Abstract
In the last 2 decades, advances in genomic technologies and molecular biology have accelerated the identification of multiple genetic loci that confer risk for cutaneous melanoma. The risk alleles range from rarely occurring, high-risk variants with a strong familial predisposition to low-risk to moderate-risk variants with modest melanoma association. Although the high-risk alleles are limited to the CDKN2A and CDK4 loci, the authors of recent genome-wide association studies have uncovered a set of variants in pigmentation loci that contribute to low risk. A biological validation of these new findings would provide greater understanding of the disease. In this review we describe some of the important risk loci and their association to risk of developing cutaneous melanoma and also address the current clinical challenges in CDKN2A genetic testing.
During the last 3 decades, the incidence of cutaneous melanoma (CM) has increased largely in Caucasian populations around the world.1–3 In the United States, the estimated number of new cases of invasive melanoma in 2010 is 62,480, of which an annual estimated mortality rate is at 2.3% for all races and 2.8% in white populations alone.1 The outcome for melanoma is highly dependent on the stage of the disease.4 On one end of the spectrum is the primary CM, which when excised appropriately, is highly curable. However, by contrast, metastatic melanoma comes with a very poor prognosis despite treatment,5–6 and patients with stage IV melanoma have an expected 9% to 15% rate of survival4 at 5 years. Because early disease is significantly more favorable in prognosis, there has been much focus on the need to identify those at greatest risk for melanoma. Recent advances in technology and molecular biology have enhanced our ability to more accurately personalize melanoma risk assessment.7,8
Cutaneous melanoma is a cancer in which a combination of genetic and environmental factors clearly influences the onset of disease.9 High-risk alleles often are expressed as familial clusters, whereas lower-risk alleles result in sporadic cases.10–12 The purpose of this review is to discuss current knowledge about the individual risk loci that modulate risk for melanoma. Also, we have addressed some of the current progress and limitations of genetic testing and the strategies for establishing risk intervention programs to promote effective risk management recommendations.
High-Risk Alleles in Melanoma Predisposition
Hereditary melanoma typically arises in an autosomal-dominant pattern within multiplex families. Often, there is an association with clinically atypical nevi, although patients may harbor nevi without melanoma and vice versa.13,14 Progress in genetic methodology in the late 1980s and early 1990s led to the identification of germ line mutations in p16Ink4a (now CDKN2A), which is located on chromosome 9p21.15,16 Shortly thereafter, germline mutations of CDK4 were also recognized in certain kindreds that lacked heritable CDKN2A alterations.17,18 Thus, within a few short years in the mid-1990s, the molecular basis of hereditary melanoma was quickly established in a subset of melanoma-prone kindreds. These loci account for approximately 20% to 57% of disease susceptibility,19 thereby suggesting that other high-risk loci still exist. At the tail end of the successful completion of the Human Genome Project, there has been intense interest in enumerating lower-risk disease alleles through genome-wide association studies (GWAS). As outlined in this article, the combination of familial and population genetics have led to a much larger and richer view of melanoma predisposition.
CDKN2A
The CDKN2A locus constitutes 4 exons that encode 2 functionally distinct proteins through alternate splicing events, leading to 2 very potent tumor suppressors: p16Ink4a and p14Arf (Fig. 1).15,16 Tumor-derived alleles of p16Ink4a result in mutant proteins that are no longer able to inhibit cyclin-dependent protein kinases (Cdks)-4 and -6 and are thus no longer able to suppress the phosphorylation of the retinoblastoma protein.20,21 When the Rb protein becomes hyperphosphorylated, the cell transitions from G1 to the S-phase and begins to proliferate. p14Arf binds to Hdm2 and promotes rapid degradation of Hdm2, thus stabilizing p5322–25; thus, in the context of tumors, the loss of p14Arf results in an indirect loss of p53 function and deregulation of cell cycling and DNA damage signaling.26 A vast array of CDKN2A mutations have been identified in families throughout the world, and most occur in exons 1-alpha and 2.17,27,28 Interestingly, the large number of mutations in exon 1-alpha suggests that p16Ink4a and not p14Arf18 may be preferentially targeted. However, mutations in the p14Arf-specific exon 1-beta locus have been described, thereby indicating that p14Arf is a bona-fide melanoma risk gene.29–31 In a recent study, the rate of germline p14Arf mutations without coinheritance of p16Ink4a and Cdk4 mutations was estimated to be 3.2%,32 which is consistent with a previous multicenter study.33 Two of the reported families shared the g.193 + 1A > G splice-site mutation, and one family had a novel missense mutation g.161G > A, which causes the substitution of the arginine in position 54 with a histidine. A CDKN2A locus-specific database that lists all common germ-line variants is publicly available (https://biodesktop.uvm.edu/perl/p16).
Figure 1.
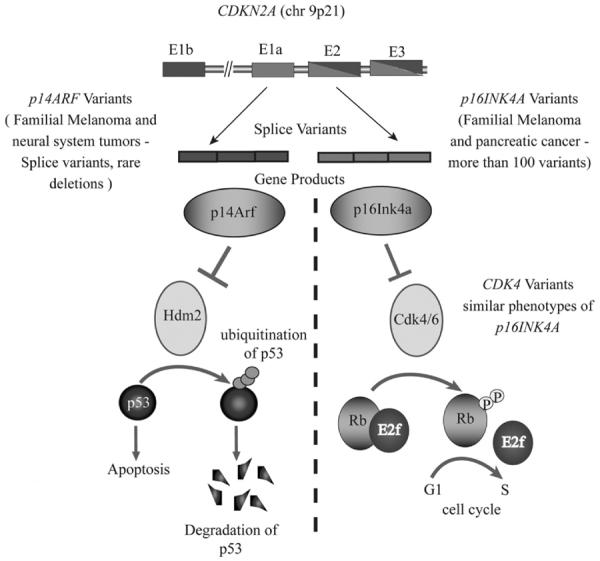
High-risk melanoma predisposition loci. The schematic represents the signaling pathways controlled by CDKN2A loci. The 2 different protein products encoded by the splice variants - p14Arf and p16Ink4a, converge on the same cellular function—cell-cycle regulation. The p14Arf regulates the p53 tumor suppressor network and the P16Ink4a regulates the Rb network. Variants in p14Arf, p16Ink4a, Cdk4, and Rb contribute to deregulated signaling and uncontrolled cell proliferation.
Investigators in several studies have attempted to estimate CDKN2A mutation penetrance, defined as the observed risk of developing melanoma over time for a mutation carrier. A large GenoMEL (The International Melanoma Genetics Consortium) study, whose authors analyzed 80 families from 3 continents, estimated CDKN2A mutation penetrance to be 0.30 (95% confidence interval (CI) = 0.12–0.62) by age 50 and 0.67 (95% CI = 0.31–0.96) by age 80. There was significant variation in the mutation rate on the basis of population geography (P = 0.003). By age 50 years, CDKN2A mutation penetrance reached 0.13 in Europe, 0.50 in the United States, and 0.32 in Australia; by age 80 years it was 0.58 in Europe, 0.76 in the United States, and 0.91 in Australia. The variation as the result of geographic location suggests that other factors, such as degree of sun exposure or other coinherited gene modifiers, also contribute to the overall risk.
Unbiased estimates of CDKN2A penetrance have also been performed on the general CM population. The relative risk of a subsequent melanoma among patients with the mutations in CDKN2A with previously diagnosed primary melanoma is estimated to be 4.3 (95% CI = 2.3–7.7).34,35 Population-based ascertainment leads to lower penetrance estimates (0.14 [95% CI = 8%–22%] by age 50, 0.24 [95% CI =15%–34%] by age 70, and 0.28 [95% CI =18%–40%] by age 80) compared with familial-based ascertainment, most likely because of shared genetic modifiers and exposures within families.27 Some of the earlier studies have also shown that CDKN2A mutations are predominantly found in high-risk families (9 of 87%, 10.3%), thus, the contribution of such a mutation to the overall burden of CM is only 0.2%.2,36 The melanoma penetrance is also greater (1.2%) in familial clusters with multiple primary melanoma (MPM) when compared with those with single primary melanoma.27 The occurrence of CDKN2A mutations in patients with MPM regardless of familial history, albeit less frequent (8%–15%), could be a significant predictor of genetic susceptibility to melanoma during melanoma risk assessments.
Mutations in CDKN2A have also been associated with an increased risk of pancreatic cancer.37 In the GenoMEL analysis of 385 families, 72% of families with 1 reported patient with pancreatic cancer had mutations (31/43) and 81% of families with ≥2 patients with pancreatic cancer had mutations (13/16).19 The role of screening in the prevention of pancreatic cancer is still unclear, and thus, the impact of CDKN2A testing on pancreatic cancer management has not yet established.
CDK4
Cyclin-dependent kinase 4 (Cdk4), which physically and genetically interacts with the p16Ink4a (Fig. 1), is another important high-risk locus that that been implicated in melanoma risk. A small number of melanoma-prone families harbor mutations in CDK4. Both somatic and germline mutations of CDK4 have been identified in melanoma cell lines and in families,18,38 respectively. Recurrent mutations at codon 24 (Arg24His and Arg24 Cys) result in an autosomal-dominant oncogenic form of Cdk4, which is resistant to p16Ink4a inhibition.17,18,39,40 Because the 2 proteins directly interact, the consequences of p16Ink4a loss and Cdk4 activation are similar; thus, carriers of CDKN2A and CDK4 mutations also share a similar phenotype.41 The Rb protein, which is downstream of p16Ink4a and Cdk4, is an essential gatekeeper for hereditary retinoblastoma. In patients cured of bilateral retinoblastoma (presumed RB1 mutation carriers), the estimates of increased lifetime risk of melanoma range from 4-fold to 80-fold (reviewed in Chin et al42). Hence, mutations in the entire CDKN2A/CDK4/Rb pathway appear to play a significant role in melanoma pathogenesis.
Low- to Moderate-Risk Susceptibility Alleles
With the Human Genome Project, it quickly became clear that single nucleotide changes, or polymorphisms (SNPs), occur throughout the human genome. These millions of variants have been recently leveraged in a host of GWAS. During the past few years, many GWAS analyses have led to the discovery of chromosomal regions linked to more common diseases and traits, including melanoma, pigmentation and nevus density. These GWAS have propelled the field forward by identifying many low to moderate risk loci that underlie sporadic CM formation (reviewed by Udayakumar and Tsao43).
MC1R
There are more than 120 genes that influence pigmentation. Recent comprehensive meta-analyses have directly linked pigmentation traits with the risk of developing melanoma.44,45 A key pigmentation gene, melanocortin-1-receptor (MC1R), has been recently implicated as a low to moderate melanoma predisposition gene (reviewed in43,46–49). It is a 7-transmembrane protein that belongs to the family of G protein-coupled receptors (Fig. 2). When alpha-melanocyte stimulating hormone (alpha-MSH) binds to its receptor, Mc1r, it activates a G protein, which in turn stimulates the synthesis of cAMP through adenylate cyclase (AC). The microphthalmia transcription factor (Mitf) is subsequently induced, which leads to up-regulation of the pigment synthetic genes and eumelanin synthesis. Mutations in MC1R abrogate this pathway; therefore, melanocytes are enriched for pheomelanin, and patients exhibit increased freckling, red hair color (RHC), and enhanced photosensitivity.50,51 A systematic study conducted by Raimondi and colleagues51 investigated all MC1R variants and classified them based on degree of association with the RHC phenotype:
Strong association with more frequency: 4 “R” MC1R variants (p.Asp84Glu, p.Arg151Cys, p.Arg160Trp, and p.Asp294His)
Familial association with fewer frequency: 2 “R” alleles (p.Arg142His and p.Ile155Thr)
Weak Association: 3 “r” alleles (p.Val60Leu, p.Val92Met, and p.Arg163Gln)
Except for p.Val60Leu and p.Val92Met, both of which showed lack of significant association to melanoma risk, all other variants appear significantly associated with melanoma risk, with odds ratios ranging from 1.42 for p.Arg163Gln to 2.45 for p.Ile155Thr. Some MC1R variants that correlate well with skin/hair color and poor tanning ability (80% in patients with RHC and/or fair skin, 20% in patients with brown or black hair) show up to a 3.9-fold increase in CM risk.52 Interestingly, the authors of several other studies have uncovered risk variants that show more complex genetic interactions. Some MC1R variants conferred little or no increased risk of CM among one cohort versus another cohort from different geographic location.53 Some variants conferred melanoma risk and no association with pigmentation phenotypes.54 This complexity may reflect significant environmental inputs that have not been fully annotated in all studies. The presence of multiple MC1R variants has been significantly associated with melanoma especially among individuals with Multiple Primary Melanomas (MPMs). In addition, an inverse relationship has been observed between the number of MC1R variants with decreased median age at diagnosis.55 Lastly, MC1R variants can also modify CDKN2A penetrance. In one study, the presence of MC1R variants Arg151Cys, Arg160Trp and Asp294His, in addition to CDKN2A mutation, increased the penetrance of CDKN2A mutations from 50% to 84% with a mean onset age of 37.8 years.56
Figure 2.
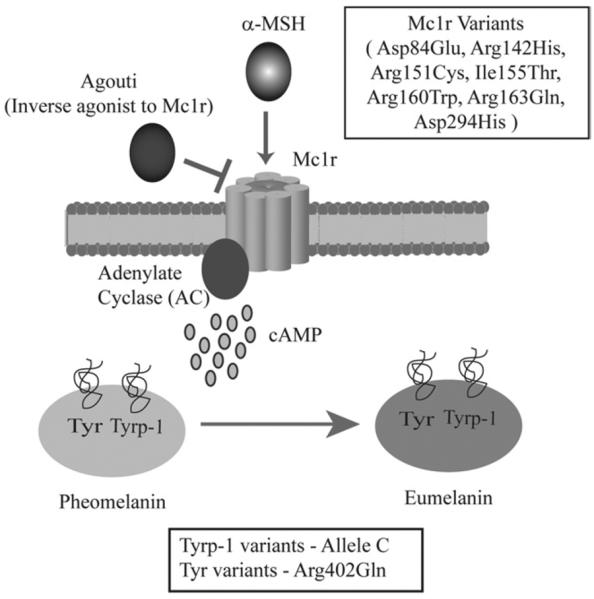
Mc1r pigmentation pathway implicated in melanoma risk. Alpha-melanocyte stimulating hormone (α-MSH) serves as a ligand of Mc1r. Agouti protein antagonizes this interaction. Binding of MSH to Mc1r stimulates adenylate cyclase to generate cAMP. cAMP is one of the key mediators of a series of events that leads to the conversion of pheomelanin to a photoprotective eumelanin during melanogenesis. The tyrosinase (Tyr) and the tyrosine-related protein kinase 1 (Tyrp-1) are the key enzymes that regulate this process. The variants of the MC1R, TYR, and TYRP-1, presumably lead to pigmentation differences, photosensitivity, and predisposition to cutaneous melanoma.
Other Low-Risk Alleles
In recent years, several groups have isolated many novel variants associated with hair and eye pigmentation, skin sensitivity to sun, and freckling.53,57–59 Some of the significant loci identified by several studies include SLC24A4, KITLG, 6p25.3, TYR, OCA2, TPCN2, ASIP, and TYRP1.43 The ASIP locus (P =1.2 × 10−9; OR = 1.45), coding variant in TYR (P = 2.8 × 10−7; combined or =1.21) and a noncoding SNP at the tyrosinase-related protein 1 (TYRP1) locus are significantly associated with melanoma risk.58 Larger population-based GWAS studies identified other SNPs that show significant association with melanoma risk.54,60–62 Others have reported low-risk variants, including methylthioadenosine phosphorylase (MTAP) gene that flanks the CDKN2A region, polymorphisms in the EGF, glutathione S-transferase (GSTM1), cytochrome P450 debrisoquine hydroxylase (CYP2D6), and vitamin D receptor gene (VDR) locus.63–66 Further studies are needed to biologically validate these loci in the pathogenesis of melanoma.
Genetic Testing and Risk Management
Genetic testing for CDKN2A mutations is available clinically and on a research basis. Currently, the Melanoma Genetics Consortium and the American Society of Clinical Oncologists caution against the routine use of clinical CDNK2A testing for patients with hereditary melanoma because of the difficulty in interpreting test results, the potentially limited impact of the result on clinical management, and concerns for genetic discrimination.36,67,68 Although, as discussed previously, mutations in this gene are associated with a significant risk for melanoma, it has also been noted that both families who test negative for a CDKN2A gene mutation as well as patients who test negative for a familial CDKN2A gene mutation remain at a risk greater than that of the general population to develop melanoma.10,69
The following features have been associated with a moderate to high risk for a hereditary melanoma predisposition syndrome: (1) multiple primary melanomas27,70,71; (2) family history of melanoma71–73; and/or (3) the presence of melanoma in addition to another related cancer, such as pancreatic cancer, on the same side of the family.33,74–76 Early age of melanoma onset in patients with a negative family history has not been shown to increase the risk for a CDKN2A mutation.27,71,73 Recently, MelaPRO, a risk-assessment model, was developed to assess the likelihood of a CDKN2A mutation in a person on the basis of personal and family history, as well as geographic penetrance. MelaPRO and other such models aid in the identification of patients and families at significant risk for melanoma who may benefit from further discussion regarding the option of genetic testing.77
Patients considering genetic testing for CDKN2A gene mutations should be informed of the possible test results, including positive, negative, and variant of uncertain significance, and the potential implications the test result could have on medical management. Informed consent for CDKN2A testing should also include a discussion regarding the cost of testing, genetic discrimination, and the possible psychological impact of testing.68 In addition, it is important that individuals who opt for CKDN2A testing are aware of the increased risk for pancreatic cancer in some families, as well as the current lack of effective screening guidelines for pancreatic cancer.37,74,75
Conclusions
In this review we have discussed some of the very important loci reported to be involved in melanoma susceptibility and discussed other loci that have been implicated in the melanoma susceptibility process either as an independent risk factor or as a modifier locus. Although the identification of these low-risk alleles has stimulated much enthusiasm in the field, an understanding of the biological consequences and clinical utility of these findings remains to be established.
Acknowledgments
This writing of this manuscript was supported by grants from the American Cancer Society (RSG MGO-112970 to H.T.) and the National Institutes of Health (K24 CA149202-01 and P50 CA-93683, both to H.T.).
Footnotes
The authors declare no conflict of interest pertaining to this work.
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Aitken J, Welch J, Duffy D, et al. CDKN2A variants in a population-based sample of Queensland families with melanoma. J Natl Cancer Inst. 1999;91:446–452. doi: 10.1093/jnci/91.5.446. [DOI] [PubMed] [Google Scholar]
- 3.Lag R, Krapcho M, Stinchcomb DG, et al., editors. SEER Cancer Statistics Review, 1975–2005. National Cancer Institute; Bethesda, MD: 2008. [Google Scholar]
- 4.Balch CM, Buzaid AC, Soong SJ, et al. Final version of the American Joint Committee on Cancer staging system for cutaneous melanoma. J Clin Oncol. 2001;19:3635–3648. doi: 10.1200/JCO.2001.19.16.3635. [DOI] [PubMed] [Google Scholar]
- 5.Sekulic A, Haluska P, Jr, Miller AJ, et al. Malignant melanoma in the 21st century: the emerging molecular landscape. Mayo Clin Proc. 2008;83:825–846. doi: 10.4065/83.7.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsao H, Atkins MB, Sober AJ. Management of cutaneous melanoma. N Engl J Med. 2004;351:998–1012. doi: 10.1056/NEJMra041245. [DOI] [PubMed] [Google Scholar]
- 7.de Snoo FA, Hayward NK. Cutaneous melanoma susceptibility and progression genes. Cancer Lett. 2005;230:153–186. doi: 10.1016/j.canlet.2004.12.033. [DOI] [PubMed] [Google Scholar]
- 8.Lin J, Hocker TL, Singh M, et al. Genetics of melanoma predisposition. Br J Dermatol. 2008;159:286–291. doi: 10.1111/j.1365-2133.2008.08682.x. [DOI] [PubMed] [Google Scholar]
- 9.Roderick McLeod G, Davis NC, Sober AJ. A history of melanoma: from hunter to Clark. In: Balch CMH, et al., editors. Cutaneous Melanoma. 4 ed Quality Medical Publishing; St Louis, MO: 2003. [Google Scholar]
- 10.Bishop DT, Demenais F, Goldstein AM, et al. Geographical variation in the penetrance of CDKN2A mutations for melanoma. J Natl Cancer Inst. 2002;94:894–903. doi: 10.1093/jnci/94.12.894. [DOI] [PubMed] [Google Scholar]
- 11.Bishop JN, Harland M, Bishop DT. The genetics of melanoma. Br J Hosp Med (Lond) 2006;67:299–304. doi: 10.12968/hmed.2006.67.6.21288. [DOI] [PubMed] [Google Scholar]
- 12.Fargnoli MC, Argenziano G, Zalaudek I, et al. High- and low-penetrance cutaneous melanoma susceptibility genes. Expert Rev Anticancer Ther. 2006;6:657–670. doi: 10.1586/14737140.6.5.657. [DOI] [PubMed] [Google Scholar]
- 13.Clark WH, Jr, Reimer RR, Greene M, et al. Origin of familial malignant melanomas from heritable melanocytic lesions. “The B-K mole syndrome.” Arch Dermatol. 1978;114:732–738. [PubMed] [Google Scholar]
- 14.Lynch HT, Frichot BC, 3rd, Lynch JF. Familial atypical multiple mole-melanoma syndrome. J Med Genet. 1978;15:352–356. doi: 10.1136/jmg.15.5.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hussussian CJ, Struewing JP, Goldstein AM, et al. Germline p16 mutations in familial melanoma. Nat Genet. 1994;8:15–21. doi: 10.1038/ng0994-15. [DOI] [PubMed] [Google Scholar]
- 16.Kamb A, Shattuck-Eidens D, Eeles R, et al. Analysis of the p16 gene (CDKN2) as a candidate for the chromosome 9p melanoma susceptibility locus. Nat Genet. 1994;8:23–26. doi: 10.1038/ng0994-22. [DOI] [PubMed] [Google Scholar]
- 17.Soufir N, Avril MF, Chompret A, et al. Prevalence of p16 and CDK4 germline mutations in 48 melanoma-prone families in France. The French Familial Melanoma Study Group. Hum Mol Genet. 1998;7:209–216. doi: 10.1093/hmg/7.2.209. [DOI] [PubMed] [Google Scholar]
- 18.Zuo L, Weger J, Yang Q, et al. Mutations in the p16INK4a binding domain of CDK4 in familial melanoma. Nat Genet. 1996;12:97–99. doi: 10.1038/ng0196-97. [DOI] [PubMed] [Google Scholar]
- 19.Goldstein AM, Chan M, Harland M, et al. Features associated with germline CDKN2A mutations: a GenoMEL study of melanoma-prone families from three continents. J Med Genet. 2007;44:99–106. doi: 10.1136/jmg.2006.043802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koh J, Enders GH, Dynlacht BD, et al. Tumour-derived p16 alleles encoding proteins defective in cell-cycle inhibition. Nature. 1995;375:506–510. doi: 10.1038/375506a0. [DOI] [PubMed] [Google Scholar]
- 21.Lukas J, Parry D, Aagaard L, et al. Retinoblastoma-protein-dependent cell-cycle inhibition by the tumour suppressor p16. Nature. 1995;375:503–506. doi: 10.1038/375503a0. [DOI] [PubMed] [Google Scholar]
- 22.Kamijo T, Weber JD, Zambetti G, et al. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci U S A. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pomerantz J, Schreiber-Agus N, Liegeois NJ, et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 24.Stott FJ, Bates S, James MC, et al. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 26.Chin L, Pomerantz J, Polsky D, et al. Cooperative effects of INK4a and ras in melanoma susceptibility in vivo. Genes Dev. 1997;11:2822–2834. doi: 10.1101/gad.11.21.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Begg CB, Orlow I, Hummer AJ, et al. Lifetime risk of melanoma in CDKN2A mutation carriers in a population-based sample. J Natl Cancer Inst. 2005;97:1507–1515. doi: 10.1093/jnci/dji312. [DOI] [PubMed] [Google Scholar]
- 28.Hayward NK. Genetics of melanoma predisposition. Oncogene. 2003;22:3053–3062. doi: 10.1038/sj.onc.1206445. [DOI] [PubMed] [Google Scholar]
- 29.Harland M, Taylor CF, Chambers PA, et al. A mutation hotspot at the p14ARF splice site. Oncogene. 2005;24:4604–4608. doi: 10.1038/sj.onc.1208678. [DOI] [PubMed] [Google Scholar]
- 30.Hewitt C, Lee Wu C, Evans G, et al. Germline mutation of ARF in a melanoma kindred. Hum Mol Genet. 2002;11:1273–1279. doi: 10.1093/hmg/11.11.1273. [DOI] [PubMed] [Google Scholar]
- 31.Garcia-Casado Z, Nagore E, Fernandez-Serra A, et al. A germline mutation of p14/ARF in a melanoma kindred. Melanoma Res. 2009;19:335–337. doi: 10.1097/CMR.0b013e32832dd2d4. [DOI] [PubMed] [Google Scholar]
- 32.Binni F, Antigoni I, De Simone P, et al. Novel and recurrent p14(ARF) mutations in Italian familial melanoma. Clin Genet. 2010;77:581–586. doi: 10.1111/j.1399-0004.2009.01298.x. [DOI] [PubMed] [Google Scholar]
- 33.Goldstein AM, Chan M, Harland M, et al. High-risk melanoma susceptibility genes and pancreatic cancer, neural system tumors, and uveal melanoma across GenoMEL. Cancer Res. 2006;66:9818–9828. doi: 10.1158/0008-5472.CAN-06-0494. [DOI] [PubMed] [Google Scholar]
- 34.Berwick M, Orlow I, Mahabir S, et al. Estimating the relative risk of developing melanoma in INK4A carriers. Eur J Cancer Prev. 2004;13:65–70. doi: 10.1097/00008469-200402000-00010. [DOI] [PubMed] [Google Scholar]
- 35.Blackwood MA, Holmes R, Synnestvedt M, et al. Multiple primary melanoma revisited. Cancer. 2002;94:2248–2255. doi: 10.1002/cncr.10454. [DOI] [PubMed] [Google Scholar]
- 36.Kefford R, Bishop JN, Tucker M, et al. Genetic testing for melanoma. Lancet Oncol. 2002;3:653–654. doi: 10.1016/s1470-2045(02)00894-x. [DOI] [PubMed] [Google Scholar]
- 37.Goldstein AM, Fraser MC, Struewing JP, et al. Increased risk of pancreatic cancer in melanoma-prone kindreds with p16INK4 mutations. N Engl J Med. 1995;333:970–974. doi: 10.1056/NEJM199510123331504. [DOI] [PubMed] [Google Scholar]
- 38.Tsao H, Benoit E, Sober AJ, et al. Novel mutations in the p16/CDKN2A binding region of the cyclin-dependent kinase-4 gene. Cancer Res. 1998;58:109–113. [PubMed] [Google Scholar]
- 39.Ranade K, Hussussian CJ, Sikorski RS, et al. Mutations associated with familial melanoma impair p16INK4 function. Nat Genet. 1995;10:114–116. doi: 10.1038/ng0595-114. [DOI] [PubMed] [Google Scholar]
- 40.Reymond A, Brent R. p16 proteins from melanoma-prone families are deficient in binding to Cdk4. Oncogene. 1995;11:1173–1178. [PubMed] [Google Scholar]
- 41.Goldstein AM, Struewing JP, Chidambaram A, et al. Genotype-phenotype relationships in U.S. melanoma-prone families with CDKN2A and CDK4 mutations. J Natl Cancer Inst. 2000;92:1006–1010. doi: 10.1093/jnci/92.12.1006. [DOI] [PubMed] [Google Scholar]
- 42.Chin L, Garraway LA, Fisher DE. Malignant melanoma: genetics and therapeutics in the genomic era. Genes Dev. 2006;20:2149–2182. doi: 10.1101/gad.1437206. [DOI] [PubMed] [Google Scholar]
- 43.Udayakumar D, Tsao H. Moderate-to low-risk variant alleles of cutaneous malignancies and nevi: lessons from genome-wide association studies. Genome Med. 2009;1:95. doi: 10.1186/gm95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gandini S, Sera F, Cattaruzza MS, et al. Meta-analysis of risk factors for cutaneous melanoma. I. Common and atypical naevi. Eur J Cancer. 2005;41:28–44. doi: 10.1016/j.ejca.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 45.Gandini S, Sera F, Cattaruzza MS, et al. Meta-analysis of risk factors for cutaneous melanoma. III. Family history, actinic damage and phenotypic factors. Eur J Cancer. 2005;41:2040–2059. doi: 10.1016/j.ejca.2005.03.034. [DOI] [PubMed] [Google Scholar]
- 46.Sturm RA. Skin colour and skin cancer—MC1R, the genetic link. Melanoma Res. 2002;12:405–416. doi: 10.1097/00008390-200209000-00001. [DOI] [PubMed] [Google Scholar]
- 47.Sturm RA, Duffy DL, Box NF, et al. The role of melanocortin-1 receptor polymorphism in skin cancer risk phenotypes. Pigment Cell Res. 2003;16:266–272. doi: 10.1034/j.1600-0749.2003.00041.x. [DOI] [PubMed] [Google Scholar]
- 48.Sturm RA, Duffy DL, Box NF, et al. Genetic association and cellular function of MC1R variant alleles in human pigmentation. Ann NY Acad Sci. 2003;994:348–358. doi: 10.1111/j.1749-6632.2003.tb03199.x. [DOI] [PubMed] [Google Scholar]
- 49.Rouzaud F, Kadekaro AL, Abdel-Malek ZA, et al. MC1R and the response of melanocytes to ultraviolet radiation. Mutat Res. 2005;571:133–152. doi: 10.1016/j.mrfmmm.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 50.Garcia-Borron JC, Sanchez-Laorden BL, Jimenez-Cervantes C. Melanocortin-1 receptor structure and functional regulation. Pigment Cell Res. 2005;18:393–410. doi: 10.1111/j.1600-0749.2005.00278.x. [DOI] [PubMed] [Google Scholar]
- 51.Raimondi S, Sera F, Gandini S, et al. MC1R variants, melanoma and red hair color phenotype: a meta-analysis. Int J Cancer. 2008;122:2753–2760. doi: 10.1002/ijc.23396. [DOI] [PubMed] [Google Scholar]
- 52.Valverde P, Healy E, Jackson I, et al. Variants of the melanocyte-stimulating hormone receptor gene are associated with red hair and fair skin in humans. Nat Genet. 1995;11:328–330. doi: 10.1038/ng1195-328. [DOI] [PubMed] [Google Scholar]
- 53.Sulem P, Gudbjartsson DF, Stacey SN, et al. Two newly identified genetic determinants of pigmentation in Europeans. Nat Genet. 2008;40:835–837. doi: 10.1038/ng.160. [DOI] [PubMed] [Google Scholar]
- 54.Kennedy C, Ter Huurne J, Berkhout M, et al. Melanocortin 1 receptor (MC1R) gene variants are associated with an increased risk for cutaneous melanoma which is largely independent of skin type and hair color. J Invest Dermatol. 2001;117:294–300. doi: 10.1046/j.0022-202x.2001.01421.x. [DOI] [PubMed] [Google Scholar]
- 55.Goldstein AM, Landi MT, Tsang S, et al. Association of MC1R variants and risk of melanoma in melanoma-prone families with CDKN2A mutations. Cancer Epidemiol Biomarkers Prev. 2005;14:2208–2212. doi: 10.1158/1055-9965.EPI-05-0321A. [DOI] [PubMed] [Google Scholar]
- 56.Box NF, Duffy DL, Chen W, et al. MC1R genotype modifies risk of melanoma in families segregating CDKN2A mutations. Am J Hum Genet. 2001;69:765–773. doi: 10.1086/323412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brown KM, Macgregor S, Montgomery GW, et al. Common sequence variants on 20q11.22 confer melanoma susceptibility. Nat Genet. 2008;40:838–840. doi: 10.1038/ng.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gudbjartsson DF, Sulem P, Stacey SN, et al. ASIP and TYR pigmentation variants associate with cutaneous melanoma and basal cell carcinoma. Nat Genet. 2008;40:886–891. doi: 10.1038/ng.161. [DOI] [PubMed] [Google Scholar]
- 59.Sulem P, Gudbjartsson DF, Stacey SN, et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat Genet. 2007;39:1443–1452. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- 60.Bishop DT, Demenais F, Iles MM, et al. Genome-wide association study identifies three loci associated with melanoma risk. Nat Genet. 2009;41:920–925. doi: 10.1038/ng.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Han J, Kraft P, Colditz GA, et al. Melanocortin 1 receptor variants and skin cancer risk. Int J Cancer. 2006;119:1976–1984. doi: 10.1002/ijc.22074. [DOI] [PubMed] [Google Scholar]
- 62.Han J, Kraft P, Nan H, et al. A genome-wide association study identifies novel alleles associated with hair color and skin pigmentation. PLoS Genet. 2008;4:e1000074. doi: 10.1371/journal.pgen.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hutchinson PE, Osborne JE, Lear JT, et al. Vitamin D receptor polymorphisms are associated with altered prognosis in patients with malignant melanoma. Clin Cancer Res. 2000;6:498–504. [PubMed] [Google Scholar]
- 64.Kanetsky PA, Swoyer J, Panossian S, et al. A polymorphism in the agouti signaling protein gene is associated with human pigmentation. Am J Hum Genet. 2002;70:770–775. doi: 10.1086/339076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Strange RC, Ellison T, Ichii-Jones F, et al. Cytochrome P450 CYP2D6 genotypes: association with hair colour, Breslow thickness and melanocyte stimulating hormone receptor alleles in patients with malignant melanoma. Pharmacogenetics. 1999;9:269–276. [PubMed] [Google Scholar]
- 66.Wolf CR, Smith CA, Gough AC, et al. Relationship between the debrisoquine hydroxylase polymorphism and cancer susceptibility. Carcinogenesis. 1992;13:1035–1038. doi: 10.1093/carcin/13.6.1035. [DOI] [PubMed] [Google Scholar]
- 67.Kefford RF, Newton Bishop JA, Bergman W, et al. Counseling and DNA testing for individuals perceived to be genetically predisposed to melanoma: A consensus statement of the Melanoma Genetics Consortium. J Clin Oncol. 1999;17:3245–3251. doi: 10.1200/JCO.1999.17.10.3245. [DOI] [PubMed] [Google Scholar]
- 68.Robson ME, Storm CD, Weitzel J, et al. Policy statement update: genetic and genomic testing for cancer susceptibility. J Clin Oncol. 2010;28:893–901. doi: 10.1200/JCO.2009.27.0660. [DOI] [PubMed] [Google Scholar]
- 69.Udayakumar D, Tsao H. Melanoma genetics: an update on risk-associated genes. Hematol/Oncol Clin North Am. 2009;23:415–429. vii. doi: 10.1016/j.hoc.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 70.Goggins WB, Tsao H. A population-based analysis of risk factors for a second primary cutaneous melanoma among melanoma survivors. Cancer. 2003;97:639–643. doi: 10.1002/cncr.11116. [DOI] [PubMed] [Google Scholar]
- 71.Leachman SA, Carucci J, Kohlmann W, et al. Selection criteria for genetic assessment of patients with familial melanoma. J Am Acad Dermatol. 2009;61:e1–14. doi: 10.1016/j.jaad.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Holland EA, Schmid H, Kefford RF, et al. CDKN2A (P16(INK4a)) And CDK4 mutation analysis in 131 Australian melanoma probands: effect of family history and multiple primary melanomas. Genes Chromosomes Cancer. 1999;25:339–348. [PubMed] [Google Scholar]
- 73.Tsao H, Zhang X, Kwitkiwski K, et al. Low prevalence of germline CDKN2A and CDK4 mutations in patients with early-onset melanoma. Arch Dermatol. 2000;136:1118–1122. doi: 10.1001/archderm.136.9.1118. [DOI] [PubMed] [Google Scholar]
- 74.Bartsch DK, Sina-Frey M, Lang S, et al. CDKN2A germline mutations in familial pancreatic cancer. Ann Surg. 2002;236:730–737. doi: 10.1097/00000658-200212000-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.de Snoo FA, Bishop DT, Bergman W, et al. Increased risk of cancer other than melanoma in CDKN2A founder mutation (p16-Leiden)-positive melanoma families. Clin Cancer Res. 2008;14:7151–7157. doi: 10.1158/1078-0432.CCR-08-0403. [DOI] [PubMed] [Google Scholar]
- 76.Sinilnikova OM, Egan KM, Quinn JL, et al. Germline BRCA2 sequence variants in patients with ocular melanoma. Int J Cancer. 1999;82:325–328. doi: 10.1002/(sici)1097-0215(19990730)82:3<325::aid-ijc3>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 77.Wang W, Niendorf KB, Patel D, et al. Estimating CDKN2A carrier probability and personalizing cancer risk assessments in hereditary melanoma using MelaPRO. Cancer Res. 2010;70:552–559. doi: 10.1158/0008-5472.CAN-09-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]


