Abstract
Muscarinic receptor-based designer receptors have as powerful novel tools to study G-proteincoupled receptor (GPCR) signaling and physiology. These new designer GPCRs, which are most frequently referred to as DREADDs (designer receptors exclusively activated by designer drug), are unable to bind acetylcholine, the endogenous muscarinic receptor agonist, but can be activated by clozapine-N-oxide (CNO), an otherwise pharmacologically inert compound, with high potency and efficacy. The various DREADDs differ primarily in their G protein coupling preference. More recently, an arrestin-biased DREADD has also been developed. The expression of DREADDs in distinct tissues or cell types has enabled researchers to study the outcome of selective stimulation of distinct GPCR (or arrestin) signaling pathways in a temporally and spatially controlled fashion in vivo. In this review, we provide an up-to-date snapshot of where this field currently stands and which important novel insights have been gained using this new technology.
Development of a novel class of designer GPCRs
Members of the superfamily of GPCRs modulate the activity of virtually every cell in the body. As a result, GPCRs are the target of a very large number of drugs in current clinical use [1]. Typically, a specific GPCR (excluding the large family of odorant GPCRs expressed by the nasal epithelium) is expressed in multiple tissues and cell types [2]. Consequently, the physiological effects observed after systemic administration of a particular GPCR ligand represent the integrated outcome of the actions of the ligand on multiple organs or tissues. Thus, to better understand the physiological and, potentially, pathophysiological roles of specific GPCR signaling pathways, an experimental system that allows the activation (in vivo) of a particular GPCR in a cell type- or tissue-specific fashion would be highly desirable.
In this regard, the development of designer GPCRs referred to as RASSLs (receptors activated solely by synthetic ligand) has provided important novel tools to study the in vivo relevance of distinct GPCR signaling pathways [3,4]. In most cases, these first-generation RASSLs represent engineered GPCRs that can be efficiently activated by a preexisting synthetic drug but are much less sensitive to activation by its endogenous ligand. Phenotypic analysis of transgenic mice expressing particular RASSLs in a cell type-specific fashion led to important novel insights into the physiological and pathophysiological roles of various GPCR signaling cascades [3,4]. However, first-generation RASSLs are usually endowed with a significant degree of constitutive activity, and the most commonly used RASSL ligands retain considerable activity at endogenous GPCRs [4], complicating the interpretation of mouse phenotyping studies.
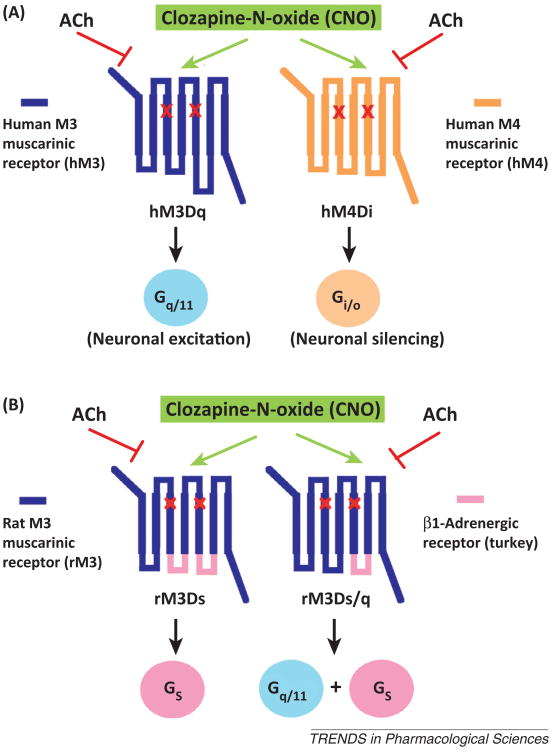
To overcome the pitfalls associated with the use of first-generation RASSLs, Bryan Roth and colleagues recently developed a series of mutationally modified muscarinic receptors that can be activated by clozapine-N-oxide (CNO) with high potency and efficacy (Figure 1A) [5]. CNO is a pharmacologically inert metabolite of clozapine, an antipsychotic drug (Figure 2A). Importantly, these new designer receptors cannot be activated by acetylcholine, the endogenous muscarinic receptor agonist (Figure 1A) [5,6]. To distinguish this new class of designer receptors from the first-generation RASSLs, most researchers in the field now refer to these CNO-sensitive mutant muscarinic receptors as DREADDs (designer receptors exclusively activated by designer drug) [5]. With only one exception [6], expression of different types of DREADDs in vivo in mice or rats did not result in constitutive signaling in the absence of CNO.
Figure 1.
Schematic representation of designer receptors exclusively activated by designer drug (DREADDs) endowed with different G protein coupling properties. All depicted DREADDs contain the Y3.33C and A5.46G point mutations in TM3 and TM5, respectively (indicated by the red × marks). The resulting designer receptors are unable to bind acetylcholine (ACh), the endogenous muscarinic receptor agonist, but can be activated by clozapine-N-oxide (CNO) with high potency and efficacy. (A) Structure and G protein coupling properties of the hM3Dq and hM4Di DREADDs [5]. CNO stimulation of hM3Dq- or hM4Di-expressing neurons leads to neuronal excitation or inhibition, respectively (see text for details). (B) Structure and G protein coupling properties of the rM3Dq and rM3Ds/q DREADDs ([6]; Guettier and Wess, unpublished data). In these two DREADDs, distinct intracellular loops of the rat M3 muscarinic receptor were replaced with the corresponding sequences derived from the turkey β1-adrenoceptor. Note that CNO binds to the extracellular surface of the receptors while the intracellular side of the receptors interacts with heterotrimeric G proteins.
Figure 2.
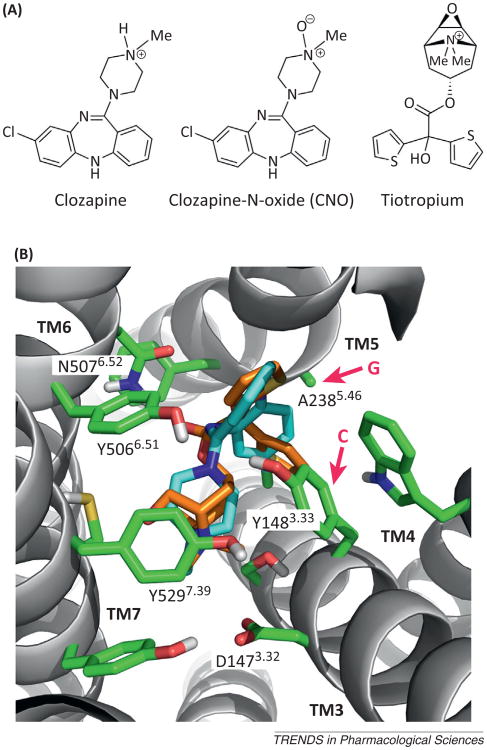
Docking of clozapine-N-oxide (CNO) into the ligand binding pocket of the rat M3 muscarinic receptor (M3R). (A) Chemical structures of clozapine, CNO, and tiotropium. Note that CNO differs from clozapine only in the presence of the N-oxide moiety. The structure of tiotropium, a muscarinic antagonist/inverse agonist, is shown for comparison because of the availability of a high-resolution X-ray structure of the tiotropium–rat M3R complex [17]. (B) Extracellular view of the tiotropium binding pocket [17]. Key amino acids that are of particular importance for tiotropium binding are highlighted. Both Y3.33 and A5.46 (red arrows) are predicted to contact the tiotropium ligand (see the text for details; note that all muscarinic receptor-based DREADDs contain the Y3.33C and A5.46G point mutations). The amino acids that replace Y3.33 and A5.46 in the DREADD constructs are indicated (C and G at the beginning of the red arrows). CNO can be docked into the M3R binding pocket in a pose very similar to that of tiotropium, without any obvious structural clashes (the two compounds are similar in size and overall shape; CNO, light blue; tiotropium, orange; Dahlia R. Weiss and Brian K. Shoichet, personal communication).
In a series of elegant studies, Armbruster et al. demonstrated that simultaneous introduction of the Y3.33C and A5.46G point mutations into the M1–M5 muscarinic receptors resulted in DREADDs that show the same G proteincoupling preference as their parent receptors (M1, M3, and M5 forGq/11;M2 and M4 forGi/o; shown for the human M1 and M3 muscarinic receptors in Figure 1A) [5]. More recently, Guettier et al. described a Gs-coupled DREADD that represents an M3 muscarinic/β1-adrenergic hybrid receptor in which the second and third intracellular loops of the M3 muscarinic receptor (M3R) have been replaced with the corresponding β1-adrenoceptor sequences (Figure 1B) [6]. Interestingly, an M3R DREADD in which only the third intracellular loop contains β1-adrenoceptor sequence (Figure 1B) shows promiscuous G protein-coupling properties; it is able to efficiently activate both Gs and Gq/11 in a CNO-dependent fashion (Guettier and Wess, unpublished observation). The generation of an arrestin-biased DREADD as a novel tool to study the in vivo role of G protein-independent signaling pathways is discussed below.
The development and initial experimental applications of DREADD technology have been covered in several excellent reviews [4,7–9]. Thus, the primary focus of the present review is on more recent findings in this very promising new area of research.
DREADD nomenclature
Different authors have used different acronyms to refer to the various DREADDs developed during the past few years (Table 1). As discussed above [4,5], the consistent use of a common DREADD nomenclature would avoid potential confusion about the molecular nature of a designer receptor used in a particular study. Similar to earlier proposals [4,5], we suggest the use of the DREADD terminology shown in Table 1. For example, in the term hM3Dq, M3 denotes the receptor subtype into which the DREADD mutations were introduced (the prefix h refers to the species, in this case human), D stands for DREADD, and q for the G protein coupling preference of this particular designer receptor (Gq/11). It should be noted, however, that some of the DREADDs represent hybrid receptors containing receptor sequences from different species (e.g., rM3Ds). In this case, the species designation refers to the receptor from which most of the sequence is derived. Moreover, the development of novel classes of DREADDs that preferentially signal via arrestin dependent (G protein-independent) pathways or stimulate G proteins without recruiting arrestins requires further refinements of the nomenclature scheme. Although the proposed nomenclature contains information about the G protein coupling preference of a particular DREADD, it has been demonstrated that the hM3Dq [10] and rM3Dq [11] constructs are also able to recruit proteins of the arrestin family (arrestin-2 and -3), similar to the wild type (WT) receptors from which they are derived. The same probably also holds true for other functional classes of DREADDs. Moreover, the different DREADDs, like their parental WT GPCRs, may also interact with various other GPCR-associated proteins. Thus, although the proposed nomenclature is far from perfect, its general adoption would simplify the identification of a specific DREADD used in a particular study.
Table 1. Examples of the proposed DREADD terminology.
| Designer receptora | Previous names (examples) | Coupling preference | Comments | Refs |
|---|---|---|---|---|
| hM3Dq | hM3-RASSL, Gq-DREADD, Sy-Rq | Gq/11 | – | [5] |
| rM3Dq | Rq | Gq/11 | Rat–human M3R hybrid construct | [6] |
| hM4Di | Gi-RASSL, M4-DREADD | Gi/o | – | [5] |
| rM3Ds | Rs, Gs-DREADD | Gs/olf | Rat M3R–turkey β1-receptor hybrid construct | [6] |
| rM3Ds/q | Gs/olf; Gq/11 | – | Unpublished b | |
| rM3Darr | Rq(R165L) | Arrestin-2, -3 | Rat/human M3R hybrid construct | [11] |
All constructs contain the Y3.33C and A5.46G point mutations.
In the rM3Ds/q construct, the third intracellular loop (i3) of the rat M3R (residues 253–489) was replaced with the corresponding turkey β1-receptor sequence (residues 230–289) (Guettier and Wess, unpublished data).
Molecular basis of CNO–DREADD interactions
The M3R binds clozapine with relatively high affinity (20–30 nM) [12,13]. Compared to clozapine, CNO binds to the M3R with ∼1000-fold lower affinity [6]. Structurally, CNO differs from clozapine only by the presence of the N-oxide group (Figure 2A). It is most likely that the negatively charged oxygen of the N-oxide moiety interferes, via electrostatic repulsion, with the proper formation of the salt bridge between the positively charged nitrogen of the ligand and the negatively charged D3.32 side chain (note that this interaction is conserved among all biogenic amine GPCRs; corresponding to D1473.32 in Figure 2B).
All muscarinic receptor-based DREADDs contain the Y3.33C and A5.46G point mutations (corresponding toY148C and A238G in the rat M3R; Figures 1 and 2B). Site-directed mutagenesis studies have shown that introduction of the Y3.33C point mutation into the M3R or the structurally closely related M1R leads to a pronounced decrease in carbachol or acetylcholine binding affinity and a loss or dramatic reduction of agonist (acetylcholine or carbachol) efficacy [14,15]. Similarly, introduction of the A5.46G point mutation into the M1R results in marked decreases in acetylcholine binding affinity, potency, and efficacy [16]. These findings explain why muscarinic receptor-based DREADDs containing both the Y3.33C and A5.46G point mutations show minimal acetylcholine binding affinity and efficacy.
The X-ray structure of the rat M3R indicates that both Y3.33 and A5.46 contact the co-crystallized ligand, tiotropium, a muscarinic antagonist/inverse agonist (Figure 2B) [17]. Y3.33 is part of an aromatic cage that surrounds the positively charged ammonium group of the ligand. The side chain of A5.46 makes contact with one of the two aromatic rings of tiotropium (Figure 2B). CNO can be docked into the M3R binding pocket in a pose very similar to that of tiotropium, without any obvious structural clashes (Figure 2B; Dahlia R. Weiss and Brian K. Shoichet, personal communication). It therefore remains unclear why CNO shows a much higher affinity for the M3R DREADD than for the WT M3R and why CNO is an agonist at the M3R DREADD but not at the WT M3R. One possibility is that that the Y3.33C point mutation causes minor conformational rearrangements of the so-called aromatic lid [17] that lead to changes in the kinetics of CNO binding, resulting in an increase in CNO binding affinity. Moreover, it is possible that the A5.46G point mutation increases the conformational flexibility of TM5, thus facilitating a CNO-dependent inward movement of TM5. In the β2-adrenoceptor, an agonist-dependent inward bulge of TM5 centered around S5.46 is a crucial structural feature of the active state of this receptor [18].
Pharmacokinetic properties of CNO
After a single intraperitoneal (i.p.) injection of CNO (1 mg/kg) into mice, CNO plasma levels peaked at 15 min and were very low after 2 h [6]. In these acute CNO injection experiments, back-transformation of CNO to clozapine was not detectable, consistent with the outcome of similar experiments performed in rats [19]. However, acutely administered CNO can be metabolically converted to clozapine in other species such as human and guinea-pig [19]. The metabolites that may form after chronic administration of CNO to DREADD-expressing mice (or other species) have not been studied systematically. However, even if back-transformation to clozapine occurs after chronic CNO administration, it should be noted that clozapine is a more potent (by ∼10-fold) DREADD agonist than CNO itself [5]. Moreover, confounding biological effects of potential CNO metabolites can be easily identified by including both saline- and CNO-treated WT animals in a particular DREADD study.
Despite the short plasma half-life of CNO in mice [6], the biological effects that have been described after acute treatment of DREADD-expressing experimental animals are usually much longer (6–10 h) [6,20]. One possibility is that CNO tends to accumulate in tissues, although other scenarios are also feasible.
Many studies have shown that systemic administration of CNO (i.p. or subcutaneous) causes pronounced central effects in DREADD-expressing experimental animals (Table 2), indicating that sufficient amounts of CNO penetrate the blood–brain barrier. Moreover, oral CNO administration via drinking water to mice expressing rM3Dq in pancreatic β-cells led to robust metabolic phenotypes [21], indicating that significant amounts of CNO are absorbed from the gastrointestinal tract.
Table 2. Summary of studies that used DREADD technology in vivo.
| DREADD-expressing cell type or tissue | DREADD | Expression system | CNO dose | Major phenotypes | Refs |
|---|---|---|---|---|---|
| Principal neurons of the forebrain (hippocampus, cortex) | hM3Dq | Double transgenic mice (CaMKIIα-tTA; Tet-off system) | 0.1–5 mg/kg i.p. | Increases in locomotion and stereotypy, limbic seizures | [20] |
| Pancreatic β-cells | rM3Dq rM3Ds | Transgenic mice (use of the ripII promoter) | 0.001–10 mg/kg i.p. | Increases in insulin release and β-cell mass; improved glucose tolerance in obese mice | [6] |
| Striatonigral (Dyn neurons) and striatopallidal MSNs(Enk neurons) | hM4Di | HSV vector; stereotactic injection into the striatum of rats; hM4Di expression is driven by the Dyn or Enk promoter | 1 and 3 mg/kg i.p. | Enhanced behavioral desensitization to amphetamine (Enk neurons); impaired persistence of behavioral desensitization to amphetamine (Dyn neurons) | [25] |
| Orexin-containing neurons of the lateral hypothalamus | hM3Dq hM4Di | Use of rAAV vectors; stereotactic injection into the lateral hypothalamus of orexin-cre transgenic mice (FLEX switch technology) | 5 mg/kg i.p. | Increased wakefulness in hM3Dq-expressing mice; decreased wakefulness in hM4Di-expressing mice | [27] |
| Agrp-containing neurons of the arcuate nucleus | hM3Dq hM4Di | Use of rAAV vectors; stereotactic injection into the arcuate nucleus of agrp-cre transgenic mice (FLEX switch technology) | 0.3 mg/kg i.p. | Increased food intake and decreased energy expenditure in hM3Dq-expressing mice; decreased food intake in hM4Di-expressing mice | [28] |
| Serotonergic neurons | hM4Di | Double transgenic mice (RC∷PDi knock-in mice that require Cre for hM4Di expression crossed with Slc6a4-cre transgenic mice) | 10 mg/kg i.p. | Attenuation of the hypercapnia-induced respiratory chemoreflex; pronounced reduction incore body temperature at RT | [23] |
| Neurons in which the c-fos promoter is active | hM3Dq | Double transgenic mice (use of a c-fos promoter-driven tTA transgene; Tet-off system) | 0.5 mg/kg i.p. | Activation of a specific ensemble of neurons during fear conditioning; spatial pattern of activity at the time of learning and retrieval must match for proper recall | [22] |
| Agrp- or Pomc-expressing neurons of the arcuate nucleus; Sim1-expressing neurons of the PVH | hM3Dq hM4Di | Use of rAAV vectors; stereotactic injection into the arcuate nucleus or PVH of agrp-cre, pomc-cre or sim1-cre transgenic mice (FLEX switch technology) | 0.3 or 5 mg/kg i.p. | Increased food intake in mice expressing hM3Dq in Agrp neurons; regulation of long-term food intake in mice expressing hM4Di in Pomc neurons; stimulation of food intake in mice expressing hM4Di in Sim1 neurons | [34] |
| Striatonigral MSNs, striatopallidal MSNs | hM4Di | Use of rAAV vector; stereotactic injection into the striatum of D1-cre, D2-cre, or adora2a-cre transgenic mice (FLEX switch technology) | 1 mg/kg s.c. for several days (neonatal mice) | Pronounced effects on striatal excitatory synaptogenesis | [30] |
| Neurons of the arcuate nucleus in which the ripII promoter is active | hM3Dq | Use of an rAAV vector; stereotactic injection into the arcuate nucleus of ripII-cre transgenic mice (FLEX switch technology) | 0.3 mg/kg i.p. | Increased energy expenditure and brown fat activity | [31] |
| Brainstem Egr2 neurons | hM4Di | Double transgenic mice (RC∷PDi knock-in mice that require Cre for hM4Di expression were crossed with egr2-cre transgenic mice) | 10 mg/kg i.p. | Impairment of the hypercapnia-induced respiratory chemoreflex | [24] |
| Lateral habenula (LHb) | hM4Di | HSV vector; stereotactic injection into the LHb of rats | 1 mg/kg i.p. | Increased swim time and decreased immobile time in a forced swim test (model of behavioral depression) | [26] |
| Striatopallidal MSNs | rM3Ds | Transgenic mice (use of the adora2a promoter) | 0.01–5 mg/kg i.p. | Increased DARP-32 phosphorylation; decreased spontaneous and novelty-induced locomotor activity; prevention of behavioral sensitization to amphetamine | [46] |
| Medio-dorsal thalamus (MD) | hM4Di | Use of an rAAV; stereotactic injection into the MD of mice | 2 mg/kg i.p. | Selective impairments in prefrontal-dependent cognitive tasks; disruption of task-dependent modulation of MD–PFC synchrony | [47] |
| MRGPRB4 + sensory neurons | hM3Dq | Use of an rAAV; injections into Mrgprb4-cre transgenic mice (FLEX switch technology) | 5 mg/kg i.p. | Promotion of conditioned place preference | [32] |
| Somatostatin-positive neurons of the lateral subdivision of the central amygdala | hM4Di | Use of an rAAV; stereotactic injection into som-IRES-cre transgenic mice (FLEX switch technology) | 10 mg/kg i.p. | Marked impairment in fear memory | [33] |
| Pancreatic β-cells | rM3Dq | Transgenic mice (use of the ripII promoter) | 1 mg/kg i.p. CNO drinking water (0.25 mg/ml) treatment for several weeks | Improved β-cell function; prevention of experimentally induced diabetes | [21] |
Strategies used for expressing DREADDs in a cell type-or tissue-specific fashion
DREADD technology has provided neuroscientists with powerful novel tools to map neuronal circuits underlying a large number of central nervous system (CNS) functions, including, for example, memory formation, regulation of food intake, and wakefulness (Table 2). Importantly, CNO is able to silence or reduce the activity of neurons that express hM4Di but leads to excitation of neurons that express hM3Dq (Figure 1A) [9]. Roth and colleagues have shown that hM4Di-mediated neuronal silencing involves the Gβγ-mediated activation of G protein-coupled inwardly-rectifying potassium (GIRK) channels [5], whereas hM3Dq-mediated neuronal excitation depends on the activation of phospholipase C [20]. Thus, most neuroscientists using DREADD approaches are less concerned about activating specific GPCR signaling pathways but use DREADD technology primarily to turn on and off specific sets of neurons in a temporally and spatially controlled fashion.
In many of the studies listed in Table 2, DREADD technology was used in combination with optogenetic approaches (for a detailed discussion of the specific features of optogenetic techniques compared to DREADD-based approaches, see [9]). The cell type- or tissue-specific expression of DREADDs in vivo has been achieved using different experimental strategies (Table 2). Such approaches include the generation of transgenic mice in which DREADD expression can be turned on in a temporally and spatially controlled fashion by the use of Tet-off technology and tissue-specific promoters [20,22]. The Dymecki group has developed a knock-in mouse model in which the expression of hM4Di is dependent on Cremediated removal of a floxed stop sequence preceding the hM4Di coding sequence [23,24]. More recently, stereotactic injection of recombinant viruses [herpes simplex virus (HSV) or adeno-associated virus (AAV)] has been widely used to achieve DREADD expression in distinct neuronal subpopulations or parts of the brain. Viral delivery techniques offer the advantage that they do not require the time-consuming development of novel transgenic mouse lines. For example, DREADD-encoding HSVs containing a cell-type-specific promoter have been injected into distinct brain regions of the rat [25,26]. More recently, recombinant AAVs have been used that contain the coding sequence of a particular DREADD in an inverted fashion, surrounded by two pairs of heterotypic, antiparallel loxP sites [27–34]. After stereotactic injection of these viruses into a particular brain region of a specific Cre mouse driver line, Cre recombinase (whose expression is restricted to specific neurons) restores proper orientation of the DREADD coding sequence, resulting in expression of the designer receptor. This strategy is usually referred to as a FLEX switch [29] or double-floxed inverted open reading frame (DIO) technology [35].
CNO is a DREADD agonist that shows no obvious functional bias
One important question for the use of DREADD technology is whether a synthetic ligand (CNO) acting on engineered GPCRs can cause signaling outcomes that are similar to activation of the native GPCRs from which the various DREADDs are derived. To address this issue, Alvarez-Curto et al. compared the signaling properties of the hM3Dq construct (agonist CNO) with those of the WT hM3R (agonist acetylcholine) expressed in cultured cells [10]. The authors demonstrated that CNO treatment of hM3Dq-expressing cells caused functional outcomes very similar to those of acetylcholine acting on cells expressing the WT hM3R. Such effects included early conformational changes following receptor activation, receptor-dependent ERK1/2 phosphorylation and arrestin recruitment, receptor internalization, and the pattern of receptor phosphorylation. Thus, these data indicate that CNO does not act as a biased ligand at the hM3Dq receptor. It remains to be explored whether other DREADDs show similar properties to their corresponding native GPCRs. However, even if a particular DREADD shows functional selectivity for a certain signaling pathway, such a construct could become a very valuable tool for exploring the physiological roles of this specific signaling cascade.
Development of an arrestin-biased DREADD as a novel experimental tool
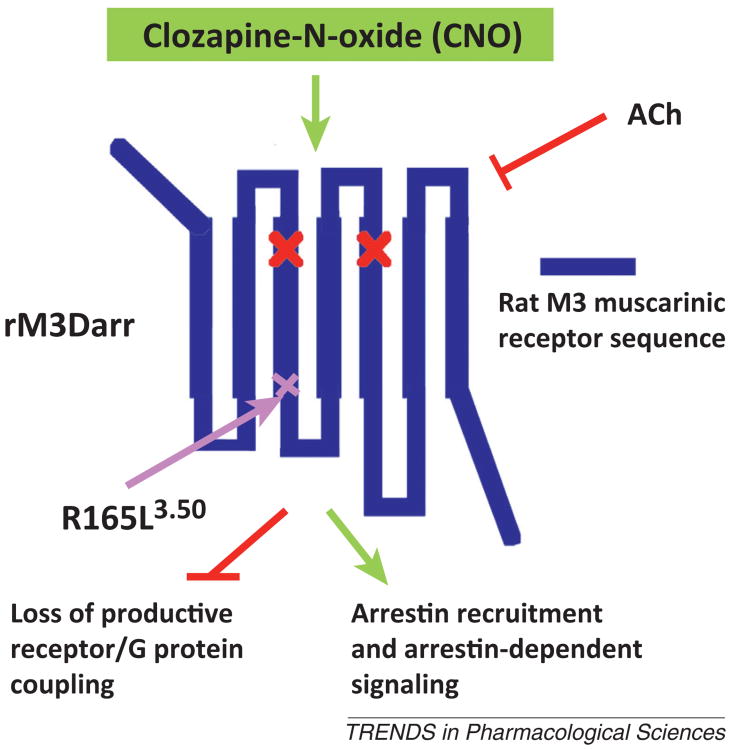
As is the case for most endogenous GPCRs, CNO-activated DREADDs not only couple to heterotrimeric G proteins but can also recruit proteins of the arrestin family (arrestin-2 and -3) to the activated receptors. For example, two recent studies have shown that CNO activation of hM3Dq- or rM3Dq-expressing cells promotes the recruitment of arrestin-2 and -3 [10,11]. It is now well established that arrestins can act as scaffolding proteins to promote signaling through G-protein-independent signaling pathways [36,37]. To generate a novel tool useful for studying the physiological relevance of such arrestin-dependent signaling pathways, Nakajima and Wess recently described the development of an arrestin-biased DREADD (rM3Darr according to the nomenclature suggested in Table 1) [11]. This novel rM3Dq-based DREADD contains the additional R3.50L point mutation (R165L in the rat M3R sequence) (Figure 3). CNO treatment of cultured cells expressing the rM3Darr construct did not result in any changes in intracellular calcium, inositol phosphate, or cAMP levels, suggesting that this DREADD is unable to activate heterotrimeric G proteins such as Gq/11, Gs, and Gi/o. However, rM3Darr was able to recruit arrestins and promote ERK1/2 phosphorylation in a CNO- and arrestin-dependent fashion. Interestingly, CNO treatment of MIN6 mouse insulinoma cells expressing the rM3Darr construct resulted in robust stimulation of insulin release. This response could be greatly reduced by pretreating cells with arrestin-2 or -3 siRNA [11]. These data support the concept that arrestin-dependent signaling plays a role in the regulation of insulin release [38]. Expression of rM3Darr in specific tissues or cell types from mice (or other experimental animals) should provide novel information about the physiological and pathophysiological roles of arrestin signaling pathways.
Figure 3.
Structure of a designer receptor exclusively activated by designer drug (DREADD; rM3Darr) that cannot activate heterotrimeric G proteins but is able to initiate arrestin-mediated signaling. Like all other muscarinic receptor-based DREADDs, the rM3Darr designer receptor contains the Y3.33C and A5.46G point mutations in TM3 and TM5, respectively (indicated by the red × marks) and is unable to bind acetylcholine (Ach) [11]. In addition, the rM3Darr construct contains the R165L3.50 point mutation at the bottom of TM3 within the highly conserved DRY motif. A previous study has shown that introduction of this point mutation into the wild type rat M3R virtually abolishes productive receptor–G protein coupling [48]. As a result, clozapine-N-oxide (CNO) treatment of rM3Darr-expressing cells does not lead to activation of heterotrimeric G proteins. However, in the presence of CNO, the rM3Darr designer receptor retains the ability to recruit arrestin-2 and -3 and to mediate arrestin-dependent downstream signaling [11]. CNO binds to the extracellular surface of rM3Darr, whereas the intracellular side of rM3Darr is predicted to interact with arrestins.
Identification of novel signal pathways using DREADD technology
DREADD-based techniques have also proven useful in identifying novel signaling pathways that are of great physiological importance. For example, extensive analysis of transgenic mice expressing the rM3Dq DREADD in pancreatic β-cells has revealed novel pathways crucial for the regulation of β-cell function [6,21]. Strikingly, chronic CNO treatment of these mutant mice resulted in pronounced improvements in β-cell function, including the upregulation of many genes critical for β-cell function, maintenance, and replication [21]. Moreover, chronic activation of β-cell rM3Dq signaling in this mouse strain effectively prevented streptozotocin-induced diabetes and greatly ameliorated the detrimental metabolic effects associated with consumption of a high-fat diet [21]. Additional in vivo and in vitro studies strongly suggest the existence of a novel signaling pathway through which activation of β-cell Gq triggers enhanced expression and function of insulin receptor substrate 2 (IRS2) and that IRS2-dependent downstream signaling plays a key role in mediating the improved β-cell function observed after chronic activation of rM3Dq [21]. These findings strongly suggest that therapeutic strategies to enhance signaling through β-cell Gq should prove useful for the treatment of diabetes.
DREADD studies with cultured cells
Although DREADD technology has been used most often for in vivo studies, experiments with cultured cells expressing specific DREADDs have also provided many important new mechanistic insights. For example, the latter approach has led to novel findings about the action of allosteric GPCR ligands [39,40], the molecular mechanisms involved in GPCR oligomerization [41], GPCR-dependent transendothelial migration of breast cancer cells [42] and mitogenic signaling [43], the modulation of Wnt–β-catenin signaling [44], and transcriptional regulation of immediate early genes [45].
Caveats for the use of DREADD technology
As summarized in Table 2, the cell type- or tissue-selective expression of DREADDs usually relies on the use of specific promoters. However, aberrant DREADD expression may sometimes occur, for example when a mouse Cre driver line has not been characterized in sufficient detail or when traditional transgenic approaches are used. To allow for a straightforward interpretation of experimental data, it is therefore essential to confirm that DREADD expression is restricted only to those tissues or cells that are under investigation. Another issue involves actual DREADD expression levels that are achieved in target cells. If at all possible, attempts should be made to quantify DREADD expression levels and compare these with the levels of endogenous GPCRs endowed with similar signaling properties. The expression of DREADDs at nonphysiologically high levels may lead to striking phenotypes but the outcome of such studies may not be relevant for targeting endogenous GPCRs for therapeutic purposes. The latter two issues are less critical for studies using DREADD technology for turning on or off distinct sets of neurons.
Concluding remarks
DREADD technology, despite its rather recent development, is now widely used in the neuroscience field to map neuronal circuits controlling many fundamental functions of the brain. In addition, studies with mutant mice expressing DREADDs in various peripheral tissues or cell types offer unprecedented new possibilities to explore the in vivo effects of activating specific peripheral GPCR signaling pathways. It is likely that the novel information that will emerge from these studies will lead to new strategies aimed at improving the therapy of many important pathophysiological conditions.
As discussed above, our group has recently designed and characterized an arrestin-biased DREADD. However, a G-protein-biased DREADD that is no longer able to interact with arrestins remains to be developed. Moreover, it should be possible to engineer DREADDs that display other specialized features, for example, DREADDs that differ in their subcellular localization or that show altered internalization, desensitization, or phosphorylation patterns. The use of such new designer receptors would prove useful in understanding the physiological roles of these and other processes in an in vivo setting. DREADDs that are selective for proteins of the Gq/11, Gi/o, or Gs families are currently used by many laboratories. However, a DREADD that can selectively activate G12/13, another major family of heterotrimeric G proteins, remains to be developed. Another area of future research involves the design of nonmuscarinic receptor-based DREADDs that can be activated by ligands different from CNO. The coexpression of such second-generation DREADDs and CNO-sensitive DREADDs would give researchers more sophisticated control over regulating the activity of specific cell types in vivo.
Although CNO represents an excellent DREADD ligand endowed with high potency, efficacy, and selectivity, systematic structural modification of this molecule may yield DREADD agonists endowed with altered (improved) pharmacological or pharmacokinetic properties. Clearly, the development of CNO derivatives endowed with a functional bias for either G-protein- or arrestin-dependent signaling pathways would represent another breakthrough in the DREADD field. A CNO analog that does not cross the blood–brain barrier may prove beneficial in studies in which activation of DREADDs expressed in the brain is not desirable. By contrast, a CNO-derived drug with enhanced lipophilicity that can enter the CNS more rapidly than CNO itself should make it possible to detect DREADD-mediated CNS responses in vivo more easily. Finally, a metabolically stable derivative of CNO would be useful in applying DREADD technology to other species, including humans (e.g., for potential gene therapy trials), in which CNO undergoes significant metabolic conversion.
Acknowledgments
Our research covered in this review was supported by the Intramural Research Program of the NIH, NIDDK. We thank Drs Dahlia R. Weiss and Brian K. Shoichet (University of California, San Francisco) for preparing Figure 2B, Dr Daniel Appella (NIH, NIDDK) for his help in generating Figure 2A, and Mr Andrew Kruse (University of California, San Francisco) for helpful discussions.
References
- 1.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 2.Regard JB, et al. Anatomical profiling of G protein-coupled receptor expression. Cell. 2008;135:561–571. doi: 10.1016/j.cell.2008.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scearce-Levie K, et al. Engineering receptors activated solely by synthetic ligands (RASSLs) Trends Pharmacol Sci. 2001;22:414–420. doi: 10.1016/s0165-6147(00)01743-0. [DOI] [PubMed] [Google Scholar]
- 4.Conklin BR, et al. Engineering GPCR signaling pathways with RASSLs. Nat Methods. 2008;5:673–678. doi: 10.1038/nmeth.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Armbruster BN, et al. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc Natl Acad Sci USA. 2007;104:5163–5168. doi: 10.1073/pnas.0700293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guettier JM, et al. A chemical-genetic approach to study G protein regulation of beta cell function in vivo. Proc Natl Acad Sci USA. 2009;106:19197–19202. doi: 10.1073/pnas.0906593106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong S, et al. A chemical-genetic approach for precise spatiotemporal control of cellular signaling. Mol Biosyst. 2010;6:1376–1380. doi: 10.1039/c002568m. [DOI] [PubMed] [Google Scholar]
- 8.Dong S, et al. Directed molecular evolution of DREADDs: a generic approach to creating next-generation RASSLs. Nat Protoc. 2010;5:561–573. doi: 10.1038/nprot.2009.239. [DOI] [PubMed] [Google Scholar]
- 9.Rogan SC, Roth BL. Remote control of neuronal signaling. Pharmacol Rev. 2011;63:291–315. doi: 10.1124/pr.110.003020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alvarez-Curto E, et al. Developing chemical genetic approaches to explore G protein-coupled receptor function: validation of the use of a receptor activated solely by synthetic ligand (RASSL) Mol Pharmacol. 2011;80:1033–1046. doi: 10.1124/mol.111.074674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakajima K, Wess J. Design and functional characterization of a novel, arrestin-biased designer G proteincoupled receptor. Mol Pharmacol. 2012;82:575–582. doi: 10.1124/mol.112.080358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olianas MC, et al. Mixed agonist-antagonist properties of clozapine at different human cloned muscarinic receptor subtypes expressed in Chinese hamster ovary cells. Neuropsychopharmacology. 1999;20:263–270. doi: 10.1016/S0893-133X(98)00048-7. [DOI] [PubMed] [Google Scholar]
- 13.Johnson DE, et al. Inhibitory effects of antipsychotics on carbachol-enhanced insulin secretion from perifused rat islets: role of muscarinic antagonism in antipsychotic-induced diabetes and hyperglycemia. Diabetes. 2005;54:1552–1558. doi: 10.2337/diabetes.54.5.1552. [DOI] [PubMed] [Google Scholar]
- 14.Han SJ, et al. Identification of an agonist-induced conformational change occurring adjacent to the ligand-binding pocket of the M3 muscarinic acetylcholine receptor. J Biol Chem. 2005;280:34849–34858. doi: 10.1074/jbc.M506711200. [DOI] [PubMed] [Google Scholar]
- 15.Lu ZL, Hulme EC. The functional topography of transmembrane domain 3 of the M1 muscarinic acetylcholine receptor, revealed by scanning mutagenesis. J Biol Chem. 1999;274:7309–7315. doi: 10.1074/jbc.274.11.7309. [DOI] [PubMed] [Google Scholar]
- 16.Allman K, et al. Scanning mutagenesis identifies amino acid side chains in transmembrane domain 5 of the M1 muscarinic receptor that participate in binding the acetyl methyl group of acetylcholine. Mol Pharmacol. 2000;58:175–184. doi: 10.1124/mol.58.1.175. [DOI] [PubMed] [Google Scholar]
- 17.Kruse AC, et al. Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature. 2012;482:552–556. doi: 10.1038/nature10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rasmussen SG, et al. Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jann MW, et al. Rapid formation of clozapine in guinea-pigs and man following clozapine-N-oxide administration. Arch Int Pharmacodyn Ther. 1994;328:243–250. [PubMed] [Google Scholar]
- 20.Alexander GM, et al. Remote control of neuronal activity in transgenic mice expressing evolved G protein-coupled receptors. Neuron. 2009;63:27–39. doi: 10.1016/j.neuron.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jain S, et al. Chronic activation of a designer Gq-coupled receptor improves β-cell function. J Clin Invest. 2013;123:1750–1762. doi: 10.1172/JCI66432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garner AR, et al. Generation of a synthetic memory trace. Science. 2012;335:1513–1516. doi: 10.1126/science.1214985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ray RS, et al. Impaired respiratory and body temperature control upon acute serotonergic neuron inhibition. Science. 2011;333:637–642. doi: 10.1126/science.1205295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ray RS, et al. Egr2-neurons control the adult respiratory response to hypercapnia. Brain Res. 2012 doi: 10.1016/j.brainres.2012.12.017. http://dx.doi.org/10.1016/j.brainres.2012.12.017. [DOI] [PMC free article] [PubMed]
- 25.Ferguson SM, et al. Transient neuronal inhibition reveals opposing roles of indirect and direct pathways in sensitization. Nat Neurosci. 2011;14:22–24. doi: 10.1038/nn.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nair SG, et al. DREADDing the lateral habenula: a review of methodological approaches for studying lateral habenula function. Brain Res. 2012 doi: 10.1016/j.brainres.2012.10.011. http://dx.doi.org/10.1016/j.brainres.2012.10.011. [DOI] [PMC free article] [PubMed]
- 27.Sasaki K, et al. Pharmacogenetic modulation of orexin neurons alters sleep/wakefulness states in mice. PLoS ONE. 2011;6:e20360. doi: 10.1371/journal.pone.0020360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krashes MJ, et al. Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. J Clin Invest. 2011;121:1424–1428. doi: 10.1172/JCI46229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Atasoy D, et al. A FLEX switch targets channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J Neurosci. 2008;28:7025–7030. doi: 10.1523/JNEUROSCI.1954-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozorovitskiy Y, et al. Recurrent network activity drives striatal synaptogenesis. Nature. 2012;485:646–650. doi: 10.1038/nature11052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kong D, et al. GABAergic RIP-Cre neurons in the arcuate nucleus selectively regulate energy expenditure. Cell. 2012;151:645–657. doi: 10.1016/j.cell.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vrontou S, et al. Genetic identification of C fibres that detect massage-like stroking of hairy skin in vivo. Nature. 2013;493:669–673. doi: 10.1038/nature11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, et al. Experience-dependent modification of a central amygdala fear circuit. Nat Neurosci. 2013;16:332–339. doi: 10.1038/nn.3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Atasoy D, et al. Deconstruction of a neural circuit for hunger. Nature. 2012;488:172–177. doi: 10.1038/nature11270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cardin JA, et al. Driving fast-spiking cells induces gamma rhythm and controls sensory responses. Nature. 2009;459:663–667. doi: 10.1038/nature08002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rajagopal S, et al. Teaching old receptors new tricks: biasing seven-transmembrane receptors. Nat Rev Drug Discov. 2010;9:373–386. doi: 10.1038/nrd3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shukla AK, et al. Emerging paradigms of b-arrestin-dependent seven transmembrane receptor signaling. Trends Biochem Sci. 2011;36:457–469. doi: 10.1016/j.tibs.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kong KC, et al. M3-muscarinic receptor promotes insulin release via receptor phosphorylation/arrestin-dependent activation of protein kinase D1. Proc Natl Acad Sci USA. 2010;107:21181–21186. doi: 10.1073/pnas.1011651107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nawaratne V, et al. New insights into the function of M4 muscarinic acetylcholine receptors gained using a novel allosteric modulator and a DREADD (designer receptor exclusively activated by a designer drug) Mol Pharmacol. 2008;74:1119–1131. doi: 10.1124/mol.108.049353. [DOI] [PubMed] [Google Scholar]
- 40.Abdul-Ridha A, et al. Allosteric modulation of a chemogenetically modified G protein-coupled receptor. Mol Pharmacol. 2013;83:521–530. doi: 10.1124/mol.112.083006. [DOI] [PubMed] [Google Scholar]
- 41.Alvarez-Curto E, et al. Ligand regulation of the quaternary organization of cell surface M3 muscarinic acetylcholine receptors analyzed by fluorescence resonance energy transfer (FRET) imaging and homogeneous time-resolved FRET. J Biol Chem. 2010;285:23318–23330. doi: 10.1074/jbc.M110.122184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yagi H, et al. A synthetic biology approach reveals a CXCR4-G13–Rho signaling axis driving transendothelial migration of metastatic breast cancer cells. Sci Signal. 2011;4:ra60. doi: 10.1126/scisignal.2002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vaqué JP, et al. A genome-wide RNAi screen reveals a Trioregulated Rho GTPase circuitry transducing mitogenic signals initiated by G protein-coupled receptors. Mol Cell. 2013;49:94–108. doi: 10.1016/j.molcel.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Regard JB, et al. Wnt/β-catenin signaling is differentially regulated by Gα proteins and contributes to fibrous dysplasia. Proc Natl Acad Sci USA. 2011;108:20101–20106. doi: 10.1073/pnas.1114656108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaufmann A, et al. Regulation of immediate-early gene transcription following activation of Gαq-coupled designer receptors. J Cell Biochem. 2013;114:681–696. doi: 10.1002/jcb.24410. [DOI] [PubMed] [Google Scholar]
- 46.Farrell MS, et al. A Gαs DREADD mouse for selective modulation of cAMP production in striatopallidal neurons. Neuropsychopharmacology. 2013;38:854–862. doi: 10.1038/npp.2012.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parnaudeau S, et al. Inhibition of medio–dorsal thalamus disrupts thalamo–frontal connectivity and cognition. Neuron. 2013;77:1151–1162. doi: 10.1016/j.neuron.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li B, et al. Random mutagenesis of the M3 muscarinic acetylcholine receptor expressed in yeast: identification of second-site mutations that restore function to a coupling-deficient mutant M3 receptor. J Biol Chem. 2005;280:5664–5675. doi: 10.1074/jbc.M411623200. [DOI] [PubMed] [Google Scholar]