No association between common polymorphisms in genes of folate and homocysteine metabolism and the risk of Down’s syndrome among French mothers. Br J Nutr. 2005;94:166–9. doi: 10.1079/bjn20051490. [DOI] [PubMed] [Google Scholar]
- 35.Sheth JJ, Sheth FJ. Gene polymorphism and folate metabolism: A maternal risk factor for Down syndrome. Indian Pediatr. 2003;40:115–23. [PubMed] [Google Scholar]
- 36.Lakshmi SV, Naushad SM, Ruparsree Y, Rao DS, Kutala VK. Interactions of 5ÚTR thymidine synthase polymorphism with 677C ->T methylene tetrahydrofolate reductase and 66A ->G methyleneteterahydrofolate homocysteine methyl-transferase reductase polymorphisms determne susceptibility to coronary artery disease. J Atheroscler Thromb. 2010;18:56–64. doi: 10.5551/jat.5702. [DOI] [PubMed] [Google Scholar]
- 37.Rai V, Yadav U, Kumar P, Gupta S. 2011.Methionine synthase reductase (MTRR) A66G polymorphism in rural population of Uttar Pradesh (India) Biotechnology. 2011;10:220–3. [Google Scholar]
- 38. Available from: http://webdev.nccd.cdc.gov/genomics/population/genvar/frequencies/MTRR.htm .
- 39. Available from: http://www.hapmap.org/cgi-perl/gbrowse/


