Summary
The human fungal pathogen Candida albicans has at least two types of morphological transitions: white to opaque cell transitions and yeast to hyphal transitions. Opaque cells have historically not been known to undergo filamentation under standard filament-inducing conditions. Here we find that Bcr1 and its downstream regulators Cup9, Nrg1 and Czf1 and the cAMP-signaling pathway control opaque cell filamentation in C. albicans. We have shown that deletion of BCR1, CUP9, NRG1 and CZF1 results in opaque cell filamentation under standard culture conditions. Disruption of BCR1 in white cells has no obvious effect on hyphal growth, suggesting that Bcr1 is an opaque-specific regulator of filamentation under the conditions tested. Moreover, inactivation of the cAMP-signaling pathway or disruption of its downstream transcriptional regulators, FLO8 and EFG1, strikingly attenuates filamentation in opaque cells of the bcr1/bcr1 mutant. Deletion of HGC1, a downstream gene of the cAMP-signaling pathway encoding G1 cyclin-related protein, completely blocks opaque cell filamentation induced by inactivation of BCR1. These results demonstrate that Bcr1 regulated opaque cell filamentation is dependent on the cAMP-signaling pathway. This study establishes a link between the white-opaque switch and the yeast-filamentous growth transition in C. albicans.
Keywords: Candida albicans, Bcr1, opaque cell filamentation, cAMP signaling pathway
Introduction
Fluctuations in environmental conditions often lead to increased phenotypic plasticity and functional robustness of living organisms. Candida albicans, a major fungal pathogen of humans, has two known phenotypic transition systems, the “white” and “opaque” morphological switch, and the “yeast” and “filamentous” growth transition (Huang, 2012). Integration of these two systems could lead to increases in phenotypic plasticity and thus considerably diversify the niches that this pathogen colonizes.
White and opaque cell types of C. albicans display a number of distinct features including colony and cellular appearances and gene expression profiles (Anderson and Soll, 1987; Tuch et al., 2010; Huang, 2012). White cells are relatively round and small and form white colonies, while opaque cells are bean-shaped and form gray or “opaque” colonies on nutrient-agar surfaces; the cell wall surface of opaque cells is pimpled (Anderson and Soll, 1987). White and opaque cells also differ in mating competency and virulence (Huang, 2012). In order to mate, white cells must first switch to opaque, and only opaque cells have been reported to mate efficiently (Miller and Johnson, 2002).
Numerous studies indicate that genetic and epigenetic regulation of gene expression and environmental cues control white-opaque switching in C. albicans (Huang, 2012). A variety of overlapping environmental cues and genes regulate white-opaque switching as well as filamentous growth (Huang, 2012). The ability to undergo filamentation is a defining feature of virulence in C. albicans (Whiteway and Bachewich, 2007; Lo et al., 1997); filamentous cells are more invasive and better at tissue penetration than yeast-form cells. The central pathway controling this process is the cAMP-signaling pathway (Biswas et al., 2007; Whiteway and Bachewich, 2007; Huang, 2012). Inactivation of the cAMP pathway by deletion of CYR1, encoding the only adenylyl cyclase in C. albicans, blocks filamentous growth under a variety of conditions (Rocha et al., 2001). Consistently, the downstream transcriptional regulators of the cAMP pathway, Flo8 and Efg1, are required for filamentous development and virulence (Lo et al., 1997; Cao et al., 2006; Huang, 2012). Hgc1, a G1 cyclin-related protein, is enriched in filamentous cells and regulates hyphal morphogenesis; HGC1 is transcriptionally regulated by the cAMP pathway (Zheng and Wang, 2004). The HGC1 transcript is not detectable in the cyr1/cyr1, efg1/efg1 or flo8/flo8 mutants under filament-inducing conditions (Zheng and Wang, 2004; Cao et al., 2006). pH signaling and MAPK pathways are also involved in the regulation of hyphal development (Biswas et al., 2007; Huang, 2012). Acidic pH conditions inhibit filamentous growth, while neutral and basic pH conditions favor filamentation (Biswas et al., 2007; Huang, 2012). The general transcriptional repressors Tup1, Nrg1 and Rfg1 are repressors of the yeast-to-filament transition in C. albicans (Braun and Johnson, 1997; Braun et al., 2001; Kadosh and Johnson, 2001; Murad et al., 2001; Huang, 2012). Deletion of TUP1, NRG1 or RFG1 leads to constitutive filamentous growth under a number of different culture conditions. Tup1 also plays a role in white-opaque switching and the regulation of phase-specific gene expression (Zhao et al., 2002; Park and Morschhauser, 2005a). Filamentous cells formed by the MTLα tup1/tup1 mutant are mating-competent, indicating that they are functionally opaque (Park and Morschhauser, 2005a). Nrg1 is a zinc-finger transcription factor that is typically associated with Tup1 (Huang, 2012). Rfg1 is an HMG-domain protein that transcriptionally regulates filamentous genes via Tup1-dependent or independent pathways (Kadosh and Johnson, 2005).
Both MTL homozygous and MTL heterozygous white cells can undergo filamentation under standard filament-inducing laboratory conditions. Opaque cells, on the other hand, have not been known to form filaments under these conditions (Anderson et al., 1989), although they have been found to undergo filamentation when grown in a perfusion chamber or on a monolayer of human epithelial skin cells (Anderson et al., 1989). Therefore, once the MTL homozygous cells differentiate into the specialized opaque type, they lose the ability to filament under standard laboratory culture conditions and acquire a number of opaque-specific attributes, such as the ability to mate and colonize the skin (Huang, 2012). Although several studies have focused on the mechanisms of filamentous growth regulation in MTL heterozygous cells (or white cells) of C. albicans (see Whiteway and Bachewich, 2007 for a review), little is known about the molecular mechanisms involved in the regulation of filament development in opaque cells.
Recently, Si et al. (2013) have found that the cAMP signaling pathway and some general transcriptional regulators including Nrg1, Ume6 and Rfg1 are involved in the regulation of opaque cell filamentation under several cultural conditions (Si et al., 2013). These regulators may play a general activating or repressing role in the regulation of filamentation both in white cells and in opaque cells. In this study, we have identified Bcr1 as a unique and key regulator of opaque cell filamentation.
Although opaque cells can not undergo filamentation under standard filament-inducing laboratory conditions (Anderson et al., 1989; Si et al., 2013), Si et al. (2013) have reported that SOR medium (synthetic complete dextrose medium supplemented with 1 M sorbitol) and MIN medium (synthetic medium lacking amino acids) can efficiently induce opaque cell filamentation (Si et al., 2013). The growth of C. albicans cells with the latter two media may represent two “stressful” conditions (high osmotic pressure for the SOR medium and starvation stress for the MIN medium, respectively). In this study, we have identified two pathways that control filament development in the opaque cells of C. albicans under standard laboratory inducing conditions: a positive and a negative pathway. The positive pathway is the cAMP-signaling pathway, in which the adenylyl cyclase Cyr1, the transcription factors Flo8, Efg1, and the G1 cyclin-related protein Hgc1, are required for opaque cell filamentation. These findings are consistent with the recently published paper (Si et al., 2013). We have discovered the negative pathway mediated by the Bcr1 transcription factor and its downstream regulators, including Nrg1, Cup9 and Czf1. We find that the central regulator, Bcr1, is an opaque-phase-specific repressor of filamentation in C. albicans under the conditions tested.
Results
Identification of Bcr1, Cup9 and Czf1 as opaque cell filamentation regulators by screening a library of transcription factor deletion mutants
Opaque cells of C. albicans are unable to form hyphae under most standard laboratory conditions (Anderson et al., 1989 and our unpublished data). To identify the regulators of opaque cell filamentation, we screened a library containing about 160 transcription factor gene deletion mutants, which were homozygous at the MTL locus (Du et al., 2012a). The white and opaque cells of the mutants were plated onto Lee’s glucose medium plates and incubated at 25°C in air for 5 days. The WT control and most mutants of the library formed typical “white” (white and smooth) and/or “opaque” (red, rough and flat) colonies on Lee’s glucose medium containing the red dye phloxine B. The white colonies of two strains (cup9/cup9 and rfg1/rfg1) were wrinkled. The opaque colonies of three mutants showed obvious wrinkled (czf1/czf1) or fuzzy (bcr1/bcr1 and cup9/cup9) appearances (Fig. 1A). The three mutants could also undergo filamentation in liquid cultures at 25°C (Fig. 1B). Of the three mutants identified, the opaque cells of the bcr1/bcr1 mutant were able to undergo the most robust filamentation (hereafter, we will refer to these cells as filamentous opaque cells), while its white cells remained in yeast form under the same culture condition, suggesting that Bcr1 is an opaque-specific regulator of filamentation. The white colonies of the cup9/cup9 mutant were wrinkled, while the opaque colonies showed filamentous growth at their periphery and top regions. The cellular morphology verified that both white and opaque cells were able to undergo filamentation, indicating that Cup9 regulates filamentous growth in opaque and white cells. The opaque colonies of the czf1/czf1 mutant formed slightly wrinkled colonies by day 5, and became more wrinkled and fuzzy at day 10. Aerial hyphae were also observed in the opaque colonies of the czf1/czf1 mutant. The white colonies remained smooth and contained only yeast form cells after incubation for 5 to 10 days (Fig. 1A). These results suggest that Czf1 is a relatively weak regulator of opaque cell filamentation dependent on culture time period or cell aging.
Fig. 1.
Identification of Bcr1, Cup9 and Czf1 as opaque cell filamentation regulators by screening a library of transcription factor deletion mutants. A. Colony morphology of the WT (SN250α), bcr1/bcr1, cup9/cup9 and czf1/czf1 mutants. Cells were plated onto Lee’s glucose medium plates and cultured at 25°C for 5 days. Incubation of the czf1/czf1 mutant was extended to 10 days. B. Cellular images of the growth cultures in liquid Lee’s glucose medium at 25°C. Opaque colonies were inoculated into liquid medium. Cells were grown to the mid-exponential phase and imaged. Scale bar, 10 μm.
Spontaneous switching between the white and opaque cell types is an important feature of the white-opaque transition in C. albicans. Indeed, both the white and filamentous opaque types of the bcr1/bcr1, cup9/cup9 and czf1/czf1 mutants were capable of switching back and forth at 25°C. The white-to-opaque switching frequencies of these mutants are shown in Table S1.
Bcr1 is a key opaque-specific regulator of filamentation
Of the mutants identified in our screen, the bcr1/bcr1 mutant was able to undergo the most robust opaque cell filamentation. Therefore, we further extensively investigated the role of Bcr1 in this process. The bcr1/bcr1 mutant formed two types of colonies: typical “white” and “fuzzy” red filamentous opaque colonies (Fig. 2A). The cellular morphology indicated that the WT white colonies contained white yeast cells, while WT opaque colonies formed typical elongated opaque cells. The white colonies of the bcr1/bcr1 mutant also contained only white yeast cells, while the red “fuzzy” colonies were composed of highly filamentous cells (Fig. 2A). One of the defining features of opaque cells is their uniquely pimpled cell surface (Anderson and Soll, 1987). To confirm that the cell identity of the “fuzzy” colonies was opaque, the cell surface was examined by scanning electron microscopy (SEM). As shown in Fig. S1, the surface of some, but not all, filamentous cells of the bcr1/bcr1 mutant contained obvious pimples as did that of the WT opaque, while the white cells of both the WT and bcr1/bcr1 mutant were smooth.
Fig. 2.
Deletion of BCR1 results in filamentous and invasive growth of opaque cells. A. Colony and cellular images of the WT (SN250α), bcr1/bcr1 mutant and bcr1/bcr1+BCR1p-BCR1 reconstituted strain. Wh, white phase; op, opaque phase. Scale bar, 10 μm. B. Invasive growth of the WT (SN250α), bcr1/bcr1 mutant and bcr1/bcr1+BCR1p-BCR1 reconstituted strain. White and opaque cells (~6 × 104) were spotted on Lee’s glucose medium plates and incubated at 25°C for 3 days. C. Expression of opaque (WOR1 and OP4) or filamentous (HWP1 and ECE1) enriched genes in the bcr1/bcr1 mutant analyzed by Northern blot.
To confirm that deletion of BCR1 in C. albicans resulted in robust filamentation of opaque cells, a fragment containing the BCR1 gene was introduced back into the genome and integrated into the original BCR1 locus. We found that either white or opaque cells of the bcr1/bcr1+BCR1p-BCR1 reconstituted strain showed similar colony and cellular phenotypes to those of the WT (Fig. 2A). The phenotypes of filamentation in opaque cells were verified in an MTLa/a strain of an independent bcr1/bcr1 mutant (CJN702) (Nobile and Mitchell, 2005). To verify that opaque cell filamentation of the bcr1/bcr1 mutant is not specific to Lee’s medium, we also performed filamentation experiments in YPD, Spider, and synthetic complete defined (SCD) medium. As shown in Fig. S2, opaque cells of the bcr1/bcr1 mutant were able to undergo obvious filamentation in all three media, while white cells remained in the yeast form. These results demonstrate that Bcr1 functions as an opaque-specific repressor of filamentous growth in C. albicans.
The bcr1/bcr1 mutant filamentous opaque cells undergo invasive growth
Invasive growth of C. albicans is important for invading and colonizing host tissues (Biswas et al., 2007; Huang, 2012). To investigate whether the bcr1/bcr1 mutant could undergo invasive growth, white and opaque cells of the WT, bcr1/bcr1 mutant and bcr1/bcr1+BCR1p-BCR1 reconstituted strain were spotted onto Lee’s glucose medium plates. After 3 days of growth at 25°C, the plates were imaged before and after being washed with water. As shown in Fig. 2B, only filamentous opaque cells of the bcr1/bcr1 mutant were able to undergo strong invasive growth. White cells of the three strains and opaque cells of the WT and BCR1 complemented strain were almost completely washed away.
We next tested whether the filamentous cells of the bcr1/bcr1 mutant contain a combination of filamentous and opaque features. Northern blot analysis showed that both opaque-enriched and filamentous-enriched genes (OP4, WOR1, HWP1 and ECE1) were exclusively expressed in filamentous cells of the bcr1/bcr1 mutant (Fig. 2C) (Birse et al., 1993; Morrow et al., 1993; Srikantha et al., 1995; Staab et al., 1999; Huang et al., 2006; Srikantha et al., 2006; Zordan et al., 2006).
Czf1 and Cup9 are opaque cell filamentation regulators downstream of Bcr1
CUP9 is downregulated in response to hyphal growth inducers (Nantel et al., 2002; Garcia-Sanchez et al., 2005). Deletion of CUP9 resulted in filamentation in white cells as well as in opaque cells (Fig. 1), suggesting that Cup9 is a general repressor of filamentation. The surface of filamentous opaque cells of the mutant was obviously pimpled, while the surface of white cells was smooth (Fig. S1). We found that Bcr1 binds to the promoter of CUP9 both in white and opaque cells (Fig. 3).
Fig. 3.

ChIP-PCRs of the promoter regions of Bcr1-regulated genes. EFG1(b1), Bcr1 binding site 1 (−7188 to −7422); EFG1(b2), Bcr1 binding site 2 (−2220 to −1886). PCR products were detected after 27 cycles. The primers were designed according to the Bcr1 binding sites reported by Nobile et al., 2012. The ChIP-PCR of the HGC1 promoter, which was known not to be bound by Bcr1 (Nobile et al., 2012), served as the negative control. White and opaque cells of the strain CJN1787a (MTLa/α Δ::SAT1 BCR1/BCR1-Myc) were used for the experiments.
Czf1 is a transcriptional regulator of white-opaque switching and hyphal development under certain conditions, such as embedded cultures (Brown et al., 1999; Zordan et al., 2007). Deletion of CZF1 had a moderate effect on the induction of opaque cell filamentation, but not on white cell filamentation (Fig. 1). ChIP-PCR experiments demonstrated that Bcr1 binds to the promoter of CZF1 both in white and in opaque cells, although the binding signal in white cell was relatively weak (Fig. 3). Consistently, Bcr1 also binds to the promoter of CZF1 in MTL heterozygous cells (Nobile et al., 2012). This suggests that Czf1 is downstream of Bcr1 in the regulation of opaque cell filamentation.
Filamentous opaque cells of the bcr1/bcr1 and cup9/cup9 mutants are mating competent
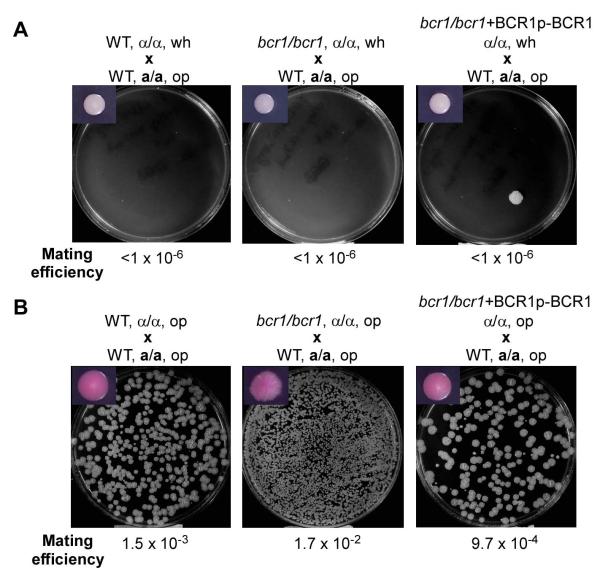
Mating competency is one of the most important characteristics of opaque cells, which mate much more efficiently than white cells (Miller and Johnson, 2002). To further confirm that the filamentous cells of the bcr1/bcr1 and cup9/cup9 mutants were functionally opaque, a quantitative mating assay was performed. WT opaque cells (MTLa/a) were used as the mating tester. We found that the mating efficiency of filamentous opaque cells was over 1,000 fold higher than that of the white cell counterparts of the bcr1/bcr1 and cup9/cup9 mutants (Fig. 4 and Table S2). As expected, the mating efficiency of opaque cells was also 1,000 times higher than that of white cells of the WT and reconstituted bcr1/bcr1+BCR1p-BCR1 reference strains (Fig. 4). Taken together, this data strongly suggests that the filamentous opaque cells are mating-competent.
Fig. 4.
Quantitative mating assay indicates that filamentous opaque cells of the bcr1/bcr1 mutant are mating-competent. MTLa/a and MTLα/α cells in white or opaque phase were mixed and cultured on Lee’s glucose plates for 4 days at 25°C. About 1 × 106 cells of each mating mixture were resuspended and plated onto selectable SCD plates. A representative selectable plate (depleted with uridine and arginine) of each cross was imaged. The mating efficiencies were calculated and shown below the images. A colony image indicating the phase of experimental cells used for mating was also shown (top left corner). A. Mating between the WT opaque tester (GH1012, MTLa/a, ura3-) and white cells of the WT (MTLα/α, arg4-), bcr1/bcr1 mutant (MTLα/α, arg4-) and bcr1/bcr1+BCR1p-BCR1 reconstituted strain (MTLα/α, arg4-). B. Mating between the WT opaque tester (GH1012, MTLa/a, ura3-) and opaque cells of the WT (MTLα/α, arg4-), bcr1/bcr1 mutant (MTLα/α, arg4-) and bcr1/bcr1+BCR1p-BCR1 reconstituted strain (MTLα/α, arg4-).
High temperature (37°C) induces filamentous opaque cells of the bcr1/bcr1 and cup9/cup9 mutants to switch to the white phase
Opaque cells are sensitive to temperature changes. In vitro experiments have shown that high temperature (37°C) induces a mass conversion from the opaque to white phase in WT strains (Anderson and Soll, 1987) (Fig. 5). To investigate whether the filamentous opaque phenotypes of the bcr1/bcr1 and cup9/cup9 mutants are unstable at high temperature, the cells were plated onto nutrient agar plates at 37°C for a series of time periods (0 to 5 days), and then transferred to 25°C for an additional incubation (5 to 0 days). The colony and cellular morphology was examined after a total culture time of 5 days at both temperatures. As expected, when cultured at 25°C, the majority of colonies of either the WT or the two mutants were opaque or filamentous opaque. Most colonies of the WT became white after incubation at 37°C for 2 days and 25°C for 3 days (Fig. 5), suggesting that 37°C induced a mass conversion to white. At this culture condition, most colonies of the bcr1/bcr1 and cup9/cup9 mutant strains become white, white-sectored, or mixed. Extended treatments (3 and 5 days) at 37°C led to a gradual increase in white or white filamentous growth (Fig. 5). The bcr1/bcr1 mutant cultures of 2 or 3 days treatment at 37°C formed mostly white colonies, containing a majority of yeast cells and a minority of filaments. After incubating at 37°C for 5 days, the bcr1/bcr1 mutant formed wrinkled filamentous colonies. It is clear that high temperature induced an opaque to white (yeast form) switch followed by filamentation of the switched white cells in the WT and bcr1/bcr1 mutant (Fig. 5). Filamentous white and opaque colonies of the bcr1/bcr1 and cup9/cup9 mutants were morphologically distinct. Filamentous white colonies of the WT, bcr1/bcr1 and cup9/cup9 mutants were wrinkled, while opaque filamentous colonies of the bcr1/bcr1 and cup9/cup9 mutants were fuzzy. To confirm their cell phase, mating competencies of the filamentous white and opaque cells were tested (data not shown). This data also suggests that, similar to the WT opaque cells, high temperature induced the switch from filamentous opaque cells to white (or filamentous white) phase in the bcr1/bcr1 and cup9/cup9 mutants (Fig. 5). These results provide additional evidence that the filamentous opaque cells of the mutants are functionally opaque.
Fig. 5.
High temperature (37°C) induces filamentous opaque cells switching to white phase. Opaque cells of the WT (SN250α), bcr1/bcr1 and cup9/cup9 mutants were plated onto Lee’s glucose plates and cultured at the indicated temperatures for 5 days. Colony morphology of the WT (SN250α), bcr1/bcr1 and cup9/cup9 mutants is shown. Wh, white colonies; Op, opaque colonies; Mix, white and opaque mixed or sectored colonies.
Nrg1 regulates both white and opaque cell filamentation
We did not identify Nrg1 as a regulator of opaque cell filamentation in our library screen because the nrg1/nrg1 mutant was able to undergo hyper-filamentation under the culture condition of our screen, which interfered with our ability to assess the transition between white and opaque cell phases. We have found that Bcr1 binds to the promoter of NRG1 in white and opaque cells of an MTL homozygous strain (Fig. 3). Consistently, Nobile et al. also recently reported that Bcr1 binds to the promoter of NRG1 in MTLa/α cells (Nobile et al., 2012). We, therefore, predicted that Nrg1 may also be involved in the regulation of opaque cell filamentation. To test this hypothesis, we deleted NRG1 in GH1013, a characterized MTLa/a strain (Huang et al., 2009). The nrg1/nrg1 mutant spontaneously switched between two cell types of filamentous colonies (white and opaque) on Lee’s glucose medium plates (pH 6.8, Fig. 6). Similar to the bcr1/bcr1 and cup9/cup9 mutants, the surface of opaque cells of the nrg1/nrg1 mutant was also pimpled, while the surface of its respective white cells was smooth (Fig. S1). A quantitative mating assay further confirms that the filamentous opaque cells of the nrg1/nrg1 mutants are functionally opaque (Table S2). These results indicate that Nrg1 is also involved in opaque cell filamentation.
Fig. 6.
Regulation of white and opaque cell filamentation by Bcr1, Cup9, Nrg1, Czf1 and Rfg1. White and opaque cells were plated onto Lee’s glucose medium with different pH (4.0 to 8.0). The colony and cellular images of the WT (SN250α), bcr1/bcr1, cup9/cup9, nrg1/nrg1 and rfg1/rgf1 were taken after incubation at 25°C for 5 days. The czf1/czf1 mutant was incubated at 25°C for 8 days and imaged. Wh, white colonies; Op, opaque colonies. Cellular morphology of the mutants is shown in Fig. S3.
Regulation of white and opaque cell filamentation by Bcr1, Cup9, Nrg1, Czf1 and Rfg1 under pH 4.0 to 8.0 at 25°C
With the exception of Bcr1, the transcription factors Cup9, Nrg1, Czf1 and Rfg1 have been well characterized in the regulation of white cell filamentation (Nantel et al., 2002; Garcia-Sanchez et al., 2005; Biswas et al., 2007; Whiteway and Bachewich, 2007; Huang, 2012). It has been shown that Bcr1 directly binds to the promoter regions of CUP9, NRG1, CZF1 and RFG1 genes in MTLa/α cells of C. albicans (Nobile et al., 2012). Chromatin immunoprecipitation PCR (ChIP-PCR) confirmed that Bcr1 also binds to the promoter regions of CUP9, NRG1 and CZF1 in MTL homozygous strains (Fig. 3). pH is a critical environmental cue regulating white cell filamentation in C. albicans (Davis, 2003; Biswas et al., 2007). We, therefore, tested the filamentous growth ability of white and opaque cells of the bcr1/bcr1, cup9/cup9, nrg1/nrg1, czf1/czf1 and rfg1/rfg1 mutants under different pH conditions. As expected, both white and opaque cells of the WT control did not form hyphae under all pH conditions tested (Fig. 6). White cells of the bcr1/bcr1 mutant also did not undergo filamentation, while opaque cells of the bcr1/bcr1 mutant were able to undergo robust filamentation under all pH conditions (pH 4.0 to 8.0). White cells of the cup9/cup9 mutant formed filamentous colonies at pH 6.8 and 8.0, but not at acidic pHs (4.0 to 6.0). Although the opaque colonies of the cup9/cup9 mutant contained less filamentous cells at acidic pHs (4.0 to 6.0) compared to pH 6.8 and 8.0, acidic pH did not completely block filamentation in opaque cells. White cells of the nrg1/nrg1 mutant developed obvious filamentous colonies at pH 6.0 to 8.0, but not at pH 4.0 and 5.0. Similar to the bcr1/bcr1 mutant, opaque cells of the nrg1/nrg1 mutant formed robust filamentous colonies at pH4.0 to 8.0. White cells of the czf1/czf1 mutant only grew as yeast form, while opaque cells of the czf1/czf1 mutant were able to undergo moderate filamentation at all pH conditions. Deletion of RFG1 induced filamentous development of white cells, but had no obvious effect on opaque cell filamentation. The colony and cellular morphologies of the mutants are shown in Fig. 6 and Fig. S3, respectively. The robustness of filamentation of all the mutants tested at pH 4.0 to 8.0 is summarized in Table 1. Taken together, these results demonstrate that opaque cell filamentation is less dependent on the environmental pH signal than white cell filamentation.
Table 1.
pH regulates white and opaque cell filamentation.
| Strain |
Cell
phase |
pH
|
||||
|---|---|---|---|---|---|---|
| 4.0 | 5.0 | 6.0 | 6.8 | 8.0 | ||
| WT | Wh | − | − | − | − | − |
| Op | − | − | − | − | − | |
|
| ||||||
| bcr1/bcr1 | Wh | − | − | − | − | − |
| Op | +++++ | +++++ | +++++ | +++++ | +++++ | |
|
| ||||||
| cup9/cup9 | Wh | − | − | − | +++ | ++++ |
| Op | ++ | ++ | +++ | ++++ | ++++ | |
|
| ||||||
| nrg1/nrg1 | Wh | + | ++ | ++++ | +++++ | +++++ |
| Op | ++++ | +++++ | +++++ | +++++ | +++++ | |
|
| ||||||
| czf1/czf1 | Wh | − | − | − | − | − |
| Op | ++ | ++ | ++ | ++ | ++ | |
|
| ||||||
| rfg1/rfg1 | Wh | +++ | ++ | + | ++++ | ++++ |
| Op | − | − | − | − | − | |
Cells were grown on Lee’s glucose plates of pH4.0 to 8.0 as indicated for 5 days at 25°C. The robustness of filamentation was evaluated by examining the percentage and length of filamentous cells in a representative colony. Wh, white cells; op, opaque cells. “−” indicated no filamentous cells were observed. The number of “+” indicated the robustness of filamentation.
To verify the cell identity of white and opaque cells of the mutants tested in the pH experiments (Fig. 6 and Fig. S3), the mating competency was tested under these conditions (data not shown). Opaque or filamentous opaque cells of all the mutants were mating-competent, while white or filamentous white cells of all the mutants were mating-incompetent (data not shown).
cAMP signaling is required for Bcr1-controlled filamentation of opaque cells
cAMP signaling regulates not only filamentous growth but also white-opaque switching in C. albicans (Rocha et al., 2001; Huang et al., 2010). Deletion of CYR1, encoding the only adenylyl cyclase in C. albicans, completely blocks filamentous growth under a variety of culture conditions (Rocha et al., 2001). Inactivation of the cAMP pathway leads to dramatically reduced white to opaque switch frequencies in GlcNAc containing medium, while activation of this pathway by deletion of PDE2, encoding a high affinity cyclic nucleotide phosphodiesterase, results in hypersensitivity to GlcNAc in C. albicans (Huang et al., 2010). We next investigated whether cAMP signaling is required for filamentation in opaque cells. Opaque cells of the WT and cyr1/cyr1 mutant did not undergo filamentous growth (Fig. 7A), suggesting that deletion of CYR1 does not increase filamentation. A cyr1/cyr1 bcr1/bcr1 double mutant was then generated. We found that, compared with the bcr1/bcr1 single mutant, filamentation of opaque cells of the cyr1/cyr1 bcr1/bcr1 double mutant was reduced (Fig. 7A). Supplementation of dibutyryl-cAMP (dbcAMP) to the cyr1/cyr1 bcr1/bcr1 double mutant increased filamentation of opaque cells. However, equal amounts of dbcAMP added to the cyr1/cyr1 opaque culture had no obvious effects on filamentous growth (Fig. 7B). These results indicate that cAMP signaling plays a critical role in Bcr1-controlled filamentation in opaque cells.
Fig. 7.
cAMP signal plays a major role in Bcr1-regulated filamentation of opaque cells. A. Opaque cells of the cyr1/cyr1 bcr1/bcr1double mutant showed reduced filamentation. Cells were cultured on Lee’s glucose medium plates. B. Dibutyryl-cAMP (dbcAMP) recovered the robust filamentation ability of opaque cells of the bcr1/bcr1 cyr1/cyr1 double mutant. The liquid cultures of the cyr1/cyr1 mutant and bcr1/bcr1 cyr1/cyr1 double mutant in the presence and absence of dbcAMP or not were incubated at 25°C for 36 hours with shaking.
Role of Efg1, Flo8 and Hgc1 in opaque cell filamentation of the bcr1/bcr1 mutant
The cAMP signaling downstream regulators, Efg1 and Flo8, function coordinately to control filamentous growth in C. albicans (Noffz et al., 2008; Cao et al., 2006; Du et al., 2012a). They also play a central role in the regulation of white-opaque switching (Huang, 2012). To test their roles in filamentation of opaque cells, we generated the efg1/efg1 bcr1/bcr1 and flo8/flo8 bcr1/bcr1 double mutants in C. albicans. Compared with the bcr1/bcr1 single mutant, the ability to undergo filamentous growth in the efg1/efg1 bcr1/bcr1 double mutant opaque cells was dramatically reduced (Fig. 8A), although extended culturing (for more than 7 days) induced weak filamentation. Since deletion of FLO8 “locks” C. albicans in the white phase in Lee’s glucose medium, and since the mutant of flo8/flo8 can only form opaque colonies in Lee’s GlcNAc medium (Du et al., 2012a), we grew the flo8/flo8 mutant and flo8/flo8 bcr1/bcr1 double mutant in both media. As shown in Fig. 8B, neither the flo8/flo8 single mutant nor the flo8/flo8 bcr1/bcr1 double mutant was able to filament on Lee’s glucose medium. On Lee’s GlcNAc medium plates, the flo8/flo8 single mutant formed opaque colonies containing only yeast form cells, while the flo8/flo8 bcr1/bcr1 double mutant formed weakly wrinkled colonies (Fig. 8B). Although the filamentation ability of the flo8/flo8 bcr1/bcr1 double mutant was weaker than that of the bcr1/bcr1 single mutant, cellular morphology confirmed the existence of typical opaque and filamentous cells in the wrinkled colonies (Fig. 8B). These results suggest that Efg1 and Flo8 contribute to filamentation of opaque cells, yet inactivation of these two transcription factors does not completely block Bcr1-controlled filamentous growth.
Fig. 8.
Roles of Efg1, Flo8, Hgc1, Tec1, and Brg1, in Bcr1-controlled filamentation in opaque cells. Cells were plated onto Lee’s glucose or Lee’s GlcNAc plates and cultured at 25°C in air for 5 days. Colonies were imaged at the 5th day after plating and cellular morphology of a representative colony is shown. A. Filamentous growth of opaque cells of the WT (SN250α), bcr1/bcr1, efg1/efg1, bcr1/bcr1 efg1/efg1 double mutant, hgc1/hgc1, bcr1/bcr1 hgc1/hgc1 double mutant, tec1/tec1, bcr1/bcr1 tec1/tec1 double mutant, brg1/brg1, bcr1/bcr1 brg1/brg1 double mutant, cph1/cph1 and cph1/cph1 brg1/brg1 double mutant on Lee’s glucose plates. Scale bar, 10 μm. B. Filamentous growth of the bcr1/bcr1 flo8/flo8 double mutant on Lee’s glucose and Lee’s GlcNAc plates.
Hgc1 is transcriptionally regulated by Efg1 and Flo8 and is downstream of the cAMP-signaling pathway (Zheng and Wang, 2004; Cao et al., 2006). We, therefore, generated an hgc1/hgc1 bcr1/bcr1 double mutant. While opaque cells of the bcr1/bcr1 single mutant were able to undergo robust filamentation, hyphal development of the hgc1/hgc1 bcr1/bcr1 double mutant opaque cells was completely blocked (Fig. 8A), suggesting that Hgc1 is essential for opaque cell filamentation induced by inactivation of Bcr1.
Overexpression of white cell hyphal-inducing gene HGC1, BRG1, and TEC1, promotes opaque cell filamentation
Hgc1, Flo8, Efg1, Tec1 and Brg1 are well characterized regulators of white cell filamentation (Homann et al., 2009; Du et al., 2012b; Huang, 2012). Tec1, Brg1 and Bcr1 are members of the master transcriptional network of biofilm formation and bind to the promoters of each other (Nobile et al., 2012). To identify activators of opaque cell filamentation, we overexpressed HGC1, FLO8, EFG1, TEC1 and BRG1 in white and opaque cells of a WT strain (GH1013, a derivative of SC5314) using a tetracycline-controlled promoter (TETp) (Park and Morschhauser, 2005b; Du et al., 2012a; Du et al., 2012b). The activity of the TET-On promoter of the plasmid pNIM1 is considerably lower in opaque cells because the Tet-On system is based on expression of cartTA from the ADH1 promoter, which is less active in opaque cells than in white cells. To increase the ectopic expression levels of these genes, a copy of the cartTA gene was first integrated into the OP4 (an opaque cell enriched gene) locus of GH1013 (WT+OP4p-cartTA). To test whether this engineered overexpression system works, we examined the expression of GFP protein with fluorescence microscopy assays. Under the inducing condition, visible GFP fluorescence could only be observed in a few white cells, but not opaque cells, of the WT+pNIM1 strain. However, the expression level of the GFP reporter in opaque cells of the WT+OP4p-cartTA+pNIM1 strain was significantly higher than that in opaque cells of the WT+pNIM1 strain under the inducing condition (Fig. 9A). Cells of the overexpression strains were grown on Lee’s glucose plates with or without 40 μg ml−1 doxycycline at 25°C. 40μg ml−1 doxycycline was used for inducing ectopic expression because high concentrations (>50 μg ml−1) of this chemical could have an inhibitory effect on filamentation (Park and Morschhauser, 2005b). As shown in Fig. 9B, overexpression of HGC1, TEC1, or BRG1 in opaque cells induced filamentous growth, while overexpression of FLO8 had no obvious effect. Overexpression of the white cell enriched gene EFG1 indeed induced filamentation, but we observed that EFG1 first induced an opaque-to-white conversion and resulted in filamentation of the converted white cells (data not shown).
Fig. 9.
Roles of overexpression of FLO8, HGC1, TEC1, and BRG1 genes in opaque cell filamentation. WT, GH1013, an MTLa/a strain of SC5314 background. A. Expression of GFP protein in the WT+pNIM1 and WT+OP4p-cartTA+pNIM1 strains. To generate the WT+OP4p-cartTA+pNIM1 strain, a copy of the cartTA gene was integrated into the OP4 (an opaque cell enriched gene) locus of GH1013. B. Cellular images of overexpression strains grown in inducing (with doxycycline) or non-inducing (without doxycycline) liquid Lee’s glucose medium plates at 25°C for 36 hours. 40 μg ml−1 doxycycline was added to the inducing medium. Scale bar, 10μm.
Deletion of TEC1, BRG1 and CPH1 in the bcr1/bcr1 mutant does not affect opaque cell filamentation
Since overexpression of TEC1 and BRG1 in opaque cells induced filamentous growth, we subsequently tested whether TEC1 and BRG1 are required for filamentation in the bcr1/bcr1 mutant. As shown in Fig. 8A, the double mutants of tec1/tec1 bcr1/bcr1 and brg1/brg1 bcr1/bcr1 underwent robust filamentation, suggesting that deletion of these two genes has no obviously effect on Bcr1-regulated filamentous growth. Consistently, Si et al. also found that Tec1 is not required for environmental cues induced opaque cell filamentation (Si et al., 2013). We also found that deletion of CPH1 in the bcr1/bcr1 mutant did not affect opaque cell filamentation (Fig. 8A).
Deletion of TEC1, BRG1, ROB1, EFG1 and NDT80 does not induce opaque cell filamentation
The transcription factors Bcr1, Tec1, Brg1, Rob1, Efg1 and Ndt80 comprise a master circuit that controls biofilm formation and form an elaborate, interconnected transcriptional network in C. albicans (Nobile et al., 2012). We, therefore, examined whether Tec1, Brg1, Rob1, Efg1 and Ndt80 were also involved in repressing opaque cell filamentation. As shown in Fig. S4 and Fig. 8A, deletion of TEC1, BRG1, ROB1, EFG1 or NDT80 did not induce filamentous growth of opaque cells, suggesting that a different transcriptional network is involved in the regulation of opaque cell filamentation.
Discussion
Eukaryotic cells become morphologically and functionally specialized through cell differentiation. C. albicans can undergo a phenotypic switch between white and opaque cell types, which are functionally specialized in the processes of filamentation, mating and infection of different host tissues (Lohse and Johnson, 2009; Soll, 2009). White cells can undergo filamentation, whereas opaque cells appear to lose the capacity to grow as a filamentous form under most inducing conditions ((Anderson et al., 1989; Si et al., 2013); and our unpublished data). White-opaque switching is a bistable and heritable epigenetic event, where both cell types are maintained for many generations (Slutsky et al., 1987). However, the yeast-filamentous growth transition is apparently non-heritable and highly environment-dependent (Huang, 2012). Despite many shared features of the white-opaque switch and the yeast-filamentous growth transition (Huang, 2012), they have traditionally been thought to exist as two separate phenotypic switching systems. Previous studies demonstrate that opaque cells indeed form filaments when grown on the surface of a perfusion chamber or on a monolayer of human epithelial skin cells (Anderson et al., 1989), suggesting that a cryptic program of filamentous growth exists in opaque cells. In this study, we report that both cAMP-signaling and Bcr1-mediated pathways are involved in the regulation of filamentation of opaque cells in C. albicans. Numerous studies have linked filamentous growth with pathogenesis in MTL heterozygous strains of C. albicans (Huang, 2012; Whiteway and Bachewich, 2007). One hypothesis is that the combination of the ability to filament with the ability to mate as an opaque cell (in addition to other opaque cell features), may be adaptive by permitting C. albicans to persist in the human host, providing advantages over other phenotypes in specific host niches.
In this study, we have identified Bcr1 as a central regulator of opaque cell filamentation. We find that Bcr1-regulated opaque cell filamentation is dependent on the cAMP signaling pathway. These findings provide new insights into a longstanding question of why opaque cells have historically been unable to undergo filamentation under standard hyphal-inducing conditions. A recent study by Si et al. reports that distinctive pathways are involved in filamentous growth in white and opaque cells (Si et al., 2013).While our study and the Si et al. study both focus on a similar topic, they have diverse and complementary features. For example, most culture media used in our study are standard laboratory media (e.g. Lee’s medium and YPD), whereas SOR (with 1 M sorbitol) and MIN (without amino acids) media were used for opaque cell filamentation in the Si paper (Si et al., 2013). Different culture conditions may represent different natural niches for C. albicans in vivo.
Bcr1, the central regulator of opaque cell filamentation
Bcr1 is the first transcription factor that has been reported to be required for biofilm development in vitro and in vivo, but is not required for filamentous growth in MTL heterozygous C. albicans isolates (Nobile and Mitchell, 2005). Bcr1 regulates the expression of a subset of cell surface genes including HWP1, ECE1, RBT5 and ALS3 (Nobile et al., 2006), which are also enriched during filamentation (Huang, 2012). The expression of BCR1 is partly dependent on the hyphal regulator Tec1 and is repressed by the general transcription repressor Tup1 (Kadosh and Johnson, 2005; Nobile and Mitchell, 2005), although Bcr1 also transcriptionally regulates TEC1 through an interwoven network during biofilm formation (Zordan et al., 2007; Nobile et al., 2012). Recently, Fanning et al. reported that Bcr1 divergently controls the expression of many genes during the process of biofilm formation in vitro and in vivo (Fanning et al., 2012). Moreover, Bcr1 and Wor1 bind to each other’s promoters (Zordan et al., 2007; Nobile et al., 2012). These findings are consistent with the idea that Bcr1 is linked to filamentation and white-opaque switching and can act differentially in “fine-tuning” the regulation of gene expression under different circumstances. In the present study, we have discovered a novel role for Bcr1 in the regulation of opaque filamentous growth. Disruption of BCR1 in opaque cells leads to hyper-filamentous and hyper-invasive growth, demonstrating that Bcr1 is a powerful repressor of opaque cell filamentation. We showed that the physiological temperature of the human host (37°C) induces opaque to white switching followed by filamentation after a prolonged incubation period (Fig. 5). However, the latter filaments are present both in the WT and in the bcr1/bcr1 mutant. Thus, Bcr1 does not repress filamentous growth in white cells under the condition tested. Nobile et al. has also found that white cells of the bcr1/bcr1 mutant and the WT control produced hyphae to comparable extents under a variety of culture conditions (Nobile et al., 2012). However, we cannot exclude the possibility that Bcr1 might play a role in white cell filamentation under some un-discovered condition, such as a specific host niche.
Filamentous opaque cells of the bcr1/bcr1 mutant are mating competent, confirming that their cell identity is opaque. Notably, the mating efficiency of the bcr1/bcr1 mutant opaque cell type is about 10 times higher than that of the WT opaque cell type. One potential explanation for this observation is that the larger length of the filamentous opaque cells of the bcr1/bcr1 mutant may increase the opportunity for interactions between mating partners by providing a larger surface area. Another explanation is that filamentous opaque cells may more easily form elongated mating projections, a prerequisite of mating, since they are already in an elongated form.
Role of previously characterized hyphal growth regulators (Czf1, Cup9, Nrg1, Rfg1, Tup1, Brg1, and Tec1) in opaque cell filamentation
The transcription factors Czf1, Cup9 and Nrg1 are also involved in the regulation of opaque cell filamentation (Table 1, Fig. 1, 6 and Fig. S3). Bcr1 directly binds to the promoter regions of these genes in MTL heterozygous as well MTL homozygous cells (Nobile et al., 2012 and our unpublished data). Deletion of CZF1 has only a moderate inducing effect on the development of filaments in aged opaque cells (Fig. 1 and Fig. 6). Consistent with this, Zordan et al. has reported that opaque colonies of the czf1/czf1 mutant are rougher than those of WT strains (Zordan et al., 2007). Cup9 and Nrg1 are involved in opaque cell filamentation as well as white cell filamentation, indicating that they are not opaque cell specific regulators of filamentous growth, but general transcriptional repressors. We have also identified a white cell specific regulator, Rfg1, an HMG-domain-containing DNA binding protein acting in Tup1-dependent and -independent pathways (Kadosh and Johnson, 2001). Deletion of RFG1 affects white cell filamentation but not opaque cell filamentation (Fig. 6 and Fig. S3). This result is consistent with the recent study reporting that Rfg1 represses filamentation in white cells but not in opaque cells, and further demonstrates that Rfg1 functions as a positive regulator of opaque cell filamentation (Si et al., 2013). Environmental pH plays an important role in the regulation of hyphal development in MTL heterozygous and MTL homozygous white cells (Davis, 2003; Biswas et al., 2007) (Fig. 6 and Fig. S3). However, alterations in pH appear to have no obvious effect on opaque cell filamentation in the bcr1/bcr1, nrg1/nrg1 and czf1/czf1 mutants.
Bcr1 directly binds to the promoter regions of NRG1 and CUP9 and may control their transcription ((Nobile et al., 2012) and Fig. 3). The major machinery of opaque cell filamentation, including the conserved and central cAMP-signaling pathway, the negative regulators of filamentation (such as Tup1, Nrg1, Czf1 and Cup9) and the cell wall components, are also used in the regulation of white cell filamentation. A slight modification of these pathways via Bcr1 may have permitted the reprogramming of a new biological process.
The MTL homozygous mutant of the repressor TUP1 can switch between four different colony types (Zhao et al., 2002; Park and Morschhauser, 2005a). The regulation of filamentous growth by Tup1 is more general in C. albicans since Tup1 is a global and highly pleiotropic transcriptional regulator. Disruption of TUP1 in MTL heterozygous strains also leads to hyper-filamentous growth under all conditions tested (Braun and Johnson, 1997). The Tup1 regulator is highly conserved and functions similarly in other yeasts. Tup1 and its associated transcription factor Nrg1 may also repress opaque cell filamentation via a pathway independent of Bcr1. Bcr1, Tec1, and Brg1, coordinate to regulate biofilm formation in C. albicans (Nobile et al., 2012). Of note, both TEC1 and BRG1 play a critical role in the regulation of white cell filamentation (Du et al., 2012b). Although deletion of TEC1 and BRG1 had no effect on the induction of opaque cell filamentation, overexpression of TEC1 and BRG1 obviously induced filamentous growth in opaque cells (Fig. 9). The regulatory pathways of opaque cell filamentation are shown in Fig. 10A.
Fig. 10.
Model of the regulation of opaque cell filamentation in C. albicans. A. Pathways involved in the regulation of opaque cell filamentation. Environmental cues signal the positive cAMP pathway and the negative pathway mediated by the key regulator Bcr1 to control the expression of filamentous specific genes. B. Models of separate and integrated phenotypic switching systems. Integration of the white-opaque switching and yeast-filamentous transition systems increases the complexity and plasticity of phenotypic switching in C. albicans. Solid lines represent confirmed phenotypic switches. Dashed lines represent unconfirmed switches. Wh, white cell phase; Op, opaque cell phase; Fila. wh, filamentous white cell phase; Fila. op, filamentous opaque cell phase.
The cAMP signaling is required for opaque cell filamentation
The adenylyl cyclase Cyr1 occupies the central position of the regulatory network of filamentation in MTL heterozygous isolates of C. albicans (Huang, 2012). We set out to explore the role of cAMP signaling in filamentous growth of opaque cells. Inactivating cAMP signaling by disruption of CYR1 results in a significant decrease in filamentation in the bcr1/bcr1 mutant opaque cells. Of note, deletion of CYR1 results in the lack of the basal level of cAMP, which may affect a range of cellular functions including cell morphogenesis and growth rate. Therefore, to some extent, the defect of filamentation in the bcr1/bcr1 cyr1/cyr1 double mutant may also be a secondary effect.
Consistent with the phenotype observed in the bcr1/bcr1 cyr1/cyr1 double mutant, two major transcription regulators downstream of cAMP signaling, Efg1 and Flo8, are also required for hyper-filamentation of the bcr1/bcr1 mutant. Filamentous-specific G1 cyclin-related protein Hgc1 is transcriptionally regulated by the cAMP pathway and the Efg1 and Flo8 transcription regulators (Zheng and Wang, 2004; Cao et al., 2006). HGC1 is essential for opaque cell filamentation in the bcr1/bcr1 mutant (Fig. 8A). Deletion of EFG1 or FLO8 partially suppresses filamentous growth, while deletion of HGC1 completely blocks opaque cell filamentation in the bcr1/bcr1 mutant. The transcriptional level of HGC1 in opaque cells of the bcr1/bcr1 mutant is 4 fold higher than that in opaque cells of the WT control (data not shown). Consistently, overexpression of HGC1 promotes filamentous development in opaque cells. These results suggest that the regulation of Bcr1-controlled filamentous growth in opaque cells is regulated by the major cAMP pathway and a minor unidentified pathway, which converge on Hgc1.
It is possible that the specialization of Bcr1 and cAMP signaling in the gene regulatory network of filamentation in opaque cells may have been gained after the evolution of white-opaque switching in C. albicans. Filamentous enriched genes HWP1 and ECE1 are expressed in filamentous opaque cells of the bcr1/bcr1 mutant (Fig. 2C), suggesting that downstream functional components of filamentation of white cells are harnessed by the mutant. The central regulatory pathway of filamentous growth in white cells, cAMP signaling and its downstream regulators Efg1, Flo8 and Hgc1 are also utilized by opaque cells. Disruption of a single gene, BCR1, is able to reprogram the regulatory circuitry of filamentation, emphasizing the importance of Bcr1 in opaque cells. Environmental cues could signal opaque cells to undergo filamentation via the positive pathway of cAMP signaling as well as the Bcr1-mediated negative transcriptional circuit.
Opaque cell filamentation may increase phenotypic plasticity of C. albicans
Evolution increases phenotypic plasticity in living organisms, where new phenotypes may arise as an adaptation to changing environmental conditions or via integration of existing regulatory networks through genetic drift. For the human fungal pathogen C. albicans, the traditionally separate white-opaque and yeast-filamentous circuits have been regarded as separate systems (Fig. 10B). Integration of the opaque and filamentous circuits into a new filamentous opaque network provides an additional level of phenotypic regulation, which results in a complex multiple-component switching system (Fig. 10B). Compared to the former systems, the latter has increased phenotypic flexibility, which is important for the pathogen to better adapt to fluctuations in the host environment. Moreover, our study of reprogramming typical opaque cells to filamentous opaque cells by disruption of BCR1 provides an example of a simple cell differentiation system in C. albicans that may serve as a model for the study of other developmental processes.
Experimental procedures
Culture conditions, strains and plasmids
YPD (1% yeast extract, 2% peptone, 2% glucose) and modified Lee’s glucose medium were used for routine culture of C. albicans cells (Huang et al., 2010). Lee’s medium is a chemically defined medium, which was developed based on the aminopeptidases of C. albicans (Lee et al., 1975). 0.07 grams per liter of L-arginine was added to the modified Lee’s medium. Modified Lee’s medium with 1.25 % (w v−1) glucose as a carbon source is referred to as Lee’s glucose medium. Modified Lee’s medium with GlcNAc 1.25 % (w v−1) as a carbon source is referred to as Lee’s GlcNAc medium. YPmal medium (1% yeast extract, 2% peptone, 2% maltose) was used for FLP-mediated excision of the SAT1/flipper cassette.
The MTLa/α strains of C. albicans were converted to MTL homozygotes by selection in YPS (1% yeast extract, 2% peptone and 10% sorbose) medium as previously described (Du et al., 2012a). To verify the phenotypes of C. albicans MTL homozygotes, the a or α locus was also deleted in some MTLa/α strains by targeted disruption with the plasmid pSFS2a-MTLKO to generate MTLa/Δ or MTLΔ/α isolates (Thyagarajan and Soll, unpublished). To generate the bcr1/bcr1+BCR1p-BCR1 reconstituted strain, the same strategy was used as we reported for the construction of a BRG1 reconstituted strain (Du et al., 2012b). The construct contained two fragments (one 3.1 kb fragment with the sequence of BCR1 ORF, 5′-UTR and 3′-UTR; the other 0.5 kb fragment contained the 3′-UTR of BCR1). The construct was integrated into the original BCR1 locus. The strains used in this study are listed in supplemental material Table S3.
To construct the BCR1 knockout plasmid pSFS2a-BCR1KO, two BCR1 flanking fragments were inserted into the ApaI/XhoI and SacII/SacI sites of the plasmid pSFS2A (Reuss et al., 2004).
The cyr1/cyr1 mutant, GH1109, was used to generate the cyr1/cyr1 bcr1/bcr1 double mutant. To delete the first copy of BCR1, GH1109 was transformed with a fragment of the ApaI/SacI linearized pSFS2a-BCR1KO, containing a SAT1/flipper cassette. The resulting strain bcr1/bcr1 BCR1/bcr1::SAT1-FLIP was then grown in YPmal medium for FLP-mediated excision of the SAT1/flipper cassette generating the strain cyr1/cyr1 BCR1/bcr1::FRT. To delete the second copy of BCR1, the strain cyr1/cyr1 BCR1/bcr1::FRT was transformed with the fusion PCR product of the URA3 gene flanked by the BCR1 gene 5′ and 3′ fragments. The plasmid pGEM-URA3 was used as a PCR template (Wilson et al., 1999). The fusion PCR assay was performed as previously reported (Hernday et al., 2010).
The flo8/flo8 mutant, GH1296, was used to generate the flo8/flo8 bcr1/bcr1 double mutant. To delete the first copy of BCR1, GH1296 was transformed with a fragment of the ApaI/SacI linearized pSFS2a-BCR1KO. The resulting strain flo8/flo8 BCR1/bcr1::SAT1-FLP was grown in YPmal medium for FLP-mediated excision of the SAT1/flipper cassette generating the strain flo8/flo8 BCR1/bcr1::FRT, and then grown on 5-fluoroorotic acid (5-FOA) containing medium for counter-selection of ura3− isolates. Similarly as in the generation of the cyr1/cyr1 bcr1/bcr1 double mutant, the second copy of BCR1 was deleted using the fusion PCR method. The plasmid pGEM-URA3 was used as a PCR template (Wilson et al., 1999).
The efg1/efg1 mutant, MMY620, and the cph1/cph1 mutant, JKC18a, were used to generate the efg1/efg1 bcr1/bcr1 double mutant. JKC18a (MTLa/a) was derived from JKC18 by growing the strain in sorbose-containing medium (Liu et al., 1994). The two copies of BCR1 were disrupted in the efg1/efg1 mutant and the cph1/cph1 mutant with the same method used in generating the flo8/flo8 bcr1/bcr1 double mutant.
The hgc1/hgc1 mutant, GH1281, was used to generate the hgc1/hgc1 bcr1/bcr1 double mutant. To delete the first copy of BCR1, GH1281 was transformed with a fragment of the ApaI/SacI linearized pSFS2a-BCR1KO, containing a SAT1/flipper cassette. To delete the second copy of BCR1, the strain hgc1/hgc1 BCR1/bcr1::FLP was transformed with the fusion PCR product of the URA3 gene flanked by BCR1 gene 5′ and 3′ fragments. The plasmid pGEM-URA3 was used as a PCR template. All primers used are listed in Table S4.
TF22 (brg1/brg1 mutant, MTLa/α) of the transcription factor mutant library was first converted to an MTLΔ/α strain by targeted disruption with the plasmid pSFS2a-MTLKO to generate MTLΔ/α isolate (TF22α). TF22α was then grown in YPmal medium for FLP-mediated excision of the SAT1/flipper cassette to generate the strain TF22α1. To delete the first copy of BCR1 in TF22α1, a fragment of the ApaI/SacI linearized pSFS2a-BCR1KO was used for disruption. To delete the second copy of BCR1, cells were transformed with the fusion PCR products of the ARG4 gene and two fragments (of the 5′ and 3′ terminals) of the BCR1 gene. Genomic DNA of SC5314 and the plasmid pRS-ARG4 was used as initial PCR templates.
A similar strategy to the deletion of BCR1 in TF22α1 was used to generate the tec1/tec1 bcr1/bcr1 double mutant. The two alleles of BCR1 were subsequently disrupted in the tec1/tec1 mutant TF115α with the ApaI/SacI linearized pSFS2a-BCR1KO plasmid and the fusion PCR products of the ARG4 gene and two fragments (of the 5′ and 3′ terminals) of the BCR1 gene.
At least three independent isolates of each double mutant were used for the phenotypic transition analysis in order to exclude any effects induced by non-specific mutations (e.g. 5-FOA induced spontaneous mutations).
The strain E5.3I-3 of the czf1/czf1 mutant was used in Fig. 1, 6 and S3. The phenotype of the czf1/czf1 mutant was verified in the strain WCZF1M4B, a czf1/czf1 mutant generated in a different background strain, WO-1 (Ramirez-Zavala et al., 2008).
Northern blot analysis
Northern blot analysis was performed according to our previous studies (Huang et al., 2009). Purified PCR products were used to make probes. Primers used for the PCR reactions are listed in Table S4.
Chromatin Immunoprecipitation PCR (ChIP-PCR)
The myc-tagged strain CJN1787 (MTLa/α BCR1/BCR1-Myc) was converted to an MTLa/Δ strain by deletion of the α locus with the plasmid pSFS2a-MTLKO. The resulting strain was named as CJN1787a (MTLa/Δ::SAT1 BCR1/BCR1-Myc). To test whether Bcr1 binds to the candidate targets, white and opaque cells of the strain CJN1787a were lysed and the supernatant was subjected to sonication to obtain sheared chromatin averaging 500 to 1000 bp in length. The ChIP assay was conducted as described previously (Hernday et al., 2010; Nobile et al., 2012). Anti-Myc polyclonal antibody (sc-789) (Santa Cruz Biotechnology, Inc.) was used. Immunoprecipitates were recovered and DNA was then extracted for PCR amplification.
Microscopy
Cells collected from liquid cultures or from nutrient agar plates as indicated in the text were used for morphological analysis. Differential interference contrast (DIC) optics was used for standard cell morphology detection. To discriminate hyphae and pseudohyphae, cells were dyed with Calcofluor white, which stains septa and chitin in the cell wall. Fluorescence microscopy was used to detect septa. Calcofluor white staining was performed as described previously (Wang et al., 2011). Scanning electron microscopy (SEM) was performed as previously described (Du et al., 2012b).
Mating assay
Mating experiments were conducted on Lee’s glucose plates as previously reported (Miller and Johnson, 2002; Huang et al., 2009). Briefly, the experimental cell samples were collected from the colonies of the right phase phenotype. The tester opaque cells were collected from a liquid culture in middle exponential phase. 1 × 106 of MTLa/a cells and 1 × 106 of MTLα/α cells were mixed in Lee’s glucose medium and spotted onto Lee’s glucose medium plates. After 4 days of growth at 25°C, the mating mixture was resuspended and plated onto selectable plates (depleted of uridine, or Arginine, or uridine/Arginine). Colonies grown on the three types of plates were counted and mating efficiencies were calculated.
Filamentous and invasive growth assays
YPD, spider and SCD medium were used in Fig. S2. Spider medium was made according to Liu et al. (1994) (Liu et al., 1994). SCD medium (pH6.8) was buffered with K2HPO4. For Fig. 8B, the flo8/flo8 and flo8/flo8 bcr1/bcr1 mutants were cultured on both Lee’s glucose and Lee’s GlcNAc media.
To examine the ability of invasive growth, 3 μl of liquid medium containing about 6 × 104 cells was spotted onto the Lee’s glucose medium plates and incubated for 3 days at 25 °C. The plates were imaged before and after washing with H2O.
Supplementary Material
Acknowledgments
The authors are indebted to Drs. David Soll, Alexander D. Johnson and Joachim Morschhäuser for the generous gifts of plasmids and strains. This work was supported by the “100 Talent Program” grant from the Chinese Academy of Sciences and grant 31170086 from the Chinese National Natural Science Foundation to G.H. C.J.N. was supported by NIH grants K99AI00896 and R01AI049187.
Footnotes
Supporting Information Additional supporting information may be found in the online version of this article.
References
- Anderson J, Cundiff L, Schnars B, Gao MX, Mackenzie I, Soll DR. Hypha formation in the white-opaque transition of Candida albicans. Infect Immun. 1989;57:458–467. doi: 10.1128/iai.57.2.458-467.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JM, Soll DR. Unique phenotype of opaque cells in the white-opaque transition of Candida albicans. J Bacteriol. 1987;169:5579–5588. doi: 10.1128/jb.169.12.5579-5588.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birse CE, Irwin MY, Fonzi WA, Sypherd PS. Cloning and characterization of ECE1, a gene expressed in association with cell elongation of the dimorphic pathogen Candida albicans. Infect Immun. 1993;61:3648–3655. doi: 10.1128/iai.61.9.3648-3655.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S, Van Dijck P, Datta A. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol Mol Biol Rev. 2007;71:348–376. doi: 10.1128/MMBR.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun BR, Johnson AD. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science. 1997;277:105–109. doi: 10.1126/science.277.5322.105. [DOI] [PubMed] [Google Scholar]
- Braun BR, Kadosh D, Johnson AD. NRG1, a repressor of filamentous growth in C.albicans, is down-regulated during filament induction. EMBO J. 2001;20:4753–4761. doi: 10.1093/emboj/20.17.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DH, Jr., Giusani AD, Chen X, Kumamoto CA. Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Mol Microbiol. 1999;34:651–662. doi: 10.1046/j.1365-2958.1999.01619.x. [DOI] [PubMed] [Google Scholar]
- Cao F, Lane S, Raniga PP, Lu Y, Zhou Z, Ramon K, et al. The Flo8 transcription factor is essential for hyphal development and virulence in Candida albicans. Mol Biol Cell. 2006;17:295–307. doi: 10.1091/mbc.E05-06-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis D. Adaptation to environmental pH in Candida albicans and its relation to pathogenesis. Curr Genet. 2003;44:1–7. doi: 10.1007/s00294-003-0415-2. [DOI] [PubMed] [Google Scholar]
- Du H, Guan G, Xie J, Cottier F, Sun Y, Jia W, et al. The transcription factor Flo8 mediates CO2 sensing in the human fungal pathogen Candida albicans. Mol Biol Cell. 2012;23:2692–2701. doi: 10.1091/mbc.E12-02-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Guan G, Xie J, Sun Y, Tong Y, Zhang L, Huang G. Roles of Candida albicans Gat2, a GATA-type zinc finger transcription factor, in biofilm formation, filamentous growth and virulence. PLoS One. 2012;7:e29707. doi: 10.1371/journal.pone.0029707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning S, Xu W, Solis N, Woolford CA, Filler SG, Mitchell AP. Divergent Targets of Candida albicans Biofilm Regulator Bcr1 In Vitro and In Vivo. Eukaryot Cell. 2012;11:896–904. doi: 10.1128/EC.00103-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sanchez S, Mavor AL, Russell CL, Argimon S, Dennison P, Enjalbert B, Brown AJ. Global roles of Ssn6 in Tup1- and Nrg1-dependent gene regulation in the fungal pathogen, Candida albicans. Mol Biol Cell. 2005;16:2913–2925. doi: 10.1091/mbc.E05-01-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernday AD, Noble SM, Mitrovich QM, Johnson AD. Genetics and molecular biology in Candida albicans. Methods Enzymol. 2010;470:737–758. doi: 10.1016/S0076-6879(10)70031-8. [DOI] [PubMed] [Google Scholar]
- Homann OR, Dea J, Noble SM, Johnson AD. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet. 2009;5:e1000783. doi: 10.1371/journal.pgen.1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G. Regulation of phenotypic transitions in the fungal pathogen Candida albicans. Virulence. 2012:3. doi: 10.4161/viru.20010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Srikantha T, Sahni N, Yi S, Soll DR. CO2 regulates white-to-opaque switching in Candida albicans. Curr Biol. 2009;19:330–334. doi: 10.1016/j.cub.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Yi S, Sahni N, Daniels KJ, Srikantha T, Soll DR. N-acetylglucosamine induces white to opaque switching, a mating prerequisite in Candida albicans. PLoS Pathog. 2010;6:e1000806. doi: 10.1371/journal.ppat.1000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang GH, Nie XY, Chen JY. CaMac1, a Candida albicans copper ion-sensing transcription factor, promotes filamentous and invasive growth in Saccharomyces cerevisiae. Acta Biochim Biophys Sin (Shanghai) 2006;38:213–217. doi: 10.1111/j.1745-7270.2006.00146.x. [DOI] [PubMed] [Google Scholar]
- Kadosh D, Johnson AD. Rfg1, a protein related to the Saccharomyces cerevisiae hypoxic regulator Rox1, controls filamentous growth and virulence in Candida albicans. Mol Cell Biol. 2001;21:2496–2505. doi: 10.1128/MCB.21.7.2496-2505.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh D, Johnson AD. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol Biol Cell. 2005;16:2903–2912. doi: 10.1091/mbc.E05-01-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KL, Buckley HR, Campbell CC. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia. 1975;13:148–153. doi: 10.1080/00362177585190271. [DOI] [PubMed] [Google Scholar]
- Liu H, Kohler J, Fink GR. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- Lohse MB, Johnson AD. White-opaque switching in Candida albicans. Curr Opin Microbiol. 2009;12:650–654. doi: 10.1016/j.mib.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MG, Johnson AD. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell. 2002;110:293–302. doi: 10.1016/s0092-8674(02)00837-1. [DOI] [PubMed] [Google Scholar]
- Morrow B, Srikantha T, Anderson J, Soll DR. Coordinate regulation of two opaque-phase-specific genes during white-opaque switching in Candida albicans. Infect Immun. 1993;61:1823–1828. doi: 10.1128/iai.61.5.1823-1828.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murad AM, Leng P, Straffon M, Wishart J, Macaskill S, MacCallum D, et al. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 2001;20:4742–4752. doi: 10.1093/emboj/20.17.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nantel A, Dignard D, Bachewich C, Harcus D, Marcil A, Bouin AP, et al. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol Biol Cell. 2002;13:3452–3465. doi: 10.1091/mbc.E02-05-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Mitchell AP. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr Biol. 2005;15:1150–1155. doi: 10.1016/j.cub.2005.05.047. [DOI] [PubMed] [Google Scholar]
- Nobile CJ, Andes DR, Nett JE, Smith FJ, Yue F, Phan QT, et al. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2006;2:e63. doi: 10.1371/journal.ppat.0020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, Hernday AD, et al. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell. 2012;148:126–138. doi: 10.1016/j.cell.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noffz CS, Liedschulte V, Lengeler K, Ernst JF. Functional mapping of the Candida albicans Efg1 regulator. Eukaryot Cell. 2008;7:881–893. doi: 10.1128/EC.00033-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YN, Morschhauser J. Candida albicans MTLalpha tup1Delta mutants can reversibly switch to mating-competent, filamentous growth forms. Mol Microbiol. 2005;58:1288–1302. doi: 10.1111/j.1365-2958.2005.04898.x. [DOI] [PubMed] [Google Scholar]
- Park YN, Morschhauser J. Tetracycline-inducible gene expression and gene deletion in Candida albicans. Eukaryot Cell. 2005;4:1328–1342. doi: 10.1128/EC.4.8.1328-1342.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Zavala B, Reuss O, Park YN, Ohlsen K, Morschhauser J. Environmental induction of white-opaque switching in Candida albicans. PLoS Pathog. 2008;4:e1000089. doi: 10.1371/journal.ppat.1000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuss O, Vik A, Kolter R, Morschhauser J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene. 2004;341:119–127. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- Rocha CR, Schroppel K, Harcus D, Marcil A, Dignard D, Taylor BN, et al. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol Biol Cell. 2001;12:3631–3643. doi: 10.1091/mbc.12.11.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si H, Hernday AD, Hirakawa MP, Johnson AD, Bennett RJ. Candida albicans White and Opaque Cells Undergo Distinct Programs of Filamentous Growth. PLoS Pathog. 2013;9:e1003210. doi: 10.1371/journal.ppat.1003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slutsky B, Staebell M, Anderson J, Risen L, Pfaller M, Soll DR. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J Bacteriol. 1987;169:189–197. doi: 10.1128/jb.169.1.189-197.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll DR. Why does Candida albicans switch? FEMS Yeast Res. 2009;9:973–989. doi: 10.1111/j.1567-1364.2009.00562.x. [DOI] [PubMed] [Google Scholar]
- Srikantha T, Chandrasekhar A, Soll DR. Functional analysis of the promoter of the phase-specific WH11 gene of Candida albicans. Mol Cell Biol. 1995;15:1797–1805. doi: 10.1128/mcb.15.3.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantha T, Borneman AR, Daniels KJ, Pujol C, Wu W, Seringhaus MR, et al. TOS9 regulates white-opaque switching in Candida albicans. Eukaryot Cell. 2006;5:1674–1687. doi: 10.1128/EC.00252-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staab JF, Bradway SD, Fidel PL, Sundstrom P. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science. 1999;283:1535–1538. doi: 10.1126/science.283.5407.1535. [DOI] [PubMed] [Google Scholar]
- Tuch BB, Mitrovich QM, Homann OR, Hernday AD, Monighetti CK, De-La-Vega FM, Johnson AD. The transcriptomes of two heritable cell types illuminate the circuit governing their differentiation. PLoS Genet. 2010;6:e1001070. doi: 10.1371/journal.pgen.1001070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HX, Douglas LM, Aimanianda V, Latge JP, Konopka JB. The Candida albicans Sur7 protein is needed for proper synthesis of the fibrillar component of the cell wall that confers strength. Eukaryot Cell. 2011;10:72–80. doi: 10.1128/EC.00167-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteway M, Bachewich C. Morphogenesis in Candida albicans. Annu Rev Microbiol. 2007;61:529–553. doi: 10.1146/annurev.micro.61.080706.093341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RB, Davis D, Mitchell AP. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol. 1999;181:1868–1874. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Lockhart SR, Daniels K, Soll DR. Roles of TUP1 in switching, phase maintenance, and phase-specific gene expression in Candida albicans. Eukaryot Cell. 2002;1:353–365. doi: 10.1128/EC.1.3.353-365.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Wang Y. Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J. 2004;23:1845–1856. doi: 10.1038/sj.emboj.7600195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zordan RE, Galgoczy DJ, Johnson AD. Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc Natl Acad Sci U S A. 2006;103:12807–12812. doi: 10.1073/pnas.0605138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zordan RE, Miller MG, Galgoczy DJ, Tuch BB, Johnson AD. Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS Biol. 2007;5:e256. doi: 10.1371/journal.pbio.0050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.