Abstract
Frizzled homolog 1 (FZD1) is a transmembrane receptor that mediates Wnt signaling. The transcriptional regulation of FZD1 and the role of FZD1 in osteoblast biology are not well understood. We examined the role of E2F1 in FZD1 promoter activation and osteoblast differentiation and mineralization. A putative E2F1 binding site in the FZD1 promoter region was initially identified in silico and characterized further in Saos2 cells in vitro by chromatin immunoprecipitation (ChIP), electrophoretic mobility shift (EMSA) and promoter reporter assays. Over-expression of E2F1 transactivated the FZD1 promoter and increased endogenous FZD1 mRNA and protein levels in Saos2 cells. Over-expression of E2F1 in Saos2 cells up-regulated osteoblast differentiation markers alkaline phosphatase (ALP), type I collagen α (COL1A), and osteocalcin (OCN). Furthermore, E2F1 over-expression enhanced mineralization of differentiated Saos2 cells, whereas siRNA knockdown of FZD1 diminished the effects of E2F1 on osteoblast mineralization. The effects of E2F1 on FZD1 expression and osteoblast mineralization were further confirmed in normal human FOB osteoblasts. Taken together, our experiments demonstrate a role of E2F1 in osteoblast differentiation and mineralization and suggest that FZD1 is required, in part, for E2F1 regulation of osteoblast mineralization.
Keywords: Frizzled homology1 (FZD1), E2F1, Osteoblasts, Mineralization
Introduction
Wnt signaling is a key regulator of osteoblast biology and bone mass in humans [1-3]. Frizzled homolog 1 (FZD1), a member of the G-protein-coupled receptor superfamily, is expressed in osteoblasts [4] and mediates Wnt signaling by serving as a co-receptor with LRP-5 for Wnt ligands. We have recently reported genetic associations of FZD1 promoter variants with bone mineral density (BMD) in humans [5,6] but the regulation of FZD1 mRNA and protein in osteoblasts and its potential role in osteoblast biology and bone mineralization are poorly understood.
The E2F family of transcription factors plays a role in cell differentiation, proliferation, and apoptosis [7-9]. E2F1, a member of the E2F family, binds to retinoblastoma protein pRb in a cell-cycle dependent manner to mediate cellular proliferation and apoptosis. E2F1 is expressed in calvaria and involved in osteoblast differentiation mediated by pRb [10]. Abnormal activation of E2F1 contributes to the inability of pRb-deficient osteoprogenitors to properly exit the cell cycle and leads to aberrant osteoblast differentiation [10]. In addition, over expression of E2F1 alters the onset of endochondral bone formation [11].
In the present study, we identified an E2F1 binding element in the FZD1 promoter and demonstrated that E2F1 is a novel transcriptional activator of FZD1 in human osteoblastic cells. Further, over-expression of E2F1 in osteoblastic cells increases mineralization in vitro whereas knockdown of FZD1 diminishes this effect.
Material and methods
Cell culture and E2F1 over-expression
Saos2 cells were cultured as described [5,6]. FOB cells were cultured in a medium containing 1:1 volume of DMEM and F12 supplemented with 10% fetal bovine serum. When performing cell transfection experiments for luciferase assays, real time PCR and Western blotting, Saos2 and FOB cells were plated at a density of 4 × 105 in a 35 mm diameter culture plate. On the next day, Saos2 cells were transfected with 0.25, 0.5 and 1 μg pCMV/E2F1 using Lipofectamine 2000 (Invitrogen). Cells were harvested at 48 h post transfection for RNA and protein preparation for real-time PCR, Western blot and luciferase analysis. In all transfection experiments, the amount of plasmid DNA was balanced as necessary with β-gal expression plasmid such that the total DNA amount was constant in each group. When assessing the effect of E2F1 on mineralization, Saos2 and FOB cells were infected by 5 × 106 pfu of adenovirus expressing E2F1 (a generous gift of Dr. Mien-Chie Hung, UT MD Anderson Cancer Center) [12] and β-gal and cultured in differentiation medium. Media were changed every 3 days and 2.5 × 106 pfu of E2F1 adenovirus was added at the same time. For nuclear protein extraction, cells were plated in 10 cm diameter culture plates at a density of 3.2 × 106.
Knockdown of FZD1 in Saos2 and E2F1in FOB
For siRNA knockdown of FZD1 in Saos2 and E2F1 in FOB, cells were seeded at a density of 2 × 105 in a 35 mm diameter culture plate and transfected with siRNA as described above. Cells were cultured for 48 h after transfection and harvested for RNA preparation and real time PCR. Experiments were performed in triplicate and repeated 2–3 times.
Identification of a putative E2F1 binding site in the FZD1 promoter
In order to identify potential binding sites for E2F1 in the FZD1 promoter in silico, we used the Transcription Element Search Software (TESS) (http://www.cbil.upenn.edu/cgi-bin/tess/tess) to screen the promoter region of FZD1 from −655 to +71 bp of the translation start site. Our analysis identified a putative E2F1 binding site (GCGCGA) at positions +47 to +52 bp downstream of the FZD1 translation start site.
Plasmid construction
To fine-map the region of the FZD1 promoter necessary for E2F1 transactivation, we made truncated promoter clones containing −175 to +75 and specific alleles of the promoter polymorphism rs2232158 using the full length promoter clones as reported [6]. The original promoter clones were digested with KpnI and SacII and followed by blunt-end ligation. Mutagenesis of the putative E2F binding site at position +47 to +54 bp of the wild type clone was performed using the Quikchange lightning site directed mutagenesis kit according to the manufacturer's instructions (Agilent Technologies) and the mutation primer pair 5′-CCCGGGCCGCCGGCGGTGGCTTGAGCTGGGAACTTTGTG CCG-3′ and 5′-CGGCACAAAGTTCCCAGCTCAAGCCACCGCCGGCGGCCCG GG-3′. DNA sequences were confirmed using direct DNA sequencing as described [5].
Luciferase assay
Cultured Saos2 cells were transfected with 0.2 μg/each pGL3-basic-FZD1 promoter constructs using Lipofectamine 2000 following the manufacturer's instructions (Invitrogen). pRL-null luciferase reporter was used as an internal control. At 48 h post transfection, cells were lysed in 1 × passive lysis buffer and centrifuged for 10 min at 4 °C. Luciferase activity of the cell lysis supernatant was determined using the Dual Luciferase reporter assay system (Promega).
Total RNA isolation and real time PCR
Total RNA was extracted from Saos2 cells using Trizol reagent. One microgram of RNA was used to reverse transcribe the cDNA (Applied Biosystems) in a total volume of 20 μl. A portion (0.25 μl) of the cDNA was added to a master mix containing SYBR green and primer mixture at a concentration of 5 μM. Real time quantitative PCR was performed using the following cycling conditions: 50 °C for 2 min, 95 °C for 10 min, 40 cycles of 15 s at 95 °C and 1 min at 60 °C using an ABI 7900HT DNA analyzer (Applied Biosystems). The hypoxanthine-guanine phosphoribosyltransferase (HPRT ) gene was used as an internal control to show the relative mRNA expression of analyzed genes. Relative expression levels of experimental samples were calculated by using the values of the HPRT internal control as one. In the real time PCR analysis, gene specific primer pairs were used for alkaline phosphatase (ALP, 5′-GGCTCCAGGGATAAAGCAGGTC-3′ and 5′-GACACCCATCCCATCTCCCA-3′), osteocalcin (OCN, 5′-GAGAGCCCTCACACTCCTCG-3′ and 5′-GTCTCTTCACTACCTCGCTGCC-3′), collagen 1A (COLA1, 5′-ACCGCCCTCCTGACGC-3′ and 5′-CGTTGTCGCAGACGCAGAT-3′), E2F1 (5′-CGCATCTATGACATCACCAACG-3′ and 5′-GTCGGAGGTCCTGGGTCAAC-3′), FZD1 (5′-CCAAGAGAGGAGCCGAGA-3′ and 5′-CGGCACAAAGTTCCCAG-3′), receptor activator of nuclear factor kappa-B ligand (RANKL, 5′-TGCCACCGACATCCCATCTG-3′ and 5′-CGAAAGCAAATGTTGGCATACAGGT-3′) and HPRT (5′-GGCTTCCTCCTCCTGAGCAGTC-3′ and 5′-ACACCCTTTCCAAATCCTCAGCA-3′).
Western blot analysis
Protein lysates were prepared by lysing the Saos2 cells at sub confluence in 1 × RIPA buffer (Santa Cruz) on ice for 15 min. Equal volumes of 2 × electrophoresis loading buffer (Santa Cruz) was added into the protein lysate prior to heat inactivation at 95 °C for 5 min. Twenty micrograms of total protein was resolved by SDS-PAGE, transferred to nitrocellulose membrane and analyzed by standard immunoblotting with antibodies specific for FZD1 (rabbit polyclonal, Abgent), E2F1 (rabbit polyclonal, Santa Cruz), and β-actin (mouse monoclonal, Sigma). At least two different experiments were performed and representative blots are shown.
Chromatin immunoprecipitation (ChIP) assay
The ChIP analyses were performed using a standard protocol. Briefly, Saos2 cells were grown on 100-mm tissue culture dishes for 2 days. Cells were cross-linked with 1% formaldehyde for 10 min and harvested for fragmentation using sonication. The chromatin fragments were immunoprecipitated with 3 μg of the indicated antibodies for E2F1 (Santa Cruz) and mouse normal IgG (Santa Cruz). The precipitated fragments were washed 5 times and analyzed by PCR using 2 pairs of primers of two forward primers and one common reverse primer (F1: 5′-CCAAGAGAGGAGCCGAGAAA-3′, F2: 5′-AGGAAGGCGGGACACGAC-3′ and R1: 5′-GCCCCGGCACAAAGTTC-3′) spanning the putative E2F1 binding site on the FZD1 promoter. ChIP assay was performed with mouse normal IgG as a negative control [13,14].
Electrophoretic mobility shift assay (EMSA)
EMSA was performed using the Lightshift Chemiluminescent EMSA kit following the manufacturer's instructions (Pierce, Rockford IL). Nuclear protein was extracted from Saos2 cells over-expressing E2F1 using the Nuclear Extract Kit (Active Motif). E2F-specifc binding oligonucleotide (5′-CCGGCGGTGGCGCGAGCTGGGAAC-3′) of the FZD1 promoter was labeled with biotin at the 3′ end using a DNA biotin labeling kit (Pierce). Three micrograms of the nuclear extract was incubated with an excess of biotin labeled ds-DNA probe. The binding reaction (20 μl) was carried out at room temperature for 30 min in binding buffer and 0.1 μg of nonspecific competitor poly-dIdC (Pierce). Monoclonal E2F1 antibody (1 μl/reaction, KH95 X, Santa Cruz Biotechnology) was used in E2F1 super-shift experiments. Complexes were resolved on a 5% polyacrylamide gel, transferred to nylon membrane and visualized by Chemiluminescent Nucleic Acid Detection Module kit (Pierce) [13,15].
Mineralization and alizarin red staining
Saos-2 cells were cultured in DMEM medium supplemented with 100 U/ml penicillin, 100 mg/ml streptomycin (Sigma), and 15% fetal bovine serum (FBS, Gibco). Mineralization was induced by culturing confluent cells in growth medium supplemented with 50 mg/ml ascorbic acid (Sigma) and 7.5 mM β-glycerophosphate (Sigma). Cells were also cultured in growth media without the supplements in parallel as a control. After 10 days, cultured cells were washed with phosphate-buffered saline (PBS) and stained with 0.5% alizarin red-S (pH 4.2) for 10 min at room temperature. Free calcium ions were removed by washing with PBS four times. The culture dishes with stained cells were photographed and cells were detained with 10% cetylpyridinium chloride in PBS pH 7.0 for 20 min at room temperature. Alizarin red-S concentration was determined by measuring the absorbance at 562 nm [16].
Statistical analysis
All transfection and mineralization studies were conducted 2–3 times. For each of the experiments, samples were executed in triplicate for all testing conditions. Tests of statistical significance for transfection and mineralization assays were conducted using the Student's t-test. A p value <0.05 was considered statistically significant. Analyses were conducted using both graph pad and excel.
Results
E2F1 up-regulates FZD1 gene and protein expression
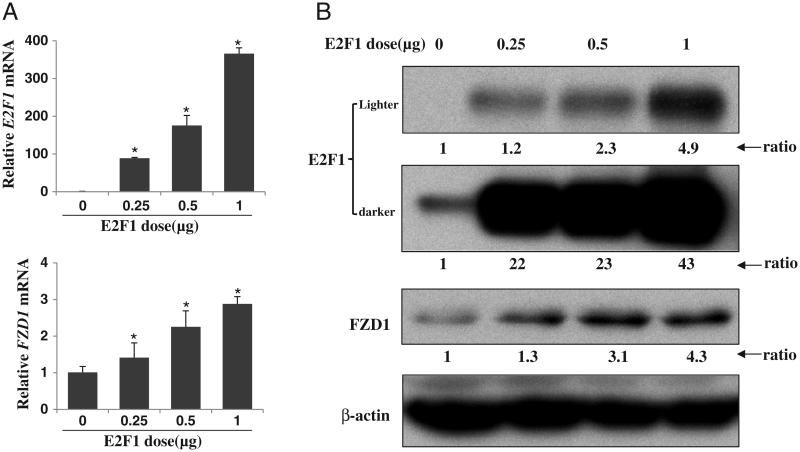
We previously reported that the FZD1 promoter polymorphisms rs2232157 and rs2232158 were associated with BMD and allele and haplotype specific promoter activity [5,6]. Based on an initial bioinformatic analysis, the G to T transversion polymorphism (rs2232157) at position−224 bp in the FZD1 promoter creates a putative E2F1 binding site [5], suggesting a role of E2F1 in FZD1 regulation. To determine if E2F1 affects the expression of FZD1 in osteoblastic cells, we analyzed FZD1 expression in Saos2 cells transiently expressing E2F1 protein. The dose-dependent over-expression of E2F1 mRNA and protein was confirmed in pCMV/E2F1 transfected Saos2 cells. Similarly, endogenous FZD1 gene expression and protein levels in these cells were up-regulated by E2F1 in a dose dependent manner (Figs. 1A and B). The FZD1 protein expression was increased 1.3, 3.1 and 4.3 fold in cells transfected with 0.25, 0.5 and 1 μg of pCMV/E2F1 plasmid, respectively.
Fig. 1.
E2F1 positively regulates expression of FZD1 in Saos2 cells. (A) Dose-dependent up-regulation of FZD1 mRNA by E2F1. Saos2 cells were transiently transfected with the indicated amounts of pCMV/E2F1 plasmid. After 36 h, cells were harvested for real time PCR analysis of both E2F1 and FZD1. (B) Level of FZD1 protein was up-regulated by E2F1. Saos2 cells were treated the same as above. After 36 h, whole cell lyses were harvested for Western blot analysis. The relative expression level of target protein related to untreated Saos2 cells are shown underneath and obtained by first calculating the expression ratio to β-actin followed by normalization to the ratio of untreated Saos2 cells.
E2F1 transactivates the FZD1 promoter
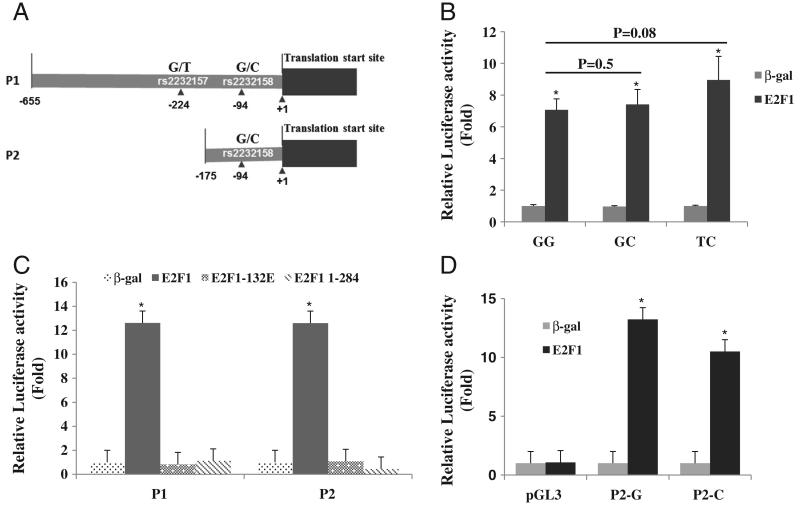
To determine if the up-regulation of FZD1 by E2F1 is due to transcriptional activation of the FZD1 promoter and if this transactivation is allele (rs2232157, rs2232158) and haplotype (rs2232157/rs2232158) specific, FZD1 promoters were co-transfected with or without pCMV/E2F1 plasmid using Saos2 cells. After 48 h of transfection, luciferase activities of the transfected cells were analyzed. Compared to the wild type GG haplotype (rs2232157G/rs2232158G), a higher transactivation was observed for the TC haplotype consisting of the rare alleles of rs2232157 and rs2232158 (Fig. 2B, 7.1 vs 9.0 fold) although this difference did not achieve statistical significance (p = 0.08). The level of transactivation for the GC haplotype (rs2232157G and rs2232158C) was similar to the GG haplotype (7.4 vs 7.1 fold). To determine if the region containing rs2232157 was essential for E2F1 transactivation, a plasmid spanning −175 to +71 of the FZD1 promoter was constructed and designated as P2. Deletion of the upstream sequences (−655 to −174) did not affect the ability of E2F to transactivate the FZD1 promoter since similar levels of promoter activity were observed for both the full-length promoter (P1) and the truncated promoter P2 (Fig. 2C, 12.6-fold relative to β-gal for both). Furthermore, we did not observe an effect of rs2232158 alleles on FZD1 activation by E2F1 in Saos2 cells (Fig. 2D).
Fig. 2.
E2F1 up-regulates FZD1 promoter activity in Saos2 cells. (A) Schematic of the FZD1 5′ UTR region and the DNA sequence locations of the P1 (−655 to +71) and P2 (−175 to +71) promoter clones. (B) Haplotype-specific E2F1 transactivation of the P1 FZD1 promoter. Saos2 cells were transfected with 0.2 μg wild type (GG), GC or TC haplotype specific FZD1 promoters in the presence or absence of 0.5 μg pCMV/E2F1 expression plasmid. After 36 h, cells were harvested for the dual luciferase assay. The p-values for direct comparisons of promoter activation by E2F1 between haplotypes are shown. (C) The effects of mutant E2F1 proteins on FZD1 promoter activity. P1 and P2 WT promoters were co-transfected with E2F1, E2F1-132E, or E2F1-1-284 expression plasmids in Saos2 cells. Dual luciferase assay was processed. (D) E2F1 activation of P2 FZD1 promoter is not influenced by rs2232158 alleles. Saos2 cells were transfected with 0.2 μg P2-G or P2-C promoter with or without 0.5 μg pCMV/E2F1 expression plasmid. After 36 h, cells were harvested for the dual luciferase assay. For all experiments, 10 ng pRL-null reporter was used as an internal control. The fold changes relative to the β-gal transfected cells are shown for panels B, C and D. * indicates significant changes in promoter activity between E2F1 and β-gal (p < 0.01).
To further confirm if E2F1 transactivates the FZD1 promoter in Saos2 cells, we co-transfected plasmids expressing mutated E2F1, E2F1-132E and E2F1-1-284. Both of the E2F1 mutants affected the binding of E2F1 to its DNA target sequence [17]. Neither of the E2F1 mutants was able to transactivate P1 or P2 FZD1 promoters (Fig. 2C), suggesting that E2F1 directly binds to the FZD1 promoter and is necessary for its transactivation.
E2F1 directly binds to the FZD1 promoter
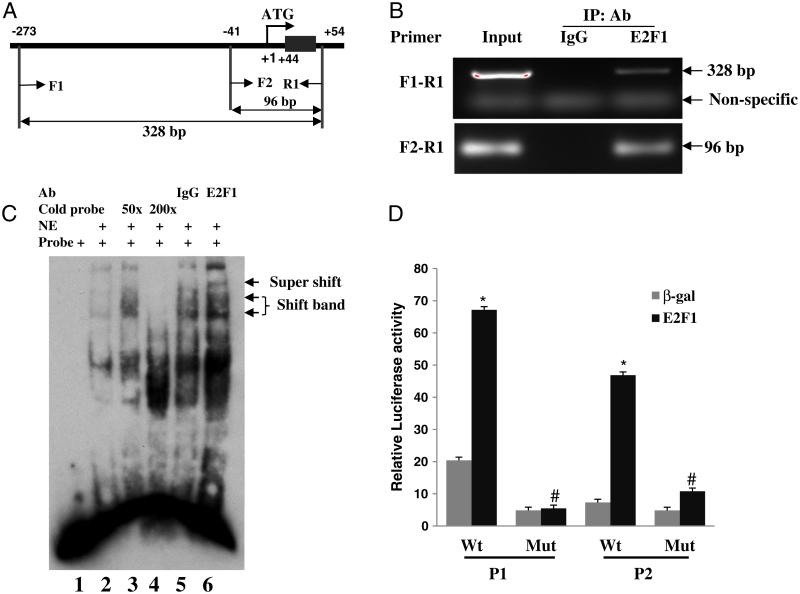
The similar transactivation of full-length and truncated FZD1 promoters by E2F1 suggests that the E2F1 binding site may be located between the region of −175 and +71 of the FZD1 promoter. To demonstrate that E2F1 directly binds to the endogenous FZD1 promoter, ChIP assay was performed on formalin fixed Saos2 using an E2F1 specific antibody. We were able to amplify the FZD1 promoter with primers spanning the region of −273 to +54 bp using the ChIP product with E2F1 antibody, while no FZD1 promoter was amplified using ChIP product of a negative IgG control (Fig. 3B). The positive amplification of both primer pairs further narrowed the E2F1 binding site to a 96 bp region (−41 to +54 bp). Consistent with the ChIP experiments, we identified a putative E2F1 consensus binding site (GCGCGA) at positions +47 to +52 bp from the FZD1 translation start site when we performed a bioinformatic analysis using wild type DNA sequence of the full-length promoter (−655 to +71 bp) and the Transcription Element Search Software (TESS). Direct binding of E2F1 to this putative binding site was subsequently demonstrated by EMSA using a labeled DNA probe containing the putative E2F1 binding site and nuclear extract from the Saos2 cells. As shown in Fig. 3C, nuclear extract formed complexes with the labeled probes (lane 2) whereas these complexes were completely competed out by 200-fold of unlabeled probe (lane 4). The existence of E2F1 in the shifted DNA-protein complexes was confirmed by supershifting the nuclear complexes with an E2F1 specific antibody (lane 6). No supershifted complexes were detected in the control reaction with non-specific rabbit IgG (lane 5). To further confirm that the putative consensus site is essential for the E2F1-dependent transactivation of FZD1, we mutated the binding site and tested its transactivation by E2F1. E2F1 mediated transactivation of the mutant FZD1 promoters was reduced 77% and 92% compared to their wild type promoter P1 and P2, respectively (Fig. 3D), further demonstrating that the putative site is a functional binding site for E2F1.
Fig. 3.
E2F1 binding to the FZD1 promoter. (A and B) ChIP assay of FZD1 promoter with antibody specific for E2F1 using Saos2 cells. A schematic representation of the relevant region of the human FZD1 promoter is shown. F1, F2 and R1 indicate PCR primers used to amplify FZD1 specific promoter sequences using ChIP DNA. (C) EMSA with probe specific for the FZD1 putative E2F1 binding site identified in silico. Labeled FZD1 DNA probe was incubated with 3 μg nuclear extracts (NE) from Saos2 cells over-expressing E2F1. For super gel shift assay, labeled FZD1 DNA probe was incubated with 3 μg NE from Saos2 cells in the presence of E2F1 antibody (lane 6) or control IgG (lane 5). The locations of shifted complex bands and E2F1 supershifted complex band are indicated. (D) Saos2 cells transfected with P1 WT or P2-WT promoter, or the same plasmid containing a 2-bp substitution mutation (Mut) in the putative E2F1-binding site with or without E2F1 expression plasmid. * indicates significant changes in promoter activity between E2F1 and β-gal (p < 0.01). # indicates significant changes in P1 or P2 promoter activity between WT and E2F1 binding site Mut reporter (p < 0.05).
E2F1-over-expressing Saos2 cells show enhanced osteoblastic differentiation
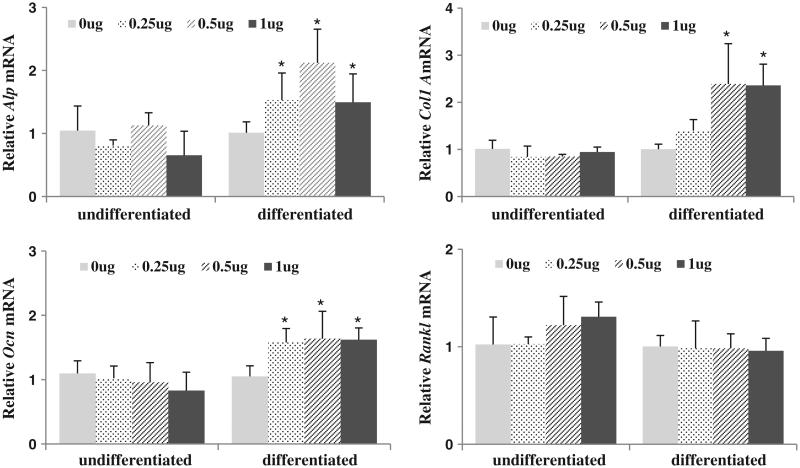
We have shown that FZD1 plays an important role in osteoblastic differentiation and mineralization of osteoblastic cells (Zhang et al, submitted). To determine if E2F1 expression is also important in osteoblastic differentiation, pCMV/E2F1 was transfected in Saos2 cells at different concentrations and the cells were treated with or without differentiation media for 48 h. Expression of genes associated with osteoblastic differentiation including ALP, COL1A, OCN and RANKL were analyzed in both differentiated and undifferentiated Saos2 cells. As shown in Fig. 4, over-expression of E2F1 led to an up-regulation of ALP, COL1A and OCN in differentiated Saos2 cells. Dose-dependent effects of E2F1 were observed for ALP and COL1A. Interestingly, with 1 μg of E2F1 expression plasmid, the expression level of ALP was lower than that observed with 0.5 μg of E2F1 plasmid, while no further increases were observed for COL1A and OCN. In undifferentiated cells, E2F1 did not significantly influence ALP, COL1A or OCN expression. In contrast, E2F1 did not influence the expression of RANKL, another osteoblast related gene, in either undifferentiated or differentiated Saos2 cells. These results suggest that ALP, COL1A and OCN are specifically regulated by E2F1 in differentiated Saos2 cells.
Fig. 4.
Over-expression of E2F1 promotes expression of genes associated with osteoblast differentiation. Saos2 cells transiently expressing E2F1 cDNA were cultured with or without ascorbic acid and β-glycophosphate for 48 h to induce osteoblast differentiation. The expression levels of ALP, Col1A, OCN and RankL were measured by real-time PCR. Results are normalized to HPRT. * indicates significant differences between the indicated groups (p < 0.05).
FZD1 plays a role in E2F1 mediated enhancement of osteoblast mineralization
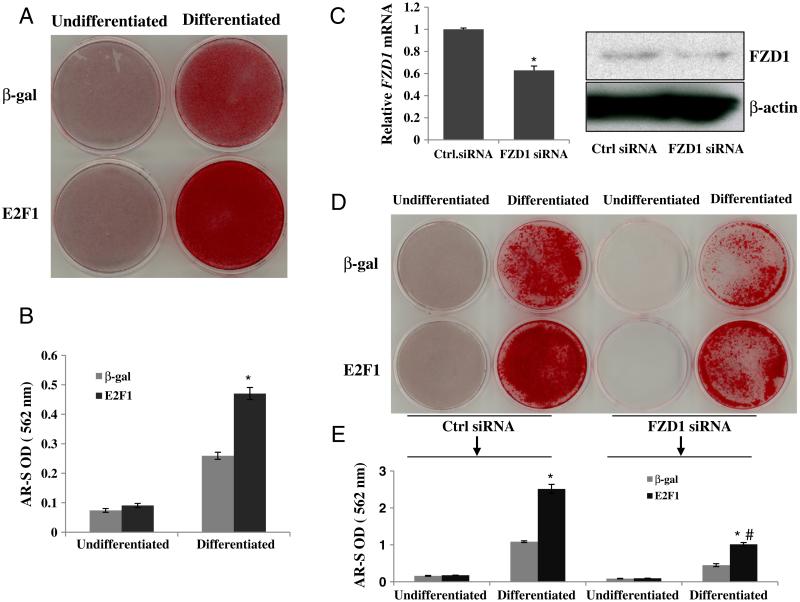
Because over-expression of E2F1 increased osteoblastic gene expression in our experiments, we hypothesized that Saos2 cells over-expressing E2F1 would also exhibit an enhanced ability to produce mineralized matrix. To test this hypothesis, we performed an initial mineralization assay in Saos2 cells infected with an E2F1 specific adenovirus (Ad-E2F1) or Ad-β-gal as a control, and the cells cultured in osteoblast differentiation medium (DMEM containing 50 mg/ml ascorbic acid and 7.5 mM β-glycerophosphate) for 10 days. Calcium nodules of osteoblastic mineralization were detected by alizarin red-S staining. Significantly stronger mineralization was detected in cultures of Saos2 cells over-expressing E2F1 compared to β-gal control (Figs. 5A and B).
Fig. 5.
E2F1 enhances osteoblast mineralization. (A) Alizarin red-S (AR-S) staining for mineralization of Saos2 cells. Saos2 cells infected with or without Ad-E2F1 were cultured in the presence or absence of osteoblast differentiation medium for 10 days, then cells were fixed and stained with AR-S. (B) Quantitative result of AR-S staining. AR-S was solubilized in cell cultures of all groups by 10% cetylpyridinium chloride and quantified at 562 nm (results are mean ± SD, n = 3). (C) FZD1 knockdown in Saos2 cells. Saos2 cell were transfected with 20 nM FZD1 siRNA or scramble siRNA. After 36 h, RNA and protein were prepared for real time PCR and Western blot, respectively. (D) AR-S staining for Saos2 cells with FZD1 knockdown and E2F1 over-expression. After treating with FZD1 siRNA for 36 h, Saos2 cells infected with or without adeno-E2F1 were cultured in the presence or absence of osteoblast differentiation medium for 10 more days, then cells were fixed and stained with AR-S. (E) Quantitative result of AR-S staining for panel D. * indicates significant changes in mineralization between E2F1 and β-gal treated Saos2 cells (p < 0.01). # indicates significant changes in mineralization between WT and FZD1 knockdown Saos2 cells (p < 0.05).
To determine if FZD1 is required for the effects of E2F1 on Saos2 mineralization, we performed FZD1 knock down experiments using FZD1 siRNA. The siRNA treatment decreased FZD1 gene and protein expression by 42%–50% (Fig. 5C). We found that FZD1 knockdown reduced Saos2 mineralization by 44% in the absence of E2F1 over-expression (Fig. 5D). Similarly, knockdown of FZD1 partially abolished the stimulatory effects of E2F1 on Saos2 mineralization (60% reduction, Figs. 5D and E). These experiments suggest that E2F1 may play a role in Saos2 osteoblastic mineralization and that this effect is mediated in part by FZD1.
E2F1 enhances FZD1 expression and osteoblast mineralization of human FOB cells
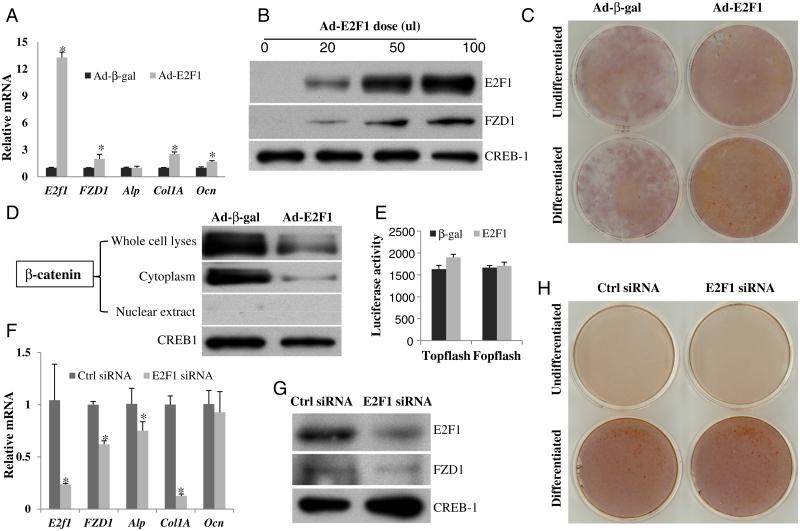
Given that the Saos2 cell line is derived from an osteosarcoma and has a major defect in the pRb/E2F1 pathway [18], we further tested the effects of E2F1 on FZD1 expression and mineralization in FOB, a normal human osteoblast cell line. Both mRNA and protein levels of FZD1 were increased by E2F1 in a dose dependent manner (Figs. 6A and B) after 36 h infection with Ad-E2F1. Additionally, relative gene expression levels of Col1A and OCN were increased by E2F1 (Fig. 6A). Furthermore, Ad-E2F1 enhanced mineralization of the FOB cells after 10 days of differentiation (Fig. 6C).
Fig. 6.
E2F1 positively regulates FZD1 expression and mineralization in human FOB cells. (A) FZD1, ALP, Col1A and OCN mRNA were up-regulated by E2F1. Human FOB cells were infected with Ad-E2F1 or Ad-β-gal. After 36 h, cells were harvested for real time PCR. The * indicates significant changes in mRNA between E2F1 and β-gal (p < 0.05). (B) Dose-dependent up-regulation of FZD1 protein by E2F1. Human FOB cells were infected with the indicated amounts of Ad-E2F1. Ad-β-gal was used to make the adenovirus concentration consistent in all treatments. After 36 h, whole cell lyses were harvested for Western blot analysis. (C) E2F1 enhances human FOB cell mineralization. FOB cells were infected with or without Ad-E2F1 for 36 h, followed by further culturing in the presence or absence of osteoblast differentiation medium for 10 more days and then cells were fixed and stained with AR-S. (D) E2F1 significantly decreases cytoplasmic β-catenin in FOB cells. (E) E2F1 has no effect on Topflash luciferase activity in FOB cells. FOB cell were co-transfected with pCMV/E2F1 and Topflash plasmids. After 36 h, protein was prepared for dual luciferase activity assay. (F) Real time PCR in human FOB cells with E2F1 knockdown. Human FOB cells were transfected with 25 nM E2F1 siRNA or scramble siRNA. After 36 h, RNA was prepared for real time PCR. The * indicates significant changes in mRNA between E2F1 and β-gal (p < 0.05). (G) Western blot in human FOB cells with E2F1 knockdown. Human FOB cells were treated the same as above, and protein was prepared for Western blot analysis. (H) E2F1 siRNA knockdown does not affect human FOB cell mineralization. FOB cells were transiently transfected with E2F1 siRNA for 36 h. Cells were cultured in the presence or absence of osteoblast differentiation medium for 10 more days and were subsequently fixed and stained with AR-S.
The effects of E2F1 on FZD1 expression and FOB mineralization were further analyzed using a loss-of-function experiment. Knockdown of E2F1 by siRNA resulted in the down-regulation of FZD1, ALP and Col1 A gene expression in FOB cells (Figs. 6F and G). However, we did not detect changes in mineralization by alizarin red staining for differentiated FOB cells treated with E2F1 siRNA (Fig. 6H).
To determine if E2F1 activates canonical Wnt signaling, we analyzed the levels of β-catenin protein and Topflash luciferase activity. We found that E2F1 significantly decreased the level of β-catenin protein in the cytoplasm, but it had a relatively smaller effect on nuclear β-catenin (Fig. 6D). Ad-E2F1 treatment had no significant effect on the Topflash luciferase activity in FOB cells (Fig. 6E).
Discussion
The Wnt pathway has emerged as a potential therapeutic target in metabolic bone diseases and Wnt signaling is mediated through the frizzled family of co-receptors [2]. The regulation of FZD1 mRNA and protein in osteoblasts and the role of FZD1 in bone biology are not well understood. In this study, we identified a putative E2F1 transcription factor binding element in the FZD1 promoter and characterized its stimulatory effects on endogenous FZD1 gene expression and protein levels in Saos2 osteoblastic cells. We also demonstrated that E2F1 increases differentiation and mineralization of Saos2 in vitro and that the stimulation of Saos2 mineralization by E2F1 was diminished when FZD1 was knocked down by siRNA. Up-regulation of FZD1 expression and osteoblast mineralization by E2F1 was further validated in normal human osteoblasts using the FOB cell line. Taken together, these data demonstrate a significant role of E2F1 in osteoblast differentiation and mineralization which appears to be mediated, at least in part, by up-regulation of FZD1.
E2F1 is a crucial transcription factor in cell cycle control and apoptosis. The role of E2F1 in FZD1 expression and osteoblast mineralization was analyzed in both Saos2 and FOB cells. In agreement with previous studies [19-21], our data show that E2F1 induces Saos2 cell apoptosis when it was transfected into cells at a high dosage (data not shown). In contrast, expression of E2F1 at a low level induces Saos2 differentiation as demonstrated by the increased expression of osteoblast differentiation marker genes ALP, COL1A and OCN at a later stage of differentiation (differentiating for 7 days), but not at an earlier undifferentiating stage (Fig. 4). More importantly, E2F1 dramatically increases Saos2 and FOB cell mineralization (Figs. 5 and 6). Our data support the concept that E2F1 plays a dose-dependent role in cell growth and survival [20,22]. The delicate balance between the expression of E2F1 target genes may be an important determinant of whether E2F1 will increase cell differentiation or induce apoptosis [20,22]. Transient expression of the highest dose (1 μg) of E2F1 did not further stimulate the expression of osteoblast marker genes (Fig. 4) but rather induced cell death in our experiments (data not shown). Many pathways, including the PI3K/Akt and p53 pathways, are involved in the regulation of gene expression by E2F1 in balancing the cellular decision of proliferation and apoptosis [20,22]. In addition, the differential effects of E2F1 on cell survival or cell death probably depends on the stage of growth and differentiation of the target cells (Fig. 4).
We have reported associations of FZD1 promoter polymorphisms with FZD1 gene expression in vitro and with BMD in population studies [5,6]. The single nucleotide polymorphism rs2232157 in the promoter region of FZD1 creates a putative E2F1 binding site [5]. We showed drastic stimulation of the FZD1 promoter by E2F1 in the current study. However, transactivation of FZD1 by E2F1 was only minimally influenced by the rs2232157T allele. Rather, E2F1 exerted its effects on FZD1 mRNA and protein expression through a novel binding site located between nucleotides +47 and +52 from the translation start site. Genome-wide analysis reveals that E2F1 binds to a large number of genomic regions and most of the binding sites are at core promoters with the distance of E2F1 sites relative to transcription start site peaking at −250 to +200 bp [23]. In addition, activation of E2F1 frequently needs the collaboration of a close SP1 binding site in the same region of the promoter [23]. There is a putative SP1 binding site at nucleotide +64 to +68 bp of the FZD1 promoter. Therefore, the novel E2F1 binding site in the FZD1 promoter fulfills the above criteria. In addition, we have demonstrated that E2F1 binds to the FZD1 promoter using both ChIP assay and EMSA and over-expression of E2F1 increases both FZD1 mRNA and protein levels in Saos2 cells (Fig. 3). Furthermore, FZD1 knockdown significantly abolished the stimulatory effects of E2F1 on Saos2 cell differentiation and mineralization (Fig. 5). Together, these results demonstrate that E2F1 increases Saos2 differentiation and mineralization, at least in part, through FZD1.
E2F1 decreased the level of β-catenin in the cytoplasm of FOB cells. This is in agreement with the reported down-regulation of β-catenin by E2F1 in Saos2 cells [18]. Consistent with the relatively small change in nuclear compared to cytoplasm β-catenin protein levels, the Topflash luciferase activity is not affected by E2F1 over-expression in FOB cells. In addition to the canonical Wnt β-catenin pathway, other signaling pathways mediated by FZD1 may also be involved in the E2F1-dependent activation of FZD1 and osteoblast mineralization. Both canonical and non-canonical Wnt signaling pathways are important for osteoblast differentiation, matrix mineralization and bone formation. For example, Wnt7b, a non-canonical Wnt pathway ligand, induces osteoblast differentiation and mineralization in vitro via the PKCδ-mediated pathway and ablation of Wnt7b in skeletal progenitors results in less bone in the mouse embryo [24]. Ror2, an orphan receptor tyrosine kinase, promotes osteoblast differentiation and enhances ex vivo bone formation through both canonical Wnt signaling dependent and independent pathways [25,26]. Caverzasio et al. also reported that Wnt3a stimulated ALP activity and mineralization of osteoblasts through the MAPK p38 pathway [27]. Non-canonical Wnt signaling also increased osteoblast maturation on microstructured titanium surfaces, in which up-regulation of FZD1, ALP activity and OCN expression were involved [28]. Over-expression of WNT3a or WNT7b resulted in the up-regulation of bone morphogenetic protein 2 (BMP2) gene expression in both FOB and Saos2 cells. The expression level of FZD1 was not affected by either of the wnt ligands (data not shown). BMP2 plays important roles in osteoblast differentiation and bone mineralization and is activated by Wnt signaling [29-31]. These findings support the fact that both canonical and non-canonical Wnt pathways are important in osteoblast differentiation and bone mineralization. Our present data, together with the aforementioned studies, indicates that E2F1 has the ability to both down-regulate cytoplasm β-catenin protein levels of the canonical Wnt pathway and potentiate non-canonical Wnt signaling. Additional studies are warranted to unravel the effects of E2F1 on non-canonical Wnt signaling in osteoblast differentiation and function.
In summary, the present study demonstrates that E2F1 is an important transcription factor for osteoblast differentiation and mineralization. FZD1 is a target of E2F1 and plays an important role in E2F1 mediated enhancement of osteoblast mineralization.
Footnotes
This study was supported by grants R01-AG033618 from the National Institute on Aging, and R01-AR049747 from the National Institute of Arthritis and Musculoskeletal and Skin Diseases and funding from the National Osteoporosis Foundation and Western Pennsylvania Arthritis Foundation. Laura M. Yerges-Armstrong was supported by National Institute on Aging grant T32-AG00181.
References
- 1.Baron R, Rawadi G, Roman-Roman S. Wnt signaling: a key regulator of bone mass. Curr Top Dev Biol. 2006;76:103–27. doi: 10.1016/S0070-2153(06)76004-5. [DOI] [PubMed] [Google Scholar]
- 2.Krishnan V, Bryant HU, Macdougald OA. Regulation of bone mass by Wnt signaling. J Clin Invest. 2006;116:1202–9. doi: 10.1172/JCI28551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Westendorf JJ, Kahler RA, Schroeder TM. Wnt signaling in osteoblasts and bone diseases. Gene. 2004;341:19–39. doi: 10.1016/j.gene.2004.06.044. [DOI] [PubMed] [Google Scholar]
- 4.Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, et al. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci U S A. 2002;99:4465–70. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y, Kuipers AL, Yerges-Armstrong LM, Nestlerode CS, Jin Z, Wheeler VW, et al. Functional and association analysis of frizzled 1 (FZD1) promoter haplotypes with femoral neck geometry. Bone. 2010;46:1131–7. doi: 10.1016/j.bone.2009.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yerges LM, Zhang Y, Cauley JA, Kammerer CM, Nestlerode CS, Wheeler VW, et al. Functional characterization of genetic variation in the frizzled 1 (FZD1) promoter and association with bone phenotypes: more to the LRP5 story? J Bone Miner Res. 2009;24:87–96. doi: 10.1359/JBMR.080816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muller H, Bracken AP, Vernell R, Moroni MC, Christians F, Grassilli E, et al. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 2001;15:267–85. doi: 10.1101/gad.864201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iaquinta PJ, Lees JA. Life and death decisions by the E2F transcription factors. Curr Opin Cell Biol. 2007;19:649–57. doi: 10.1016/j.ceb.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lees JA, Saito M, Vidal M, Valentine M, Look T, Harlow E, et al. The retinoblastoma protein binds to a family of E2F transcription factors. Mol Cell Biol. 1993;13:7813–25. doi: 10.1128/mcb.13.12.7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berman SD, Yuan TL, Miller ES, Lee EY, Caron A, Lees JA. The retinoblastoma protein tumor suppressor is important for appropriate osteoblast differentiation and bone development. Mol Cancer Res. 2008;6:1440–51. doi: 10.1158/1541-7786.MCR-08-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scheijen B, Bronk M, van der Meer T, Bernards R. Constitutive E2F1 overexpression delays endochondral bone formation by inhibiting chondrocyte differentiation. Mol Cell Biol. 2003;23:3656–68. doi: 10.1128/MCB.23.10.3656-3668.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunt KK, Deng J, Liu TJ, Wilson-Heiner M, Swisher SG, Clayman G, et al. Adenovirus-mediated overexpression of the transcription factor E2F-1 induces apoptosis in human breast and ovarian carcinoma cell lines and does not require p53. Cancer Res. 1997;57:4722–6. [PubMed] [Google Scholar]
- 13.Yu S, Franceschi RT, Luo M, Fan J, Jiang D, Cao H, et al. Critical role of activating transcription factor 4 in the anabolic actions of parathyroid hormone in bone. PLoS One. 2009;4:e7583. doi: 10.1371/journal.pone.0007583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu S, Jiang Y, Galson DL, Luo M, Lai Y, Lu Y, et al. General transcription factor IIA-gamma increases osteoblast-specific osteocalcin gene expression via activating transcription factor 4 and runt-related transcription factor 2. J Biol Chem. 2008;283:5542–53. doi: 10.1074/jbc.M705653200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu B, Zhao X, Yang C, Crane J, Xian L, Lu W, et al. Parathyroid hormone induces differentiation of mesenchymal stromal/stem cells by enhancing bone morphogenetic protein signaling. J Bone Miner Res. 2001;27:14. doi: 10.1002/jbmr.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu S, Zhu K, Lai Y, Zhao Z, Fan J, Im HJ, et al. ATF4 promotes beta-catenin expression and osteoblastic differentiation of bone marrow mesenchymal stem cells. Int J Biol Sci. 9:256–66. doi: 10.7150/ijbs.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cress WD, Johnson DG, Nevins JR. A genetic analysis of the E2F1 gene distinguishes regulation by Rb, p107, and adenovirus E4. Mol Cell Biol. 1993;13:6314–25. doi: 10.1128/mcb.13.10.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morris EJ, Ji JY, Yang F, Di Stefano L, Herr A, Moon NS, et al. E2F1 represses beta-catenin transcription and is antagonized by both pRB and CDK8. Nature. 2008;455:552–6. doi: 10.1038/nature07310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsieh JK, Fredersdorf S, Kouzarides T, Martin K, Lu X. E2F1-induced apoptosis requires DNA binding but not transactivation and is inhibited by the retinoblastoma protein through direct interaction. Genes Dev. 1997;11:1840–52. doi: 10.1101/gad.11.14.1840. [DOI] [PubMed] [Google Scholar]
- 20.Li Z, Kreutzer M, Mikkat S, Mise N, Glocker MO, Putzer BM. Proteomic analysis of the E2F1 response in p53-negative cancer cells: new aspects in the regulation of cell survival and death. Proteomics. 2006;6:5735–45. doi: 10.1002/pmic.200600290. [DOI] [PubMed] [Google Scholar]
- 21.Wang C, Chen L, Hou X, Li Z, Kabra N, Ma Y, et al. Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nat Cell Biol. 2006;8:1025–31. doi: 10.1038/ncb1468. [DOI] [PubMed] [Google Scholar]
- 22.Hallstrom TC, Mori S, Nevins JR. An E2F1-dependent gene expression program that determines the balance between proliferation and cell death. Cancer Cell. 2008;13:11–22. doi: 10.1016/j.ccr.2007.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao AR, Rabinovich R, Xu M, Xu X, Jin VX, Farnham PJ. Genome-wide analysis of transcription factor E2F1 mutant proteins reveals that N- and C-terminal protein interaction domains do not participate in targeting E2F1 to the human genome. J Biol Chem. 2011;286:11985–96. doi: 10.1074/jbc.M110.217158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tu X, Joeng KS, Nakayama KI, Nakayama K, Rajagopal J, Carroll TJ, et al. Noncanonical Wnt signaling through G protein-linked PKCdelta activation promotes bone formation. Dev Cell. 2007;12:113–27. doi: 10.1016/j.devcel.2006.11.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Billiard J, Way DS, Seestaller-Wehr LM, Moran RA, Mangine A, Bodine PV. The orphan receptor tyrosine kinase Ror2 modulates canonical Wnt signaling in osteoblastic cells. Mol Endocrinol. 2005;19:90–101. doi: 10.1210/me.2004-0153. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, Bhat RA, Seestaller-Wehr LM, Fukayama S, Mangine A, Moran RA, et al. The orphan receptor tyrosine kinase Ror2 promotes osteoblast differentiation and enhances ex vivo bone formation. Mol Endocrinol. 2007;21:376–87. doi: 10.1210/me.2006-0342. [DOI] [PubMed] [Google Scholar]
- 27.Caverzasio J, Manen D. Essential role of Wnt3a-mediated activation of mitogen-activated protein kinase p38 for the stimulation of alkaline phosphatase activity and matrix mineralization in C3H10T1/2 mesenchymal cells. Endocrinology. 2007;148:5323–30. doi: 10.1210/en.2007-0520. [DOI] [PubMed] [Google Scholar]
- 28.Olivares-Navarrete R, Hyzy SL, Hutton DL, Dunn GR, Appert C, Boyan BD, et al. Role of non-canonical Wnt signaling in osteoblast maturation on microstructured titanium surfaces. Acta Biomater. 7:2740–50. doi: 10.1016/j.actbio.2011.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katagiri T, Yamaguchi A, Komaki M, Abe E, Takahashi N, Ikeda T, et al. Bone morphogenetic protein-2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. J Cell Biol. 1994;127:1755–66. doi: 10.1083/jcb.127.6.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wozney JM, Rosen V. Bone morphogenetic protein and bone morphogenetic protein gene family in bone formation and repair. Clin Orthop Relat Res. 1998:26–37. [PubMed] [Google Scholar]
- 31.Zhang R, Oyajobi BO, Harris SE, Chen D, Tsao C, Deng HW, et al. Wnt/beta-catenin signaling activates bone morphogenetic protein 2 expression in osteoblasts. Bone. 52:145–56. doi: 10.1016/j.bone.2012.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]