Abstract
Neurons have highly specialized intracellular compartments that facilitate the development and activity of the nervous system. Ubiquitination is a post-translational modification that controls many aspects of neuronal function by regulating protein abundance. Disruption of this signaling pathway has been demonstrated in neurological disorders such as Parkinson’s disease, Amyotrophic Lateral Sclerosis and Angleman Syndrome. Since many neurological disorders exhibit ubiquitinated protein aggregates, the loss of neuronal ubiquitin homeostasis may be an important contributor of disease. This review discusses the mechanisms utilized by neurons to control the free pool of ubiquitin necessary for normal nervous system development and function as well as new roles of protein ubiquitination in regulating synaptic activity.
Introduction
While many of the processes that require ubiquitin are common to all cell types, ubiquitin also has distinct roles in highly specialized and structurally unique cells like neurons [1]. During neuronal development, synapse formation [2] and synaptic pruning [3] are regulated by the ubiquitin proteasome system, as are excitatory [4,5] and inhibitory [5] transmission in the mature brain. Although protein degradation is the most well-studied aspect of the ubiquitin proteasome system, protein ubiquitination is also responsible for regulating cell signaling by controlling the endocytosis of plasma membrane receptors [1]. The complex morphology of neurons makes the regulation of ubiquitin levels more challenging than in morphologically simpler cell types [6].
Ubiquitin, a highly conserved 76-amino acid protein, was originally described in 1975 in studies aimed at discovering hormones produced by the thymus [7]. It was later shown to be identical to ATP-dependent proteolysis factor 1 [8], which was implicated in protein degradation by the pioneering work of Aaron Ciechanover, Avram Hershko, and Irwin Rose [9,10]. Ubiquitin has since been identified in all eukaryotic cells and, although it was first studied for its role in tagging proteins for degradation by the proteasome, it is now known to be involved in processes as varied as signal transduction, endocytosis, and DNA repair.
The levels of free ubiquitin in a cell are determined by the rates of ubiquitin synthesis, polyubiquitin chain formation, polyubiquitin chain disassembly, and ubiquitin degradation. We use to the term “free” ubiquitin to designate the unconjugated pool of ubiquitin and “conjugated” to refer to ubiquitin that has been covalently attached to substrates of the ubiquitin proteasome system. In mouse brain, 60% of the processed ubiquitin is found as a free monomer and 40% is conjugated onto substrates [11]. Of the conjugated ubiquitin, approximately 90% is found on mono-ubiquitinated substrates and 10% is found on polyubiquitinated substrates. The high levels of free ubiquitin found in neurons may serve as a reservoir to allow for rapid responses to cell stimulation or stress. In this review, we will discuss the mechanisms utilized by neurons to control ubiquitin levels, the consequences of altering ubiquitin homeostasis, and novel roles for protein ubiquitination in regulating synaptic activity.
De novo ubiquitin synthesis
Two classes of genes encode ubiquitin in the mammalian genome: the ribosomal fusion proteins Rsp27a and Uba52, which are constitutively expressed, and the polyubiquitin genes Ubb and Ubc, which are induced by various forms of cellular stress [12,13]. Rsp27a and Uba52 each encode a single ubiquitin moiety fused to a small ribosomal protein, whereas Ubb and Ubc encode 3 and 9 tandem ubiquitin repeats, respectively. In all cases, the generation of monomeric ubiquitin requires post-translational cleavage of fusion proteins by ubiquitin-specific proteases, notably Usp5 [14].
Although both Ubb and Ubc are transcribed at increased rates in response to cell stress [12,13], both ubiquitin poly-proteins also appear to contribute to basal ubiquitin levels. For instance, in mice, Ubb accounts for more than 60% of the total ubiquitin transcripts in the testes and nearly 40% in brain [15]. Although the redundancy in ubiquitin genes should, presumably, allow for an increase in transcription of the remaining genes to compensate for loss of Ubb, ubiquitin levels are significantly decreased in both the gonads and the hypothalamus of Ubb knockout mice [15–17]., Ubb knockout mice have a developmental arrest of spermatocytes and oocytes prior to the first meiotic division, hypogonadism, and late onset obesity due to hypothalamic dysfunction. Similarly, mouse embryonic fibroblasts that lack Ubc show a significant decrease in ubiquitin. Expression of Ubc appears to be especially important in liver development, as knockout of Ubc in mice is embryonically lethal due to reduced proliferation of fetal liver cells [16,18].
Transport of ubiquitin in neurons
Following the de novo generation of ubiquitin in the cell body, ubiquitin is transported from the soma to distant locals like axons and dendrites. A single study in the literature indicates that ubiquitin is trafficked via slow axonal transport down the rat optic nerve [6]. This transport proceeds at a rate of approximately 3 mm/day, indicating that the length of time required for newly generated ubiquitin to reach synaptic terminals is on the order of days, or even weeks, in some neurons. As ubiquitin is a component of the cellular response to heat shock and other stressors, the slow rate of transport may therefore make distal axons and dendrites particularly vulnerable to stress. For example, the accumulation of ubiquitin-positive deposits in axons and dendrites of diseased neurons demonstrates that protein degradation is compromised in these compartments. Sequestration of ubiquitin in these aggregates may contribute to a local depletion in free ubiquitin that can only be replenished by ubiquitin synthesized in the soma.
Conjugation of ubiquitin to target proteins
Rates of ubiquitin conjugation and deconjugation can directly influence the steady-state levels of free ubiquitin. For example, proteasome inhibition results in a shift in ubiquitin from the free pool to the conjugated pool. Ubiquitin is activated for conjugation by an E1 enzyme that harnesses ATP hydrolysis to adenlyate ubiquitin’s C-terminal glycine [19]. This high-energy intermediate is subject to rapid nucleophilic attack by the E1 enzyme’s active site cysteine, forming a high-energy thioester bond. Activated ubiquitin is then passed to the active-site cysteine of an E2 enzyme, which interacts with the ubiquitin-charged E1 enzyme with high affinity [20,21]. Upon dissociation of the uncharged E1 activating enzyme [22], the E2 enzyme interacts with an E3 enzyme that is associated with the protein to be ubiquitinated, resulting in an isopeptide bond between ubiquitin’s C-terminal glycine and the ε-amino group in a substrate’s lysine residue.
Conjugation of a single ubiquitin moiety to a protein, termed monoubiquitination, is an important signaling event in endocytosis and transcriptional regulation [23]. However, because ubiquitin itself contains 7 lysines, ubiquitin chains can be formed by ligation of the C-terminal glycine residue to a lysine residue in another ubiquitin monomer [24]. The most well characterized ubiquitin chain is the K48-linkage, which targets proteins to the proteasome for degradation [24–26]. K63-chains are involved in signal transduction, scaffolding [27], and DNA repair [28]. Although monoubiquitination has been shown, in vitro, to be sufficient for receptor endocytosis [29,30], many subsequent studies show that K63-linked chains lead to more efficient processing through the endosome-lysosome pathway. However, recent studies indicate that ubiquitin signaling is significantly more complicated than previously appreciated and, although these generalizations form a useful framework for introducing the biological consequences of various ubiquitin linkages, functional categorization of specific ubiquitination events is far from straightforward [31].
Ubiquitin deposition in protein aggregates
An increase in ubiquitin conjugates is associated with several neurodegenerative disorders [32–34]. In Parkinson’s disease, focal accumulations of ubiquitinated α-synuclein can be found in Lewy-bodies. Ubiquitinated protein aggregates are also found in neurofibrillary tangles from post-mortem brains of patients with Alzheimer’s disease. While the role that these aggregates play in actually causing disease is controversial, the formation of these aggregates could result in ‘trapping’ ubiquitin molecules, lowering the free ubiquitin pool, and decreasing ubiquitin-dependent proteolysis. Consistent with this idea, mouse models of ubiquitin deficiency due to either loss of ubiquitin expression or increased ubiquitin turnover result in neurodegenerative disease [15–17,35,36].
Post-translational stability of ubiquitin in neurons
An under-appreciated consequence of protein turnover at the proteasome is the destruction of ubiquitin. In vitro studies demonstrate that ubiquitin can enter the proteasome either as free monomers or conjugated to substrates [37]. This finding indicates that, although the proteasome contains several deubiquitinating enzymes to facilitate ubiquitin recycling, there is continual degradation of ubiquitin by the proteasome. Since the consequences of ubiquitin depletion can be dire, especially in neurons, the advantages of constitutive ubiquitin degradation by the proteasome are intriguing. Ubiquitin turnover could serve to eliminate damaged ubiquitin molecules to maintain a functional pool of ubiquitin or allow for acute changes in ubiquitin levels to modulate ubiquitin-signaling events. The following section will address how changes in deubiquitinating enzymes can alter the levels of ubiquitin in neurons and discuss how these changes in ubiquitin levels affect synaptic development and function.
Sequestration of ubiquitin pools by UCHL1
Ubiquitin C-terminal hydrolase 1 (UCHL1) is a highly abundant protein in neurons and is thought to play an important role in ubiquitin homeostasis. Interest in the role of UCHL1 in the nervous system was sparked when a German family with Parkinson’s disease was shown to have a missense mutation in Uchl1 that reduced its protease activity [38]. This finding implicated a possible connection between defective ubiquitin-dependent protein turnover and Parkinson’s disease. Shortly after this study was published, the spontaneously occurring gracile axonal dystrophy (gad) mouse mutation was shown to be in the Uchl1 gene [39]. The gad mice display a late onset motor ataxia that correlates with the accumulation of ubiquitinated proteins and nerve terminal swellings [39]. A first indication of UCHL1’s involvement in ubiquitin homeostasis was the finding that monomeric ubiquitin levels are significantly reduced in brain extracts from gad mice [35]. This decrease in monomeric ubiquitin levels was not associated with an up-regulation of Ubb or Ubc transcription. The lack of a ubiquitin stress response suggests that loss of UCHL1 may result in compartment-specific destabilization of ubiquitin and that the ubiquitin present in other compartments prevents an increase in Ubb and Ubc transcription. Since UCHL1 has a high affinity for ubiquitin, it has been suggested that UCHL1 may serve to sequester ubiquitin and prevent its degradation. Consistent with this idea, transgenic overexpression of UCHL1 results in increased levels of free ubiquitin [35].
The postsynaptic compartment of a neuron is one region that is sensitive to loss of UCHL1. Rapid ubiquitin-dependent proteasomal protein turnover occurs in the postsynaptic compartment following synaptic activity, and this regulated protein degradation is critical to activity-dependent strengthening and weakening of synaptic connections [40]. Genetic deletion of UCHL1 reduces long-term potentiation (a long-term increase in synaptic strength) [41], and UCHL1 inhibition leads to an increase in the size of dendritic spines and a decrease in their density [42], demonstrating the importance of UCHL1 in the postsynaptic compartment. Interestingly, these changes in spine morphology are reversed by ectopic expression of ubiquitin, arguing that a critical role of UCHL1 in the post-synapse is the maintenance of ubiquitin pools.
USP14 prevents proteasomal degradation of ubiquitin
Ubiquitin-specific protease 14 (USP14) is a deubiquitinating enzyme that dynamically associates with the proteasome to stabilize ubiquitin levels [36,43]. While unbound USP14 is catalytically inactive, association of USP14 with the proteasome stimulates its ubiquitin hydrolase activity [36,44]. By controlling the association of USP14 with the proteasome, the cell is therefore able to restrict USP14’s deubiquitinating activity to substrates bound to the proteasome. In vitro studies have also shown that the loss of USP14 leads to enhanced turnover of ubiquitinated proteins by the proteasome [45]. These findings indicate that USP14 is tightly regulated by the proteasome, and changes in the activity of USP14 can alter ubiquitin-dependent protein turnover by the proteasome.
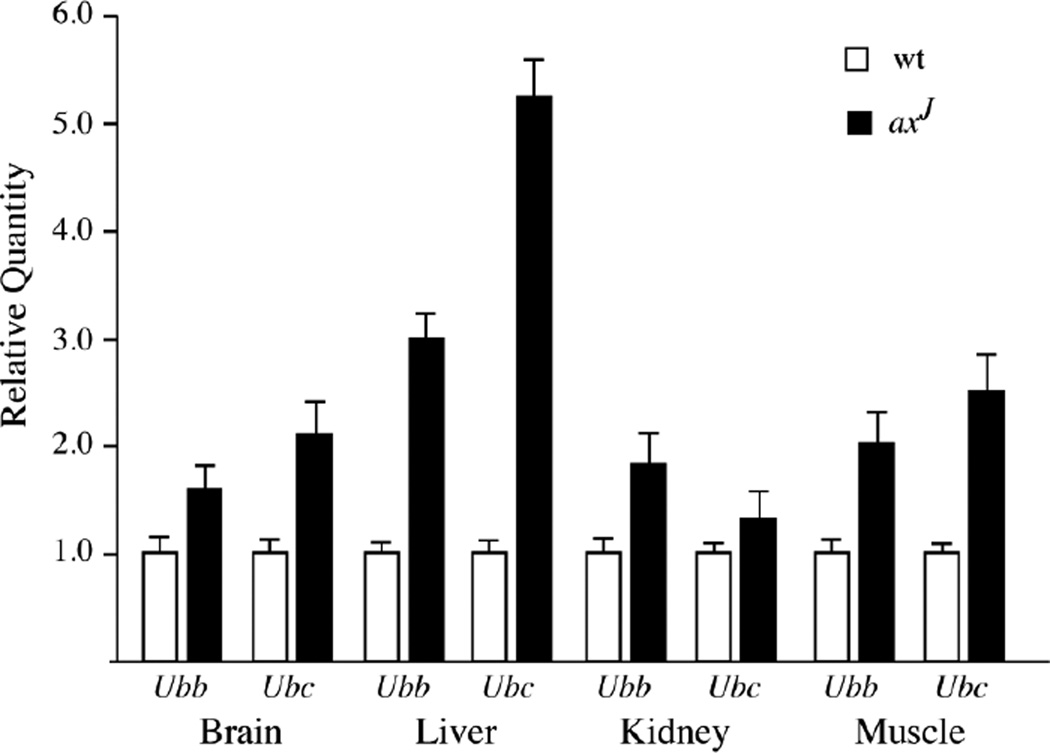
The ataxia (axJ) mice have been used as an animal model to investigate the in vivo effects of Usp14 deficiency on neuronal function [46]. In the axJ mice, there is a 40% loss of monomeric ubiquitin in all tissues, which results in a “ubiquitin-stress” response that is measured by 2 to 5-fold increases in Ubb and Ubc transcripts, indicating that USP14 plays an important role in stabilizing ubiquitin pools (Fig. 1). The greatest levels of ubiquitin loss are observed at synaptic terminals, demonstrating the importance of USP14 for ubiquitin stability at synapses [47]. These changes in free ubiquitin correlate with the accumulation of neurofilaments and nerve terminal swellings at the axJ neuromuscular junctions [47]. In addition to the structural changes, the axJ neuromuscular junctions are functionally compromised and exhibit giant mini-endplate potentials, reduced mini-endplate frequencies, reduced quantal content, and synaptic failures. These structural and functional deficits are corrected by neuronal-specific transgenic expression of Usp14 in the axJ mice, demonstrating that USP14 has a critical presynaptic function in neurons [48]. This function appears to be in maintaining ubiquitin stability, as transgenic expression of ubiquitin also corrected the synaptic transmission and structural deficits at the axJ neuromuscular junction [49]. Since no postsynaptic deficits have been observed in the axJ mice, USP14 may have a primary function at the presynaptic terminal. As the motor neuron terminals are located great distances from the site of ubiquitin synthesis, the local levels of ubiquitin may be highly dependent on USP14 to maintain the ubiquitin pools required by the ubiquitin proteasome system in the presynaptic compartment.
Figure 1.
Effect of USP14 deficiency on ubiquitin gene transcription in neuronal and non-neuronal tissues. Quantitative PCR was performed on reverse-transcribed RNA isolated from the livers, kidneys, muscles and brains of 6-week-old wild type (wt) and USP14-deficient axJ mice. Gapdh was used as control. Data are shown as mean ± SE, where n = 3 animals per genotype.
Ubiquitin over-expression leads to neurological abnormalities
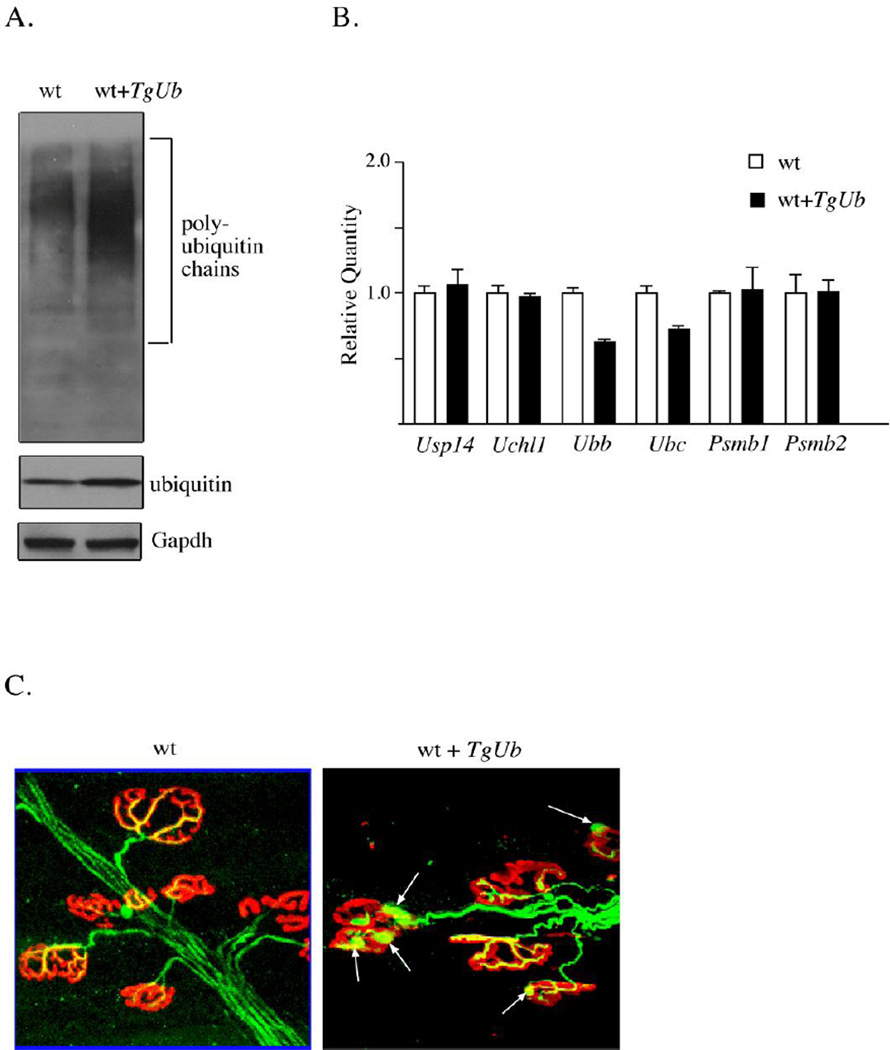
Although studies have investigated how loss of ubiquitin can contribute to neurological disease, there have not been any reports on the effect of ubiquitin overexpression on the nervous system. Research on the yeast orthologue of USP14, Ubp6, indicated that the expression of a deubiquitinating enzyme can be tightly coupled to ubiquitin levels [50]. In mice, ubiquitin-depletion by loss of UCHL1 did not affect the neuronal expression of Usp14, and loss of Usp14, did not affect the levels of UchL1. Transgenic mice that overexpress ubiquitin have also been developed to determine the interplay of ubiquitin and USP14 in the mammalian nervous system [49]. Overexpression of ubiquitin 2 to 3-fold in the nervous system did not alter the transcription of Usp14, showing that Usp14 expression is not controlled by ubiquitin levels in neurons (Fig. 2A and B). Interestingly, increasing neuronal ubiquitin levels results in a significant increase in the levels of conjugated ubiquitin, indicating that modulation of local ubiquitin levels can alter the landscape of ubiquitin protein conjugates and affects ubiquitin signaling. Consistent with a cellular attempt to maintain ubiquitin levels at a specific set point, expression of endogenous Ubb and Ubc significantly decreased in the ubiquitin-overexpressing mice. In contrast, the transcript levels of Psmb1, Psmb2, Uch37, Uchl3 and Uchl1 did not appear to be altered in the brains of mice overexpressing ubiquitin. While ubiquitin overexpression did not have broad effects on ubiquitin proteasome system gene expression, transgenic mice that overexpressed ubiquitin in neurons exhibited structural defects at the neuromuscular junction. These alterations included motor endplate swellings and decreased acetylcholine receptor arborization (Fig. 2B). Surprisingly, the effects of ubiquitin overexpression on the morphology of the neuromuscular junction were very similar to those seen in ubiquitin-deficient Usp14 and Uchl1-deficient mice [47]. In addition to the structural deficits, the effect of increased ubiquitin expression can also be seen functionally at the neuromuscular junction. Increasing free ubiquitin levels in Usp14-deficient mice stimulates mini-endplate frequency [49]. Given the far-reaching effects of ubiquitin in the cell, it is not clear if ubiquitin complementation corrects the initial mechanism of disease in axJ mice or simply masks it. However, these results suggest that ubiquitination is coupled to synaptic transmission, and the regulation of ubiquitin levels must therefore be tightly controlled to allow for proper synapse development and function.
Figure 2.
Effects of ubiquitin overexpression on the mouse nervous system. (A) Representative immunoblot of ubiquitin levels from the brains of 6-week-old wild type mice (wt) and wild type mice containing the Thy1-ubiquitin transgene (wt + TgUb). Gapdh was used as a loading control. (B) Quantitative PCR was performed on reversetranscribed RNA isolated from brains of 6-week-old wild type (wt) and wild type ubiquitin transgenic mice (wt+TgUb). Data are shown as mean ± SE, where n = 3 animals per genotype. (C) Tibialis anterior muscles were prepared from 6-week-old wild type mice (wt) and wild type mice containing the Thy1-ubiquitin transgene (wt + TgUb). Muscles were stained for neurofilaments (green) and acetylcholine receptors (red). Arrows indicate neurofilament accumulations.
Roles for ubiquitination at the synapse
Since the original description of the ubiquitin proteasome system by Ciechanover, Hershko, and Rose in the late 1970’s and early 1980’s, numerous studies have linked ubiquitin to neuronal function [51]. While the contribution of protein ubiquitination in regulating synaptic protein abundance is well documented, there are hints in the literature of additional roles for ubiquitin at the synapse, which may be independent of protein degradation. For example, in isolated synaptic terminals, calcium entry mediates a rapid decrease in ubiquitin conjugates that is reversed within seconds [52]. As the reversibility of this phenomenon suggests, the decrease in ubiquitinated proteins is not blocked by proteasome inhibition and therefore does not appear to result from increased protein turnover. Instead, it may result from calcium-mediated stimulation of deubiquitinating enzymes or, as the authors suggest, inhibition of on-going protein ubiquitination. Although the functional consequences of the decrease in ubiquitin conjugates are not known, it is certainly suggestive that the same stimulus that results in neurotransmitter release also decreases protein ubiquitination. It may be that ubiquitination, or lack thereof, transiently changes protein activity, and these changes favor release of neurotransmitter from the synapse.
A later study by Felix Schweizer’s group further implicates acute ubiquitination events in neurotransmitter release [53]. These researchers demonstrate that proteasome inhibition in cultured hippocampal neurons increases the frequency of miniature synaptic events (mEPSCs and mIPSCs) without changing their amplitude or kinetics, which is suggestive of a presynaptic change in the probability of neurotransmitter release. The increase is independent of calcium, which implicates a change in activity of the presynaptic release machinery downstream of calcium entry. Surprisingly, an increase in ubiquitinated proteins is not observed during this same time frame, arguing against simple stabilization of the release machinery or other pro-release proteins. Inhibition of the E1 activating enzyme, which should affect the global level of ubiquitinated proteins in the opposite direction of proteasome inhibition, yields a similar increase in miniature postsynaptic potentials [53]. The authors hypothesize that the increased neurotransmitter release is derived from altering the state of a group of dynamically ubiquitinated proteins. This change can be brought about by reducing protein ubiquitination or by inhibiting the proteasome and sequestering ubiquitin on proteins bound for degradation. This study, together with the work of Chen et al (2003), suggests that dynamic ubiquitination events may be important to presynaptic function, although the nature of these events and their substrates remain unknown.
Conclusions
Significant progress has been made in our understanding of how cells utilize ubiquitin to regulate cellular pathways. The structure and function of the nervous system is dependent on regulated protein turnover by the proteasome, and changes in protein stability are thought to contribute to many chronic neurological diseases. While loss of ubiquitin modifying enzymes can disrupt neuronal function by altering the stability of individual proteins, loss of ubiquitin homeostasis is also a mechanism that can lead to neuronal dysfunction. Surprisingly, neurons seem to have a restricted range in which ubiquitin levels must be maintained, as either a decrease or increase in ubiquitin expression can negatively impact neuronal structure and function. The highly compartmentalized organization of neurons is likely an important contributor to their selective sensitivity to changes in ubiquitin pools. Future work will likely uncover new targets of regulated protein degradation at synapses as well as identify additional roles for protein ubiquitination in regulating synaptic plasticity.
Acknowledgements
This research was supported by the Evelyn F. McKnight Brain Institute, NIH/NINDS Grants NS047533 and NS074456 (S.M.W).
References
- 1.Yi JJ, Ehlers MD. Emerging roles for ubiquitin and protein degradation in neuronal function. Pharmacol Rev. 2007;59:14–39. doi: 10.1124/pr.59.1.4. [DOI] [PubMed] [Google Scholar]
- 2.DiAntonio A, Haghighi AP, Portman SL, Lee JD, Amaranto AM, et al. Ubiquitination-dependent mechanisms regulate synaptic growth and function. Nature. 2001;412:449–452. doi: 10.1038/35086595. [DOI] [PubMed] [Google Scholar]
- 3.Ding M, Chao D, Wang G, Shen K. Spatial regulation of an E3 ubiquitin ligase directs selective synapse elimination. Science. 2007;317:947–951. doi: 10.1126/science.1145727. [DOI] [PubMed] [Google Scholar]
- 4.Burbea M, Dreier L, Dittman JS, Grunwald ME, Kaplan JM. Ubiquitin and AP180 regulate the abundance of GLR-1 glutamate receptors at postsynaptic elements in C-elegans. Neuron. 2002;35:107–120. doi: 10.1016/s0896-6273(02)00749-3. [DOI] [PubMed] [Google Scholar]
- 5.Colledge M, Snyder EM, Crozier RA, Soderling JA, Jin Y, et al. Ubiquitination regulates PSD-95 degradation and AMPA receptor surface expression. Neuron. 2003;40:595–607. doi: 10.1016/s0896-6273(03)00687-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bizzi A, Schaetzle B, Patton A, Gambetti P, Autilio-Gambetti L. Axonal transport of two major components of the ubiquitin system: free ubiquitin and ubiquitin carboxyl-terminal hydrolase PGP 9.5. Brain Res. 1991;548:292–299. doi: 10.1016/0006-8993(91)91135-n. [DOI] [PubMed] [Google Scholar]
- 7.Goldstein G, Scheid M, Hammerling U, Schlesinger DH, Niall HD, et al. Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells. Proc Natl Acad Sci U S A. 1975;72:11–15. doi: 10.1073/pnas.72.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilkinson KD, Urban MK, Haas AL. Ubiquitin is the ATP-dependent proteolysis factor I of rabbit reticulocytes. J Biol Chem. 1980;255:7529–7532. [PubMed] [Google Scholar]
- 9.Hershko A, Eytan E, Ciechanover A, Haas AL. Immunochemical analysis of the turnover of ubiquitin-protein conjugates in intact cells. Relationship to the breakdown of abnormal proteins. J Biol Chem. 1982;257:13964–13970. [PubMed] [Google Scholar]
- 10.Ciehanover A, Hod Y, Hershko A. A heat-stable polypeptide component of an ATP-dependent proteolytic system from reticulocytes. Biochem Biophys Res Commun. 1978;81:1100–1105. doi: 10.1016/0006-291x(78)91249-4. [DOI] [PubMed] [Google Scholar]
- 11.Kaiser SE, Riley BE, Shaler TA, Trevino RS, Becker CH, et al. Protein standard absolute quantification (PSAQ) method for the measurement of cellular ubiquitin pools. Nat Methods. 2011;8:691–696. doi: 10.1038/nmeth.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fornace AJ, Jr, Alamo I, Jr, Hollander MC, Lamoreaux E. Ubiquitin mRNA is a major stress-induced transcript in mammalian cells. Nucleic Acids Res. 1989;17:1215–1230. doi: 10.1093/nar/17.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bond U, Schlesinger MJ. The chicken ubiquitin gene contains a heat shock promoter and expresses an unstable mRNA in heat-shocked cells. Mol Cell Biol. 1986;6:4602–4610. doi: 10.1128/mcb.6.12.4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilkinson KD, Tashayev VL, O'Connor LB, Larsen CN, Kasperek E, et al. Metabolism of the polyubiquitin degradation signal: structure, mechanism, and role of isopeptidase T. Biochemistry. 1995;34:14535–14546. doi: 10.1021/bi00044a032. [DOI] [PubMed] [Google Scholar]
- 15.Ryu KY, Garza JC, Lu XY, Barsh GS, Kopito RR. Hypothalamic neurodegeneration and adult-onset obesity in mice lacking the Ubb polyubiquitin gene. Proc Natl Acad Sci U S A. 2008;105:4016–4021. doi: 10.1073/pnas.0800096105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryu KY, Maehr R, Gilchrist CA, Long MA, Bouley DM, et al. The mouse polyubiquitin gene UbC is essential for fetal liver development, cell-cycle progression and stress tolerance. Embo Journal. 2007;26:2693–2706. doi: 10.1038/sj.emboj.7601722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryu KY, Sinnar SA, Reinholdt LG, Vaccari S, Hall S, et al. The mouse polyubiquitin gene Ubb is essential for meiotic progression. Mol Cell Biol. 2008;28:1136–1146. doi: 10.1128/MCB.01566-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ryu KY, Park H, Rossi DJ, Weissman IL, Kopito RR. Perturbation of the hematopoietic system during embryonic liver development due to disruption of polyubiquitin gene Ubc in mice. PLoS One. 2012;7:e32956. doi: 10.1371/journal.pone.0032956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haas AL, Warms JV, Hershko A, Rose IA. Ubiquitin-activating enzyme. Mechanism and role in protein-ubiquitin conjugation. J Biol Chem. 1982;257:2543–2548. [PubMed] [Google Scholar]
- 20.Hershko A, Heller H, Elias S, Ciechanover A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J Biol Chem. 1983;258:8206–8214. [PubMed] [Google Scholar]
- 21.Pickart CM, Rose IA. Functional heterogeneity of ubiquitin carrier proteins. J Biol Chem. 1985;260:1573–1581. [PubMed] [Google Scholar]
- 22.Eletr ZM, Huang DT, Duda DM, Schulman BA, Kuhlman B. E2 conjugating enzymes must disengage from their E1 enzymes before E3-dependent ubiquitin and ubiquitin-like transfer. Nature Structural & Molecular Biology. 2005;12:933–934. doi: 10.1038/nsmb984. [DOI] [PubMed] [Google Scholar]
- 23.Clague MJ, Liu H, Urbe S. Governance of endocytic trafficking and signaling by reversible ubiquitylation. Dev Cell. 2012;23:457–467. doi: 10.1016/j.devcel.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 24.Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, et al. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 25.Finley D, Sadis S, Monia BP, Boucher P, Ecker DJ, et al. Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Mol Cell Biol. 1994;14:5501–5509. doi: 10.1128/mcb.14.8.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. Embo J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abbott DW, Yang Y, Hutti JE, Madhavarapu S, Kelliher MA, et al. Coordinated regulation of Toll-like receptor and NOD2 signaling by K63-linked polyubiquitin chains. Mol Cell Biol. 2007;27:6012–6025. doi: 10.1128/MCB.00270-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan NL, Hill CP. Defining polyubiquitin chain topology. Nat Struct Biol. 2001;8:650–652. doi: 10.1038/90337. [DOI] [PubMed] [Google Scholar]
- 29.Reggiori F, Pelham HRB. Sorting of proteins into multivesicular bodies: ubiquitin-dependent and -independent targeting. Embo Journal. 2001;20:5176–5186. doi: 10.1093/emboj/20.18.5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sigismund S, Woelk T, Puri C, Maspero E, Tacchetti C, et al. Clathrin-independent endocytosis of ubiquitinated cargos. Proc Natl Acad Sci U S A. 2005;102:2760–2765. doi: 10.1073/pnas.0409817102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kravtsova-Ivantsiv Y, Ciechanover A. Non-canonical ubiquitin-based signals for proteasomal degradation. J Cell Sci. 2012;125:539–548. doi: 10.1242/jcs.093567. [DOI] [PubMed] [Google Scholar]
- 32.Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 33.Upadhya SC, Hegde AN. Role of the ubiquitin proteasome system in Alzheimer's disease. BMC Biochem. 2007;8(Suppl 1):S12. doi: 10.1186/1471-2091-8-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dawson TM, Dawson VL. Molecular pathways of neurodegeneration in Parkinson's disease. Science. 2003;302:819–822. doi: 10.1126/science.1087753. [DOI] [PubMed] [Google Scholar]
- 35.Osaka H, Wang YL, Takada K, Takizawa S, Setsuie R, et al. Ubiquitin carboxy-terminal hydrolase L1 binds to and stabilizes monoubiquitin in neuron. Hum Mol Genet. 2003;12:1945–1958. doi: 10.1093/hmg/ddg211. [DOI] [PubMed] [Google Scholar]
- 36.Anderson C, Crimmins S, Wilson JA, Korbel GA, Ploegh HL, et al. Loss of Usp14 results in reduced levels of ubiquitin in ataxia mice. J Neurochem. 2005;95:724–731. doi: 10.1111/j.1471-4159.2005.03409.x. [DOI] [PubMed] [Google Scholar]
- 37.Shabek N, Ciechanover A. Degradation of ubiquitin: the fate of the cellular reaper. Cell Cycle. 2010;9:523–530. doi: 10.4161/cc.9.3.11152. [DOI] [PubMed] [Google Scholar]
- 38.Leroy E, Boyer R, Auburger G, Leube B, Ulm G, et al. The ubiquitin pathway in Parkinson's disease. Nature. 1998;395:451–452. doi: 10.1038/26652. [DOI] [PubMed] [Google Scholar]
- 39.Saigoh K, Wang YL, Suh JG, Yamanishi T, Sakai Y, et al. Intragenic deletion in the gene encoding ubiquitin carboxy-terminal hydrolase in gad mice. Nat Genet. 1999;23:47–51. doi: 10.1038/12647. [DOI] [PubMed] [Google Scholar]
- 40.Ehlers MD. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat Neurosci. 2003;6:231–242. doi: 10.1038/nn1013. [DOI] [PubMed] [Google Scholar]
- 41.Sakurai M, Sekiguchi M, Zushida K, Yamada K, Nagamine S, et al. Reduction in memory in passive avoidance learning, exploratory behaviour and synaptic plasticity in mice with a spontaneous deletion in the ubiquitin C-terminal hydrolase L1 gene. Eur J Neurosci. 2008;27:691–701. doi: 10.1111/j.1460-9568.2008.06047.x. [DOI] [PubMed] [Google Scholar]
- 42.Cartier AE, Djakovic SN, Salehi A, Wilson SM, Masliah E, et al. Regulation of synaptic structure by ubiquitin C-terminal hydrolase L1. J Neurosci. 2009;29:7857–7868. doi: 10.1523/JNEUROSCI.1817-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Borodovsky A, Kessler BM, Casagrande R, Overkleeft HS, Wilkinson KD, et al. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. Embo J. 2001;20:5187–5196. doi: 10.1093/emboj/20.18.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu M, Li P, Song L, Jeffrey PD, Chenova TA, et al. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. Embo J. 2005;24:3747–3756. doi: 10.1038/sj.emboj.7600832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson SM, Bhattacharyya B, Rachel RA, Coppola V, Tessarollo L, et al. Synaptic defects in ataxia mice result from a mutation in Usp14, encoding a ubiquitin-specific protease. Nat Genet. 2002;32:420–425. doi: 10.1038/ng1006. [DOI] [PubMed] [Google Scholar]
- 47.Chen PC, Qin LN, Li XM, Walters BJ, Wilson JA, et al. The proteasome-associated deubiquitinating enzyme Usp14 is essential for the maintenance of synaptic ubiquitin levels and the development of neuromuscular junctions. J Neurosci. 2009;29:10909–10919. doi: 10.1523/JNEUROSCI.2635-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crimmins S, Jin Y, Wheeler C, Huffman AK, Chapman C, et al. Transgenic rescue of ataxia mice with neuronal-specific expression of ubiquitin-specific protease 14. J Neurosci. 2006;26:11423–11431. doi: 10.1523/JNEUROSCI.3600-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen PC, Bhattacharyya BJ, Hanna J, Minkel H, Wilson JA, et al. Ubiquitin homeostasis is critical for synaptic development and function. J Neurosci. 2011;31:17505–17513. doi: 10.1523/JNEUROSCI.2922-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hanna J, Meides A, Zhang DP, Finley D. A ubiquitin stress response induces altered proteasome composition. Cell. 2007;129:747–759. doi: 10.1016/j.cell.2007.03.042. [DOI] [PubMed] [Google Scholar]
- 51.Bingol B, Sheng M. Deconstruction for reconstruction: the role of proteolysis in neural plasticity and disease. Neuron. 2011;69:22–32. doi: 10.1016/j.neuron.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 52.Chen H, Polo S, Di Fiore PP, De Camilli PV. Rapid Ca2+-dependent decrease of protein ubiquitination at synapses. Proc Natl Acad Sci U S A. 2003;100:14908–14913. doi: 10.1073/pnas.2136625100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rinetti GV, Schweizer FE. Ubiquitination Acutely Regulates Presynaptic Neurotransmitter Release in Mammalian Neurons. Journal of Neuroscience. 2010;30:3157–3166. doi: 10.1523/JNEUROSCI.3712-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]