Abstract
β-Lactamases produced by pathogenic bacteria cleave β-lactam antibiotics and render them ineffective. Understanding the principles that govern the structural stability of β-lactamases requires elucidation of the nature of the interactions that are involved in stabilization. In the present study, we systematically analyze the influence of CH...O interactions on determining the specificity and stability of β-lactamases in relation to environmental preferences. It is interesting to note that all the residues located in the active site of β-lactamases are involved in CH...O interactions. A significant percentage of CH...O interactions have a higher conservation score and short-range interactions are the predominant type of interactions in β-lactamases. These results will be useful in understanding the stability patterns of β-lactamases.
Keywords: β-lactamases, CH...O interactions, Stabilization centers, Conservation, Solvent accessibility, Secondary structure
Introduction
The evolution and spread of β-lactamase against β-lactam antibiotics cause a serious threat to antimicrobial treatment [1]. Bacteria may become resistant to the antibiotic by genetic mutation or by acquiring resistant genes from other bacteria. The genes encoding proteins, which counteract the effects of antibiotics, may be found on bacterial chromosomes or plasmids [2]. Selective pressure created by the use of antibacterial agents increases the emergence of resistant strains [3]. Therefore, a detailed study of β-lactamases is essential for the design of novel lactamase inhibitors.
The hydrogen bond plays an important role in the structure and function of proteins and occurs when a proton is shared by two electronegative atoms [4, 5]. Non-canonical interactions, such as C-H⋯O interactions, have gained much attention, with a number of studies emphasizing the importance of these unconventional hydrogen bonds in stabilizing the structure of proteins [6]. The importance of C H⋯O interactions in biological structures is also evident from the early analysis of C H⋯O interactions in glycoproteins [7], RNA binding proteins [8], protein–DNA recognition [9], and carbohydrate–lectin interactions [10]. In 1996, Bella and Berman identified the importance of C H⋯O interactions in a collagen triplex helix [11].
Mutational analysis of protein-ligand complexes also reveals the importance of C-H...O interactions [12–14]. Other studies demonstrating the role played by C-H⋯O interactions in determining molecular packing and conformation [15], RNA structures [16], minor groove of A-tracts in DNA double helices [17], and organic crystal structures [18] also reveal the importance of the C-H...O hydrogen bond. Furthermore, C-H...O hydrogen bonds also play a key role in structural implications and supramolecular design [19]. The C-H...O hydrogen bond is the second most important group to be widely distributed among protein structures, constituting 20–25 % of total hydrogen bonds [20–22]. However, there are no reports on the role of C-H...O interactions in β-lactamases and we assume ours is the first such report.
Materials and methods
We selected a non-redundant data set of 95 β-lactamases from the Protein Data Bank (PDB) [23] for our analysis. The criteria used for the selection of the non-redundant data set were as follows: (i) the structures were solved with <2.5 Å resolution, (ii) the sequence identity among the proteins in the data set was less than 40 %. If the protein contains more than one chain, then chain A was considered for analysis. The PDB codes of the β-lactamases investigated in the present study are as follows:
1BC2_A,1BMC_A, 1BSG_A, 1DD6_A, 1DDK_A, 1DY6_A, 1E25_A, 1FOF_B, 1FR6_A, 1GCE_A, 1GHI_A, 1H8Z_A, 1HTZ_A, 1HZO_A, 1I2S_A, 1IYS_A, 1JJE_A, 1JTD_A, 1JTG_A, 1K38_A, 1K4F_A, 1L9Y_A, 1M2X_A, 1M6K_A, 1MBL_A, 1N4O_ A, 1O7E_A, 1PIO_A, 1Q2Q_A, 1RGY_A, 1S0W_A, 1S6R_A, 1SHV_A, 1W7F_A, 1WUP_A, 1Y54_A, 1ZC2_A, 1ZG4_A, 1ZKP_B, 2A3U_A, 2A49_A, 2BG8_A, 2CC1_ A, 2DKF_A, 2FHX_A, 2FU6_A, 2G2W_A, 2GDN_A, 2GMN_A, 2P4Z_A, 2QZ6_A, 2V20_A, 2WGI_A, 2WRS_A, 2XR0_A, 2Y87_A, 2YZ3_A, 2ZJ9_A, 2ZO4_A, 3BFD_A, 3BYD_A, 3C5A_A, 3C7V_A, 3CJM_A, 3DTM_A, 3DW0_A, 3GMV_X, 3GMW_ A, 3GSG_A, 3H3E_A, 3IF6_A, 3IWI_A, 3L6N_A, 3LVZ_A, 3LY3_A, 3M2K_A, 3OPL_A, 3OZH_A, 3P09_A, 3P98_A, 3PAE_A, 3PG4_A, 3PY6_A, 3Q6X_A, 3QHY_ A, 3QI0_A, 3QNC_A, 3RKK_A, 3S0Z_A, 3SIY_A, 3SH8_A, 3SPU_A, 3ZR9_A, 4BLM_A, 4ZNB_A.
C H⋯O interactions
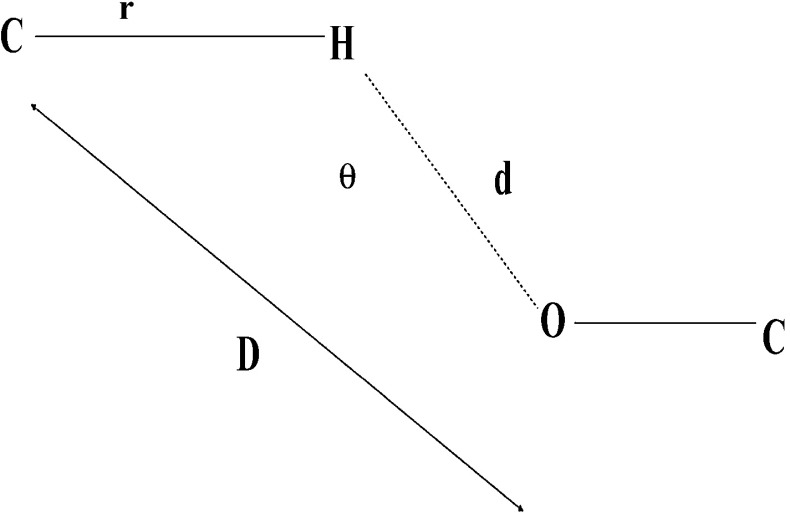
C H⋯O interactions frequently occur in protein structures and play a central role in the stability of proteins [24]. The C H⋯O interactions in the data set were identified using the Hydrogen Bond Analysis Tool (HBAT) program, which requires structural information in PDB format [25]. The standard H-bonding criteria for C–H⋯O interactions are d (H⋯O) ≤3.0 Å and θ (C–H⋯O) ≥90°; where d is the distance between the H atom and the O atom; θ is the angle between the C–H bond and the center of the acceptor atom. These parameters are represented in Fig. 1 [26]. The C-H...O interactions considered here were between the entire possible donor (CαH, Cali − H and Caro-H) and the oxygen atoms in the proteins were of hydroxyl, carbonyl, and carboxyl type. The C H⋯O interactions are represented by a two-letter code in which the first letter indicates the donor atom and the second letter indicates the acceptor [8]. C-H...O interactions were classified into four types, namely, main-chain to main-chain C-H...O interactions (MM-C-H...O), main-chain to side-chain C-H...O interactions (MS-CH...O), side-chain to main-chain C-H...O interactions (SM-C-H...O) and side-chain to side-chain C-H...O interactions (SS-C-H...O) [7].
Fig. 1.
Parameters of C-H⋯O interacting pairs in HBAT [26]. Parameters are r, distance between C and H atom; d, distance between the H atom and the O atom; D, distance between C and O atom; θ, defined as the angle between the C–H bond and the center of the acceptor atom
Secondary structure preferences
As the name implies, secondary structure constitutes the second level of the protein structure and is an important determinant of protein structure and function [27]. In order to understand the occurrence of C-H...O interaction forming residues in different secondary structures, we performed a systematic investigation based on the information available in PDB [23] and by using letters we denoted H for helix, T for turn and S for strand [28].
Computation of solvent accessibility
Interactions with a surrounding aqueous environment are important factors for the structure and dynamic properties of biological macromolecules. An important element in the elucidation of such interactions is the analysis of the solvent-accessible surface area. The solvent accessibility pattern of residues involved in C-H...O interactions was analyzed by using the ASA-View program [29]. These residues were classified into buried, partially buried, and exposed, indicating minimal, moderate, and high accessibility of the amino acid residues to the solvent [28].
Sequential separation
The composition of the surrounding residues associated with the given residue was calculated for a sphere of radius 8 Å [30]. The contribution from < ±4 were treated as short-range contacts, ±4 to ±10 as medium-range contacts and > ±10 were treated as long-range contacts. The definition of short, medium, and long range in amino acid residues was based on their respective locations in the sequence. This classification allows us to evaluate the contribution of short-range, medium-range, and long-range contacts in the formation of C-H...O interactions [31].
Stabilization centers
Identification of the residues, which plays a key role in the stabilization of proteins, leads to a better understanding of the mechanism of stabilization in β-lactamases. We used the SCide server [32] for computing the stabilization centers in β-lactamases. These are residues involved in long-range contacts and play an important role in maintaining the flexibility and stability of a protein.
Conservation score
We computed the conservation score of C-H⋯O interacting residues using the ConSurf program [33, 34], which provides evolutionary conservation profiles for proteins of known structures in the PDB. The evolutionary conservation of each amino acid position in the alignment was calculated using the Rate4Site algorithm, which assigns a conservation level for each residue using an empirical Bayesian inference [35]. The conservation scores were divided into distinct scales of nine grades; residues with a score of 1 were considered highly variable and residues with a score of 9 were considered highly conserved. A conservation score of 6 was the cut-off value used to identify the stabilizing residues [36].
C-H⋯O interacting residues in the binding site of β-lactamases
The importance of C-H⋯O interacting residues in the binding site of β-lactamases was analyzed using the Ligplot program, which generates 2D schematic diagrams of protein–ligand interactions from the 3D coordinates of a given PDB file in order to generate diagrams of binding sites [37].
Results
C-H⋯O interactions
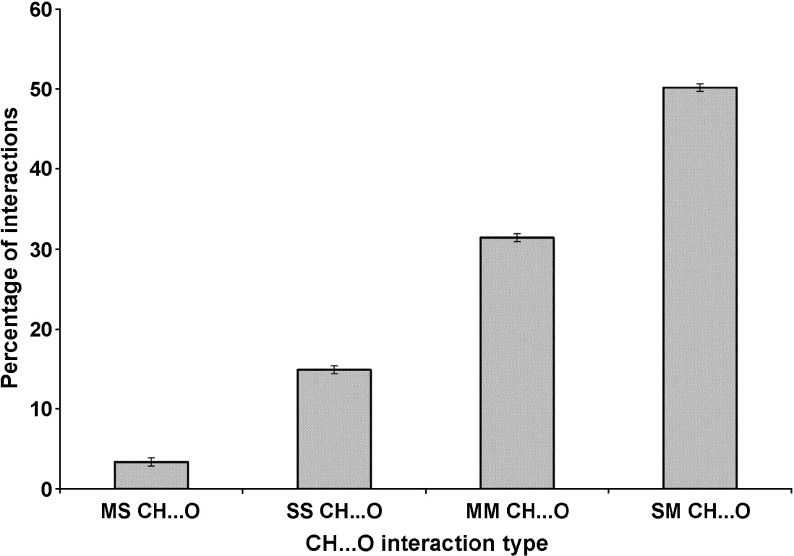
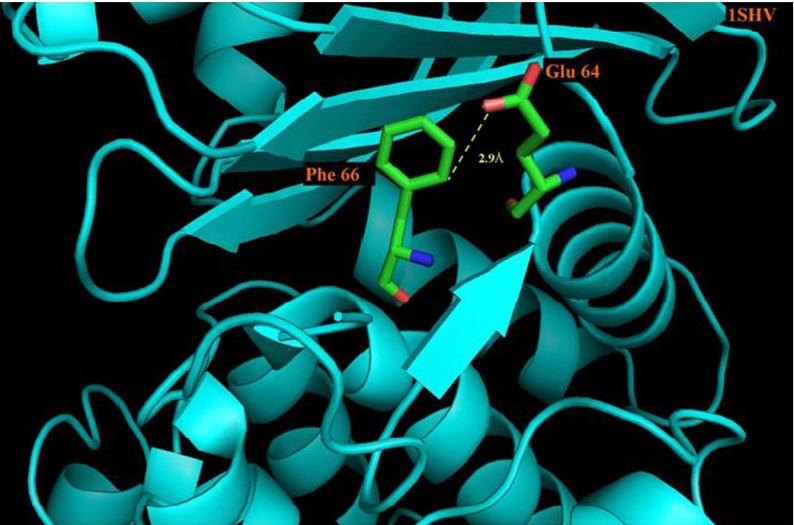
The C-H⋯O interaction types in the β-lactamases studies are depicted in Fig. 2. We find that 50.2 % are SM C-H⋯O interactions, 31.4 % are MM C-H⋯O interactions, 14.9 % are SS C-H⋯O interactions and the remaining 3.4 % are MS C-H⋯O interactions. An illustrative picture of C-H⋯O interactions between Glu 64 and Phe 66 of PDB Code 1SHV is shown in Fig. 3. The major contribution to C-H⋯O interactions is mainly from SM C-H⋯O interactions. The contributions of SS C-H⋯O interactions, MM C-H⋯O interactions and MS C-H⋯O interactions are minimal. From these observations, we consider that the contribution of SM C-H⋯O interactions is significant and hence may play an important role in the structure and stability of β-lactamases.
Fig. 2.
C-H⋯O interaction types in β-lactamases
Fig. 3.
PyMOL view of Glu 64 - Phe 66 C-H⋯O interacting pairs in PDB ID-1SHV
Secondary structure preferences
We calculated the occurrence of C-H⋯O interacting residues in different secondary structures in our data set and the results are presented in Table 1. Secondary structure preference of C-H⋯O interacting residues shows that Ala, Asn, Asp, and Glu prefer to be helix, while Gly, Val, and Tyr were found to prefer strand conformation. It is interesting to note that a significant percentage of Met residues favored helix and strand conformation. From these observations, we infer that Met residues might stabilize helices and strands by C-H⋯O bonding in β-lactamases.
Table 1.
Frequency occurrence of C-H⋯O interaction forming residues in different secondary structures
| Residues | Helix (%) | Strand (%) | Turn (%) |
|---|---|---|---|
| A | 53.7 | 33.5 | 7.8 |
| R | 38 | 41.5 | 21.3 |
| N | 29.6 | 29.6 | 27.2 |
| D | 37.8 | 30.5 | 31.7 |
| C | 44.8 | 37.5 | 17.5 |
| Q | 51.1 | 40.8 | 20.2 |
| E | 49.3 | 30.1 | 15.5 |
| G | 21.5 | 47.4 | 30.9 |
| H | 38.5 | 38.5 | 22.9 |
| L | 52.9 | 33.7 | 13.2 |
| I | 37.6 | 50.5 | 12 |
| K | 48 | 36.9 | 14.9 |
| M | 48.5 | 45.7 | 5.7 |
| F | 34 | 55.2 | 10.6 |
| P | 33.1 | 33.8 | 33.1 |
| S | 44.1 | 38 | 16.4 |
| T | 38.3 | 47.6 | 13.8 |
| W | 32 | 50.9 | 17 |
| V | 29.5 | 60.9 | 10 |
| Y | 29.1 | 53.2 | 17.5 |
Computations of solvent accessibility pattern
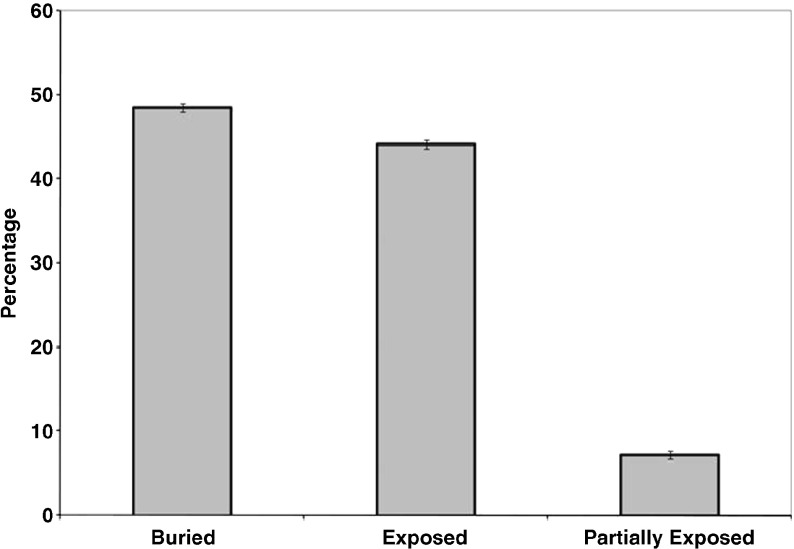
We carried out a systematic analysis of the solvent accessibility patterns of the C-H⋯O interacting residues and the results are shown in Fig. 4. The solvent accessibility of amino acid residues was categorized into buried, partially buried, and exposed, for different ranges of solvent accessibility values: <10, 10–15, >15, respectively. From our analysis we find that 48.5 % of C-H⋯O interacting residues preferred to be in a buried region, 44.1 % of residues in an exposed region and the remaining in a partially buried region. Hence, C-H⋯O interactions are important in stabilizing core regions in β-lactamases.
Fig. 4.
Solvent accessibility pattern of C-H⋯O interacting residues in β-lactamases
Sequential separation between the residues that form C-H⋯O interactions
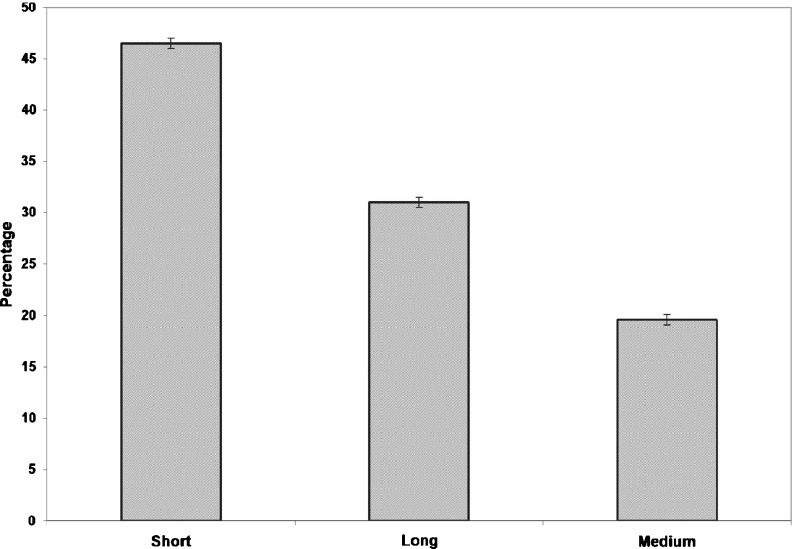
The contribution of C-H⋯O interactions in the local or global stability of β-lactamases is evaluated and the results are shown in Fig. 5. Short-range interactions are the predominant type of interaction in the β-lactamases. These results indicate that C-H⋯O interactions might significantly contribute to the local conformation stability of secondary structural elements in β-lactamases.
Fig. 5.
Sequential separation of C-H⋯O interacting residues in β-lactamases
Stabilization centers
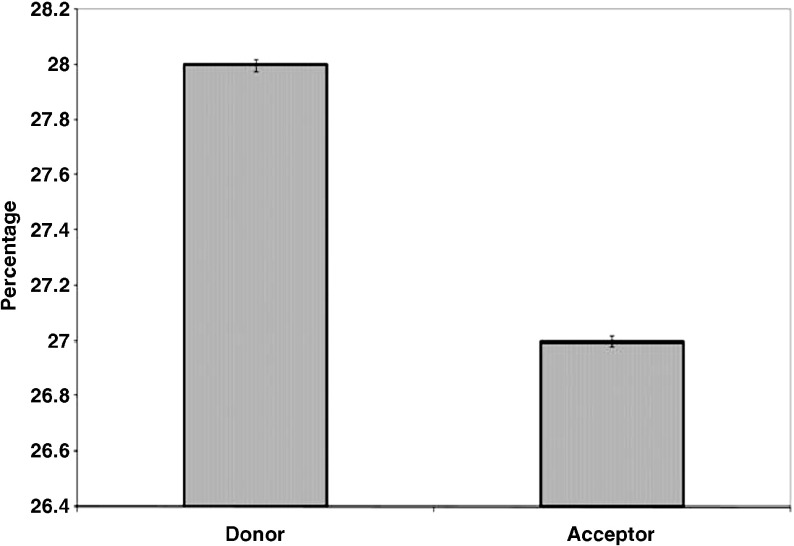
Stabilization centers in β-lactamases are computed and the results are presented in Fig. 6. We find that 28 % of the amino acid residues that contribute donor atoms and 27 % of the amino acid residues that contribute acceptor atoms to C-H⋯O interactions had one or more stabilization centers. From our analysis, we infer that these residues might contribute additional stability to the β-lactamases in addition to their participation in C-H⋯O interactions.
Fig. 6.
Stabilization centers in β-lactamases
Conservation score
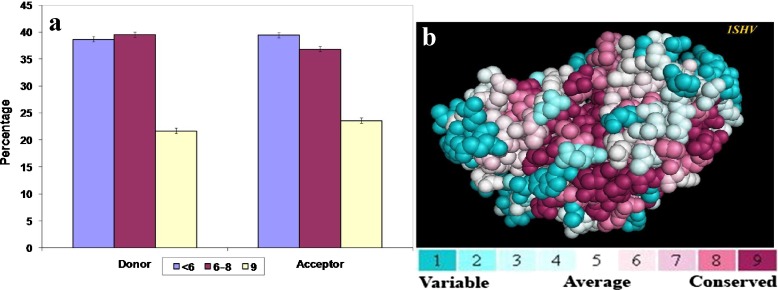
The conservation scores of the amino acid residues involved in C-H⋯O interactions are computed and the results are shown in Fig. 7a. Considering donor residues we find that 21.7 % of the residues have the highest conservation score of 9, while 39.6 % of the residues are in the range of 6–8. In the case of acceptor residues we find that 23.6 % of residues have the highest conservation score of 9 and 36.9 % of the residues are in the range of 6–8. Thus 61.3 % of donor residues and 60.5 % of acceptor residues have a higher conservation score of 6 and above (cut off score for conserved residues). From these observations, we infer that a considerable number of the amino acid residues involved in C-H⋯O interactions are evolutionarily conserved in β-lactamases. As a representative picture, the amino acids of the PDB ID -1SHV colored by their conservation grades are shown in Fig. 7b.
Fig. 7.
a Conservation patterns of C-H⋯O interacting residues in β-lactamases and b PyMOL view of the PDB ID-1SHV colored by their conservation grades using the color-coding bar, with turquoise-through-maroon indicating variable-conserved regions
C-H⋯O-interacting residues in the binding site of protein
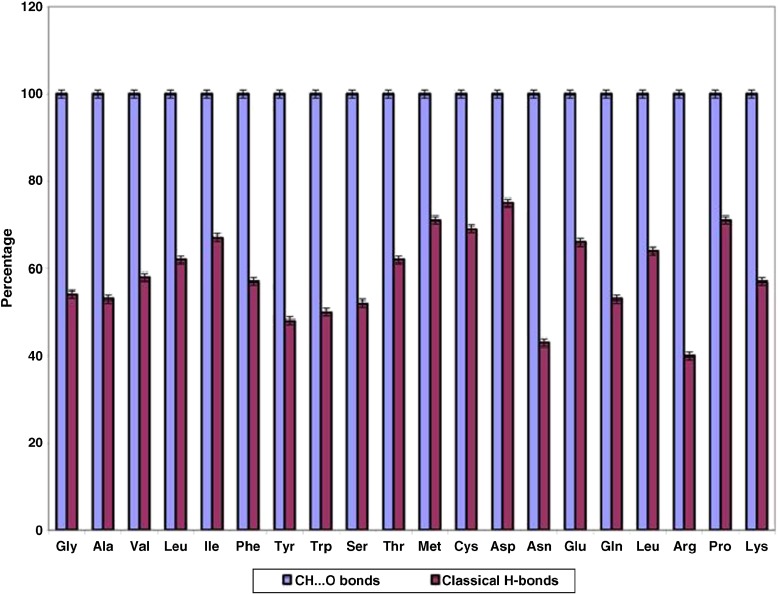
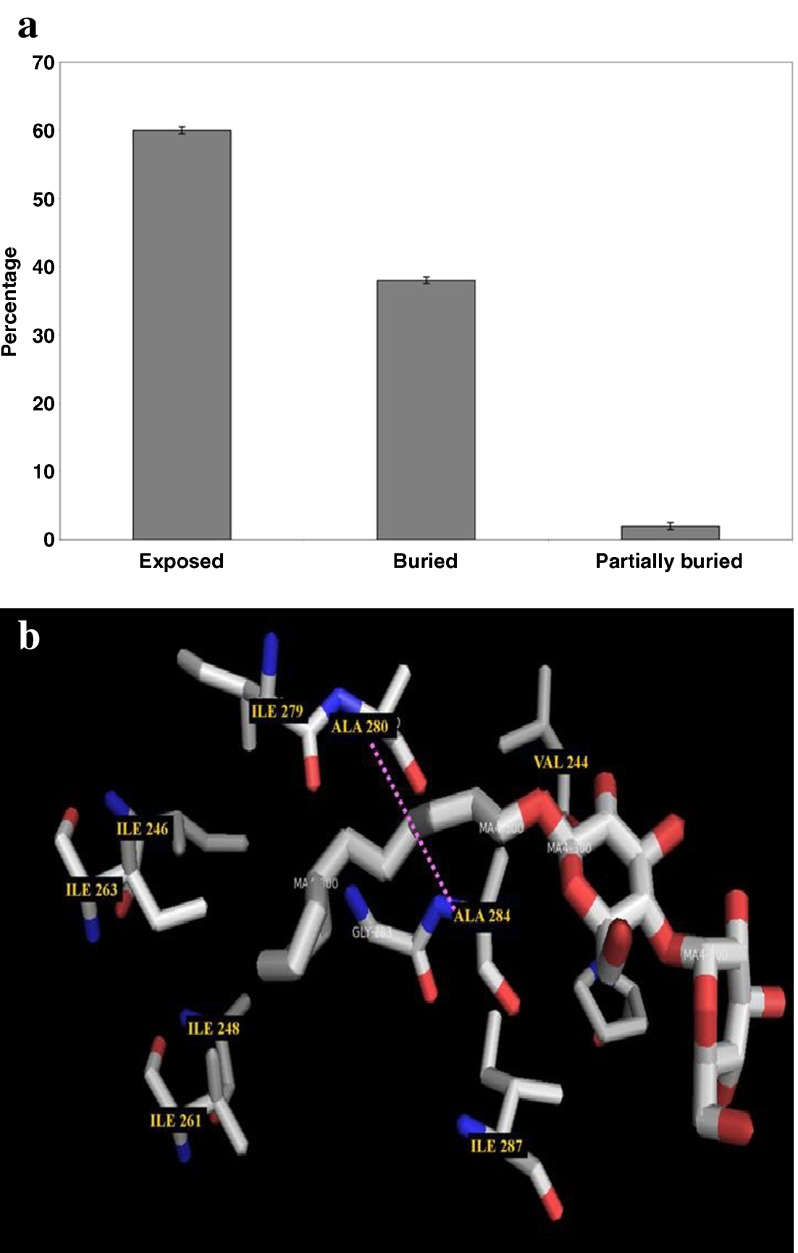
Binding site information is only available for 65 β-lactamases and the results are shown in Table 2. It is interesting to note that all the residues located in the binding pockets of β-lactamases are involved in C-H⋯O interactions, while in the case of classical H-bonds it is found that only 60 % of residues located in the binding sites of β-lactamases are involved; the results are shown in Fig. 8. After further analyzing the solvent accessibility pattern of active site residues, it is interesting to note that the majority of the active site residues are located in the exposed region of the proteins, which is shown in Fig. 9a. Hence, we conclude that C-H⋯O interacting residues play an important role in the functional specificity of β-lactamases. A PyMOL view of C-H⋯O interacting residues in the binding pockets of PDB ID-1SHV was obtained using the LigPlot+ program [38], which is shown in Fig. 9b.
Table 2.
C-H⋯O interacting residues in the active site of β-lactamases
| PDB ID | Active site residues |
|---|---|
| 3P09-A | Asp 192 Lys 193 Lys 176 Tyr 39 Asn 48 Lys 50 Leu 137 Gly 136 Gln 138 Gly 228 Thr 229 Ser 61 Ser 122 Thr 227 Asn 150 Lys 35 |
| 3GSG-A | Leu 157 Leu 161 Trp 93 Lys 164 Gly 81 Ala 79 Val 308 Arg 309 Ala 307 Ser 64 Gly 317 Thr 316 Ala 318 Tyr 221 Asn 152 |
| 1 W7F-A | Thr 235 Thr 216 Ser 130 Lys 73 Tyr 105 Ser 70 Ala 237 Ala 69 Gly 236 |
| 1I2S-A | Ser 130 Ser 70 Thr 235 Arg 244 Gly 236 Ala 237 Thr 216 Thr 274 |
| 3S1Y-A | Asn 373 Tyr 177 Ala 319 Gly 344 Gly 89 Thr 346 Val 239 Pro 241 Gly 242 Tyr 249 Gln 146 Lys 342 Thr 343 Thr 316 Leu 320 Ala 319 Gln 321 Pro 322 Phe 147 |
| 1FR6-A | Thr 316 Tyr 150 Gln 120 Leu 119 Asp 123 Pro 122 Asp 124 Tyr 221 Asn 152 Thr 319 Gly 63 Gly 317 Lys 315 |
| 1JTG-A | Arg 144 Asp 135 Asp 133 |
| 1JTD-A | Glu 37 |
| 4BLM-A | Ser 70 Ser 130 Tyr 105 |
| 3BFD-A | Ser 130 Ser 70 Trp 105 Ala 237 |
| 1SHV-A | Val 224 Ala 280 Ile 279 Val 261 Gly 283 Ile 263 Ile 246 Ala 248 Ile 287 Ala 284 Pro 226 |
| 3L6N-A | Thr 80 Lys 23 Lys 83 His 228 Val 229 Ile 216 Gly 226 Gly 227 His 98 His 96 His 159 Cys 178 His 220 Asp 100 |
| 1ZKP-B | Ala 237 Ser 130 Ser 70 Ser 235 Arg 244 Met 69 Tyr 105 Val 216 Gly 236 |
| 3PY6-A | Arg 222 Leu 225 |
| 3OPL-A | Gln 277 Ile 246 Ala 280 Ala 284 Gly 281 Ile 231 Gly 283 Ala 248 Leu 250 Val 224 Ile 279 Arg 244 Tyr 105 Gly 236 Ser 70 Ser 130 Val 216 Lys 234 Thr 235 |
| 3RKK-A | His 189 Lys 211 His 250 Gly 219 Ser 251 His 250 Asp 212 Asp 212 Lys 211 Arg264 Arg 234 Gln 44 Gln 107 Phe 46 Gln 53 Leu 54 Ala 55 Gly 200 Thr 201 Pro 56 Asn 57 Ser 249 His 250 Asp 124 Lys 125 His 120 Cys 208 |
| 3BYD-A | Thr 238 Ser 73 Ser 133 Gly 239 Ala 240 Trp 232 His 257 Glu 255 Ser 231 Asn 256 Pro 254 Gly 221 Lys 180 Arg 68 Arg 181 |
| 1GHI-A | Ser 70 Ser 130 Ser 235 |
| 3SH8-A | Asn 104 Asn 132 Ser 70 Glu 168 Ala 69 Ser 130 Gly 236 Thr 235 Arg 244 Ala 237 |
| 1K4F-A | Arg 160 Thr 206 Ser 115 Ser 67 Arg 250 Phe 208 Gly 207 |
| 3CJM-A | Ser 108 Ala 107 Lys 111 Lys 174 Ile 110 Gln 193 Glu 190 Arg 189 Lys 77 Val 76 Gln 193 Glu 246 Gln 251 Thr 247 Pro 248 Glu 243 Asn 288 Ala 292 Glu 243 Ile 258 Phe 241 |
| 1K38-A | Arg 244 Ser 115 Thr 206 Gly 207 Trp 208 |
| 3M2K-A | Ser 70 Asn 132 Lys 73 Ser 130 Ala 238 Gly 236 Ala 237 Thr 235 |
| 2FU6-A | Thr 181 Arg 172 Ile 180 Lys 297 Asn 210 Arg 252 Asp 152 Arg 148 Arg 130 Thr 181 Arg 172 Ile 180 Arg 110 |
| 3QI0-A | Ala 99 Pro 98 Ala 60 Gly 48 Trp 47 Asn 90 Ala 89 Ala 177 Pro 176 Ala 216 Pro 215 Arg 286 Gly 49 Gly 48 |
| 1S6R-A | Ser 64 Tyr 150 Gln 120 Ser 318 Gly 317 Gly 63 Tyr 221 Asn 152 |
| 1BC2-A | Ser 172 Glu 212 Ser 174 Thr 173 Gly 211 |
| 1ZC2-A | Ser 64 Ser 318 Asn 346 Lys 315 Thr 316 Gly 317 Ala 292 Leu 293 |
| 1IYS-A | Ser 237 Thr 235 Ser 70 Ser 130 Gly 236 Arg 276 Arg 274 Glu 273 Arg 276 Asp 277 Arg 275 Arg 178 Gln 270 Glu 269 Trp 229 Glu 254 Asn 255 Pro 252 Phe 290 His 256 Asn 92 His 89 His 112 Lys 111 Ile 108 |
| 2YZ3-A | Ser 17 Thr 15 Val 16 Asp 43 |
| 1WUP-A | Lys 161 Glu 81 His 139 Asn 167 Cys 158 |
| 1FOF-B | Thr 206 Lys 205 Arg 250 Ser 67 Ser 115 Gly 207 |
| 1N4O-A | Ser 237 Ser 70 Thr 235 Gly 236 Ser 130 Arg 178 |
| 1Q2Q-A | Asn 152 Ser 321 Gln 120 Ser 64 Thr 322 Tyr 224 Gly 320 Leu 119 Asp 248 Ala 247 Val 246 Leu 182 Pro 181 |
| 1RGY-A | Tyr 150 Ser 64 Asn 152 Gln 120 Ser 318 Gly 317 Gly 63 Val 211 Tyr 221 Thr 319 |
| 1H8Z-A | Thr 206 Arg 250 Gly 207 Lys 182 Asn 176 Asn 85 |
| 1MBL-A | Arg 244 Ser 130 Thr 235 Ser 70 Gly 236 |
| 2A3U-A | Asp 104 Tyr 105 Asn170 Asn 132 Ser 70 Ala 237 Thr 167 Met 69 Met 69 Ala 284 Ile 279 Ile 287 Ala 280 Ile 263 Ile 246 Val 261 Ala 248 Ala 280 Ile 221 Pro 226 Val 224 Ala 280 Asn 276 Ile 279 Ile 246 Thr 235 Arg 244 Thr 235 Ser 70 Ser 130 Lys 234 Arg 244 Gly 236 Lys 234 |
| 1Y54-A | Ser 64 Asn 154 Leu 119 Tyr 221 Gly 317 |
| 2BG8-A | Asp 89 Asp 88 Lys 90 Gly 193 Tyr 191 Lys 194 Gln 210 Lys 186 Asn 75 Lys 106 Tyr 191 Gly 193 Pro 192 Asn 248 Thr 176 Lys 148 Ser 227 Ser 225 Glu 265 Gly 264 Thr 226 Arg 131 Lys 93 Glu 97 Lys 147 Asn 233 |
| 1DD6-A | Arg 266 Glu 23 Phe 51 Val 25 Lys 33 His 79 His 197 Asp 81 Cys 158 Val 31 Trp 28 Gly 164 Leu 165 His 139 Asn 167 Gly 166 |
| 1M2X-A | Lys 197 Asp 119 Tyr 233 His 118 Asp120 His 196 His 263 Ala 237 Tyr 238 Glu 236 |
| 1JJE-A | Pro 32 Glu 199 Phe 22 Thr 20 His 34 Lys 8 His 79 Asp 170 His 139 Asn 167 His 197 Asp 81 Lys 161 Asn 167 Cys 158 Gly 166 Trp 28 Val 25 Ser 80 |
| 2A49-A | Ser 70 Ala 237 Met 272 Gly 236 Asn 170 Met 69 Val 224 Glu 288 Pro 226 Ala 284 Ile 221 Ile 279 Ala 280 Ile 263 Ile 231 Ala 248 Val 261 Leu 220 Ala 280 Ile 279 Asn 276 Arg 244 Thr 235 Ser 130 Ser 70 Thr 235 Arg 244 Val 216 |
| 1BSG-A | Lys 290 Gly 291 Ala 287 |
| 2WRS-A | Trp 87 Asp 118 Phe 62 Gly 209 His 179 His 114 His 240 Asn 210 Arg 205 Gly 168 Arg 109 Phe 167 Arg 166 Ala 132 Gly 130 Arg 127 His 153 Thr 152 |
| 1E25-A | Gly 236 Thr 235 Ser 130 Thr 237 Ser 70 |
| 2V20-A | Arg 220 Arg 219 Lys 214 Ser 215 Ser 110 Arg 223 Val 196 Ser 50 Gly 216 |
| 1DDK-A | Asp 81 His 197 |
| 2ZJ9-A | Tyr 266 Arg 148 Ile 291 Met 265 Glu 272 Gln 139 Gln 100 Trp 138 |
| 1L9Y-A | Lys 100 Phe 106 Ser 107 Lys 105 Asp 67 Asp 120 His 263 His 196 Asn 225 Lys 291 Lys 212 Tyr 214 Arg 78 Lys 172 Glu 181 Arg 179 Met 137 Val 180 His 190 Thr 204 Met 254 Gln 215 Ser 252 Met 205 Lys 206 Thr 188 His 118 His 196 His 116 His 121 Asp 120 Asp 86 His 263 |
| 1HZO-A | Ser 130 Ser 237 Lys 234 Thr 235 Tyr 105 Gly 236 Ser 70 |
| 3GMV-X | Pro 114 Trp 65 Thr 59 |
| 1O7E-A | Val 141 Gly 140 Ser 130 Ser 70 Thr 235 Ser 237 Gly 226 Arg 178 Lys 38 Lys 38 Gly 175 Lys 174 Arg 65 Arg 153 Arg 204 Gln 205 |
| 1M6K-A | Ala 215 Val 117 Trp 102 Ser 115 Leu 255 Glu 122 Lys 126 Ile 82 Phe 114 Ile 101 Glu 196 Asn 193 Ser 189 Glu 192 Pro 185 Leu 184 Asn 183 Asn 188 Val 186 Lys 187 |
| 3IWI-A | His 186 Asp 232 |
| 2Y87-A | His 196 His 116 His 118 His 196 Asp 236 Cys 221 His 263 Asp 120 Cys 221 |
| 2WGI-A | Ser 67 Ser 115 Arg 250 Thr 206 Val 117 Leu 155 Ala 66 |
| 3IF6-A | Lys 75 Ser 72 Lys 210 Thr 211 Arg 249 Asn 119 Ser 120 Arg 242 Gly 212 Ile 198 Arg 118 Thr 199 Met 117 Lys 210 Leu 196 Met 197 |
| 3C5A-A | Gly 236 Thr 237 Thr 235 Thr 216 Ser 130 Ser 70 |
| 2FHX-A | Tyr 67 Lys 224 Gly 232 Tyr 233 His 196 |
| 3PAE-A | Leu 168 Ala 80 Val 130 Leu 127 Ser 128 Met 114 Trp 115 Tyr 112 Gly 258 Gly 220 Trp 221 Glu 143 Gly 141 Leu 142 |
| 3Q6X-A | Asn 220 His 189 Trp 93 His 122 Met 67 Leu 65 Gly 219 Asp 124 Cys208 Ile 203 Ala 243 Lys 242 Phe 240 Asp 199 Ile 198 Thr 201 |
| 3QNC-A | Tyr 208 Gly 207 Arg 250 Thr 206 Lys 205 Ser 115 Ser 67 Lys 134 Lys 138 Lys 49 Gly 20 |
| 3GMW-A | Ser 140 Gln 109 His 142 Lys 125 Tyr 126 |
Fifth letter of the PDB code indicates the chain
Fig. 8.
Percentage of binding site residues involved in CH...O bonds and classical H-bonds
Fig. 9.
a Solvent accessibility pattern of binding site residues involved in C-H⋯O interactions in β-lactamases. b PyMOL view of C-H⋯O interacting residues in the binding pockets of β-lactamase (PDB ID-1SHV)
Discussion and conclusions
In the present study, we have analyzed the roles played by C-H⋯O interactions in the structural stability of β-lactamases. Ab initio calculations state that the energy of these C-H...O interactions is less than the energy of a conventional hydrogen bond, which estimates the C-H...O bond energy as −2 to −3Kcal/mol [9, 39–42]. From our analysis we found an average of 605 C–H...O interactions per β-lactamase. Therefore the minimum energy contribution by the C–H...O interaction is around −1,210 kcal/mol, whereas the maximum energy is −1,815 kcal/mol, which makes the C–H...O interaction a potentially important contributor to the overall stability of β-lactamases. The donor atom contribution to C-H⋯O interactions was mainly from aliphatic residues, while Asp, Glu, Leu, and Thr were involved as acceptor residues in all the four types of C-H⋯O interactions. SM C-H⋯O interactions are the predominant type of interactions in β-lactamases and thus we conclude that a contribution of SM-mediated interactions might be significant in C-H⋯O interactions. It is interesting to note that these results are different form those observed with RNA binding proteins [8]. Conformation of an amino acid residue in a protein structure is primarily determined by short-range interactions [43]. The geometry of these interactions reveals that C-H⋯O interactions prefer to be in short-range contacts and thus these residues might contribute to the local conformational stability of β-lactamases. A decrease in the surface contact with a solvent increases the hydrophobic effect. Previous studies have stated that when a protein folds into a well-defined three-dimensional structure, the majority of non-polar residues are excluded from water and cluster together within the core of the protein [44–46]. Thus, the energy generated by the exclusion of hydrophobic side chains from an aqueous environment helps to maintain the three-dimensional structure of the protein [47]. Our analysis correlates with the previous studies as most of the hydrophobic amino acids are located in the core region of the protein, where they are shielded from water. It is also interesting to note that a considerable number of the C-H...O interacting polar residues are located in the core region of the protein. The contribution of buried polar residues in the structure and stability of proteins has been reported by many investigators [48–51]. These studies provide considerable evidence for the importance of buried polar residues for protein stability compared to non-polar groups. The solvent accessibility pattern of β-lactamases reveals that most of the residues prefer to be in buried regions, thus these residues might play an important role in the stability of the core region of β-lactamases. Previous studies have shown that different amino acids have a distinct tendency for the adoption of helical, strand, and random coil conformations. Our analysis of the secondary structure preference of C-H...O interacting residues shows that most amino acids have a clear preference to participate in a certain secondary structure type, which is consistent with previous studies. Amino acids belonging to the same group behave in a similar manner to certain secondary structure types [52–56]. The level of conservation depends on the biological significance of the amino acids in the protein [34]. Previous studies have suggested that biologically important sites are extremely conserved and evolutionarily favored [57]; the rate of evolution at the amino acid site is inversely proportional to the degree of its conservation [33]. Previously, it has been reported that the residues located in the core region of proteins are highly conserved compared to rim residues [58]. Residues that are critical for the stability of the protein are often clustered together in the core regions of the proteins. The evolutionary pressure for both stability and function could lead to clustering of conserved residues. Non-polar residues located in the core region of proteins provide resistance against thermal and chemical denaturation, thereby contributing to the stability of proteins. Our analysis of C-H...O interacting residues shows that most of the non-polar residues located in the core region of the protein show a relatively higher conservation score. Hence, we propose the importance of these residues in maintaining the structure and stability of proteins; these results correlate with previous studies [59–62]. Stabilization centers are formed by supportive long-range interactions, which protect the structure of proteins from thermal fluctuations [63]. The biological importance of these centers is not only demonstrated by their long-range interactions but also by their evolutionarily conservative characters. Analysis of a stabilization center in structurally similar proteins provides valuable information regarding their evolutionary relationship [64]. A significant percentage of the C-H⋯O interacting residues had one or more stabilization centers and these residues might contribute additional stability to β-lactamases. Recognition of binding site residues is an important task for a structure-based drug design and functional annotation. Cellular processes highly depend on interaction with other molecules to carry out their biological functions, and thus knowing the location of the ligand binding sites makes it possible to design inhibitors or antagonists [65–69]. Analysis of these binding site residues reveals that all the residues located in the binding site of β-lactamases are involved in C-H⋯O interactions, which shows the significance of C-H⋯O interactions in determining the stability of β-lactamases. After further analyzing the solvent accessibility pattern of active site residues it is interesting to note that the majority of the active site residues are located in the exposed region of the protein. These results show the importance of C-H·O interactions in the specificity and stability of β-lactamases.
Acknowledgements
Dr. Anand Anbarasu gratefully acknowledges the Indian Council of Medical Research (ICMR) and the Government of India Agency for the research grant [IRIS ID: 2011–03260] to carry out this research. P. Lavanya thanks ICMR for the Research Fellowship through ICMR grant IRIS ID: 2011–03260. We would like to thank the management of VIT University for providing us the necessary facilities to carry out this research project.
Conflict of interest
The authors declare that there are no conflicts of interest.
References
- 1.Mandage R, Kamath P, Wakle M, Momin A. Discovery of β-lactam antibiotic resistance specific functional residues: a bioinformatics approach. eJBio. 2012;8(1):15–18. [Google Scholar]
- 2.Petrosino J, Cantu C, Palzkill T. Beta-lactamases: protein evolution in real time. Trends Microbiol. 1998;6(8):322–327. doi: 10.1016/S0966-842X(98)01317-1. [DOI] [PubMed] [Google Scholar]
- 3.Tenover FC. Mechanisms of antimicrobial resistance in bacteria. Am. J. Med. 2006;119(6):3–10. doi: 10.1016/j.amjmed.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 4.Baker EN, Hubbard RE. Hydrogen bonding in globular proteins. Prog. Biophys. Mol. Biol. 1984;44(2):97–179. doi: 10.1016/0079-6107(84)90007-5. [DOI] [PubMed] [Google Scholar]
- 5.Jeffery GA, Saenger W. Hydrogen Bonding in Biological Structures. New York: Springer; 1991. [Google Scholar]
- 6.Steiner T, Saenger W. Lengthening of the covalent O-H bond in O-H...O hydrogen bonds re-examined from low-temperature neutron diffraction data of organic compounds. Acta Crystallogr. B. 1994;50:348–357. doi: 10.1107/S0108768193011966. [DOI] [Google Scholar]
- 7.Anand S, Anbarasu A, Sethumadhavan R. Exploring the C-H...O interactions in glycoproteins. Appl. Biochem. Biotechnol. 2009;159:343–354. doi: 10.1007/s12010-008-8518-3. [DOI] [PubMed] [Google Scholar]
- 8.Anbarasu A, Anand S, Sethumadhavan R. Investigations on unconventional hydrogen bonds in RNA binding proteins: the role in CH⋯O C interactions. BioSystems. 2007;90:792–801. doi: 10.1016/j.biosystems.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 9.Mandel-Gutfreund Y, Margalit H, Jernigan H, Zhurkin VB. A role for in CH⋯O C interactions in protein-DNA recognition. J. Mol. Biol. 1998;277:1129–1140. doi: 10.1006/jmbi.1998.1660. [DOI] [PubMed] [Google Scholar]
- 10.Swaminathan CP, Gupta A, Surolia NA. Plasticity in the primary binding site of galactose/N-acetyl galactosamine-specific lectins. Implication of the C-H...O hydrogen bond at the specificity-determining C-4 locus of the saccharide in 4-methoxygalactose recognition by jacalin and winged bean (basic) agglutinin. J. Biol. Chem. 2000;275(37):28483–28487. doi: 10.1074/jbc.M004685200. [DOI] [PubMed] [Google Scholar]
- 11.Bella J, Berman HM. Crystallographic evidence for Cα-H⋯O=C hydrogen bonds in a collagen triple helix. J. Mol. Biol. 1996;264:734–742. doi: 10.1006/jmbi.1996.0673. [DOI] [PubMed] [Google Scholar]
- 12.Wahl MC, Sundaralingam M. C–H...O hydrogen bonding in biology. Trends Biochem. Sci. 1997;22:97–102. doi: 10.1016/S0968-0004(97)01004-9. [DOI] [PubMed] [Google Scholar]
- 13.Musah RA, Jensen GM, Rosenfeld RJ, McRee DE, Goodin DB, Bunte SW. Variation in strength of an unconventional C–H...O hydrogen bond in an engineered protein cavity. J. Am. Chem. Soc. 1997;119:9083–9084. doi: 10.1021/ja9716766. [DOI] [Google Scholar]
- 14.Baures PW, Wiznycia A, Beatty AM. Hydrogen bonding isosteres: biomolecular carboxylic acid and amine-N-oxide interactions mediated via CH...O hydrogen bonds. Bioorg. Med. Chem. 2000;8(7):1599–1605. doi: 10.1016/S0968-0896(00)00090-0. [DOI] [PubMed] [Google Scholar]
- 15.Yellin ZB, Leiserowitz L. The role played by C-H...O and C-H...N interactions in determining molecular packing and conformation. Acta Crystallogr. 1984;40:159–165. doi: 10.1107/S0108768184001919. [DOI] [Google Scholar]
- 16.Brandl M, Lindauer MM, Sühnel J. C-H...O and C-H...N interactions in RNA structures. Theor. Chem. Acc. 1999;101:103–113. doi: 10.1007/s002140050415. [DOI] [Google Scholar]
- 17.Ghosh A, Bansal M. Three-centre C-H—O hydrogen bonds in the DNA minor groove: analysis of oligonucleotide crystal structures. Acta Crystallogr. D Biol. Crystallogr. 1999;55:2005–2012. doi: 10.1107/S0907444999012858. [DOI] [PubMed] [Google Scholar]
- 18.Taylor R, Kennard O. Crystallographic evidence for the existence of C-H O, C-H N, and C-H Cl hydrogen bonds. J. Am. Chem. Soc. 1982;104:5063–5070. doi: 10.1021/ja00383a012. [DOI] [Google Scholar]
- 19.Desiraju GR. The C-H⋯O hydrogen bond: structural implications and supramolecular design. Acc. Chem. Res. 1996;29(9):441–449. doi: 10.1021/ar950135n. [DOI] [PubMed] [Google Scholar]
- 20.Derewenda ZS, Lee L, Derewenda U. The occurrence of C-H...O hydrogen bonds in proteins. J. Mol. Biol. 1995;252:248–262. doi: 10.1006/jmbi.1995.0492. [DOI] [PubMed] [Google Scholar]
- 21.Fabiola GF, Krishnaswamy S, Nagarajan V, Pattabhi V. C-H...O hydrogen bonds in beta sheets. Acta Crystallogr. D. 1997;53:316–320. doi: 10.1107/S0907444997000383. [DOI] [PubMed] [Google Scholar]
- 22.Weiss MS. More hydrogen bonds for the (structural) biologist. Trends Biochem. Sci. 2001;26:521–523. doi: 10.1016/S0968-0004(01)01935-1. [DOI] [PubMed] [Google Scholar]
- 23.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyallow IN, Bourne PE. The Protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang L, Lai L. CH⋯O Hydrogen bonds at protein–protein interfaces. J. Biol. Chem. 2002;277(40):37732–37740. doi: 10.1074/jbc.M204514200. [DOI] [PubMed] [Google Scholar]
- 25.Panigrahi SK, Desiraju GR. Strong and weak hydrogen bond in drug-DNA complexes: a statistical analysis. J. Bio. Sci. 2007;32:677–691. doi: 10.1007/s12038-007-0068-2. [DOI] [PubMed] [Google Scholar]
- 26.Tiwari A, Panigrahi SK, HBAT a complete package for analysing strong and weak hydrogen bonds in macromolecular crystal structures. In Silico Biol. 2007;7:651–661. [PubMed] [Google Scholar]
- 27.Malkov SN, Zivkovic MV, Beljanski MV, Hall MB, Zaric SD. A reexamination of the propensities of amino acids towards a particular secondary structure: classification of amino acids based on their chemical structure. J. Mol. Model. 2008;14(8):769–775. doi: 10.1007/s00894-008-0313-0. [DOI] [PubMed] [Google Scholar]
- 28.Gromiha MM. Influence of cation-π interactions in different folding types of membrane proteins. Biophys. Chem. 2003;25:251–258. doi: 10.1016/S0301-4622(02)00318-6. [DOI] [PubMed] [Google Scholar]
- 29.Ahmad S, Gromiha M, Fawareh H, Sarain A. ASA view: database and tool for solvent accessibility representations in proteins. BMC Bioinforma. 2004;5:51. doi: 10.1186/1471-2105-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manavalan P, Ponnuswamy PK. A study of the preferred environment of amino acid residues in globular proteins. Arch. Biochem. Biophys. 1977;184(2):476–487. doi: 10.1016/0003-9861(77)90457-X. [DOI] [PubMed] [Google Scholar]
- 31.Gromiha MM, Selvaraj S. Inter-residue interactions in protein folding and stability. Prog. Biophys. Mol. Biol. 2004;56(2):235–237. doi: 10.1016/j.pbiomolbio.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 32.Dostanyi Z, Magyar C, Tusnady G, Simon I. SCide: identification of stabilization of stabilization centers in proteins. Bioinformatics. 2003;19:899–900. doi: 10.1093/bioinformatics/btg110. [DOI] [PubMed] [Google Scholar]
- 33.Glaser F, Pupko T, Paz I, Bell RE, Bechor D, Martz E, Tal NB. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19:163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- 34.Landau M, Maryrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Tal NB. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:299–302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goldenberg O, Erez E, Nimrod GM, Tal NB. The ConSurf-DB: pre-calculated evolutionary conservation profiles of protein structures. Nucleic Acids Res. 2000;37:323–327. doi: 10.1093/nar/gkn822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gromiha MM, Pujadas G, Magyar C, Selvaraj S, Simon I. Locating the stabilizing residues in (alpha/beta)8 barrel proteins based on hydrophobicity, long-range interactions, and sequence conservation. Proteins. 2004;55(2):316–329. doi: 10.1002/prot.20052. [DOI] [PubMed] [Google Scholar]
- 37.Wallace AC, Laskowski RA, Thornton JM. Ligplot: a program to generate schematic diagrams of protein–ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 38.Laskowski RA, Swindells MB. LigPlot+: Multiple ligand–protein interaction. Diagrams for drug discovery. J. Chem. Inf. Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 39.Levitt M, Perutz MF. Aromatic rings act as hydrogen bond acceptors. J. Mol. Biol. 1988;201:751–754. doi: 10.1016/0022-2836(88)90471-8. [DOI] [PubMed] [Google Scholar]
- 40.Scheiner S, Kar T, Gu Y. Strength of the C H⋯O hydrogen bond of amino acid residues. J. Biol. Chem. 2001;276:9832–9837. doi: 10.1074/jbc.M010770200. [DOI] [PubMed] [Google Scholar]
- 41.Vargas R, Garza J, Dixon DA, Hay BP. How strong is the C H⋯O C hydrogen bond? J. Am. Chem. Soc. 2000;122:4750–4755. doi: 10.1021/ja993600a. [DOI] [Google Scholar]
- 42.Duan G, Smith VH, Jr, Weaver DF. An ab initio and data mining study on aromatic–amide interactions. Chem. Phys. Lett. 1999;310:323–332. doi: 10.1016/S0009-2614(99)00804-0. [DOI] [Google Scholar]
- 43.Ponnuswamy PK, Warme PK, Scheraga HA. Role of medium-range interactions in proteins. Proc. Natl. Acad. Sci. 1973;70(3):830–833. doi: 10.1073/pnas.70.3.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koehl P, Delarue M. Polar and non polar atomic environments in the protein core: implication for folding and binding. Proteins. 1994;20(3):264–278. doi: 10.1002/prot.340200307. [DOI] [PubMed] [Google Scholar]
- 45.Miller S, Lesk AM, Janin J, Chothia C. The accessible surface area and stability of oligomeric proteins. Nature. 1987;328(6133):834–836. doi: 10.1038/328834a0. [DOI] [PubMed] [Google Scholar]
- 46.Tejedor SA, Abian O, Sancho J. Underexposed polar residues and protein stabilization. Protein Eng. Des. Sel. 2001;24(1–2):171–177. doi: 10.1093/protein/gzq072. [DOI] [PubMed] [Google Scholar]
- 47.Pace CN. Polar group burial contributes more to protein stability than non polar group burial. Biochemistry. 2001;40(2):310–313. doi: 10.1021/bi001574j. [DOI] [PubMed] [Google Scholar]
- 48.Takano K, Yamagata Y, Yutani K. Contribution of polar groups in the interior of a protein to the conformational stability. Biochemistry. 2001;40(15):4853–4858. doi: 10.1021/bi002792f. [DOI] [PubMed] [Google Scholar]
- 49.Lins L, Thomas A, Brasseur R. Analysis of accessible surface of residues in proteins. Protein Sci. 2003;12(7):1406–1417. doi: 10.1110/ps.0304803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson RJ, Lin SR, Raines RT. Genetic selection reveals the role of buried, conserved polar residue. Protein Sci. 2007;16(8):1609–1616. doi: 10.1110/ps.072938907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moret MA, Zebende GF. Amino acid hydrophobicity and accessible surface area. Phys. Rev. E. 2007;75:11920. doi: 10.1103/PhysRevE.75.011920. [DOI] [PubMed] [Google Scholar]
- 52.Chou PY, Fasman GD. Conformational parameters for amino acids in helical, beta-sheet, and random coil regions calculated from proteins. Biochemistry. 1974;13(2):211–222. doi: 10.1021/bi00699a001. [DOI] [PubMed] [Google Scholar]
- 53.Betts, M.J., Russell, R.B.: Amino acid properties and consequences of substitutions. In: Barnes, M., Gray, I., (eds.) Bioinformatics for Geneticists, pp. 289–316. Wiley (2003)
- 54.Willis MA, Bishop B, Regan L. Dramatic structural and thermodynamic consequences of repacking a protein’s hydrophobic core. Structure. 2000;8:1319–1328. doi: 10.1016/S0969-2126(00)00544-X. [DOI] [PubMed] [Google Scholar]
- 55.Chothia C. The nature of the accessible and buried surfaces in proteins. J. Mol. Biol. 1976;105:1–14. doi: 10.1016/0022-2836(76)90191-1. [DOI] [PubMed] [Google Scholar]
- 56.Rose GR, Geselowitz AR, Lesser GJ, Lee RH, Zehfus MH. Hydrophobicity of amino acid residues in globular proteins. Science. 1985;229(4716):834–838. doi: 10.1126/science.4023714. [DOI] [PubMed] [Google Scholar]
- 57.Chakrabarti S, Lanczycki CJ. Analysis and prediction of functionally important sites in proteins. Protein Sci. 2007;16(1):4–13. doi: 10.1110/ps.062506407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guharoy M, Chakrabarti P. Conserved residue clusters at protein–protein interfaces and their use in binding site identification. BMC Bioinformatics. 2010;11:286. doi: 10.1186/1471-2105-11-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guharoy M, Chakrabarti P. Conservation and relative importance of residues across protein–protein interfaces. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15447–15452. doi: 10.1073/pnas.0505425102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Furman OS, Baker D. Conserved residue clustering and protein structure prediction. Proteins: Struct. Funct. Genet. 2003;52:225–235. doi: 10.1002/prot.10365. [DOI] [PubMed] [Google Scholar]
- 61.Zhou H, Zhou Y. Quantifying the effect of burial amino acid residues on protein stability. Proteins Struct. Funct. Bioinf. 2004;54:315–322. doi: 10.1002/prot.10584. [DOI] [PubMed] [Google Scholar]
- 62.Gutfreund YM, Gregoret LM. On the significance of alternating patterns of polar and non-polar residues in beta-strands. J. Mol. Biol. 2002;323:453–461. doi: 10.1016/S0022-2836(02)00973-7. [DOI] [PubMed] [Google Scholar]
- 63.Creighton TE. Proteins: Structures and Molecular Properties. 2. New York: Freeman; 1993. pp. 419–429. [Google Scholar]
- 64.Fuxreiter M, Simon I. Role of stabilization centers in 4 helix bundle proteins. Proteins Struct. Funct. Bioinf. 2002;48:320–326. doi: 10.1002/prot.10167. [DOI] [PubMed] [Google Scholar]
- 65.Huang B. MetaPocket: a meta approach to improve protein ligand binding site prediction. J. Integr. Biol. 2009;13:4. doi: 10.1089/omi.2009.0045. [DOI] [PubMed] [Google Scholar]
- 66.Xie ZR, Hwang MJ. Ligand binding site prediction using ligand interacting and binding site-enriched protein triangles. Bioinformatics. 2012;28(12):1579–1585. doi: 10.1093/bioinformatics/bts182. [DOI] [PubMed] [Google Scholar]
- 67.Gao J, Liu Q, Kang J, Cao Z, Zhu R. Comparison of different ranking methods in protein-ligand binding site prediction. Int. J. Mol. Sci. 2012;13:8752–8761. doi: 10.3390/ijms13078752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Laurie ATR, Jackson RM. Methods for the prediction of protein-ligand binding sites for structure based drug design and virtual ligand screening. Curr. Protein Pept Sci. 2006;7:395–406. doi: 10.2174/138920306778559386. [DOI] [PubMed] [Google Scholar]
- 69.Park K, Kim D. A method to detect important residues using protein binding site comparison. Genome Informa. 2006;17(2):216–225. [PubMed] [Google Scholar]