Abstract
Targeted nanoparticles are being pursued for a range of medical applications. Here, we utilized targeted nanoparticles (synthetic platelets) to halt bleeding in acute trauma. One of the major questions that arises in the field is the role of surface ligand density on targeted nanoparticles’ performance. We developed intravenous hemostatic nanoparticles (GRGDS-NP1), and previously demonstrated their ability to reduce bleeding following femoral artery injury and increase survival after lethal liver trauma in the rat. These nanoparticles are made from block copolymers, poly(lactic-co-glycolic acid)-b- poly-ι-lysine-b-poly(ethylene glycol). Surface-conjugated targeting ligand density can be tightly controlled with this system, and here we investigated the effect of varying density on hemostasis and biodistribution. We increased the targeting peptide (GRGDS) concentration 100-fold (GRGDS-NP100) and undertook an in vitro dose-response study using rotational thromboelastometry (ROTEM), finding GRGDS-NP100 hemostatic nanoparticles were efficacious at doses at least 10-fold lower than the GRGDS-NP1. These results were recapitulated in vivo, demonstrating efficacy at 8-fold lower concentration after lethal liver trauma. 1-hour survival increased to 92%, compared to a scrambled peptide control, 45% (OR=14.4, 95% CI=[1.36, 143]), a saline control, 47% (OR=13.5, 95% CI=[1.42, 125]), and GRGDS-NP1, 80% (OR=1.30, n.s.). This work demonstrates the impact of changing synthetic platelet ligand density on hemostasis, and lays the foundation for methods to determine optimal ligand concentration parameters.
Keywords: nanomedicine, synthetic platelets, hemorrhage, ROTEM, hemostasis, coagulation
INTRODUCTION
Targeted nanotherapeutics have been developed for a broad range of medical applications.1–3 Factors that influence targeting efficacy include ligand-receptor affinities4, heteromultivalent ligand targeting strategies5, ligand presentation (linkers)6–8, as well as targeting ligand density. Studies to determine optimal surface ligand densities have demonstrated unique challenges specific to each material, ligand and application combination.9–15 We have previously developed intravenously injectable nanoparticles that augment hemostasis after injury.16, 17 In order to further optimize these nanoparticles for platelet targeting, reliable methods are needed that allow for tuning and measuring the effect of targeting-ligand densities.
These hemostatic nanoparticles are made of biodegradable block copolymers, reducing the risk of long-term inflammatory reactions. They consist of a nanoparticle core of biodegradable block copolymer of poly(lactic-co-glycolic acid) (PLGA) and poly-ι-lysine (PLL) with poly(ethylene glycol) (PEG) arms terminated with arginine-glycine-aspardic acid (GRGDS)-based targeting ligands. GRADSP ligands are used as a scrambled peptide to control for nonspecific actions of the particles (Scrambled-NPs). For research purposes, the nanoparticles have been loaded with coumarin-6, a fluorescent dye that allows us to track their biodistribution as previously described.16, 17
We previously investigated the role of targeting peptide length and PEG arm length on the efficacy of these hemostatic nanoparticles (NP’s), but the impact of ligand density on this system has not yet been addressed.17 From the literature, ligand density is known to play a critical role in targeting of nanoparticles. 9–13 Gu et al. developed a method to precisely engineer targeting-ligand-tunable nanoparticles for prostate cancer drug delivery and identified the narrow conjugation ratio that optimized targeting (5% for this application).9 Fakhari et al. varied the ligand density of Cyclo-(1,12)-PenITDGEATDSGC (cLABL) on PLGA nanoparticles to optimize the targeting of intercellular adhesion molecule-1 (ICAM-1), and found that the optimal density was roughly (50:50), and that particles with higher conjugation density performed worse.13 In all cases, the “optimal” conjugation of targeting ligand was highly application and condition-specific. In terms of the RGD-GPIIb/IIIa interaction that our nanoparticles utilize to augment platelet-platelet aggregation, there is evidence suggesting that receptor density may play a large role in determining the nature and strength of this interaction. 15 Coller et al. found that platelet binding to high density fibrinogen prevents aggregation of platelets to a plate through “paradoxical loss of luminal receptors”.15
Several groups, including our own, have investigated the concept of hemostatic particles to mitigate complications with sourcing, storage, immunocompatibility, and administration in the field of blood-product transfusions. These particles have shown vast promise along with an array of challenges.16–24 These challenges include establishing a small homogenous particle size20, avoiding promotion of nonspecific aggregation25, making design choices about targeting ligands, and hetero-multi-functionality5, 26, optimizing targeting-linker length17, 19, and demonstrating efficacy in both in vivo16, 17, 27 and relevant in vitro models.21, 28
Trauma is the leading cause of death for individuals between ages 1–44, and uncontrolled hemorrhage accounts for nearly one third of trauma-associated mortality.29 In combat, injuries can be especially severe and can be exacerbated by a prolonged pre-hospital phase, defined as the time between injury and admission to the hospital.30, 31 Recently, due to the advancement of body armor and changing warfare paradigms, the military has observed increased numbers of extremity injuries and internal (noncompressible) injuries (e.g. junctional, blast, central nervous system).30, 32, 33
We have previously shown that intravenously administered hemostatic nanoparticles (GRGDS-NP1) reduce bleeding times after femoral artery injury and increase survival after lethal liver trauma in the rat.16, 17 Here, our challenge was to develop a method for reproducibly controlling nanoparticle surface ligand conjugation and determine the impact of this change on hemostasis. We tested this change in both an in vitro assay, rotational thromboelastometry (ROTEM), and an in vivo model of lethal liver trauma. A dose-response study was undertaken utilizing ROTEM, and then applied in a lethal liver trauma in the rat. This work demonstrates the impact of changing synthetic platelet ligand density on hemostasis, and lays the foundation for methods to determine optimal ligand concentration parameters, providing a critical step toward translation of this nanotechnology.
METHODS
Nanoparticle Synthesis
PLGA (Resomer 503H) was purchased from Evonik Industries. Poly-ι-lysine (500–4000 Da MW) and PEG (~4600 Da MW) were purchased from Sigma Aldrich. All reagents were ACS grade and were purchased from Fisher Scientific. PLGA-PLL-PEG coblock polymer was made using standard bioconjugation techniques as previously described.16, 17, 38, 39
PLGA-PLL-PEG (1 g) was dissolved in anhydrous DMSO to a concentration of 100 mg/ml. Oligopeptides (25 mg GRGDS or GRADSP) was dissolved in 1 ml DMSO and added to the stirring polymer solution. This was reacted for 3 hours, and then transferred to dialysis tubing (SpectraPor 2 kDa MWCO). Dialysis water was changed every half hour for 4 hours with Type I D.I. water. The product was then snap-frozen in liquid nitrogen and lyophilized for 2–5 days.
The resulting quadblock copolymer PLGA-PLL-PEG-GRGDS was then dissolved to a concentration of 20 mg/ml in acetonitrile (120 mg/6 ml). This solution was added dropwise to a stirring volume of PBS. Precipitated nanoparticles form as the water-miscible solvent dissipates. Particles were collected using a coacervate precipitation method. Briefly, one mass equivalent of dry poly(acrylic acid) was added to the stirring particle suspension. 15ml of 1% w/v pAA was then added slowly to the stirring suspension until flocculation occurs. After 5 minutes, the flocculated particles were collected by centrifugation at 500g, and rinsed 3 times with 1% pAA (centrifuging @ 500 g, 2m, 4C between rinses). On the final rinse, particles were resuspended with D.I. water, snap-frozen and lyophilized for 2–5 days. Particles were resuspended in PBS and briefly sonicated at 4W to a total energy of 50 J using a probe sonicator (VCX-130, Sonics & Materials, Inc.) prior to use.
Characterization
As previously demonstrated, successful conjugation of PLL, PEG and peptide ligands was confirmed using UV-spectroscopy, 1H-NMR and amino acid analysis HPLC (BioRad, Varian and Shimadzu respectively).16 Nanoparticles from 5 independent syntheses batches were characterized for size distribution using dynamic light scattering (90Plus, Brookhaven Instruments Corporation) and zeta potential (Zeta Pals, Brookhaven Instruments Corporation). Reported figures from DLS are given as number average. Scanning electron microscopy was performed to visualize particle morphology (Hitachi S4500).
Amino acid analysis (AAA) was used to quantify the GRGDS peptide conjugation to the triblock polymer PLGA-PLL-PEG. The outcome arg:lys ratio was used to measure this relative conjugation efficiency. Briefly, a 5 mg aliquot of polymer was hydrolyzed for 24 h in a hydrolysis/derivitization workstation (Eldex Laboratories, Inc., Napa, CA). The hyrolysate was then neutralized with a redrying solution (ethanol: water: triethylamine in a 2:2:1 ratio) and derivitized with 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate, using the Water’s AccQ-Tag system. These samples were run on an HPLC (Shimadzu, with Water’s PicoTag Column) and measured using a fluorescence detector. Standard addition of known quantities of arg and lys to hydrolyzed samples was used to correct for polymer hydrolysate background.
Liver Trauma Model
All animal studies were performed in accordance with the Case Western Reserve University Institutional Animal Care and Use Committee (IACUC). Sprague Dawley rats (225–275g, Charles River) were anesthetized with intraperitoneal ketamine/xylazine (90:10 mg/kg, respectively), and injured according to the previously established liver injury model.16, 40–43 In brief, after confirmation of complete anesthesia, the abdomen was accessed and the medial lobe of the liver was marked with an arch radius 1.3 cm from the suprahepatic vena cava using a handheld cautery device. Once marked, the tail vein was catheterized with a saline-flushed 24G × 3/4″ Excel Safelet Catheter. The medial liver lobe was then resected along the marked lines to create the injury. Treatments were administered via tail vein catheter immediately after injury and included saline, scrambled particles, and functionalized particles. All particle treatments were resuspended in a 0.5 ml PBS carrier solution.
The rats were allowed to bleed for 1 hour or until death, as confirmed by lack of both breathing and a palpable heartbeat. Before measuring blood loss, all rats were injected with a 1 ml lethal dose of sodium pentobarbital. The abdomen was then reopened and blood collected with pre-weighed gauze. The clot adherent to the liver was collected last as this usually caused additionally bleeding to occur. The resected liver was weighed and fixed in 10% buffered formalin solution. Remaining liver, kidney, spleen and lungs were harvested and similarly preserved in 10% buffered formalin.
Biodistribution
Liver, kidney, spleen, lung and adherent clots were harvested and lyophilized for the biodistribution assay. The dry weight of the whole organ was recorded and 100–200 mg of dry tissue was homogenized (Precellys 24) and incubated overnight in acetonitrile at 37 C. This dissolved any nanoparticles present in the tissue and left the C6 in the organic solvent solution. Tubes were then centrifuged at 15,000 g for 10 minutes to remove solid matter and supernatant was tested on the HPLC. Mobile phase was 80% acetonitrile, and 20% aqueous (8% acetic acid). Stationary phase was a Waters Symmetry C18 Column, 100Å, 5 μm, 3.9 mm × 150 mm with fluorescence detection (450/490 nm ex/em). Based on the known C6 loading and dosage, data is represented as percent (%) of particles injected.
Histology
Tissue samples from the left lobe of the liver (uninjured), medial lobe (injured) with adherent clot, lung, kidney, and spleen were fixed in formalin, soaked overnight in sucrose, frozen and cryosectioned to 20-μm thickness. Sections were imaged with an inverted fluorescence microscope (Zeiss Axio Observer.Z1). The DsRed filter was used to image tissue background fluorescence as a reference channel since staining with VectaShield DAPI, or H&E displaced nanoparticles from the tissue.
In vitro Assay
Coagulation assays, using Sprague Dawley rat blood, were performed using the ROTEM’s NATEM test in the presence of either saline, GRGDS-NPs, or scrambled GRADSP-NPs as previously described.16 The outcomes we considered include the standard ROTEM parameters clotting time (CT), clot formation time (CFT), the sum of the two (CT+CFT), and maximum clot firmness (MCF). CT is defined as the time from the start of the assay until the initial clotting is detected (thickness = 2mm). CFT is defined as the time between clot initiation until a clot thickness of 20 mm is detected. MCF is defined as the maximum thickness (in mm) that a clot reaches during the duration of the test. The dosing study was performed blood samples from 10 rats, starting with the highest concentration of particles (20 mg/ml, n=5), and titrating downward (2 mg/ml, n=7; 0.2 mg/ml, n=9; 0.02 mg/ml, n=9) until no effect was observed (0.002 mg/ml, n=3). Each sample was run in triplicate on the ROTEM, normalized to a saline control, and averaged for each rat.
Statistics
ANOVA with ad-hoc Tukey comparisons was used to analyze blood loss and ROTEM data (Minitab). 1-hour survival was analyzed with a binomial logistic regression with chi-squared tests between odds-ratios (SAS), and survival curves with a log-rank (Mantel-Cox) test. Quantification of histology was analyzed with two sample t-tests with Welch’s correction.
RESULTS
GRGDS conjugation density
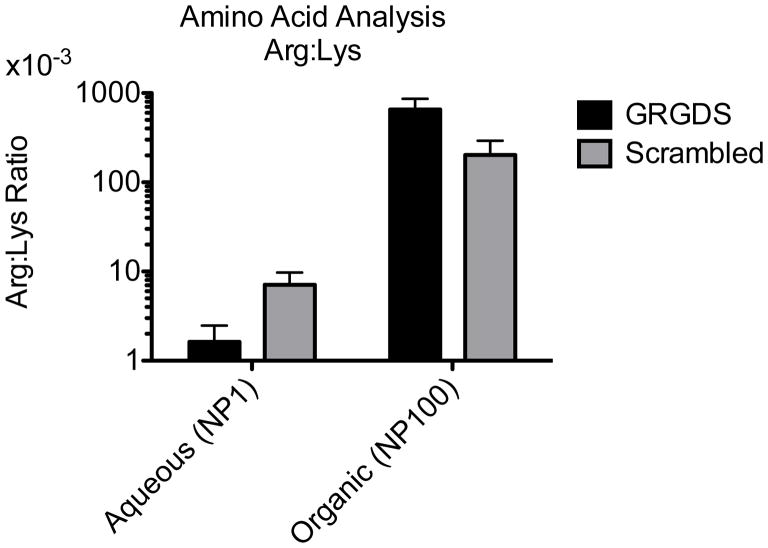
Polymer nanoparticle characteristics were the same as previously described (Table 1, Figure 1)16, except for the higher conjugation efficiency of RGD peptide to the activated PEG groups (Figure 2). This was accomplished primarily by performing the peptide conjugation before nanosphere formation and reacting in organic phase (anhydrous DMSO) rather than aqueous to form what we term the quadblock polymer (PLGA-b-p(lys)-b-PEG-b-GRGDS). After nanoparticle formation, peptide loading levels were measured with amino acid analysis and the arginine to lysine ratio was determined to obtain the percentage of polymer chains with the GRGDS peptide. Nanoparticles made from the quadblock polymer had 2 orders of magnitude more GRGDS than the nanoparticles made from triblock polymer used in the hemostatic nanoparticles tested previously (Arg:Lys ratio 4.35*10−3 compared to 0.428).16 The high-RGD-loaded nanoparticles based on the quadblock polymer are referred to as GRGDS-NP100 and Scrambled-NP100, while the previously used triblock nanoparticles are referred to as GRGDS-NP1 and Scrambled-NP1.
Table 1.
NP1 and NP100 Characterization: Size and Zeta Potential
| Nanoparticle formulation | DLS; no. avg. dia. (nm; mean ± SD) | Zeta potential (mV; mean ± SD) |
|---|---|---|
| GRGDS-NP1 | 499.4 ± 95.9 | −22.6 ± 2.1 |
| GRGDS-NP100 | 574.9 ± 196. 5 | −16.7 ± 5.5 |
| GRADSP-NP1 | 535.6 ± 133.8 | −24.8 ± 6.2 |
| GRADSP-NP100 | 611.4 ± 219.2 | −14.7 ± 2.5 |
Figure 1.
SCANNING ELECTRON MICROSCOPY. Image of nanoparticles under SEM (Hitachi S4500).
Figure 2.
AMINO ACID ANALYSIS. Peptide conjugation efficiency Arg:Lys ratio. Peptide conjugation levels are approximately 100-fold higher when the conjugation reaction is performed in DMSO instead of aqueous phase. This leads to the nomenclature, NP100 and NP1 for the organic and aqueous phase polymers respectively. Error bars denote SEM.
In vitro test of GRGDS NP100
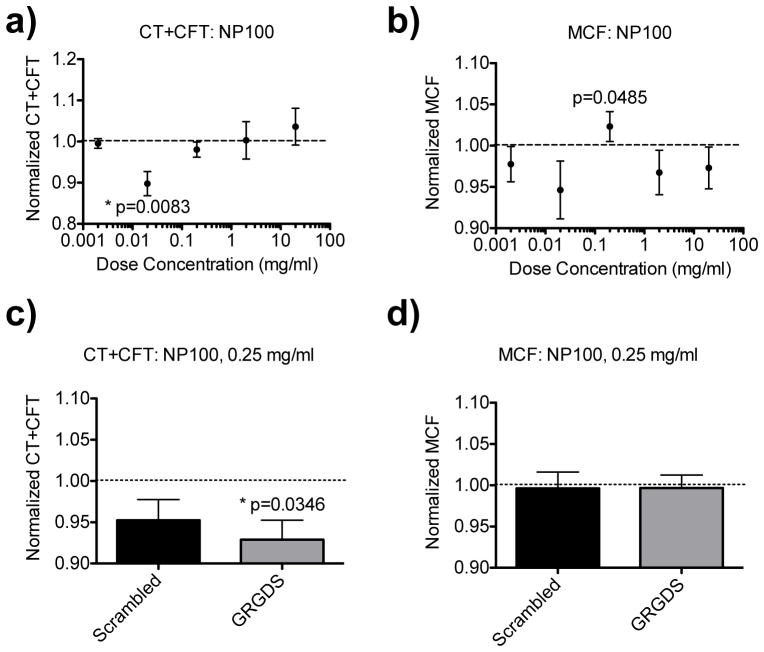
A dosing study with anticoagulated whole rat blood was performed to titrate the optimal dose of the NP100 nanoparticles (Figure 3). Rotational thromboelastometery (ROTEM) was used to determine clotting time (CT), clot formation time (CFT), and maximum clot firmness (MCF). Each sample consisted of 300 μl of anticoagulated blood, 20 μl of a particle dosing solution, and 20 μl of CaCl solution to replace the calcium in the blood and initiation coagulation. Previously, using the GRGDS-NP1 particles, we found that a particle dosing concentration of 2.5 mg/ml (blood concentration = 147 μg/ml) reduced clotting time in this in vitro model. When testing the GRGDS-NP100 particles at this same concentration, we observed a trend of increased total clotting time (CT+CFT), and a decrease in MCF, demonstrating an anticoagulant-like effect (Figure 3a). A dose response was then performed to titrate down to the optimal dose for the GRGDS-NP100 particles (Figure 3a–b). We found that the optimal dose was at least 10-fold lower, between 0.02–0.25 mg/ml (blood concentration = 1.2–14.7 μg/ml). Further testing 0.25 mg/ml at this concentration yielded a reduced total clotting time (CT+CFT, p=0.0346 versus saline), with no adverse impact on MCF (Figure 3c–d). There was no significant difference between the scrambled and GRGDS groups.
Figure 3.
IN VITRO DOSE RESPONSE GRGDS-NP100. a–b) Total clotting time (CT+CFT) and maximum clot firmness (MCF) dose-responses, recapitulated the in vivo response observed: high doses adversely impact clotting parameters (increase CT+CFT; decrease MCF). This is observed until dosing down to 0.02–0.2 mg/ml. C–D) 0.25 mg/ml was then further tested directly against a scrambled-NP100 control (n=6). CT+CFT was reduced compared to saline (p=0.0346), with no significant impact on MCF. Dotted lines denote normalization to the saline-treated controls values. Error bars denote SEM.
NP100 particles in liver injury model
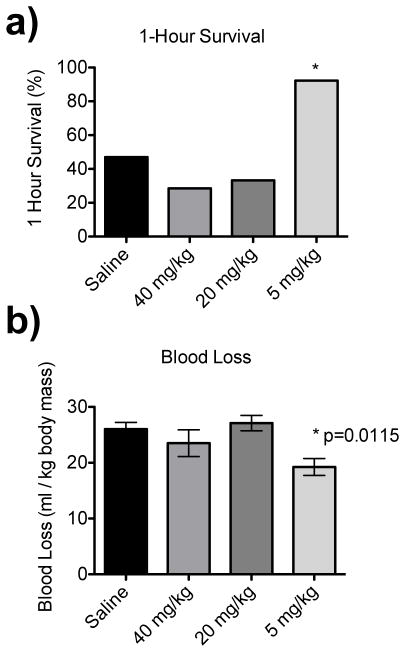
Previous experiments with the low-peptide conjugated nanoparticles (GRGDS-NP1) at 20 mg/ml concentration in a 0.5 ml carrier solution (40mg/kg) led to increased 1 hour survival (80% compared to 47% saline control) in a model of lethal liver trauma.16 When this experiment was repeated with high-peptide conjugated nanoparticles (GRGDS-NP100) at the same dose concentration (40 mg/kg) and ½ dose (20 mg/kg), survival time was drastically reduced from a mean time of 43 minutes (saline) to 28 minutes and 34 minutes, respectively, suggesting an adverse effect on the injury model. The effects of NP100 particles appeared to be harmful until dosing down to 5 mg/kg, at which, 1 hour survival increased to 100% for the pilot study with n=3 animals (Figure 4a). Blood loss was also significantly reduced compared to the saline control (p=0.0115, Figure 4b).
Figure 4.
IN VIVO DOSE RESPONSE GRGDS-NP100. Dose response with GRGDS-NP100 in rat liver injury model (n=3 for pilot study). b) Percentage of animals surviving to 1-hour is reduced in the 40 mg/kg and 20 mg/kg groups, but increased in the 5 mg/kg dose. b) Blood loss is significantly reduced in the 5 mg/kg dose, and not significantly changed with either 40 mg/kg or 20 mg/kg doses compared to the saline control. Error bars denote SEM.
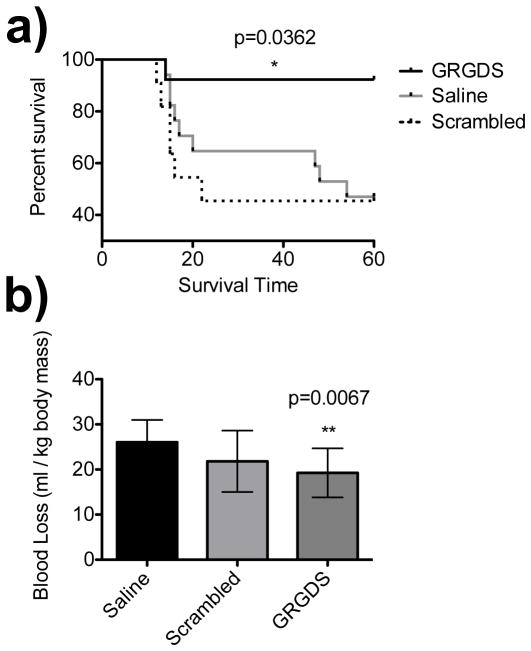
We then scaled up the study (n=13) at this new dosage, 5 mg/kg. 1-hour survival was increased to 92.3% compared to a scrambled peptide control 45% (OR=14.4, 95% CI=[1.36, 143], power=0.836) a saline control 47% (OR=13.5, 95% CI=[1.42, 125], power=0.888) and the previously reported hemostatic nanoparticles 80% (OR=1.3, n.s., Figure 5a). Blood loss, as measured by collecting intra-abdominal blood with pre-weighed gauze, was significantly decreased from a mean of 26.0 ml/kg (saline) to 19.25 ml/kg (GRGDS-NP100, p=0.0067, Figure 5b).
Figure 5.
LIVER INJURY RESULTS NP100. Rat medial liver injury model at 5 mg/kg dose. a) 1-hour (endpoint) survival was increased to 92%, compared to a scrambled peptide control, 45% (OR=14.4, 95% CI=[1.36, 143]), a saline control, 47% (OR=13.5, 95% CI=[1.42, 125]), and GRGDS-NP1, 80% (OR=1.30, n.s.). Survival curves display increased survival with GRGDS-NP100 compared to the scrambled and saline groups, log-rank (Mantel-Cox) test, p=0.0362. b) Blood loss was significantly reduced in the GRGDS-NP100 group compared to saline (p=0.0067). Error bars denote SEM.
Biodistribution
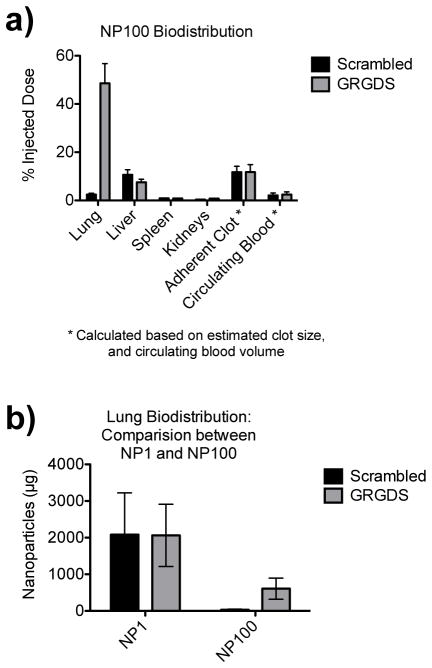
Compared to the previous liver injury study16, where 40 mg/kg of GRGDS-NP1 particles were injected, the present study only used 5 mg/kg of GRGDS-NP100 particles, 1/8 the previous mass. We find that similar proportions of the injected dose are found in the tissues, with the majority of particles being cleared through the liver (7.5–10.5%) or becoming entrapped in the clot (11%) or lungs (2–46%) as measured using an HPLC fluorescence assay for C6 (Figure 6). Less than 1% is found in the kidney and spleen, and the particles are rapidly cleared from the blood plasma, with only 2% remaining in circulation at the end of the 1-hour experiment.
Figure 6.
BIODISTRIBUTION (HPLC ASSAY). An assay for fluorescent C6 was performed using HPLC. a) There is a large proportion of nanoparticles in the lungs for the targeted GRGDS group (~50%). The liver accumulates 7.5–10.5% of the injected dose (GRGDS, scrambled respectively), while ~11% becomes entrapped in the adherent clot found in the abdominal cavity post-mortem, regardless of the peptide group. Less than 1% is found in the kidney and spleen, and the particles are rapidly cleared from the blood plasma, with only 2% remaining in circulation at the end of the 1-hour experiment. b) However, due to 8x lower dose with NP100, there are fewer nanoparticles by mass in the lungs compared to the NP1. Error bars denote SEM.
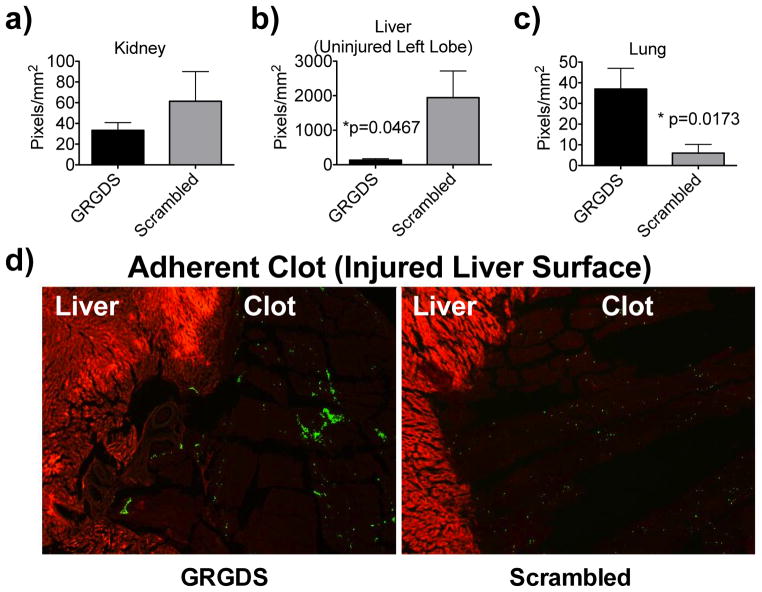
Since biodistribution within anisotropic organs can have a heterogeneous distribution, a histological investigation was conducted, looking at the kidney, capillary beds of the deep lung, the uninjured left lobe of the liver, and the adherent clot attached to the injured medial lobe of the liver (Figure 7). We found that while the proportion of particles accumulating in the clot was similar between the GRGDS and scrambled particles (by HPLC), the GRGDS particles appeared in clusters rather than individual satellite particles (by histology), suggesting that they may be actively participating in platelet aggregation (Figure 7d). The number of particles found within the clot (11% injected dose), while a small mass, represents a large number of particles, 9.2×109, or a number equivalent to ~50% of the total circulating platelets in a 250 g rat (assuming: nanoparticle density, 1.3 g/cm3; rat blood volume, 68.6 ml/kg; normal rat platelet concentration 1.180×109/ml)44, 45
Figure 7.
HISTOLOGY AND QUANTIFICATION. Histology was performed on the kidneys, uninjured left lobe of the liver, lungs, and injured medial lobe of the liver with adherent clot intact. Sections are 20 μm, and were not stained to prevent displacement of the nanoparticles. Quantification of the particles is measured in triplicate for n=3 rats per group, and represented as pixels/mm2. a) Kidneys show no significant differences between treatment groups, b) Uninjured liver (left lobe), contains higher density of particles in the scrambled group compared to GRGDS (p=0.0467). c) Lungs show a larger proportion of particles accumulating in the GRGDS group compared to scrambled, d) Clot adherent to remaining liver. Green = coumarin-6 (C6) loaded hemostatic nanoparticles; Red = Tissue background fluorescence (DsRed filter) used as reference channel. While the concentration of particles in the adherent clot is equal between groups, the particles in the GRGDS group appear as clusters, while the scrambled particles appear evenly dispersed. Error bars denote SEM.
The distribution of particles to the capillary beds of the lung was significantly smaller than expected for the GRGDS group (Figure 7b), suggesting that the large quantity of particles found in the GRGDS-NP100 group, by the HPLC assay of the whole lung tissue, is not collected in the capillary beds, and must be accumulating in higher order branches.
DISCUSSION
Internal hemorrhage is currently treated with a combination of i.v. blood products with or without additional administration of soluble clotting factors, such as fibrinogen or recombinant factor VIIa.35, 46–48 Unfortunately, resuscitative strategies involving blood products have the drawbacks necessitating donor sources and refrigeration.46 They also may carry the risk for immune responses and suffer from loss of hemostatic activity during storage.46 These issues limit the application in first responder situations, which is exceptionally important because early intervention is the best predictor of survival following trauma. We developed a synthetic substitute for platelet administration to mitigate these complications.
We previously showed that administration of GRGDS-NP1 nanoparticles significantly improve survival after lethal liver trauma.16 In this first generation system, the GRGDS peptide was crucial for function of these nanoparticles binding with the GIIb/IIIa receptor on activated platelets to form mechanically robust platelet plugs. In this work, we have gone onto investigate the role that peptide density plays in the efficacy and safety of these hemostatic nanoparticles. Here, we have increased the targeting ligand concentration on our nanoparticles 100-fold and achieved a 92% survival rate when administering GRGDS-NP100 (5 mg/kg) compared to 80% from the previous work. The higher peptide-conjugated particles, GRGDS-NP100, led to significantly increased survival and significantly reduced blood loss at a concentration 8-fold lower than the dose required for the previous GRGDS-NP1 formulation. 16 This finding is incredibly important because it suggests that peptide concentration is a critical variable, and demonstrates the significant impact on effective nanoparticle dosage by modulating targeting ligand concentration.
We also found blood loss was significantly reduced with the GRGDS-NP100 at 5 mg/kg. This is a significant finding as it continues to show that intravenous hemostatic nanoparticles can augment the clotting process, and that this can produce a large impact on survival. Furthermore, by reducing the effective dose, we also found that we can improve safety by having very few particles in non-injured tissues such as the lungs. Optimizing the targeting ligand is critical for the safety and efficacy of this system.
At higher doses (>5 mg/kg), it is evident that the GRGDS-NP100 particles have an adverse effect, substantially reducing mean survival time in the lethal liver trauma. These in vivo findings are recapitulated by our in vitro observations, that higher doses of the GRGDS-NP100 hemostatic nanoparticles actually adversely impact hemostasis. Specifically, we find that higher doses appear to inhibit the standard ROTEM parameters of clotting time and, to a lesser degree, maximum clot firmness. This sort of response, where a dose that is too high or too low is not effective, has been previously reported.15, 17
These observations are likely the result of a saturation effect of the endogenous platelets. Too many platelet-bound nanoparticles could theoretically sterically hinder or saturate the receptors on the activated platelets leading to reduced platelet-platelet interactions and inhibition of their aggregation. These findings suggest that titrating the correct dosing in vitro will be crucial as this technology moves forward into large animal and potentially clinical trials.
We previously found that accumulation of nanoparticles in the lungs was independent of peptide targeting, with ~2 mg dose accumulation in the lung for both the GRGDS-NP1 and Scrambled-NP1 particles.16 However, peptide density does appear to have an effect, with a higher percentage of injected GRGDS-NP100 accumulating in the lungs than scrambled-NP100, but at smaller total particle mass with the GRGDS-NP100 (0.57 mg) than the GRGDS-NP1 (2 mg) due to the lower effective dose. Lung accumulation was further investigated histologically and revealed extremely few, sparsely distributed nanoparticles in sections of the deep capillary beds of the lungs. Virtually no particles were found in the capillary beds of the deep lung tissue, where potential complications could arise as a result of nanoparticle aggregates.49–51 It is possible that if particles are associating with clots, as has been previously suspected16, 52, 53, and are subsequently shed from the injury site, these may be too large to reach the deep lung and are likely being incorporated in the higher order vasculature of the lung. The increased number of GRGDS-NP100 particles found in the lung in this study would therefore suggest a stronger clot-targeting effect compared to the scrambled-NP100, but has the disadvantage of accumulating in what appears to be clots in the lung. It may be possible to ameliorate these effects by changing the route of administration to one that directly feeds the injury site. Regardless, the question to answer will be whether or not these particles antagonize pulmonary function (e.g. pulmonary artery pressure) or increase risk for pulmonary embolism. It is clear that the uptake of particles in the various tissues can be widely heterogeneous, and further investigations are required to determine the impact of nanoparticles on pulmonary function and assess the risk for pulmonary embolism development.
Optimizing targeting ligand conjugation densities to increase targeting potential of nanotherapuetics is critical to efficacy and safety of these systems.7–12 It is clear that more in not always better.9, 12, 13 Here, we presented a method to tune the density of targeting ligands presented on the surface of the intravenous hemostatic nanoparticles, by producing them with blends of the GRGDS-NP100 and the nonfunctionalized pegylated polymer, PLGA-PLL-PEG. While we developed our methods independently in our lab, other groups have taken similar approaches previously.9, 54 Unsurprisingly, this method is more efficient, repeatable, and allows for greater control over peptide conjugation than other potential approaches such as tuning the stoichiometry or other reaction conditions of the conjugation chemistry.9 Interestingly the scrambled peptide, GRADSP, had a slightly higher conjugation efficiency in aqueous phase relative to GRGDS, while the GRADSP peptide had a slightly lower conjugation efficiency in organic relative to GRGDS, possibly suggesting an interaction of the additional hydrophobic proline residue during this reaction in the different phases.
One can imagine that functionalized polymer (PLGA-b-p(lys)-b-PEG-b-GRGDS) can be blended with the terminally pegylated polymer (PLGA-b-p(lys)-b-PEG), to accurately tune the peptide density. As documented by Gu et al.9 the “optimal” window can be extremely narrow, and further research is indicated to fully determine optimal blend and dose parameters. We have shown here that ROTEM is a potential assay for elucidating the effects of these changes in an in vitro system using whole blood, and that the same dose-relationship trends are observed in an in vivo model of lethal liver trauma.
CONCLUSION
Here, our challenge was to develop a method for reproducibly controlling nano-scale surface ligand conjugation and determine the impact of this change on hemostasis. We found using ROTEM the efficacious dose of GRGDS-NP100 was 10-fold lower than with previously recorded with GRGDS-NP1, and this result was recapitulated in vivo, after lethal liver trauma in the rat, showing efficacy at 8-fold lower concentration. Moving this technology toward clinical trials will require validation of the initial dose before proceeding. In vitro ROTEM testing with human blood may be one method for ascertaining an initial dosage estimate as well as provide a mechanism for optimizing surface ligand concentrations. This work demonstrates the potential utility of nanomedicine in addressing hemorrhage after lethal trauma. We have demonstrated the impact of, and the methods for, tightly control surface ligand density to optimize hemostatic efficacy.
Acknowledgments
Funding Sources
NIH Director’s New Innovator Award Grant, DP20D007338.
The authors would like to acknowledge R. Groynom, E. Shoffstall, M. Lashof-Sullivan for their contributions to this work and a NIH Director’s New Innovator Award Grant, DP20D007338.
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript. AJS performed, analyzed experiments and wrote the manuscript; AMS, LE, MV and ES performed experiments and analyzed data; JU and EL analyzed and interpreted data and contributed to the experimental designs.
References
- 1.Bhattacharyya S, Kudgus RA, Bhattacharya R, Mukherjee P. Pharm Res. 2011;28(2):237–59. doi: 10.1007/s11095-010-0318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Danhier F, Ansorena E, Silva JM, Coco R, Le Breton A, Preat V. J Control Release. 2012;161(2):505–22. doi: 10.1016/j.jconrel.2012.01.043. [DOI] [PubMed] [Google Scholar]
- 3.Prabhu P, Patravale V. J Biomed Nanotechnol. 2012;8(6):859–82. doi: 10.1166/jbn.2012.1459. [DOI] [PubMed] [Google Scholar]
- 4.Thibault G, Tardif P, Lapalme G. J Pharmacol Exp Ther. 2001;296(3):690–6. [PubMed] [Google Scholar]
- 5.Modery CL, Ravikumar M, Wong TL, Dzuricky MJ, Durongkaveroj N, Sen Gupta A. Biomaterials. 2011;32(35):9504–14. doi: 10.1016/j.biomaterials.2011.08.067. [DOI] [PubMed] [Google Scholar]
- 6.Beer JH, Springer KT, Coller BS. Blood. 1992;79(1):117–28. [PubMed] [Google Scholar]
- 7.Ng QK, Sutton MK, Soonsawad P, Xing L, Cheng H, Segura T. Mol Ther. 2009;17(5):828–36. doi: 10.1038/mt.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sancey L, Garanger E, Foillard S, Schoehn G, Hurbin A, Albiges-Rizo C, Boturyn D, Souchier C, Grichine A, Dumy P, Coll JL. Mol Ther. 2009;17(5):837–43. doi: 10.1038/mt.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu F, Zhang L, Teply BA, Mann N, Wang A, Radovic-Moreno AF, Langer R, Farokhzad OC. Proc Natl Acad Sci U S A. 2008;105(7):2586–91. doi: 10.1073/pnas.0711714105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garg A, Tisdale AW, Haidari E, Kokkoli E. Int J Pharm. 2009;366(1–2):201–10. doi: 10.1016/j.ijpharm.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gindy ME, Ji S, Hoye TR, Panagiotopoulos AZ, Prud’homme RK. Biomacromolecules. 2008;9(10):2705–11. doi: 10.1021/bm8002013. [DOI] [PubMed] [Google Scholar]
- 12.Olivier V, Meisen I, Meckelein B, Hirst TR, Peter-Katalinic J, Schmidt MA, Frey A. Bioconjug Chem. 2003;14(6):1203–8. doi: 10.1021/bc034077z. [DOI] [PubMed] [Google Scholar]
- 13.Fakhari A, Baoum A, Siahaan TJ, Le KB, Berkland C. J Pharm Sci. 2011;100(3):1045–56. doi: 10.1002/jps.22342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Modery-Pawlowski CL, Tian LL, Ravikumar M, Wong TL, Sen Gupta A. Biomaterials. 2013;34(12):3031–41. doi: 10.1016/j.biomaterials.2012.12.045. [DOI] [PubMed] [Google Scholar]
- 15.Coller BS, Kutok JL, Scudder LE, Galanakis DK, West SM, Rudomen GS, Springer KT. J Clin Invest. 1993;92(6):2796–806. doi: 10.1172/JCI116899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shoffstall AJ, Atkins KT, Groynom RE, Varley ME, Everhart LM, Lashof-Sullivan MM, Martyn-Dow B, Butler RS, Ustin JS, Lavik EB. Biomacromolecules. 2012;13(11):3850–7. doi: 10.1021/bm3013023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bertram JP, Williams CA, Robinson R, Segal SS, Flynn NT, Lavik EB. Sci Transl Med. 2009;1(11):11ra22. doi: 10.1126/scitranslmed.3000397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alving BM, Reid TJ, Fratantoni JC, Finlayson JS. Transfusion (Paris) 1997;37(8):866–76. doi: 10.1046/j.1537-2995.1997.37897424413.x. [DOI] [PubMed] [Google Scholar]
- 19.Coller BS, Springer KT, Beer JH, Mohandas N, Scudder LE, Norton KJ, West SM. J Clin Invest. 1992;89(2):546–55. doi: 10.1172/JCI115619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levi M, Friederich PW, Middleton S, de Groot PG, Wu YP, Harris R, Biemond BJ, Heijnen HF, Levin J, ten Cate JW. Nat Med. 1999;5(1):107–11. doi: 10.1038/4795. [DOI] [PubMed] [Google Scholar]
- 21.Okamura Y, Maekawa I, Teramura Y, Maruyama H, Handa M, Ikeda Y, Takeoka S. Bioconjug Chem. 2005;16(6):1589–96. doi: 10.1021/bc050178g. [DOI] [PubMed] [Google Scholar]
- 22.Okamura Y, Eto K, Maruyama H, Handa M, Ikeda Y, Takeoka S. Nanomedicine. 2010;6(2):391–6. doi: 10.1016/j.nano.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 23.Blajchman MA. Vox Sang. 2000;78(Suppl 2):183–6. [PubMed] [Google Scholar]
- 24.Blajchman MA. Transfus Clin Biol. 2001;8(3):267–71. doi: 10.1016/s1246-7820(01)00127-6. [DOI] [PubMed] [Google Scholar]
- 25.Davies AR, Judge HM, May JA, Glenn JR, Heptinstall S. Platelets. 2002;13(4):197–205. doi: 10.1080/0953371027000. [DOI] [PubMed] [Google Scholar]
- 26.Ravikumar M, Modery CL, Wong TL, Dzuricky M, Sen Gupta A. Bioconjug Chem. 2012;23(6):1266–1275. doi: 10.1021/bc300086d. [DOI] [PubMed] [Google Scholar]
- 27.Nishikawa K, Hagisawa K, Kinoshita M, Shono S, Katsuno S, Doi M, Yanagawa R, Suzuki H, Iwaya K, Saitoh D, Sakamoto T, Seki S, Takeoka S, Handa M. J Thromb Haemost. 2012;10(10):2137–48. doi: 10.1111/j.1538-7836.2012.04889.x. [DOI] [PubMed] [Google Scholar]
- 28.Ravikumar M, Modery CL, Wong TL, Gupta AS. Biomacromolecules. 2012;13(5):1495–502. doi: 10.1021/bm300192t. [DOI] [PubMed] [Google Scholar]
- 29.Kauvar DS, Lefering R, Wade CE. J Trauma. 2006;60(6 Suppl):S3–11. doi: 10.1097/01.ta.0000199961.02677.19. [DOI] [PubMed] [Google Scholar]
- 30.Holcomb JB, McMullin NR, Pearse L, Caruso J, Wade CE, Oetjen-Gerdes L, Champion HR, Lawnick M, Farr W, Rodriguez S, Butler FK. Ann Surg. 2007;245(6):986–91. doi: 10.1097/01.sla.0000259433.03754.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Champion HR, Bellamy RF, Roberts CP, Leppaniemi A. J Trauma. 2003;54(5 Suppl):S13–9. doi: 10.1097/01.TA.0000057151.02906.27. [DOI] [PubMed] [Google Scholar]
- 32.Johannigman J, Rhee P, Jenkins D, Holcomb J. Modern Combat Casualty Care. In: Mattox KL, Moore EE, Feliciano DV, editors. Trauma. 7. McGraw-Hill Professional; New York: 2012. [Google Scholar]
- 33.Kelly JF, Ritenour AE, McLaughlin DF, Bagg KA, Apodaca AN, Mallak CT, Pearse L, Lawnick MM, Champion HR, Wade CE, Holcomb JB. J Trauma. 2008;64(2 Suppl):S21–6. doi: 10.1097/TA.0b013e318160b9fb. discussion S26–7. [DOI] [PubMed] [Google Scholar]
- 34.Kauvar DS, Wade CE. Crit Care. 2005;9(Suppl 5):S1–9. doi: 10.1186/cc3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clifford CC. Mil Med. 2004;169(12 Suppl):8–10. 4. doi: 10.7205/milmed.169.12s.8. [DOI] [PubMed] [Google Scholar]
- 36.Morrison JJ, Rasmussen TE. Surg Clin North Am. 2012;92(4):843–58. doi: 10.1016/j.suc.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 37.Schrager JJ, Branson RD, Johannigman JA. Respir Care. 2012;57(8):1305–13. doi: 10.4187/respcare.01881. [DOI] [PubMed] [Google Scholar]
- 38.Lavik EB, Hrkach JS, Lotan N, Nazarov R, Langer R. J Biomed Mater Res. 2001;58(3):291–4. doi: 10.1002/1097-4636(2001)58:3<291::aid-jbm1019>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 39.Bertram JP, Jay SM, Hynes SR, Robinson R, Criscione JM, Lavik EB. Acta Biomater. 2009;5(8):2860–71. doi: 10.1016/j.actbio.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holcomb JB, McClain JM, Pusateri AE, Beall D, Macaitis JM, Harris RA, MacPhee MJ, Hess JR. J Trauma. 2000;49(2):246–50. doi: 10.1097/00005373-200008000-00010. [DOI] [PubMed] [Google Scholar]
- 41.Ryan KL, Cortez DS, Dick EJ, Jr, Pusateri AE. Resuscitation. 2006;70(1):133–44. doi: 10.1016/j.resuscitation.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 42.Matsuoka T, Wisner DH. J Trauma. 1996;41(3):439–45. doi: 10.1097/00005373-199609000-00009. [DOI] [PubMed] [Google Scholar]
- 43.Matsuoka T, Hildreth J, Wisner DH. J Trauma. 1995;39(4):674–80. doi: 10.1097/00005373-199510000-00010. [DOI] [PubMed] [Google Scholar]
- 44.Siller-Matula JM, Plasenzotti R, Spiel A, Quehenberger P, Jilma B. Thromb Haemost. 2008;100(3):397–404. [PubMed] [Google Scholar]
- 45.Probst RJ, Lim JM, Bird DN, Pole GL, Sato AK, Claybaugh JR. Journal of the American Association for Laboratory Animal Science : JAALAS. 2006;45(2):49–52. [PMC free article] [PubMed] [Google Scholar]
- 46.Nunez TC, Cotton BA. Curr Opin Crit Care. 2009;15(6):536–41. doi: 10.1097/MCC.0b013e328331575b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martinowitz U, Zaarur M, Yaron BL, Blumenfeld A, Martonovits G. Mil Med. 2004;169(12 Suppl):16–8. 4. doi: 10.7205/milmed.169.12s.16. [DOI] [PubMed] [Google Scholar]
- 48.Hess JR, Holcomb JB. Transfus Med. 2008;18(3):143–50. doi: 10.1111/j.1365-3148.2008.00855.x. [DOI] [PubMed] [Google Scholar]
- 49.Grislain L, Couvreur P, Lenaerts V, Roland M, Deprezdecampeneere D, Speiser P. Int J Pharm. 1983;15(3):335–345. [Google Scholar]
- 50.Martin FJ, Melnik K, West T, Shapiro J, Cohen M, Boiarski AA, Ferrari M. Drugs R D. 2005;6(2):71–81. doi: 10.2165/00126839-200506020-00002. [DOI] [PubMed] [Google Scholar]
- 51.Decuzzi P, Godin B, Tanaka T, Lee SY, Chiappini C, Liu X, Ferrari M. J Control Release. 2010;141(3):320–7. doi: 10.1016/j.jconrel.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 52.Guo G, Kang Y, Li X, Cai ZH, Chen JH, Wang G, Pei GX. Chin J Traumatol. 2007;10(4):237–41. [PubMed] [Google Scholar]
- 53.Knudson MM, Ikossi DG, Khaw L, Morabito D, Speetzen LS. Ann Surg. 2004;240(3):490–6. doi: 10.1097/01.sla.0000137138.40116.6c. discussion 496–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ali MM, Zale SE. US Patent 8,236,330. Cancer cell targeting using nanoparticles. 2007 Mar 30;