Abstract
Recurrent chromosomal aberrations in solid tumors can reveal the genetic pathways involved in the evolution of a malignancy and in some cases predict biological behavior. However, the role of individual genetic backgrounds in shaping karyotypes of sporadic tumors is unknown. The genetic structure of purebred dog breeds coupled with their susceptibility to spontaneous cancers provides a robust model with which to address this question. We tested the hypothesis that there is an association between breed and the distribution of genomic copy number imbalances in naturally-occurring canine tumors through assessment of a cohort of Golden Retrievers and Rottweilers diagnosed with spontaneous appendicular osteosarcoma. Our findings reveal significant correlations between breed and tumor karyotypes that are independent from gender, age at diagnosis and histological classification. These data indicate for the first time that individual genetic backgrounds, as defined by breed in dogs, influence tumor karyotypes in a cancer with extensive genomic instability.
Keywords: microarray, comparative genomic hybridization (CGH), canine, osteosarcoma, chromosome
Introduction
Appendicular osteosarcoma (OSA) in domestic dogs is a spontaneous malignancy that, unlike induced tumors of rodent model systems, shares the extensive degree of heterogeneity in histopathology, clinical presentation and outcome associated with that of its human counterpart (Mueller et al. 2007). As a consequence the dog has been used to model therapeutic development for human OSA for more than three decades (MacEwen 1990). During this time the significance of genetic factors in influencing OSA development and behavior has become increasingly well recognized (reviewed in Sandberg and Bridge 2003; Wang 2005; Kansara and Thomas 2007). Extensive studies have shown that numerous cancers are associated with non-random chromosome abnormalities (Mitelman et al. 2008), a proportion of which act as reliable indicators for detailed molecular subclassification of tumor subtypes. Unlike some tumors OSA in human patients is not associated with a small and well-defined subset of highly recurrent chromosome aberrations that indicate distinct diagnostic and/or prognostic status. Rather, human OSA typically presents with highly variable and chaotic karyotypes ranging from hypodiploidy to extreme hyperploidy, comprising numerous and diverse structural and numerical chromosome aberrations (for example Bridge et al. 1997; Ragland et al. 2002; Sandberg and Bridge 2003). This in part reflects the fact that multiple complex molecular pathways appear to be involved in the pathogenesis of OSA (reviewed in Wang 2005). A more complete understanding of the nature of these pathways, and how they interact in the development and progression of a tumor, will undoubtedly aid the development of more sophisticated diagnostic definitions, improved prognostication and novel therapies that target the key components of these molecular processes (reviewed by Boehm et al. 2000; Sandberg and Bridge 2003; Wang 2005). The vast spectrum of findings arising from the many cytogenetic studies of human OSA (for example Bridge et al. 1997; Tarkkanen et al. 1999; Boehm et al. 2000; Stock et al. 2000; Ozaki et al. 2002; Atiye et al. 2005), however, highlights the difficulty in defining the biological and clinical significance of this multitude of cytogenetic abnormalities. There is growing evidence that canine and human OSA share commonality in their molecular pathogenesis (Mendoza et al. 1998; Levine and Fleischli 2000; Levine 2002; Levine et al. 2002, reviewed in Vail and MacEwen 2000; Khanna et al. 2004; Modiano et al. 2006; Kansara and Thomas 2007), which has led to exciting advances in preclinical gene therapy approaches (for example Hemminki et al. 2003; Modiano et al. 2006; Witlox et al. 2007). Since the canine model possesses key attributes that may accelerate discovery of pathogenetically significant factors for OSA in both species this neoplasm represents an excellent example for which a comparative and translational approach to molecular oncology is warranted. Human OSA is essentially an orphan disease and thus accrual of large, homogeneous datasets is challenging (Khanna et al. 2002; Mansky et al. 2002). With approximately 10,000 cases of canine primary appendicular OSA diagnosed each year in the USA the incidence of OSA in the dog is at least 10-fold greater than its human counterpart (Withrow 2003; Khanna et al. 2006; Jemal et al. 2008) and is highly elevated in certain large and giant dog breeds, including the Rottweiler, German Shepherd Dog, Great Dane, Scottish Deerhound and Greyhound (Vail and MacEwen 2000; Lord et al. 2007; Phillips et al. 2007; Rosenberger et al. 2007 and others). The strong association between breed and predisposition to OSA suggests that genetic factors play a key role in determining disease susceptibility. These factors may contribute a more clearly defined risk of tumorigenesis in the relatively ‘genetically homogeneous’ purebred dog than is the case for more genetically diverse human populations, in which biologically significant genomic changes may lie close to the limits of detection among the high level of ‘background noise’. We, and others, therefore propose that identifying genetic factors associated with OSA susceptibility and clinical behavior in dog breeds may be simpler than in human populations (see Ostrander and Wayne 2005 for a review), particularly for highly complex genomes such as those associated with OSA. Furthermore, we propose that identification of chromosome aberrations that are evolutionarily conserved in canine and human OSA will aid identification of those genetic factors that are most intimately involved in the initiation and evolution of the tumor versus those that are secondary features of the chaotic OSA karyotypes of human patients.
We reported recently key evidence supporting a common molecular basis for canine and human tumors through identification of etiologically significant cytogenetic aberrations in lymphoma and leukemia of the dog that are orthologous to those of their human counterpart (Breen and Modiano 2008). The molecular cytogenetics of canine OSA has not yet been explored in any detail, though a comprehensive, genome-wide analysis would undoubtedly help to clarify and refine the molecular basis of this cancer. This has now become possible through development of robust resources for genomic analysis of canine tumors (Lindblad-Toh et al. 2005; Thomas et al. 2005; Karlsson et al. 2007; Thomas et al. 2007; Boggs et al. 2008). We report targeted microarray-based comparative genomic hybridization (aCGH) analysis of 38 canine OSA cases from two popular but genetically distinct dog breeds with high generalized cancer susceptibility, morbidity and mortality. The first, the Rottweiller, presents with a high relative risk of OSA (~ 12.5% incidence rate) while the second, the Golden Retriever, presents with a 2-3 fold lower relative risk of OSA (~ 5% incidence rate) but with a particularly high frequency of other cancers (Glickman et al. 2000; Cooley et al. 2002; Parker et al. 2004). We investigated the range and frequency of recurrent chromosome copy number aberrations (CNAs) and assessed evidence for genomic imbalances that were associated with the genetic background (i.e. breed) of the patient, and thus potentially with disease risk. We use these data to draw comparisons with prior cytogenetic studies of human OSA to identify genomic imbalances that appear to be conserved between species, and discuss further the relevance of the dog as a model for human OSA.
Materials and Methods
Appendicular osteosarcoma cohort
Cases of naturally occurring appendicular osteosarcoma (stage I or II) in family-owned dogs were recruited from community or institutional practices within the USA between November 1999 and January 2006. A total of 38 OSA cases were studied by aCGH, comprising 29 Rottweilers and nine Golden Retrievers. Clinical specimens were obtained during routine clinical work-up, prior to initiation of chemotherapy or radiation, and under approved protocols with informed client consent. Specimens were processed as described previously (Thomas et al. 2005). Briefly, tumor tissue was retrieved under sterile conditions as part of a diagnostic biopsy procedure, or immediately after amputation of the affected limb, and grossly normal tissue was dissected away. A representative portion of the tumor was fixed in 10% neutral-buffered formalin and used to confirm the diagnosis of OSA and to classify the tumor according to the criteria of Kirpensteijn et al. (2002). Specifically, the presence of osteoid or bony matrix was required to confirm a diagnosis of OSA, and subclassification into osteoblastic, chondroblastic, or fibroblastic OSA was based on cellular morphology and the presence of other types of matrix (chondroid or collagen) within the tumor. Signalment data for all OSA cases analyzed are shown in table 1, and are summarized in table 2.
Table 1.
Signalment data for canine appendicular OSA cases analyzed by aCGH.
| Case | Gender | Breed | Subtype | Age at Diagnosis (YR) |
|---|---|---|---|---|
| OSA-1 | F | Rottweiler | Chondroblastic | 10 |
| OSA-2 | F | Rottweiler | Osteoblastic | 9 |
| OSA-3 | M | Rottweiler | Osteoblastic | 9 |
| OSA-4 | M | Rottweiler | Osteoblastic | 10 |
| OSA-5 | F | Rottweiler | Osteoblastic | 4 |
| OSA-6 | F | Rottweiler | Osteoblastic | 6 |
| OSA-7 | M | Rottweiler | Osteoblastic | 2 |
| OSA-8 | M | Golden Retriever | Osteoblastic | 8 |
| OSA-9 | F | Rottweiler | Chondroblastic | 5 |
| OSA-10 | M | Rottweiler | Osteoblastic | 7 |
| OSA-11 | M | Golden Retriever | Osteoblastic | 7 |
| OSA-12 | F | Rottweiler | Fibroblastic | 9 |
| OSA-13 | F | Rottweiler | Osteoblastic | 6 |
| OSA-14 | M | Rottweiler | Osteoblastic | 10 |
| OSA-15 | M | Rottweiler | Fibroblastic | 9 |
| OSA-16 | M | Rottweiler | Osteoblastic | 9 |
| OSA-17 | M | Golden Retriever | Osteoblastic | 9 |
| OSA-18 | F | Golden Retriever | Osteoblastic | 3 |
| OSA-19 | F | Rottweiler | Fibroblastic | 7 |
| OSA-20 | F | Rottweiler | Osteoblastic | 7 |
| OSA-21 | M | Rottweiler | Chondroblastic | 10 |
| OSA-22 | F | Rottweiler | Osteoblastic | 12 |
| OSA-23 | M | Rottweiler | Chondroblastic | 10 |
| OSA-24 | F | Golden Retriever | Osteoblastic | 3 |
| OSA-25 | M | Rottweiler | Osteoblastic | 4 |
| OSA-26 | F | Rottweiler | Fibroblastic | ? |
| OSA-27 | F | Rottweiler | Osteoblastic | 8 |
| OSA-28 | M | Rottweiler | Osteoblastic | 7 |
| OSA-29 | F | Rottweiler | Osteoblastic | 8 |
| OSA-30 | F | Rottweiler | Osteoblastic | 9 |
| OSA-31 | M | Golden Retriever | Chondroblastic | 4 |
| OSA-32 | ? | Rottweiler | Osteoblastic | ? |
| OSA-33 | M | Golden Retriever | Osteoblastic | 8 |
| OSA-34 | M | Rottweiler | Osteoblastic | 8 |
| OSA-35 | F | Golden Retriever | Chondroblastic | 9 |
| OSA-36 | M | Rottweiler | Osteoblastic | 9 |
| OSA-37 | F | Golden Retriever | Fibroblastic | 11 |
| OSA-38 | F | Rottweiler | Fibroblastic | 8 |
Table 2.
Comparison of signalment characteristics according to breed.
| Golden Retrievers | Rottweilers | |
|---|---|---|
| Relative incidence of OSA* | ~ 5% | ~ 12.5% |
|
| ||
| Number of cases analyzed | 9 | 29 |
|
| ||
| Age** | ||
| Mean (SD) | 6.9 (2.7) | 7.5 (2.3) |
| Median | 7.8 | 7.8 |
| Range | 2.7 – 11.0 | 2.0 – 12.1 |
|
| ||
| Gender | ||
| Male (%) | 5 (55.6) | 15 (51.7) |
| Female (%) | 4 (44.4) | 14 (48.3) |
| Neutered*** (%) | 6 (66.7) | 12 (42.9) |
|
| ||
| Tumor histology | ||
| Osteoblastic (%) | 6 (66.7) | 20 (69.0) |
| Chondroblastic (%) | 2 (22.2) | 4 (13.8) |
| Fibroblastic (%) | 1 (11.1) | 5 (17.2) |
Glickman et al. 2000; Cooley et al. 2003
The age of two Rottweilers was unknown
Neutered includes castrated males and spayed females. The neuter status of one male Rottweiler was unknown.
Cytogenetic analysis of dog osteosarcomas
High molecular weight DNA was isolated from fresh tumor tissue for aCGH analysis using conventional techniques. Primary cell cultures were also initiated, and the osteoblastic origin of the isolated cells was confirmed at the third passage by expression of alkaline phosphatase and osteocalcin (IHC Services, Smithville, TX). Chromosome preparations and interphase nuclei for subsequent FISH analysis were generated by direct preparation from the primary bone tumor or from low passage (n = 3) cell lines using conventional techniques of colcemid arrest, hypotonic treatment and methanol/glacial acetic acid fixation. aCGH analysis of OSA cases was performed as described previously (Thomas et al. 2007) using a genomic microarray comprising 200 canine bacterial artificial chromosome (BAC) clones, which have been i) cytogenetically assigned to a unique chromosome location by multicolor fluorescence in situ hybridization (FISH) analysis and ii) anchored into the 7.5× dog genome assembly (Lindblad-Toh et al. 2005). The array included BAC clones representing dog orthologs of 25 human cancer-associated genes (Thomas et al. 2003). Equimolar pools of DNA isolated from 10 or more healthy, sex-matched individuals of the same breed were used as the reference sample. Hybridization and data analysis were performed as described elsewhere (Thomas et al. 2007). Briefly, tumor and reference samples were differentially labeled with Cyanine-3-dCTP and Cyanine-5-dCTP respectively, and combined in the presence of dog Cot1 competitor DNA. The combined probe mix was denatured and applied to the microarray for 48 hours, followed by a series of stringency washes. Arrays were scanned using a Perkin Elmer Scannarray Express, using the manufacturer’s software to determine the tumor DNA:reference DNA fluorescence intensity at each genomic locus. Threshold limits for detection of CNAs were set at log2 values equivalent to 1.15:1 (copy number gain) and 0.85:1 (copy number loss) of tumor DNA vs. reference DNA ratios respectively. Associations between CNAs and signalment data were then investigated using the Fisher’s exact test.
FISH analysis was performed on a representative subset of the patient population, in order to evaluate the distribution of copy number changes revealed by aCGH data within individual cells isolated from the primary tumor. For these cases multicolor FISH analysis of tumors was carried out as described elsewhere (Breen et al. 2004) using BAC clones selected from regions showing a range of normal and aberrant copy number ratios in aCGH analysis. All probes were hybridized first onto metaphase chromosome preparations from clinically healthy dogs to confirm the expected copy number (n = 2) for each probe at the expected chromosomal location. Images were acquired from a minimum of 30 representative cells in each instance. The copy number status of each probe was scored by two independent investigators with no prior knowledge of the corresponding aCGH data.
Chromosome assignments of BAC clones are reported according to the DAPI-banded dog karyotype nomenclature of Breen et al. (1999). The corresponding location of each clone within the dog genome assembly (Lindblad-Toh et al. 2005) is denoted according to the chromosome of origin and then the megabase (Mb) position on that chromosome (for example, CFA 1; 3.2Mb). Regions of conserved synteny between the dog (CanFam v2.0) and human (build 36.1) genomes are based on comparative genome sequence assembly data located at http://genome.ucsc.edu (Kent 2002).
Results
Observations of genomic organization in dog OSA
The domestic dog karyotype comprises 38 pairs of acrocentric autosomes and two metacentric sex chromosomes (Breen et al. 1999; Breen 2008). In contrast, a combination of classical banding analysis, aCGH and multicolor FISH analysis showed that appendicular OSA of the dog is a cytogenetically chaotic tumor that typically presents with extensive numerical and structural genomic rearrangements. The most striking observations in canine OSA were the formation of aberrant bi-armed derivative chromosomes through centric fusion events, with a consequent reduction in chromosome number (hypodiploidy). Chromatid breakages were evident in cells of several tumors, as were chromosomes with multiple centromeres. The observed complex nature of canine OSA karyotypes exemplified the immense challenge of identifying and characterizing recurrent chromosome aberrations using classical cytogenetic techniques.
Recurrent genomic imbalances exist in dog OSA
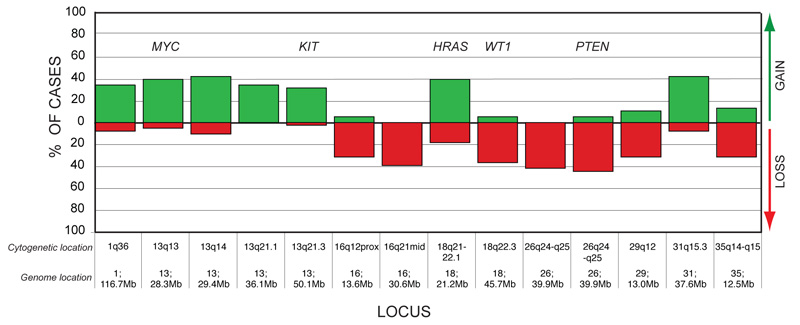
The high level of genomic rearrangement commonly observed in individual tumor metaphases was recapitulated by aCGH analysis, which revealed a wide range of non-random CNAs distributed throughout the genome. A complete listing of the aCGH data for each case is available as supplementary on line material (Supplementary Table S1). Of the loci assayed in this study, 14 loci showed either copy number gain or loss in at least 30% of the OSA cases analyzed (≥ 12/38 cases, figure 1). These 14 loci were distributed across eight different chromosomes and included regions containing five key cancer-associated genes. The most common aberration observed in the cohort was deletion of CFA 26q25 (39.9Mb), which encompasses the PTEN tumor suppressor gene locus, occurring in 42% of OSA cases (16/38 cases). Four cases, all of which were osteoblastic tumors from Rottweilers, showed log2 tumor DNA:reference DNA ratios < 1.0 at the PTEN locus, highly suggestive of homozygous deletion. Recurrent losses were also observed for regions of CFA 16, 18, 29 and 35 (figure 1). Of particular note was loss of the WT1 gene locus at CFA 18q22.3 (45.7Mb) in 37% of tumors (14/38 cases).
Figure 1.
Summary of highly-recurrent genomic imbalances in dog OSA. Genomic location (chromosome and assembly position) of loci that were aberrant in ≥ 30% of the cohort are shown on the x-axis, along with their cytogenetic location (Breen et al. 1999) and their Mb position within the dog genome sequence assembly (Lindblad-Toh et al. 2005; Thomas et al. in press). The y-axis indicates the percentage of the OSA cohort that showed copy number gain (green bar above the x-axis) or loss (red bar below the x-axis) of that locus. Regions harboring known cancer-associated genes are indicated above the corresponding bar.
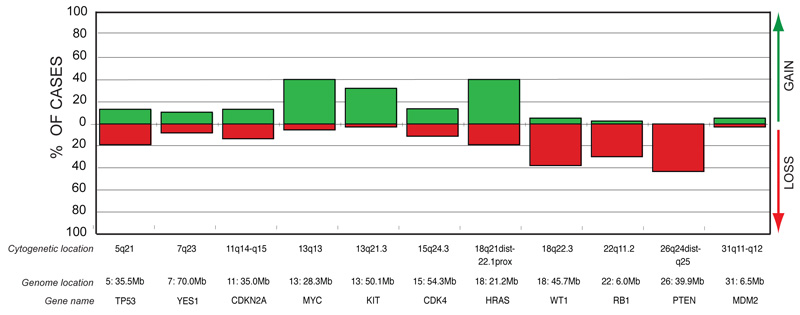
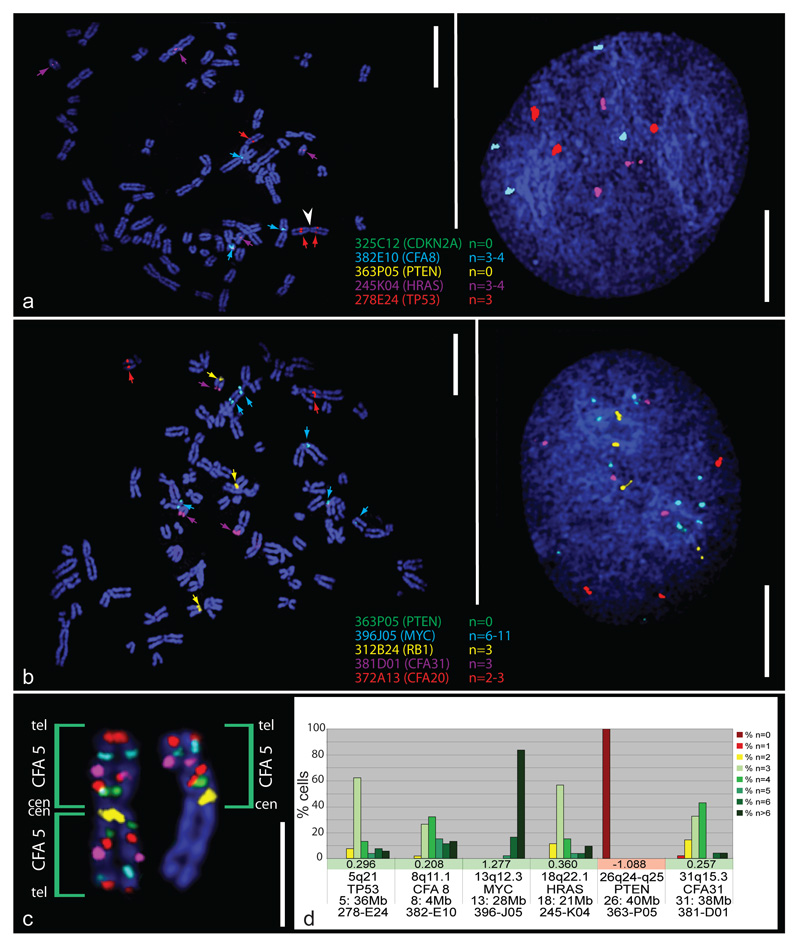
The most common copy number increases were identified for CFA 13q14 (29.4Mb) and CFA 31q15.3 (37.6Mb), both present in 42% of tumors (16/38 cases). Copy number gains of two oncogenes, MYC (CFA 13q13; 28.3Mb) and HRAS (CFA 18q21-q22.1; 21.2Mb), occurred in 40% of the cohort (15/38 cases). Three osteoblastic OSA cases (two Golden Retrievers and one Rottweiler) showed high-level amplification of MYC, with log2 tumor DNA:reference DNA ratios > 1.0. Figure 2 shows the incidence of copy number imbalance for 11 key cancer-associated genes within the OSA cohort. Figure 3 illustrates FISH analysis of one of the 38 cases of canine OSA that showed a range of copy number aberrations, including both high level gain of MYC and homozygous deletion of PTEN.
Figure 2.
Copy number imbalances for 11 cancer-associated genes within the OSA cohort. Cytogenetic and genome locations are indicated above each gene name
Figure 3.
aCGH-directed FISH analysis of dog OSA. a) and b) show multicolor FISH analysis of 10 BAC clones on metaphase chromosomes (left) and interphase nuclei (right) from a case of osteoblastic osteosarcoma in a female Rottweiler. Numerous aberrant bi-armed chromosomes are apparent, resulting from fusion events between acrocentric chromosomes. FISH analysis supported genomic copy number aberrations identified in prior aCGH-analysis, involving a series of known oncogenes and tumor suppressor genes, including homozygous deletion of PTEN and CDKN2A, and high-level amplification of MYC. Additionally, targeted FISH analysis revealed structural chromosome aberrations that are intractable to aCGH analysis. For example, a) shows a metaphase spread with three copies of the TP53 marker (CFA 5q21, red probe). One hybridization signal was located on each arm of a bicentric chromosome structure (white arrowhead), suggestive of an isochromosome of CFA 5q. The third TP53 signal was located on another biarmed structure, consistent with CFA 5 fused with another chromosome. The extent of CFA 5 DNA sequence represented by these aberrant chromosomes was then investigated by hybridization of a panel of six BAC clones selected at intervals along the full length of the CFA 5 sequence assembly. These six clones are located on CFA 5 at 3Mb (yellow signal); 24Mb (green signal); 36Mb (TP53, red signal), 53Mb (purple signal), 74Mb (aqua signal) and 92Mb (red signal) (Thomas et al. 2007). The resulting pattern of probe hybridization (c) supported the presence of an isochromosome of CFA 5 (left homologue). d) Summary of enumeration data for selected BAC probes in a) and b) within ≥ 30 cells from the OSA case. These data also show the log2 ratios from the BAC array for these loci and indicate that the FISH probe enumeration data and aCGH data are mutually supportive. Scale bars in a) and b) represent 10μm and the scale bar in c) represents 5μm.
Identification of recurrent breed-associated genomic imbalances in dog OSA
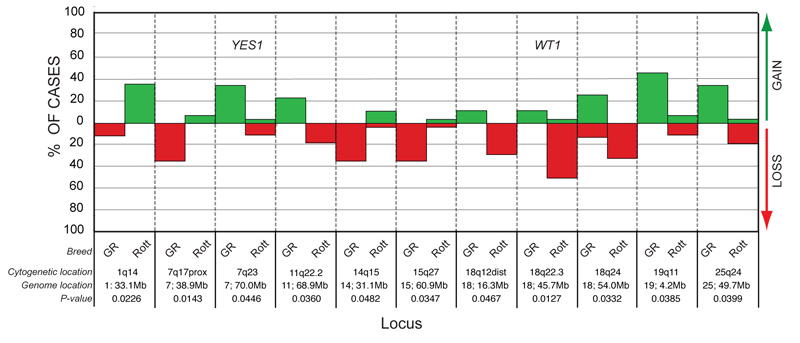
Comparison of signalment data for OSA cases showed no significant differences between Golden Retrievers and Rottweilers based upon the mean age of onset, the distribution of cases by gender or the distribution of cases by histological subtype (p > 0.05). A Fisher’s exact test was performed to test the association between the copy number status of each genomic locus from the microarray and the breed of the patient. Eleven loci (from eight different chromosomes) showed a significant (p < 0.05) difference in the distribution of DNA copy number imbalances between tumors from Golden Retrievers as compared to those from Rottweilers (figure 4). The most significant of these (p = 0.0127) was deletion of the WT1 gene (CFA 18q22.3, 45.7Mb), which occurred in 14/29 Rottweiler OSA cases (48%), but which was not observed in any of the nine Golden Retrievers. Deletions of chromosomal regions both proximal and distal to the WT1 locus on CFA 18 also were significantly more common in tumors from Rottweilers (p = 0.0127 to 0.0467), while tumors from Golden Retrievers tended towards genomic balance in these regions. Loss of CFA 7q17 (38.9Mb) was significantly more common in tumors from Golden Retrievers than Rottweilers (p = 0.0143). In contrast, Golden Retrievers showed a significant (p = 0.0446) association with genomic gain of a more distal region of the same chromosome, CFA 7q23 (70.0Mb), which contains the YES1 locus. Gains of CFA 19q11 (4.2Mb) and CFA 25q24 (49.7Mb), and loss of CFA 14q15 (31.1Mb), were also more frequently associated with OSA cases from Golden Retrievers (p = 0.0385, p = 0.0399 and p = 0.0482, respectively), while gains of CFA 1q14 (33.1Mb) were restricted to Rottweilers (p = 0.0226). Finally, when breed comparisons were restricted to osteoblastic OSA cases, a significant association was detected between gain of the region containing the KIT oncogene (CFA 13q21.3) and the Rottweiler breed (observed in 8/20 Rottweiler tumors but 0/6 Golden Retriever tumors, p = 0.037). The numbers of chondroblastic and fibroblastic cases were insufficient for this analysis.
Figure 4.
Breed-associated chromosome copy number changes in OSA. Eleven genomic regions showed a significant association between copy number status and the breed of the patient. Regions harboring known cancer-associated genes are indicated in the corresponding bar. The p-value denoting the significance of breed-association is listed against each locus.
Discussion
Canine and human osteosarcoma share common genomic imbalances
We present a preliminary assessment of gross genomic aneuploidy in 38 OSA cases from two dog breeds with differing relative risk of this cancer. Our hypothesis was that there is a direct correlation between breed and cytogenetic aberrations in spontaneous canine OSA. If supported, this would allow us to leverage the power of this model to identify evolutionarily conserved, heritable abnormalities that might influence risk and progression of this disease in both dogs and humans.
In testing our hypothesis we have investigated whether dog OSA harbors recurrent cytogenetic aberrations that are evolutionarily consistent with those previously reported in human OSA. Our findings demonstrate that, as with its human counterpart, dog OSA has a tendency towards highly complex and chaotic karyotypes comprising a wide range of both structural and numerical aberrations, including gene dosage imbalances of known oncogenes and tumor suppressor genes (figure 2). Copy number gain of CFA 13, including both the MYC and KIT oncogenes, was among the most frequent CNAs identified in our dog OSA cohort (figure 1). Gain of CFA 13 is also common within a broad spectrum of other dog cancers, including lymphoma, glial tumors, histiocytic sarcomas and prostate tumors (Hahn et al. 1994; Dunn et al. 2000; Thomas et al. 2005; Winkler et al. 2006; Kisseberth et al. 2007 and our unpublished observations). Our observations are consistent with the fact that genomic gains of HSA 8q24 and HSA 4p, the human chromosome regions orthologous to CFA 13, are among the most common numerical aberrations encountered in human OSA, in which they are predictive of outcome (Tarkkanen et al. 1999; Boehm et al. 2000; Stock et al. 2000; Entz-Werle et al. 2005; Entz-Werle et al. 2007 and others, reviewed in Sandberg and Bridge 2003). Interestingly the KIT oncogene has recently been proposed as a novel target for therapy in pediatric OSA (Entz-Werle et al. 2007). The identification of frequent KIT copy number increases in dog OSA suggests that this approach may also be clinically effective in a veterinary setting. Genomic amplification of the HRAS oncogene is also commonly encountered in human OSA (reviewed in Ragland et al. 2002), and similarly was among the most frequent observations in our dog OSA cohort (CFA 18q22.1) (figure 1).
We also assessed the copy number status of key tumor suppressor genes in dog OSA (figures 1 and 2). Deletion of the PTEN tumor suppressor gene (CFA 26q24-q25) was the most common copy number loss in our dog OSA cases (41% of the cohort). This is consistent with an earlier study in which reduction of PTEN protein, coupled with deletions involving the gene itself, was reported in canine OSA cell lines (Levine et al. 2002). Similarly, genomic loss of PTEN, and a consequent decrease in PTEN gene expression, is a frequent event in human OSA, which often exhibits homozygous gene deletion resulting in complete loss of PTEN gene expression (Freeman et al. 2008). Loss of heterozygosity of regions harboring RB1 and TP53 (HSA 13q14 and HSA 17p13 respectively) is also common in human OSA and indeed inactivation of Rb and p53 are both among the most frequently encountered molecular abnormalities in human OSA (reviewed in Sandberg and Bridge 2003). A recent conditional mouse model showed that inactivation of p53 is essential for OSA development, and that the process is significantly accelerated by loss of Rb (Walkley et al. 2008). Curiously, OSA in these mice developed only in the axial skeleton (mostly in the skull), emphasizing differences in susceptibility for appendicular OSA between rodents (where this condition is difficult to recapitulate outside the orthotopic transplantable tumor setting) and human and canine OSA, which occur spontaneously. Intriguingly, the dog RB1 gene (CFA 22q11.2), which is reportedly deleted or inactivated in more than 50% of human OSA cases (Wadayama et al. 1994), showed copy number loss in only 11/38 cases (29%) from our canine OSA cohort, while TP53 (CFA 5q21) was deleted in only 7/38 cases (18%). Preliminary data from tumor immunohistochemistry and from Western blots of cell lines established as outgrowths of a subset of the tumors analyzed in the present study show, however, that Rb expression is reduced or absent in 8/13 of the samples tested (M. Ambron, S. Fosmire and J. Modiano, unpublished data). Previous studies reporting immunohistochemical staining for p53 similarly suggest that the protein is often functionally inactivated in canine OSA (Sagartz et al. 1996; Loukopoulos et al. 2003). The relative infrequency of genomic deletions of TP53 and RB1 in our canine OSA cohort therefore does not take into account other mechanisms that can inactivate these tumor suppressor genes. It is clear that proteins within the Rb pathway operate synchronously and that abnormalities of several different genes can produce an inactive Rb phenotype (Nielsen et al. 1998; Wei et al. 1999; Lopez-Guerrero et al. 2004). For example, if we group together all canine OSA cases that showed genomic loss of RB1 itself (CFA 22q11.2) with those exhibiting copy number imbalance of one or more of regulators of RB1 (loss of the p16INKA gene encoded by CDKN2A [11q15] or gain of CDK4 [CFA 15q24.3]) 17/38 dogs (45%) show evidence of a disrupted Rb pathway. Similarly, Levine and Fleischli (2000) and Mendoza et al. (1998) reported that genomic abnormalities of TP53 in canine OSA were largely restricted to sporadic mutations (rather than genomic deletions) and that amplification of MDM2 (a negative regulator of p53) was infrequent. Our data support these general observations: genomic loss of the TP53 locus (CFA 5q21) was detected in only 7/38 of our canine OSA cases (18% of the cohort, of which two cases also showed gain of MDM2 at CFA 31q11-q12). As with the Rb pathway, a disrupted p53 phenotype may arise through other means, among which are copy number aberrations of the CDKN2A locus, which are reportedly common in canine OSA (Levine and Fleischli 2000). In addition to p16INK4A CDKN2A also encodes p14ARF, which inhibits MDM2-mediated degradation of p53. Thus, genomic deletion of CDKN2A results not only in loss of the tumor suppressor activity of p16INK4A, but also the stabilizing effect of p14ARF on p53, resulting in dysregulation of both Rb and p53 pathways. In our canine OSA cohort, genomic loss of CDKN2A occurred in an additional four cases that showed intact TP53 copy number. Therefore, when evaluated in the context of their biological pathways, a total of 11/38 cases (29% of our canine OSA cohort) may have disruption to the p53 pathway (seven cases though direct loss of TP53, and four indirectly through loss of CDKN2A). In combination these data suggest that as in human OSA, the complex interactions between the p53 and Rb gene pathways play a key role in canine OSA and that the manner in which this occurs likely involves both chromosomal imbalances and defects in gene expression.
Identification of breed-associated genomic copy number aberrations
Genomic copy number aberrations that occur at high frequency in our dog OSA cohort may represent generalized tumor- and/or OSA-associated defects that contribute to tumor initiation and progression. Although population characteristics for OSA cases were comparable for both breeds (table 2, also see McNeill et al. 2007), the differing incidence of OSA in Golden Retriever and Rottweiler populations suggests that individual genetic backgrounds may play a role in defining the risk of tumorigenesis (measured both by the occurrence and the rate of clinical progression). We have shown previously that specific genomic imbalances occurred with significantly higher frequency in lymphoid tumors from Golden Retrievers than in comparable tumors from other breeds (Modiano et al. 2005). We therefore examined whether this association of genetic background (breed) and tumor genotype extends to OSA, a non-hematopoietic tumor that originates from multipotent mesenchymal progenitors. Our hypothesis was that breed would be significantly associated with OSA karyotypes. We identified 11 genomic loci that showed significant association between copy number status and dog breed. These chromosome regions, and the genes located therein, highlight targets for extended studies of breed-associated genetic lesions in dog OSA, and ultimately may represent promising targets for individualized therapies both in dogs and in human patients.
Interestingly, deletion of the WT1 tumor suppressor gene (CFA 18q22.3), among the most common aberrations encountered in our OSA cases, was restricted solely to Rottweilers (14/29 cases). Elevated WT1 expression has been associated with poor survival of human patients with metastatic OSA (Srivastava et al. 2006), and appears to have similar prognostic value in other cancers, including soft-tissue sarcomas (Ueda et al. 2003; Sotobori et al. 2006) and leukemias (Bergmann et al. 1997). It is not yet clear whether the survival characteristics of OSA patients are breed associated, but as such data become available it will be interesting to establish whether deletions of the WT1 gene confer any prognostic advantage to the Rottweiler population. In the present study, genomic loss of TP53 and CDKN2A suppressor genes were also restricted to Rottweilers (7/29 [24%] and 5/29 cases [17%], respectively). This does not account for possible somatic mutations, but it suggests that heritable factors may influence the fragility of these loci. Overall, approximately half of the Rottweilers in this cohort (15/29 cases) showed genomic deletion of at least one of the WT1, TP53, CDKN2A, PTEN or RB1 tumor suppressor genes. In all, 22/29 (76%) of the Rottweiler tumors had a cytogenetic aberration that involved either deletion of one of the aforementioned tumor suppressor genes or amplification of the MYC or KIT oncogenes. While this corroborates the importance of these genes in sporadic appendicular osteosarcoma, it suggests that 25% of OSA cases in Rottweilers may rely on other genomic abnormalities to survive, proliferate or metastasize. Although one retrospective study did not detect differences in response to treatment between Rottweilers and other dogs with OSA (McNeill et al. 2007) it remains to be established whether breed influences survival of dogs with peculiar OSA genotypes, and if so, whether deletions of tumor suppressor genes, most notably the WT1 gene, will confer a prognostic benefit to the Rottweiler population.
Summary
We show here that, as is true for human OSA, canine OSA exhibits considerable genomic instability resulting in extensive cytogenetic disorganization. aCGH analysis revealed a subset of recurrent CNAs that may be evolutionarily conserved in OSA in both these species. These include genomic amplifications of oncogenes and deletions of tumor-suppressor genes, indicating that human and dog OSA involve disruptions of the same key signal transduction pathways. The downstream consequences of these gene dosage alterations on cell-cycle regulation and DNA repair, combined with their presence in both species, strongly suggests that they are intimately involved in the pathogenesis of this neoplasm. Perhaps more importantly, we describe dog breed-associated genomic imbalances that may contribute to, or result from, heritable risk factors in two genetically-isolated populations, namely the Golden Retriever and Rottweiler breeds. We have identified genes within these regions that will undergo detailed investigation in more extensive studies using higher-resolution cytogenetic resources and a larger sample size, comprising a wider range of breeds and histologic subtypes of OSA. While extending opportunities to enhance canine health and welfare, our observations add to the expanding pool of information supporting a comparative approach to the study of OSA (and other tumors) as a means to identify novel genomic features that influence disease risk. We anticipate that extrapolation of these data back to human populations will aid definition of new diagnostic and prognostic tools and therapeutic targets for both species.
Supplementary Material
Acknowledgements
This work was supported by a grant from the American Kennel Club Canine Health Foundation awarded to MB/JM (CHF 2254), and by charitable donations from individuals, the Startlight Fund, and the Kate Koogler Canine Cancer Research Fund. We thank Eric Seiser for assistance with statistical analysis. We thank all the owners, breeders and veterinarians who contributed samples and data to this study.
References
- Atiye J, Wolf M, Kaur S, et al. Gene amplifications in osteosarcoma-CGH microarray analysis. Genes Chromosomes Cancer. 2005;42:158–163. doi: 10.1002/gcc.20120. [DOI] [PubMed] [Google Scholar]
- Bergmann L, Miething C, Maurer U, et al. High levels of Wilms’ tumor gene (wt1) mRNA in acute myeloid leukemias are associated with a worse long-term outcome. Blood. 1997;90:1217–1225. [PubMed] [Google Scholar]
- Boehm A, Neff J, Squire J, et al. Cytogenetic findings in 36 osteosarcoma specimens and a review of the literature. Pediatric Pathology and Molecular Medicine. 2000;19:359–376. [Google Scholar]
- Boggs RM, Wright ZM, Stickney MJ, Porter WW, Murphy KE. MicroRNA expression in canine mammary cancer. Mamm Genome. 2008 doi: 10.1007/s00335-008-9128-7. [DOI] [PubMed] [Google Scholar]
- Breen M. Canine cytogenetics--from band to basepair. Cytogenet Genome Res. 2008;120:50–60. doi: 10.1159/000118740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen M, Bullerdiek J, Langford CF. The DAPI banded karyotype of the domestic dog (Canis familiaris) generated using chromosome-specific paint probes. Chromosome Res. 1999;7:401–406. doi: 10.1023/a:1009224232134. [DOI] [PubMed] [Google Scholar]
- Breen M, Hitte C, Lorentzen TD, et al. An integrated 4249 marker FISH/RH map of the canine genome. BMC Genomics. 2004;5:65–75. doi: 10.1186/1471-2164-5-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen M, Modiano JF. Evolutionarily conserved cytogenetic changes in hematological malignancies of dogs and humans - man and his best friend share more than companionship. Chromosome Res. 2008;16:145–154. doi: 10.1007/s10577-007-1212-4. [DOI] [PubMed] [Google Scholar]
- Bridge JA, Nelson M, McComb E, et al. Cytogenetic findings in 73 osteosarcoma specimens and a review of the literature. Cancer Genet Cytogenet. 1997;95:74–87. doi: 10.1016/s0165-4608(96)00306-8. [DOI] [PubMed] [Google Scholar]
- Cooley DM, Beranek BC, Schlittler DL, et al. Endogenous gonadal hormone exposure and bone sarcoma risk. Cancer Epidemiol Biomarkers Prev. 2002;11:1434–1440. [PubMed] [Google Scholar]
- Dunn KA, Thomas R, Binns MM, Breen M. Comparative genomic hybridization (CGH) in dogs--application to the study of a canine glial tumour cell line. Vet J. 2000;160:77–82. doi: 10.1053/tvjl.2000.0495. [DOI] [PubMed] [Google Scholar]
- Entz-Werle N, Marcellin L, Gaub MP, et al. Prognostic significance of allelic imbalance at the c-kit gene locus and c-kit overexpression by immunohistochemistry in pediatric osteosarcomas. J Clin Oncol. 2005;23:2248–2255. doi: 10.1200/JCO.2005.03.119. [DOI] [PubMed] [Google Scholar]
- Entz-Werle N, Marie-Pierre G, Thomas L, et al. KIT gene in pediatric osteosarcomas: Could it be a new therapeutic target? Int J Cancer. 2007 doi: 10.1002/ijc.22593. [DOI] [PubMed] [Google Scholar]
- Freeman SS, Allen SW, Ganti R, et al. Copy number gains in EGFR and copy number losses in PTEN are common events in osteosarcoma tumors. Cancer. 2008;113:1453–1461. doi: 10.1002/cncr.23782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman L, Glickman N, Thorpe R. Golden Retriever Club of America. 2000 www.grca.org/healthsurvey.pdf.
- Hahn KA, Richardson RC, Hahn EA, Chrisman CL. Diagnostic and prognostic importance of chromosomal aberrations identified in 61 dogs with lymphosarcoma. Vet Pathol. 1994;31:528–540. doi: 10.1177/030098589403100504. [DOI] [PubMed] [Google Scholar]
- Hemminki A, Kanerva A, Kremer EJ, et al. A canine conditionally replicating adenovirus for evaluating oncolytic virotherapy in a syngeneic animal model. Mol Ther. 2003;7:163–173. doi: 10.1016/s1525-0016(02)00049-7. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- Kansara M, Thomas DM. Molecular pathogenesis of osteosarcoma. DNA Cell Biol. 2007;26:1–18. doi: 10.1089/dna.2006.0505. [DOI] [PubMed] [Google Scholar]
- Karlsson EK, Baranowska I, Wade CM, et al. Efficient mapping of mendelian traits in dogs through genome-wide association. Nat Genet. 2007;39:1321–1328. doi: 10.1038/ng.2007.10. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna C, Lindblad-Toh K, Vail D, et al. The dog as a cancer model. Nat Biotechnol. 2006;24:1065–1066. doi: 10.1038/nbt0906-1065b. [DOI] [PubMed] [Google Scholar]
- Khanna C, Prehn J, Hayden D, et al. A randomized controlled trial of octreotide pamoate long-acting release and carboplatin versus carboplatin alone in dogs with naturally occurring osteosarcoma: evaluation of insulin-like growth factor suppression and chemotherapy. Clin Cancer Res. 2002;8:2406–2412. [PubMed] [Google Scholar]
- Khanna C, Wan X, Bose S, et al. The membrane-cytoskeleton linker ezrin is necessary for osteosarcoma metastasis. Nat Med. 2004;10:182–186. doi: 10.1038/nm982. [DOI] [PubMed] [Google Scholar]
- Kirpensteijn J, Kik M, Rutteman GR, Teske E. Prognostic significance of a new histologic grading system for canine osteosarcoma. Vet Pathol. 2002;39:240–246. doi: 10.1354/vp.39-2-240. [DOI] [PubMed] [Google Scholar]
- Kisseberth WC, Nadella MV, Breen M, et al. A novel canine lymphoma cell line: A translational and comparative model for lymphoma research. Leuk Res. 2007 doi: 10.1016/j.leukres.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Levine RA. Overexpression of the sis oncogene in a canine osteosarcoma cell line. Vet Pathol. 2002;39:411–412. doi: 10.1354/vp.39-3-411. [DOI] [PubMed] [Google Scholar]
- Levine RA, Fleischli MA. Inactivation of p53 and retinoblastoma family pathways in canine osteosarcoma cell lines. Vet Pathol. 2000;37:54–61. doi: 10.1354/vp.37-1-54. [DOI] [PubMed] [Google Scholar]
- Levine RA, Forest T, Smith C. Tumor suppressor PTEN is mutated in canine osteosarcoma cell lines and tumors. Vet Pathol. 2002;39:372–378. doi: 10.1354/vp.39-3-372. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Lopez-Guerrero JA, Lopez-Gines C, Pellin A, Carda C, Llombart-Bosch A. Deregulation of the G1 to S-phase cell cycle checkpoint is involved in the pathogenesis of human osteosarcoma. Diagn Mol Pathol. 2004;13:81–91. doi: 10.1097/00019606-200406000-00004. [DOI] [PubMed] [Google Scholar]
- Lord LK, Yaissle JE, Marin L, Couto CG. Results of a web-based health survey of retired racing Greyhounds. J Vet Intern Med. 2007;21:1243–1250. doi: 10.1892/07-063.1. [DOI] [PubMed] [Google Scholar]
- Loukopoulos P, Thornton JR, Robinson WF. Clinical and pathologic relevance of p53 index in canine osseous tumors. Vet Pathol. 2003;40:237–248. doi: 10.1354/vp.40-3-237. [DOI] [PubMed] [Google Scholar]
- MacEwen EG. Spontaneous tumors in dogs and cats: models for the study of cancer biology and treatment. Cancer Metastasis Rev. 1990;9:125–136. doi: 10.1007/BF00046339. [DOI] [PubMed] [Google Scholar]
- Mansky PJ, Liewehr DJ, Steinberg SM, et al. Treatment of metastatic osteosarcoma with the somatostatin analog OncoLar: significant reduction of insulin-like growth factor-1 serum levels. J Pediatr Hematol Oncol. 2002;24:440–446. doi: 10.1097/00043426-200208000-00007. [DOI] [PubMed] [Google Scholar]
- McNeill CJ, Overley B, Shofer FS, et al. Characterization of the biological behaviour of appendicular osteosarcoma in Rottweilers and a comparison with other breeds: a review of 258 dogs. Veterinary and Comparative Oncology. 2007;5:90–98. doi: 10.1111/j.1476-5829.2006.00116.x. [DOI] [PubMed] [Google Scholar]
- Mendoza S, Konishi T, Dernell WS, Withrow SJ, Miller CW. Status of the p53, Rb and MDM2 genes in canine osteosarcoma. Anticancer Res. 1998;18:4449–4453. [PubMed] [Google Scholar]
- Mitelman F, Johansson B, Mertens F. Mitelman Database of Chromosome Aberrations in Cancer. 2008. 2008.
- Modiano J, Breen M, Lana S, et al. Naturally occurring translational models for development of cancer gene therapy. Gene Therapy and Molecular Biology. 2006;10:31–40. [Google Scholar]
- Modiano JF, Breen M, Burnett RC, et al. Distinct B-cell and T-cell lymphoproliferative disease prevalence among dog breeds indicates heritable risk. Cancer Res. 2005;65:5654–5661. doi: 10.1158/0008-5472.CAN-04-4613. [DOI] [PubMed] [Google Scholar]
- Mueller F, Fuchs B, Kaser-Hotz B. Comparative biology of human and canine osteosarcoma. Anticancer Res. 2007;27:155–164. [PubMed] [Google Scholar]
- Nielsen GP, Burns KL, Rosenberg AE, Louis DN. CDKN2A gene deletions and loss of p16 expression occur in osteosarcomas that lack RB alterations. Am J Pathol. 1998;153:159–163. doi: 10.1016/S0002-9440(10)65556-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrander EA, Wayne RK. The canine genome. Genome Res. 2005;15:1706–1716. doi: 10.1101/gr.3736605. [DOI] [PubMed] [Google Scholar]
- Ozaki T, Schaefer KL, Wai D, et al. Genetic imbalances revealed by comparative genomic hybridization in osteosarcomas. Int J Cancer. 2002;102:355–365. doi: 10.1002/ijc.10709. [DOI] [PubMed] [Google Scholar]
- Parker HG, Kim LV, Sutter NB, et al. Genetic structure of the purebred domestic dog. Science. 2004;304:1160–1164. doi: 10.1126/science.1097406. [DOI] [PubMed] [Google Scholar]
- Phillips JC, Stephenson B, Hauck M, Dillberger J. Heritability and segregation analysis of osteosarcoma in the Scottish deerhound. Genomics. 2007;90:354–363. doi: 10.1016/j.ygeno.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Ragland BD, Bell WC, Lopez RR, Siegal GP. Cytogenetics and molecular biology of osteosarcoma. Lab Invest. 2002;82:365–373. doi: 10.1038/labinvest.3780431. [DOI] [PubMed] [Google Scholar]
- Rosenberger JA, Pablo NV, Crawford PC. Prevalence of and intrinsic risk factors for appendicular osteosarcoma in dogs: 179 cases (1996-2005) J Am Vet Med Assoc. 2007;231:1076–1080. doi: 10.2460/javma.231.7.1076. [DOI] [PubMed] [Google Scholar]
- Sagartz JE, Bodley WL, Gamblin RM, et al. p53 tumor suppressor protein overexpression in osteogenic tumors of dogs. Vet Pathol. 1996;33:213–221. doi: 10.1177/030098589603300211. [DOI] [PubMed] [Google Scholar]
- Sandberg AA, Bridge JA. Updates on the cytogenetics and molecular genetics of bone and soft tissue tumors: osteosarcoma and related tumors. Cancer Genet Cytogenet. 2003;145:1–30. [PubMed] [Google Scholar]
- Sotobori T, Ueda T, Oji Y, et al. Prognostic significance of Wilms tumor gene (WT1) mRNA expression in soft tissue sarcoma. Cancer. 2006;106:2233–2240. doi: 10.1002/cncr.21861. [DOI] [PubMed] [Google Scholar]
- Srivastava A, Fuchs B, Zhang K, et al. High WT1 expression is associated with very poor survival of patients with osteogenic sarcoma metastasis. Clin Cancer Res. 2006;12:4237–4243. doi: 10.1158/1078-0432.CCR-05-2307. [DOI] [PubMed] [Google Scholar]
- Stock C, Kager L, Fink FM, Gadner H, Ambros PF. Chromosomal regions involved in the pathogenesis of osteosarcomas. Genes Chromosomes Cancer. 2000;28:329–336. doi: 10.1002/1098-2264(200007)28:3<329::aid-gcc11>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Tarkkanen M, Elomaa I, Blomqvist C, et al. DNA sequence copy number increase at 8q: a potential new prognostic marker in high-grade osteosarcoma. Int J Cancer. 1999;84:114–121. doi: 10.1002/(sici)1097-0215(19990420)84:2<114::aid-ijc4>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Thomas R, Bridge W, Benke K, Breen M. Isolation and chromosomal assignment of canine genomic BAC clones representing 25 cancer-related genes. Cytogenet Genome Res. 2003;102:249–253. doi: 10.1159/000075757. [DOI] [PubMed] [Google Scholar]
- Thomas R, Duke S, Karlsson K, et al. Development of a 1Mb resolution, cytogenetically-validated genomic microarray for canine CGH analysis. Cytogenet Genome Res. (in press) [Google Scholar]
- Thomas R, Duke SE, Bloom SK, et al. A Cytogenetically Characterized, Genome-Anchored 10-Mb BAC Set and CGH Array for the Domestic Dog. J Hered. 2007;98:474–484. doi: 10.1093/jhered/esm053. [DOI] [PubMed] [Google Scholar]
- Thomas R, Scott A, Langford CF, et al. Construction of a 2-Mb resolution BAC microarray for CGH analysis of canine tumors. Genome Res. 2005;15:1831–1837. doi: 10.1101/gr.3825705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Oji Y, Naka N, et al. Overexpression of the Wilms’ tumor gene WT1 in human bone and soft-tissue sarcomas. Cancer Sci. 2003;94:271–276. doi: 10.1111/j.1349-7006.2003.tb01432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vail DM, MacEwen EG. Spontaneously occurring tumors of companion animals as models for human cancer. Cancer Invest. 2000;18:781–792. doi: 10.3109/07357900009012210. [DOI] [PubMed] [Google Scholar]
- Wadayama B, Toguchida J, Shimizu T, et al. Mutation spectrum of the retinoblastoma gene in osteosarcomas. Cancer Res. 1994;54:3042–3048. [PubMed] [Google Scholar]
- Walkley CR, Qudsi R, Sankaran VG, et al. Conditional mouse osteosarcoma, dependent on p53 loss and potentiated by loss of Rb, mimics the human disease. Genes Dev. 2008;22:1662–1676. doi: 10.1101/gad.1656808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LL. Biology of osteogenic sarcoma. Cancer J. 2005;11:294–305. doi: 10.1097/00130404-200507000-00005. [DOI] [PubMed] [Google Scholar]
- Wei G, Lonardo F, Ueda T, et al. CDK4 gene amplification in osteosarcoma: reciprocal relationship with INK4A gene alterations and mapping of 12q13 amplicons. Int J Cancer. 1999;80:199–204. doi: 10.1002/(sici)1097-0215(19990118)80:2<199::aid-ijc7>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Winkler S, Reimann-Berg N, Escobar HM, et al. Polysomy 13 in a canine prostate carcinoma underlining its significance in the development of prostate cancer. Cancer Genet Cytogenet. 2006;169:154–158. doi: 10.1016/j.cancergencyto.2006.03.015. [DOI] [PubMed] [Google Scholar]
- Withrow S. Limb Sparing Trials and Canine Osteosarcoma. In: Modiano J, editor. Genes, Dogs and Cancer; 3rd Annual Canine Cancer Conference; Ithaca NY, Seattle, WA, USA. International Veterinary Information Service; 2003. [Google Scholar]
- Witlox MA, Lamfers ML, Wuisman PI, Curiel DT, Siegal GP. Evolving gene therapy approaches for osteosarcoma using viral vectors: review. Bone. 2007;40:797–812. doi: 10.1016/j.bone.2006.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.