Abstract
The ubiquitin proteasome system (UPS) is the primary pathway responsible for the recognition and degradation of misfolded, damaged, or tightly regulated proteins in addition to performing essential roles in DNA repair, cell cycle regulation, cell migration, and the immune response. While traditional biochemical techniques have proven useful in the identification of key proteins involved in this pathway, the implementation of novel reporters responsible for measuring enzymatic activity of the UPS have provided valuable insight into the effectiveness of therapeutics and role of the UPS in various human diseases such as multiple myeloma and Huntington’s disease. These reporters, usually consisting of a recognition sequences fused to an analytical handle, are designed to specifically evaluate enzymatic activity of certain members of the UPS including the proteasome, E3 ubiquitin ligases, and deubiquitinating enzymes (DUBs). This review highlights the more commonly used reporters employed in a variety of scenarios ranging from high-throughput screening of novel inhibitors to single cell microscopy techniques measuring E3 ligase or proteasome activity. Finally, recent work is presented highlighting the development of novel degron-based substrate designed to overcome the limitations of current reporting techniques in measuring E3 ligase and proteasome activity in patient samples.
Introduction
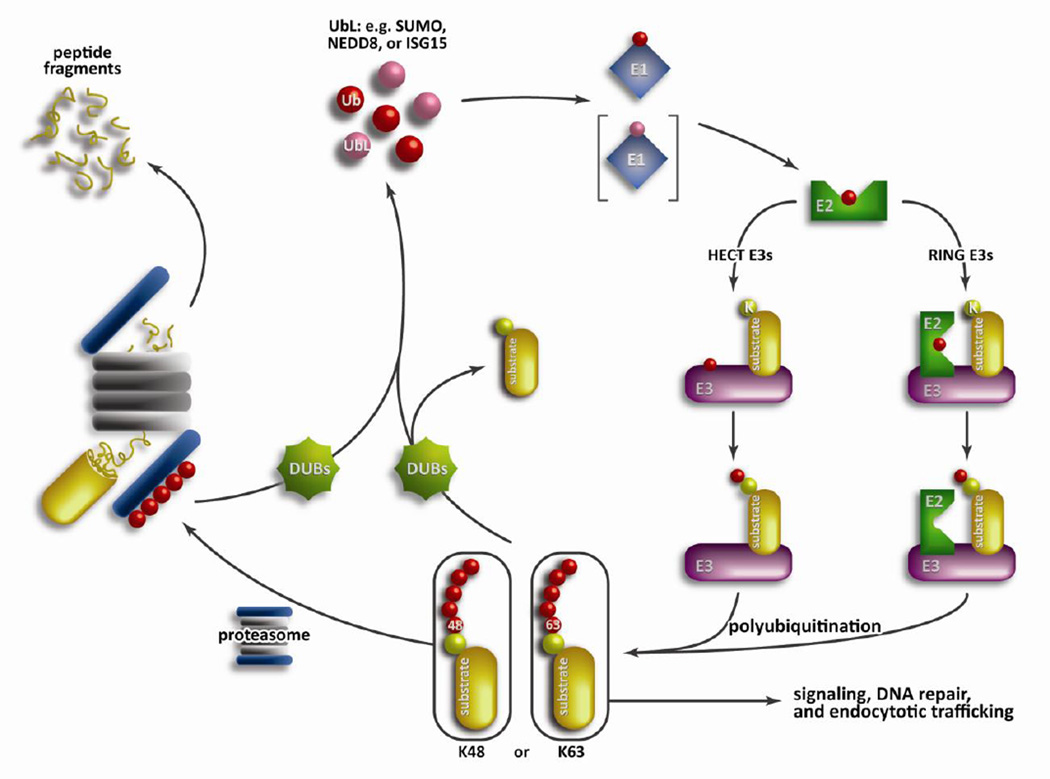
Degradation of regulatory, damaged, and misfolded proteins is essential to proper cellular homeostasis (1). In eukaryotes the targeting and controlled degradation of these proteins is governed by the ubiquitin proteasome system (UPS). Posttranslational protein modification by ubiquitin (Ub) targets the protein to the 26S proteasome, which unfolds and degrades the protein into small peptide fragments. Protein ubiquitination requires a cascade of three increasingly diverse enzymes (Figure 1). First, an E1 ubiquitin activating enzyme forms an ATP-dependent high-energy thioester bond with free ubiquitin. Next, the Ub∼E1 complex interacts with an E2 ubiquitin conjugating enzyme, transferring the ubiquitin from E1 to E2. The Ub∼E2 complex proceeds to interact with a third class of enzymes, the E3 ubiquitin ligases, which mediates either the direct (RING family E3 ligases) or indirect (HECT family E3 ligases) transfer of ubiquitin to a proximal lysine on the targeted protein (2). The recognition of the target protein occurs through a unique degradation signal, or degron. There are many different classes of degrons including phospho-degrons, oxygen-dependent degrons, and N-degrons (3). Following initial ubiquitin-protein conjugation, additional ubiquitins are added to form a polyubiquitin (polyUb) chain. This polyubiquitin chain can be linked via one of seven different lysine residues found on ubiquitin (e.g. K48, K63, or K11) or through the N-terminal methionine residue to form a linear ubiquitin chain (4). The manner in which the polyubiquitin chain is formed dictates the outcome of the protein, with K48-linked chains being targeted toward the proteasome for degradation while K63-linked chains play a role in cell signaling, DNA damage repair, and endocytic trafficking (5). Further, the number of conjugated ubiquitins (mono- vs. multiple mono- vs. poly-ubiquitination) can also decide the eventual fate of the modified protein.
Figure 1. Overview of the UPS.
Posttranslational protein modification by ubiquitin (or ubiquitin-like proteins, UbLs) requires a cascade of three increasingly diverse enzymes: an E1 ubiquitin activating enzyme, an E2 ubiquitin conjugating enzyme, and an E3 ubiquitin ligase. Protein ubiquitination starts with an E1 forming a high energy thioester bond with free ubiquitin, which is recognized and transferred to the E2 enzyme. Next, an E3 ubiquitin ligase forms a complex with the E2 enzyme to mediate the transfer of ubiquitin in either a direct (RING family) or indirect (HECT family) manner. Polyubiquitin chains are formed through linkages of one of the seven lysines present on ubiquitin with K48-linked chains being targeted towards the proteasome. The 19S cap of the 26S proteasome recognizes the polyubiquitin chain and then unfolds and degrades the protein into small peptide fragments in the 20S core particle.
A poly-ubiquitinated protein targeted for degradation is recognized by the 19S cap of the 26S proteasome, where the target protein is deubiquitinated, unfolded, and degraded by the 20S core particle (6). The barrel shaped 20S core consists of four heptameric rings, with the inner rings consisting of β-subunits, three of which are responsible for the catalytic cleavage of a protein. Control of polyubiquitin chain formation is further refined by another class of proteins, deubiquitinating enzymes (DUBs). DUBs are capable of cleaving the isopeptide bond between ubiquitin and the target protein, initiating either the rescue of a poly-ubiquitinated protein or the recycling of ubiquitin from the proteasome (Figure 1). The E1-E3 enzymatic cascade is paralleled by several ubiquitin-like proteins (UbL) (Figure 1). Examples include SUMO (small ubiquitin like modifier), ISG15 (interferon stimulated gene 15), and NEDD8 (neural precursor cell expressed developmentally down regulated protein 8) all of which are activated and conjugated to proteins using similar machinery. It has become increasingly apparent that these UbLs play an integral role in the regulation of cellular processes, including transcriptional regulation and DNA repair (7). In a process reminiscent of DUBs in the ubiquitin pathway, the isopeptide bond between an UbL and a protein is cleaved by UbL proteases allowing them to function as a reversible post-translational modification.
The unique and vital functions carried out by the UPS in regulating protein levels have made it a very attractive target for novel therapeutics. This interest has been compounded by the clinical success of the proteasome inhibitor Bortezomib in the treatment of multiple myeloma (8). Dysregulation of protein production, particularly antibodies, in this cancer can create an increased reliance on the proteasome to degrade misfolded or overproduced proteins. Incomplete inhibition of the proteasome proves too stressful for cancer cells, while proving less cytotoxic to healthy cells. This permits a more targeted treatment approach relative traditional cancer therapies and has led to the development of the next generation proteasome inhibitor Carfilzomib (9). Members of the UPS are also thought to play a direct role in other cancers and neurodegenerative disease. Though there are conflicting reports, decreased proteasome activity is suspected to play a key role in conformational diseases such as Parkinson’s and Huntington’s disease (10). These diseases are characterized by the accumulation of misfolded proteins and decreased proteasome activity, potentially implicating the proteasome as a pivotal target. Mutations of UPS components may also play a role in neurodegenerative disorders, such as the frame shifted ubiquitin UBB+1 which consists of an extra 19 amino acids on the Ub C-terminus resulting in an inability to be activated by E1 enzymes. Further, the tumor suppressor p53 and its E3 ligase Mdm2 (or Hdm2 in humans) demonstrate an additional role for the UPS in disease. Strict regulation of p53 is maintained by constant degradation in healthy cells; however, when under genotoxic stress p53 degradation is suppressed. Accumulation of p53, in turn, leads to an upregulation of genes associated with apoptosis or growth. Due to the prominent role of p53 in many cancers, the role of the UPS in its regulation has become an area of intense interest (11,12). Finally, it has been demonstrated that deregulation of DUBs is also associated with human disease (13). DUBs such as CYLD and USP9 have been found to be mutated in cancers, suggesting their potential status as oncogenes.
Due to the increased interest in targeting the UPS for clinical therapeutics, it has become necessary to create new metrics to evaluate the enzymatic activity of the members of the UPS including the proteasome, E3 ligase, and DUBs. In this review we survey common reporters and reporting techniques that have been used to detect not only enzymatic activity, but also the relative success of novel inhibitors targeting the UPS. We highlight reporters that evaluate the proteasome as well as members of the ubiquitin enzymatic cascade including E3 ligases, DUBs and polyubiquitin chain formation. Finally, we present some recent work from our lab detailing the development and characterization of a novel substrate, based on the degron from β-Catenin, as the groundwork for the next generation of E3 ligase and proteasome reporters.
Measuring proteasome activity
Several strategies have been developed to create reporters capable of directly measuring proteasome activity. The majority of these reporters consist of two major elements: a targeting sequence to recruit the reporter to the proteasome and an analytical handle that can produce a measureable signal. There are number of different schemes to target the proteasome, which is reflected in the number of successful reporters described in the literature. By far the most popular analytical handles are photometric so that microscopy-based imaging and spectrometric technologies, as well as FACS, can be used for sensor read-out. In this section we discuss these reporting methods and highlight recent work targeting a reporter to the proteasome.
Ubiquitin-dependent proteasome targeting
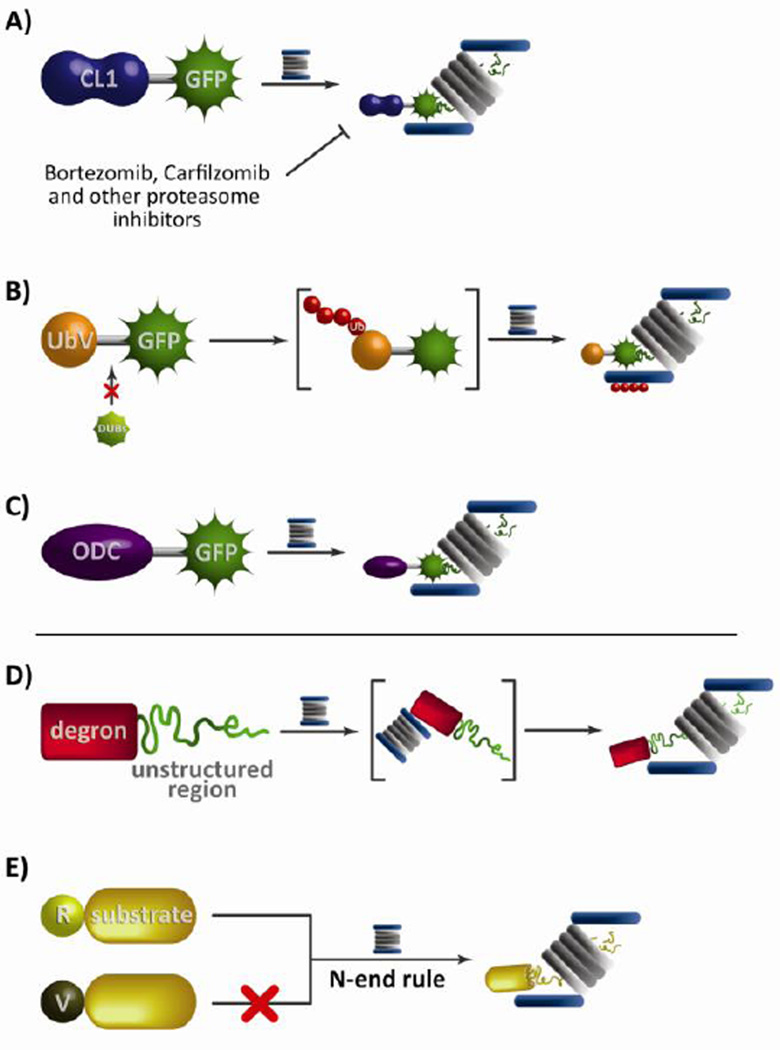
The earliest strategies for targeting a reporter to the proteasome exploited known degradation signals. Strong degrons, such as CL1, allow reporters to be ubiquitinated in an E3 ligase-mediated fashion. Indeed, one popular reporter which makes use of this approach is GFPu (Figure 2a). This reporter consists of the sixteen amino acid CL1 degron fused to green fluorescent protein (GFP) (14). Cells stably transfected with the reporter express GFPu constitutively which is immediately recruited to the proteasome for degradation after synthesis. Cellular concentrations of the reporter are low, yielding low levels of cellular fluorescence; however, inhibition of the proteasome by compounds such as MG-132 or Bortezomib causes an accumulation of GFPu resulting in increased cellular fluorescence This reporting technique has also been used to demonstrate the importance of α-Synuclein-mediated inhibition of the proteasome (15). Of particular interest are transgenic mouse models expressing GFPu, which have been used to investigate proteasome activity in neurons. One study using this model demonstrated that proteasome activity is not impaired in neurons as a function of aging (16). These models have elucidated the contribution of the proteasome to protein-conformation diseases such as Huntington’s disease and revealed that GFPu does not accumulate in mouse models of Huntington’s disease, calling into question a link between cellular proteasome activity and protein aggregation (17).
Figure 2. Measuring proteasome activity.
Proteasome reporters consist of two major elements; a targeting sequence to recruit the reporter to the proteasome (usually a degron) and an analytical handle that can produce a measureable signal (depicted here as the green fluorescent protein (GFP); however other signals such as luciferase or FLAG tags have been used). Ubiquitin-dependent proteasomal targeting uses either the 16 amino acid degron from CL1 (A) or a non-cleavable ubiquitin (B) to measure proteasome activity in response to various conditions such as drug-induced inhibition. (C) The ubiquitin-independent degron ODC is also recruited to the proteasome without requiring the E1-E3 enzymatic cascade. (D) A two component degron demonstrates the importance of an unstructured initiation region for efficient targeting and degradation of reporters. (E) The N-end rule governs N-terminal amino acids that are stabilizing (valine) or destabilizing (arginine).
Another equally important targeting mechanism fuses a non-cleavable ubiquitin (G76V) to an analytical handle such as GFP (this reporter is called UbV-GFP). The single ubiquitin promotes polyubiquitin chain formation to ultimately target the reporter to the proteasome while minimizing the threat of isopeptide bond cleavage by DUBs (Figure 2b) (18). Notably, the UbV-GFP reporter has been successfully implemented in Caenorhabditis elegans, allowing for in vivo mapping of proteasomal activity in response to elevated levels of unfolded protein or heat stress. In certain mutants, which exhibited a hypersensitivity to stress, mildly increased levels of unfolded protein resulted in an accumulation of the reporter, while heat stress resulted in enhanced proteasomal activity (19). An interesting reporter related to UbV-GFP incorporates a photoconvertible fluorescent protein, Dendra2, which could be converted from a green to red fluorescent state immediately prior to measurement. This reporter was used in C. elegans to monitor proteasome activity independent of new protein production, as newly synthesized protein would remain in a green fluorescent state (20). Ubiquitin dependent reporters have proven to be both popular and effective in monitoring overall proteasome activity; however, it is important to keep in mind when dealing with reporters that utilize the ubiquitination machinery that the E1-E3 enzymatic cascade is strongly linked to the proteasome. As such, these reporters must be carefully designed to minimize convolution of proteasome activity with other UPS activity.
Ubiquitin-independent proteasome targeting
An effective way of minimizing the potential effects of E3 ligases is to design a reporter independent of this enzymatic cascade. One such reporting strategy is the fusion of a tetra-ubiquitin chain to the enzyme luciferase. Similar to GFPu, following proteasome inhibition transfected tetra-ubiquitin-luciferase accumulates in cells catalyzing a bioluminescent reaction for quantification of proteasome activity. Reporters employing this strategy have been used to study the proteasome in high-throughput cellular assays and across several species at the organismal level. These studies have revealed a 5-fold difference between the proteasome processivity of yeast and mammals (21). This reporter has been also used successfully in one study to screen for proteasome inhibitors (22). The work examined ∼18,000 molecules and ∼15,000 plant extracts, discovering 21 molecules and 66 extracts capable of inhibiting the proteasome (23). Additionally, this reporter has been used to great effect in the imaging of the proteasome at the organismal level. Xenografted tumors in mice were shown to rapidly degrade the reporter under normal circumstances; however the effect was abrogated in a time and concentration dependent fashion when the mice were treated with a proteasome inhibitor (24). Another example of ubiquitin-independent targeting to the proteasome uses the degron from ornithine decarboxylase (ODC). A 37 amino acid sequence from the C-terminus of ODC (cODC) is recruited to the proteasome in a ubiquitin-independent manner, allowing for its use as a proteasome targeting sequence that does not require the E1-E3 enzymatic cascade (Figure 2c) (25). Stable expression of the ODC-GFP reporter in mouse models has been used to evaluate proteasome activity. This system has been shown capable of detecting Bortezomib inhibition of proteasome activity in tumor cells, indicating that it could be used in the future for library-based screens for proteasome inhibitors (26). Affinity tags combined with the ODC degron produced a reporter used in Saccharomyces cerevisiae. This tag allowed for purification and quantification of the reporter and helped identify strains of yeast with mutations affecting the proteasome (25). A recent study identified another degron that could be incorporated into a full length protein to target it for degradation, commonly referred to as a portable degron. The 21 C-terminal amino acids from the tumor suppressor NKX3.1 (termed the C21 degron) was fused to the C-terminus of GFP to initiate proteasome-dependent degradation of GFP (27). While it was not incorporated into a proteasome reporter, the C21 degron exhibits the potential for monitoring proteasome activity.
Reporters employing other targeting mechanisms have been used to monitor proteasome activity. Carefully designed small peptide reporters were able to target specific active subunits of the proteasome. These reporters use fluorophores such as 7-amino-4methylcoumarin (AMC) or 2-naphtylamine (βNA), which, when cleaved, release a highly fluorescent product (28). Unlike the previously described techniques, these reporters are not transfected into the cell; but are added immediately prior to measurement, allowing direct quantitation of proteasome activity (29). A second generation of these reporters incorporated the luciferase substrate aminoluciferin instead of a fluorophore, permitting increased sensitivity (30). FRET-based reporters have also been developed to evaluate proteasome activity. One such reporter used small peptides that, when degraded by the proteasome, would exhibit a decreased FRET-behavior and direct measurement of proteasome activity (31). Finally, a novel reporting technique incorporated a short peptide along with nuclear magnetic resonance (NMR) to detect cleavage of an isotopically labeled reporter (32). This enabled detection of proteasome activity even in optically dense environments, and has been used to discover potential proteasome inhibitors in the secretions of Photorhabdus luminescens.
The Two-Component Degron
While the aforementioned proteasome reporters containing degrons fused to various analytical handles have demonstrated success in evaluating proteasome activity, other proteasome reporters have been developed using what has been commonly referred to as the two component degron. These reporters are based on the concept that efficient proteasomal degradation requires both the degradation sequence and an unstructured region C-terminal to the degron serves as an initiation region (Figure 2d) (33). While a polyubiquitin chain consisting of at least four subunits is sufficient for targeting a protein to the proteasome, it has been demonstrated that this unstructured region acts as an initiation point for the proteasome to begin substrate degradation (34). The importance of the two-component degron has been demonstrated in mycobacterium, which used the prokaryotic ubiquitin-like protein (Pup) in proteasome-mediated degradation (35). However, the importance of initiation region length and structure was not clearly defined until recently, when researchers began to utilize novel proteasome reporters consisting of the two-component degron.
The effect of initiation region length was recently clarified using a non-cleavable tetra-ubiquitin tag (or a UbL) fused to an E. coli DHFR domain along with unstructured C-terminal tails of different lengths (36). It was determined that the initiation region needs to be located an appropriate distance relative to the proteasome-binding tag, with the distance depending upon the proteasome-targeting method. For a polyubiquitin chain, the initiation region must be adjacent to the ubiquitination site, while an UbL-targeted substrate required the initiation region to be separated from the UbL domain. The importance of protein stability was studied using a two-component degron reporter consisting of either cODC or Rpn10 as the proteasome targeting sequence coupled with either the titin I27 domain or DHFR, both of which could be switched from a folded to an unstructured state either by a point mutation or by the addition of a tight-binding ligand respectively (37). These reporters indicated that increased structural stability corresponded to a decrease in protein degradation in vivo and with purified proteasome. Another recent study found that the proteasome could be manipulated to initiate degradation in the middle of a polypeptide chain using a reporter consisting of an initiation region flanked on either side with folded domains (38). This reporter showed that while these domains do not directly interact, they do stabilize each other and effectively decrease proteasome processivity. While these and other studies have demonstrated the importance of the two-component degron, a recent study suggested that a polyubiquitin chain was not even necessary for efficient proteasomal targeting (39). This work revealed that short proteins between ∼20–150 amino acids only required mono-ubiquitination for proteasome-mediated degradation. These experiments employed multiple different reporters including non-cleavable, non-polymerizable ubiquitin fused to a poly-His tag, GFP, and shortened iterations of the DHFR domain. However, if a protein fused to the ubiquitin was larger than ∼150 amino acids it required polyubiquitination for proteasome-mediated degradation. These new results provide more information on the role of the two-component degron and the requirement for mono- vs. poly-ubiquitination to target a protein for degradation.
The N-end rule pathway
Another strategy to target a protein for degradation exploits the N-end rule, a generalized rule that governs the protein-destabilizing activity of a given amino-terminal amino acid. This pathway was initially discovered when the stability of an engineered substrate was found to be dramatically affected by the identity of its N-terminal residue (40). Following this discovery, a hierarchy of amino acids was established with certain residues, like arginine, being highly destabilizing and other residues, like valine, being fairly stable (Figure 2e). Similar to the activation of phospho-degrons by kinases, there exists a class of proteins that are involved in the creation of N-degrons, degrons containing a destabilizing amino acid at the N-terminus. These N-degrons are then recognized and targeted for degradation by N-end rule pathway ubiquitin ligases, such as UBR1 in the yeast Saccharomyces cerevisiae (41). A very comprehensive review detailing the N-end rule in various model systems including yeast, plants and mammalian cells has recently been published (42); so for the sake of this review we will only focus on some of the more recent examples of reporters using the N-end rule. One such reporter was developed based on the C-terminal proteolytic fragment of BRCA1 generated during apoptosis (43). This reporter fused ubiquitin to the N-terminus of the BRCA1 fragment such that ubiquitin could be selectively removed by addition of ubiquitin hydrolase. Removal of the ubiquitin exposed the N-terminal amino acid to allow the reporter to be targeted by the N-end rule. Different iterations of the N-terminal amino acid showed that the C-terminal fragment was in fact being degraded according to the N-end rule. Another recent study developed a short-lived reporter of transcription by using the N-end rule to control the half-life of the reporter (44). It consisted of an N-degron version of GFP with only a half-life of ∼ 7 minutes, permitting precise monitoring of gene expression in living cells. While the N-end rule is a useful method for targeting proteins for degradation, N-degrons are not normally utilized as proteasome reporters due to the requirement of an additional protein to create the N-degron, as well as the consideration that many members of the pathways are still not well characterized.
Measuring ubiquitination enzyme activity and polyubiquitin chain formation
While many of the preliminary experiments on the UPS have focused solely on protein degradation, it has become clear that the process of targeting a protein for degradation involves a substantially greater number of proteins, each with their own specificity and activity. As a result, new reporting methods were developed to measure both the activity of the enzymes controlling ubiquitination and deubiquitination as well as to find a way to quantify the formation of the various linkages of polyubiquitin chains. In this section we highlight some of the recent advances in reporting technology for E3 ligase activity, DUB inhibition, and K63-linked polyubiquitin chain formation.
E3 ligase activity
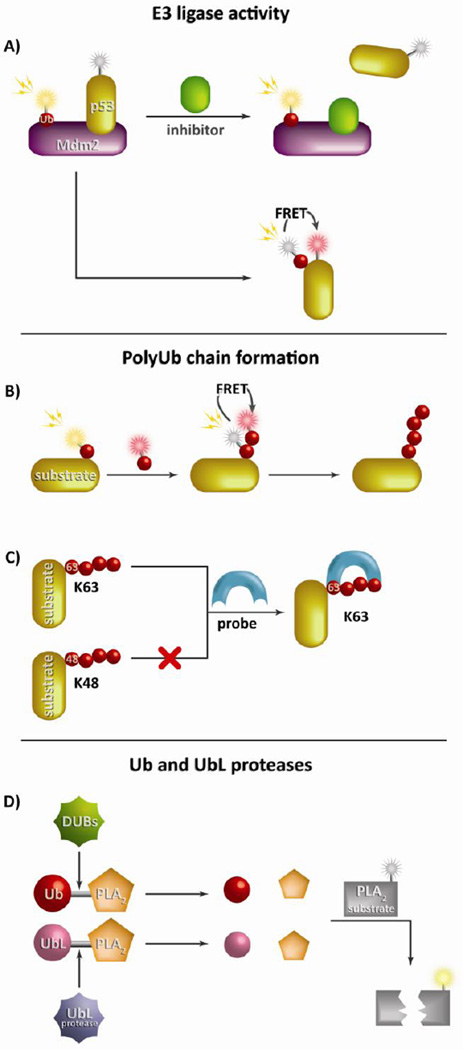
Of the three members composing the enzymatic cascade controlling protein ubiquitination, the E3 ubiquitin ligases have received the most attention with regards to measuring activity and developing inhibitors for clinical use. The desire to target this class of enzymes is due to their abundance (>600 known in humans), specificity, and known role in diseases such as cancer and neurodegenerative disorders. Many of the initial reporters for E3 ligase activity were incorporated into a high throughput screening assay in order to evaluate the effectiveness of inhibitors against the binding of p53 and Mdm2 (or Hdm2 in humans), a major mechanism of p53 inactivation in tumors. Hyperactivation of Mdm2, the E3 ligase responsible for the ubiquitination and degradation of p53, causes the accelerated clearance of p53 and the elimination of its tumor suppression capabilities. An initial assay simply utilized GST-tagged Hdm2 along with labeled ubiquitin to monitor the auto-ubiquitination of Hdm2 in the presence of inhibitors during a controlled reaction (45). While this was effective in studying Hdm2 autoubiquitination, autoubiquitination can occur in the absence of p53 and, as a result, did not provide information on the specific interaction between p53 and its E3 ligase. Recent work has focused more on the precise interaction between p53 and Hdm2, especially through the use of high throughput screening to identify compounds which can inhibit this specific interaction (Figure 3a). An assay utilizing Cy-5 labeled ubiquitin (acceptor) and europium-labeled p53 (donor) was employed to measure Mdm2-mediated ubiquitination by time resolved fluorescent resonance energy transfer (TR-FRET) (46). This TR-FRET assay was easily adapted to a high-throughput screen which was used to assess >600,000 compounds for Mdm2-p53 binding inhibition. A similar study screened inhibitors of the p53-Mdm2 interaction using mammalian two-hybrid technology in tandem with a dual-luciferase reporter system with the goal of isolating a single compound that induced growth arrest in tumor cells (47). Large screens of E3 ligase activity have also been developed for E3 ligases other than Mdm2. One report used protein microarrays to analyze cell extracts from a mitotic checkpoint system to identify possible substrates of the anaphase promoting complex (APC), a known E3 ligase essential to the cell cycle (48).
Figure 3. Measuring E3 ligase and DUB enzymatic activity and polyubiquitin chain formation.
Due to the complex nature of the ubiquitination enzymatic cascade, reporters have been developed to study a variety of key components. (A) A hallmark E3 ligase reporter was generated to monitor the interaction between p53 and its E3 ligase Mdm2 primarily using FRET pairings to evaluate substrate ubiquitination. This method is commonly employed in high throughput inhibitor screens. Reporters of polyUb chain formation can use multiple ubiquitin-conjugated fluorophores to detect chain formation by FRET (B) or specific binding domains to study precise lysine linkages such a K63 or linear polyUb chain formation (C). (D) The most common method to study Ub (or UbL) proteases such as DUBs employs Ub (or UbL) conjugated to the N-terminus of PLA2. Upon cleavage PLA2 is free is to cleave its substrate effectively liberating a detectable fluorophore.
Measuring intracellular E3 ligase activity can be difficult for a variety of reasons, including the presence of DUBs or the participation of E2 enzymes in substrate ubiquitination. To overcome the inhibitor effects of DUBs, one group sought to reconstitute the E1-E2-E3 enzymatic cascade in bacteria (49). Co-expression of affinity-tagged substrates and ubiquitin resulted in purified proteins that were used to study E3 autoubiquitination and map ubiquitination sites on the E3 ligase Mindbomb. Another study made use of a reporter consisting of a substrate tethered to an E2 enzyme in order to demonstrate that an E2 could promote ubiquitination in the absence of the E3 ligase (50). However, this ubiquitination is highly non-specific and, by using a substrate-fused to an E3 ligase reporter, it was shown that the E3 ligase actually determines mono- vs. polyubiquitination as well as the type of polyubiquitin chain linkage. Finally, a number of reporters have been developed that indirectly monitor E3 ligase activity. A recent study showed that the E3 ligase Smad ubiquitin regulatory factor-1 (Smurf1) interacts with LIM mineralization protein-1 (LMP-1) to increase the cellular responsiveness of human bone morphogenetic protein-2 (BMP-2), an essential protein used clinically to stimulate bone growth (51). Although the reporting mechanism in this work did not specifically measure E3 ligase activity, it was able to demonstrate the importance of its activity.
Polyubiquitin chain formation
The manner in which a polyubiquitin chain forms on a substrate can dramatically alter the fate of the substrate, depending on the number of ubiquitins as well as the manner in which the chain is linked. As a result, it has become important to develop reporting technologies to specifically monitor polyubiquitin chain formation. Early work in substrate ubiquitination utilized bioluminescence resonance energy transfer (BRET) to study G-protein coupled receptor (GPCR)-mediated β-arrestin 2 ubiquitination (52). The reporter system consisted of a GFP-ubiquitin fusion protein and a substrate that had been fused to Renilla luciferase (Rluc). When the GFP-Ub was conjugated to the substrate, the two moieties came into close proximity with substrate ubiquitination measured by light emission. A similar concept has been used to study the kinetics of polyubiquitin chain formation using two different fluorescently labeled ubiquitins, FAM-Ub and TAMRA-Ub, to produce a measurable FRET signal upon polyubiquitin chain formation (Figure 3b) (53). Another pairwise detection metric was used in a wheat cell-free protein synthesis system to analyze the ubiquitin pathway in Arabidopsis and identify E3 ligases (54). This reporting schemed used FLAG-tagged and biotinylated ubiquitins to produce a luminescent signal by bringing streptavidin-coated donor beads into proximity of protein A-conjugated acceptor beads during polyubiquitin chain formation. Additionally, radiolabeled 125I-ubiquitin was used to elucidate multiple aspects of the E1-E3 enzymatic cascade, such as the stoichiometric determination of E1 and E2 concentrations and a screen for E2-E3 specificity, solely by measuring polyubiquitin chain formation (55).
Recent developments in the reporting of polyubiquitin chain formation have provided insight into the role of K63-linked chains as well as the importance of the conformational state on ubiquitin recognition. Using a string of three ubiquitin-interacting motifs (UIMs) researchers created a reporter that binds to K63-polyUb chains with a 70-fold greater specificity over K48-linked chains (Figure 3c) (56). This probe was then used as a competitive inhibitor to study the role of K63-polyUb on NF-κB activation; however this method has the potential to extend beyond its use as an inhibitor. Similarly, an ubiquitin-binding domain (UBD)-based fluorescent sensor was developed to monitor linear and K63-linked chain production with confocal microscopy in response to Salmonella infection, DNA damage, and mitophagy (57). Further, a novel FRET-based reporter was designed to examine how different conformational states of ubiquitin chains can impact recognition by DUBs (58). Using FRET-labeled K63-, K48, and Met1-linked diubiquitin reporters, it was discovered that the type of lysine linkage conferred either a more ‘open’ or ‘closed’ conformation, which directly correlated with DUB recognition efficiency. While reporting substrate polyubiquitination is essential, it is also important to monitor the different pools of ubiquitin found in the cell. A new method called protein standard absolute quantification (PSAQ) was developed to measure cellular concentrations of ubiquitin species using isotope-labeled protein standards, selective pull down, and mass spectrometry (59). This assay was able to map the population of ubiquitin in free, mono-, or polyubiquitin forms under different experimental conditions such as proteasome inhibition.
Ubiquitin and UbL proteases
Control of the enzymes responsible for the cleavage of the ubiquitin-protein isopeptide bond has become a central focus in the development of novel inhibitors. As such, new reporting techniques needed to be developed to characterize the selectivity of these enzymes, as well as the effectiveness of inhibitors targeting them. The most common type of reporter consists of a single ubiquitin (or UbL) fused to the N-terminus of the enzyme phospholipase A2 (PLA2). This reporter has shown a great deal of success and works because the PLA2 amino terminus must be unobstructed for PLA2 catalytic activity. Upon cleavage of the ubiquitin by a DUB (or UbL by a corresponding protease) the free PLA2 is able to cleave the 2-acyl linkage of a fluorescent 3-sn-phosphoglyceride, effectively liberating a fluorophore from this class of molecule (Figure 3d). Ubiquitin and UbL-fused reporters can be used to discriminate between DUB, deSUMOylase, deNEDDylase, and deISGylase activities, and ultimately profile nonselective isopeptidase inhibitors (60). Highlighting the success and popularity of this reporter system, a high-throughput reporter to monitor sentrin specific proteases (SENPs) for SUMO protease activity is now commercially available (61). Additionally, an Ub-PLA2 reporter was used in a high-throughput screen to identify compounds to inhibit the DUB USP7 (62). This screen led to the discovery of a novel inhibitor that could be used to overcome Bortezomib resistance in multiple myeloma patients. Further improvements have been made on this reporting scheme enabling the creation of a multiplexed assay to ascertain inhibitor selectivity (63). By fusing ubiquitin, or a UbL, to the N-terminus of PLA2 and two other proteins, enterokinase light chain (EKL) and granzyme B (GZMB), that also require a free amino terminus for activity, researchers were able to detect three distinct protease activities simultaneously in a single well. This reporter system was ultimately used to distinguish between selective and non-selective protease inhibitors. In addition to the previously described reporters, there are other means of measuring isopeptidase activity that do not utilize the PLA2 strategy. A FRET-based reporter consisting of YFP-SUMO fused to an enhanced cyan fluorescent protein (ECFP)-peptide was used to evaluate SUMO protease activity (64). When the protease cleaved SUMO from the peptide the separation of the two fluorophores resulted in a quenching of fluorescent signal, allowing effective monitoring of protease activity. This reporting scheme was demonstrated to have applications in protease characterization, enzyme kinetic analysis, and high-throughput inhibitor screening.
Beyond reporting enzymatic activity
In addition to the development of novel reporting techniques, it is important for researchers to continue to identify new proteins regulated by the ubiquitin proteasome system. Global high-throughput screening techniques such as stable isotope labeling with amino acids in cell culture (SILAC) used mass spectrometry in tandem with specific antibodies or siRNA to identify new E3 ligases or E3 ligase targets (65–67). Conversely, the increase in computational power and expanding protein databases have allowed researchers to mine for targets of SUMO ubiquitin ligases, effectively diminishing the need for such costly high-throughput experiments (68). Additionally, novel degrons have been identified not as reporters but as controllable degradation signals. The incorporation of degrons into native proteins has permitted researchers to specifically control degradation through the incorporation of a ligand-induced degradation (LID) domain (69) or by appending a hydrophobic moiety to the surface of a protein (70). While these inducible degradation signals are not currently used for reporting proteasome or E3 ligase activity, they have the potential to be incorporated into the next round of reporters to provide more diverse evaluation of the UPS.
Development of a novel UPS substrate based on the β-Catenin degron
A wide array of methods to measure the activity of members of the ubiquitin proteasome are available and applicable to in vitro assays, single-cell, microscopy-based measurements and transgenic animal analyses. The methods have been indispensable in identifying new proteins involved in the UPS, in the discovery of new inhibitors with translational potential, and in visualizing the role the UPS plays in various diseases. However, these methods are limited when it comes to the analysis of patient samples. Clinical sample size is often limited and diseased cells are typically mixed with healthy cells, which severely limit analytical techniques. For example, in a randomized, multicenter, international phase III study comparing the clinical use of Bortezomib to dexamethasone (another multiple myeloma therapeutic) only 156 of 459 samples collected could be used for gene expression profiling due to low numbers of cells available (71). Additionally, single cells, even those isolated from the same patient or even tumor site, can exhibit differences in enzyme activity due to cell heterogeneity necessitating a need for assay systems with the sensitivity for single-cell measurements (72,73). As such, clinical reporters must be developed that are easily incorporated into single primary cells and that can be analyzed by quantitative techniques such as capillary electrophoresis or chemical cytometry. While the previously described reporters work well with cultured cell lines and transgenic animals, their ability to translate to clinical samples is impaired due to their need to be genetically engineered into cells, a technique that is at best extremely difficult with patient samples of limited size. Further, cultured cell lines have adapted themselves to growing in tissue culture and are very often not reflective of the biology of cells in vivo (74–76) . To address this need, we sought to characterize a known degron that could ultimately be incorporated into single cell reporters to evaluate either E3 ligase or proteasome activity (77). These degron-based substrates were synthesized using solid phase peptide synthesis, a non-vector based method which avoids the need for cellular genetic engineering. Previous work in the lab demonstrated the utility of this technique when evaluating enzymatic activity in single cells (78,79).
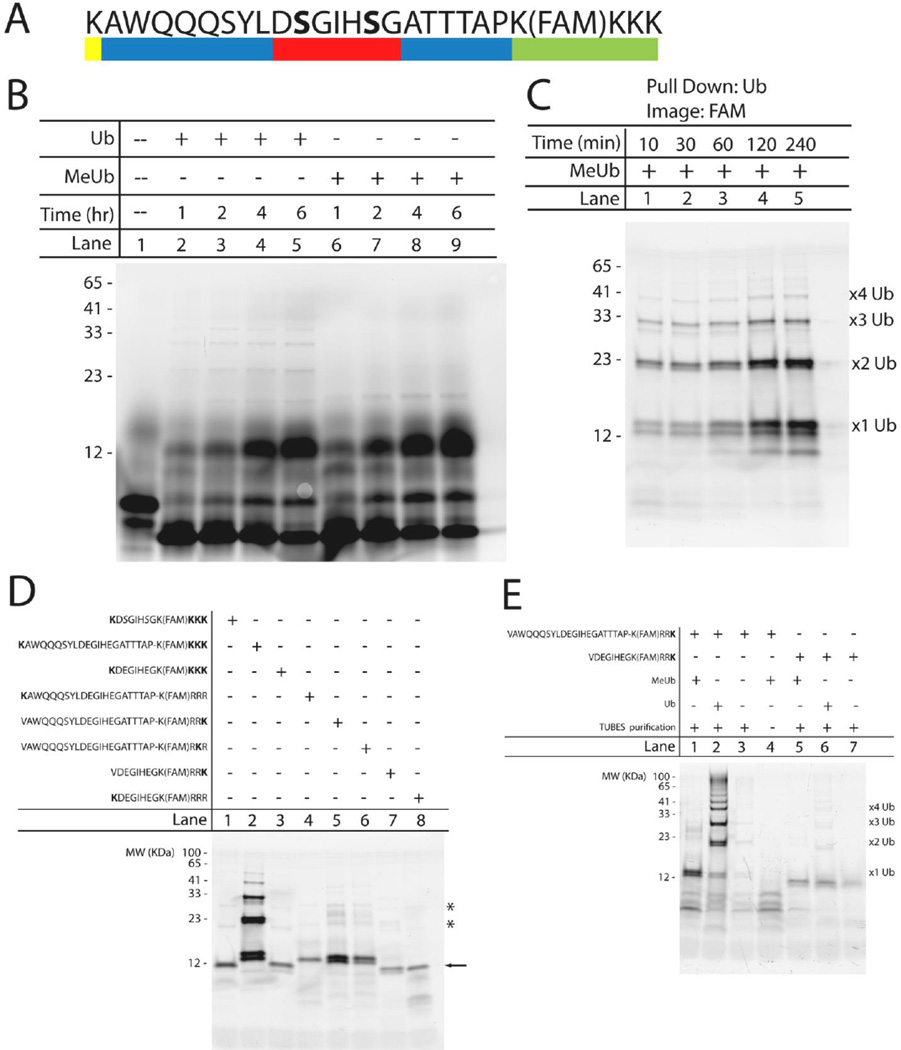
Based on the success of GFPu, the first substrate selected was the CL1 degron; however, the peptide was difficult to synthesize with sufficient yield and purity using standard Fmoc-based synthesis. Next, the phospho-degron from β-Catenin (DSGIHSG) was selected based on its success as a peptide reporter for E3 ligase-mediated ubiquitination. The two serine residues in this motif are phosphorylated (80). Based on these results, a substrate was developed based on the β-Catenin degron that included four important components: the degron (red), a ubiquitination site lysine (yellow), a spacing region to separate the degron and ubiquitin accepting lysine to avoid potential steric hindrance (blue), and a fluorescein tag (green) (Figure 4a). The incorporation of the fluorescein tag into the degron-based substrate simplified detection so that only a fluorescence measurement was needed to visualize substrate ubiquitination rather than other more complex means such as an antibody-based detection method. Further, the fluorescence tag also enables future high sensitivity detection in more precise analytical techniques such as capillary electrophoresis (10−20 to 10−21 moles of peptide or 10–100 nM peptide in a single cell) (72). During substrate design it was decided to incorporate three lysines at the C-terminus to improve peptide solubility by adding an overall positive charge to the sequence and also acted as a spacer between the bulky fluorophore and the amide resin. The substrates were generated using standard Fmoc-protected solid phase peptide synthesis (SPPS), purified by reverse-phase HPLC, and molecular weight confirmed by MALDI-TOF. Finally, a Nanodrop 2000 (Thermo Scientific) was used to measure the fluorescence intensity of all the peptides to determine both the peptide concentration and ensure that each peptide exhibited the same level of basal fluorescence.
Figure 4. Development of a novel UPS substrate based on the β-Catenin degron.
(A) Sequence of the degron-based substrate containing four essential components: the β-Catenin degron (red), an ubiquitination site lysine (yellow), amino acid spacing sequences to prevent steric hindrance (blue), and a fluorescein tag for detection (green). The bold serines are phosphorylated. (B) Time-dependent ubiquitination of the degron-based substrate using either ubiquitin (lanes 2–5) or methylated ubiquitin (lanes 6–9). Lane 1 contains unmodified substrate (∼3.1 kDa). All bands were detected using a Typhon Imager (GE Healthcare Life Sciences) to visualize the 5,6 carboxyfluorescein tag. Numbers to the left of the gel correspond to the apparent molecular weight (in kDa) determined by the known Benchmark fluorescent protein standard (Invitrogen). (C) Verification of substrate ubiquitination using TUBES to selectively pull down ubiquitinated substrate at the indicated times. Assay samples were incubated with methylated ubiquitin. Suspected mono-, di-, tri-, and tetra-ubiquitinated substrates are labeled accordingly. (D) Iterative approach to identify the importance of proximity and location of ubiquitination site lysine on substrate ubiquitination. All samples were incubated in the in vitro ubiquitination assay for 2 hours with methylated ubiquitin prior to pull down with TUBES. (E) Two degron-based substrates, containing a single C-terminal ubiquitination site lysine, were incubated in the in vitro ubiquitination assay, along with TUBES pull down, under the following conditions: methylated ubiquitin (lane 1,5), ubiquitin (lane 2,6), no exogenous MeUb or Ub (lane 3,7) or control-agarose beads instead of the agarose-TUBES (lane 4). Mono-, di-, tri, and tetra-ubiquitinated species are labeled accordingly.
To confirm that the β-Catenin degron-based substrate could be ubiquitinated, the substrate was incubated with HeLa S100 cytosolic lysates as the source of E1, E2, and E3 enzymes (81). Additionally, either exogenous ubiquitin (Ub) or methylated ubiquitin (MeUb) was incorporated into the assay. Native ubiquitin enabled the formation of polyubiquitin chains while MeUb eliminated polyubiquitin chain formation due to each lysine residue being capped with a methyl group allowing for only mono-ubiquitin chain formation. After the in vitro ubiquitination assay, experimental samples were separated and imaged using gel electrophoresis and the bands were detected using a Typhoon Imager to visualize the fluorescein tag. Substrate ubiquitination increased with time when using either Ub or MeUb, as demonstrated by the hallmark ubiquitin ladder (Figure 4b). Additionally, a pronounced band at a very low molecular weight was observed in all lanes and could potentially be due to substrate degradation by cellular peptidases, which remain active in spite of added protease inhibitors (data not shown). Although these results strongly implicated substrate ubiquitination, a ubiquitin pull down assay was performed using tandem ubiquitin binding entity (TUBE) agarose beads (LifeSensors, Malvern, PA) to confirm this (82). To obtain more quantifiable results, the ubiquitin pull down assay utilized MeUb, instead of exogenous Ub, to isolate the mono-ubiquitinated substrates at ∼12 kDa (Figure 4b, lane 6–9). These results verified that the high intensity bands observed from the ubiquitination assay did, in fact, correspond to ubiquitinated substrate (Figure 4c). Control experiments were performed to ensure that the results were not the result of non-specific binding between substrate and agarose bead or basal fluorescent levels of native proteins found in the S100 lysates (data not shown). Based on these controls, it was concluded that all the bands corresponded to specific substrate ubiquitination, with the higher molecular weight bands representing di-, tri-, and tetra-ubiquitinated substrates.
While these results do correspond to successful ubiquitination, the presence of higher molecular weight bands suggested the presence of greater than one ubiquitin per peptide even in the presence of methylated ubiquitin (Figure 4c). It was calculated that a 100-fold excess of methylated ubiquitin (45 µM) was added to the assay over endogenous ubiquitin contained in the S100 lysates (0.3 µM), which suggested that the ubiquitin ladder in the samples with methylated ubiquitin did not correspond to polyubiquitin chain formation. The most likely explanation for the higher molecular weight bands was substrate multi-monoubiquitination, where single ubiquitins are conjugated to different lysine side chains on the same substrate. Thus the E3 ligase, SCFβTrCP, might not possess absolute fidelity for the location of the ubiquitination site lysine on this non-native substrate and C-terminal lysine residues might be readily ubiquitinated in this system. To determine if the above results corresponded to multiple single ubiquitins on the same peptide, substrates were redesigned to replace the three lysines at the C-terminus with three arginines. A subsequent pull-down assay, using only MeUb to prevent polyubiquitin chain formation, demonstrated that eliminating these additional lysine residues prevented multimonoubiquitination (Figure 4d, lane 4) when compared to the initial β-Catenin substrate, which was multi-monoubiquitinated at the C-terminal lysines (Figure 4d, lane 2). Removal of the C-terminal lysines resulted in specific mono-ubiquitination of the degron-based substrates (Figure 4d, arrow). During the redesign, a phospho-mimic was also incorporated through a substitution of glutamic acid for phosphorylated serine to avoid any potential issues with substrate dephosphorylation. The pS→E substitution did not reduce substrate ubiquitination (Figure 4c, lane 4 vs. Figure 4d, lane 2), so all further peptide designs possessed this modification. Based on these results, it was next examined whether the E3 ligase possessed a preference for the location and proximity of the ubiquitination site lysine by placing the lysine at either the N- or C-terminal region on either full length or shortened substrates. An earlier study by Wu and colleagues focusing on SCFβTrCP-mediated β-Catenin ubiquitination found that the rate of ubiquitination of the degron was strongly dependent on the spacing between the ubiquitination site lysine and the degron itself (83). To address the issue of proximity between the ubiquitination site lysine and the degron, substrates were designed either with or without the ∼6–8 amino acid spacing sequences (Figure 4a, blue region) between the N- or C-terminal lysines and the degron (Figure 4d). Removal of the additional amino acids on the peptides with the triple lysine C-terminus demonstrated a pronounced lack of substrate multi-monoubiquitination, implying that there exists a spatial significance for precise substrate ubiquitination (Figure 4d lane 2 vs. lane 3). Further, when comparing full length versus shortened substrates all containing a single ubiquitination site lysine, there was a noticeable increase in substrate ubiquitination in the full length substrate (Figure 4d lane 5 vs. lane 7) further verifying the need for these ‘spacing’ amino acids between the degron and the ubiquitination site lysine. Next, the location of the ubiquitination site lysine was studied to determine the ideal position for maximal substrate ubiquitination. The results obtained indicated that a C-terminal lysine exhibited greater ubiquitination than an N-terminal lysine in both full length (Figure 4d lane 4 vs. lane 5) and shortened (Figure 4d lane 7 vs. lane 8) peptides. Additionally, it was observed that moving the ubiquitination site lysine one residue in from the C-terminus actually decreased the amount of substrate ubiquitination when compared to the C-terminal lysine (Figure 4d lane 6 vs. lane 5). Ultimately, this analysis indicated that there was a slight preference for a C-terminal ubiquitination-site lysine in the full length peptide (Figure 4d, lane 5); however, all of the peptides were ubiquitinated to various degrees which suggested that the E3 ligase did not possess an absolute requirement for a specific location of the lysine.
Finally, it is important to note the presence of the less intense bands found at ∼23 and 29 kDa in the single lysine containing substrates (Figure 4d, asterisk). To address the identity of these bands, two substrates containing a single C-terminal ubiquitination site lysine (full length or shortened) were incubated in the ubiquitin pull down assay in the presence of either MeUb, Ub, or no additional exogenous ubiquitin (Figure 4e). Incubating the two substrates with Ub resulted in the hallmark polyubiquitin ladder (Figure 4e lane 2, 6), while the same ladder was also observed in the full length substrate incubated without any exogenous MeUb or Ub (Figure 4e lane 3). This polyubiquitin ladder, in the absence of exogenous ubiquitin, is most likely due to the presence of endogenous ubiquitin found in the HeLa S100 lysates. Additionally, there was a marked increase in polyubiquitination of the full length substrate when compared with the shortened substrate, verifying what was previously observed that the ‘spacing’ amino acids are important for efficient substrate polyubiquitination (Figure 4e lane 2 vs. lane 6). This was observed further in the substrates incubated without any exogenous ubiquitin, with the shortened substrate lacking any type polyubiquitination (Figure 4e lane 7). Further, these results indicated that the higher MW bands at ∼23 and 29 kDa observed in Figure 4d are not due polyubiquitinated substrate (Figure 4e lane 1 vs. lane 2). The presence of these bands, after the ubiquitin pull down, could be attributed to a number of other issues such as non-specific binding of the MeUb-bound substrate or other modifications. Further work is underway to determine the identity of these unknown species. Finally, incubating the degron-based substrate with control agarose beads instead of the agarose-TUBES resulted in no pull down of ubiquitinated substrate. However, this did confirm that the lower MW bands (< 12 kDa) are most likely due to non-specific binding of degradation products to the agarose beads (Figure 4e lane 4).
This work highlights the isolation and characterization of a portable degron, based on the β-TrCP binding site on β-Catenin, which could ultimately be incorporated into a novel reporter to measure either E3 ligase or proteasome activity in single cells from clinical samples. It is important to note that the observed results only correspond to substrates based on the β-Catenin degron and do not translate to β-Catenin ubiquitination by SCFβ -TrCP under physiological conditions. While the N-terminal lysine found on the original substrate (Figure 4a) does correspond to the natural lysine position, the additional lysines incorporated into the C-terminus of the peptides tested are not representative of the native protein. Finally, one current limitation to this substrate is the rapid degradation in single cells by non-proteasomal systems (84). As such, current work is underway to further optimize the portable degron from β-Catenin to confer resistance to intracellular peptidases to create a more stable reporter. Eventually, these substrates based on portable degrons may exhibit the potential as new and exciting reporting tools to evaluate UPS activity in patient samples.
Outlook
After the clinical success of the proteasome inhibitor Bortezomib and the approval of the next generation inhibitor Carfilzomib (9), the development of therapeutics targeting the ubiquitin proteasome system have become a top priority. Similar to the intensive development of kinase-focused clinical drugs, inhibitors for the proteasome, E3 ligases, deubiquitinating enzymes, and UbL proteases are starting to funnel into clinical trials in greater and greater numbers. However for these inhibitors to succeed, we need to continue to expand our understanding of this essential pathway. In this review we aspire to briefly summarize some of the methods used to report the activity and performance of members of the ubiquitin proteasome system; yet, there is still much to be done in terms of reporter development. Current reporting techniques have proven successful in demonstrating the importance of the proteasome in multiple diseases ranging from cancer to neurodegenerative diseases. Ubiquitin dependent and independent reporters such as GFPu and ODC-GFP have provided preliminary information on Bortezomib effectiveness. Methods to measure E3 ligase and DUB activity have identified new potential inhibitors to complement proteasome inhibition by Bortezomib. From high-throughput screening methods to single cell microscopy these reporters have greatly advanced the understanding and selective targeting of the UPS. However, while cultured cells have been useful in studying the UPS, methods to screen patient samples for the enzymatic activity of therapeutic targets such as the proteasome would greatly benefit clinicians in the diagnosis and treatment of diseases such as multiple myeloma. As such, the next generation of UPS reporters, especially those used for drug discovery, need to be compatible with patient samples. This means reporters that are (1) easily incorporated in patient samples without complex techniques like transfection, (2) specific for the UPS system and resistant to degradation by other intracellular proteases and peptidases, and (3) compatible with single cell analysis techniques like capillary electrophoresis or mass cytometry. In particular, single-cell based technologies have the potential to reveal the full diversity and intricacies of UPS behavior in cells. Further, microscopy-based reporters need to be developed that allow for real-time, in vivo imaging of proteasome activity without the need to sacrifice the test subject. While GFPu- reporters have been useful, novel reporters have the potential to further expand on the known role of the UPS in neurodegenerative disorders such as Huntington’s and Parkinson’s. Through the development of more versatile reporters, there exists the potential to more accurately assess UPS activity in a variety of human diseases, providing the foundation for not only more effective clinical treatment but also greater successful in the discovery of novel therapeutics.
Acknowledgements
This work was supported by grants from the National Institutes of Biomedical Imaging and Bioengineering, R01EB011763 (NLA), and the National Cancer Institute, F32CA162574 (ATM).
References
- 1.Schrader E, Harstad K, Matouschek A. Targeting proteins for degradation. Nature Chemical Biology. 2009;5:815–822. doi: 10.1038/nchembio.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deshaies R, Joazeiro C. RING Domain E3 Ubiquitin Ligases. Annual Review of Biochemistry. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 3.Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nature Reviews Molecular Cell Biology. 2008;9:679–U625. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behrends C, Harper J. Constructing and decoding unconventional ubiquitin chains. Nature Structural & Molecular Biology. 2011;18:520–528. doi: 10.1038/nsmb.2066. [DOI] [PubMed] [Google Scholar]
- 5.Grabbe C, Husnjak K, Dikic I. The spatial and temporal organization of ubiquitin networks. Nature Reviews Molecular Cell Biology. 2011;12:295–307. doi: 10.1038/nrm3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maupin-Furlow J. Proteasome and protein conjugation across domains of life. Nature Reviews Microbiology. 2012;10:100–111. doi: 10.1038/nrmicro2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hickey C, Wilson N, Hochstrasser M. Function and regulation of SUMO proteases. Nature Reviews Molecular Cell Biology. 2012;13:755–766. doi: 10.1038/nrm3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tobinai K. Proteasome inhibitor, bortezomib, for myeloma and lymphoma. International Journal of Clinical Oncology. 2007;12:318–326. doi: 10.1007/s10147-007-0695-5. [DOI] [PubMed] [Google Scholar]
- 9.Kuhn D, Chen Q, Voorhees P, Strader J, Shenk K, Sun C, Derno S, Bennett M, Van Leeuwen F, Chanan-Khan A, Orlowski R. Potent activity of carfilzomib, a novel, irreversible inhibitor of the ubiquitin-proteasome pathway, against preclinical models of multiple myelorna. Blood. 2007;110:3281–3290. doi: 10.1182/blood-2007-01-065888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindsten K, Dantuma N. Monitoring the ubiquitin/proteasome system in conformational diseases. Ageing Research Reviews. 2003;2:433–449. doi: 10.1016/s1568-1637(03)00031-x. [DOI] [PubMed] [Google Scholar]
- 11.Weissman A, Shabek N, Ciechanover A. The predator becomes the prey: regulating the ubiquitin system by ubiquitylation and degradation. Nature Reviews Molecular Cell Biology. 2011;12:605–620. doi: 10.1038/nrm3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muller PAJ, Vousden KH. p53 mutations in cancer. Nature Cell Biology. 2013;15:2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- 13.Hussain S, Zhang Y, Galardy P. DUBs and cancer The role of deubiquitinating enzymes as oncogenes, non-oncogenes and tumor suppressors. Cell Cycle. 2009;8:1688–1697. doi: 10.4161/cc.8.11.8739. [DOI] [PubMed] [Google Scholar]
- 14.Bence N, Bennett E, Kopito R, Deshaies R. Application and analysis of the GFP(u) family of ubiquitin-proteasome system reporters. Ubiquitin and Protein Degradation, Pt B. 2005;399:481–490. doi: 10.1016/S0076-6879(05)99033-2. [DOI] [PubMed] [Google Scholar]
- 15.Nonaka T, Hasegawa M. A Cellular Model To Monitor Proteasome Dysfunction by alpha-Synuclein. Biochemistry. 2009;48:8014–8022. doi: 10.1021/bi900619j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cook C, Gass J, Dunmore J, Tong J, Taylor J, Eriksen J, McGowan E, Lewis J, Johnston J, Petrucelli L. Aging Is Not Associated with Proteasome Impairment in UPS Reporter Mice. Plos One. 2009;4 doi: 10.1371/journal.pone.0005888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bett J, Cook C, Petrucelli L, Bates G. The Ubiquitin-Proteasome Reporter GFPu Does Not Accumulate in Neurons of the R6/2 Transgenic Mouse Model of Huntington's Disease. Plos One. 2009;4 doi: 10.1371/journal.pone.0005128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menendez-Benito V, Heessen S, Dantuma N, Deshaies R. Monitoring of ubiquitin-dependent proteolysis with green fluorescent protein substrates. Ubiquitin and Protein Degradation, Pt B. 2005;399:490–511. doi: 10.1016/S0076-6879(05)99034-4. [DOI] [PubMed] [Google Scholar]
- 19.Segref A, Torres S, Hoppe T. A Screenable in vivo Assay to Study Proteostasis Networks in Caenorhabditis elegans. Genetics. 2011;187:1235–U1438. doi: 10.1534/genetics.111.126797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamer G, Matilainen O, Holmberg C. A photoconvertible reporter of the ubiquitin-proteasome system in vivo (vol 7, pg 473, 2010) Nature Methods. 2010;7:567–567. doi: 10.1038/nmeth.1460. [DOI] [PubMed] [Google Scholar]
- 21.Kraut D, Israeli E, Schrader E, Patil A, Nakai K, Nanavati D, Inobe C, Matouschek A. Sequence- and Species-Dependence of Proteasomal Processivity. Acs Chemical Biology. 2012;7:1444–1453. doi: 10.1021/cb3001155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blackburn C, Gigstad K, Hales P, Garcia K, Jones M, Bruzzese F, Barrett C, Liu J, Soucy T, Sappal D, Bump N, Olhava E, Fleming P, Dick L, Tsu C, Sintchak M, Blank J. Characterization of a new series of non-covalent proteasome inhibitors with exquisite potency and selectivity for the 20S beta 5-subunit (vol 430, pg 461, 2010) Biochemical Journal. 2010;431:433–433. doi: 10.1042/BJ20100383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ausseil F, Samson A, Aussagues Y, Vandenberghe I, Creancier L, Pouny I, Kruczynski A, Massiot G, Bailly C. High-throughput bioluminescence screening of ubiquitin-proteasome pathway inhibitors from chemical and natural sources. Journal of Biomolecular Screening. 2007;12:106–116. doi: 10.1177/1087057106296494. [DOI] [PubMed] [Google Scholar]
- 24.Gross S, Piwnica-Worms D. Spying on cancer: Molecular imaging in vivo with genetically encoded reporters. Cancer Cell. 2005;7:5–15. doi: 10.1016/j.ccr.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 25.Hoyt M, Zhang M, Coffino P, Deshaies R. Probing the ubiquitin/proteasome system with ornithine decarboxylase, a ubiquitin-independent substrate. Ubiquitin and Protein Degradation, Part a. 2005;398:399–413. doi: 10.1016/S0076-6879(05)98033-6. [DOI] [PubMed] [Google Scholar]
- 26.Momose I, Tatsuda D, Ohba S, Masuda T, Ikeda D, Nomoto A. In vivo imaging of proteasome inhibition using a proteasome-sensitive fluorescent reporter. Cancer Science. 2012;103:1730–1736. doi: 10.1111/j.1349-7006.2012.02352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao V, Guan B, Mutton L, Bieberich C. Proline-mediated Proteasomal Degradation of the Prostate-specific Tumor Suppressor NKX3.1. Journal of Biological Chemistry. 2012;287:36331–36340. doi: 10.1074/jbc.M112.352823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kisselev A, Goldberg A, Deshaies R. Monitoring activity and inhibition of 26S proteasomes with fluorogenic peptide substrates. Ubiquitin and Protein Degradation, Part a. 2005;398:364–378. doi: 10.1016/S0076-6879(05)98030-0. [DOI] [PubMed] [Google Scholar]
- 29.Berkers C, Verdoes M, Lichtman E, Fiebiger E, Kessler B, Anderson K, Ploegh H, Ovaa H, Galardy P. Activity probe for in vivo profiling of the specificity of proteasome inhibitor bortezomib. Nature Methods. 2005;2:357–362. doi: 10.1038/nmeth759. [DOI] [PubMed] [Google Scholar]
- 30.Moravec R, O'Brien M, Daily W, Scurria M, Bernad L, Riss T. Cell-based bioluminescent assays for all three proteasome activities in a homogeneous format. Analytical Biochemistry. 2009;387:294–302. doi: 10.1016/j.ab.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 31.Urru S, Veglianese P, De Luigi A, Fumagalli E, Erba E, Diaza R, Carra A, Davoli E, Borsello T, Forloni G, Pengo N, Monzani E, Cascio P, Cenci S, Sitia R, Salmona M. A New Fluorogenic Peptide Determines Proteasome Activity in Single Cells. Journal of Medicinal Chemistry. 2010;53:7452–7460. doi: 10.1021/jm100362x. [DOI] [PubMed] [Google Scholar]
- 32.Stein M, Beck P, Kaiser M, Dudler R, Becker C, Groll M. One-shot NMR analysis of microbial secretions identifies highly potent proteasome inhibitor. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:18367–18371. doi: 10.1073/pnas.1211423109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prakash S, Tian L, Ratliff K, Lehotzky R, Matouschek A. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nature Structural & Molecular Biology. 2004;11:830–837. doi: 10.1038/nsmb814. [DOI] [PubMed] [Google Scholar]
- 34.Takeuchi J, Chen H, Coffino P. Proteasome substrate degradation requires association plus extended peptide. Embo Journal. 2007;26:123–131. doi: 10.1038/sj.emboj.7601476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burns K, Pearce M, Darwin K. Prokaryotic Ubiquitin-Like Protein Provides a Two-Part Degron to Mycobacterium Proteasome Substrates. Journal of Bacteriology. 2010;192:2933–2935. doi: 10.1128/JB.01639-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Inobe T, Fishbain S, Prakash S, Matouschek A. Defining the geometry of the two-component proteasome degron. Nature Chemical Biology. 2011;7:161–167. doi: 10.1038/nchembio.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Henderson A, Erales J, Hoyt M, Coffino P. Dependence of Proteasome Processing Rate on Substrate Unfolding. Journal of Biological Chemistry. 2011;286:17495–17502. doi: 10.1074/jbc.M110.212027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kraut D, Matouschek A. Proteasomal Degradation from Internal Sites Favors Partial Proteolysis via Remote Domain Stabilization. Acs Chemical Biology. 2011;6:1087–1095. doi: 10.1021/cb2002285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shabek N, Herman-Bachinsky Y, Buchsbaum S, Lewinson O, Haj-Yahya M, Hejjaoui M, Lashuel H, Sommer T, Brik A, Ciechanover A. The Size of the Proteasomal Substrate Determines Whether Its Degradation Will Be Mediated by Mono- or Polyubiquitylation. Molecular Cell. 2012;48:87–97. doi: 10.1016/j.molcel.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 40.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 41.Xia Z, Webster A, Du F, Piatkov K, Ghislain M, Varshavsky A. Substrate-binding sites of UBR1, the ubiquitin ligase of the N-end rule pathway. Journal of Biological Chemistry. 2008;283:24011–24028. doi: 10.1074/jbc.M802583200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sriram S, Kim B, Kwon Y. The N-end rule pathway: emerging functions and molecular principles of substrate recognition. Nature Reviews Molecular Cell Biology. 2011;12:735–747. doi: 10.1038/nrm3217. [DOI] [PubMed] [Google Scholar]
- 43.Xu Z, Payoe R, Fahlman R. The C-terminal Proteolytic Fragment of the Breast Cancer Susceptibility Type 1 Protein (BRCA1) Is Degraded by the N-end Rule Pathway. Journal of Biological Chemistry. 2012;287:7495–7502. doi: 10.1074/jbc.M111.301002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Houser J, Ford E, Chatterjea S, Maleri S, Elston T, Errede B. An improved short-lived fluorescent protein transcriptional reporter for Saccharomyces cerevisiae. Yeast. 2012;29:519–530. doi: 10.1002/yea.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davydov I, Woods D, Safiran Y, Oberoi P, Fearnhead H, Fang S, Jensen J, Weissman A, Kenten J, Vousden K. Assay for ubiquitin ligase activity: High-throughput screen for inhibitors of HDM2. Journal of Biomolecular Screening. 2004;9:695–703. doi: 10.1177/1087057104267956. [DOI] [PubMed] [Google Scholar]
- 46.Murray M, Jurewicz A, Martin J, Ho T, Zhang H, Johanson K, Kirkpatrick R, Ma J, Lor L, Thrall S, Schwartz B. A high-throughput screen measuring ubiquitination of p53 by human mdm2. Journal of Biomolecular Screening. 2007;12:1050–1058. doi: 10.1177/1087057107308556. [DOI] [PubMed] [Google Scholar]
- 47.Li J, Zhang S, Gao L, Chen Y, Xie X. A Cell-Based High-Throughput Assay for the Screening of Small-Molecule Inhibitors of p53-MDM2 Interaction. Journal of Biomolecular Screening. 2011;16:450–456. doi: 10.1177/1087057111399191. [DOI] [PubMed] [Google Scholar]
- 48.Merbl Y, Kirschner M. Large-scale detection of ubiquitination substrates using cell extracts and protein microarrays. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2543–2548. doi: 10.1073/pnas.0812892106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keren-Kaplan T, Attali I, Motamedchaboki K, Davis B, Tanner N, Reshef Y, Laudon E, Kolot M, Levin-Kravets O, Kleifeld O, Glickman M, Horazdovsky B, Wolf D, Prag G. Synthetic biology approach to reconstituting the ubiquitylation cascade in bacteria. Embo Journal. 2012;31:378–390. doi: 10.1038/emboj.2011.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.David Y, Ternette N, Edelmann M, Ziv T, Gayer B, Sertchook R, Dadon Y, Kessler B, Navon A. E3 Ligases Determine Ubiquitination Site and Conjugate Type by Enforcing Specificity on E2 Enzymes. Journal of Biological Chemistry. 2011;286:44104–44115. doi: 10.1074/jbc.M111.234559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okada M, Sangadala S, Liu Y, Yoshida M, Reddy B, Titus L, Boden S. Development and optimization of a cell-based assay for the selection of synthetic compounds that potentiate bone morphogenetic protein-2 activity. Cell Biochemistry and Function. 2009;27:526–534. doi: 10.1002/cbf.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perroy J, Pontier S, Charest P, Aubry M, Bouvier M. Real-time monitoring of ubiquitination in living cells by BRET. Nature Methods. 2004;1:203–208. doi: 10.1038/nmeth722. [DOI] [PubMed] [Google Scholar]
- 53.Gururaja T, Pray T, Lowe R, Dong G, Huang J, Daniel-Issakani S, Payan D. A homogeneous FRET assay system for multiubiquitin chain assembly and disassembly. Methods in Enzymology. 2005;399:663–682. doi: 10.1016/S0076-6879(05)99044-7. [DOI] [PubMed] [Google Scholar]
- 54.Takahashi H, Nozawa A, Seki M, Shinozaki K, Endo Y, Sawasaki T. A simple and high-sensitivity method for analysis of ubiquitination and polyubiquitination based on wheat cell-free protein synthesis. Bmc Plant Biology. 2009;9 doi: 10.1186/1471-2229-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ronchi V, Haas A. Measuring rates of ubiquitin chain formation as a functional readout of ligase activity. Methods in Molecular Biology. 2012;832:197–218. doi: 10.1007/978-1-61779-474-2_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sims J, Scavone F, Cooper E, Kane L, Youle R, Boeke J, Cohen R. Polyubiquitin-sensor proteins reveal localization and linkage-type dependence of cellular ubiquitin signaling. Nature Methods. 2012;9:303–U122. doi: 10.1038/nmeth.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Wijk S, Fiskin E, Putyrski M, Pampaloni F, Hou J, Wild P, Kensche T, Grecco H, Bastiaens P, Dikic I. Fluorescence-Based Sensors to Monitor Localization and Functions of Linear and K63-Linked Ubiquitin Chains in Cells. Molecular Cell. 2012;47:797–809. doi: 10.1016/j.molcel.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ye Y, Blaser G, Horrocks M, Ruedas-Rama M, Ibrahim S, Zhukov A, Orte A, Kleneman D, Jackson S, Komander D. Ubiquitin chain conformation regulates recognition and activity of interacting proteins. Nature. 2012;492:266–270. doi: 10.1038/nature11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaiser S, Riley B, Shaler T, Trevino R, Becker C, Schulman H, Kopito R. Protein standard absolute quantification (PSAQ) method for the measurement of cellular ubiquitin pools. Nature Methods. 2011;8:691–U129. doi: 10.1038/nmeth.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nicholson B, Leach C, Goldenberg S, Francis D, Kodrasov M, Tian X, Shanks J, Sterner D, Bernal A, Mattern M, Wilkinson K, Butt T. Characterization of ubiquitin and ubiquitin-like-protein isopeptidase activities. Protein Science. 2008;17:1035–1043. doi: 10.1110/ps.083450408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leach C, Tian X, Mattern M, Nicholson B. Detection and characterization of SUMO protease activity using a sensitive enzyme-based reporter assay. Methods in Molecular Biology. 2009;497:269–281. doi: 10.1007/978-1-59745-566-4_18. [DOI] [PubMed] [Google Scholar]
- 62.Chauhan D, Tian Z, Nicholson B, Kumar K, Zhou B, Carrasco R, McDermott J, Leach C, Fulcinniti M, Kodrasov M, Weinstock J, Kingsbury W, Hideshima T, Shah P, Minvielle S, Altun M, Kessler B, Orlowski R, Richardson P, Munshi N, Anderson K. A Small Molecule Inhibitor of Ubiquitin-Specific Protease-7 Induces Apoptosis in Multiple Myeloma Cells and Overcomes Bortezomib Resistance. Cancer Cell. 2012;22:345–358. doi: 10.1016/j.ccr.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tian X, Isamiddinova N, Peroutka R, Goldenberg S, Mattern M, Nicholson B, Leach C. Characterization of Selective Ubiquitin and Ubiquitin-Like Protease Inhibitors Using a Fluorescence-Based Multiplex Assay Format. Assay and Drug Development Technologies. 2011;9:165–173. doi: 10.1089/adt.2010.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martin S, Hattersley N, Samuel I, Hay R, Tatham M. A fluorescence-resonance-energy-transfer-based protease activity assay and its use to monitor paralog-specific small ubiquitin-like modifier processing. Analytical Biochemistry. 2007;363:83–90. doi: 10.1016/j.ab.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 65.Emanuele M, Elia A, Xu Q, Thoma C, Izhar L, Leng Y, Guo A, Chen Y, Rush J, Hsu P, Yen H, Elledge S. Global Identification of Modular Cullin-RING Ligase Substrates. Cell. 2011;147:459–474. doi: 10.1016/j.cell.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim W, Bennett E, Huttlin E, Guo A, Li J, Possemato A, Sowa M, Rad R, Rush J, Comb M, Harper J, Gygi S. Systematic and Quantitative Assessment of the Ubiquitin-Modified Proteome. Molecular Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee K, Hammerle L, Andrews P, Stokes M, Mustelin T, Silva J, Black R, Doedens J. Ubiquitin Ligase Substrate Identification through Quantitative Proteomics at Both the Protein and Peptide Levels. Journal of Biological Chemistry. 2011;286:41530–41538. doi: 10.1074/jbc.M111.248856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun H, Hunter T. Poly-small ubiquitin-like modifier (PolySUMO)-binding proteins identified through a string search. Journal of Biological Chemistry. 2012;287:42071–42083. doi: 10.1074/jbc.M112.410985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bonger K, Chen L, Liu C, Wandless T. Small-molecule displacement of a cryptic degron causes conditional protein degradation. Nature Chemical Biology. 2011;7:531–537. doi: 10.1038/nchembio.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Neklesa T, Tae H, Schneekloth A, Stulberg M, Corson T, Sundberg T, Raina K, Holley S, Crews C. Small-molecule hydrophobic tagging-induced degradation of HaloTag fusion proteins. Nature Chemical Biology. 2011;7:538–543. doi: 10.1038/nchembio.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mulligan G, Mitsiades C, Bryant B, Zhan F, Chng W, Roels S, Koenig E, Fergus A, Huang Y, Richardson P, Trepicchio W, Broyl A, Sonneveld P, Shaughnessy J, Jr., Bergsagel L, Schenkein D, Esseltine D-L, Boral A, Anderson K. Gene expression profiling and correlation with outcome in clinical trials of the proteasome inhibitor bortezomib. Blood. 2007;109:3177–3188. doi: 10.1182/blood-2006-09-044974. [DOI] [PubMed] [Google Scholar]
- 72.Kovarik M, Allbritton N. Measuring enzyme activity in single cells. Trends in Biotechnology. 2011;29:222–230. doi: 10.1016/j.tibtech.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marusyk A, Almendro V, Polyank K. Intra-tumour heterogeneity: a looking glass for cancer. Nature Reviews Cancer. 2012;12:323–334. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- 74.Ince TA, Richardson AL, Bell GW, Saitoh M, Godar S, Karnoub AE, Iglehart JD, Weinberg RA. Transformation of different human breast epithelial cell types leads to distinct tumor phenotypes. Cancer Cell. 2007;12:160–170. doi: 10.1016/j.ccr.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 75.Jin K, Teng L, Shen Y, He K, Xu Z, Li G. Patient-derived human tumour tissue xenografts in immunodeficient mice: a systematic review. Clinical and Translational Oncology. 2010;12:473–480. doi: 10.1007/s12094-010-0540-6. [DOI] [PubMed] [Google Scholar]
- 76.Tentler JJ, Tan AC, Weekes CD, Jimeno A, Leong S, Pitts TM, Arcaroli JJ, Messersmith WA, Eckhardt SG. Patient-derived tumour xenografts as models for oncology drug development. Nature Reviews Clinical Oncology. 2012;9:338–350. doi: 10.1038/nrclinonc.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Melvin AT, Woss GS, Park JH, Waters ML, Allbritton NA. Unpublished data [Google Scholar]
- 78.Jiang D, Sims CE, Allbritton NL. Single cell analysis of phosphoinositide 3-kinase (PI3K) and phosphatase and tensins homolog (PTEN) activation. Faraday Discussions. 2011;149:187–200. doi: 10.1039/C005362G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Proctor A, Wang Q, Lawrence DS, Allbritton NL. Development of a peptidase-resistant substrate for single-cell measurements of protein kinase B activation. Analytical Chemistry. 2012;84:7195–7202. doi: 10.1021/ac301489d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pierce N, Kleiger G, Shan S, Deshaies R. Detection of sequential polyubiquitylation on a millisecond timescale. Nature. 2009;462:615–U685. doi: 10.1038/nature08595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dignam J, Lebovitz R, Roeder R. Accurate Transcription Initiation by RNA Polymerase-II in a Soluble Extract From Isolated Mammalian Nuclei. Nucleic Acids Research. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hjerpe R, Aillet F, Lopitz-Otsoa F, Lang V, England P, Rodriguez M. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. Embo Reports. 2009;10:1250–1258. doi: 10.1038/embor.2009.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu G, Xu G, Schulman B, Jeffrey P, Harper J, Pavletich N. Structure of a beta-TrCP1-Skp1-beta-catenin complex: Destruction motif binding and lysine specificity of the SCF beta-TrCP1 ubiquitin ligase. Molecular Cell. 2003;11:1445–1456. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- 84.Proctor A, Wang Q, Lawrence D, Allbritton N. Metabolism of peptide reporters in cell lysates and single cells. Analyst. 2012;137:3028–3038. doi: 10.1039/c2an16162a. [DOI] [PMC free article] [PubMed] [Google Scholar]