Abstract
Metastatic melanoma is a highly lethal type of skin cancer and is often refractory to all traditional chemo-therapeutic agents. Key insights into the genetic makeup of melanoma tumors have led to the development of promising targeted agents. An activated RAS pathway, anchored by oncogenic BRAF, appears to be the central motor driving melanoma proliferation. Although recent clinical trials have brought enormous hope to patients with melanoma, adverse effects and novel escape mechanisms of these inhibitors have already emerged. Definition of the limits of the first successful targeted therapies will provide the basis for further advances in management of disseminated melanoma. In this review, the current state of targeted therapy for melanoma is discussed, including the potent BRAFV600E inhibitor vemurafenib.
Current therapies for melanoma
The incidence of melanoma has rapidly increased over the past several decades [1]. Approximately 10% of all patients who are diagnosed with melanoma eventually die from this cancer [2]. When it becomes metastatic, melanoma often leads to death within a year [3,4], a dismal prognosis that has resulted from a lack of highly curative therapies for advanced disease [1].
The United States Food and Drug Administration (FDA) has approved few therapies for metastatic melanoma, all of which have minimal beneficial effects on patient survival [5,6]. Many of these have been immunologic in nature, including interferon (IFN)-α2b, high-dose interleu-kin (IL)-2 and, as of March 2011, ipilumimab. IFN-α2b is associated with a 10–15% reduction in the risk of relapse in the adjuvant setting, whereas IL-2 produces objective response in 15% of metastatic patients [6–10]. An older FDA-approved melanoma therapy is the alkylating agent dacarbazine (DTIC), which achieves responses in less than 10% of patients [11], a profile similar to other available agents such as carmustine (BCNU), temozolomide, tax-anes and platinum analogs [6,12–14]. In the face of these limited options, there has been a sea change in melanoma treatments ushered in by recent molecular advances.
Targeted agents aimed at oncogenic drivers that have been identified over the past decade provide an opportunity for novel melanoma therapeutics [15,16]. This review focuses on the central molecular network that fuels melanoma growth and recent drug development progress towards targeting these key proteins and signaling pathways.
The central melanoma axis and therapeutic targets
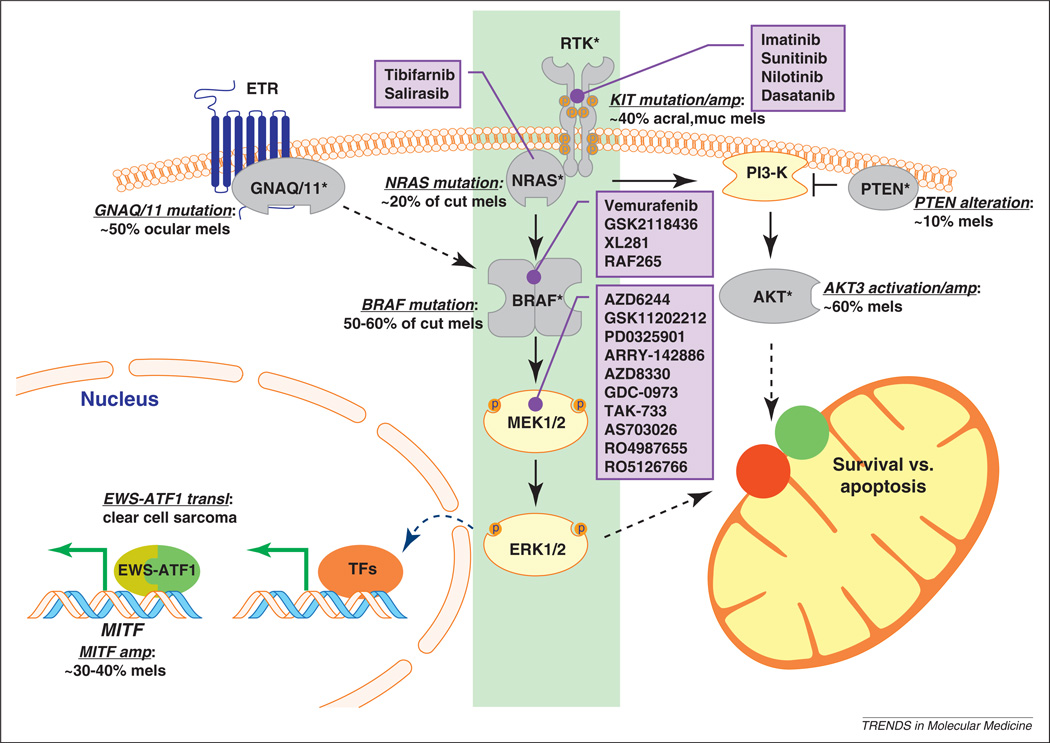
Over the past decade, much has been learned about genetic lesions that stimulate growth and signaling pathways in melanomas [17]. As shown in Figure 1, many components of the RAS pathway are either activated through oncogenic mutations or inactivated through deleterious alterations. From this composite view, activation of a KIT–NRAS–BRAF–MEK–ERK central axis (Figure 1, shaded in green) seems to be crucial in almost all forms of melanoma. Figure 1 also lists some of the drugs in the pipeline for inhibiting various components of the pathway.
Figure 1.
Key mutational and therapeutic targets in melanoma. The RAS signaling network is rife with cancer-associated mutations. BRAF is the most commonly activated oncogene in cutaneous melanomas (cut mels), followed by NRAS. Upstream of RAS, the c-KIT receptor tyrosine kinase (RTK) is amplified or activated in a substantial fraction of acral and mucosal melanomas. It thus seems that the RTK–NRAS–BRAF–MEK–ERK cascade represents a central axis (highlighted in green) that is activated in nearly all melanomas. Parallel to this axis is the PI3-K pathway, which is also activated in melanomas either through loss of PTEN or activation of AKT3. In addition, it has recently been shown that GNAQ and GNA11, which are transducers of the endothelin receptor signal (ETR), are mutated in ocular melanomas. These heightened signaling events lead to both increased transcription of survival genes (such as MITF) and enhanced pro-survival factors in mitochondria through the regulation of BCL2 family proteins (red, pro-apoptotic; green, pro-survival). MITF, which is a master regulator of melanocyte survival, is also amplified in melanomas and a target of the EWS-ATF1 fusion protein described in clear cell sarcomas (so-called melanoma of the soft parts). Drugs known to inhibit the central axis and with a potential therapeutic impact on melanoma are listed in the purple boxes.
Receptor tyrosine kinases (RTKs)
A number of growth factor RTKs such as EGFR, PDGFR and KIT are expressed in melanoma cells, although recurrent activating mutations are uncommon. One lineage-derived RTK is c-KIT, a receptor known to be crucial in melanocyte differentiation but whose expression appears to be lost in many melanomas [18,19]. A more direct role for c-KIT was recently recognized when genomic screens revealed that the KIT locus (chromosome 4q11) was amplified and/or mutated in a subset of mucosal, acral and chronically sun-damaged (CSD) melanomas (MACs) [20]. Approximately 10–20% of these melanomas harbor the same activating KIT mutations described in gastrointestinal stromal tumors (GISTs) [20–24].
The earlier successes of imatinib in c-KIT-mutated GISTs suggested that MAC melanomas may be particularly vulnerable to c-KIT inhibitors. The idea was initially bolstered by reports of several melanoma cases treated with imatinib [25,26]. These clinical results were subsequently confirmed in other melanoma cell lines sustained by an activating c-KIT mutation or an SCF–c-KIT autocrine loop [21,27]. Imatinib has minimal inhibitory effects on melanoma cell lines containing the BRAFV600E mutation despite evidence of c-KIT expression; furthermore, the mere presence of c-KIT receptor expression does not seem to predict response [28,29]. Thus, it appears that the potential clinical role of c-KIT inhibitors is probably restricted to those melanomas that have activating mutations and consequent c-KIT-dependent signaling. Interestingly, response seems to correlate with the site of mutation in c-KIT. For example, melanomas withmutations in the juxtamembrane region of c-KIT are associated with a better response to imatinib treatment [28]. Because imatinib is not c-KIT-specific, it is possible that a more selective agent could achieve a greater degree of inhibition and result in more profound responses.
Reports on two open-label Phase II trials of imatinib mesylate for KIT-mutated melanomas have recently been published. In the first trial, Carvajal et al. treated 28 patients who developed metastatic melanoma from MAC sites with 400 mg of imatinib twice daily [30]. There were 2 complete responses (CRs) lasting 94 and 95 weeks, 2 durable partial responses (PRs) lasting 53 and 89 weeks, and 2 transient PRs lasting 12 and 18 weeks among 25 evaluable patients. The median progression-free survival (PFS) was 12 weeks, with a median overall survival (OS) of 46.3 weeks. At a molecular level, 23.4% of the cases harbored either KIT mutations or amplifications, whereas 27.8% of the tumors actually contained either BRAF or NRAS mutations. The most significant responses occurred in patients with KITk642E or KITL576P variants and those with a mutant/allele ratio >1, that is, tumors with greater activated KIT dependence. In the second trial, Guo et al. treated 43 metastatic melanoma patients with 400 mg of imatinib per day unless intolerable toxicity or disease progression occurred [31]. Eligibility in the Guo trial required KIT aberrations defined as mutations in exons 9,11, 13,17, or 18 and/or increases in copy number. Overall, PRs, stable disease and progressive disease were observed in 10 patients (23.3%), 13 patients (30.2%) and 20 patients (46.5%), respectively. The 6-month PFS and 1-year OS rates were 36.6% and 51.0%, respectively. The median PFS time was 3.5 months (range 1.3–5.7 months) and the OS time was 14.0 months (range 10.8–17.2 months). There were no clear-cut associations between outcome and KIT mutation characteristics. Although the overall benefits of imatinib in these studies are encouraging, albeit modest, other RTK inhibitors (e.g. sunitinib, nilotinib and dasatanib) are emerging (Table 1) and may prove more efficacious in trials. For instance, a current Phase III trial is comparing nilotinib to dacarbazine (NCT01028222) in patients with KIT-mutated metastatic melanoma (exons 9, 11 or 13, or exon 17 mutations p.Tyr822Asp, p.Asp820Tyr, or p.Tyr823Asp). Masitinib is another potent and highly selective oral RTK inhibitor that has combined activity against both c-KIT and LYN. A recent small study showed some effect against another KIT-mediated disease, systemic mastocytosis [32], and a Phase III trial of masitinib for metastatic melanomas with juxtamembrane mutations is also currently enrolling patients (NCTO1280565).
Table 1.
Summary of RAS pathway inhibitors
| Drug | Targets | Stage of clinical development |
|---|---|---|
| Imatinib (Gleevec, STI571) | KIT, ABL, PDGFR, NQO2 [84], V-ATPase [85] | Approved for CML and GIST [86] |
| Sunitinib (SU11248) | KIT, PDGFR, VEGFR [87] | Approved for RCC and GIST [87] |
| Nilotinib | KIT, ABL, LCK, NQO2, DDR1 [84] | Approved for CML [88] |
| Dasatanib (BMS-354825) | KIT, ABL, SRC [89], DDR1, BTK, TEC [84] | Approved for CML [90] |
| Tipifarnib (R115777) | RAS and other proteins that require farnesyl transferase [91] |
Phase II/III [91] |
| Salirasib (FTS) | RAS, mTOR [92] | Phase II [43] |
| Sorafenib (BAY 43-9006) | BRAF | Approved for RCC and HCC Failed at phase II for advanced melanoma [52] |
| PLX4720 | BRAF, CRAF, VEGF, PDGF, FLT3, KIT [46] | Precursor of PLX4032 |
| Vemurafenib (PLX4032) | BRAFV600E, BRAFWT, BRK [56] | Phase III [61] |
| GSK2118436 | BRAFV600E, CRAF, BRAFWI, ARAF, ACK1, SRMS and MAP4K5 [57,58] |
Phase I/II [68] |
| PD0325901 | MEK | Phase II [75] |
| AZD6244 (ARRY-142886) | MEK | Phase II [93] |
| Anthrax lethal toxin | MEK | |
| GSK1120212 (JTP-74057) | MEK | Phase III |
Prickett and colleagues recently scanned the tyrosine kinome and identified mutations in ERBB4 in 19% of melanoma cases, although there were no mutational hot-spots [33]. The alterations were clearly oncogenic in several in vitro phenotypes, such as NIH-3T3 transformation and soft-agar growth. Furthermore, inhibition of ERBB4 by lapatinib induced apoptosis, especially in ERBB4-mu-tated cells. These recent findings have led to a Phase II trial of lapatinib in stage IV melanoma for patients with ERBB4-mutated melanomas (NCT01264081).
RAS inhibitors
NRAS is the second most commonly activated oncogene in melanoma after BRAF. Like other RAS protein members, activating changes occur on p.Glyl2 or p.Gln61. The potential of NRAS as a therapeutic target has been validated in preclinical models with siRNA [34], but potent and selective pharmacologic inhibitors are not readily available. The first group of compounds used to target RAS were farnesyl transferase inhibitors (FTIs), such as R115777 (tibifarnib; Figure 1). The FTIs were designed to block the post-translational lipid modification of RAS that is required for full RAS activity [35]. FTIs inhibit tumor growth in preclinical models [36], although their performance in the clinical trial setting has been lackluster [37]. The prevailing hypothesis for the lack of efficacy is that FTIs inhibit a large family of proteins that require farnesylation as a post-translational modification. Thus, it is not possible to achieve sufficient downregulation of RAS without profoundly impairing the function of other farnesylated proteins, a fact that is responsible for the dose-limiting toxicity of these agents. Furthermore, KRAS and NRAS can circumvent FTIs by employing geranylgeranyltransferases in cells and thereby maintaining their function in the presence of FTIs. Nevertheless, it remains possible that FTIs might still enhance the effect of other chemotherapeutic agents in melanoma, as shown in at least one preclinical study [38].
Salirasib (S-trans,trans-farnesylthiosalicylic acid, FTS) belongs to another group of RAS antagonists and mimics the carboxy-terminal farnesylcysteine that is common to all three RAS isoforms [39]. Thus, FTS competes with the active, GTP-bound forms of RAS proteins for specific binding sites on the cytoplasmic membrane [40] and inhibits melanoma cell growth in vitro and in xenograft models [41,42]. Initial results from recent clinical trials with pancreatic cancer patients support the ability of FTS to suppress RAS function and possibly mediate survival benefits [43,44]. These preliminary results require validation in larger patient populations and randomized trials. RAS inhibitors still hold unrealized potential as a therapeutic approach for melanoma, especially for the ∼20% of tumors in which activating mutations are found.
RAF inhibitors
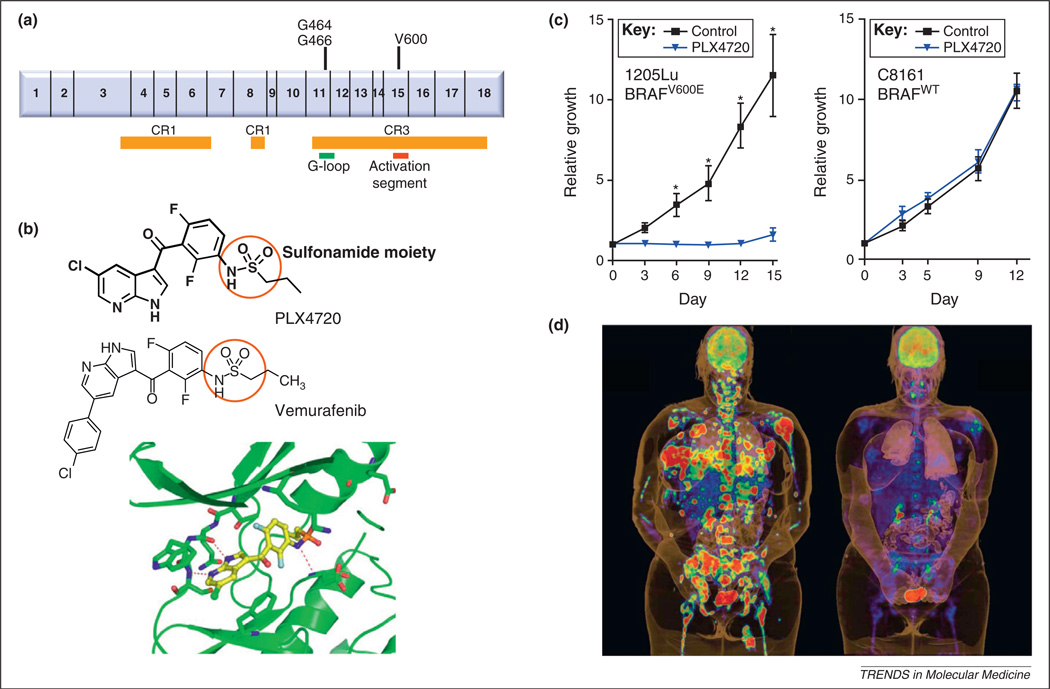
As mentioned above, the most prevalent alteration in cutaneous melanoma is activation of the serine/threonine protein kinase BRAF [45], which makes BRAF a veritable target in the therapeutic landscape. Overall, approximately 40–50% of uncultured cutaneous melanomas harbor BRAF mutations, with the p.Val600Glu missense mutation (BRAFV600E), which lies in the CR3 kinase domain (Figure 2), comprising ≥90% of reported mutations.
Figure 2.
BRAF inhibition and development of vemurafenib. (a) Domain structure of BRAF showing the CR domains and the location of the most common mutations in exons 11 and 15. (b) Chemical structure of PLX4720 and PLX4032 (vemurafenib). These compounds are competitive ATP protein kinase inhibitors (class I) selective for the BRAFV600E mutant protein. Also shown is co-structure of PLX4720 with the BRAF kinase. It has been postulated that the key to the specificity of PLX4720 lies in the differential interaction of the sulfonamide moiety (red circle) with the Asp and Phe residues of the conserved kinase Asp-Phe-Gly (DFG) sequence. In the active conformation, the nitrogen atom of the sulfonamide moiety is deprotonated and interacts with the main-chain NH group of Asp-594, whereas the oxygen atoms of the sulfonamide form hydrogen bonds to the NH group of Phe-595 and the side chain of Lys-483. Crystallographic data support a selectivity model in which the deprotonated sulfonamide interacts more favorably with the active kinase conformation that is characteristic of BRAFV600E. (c) In tumor xenografts, a BRAFV600E mutated melanoma line (1205Lu) showed significantly greater response to PLX4720 (100 mg/kg by oral gavage) compared to a BRAFWT line (C8161). (d) Dramatic tumor shrinkage after treatment with PLX4032 in a patient with metastatic BRAFV600E melanoma. Panels b and c and panel d are reproduced with permission from [56] and [83], respectively.
The first BRAF inhibitor that progressed to clinical trial was sorafenib, which targets multiple protein kinases including BRAF, CRAF and the VEGF and PDGF RTKs (Figure 1) [46]. Sorafenib has minimal activity as a single agent in melanoma, although initial results were more encouraging when it was combined with carboplatin and paclitaxel or temozolomide [47–49]. However, the activity of the combination regimen did not correlate with BRAF mutational status. A Phase III trial of sorafenib in combination with carboplatin and paclitaxel in patients with advanced melanoma failed to improve overall survival [50]. The failure of sorafenib has been attributed in part to incomplete MAP kinase inhibition at the maximum tolerated dose [51] and to compensatory RAF–MEK–ERK signaling [52] or other escape mechanisms [53,54]. A recent study challenged the entire notion that sorafenib works through BRAF in vivo. Whittaker et al. engineered cells with a gatekeeper mutation (p.Thr529Asn or p.T529N) that rendered BRAFT529N/V600E cells resistant to sorafenib in vitro and yet sensitive to sorafenib in vivo. By contrast, truly selective BRAF inhibitors (SBIs) such as PLX4720 (see below) lost their effectiveness both in vitro and in vivo when the gatekeeper mutation was introduced. This suggests that sorafenib has BRAF-independent anti-tumor activity and may also explain why sorafenib causes side effects at a dose that does not efficiently inhibit MEK signaling [55].
More selective BRAF inhibitors have been synthesized over the past few years. The first of these, PLX4720, selectively inhibits BRAFV600E (IC50=13 nM vs 160 nM for wild-type BRAF, BRAFWT) and has been thoroughly tested and validated in preclinical BRAFV600E models [56]. The clinical compound vemurafenib (PLX4032; Figure 2), which is an analog of PLX4720, shows higher selectivity for BRAFV600E (IC50=31 nM) and CRAF (IC50=48 nM) than wild-type BRAF (IC50=100 nM) [57]. These in vitro parameters, however, do not necessarily predict drug activity in vivo, where signaling networks may produce more dynamic physiologic responses. Furthermore, vemurafenib, at least in vitro, also inhibits several other protein kinases, such as ACK1, SRMS and MAP4K5 [58], with similar potency (IC50<50 nM) compared with BRAFV600E. Although speculative, suppression of these secondary targets could potentially contribute to the observed responses [58] and more investigation is needed to understand the consequences of these off-target effects.
The clinical efficacy of vemurafenib in melanoma patients with BRAFV600E mutations has been firmly established through three trials. The objective response exceeded 50% among those treated at the higher doses in the Phase I trial and an even higher response rate was obtained when a cohort of patients received the recommended Phase II dose [59,60]. Correlative studies also demonstrated that doses of vemurafenib that lead to >90% reductions in ERK phosphorylation are required to achieve a meaningful clinical response [58]. The pivotal Phase III study (BRIM-3) enrolled 675 patients with previously untreated metastatic melanoma that could not be surgically removed [61]. Patients with BRAFV600E-mutated tumors were randomly assigned to receive either vemur-afenib (960 mg orally twice daily) or dacarbazine. At the 6-month evaluation, OS was 84% in the vemurafenib group (n=336) and 64% in the dacarbazine group (n=336; hazard ratio 0.37, p<0.001). The hazard ratio for tumor progression in the vemurafenib group was 0.26 (p<0.001) and the estimated median PFS was 5.3 months in the vemurafenib group and 1.6 months in the dacarbazine group. The objective response rate for individuals treated with vemurafenib was 48% compared to 5% for dacarbazine-treated patients (p<0.001). As observed in earlier trials, kerati-nocytic neoplasms (cutaneous squamous cell carcinomas or a related entity, keratoacanthoma) developed in 18% of patients treated with vemurafenib. BRIM-3 represents the first prospective randomized molecular therapy trial in melanoma and the first to demonstrate a convincing survival benefit. Although its importance cannot be overstated, there are several lingering challenges worth addressing. First, patients with BRAFWT tumors are not eligible for vemurafenib and are in desperate need of effective agents. Second, although 48% of the patients showed an objective response, a significant fraction of BRAF-mutated tumors did not reach the RECIST criteria and therefore seem to be innately insensitive to vemurafenib; it will be important to identify these primarily resistant tumors through secondary biomarkers beyond BRAF status. Third, nearly all patients relapse with time despite ongoing treatment, and thus OS after longer follow-up will be important to establish the true rate of cure.
Secondary resistance in RAF-mutated tumors
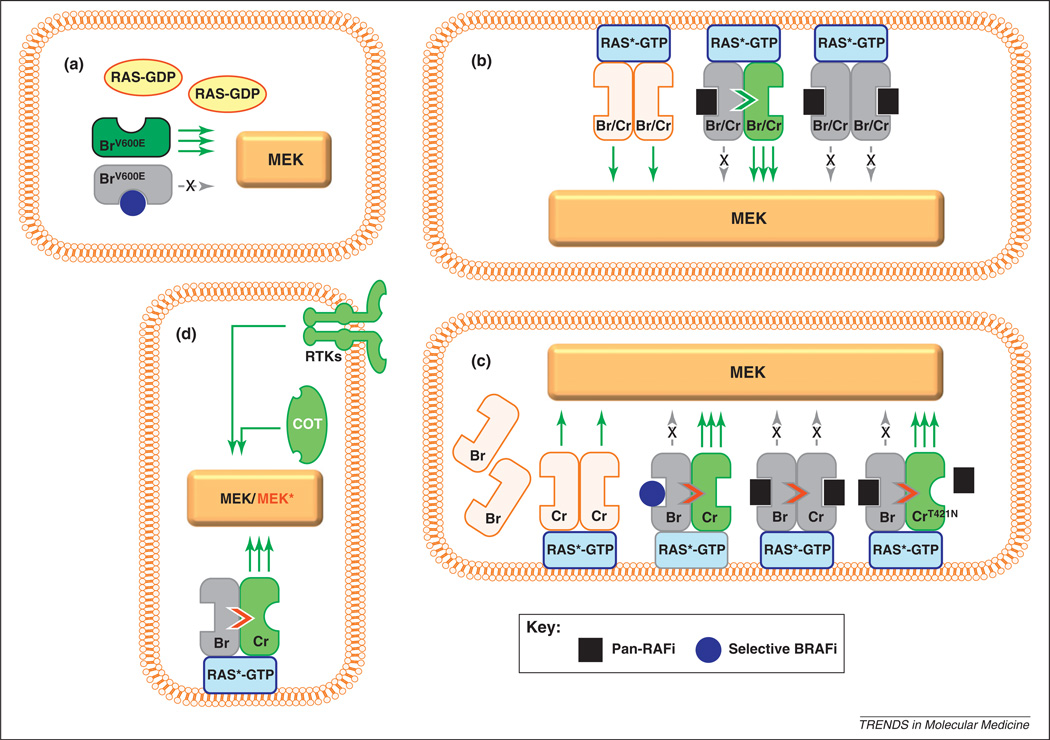
One of the most pressing and exciting areas of investigation is the elucidation of primary and secondary resistance mechanisms. Biochemical and genetic studies into the development of resistance have largely focused on two specific questions: why is MEK–ERK signaling paradoxically activated by SBIs in RAS-mutated cells, and how do BRAF-mutated cells gain resistance to SBIs amid chronic suppression? There are at least two models that address the first question and both invoke RAF dimerization, although the two models differ in their molecular details (Figure 3) [57,62]. In response to growth factor receptor activation or an oncogenic RAS mutation, the RAS protein binds GTP, is activated (RAS*-GTP; Figure 3), localizes to the plasma membrane and induces homodimerization and heterodimerization of BRAF and CRAF. In cells driven by a BRAFV600E mutation, RAS activation is bypassed and signal initiation occurs in the cytoplasm; MEK phosphorylation results from constitutive BRAF activity. Consequently, an SBI attenuates nearly all downstream MEK–ERK signal propagation. However, in BRAFWT cells, this molecular assembly at the cellular membrane is a crucial step for subsequent MEK phosphorylation and downstream signaling. One unexpected observation is the paradoxical MAPK pathway stimulation by SBIs in WT BRAF cells [57,62]. How does this occur? In one model (Figure 3b), low concentrations of a RAF inhibitor inactivate only a single monomer in the RAF dimer; dimerization still transactivates the uninhibited partner RAF molecule and triggers MAPK signaling [57]. Higher RAF inhibitor levels lead to inhibition of both RAF partners and all signaling is suppressed. In a second model (Figure 3c) [62], WT BRAF remains largely inactive in the cytoplasm of RAS*-GTP cells until it binds an SBI. The SBI-bound BRAFWT then translocates to the cell membrane where it dimerizes with CRAF, further stimulating CRAF signaling. A pan-RAF inhibitor (PRI) suppresses both BRAF and CRAF and thereby abrogates all MAPK signaling. Gatekeeper CRAF mutations, such as CRAF Thr421Asn, could disrupt binding of the PRI to CRAF and restore signaling. The implication for both models is that RAS-mutated cells may in fact be stimulated by an SBI, which could explain the observed squamous cell carcinomas that develop while on vemurafenib. Thus, the use of vemurafenib requires absolute genetic precision.
Figure 3.
Models of RAF activation and inhibition, (a) In cells with mutated BRAF (BrV600E), RAS is inactive, cytoplasmic and unable to induce RAF dimerization. MEK phosphorylation and activation occur almost exclusively via BRAFV600E and thus are effectively silenced by a selective BRAF inhibitor (blue circle). This would explain the dramatic tumor shrinkage early in treatment with vemurafenib. In some cells with wild-type BRAF, RAS is activated (RAS*-GTP), presumably by mutagenesis (e.g. NRAS mutations) or other upstream receptor events. There are at least two models of signal transduction in these RAS-stimulated tumors. In the transactivation model (b), active RAS*-GTP mobilizes to the membrane and induces homodimerization or heterodimerization of wild-type BRAF and CRAF proteins (Br/Cr), which in turn initiates signaling. At low concentrations, an ATP-competitive RAF inhibitor may occupy the active site of one of the RAF protomers. Inhibitor binding to the active site of one RAF kinase induces a conformational change that facilitates transactivation of the other RAF molecule in the dimer; therefore, there is paradoxical enhancement of MEK/ERK signaling through the uninhibited partner. At higher inhibitor concentrations, both RAF molecules are inhibited and MEK/ERK signaling is completely abolished. In the translocation model (c), wild-type BRAF (Br) remains largely inactive in the cytoplasm owing to autoinhibitory signals. Activated RAS*-GTP triggers MAPK signaling through CRAF (Cr). When wild-type BRAF binds a selective ATP-competitive BRAF inhibitor, the BRAF molecule is recruited to the membrane, thereby enhancing the RAS*-GTP/CRAF interaction and signaling (as in b). With pan-RAF inhibitors, wild-type BRAF may still enhance the RAS*-GTP/CRAF interaction, but MEK/ERK signaling is suppressed because CRAF activity itself is concurrently abrogated by inhibitor binding. In situations in which CRAF harbors a gatekeeper mutation, such as p.Thr421Asn (CrT421N), a pan-RAF agent inhibits BRAF, which then transactivates CRAFT421N; however, CRAFT421N is not inhibited by the pan-RAF agent because the mutation prevents drug binding. Both models suggest mechanisms by which BRAF inhibition may lead to increased stimulation of CRAF in RAS-mutated cells or growth-factor-stimulated cells, (d) Mechanisms of BRAF inhibitor resistance include activation of receptor tyrosine kinases (RTKs) such as PDGFR and IGF1R, CRAF and other protein kinases (e.g. COT), along with mutational activation of NRAS and MEK (MEK*).
Several recent reports have also shed light on possible mechanisms that are responsible for resistance to vemurafenib (Figure 3). For example, reactivation of the ERK pathway by an NRAS mutation confers secondary resistance to vemurafenib, as functionally shown in a relapse-derived cell line and verified in a clinically resistant melanoma sample [63]. In another study, Johannessen et al. undertook a kinome-wide screen for molecules that could confer resistance to PLX4720 and identified both CRAF and the kinase MAP3K8 (COT, TPL2) [64]. Naїve melanoma cell lines with elevated COT levels exhibit de novo resistance to PLX4720 and 2 of 3 melanoma samples taken from patients early in the course of therapy or at the time of progression also had increased COT expression. Finally, a more recent report identified a MEK1 mutation in a single tumor that had become resistant to vemurafenib [65].
Interestingly, ERK reactivation may not to be the only means of acquiring vemurafenib resistance. Nazarian et al. found that PDGFRβ upregulation can be associated with vemurafenib resistance in the absence of ostensible ERK activation [63]. There is biochemical evidence to suggest that AKT activation is correlated with heightened PDGFRβ expression, although other unidentified downstream effectors may also play a role. Similarly, Villanueva et al. reported that increased IGF receptor signaling may also be correlated with acquired SBI resistance [66].
There are several other BRAF inhibitors currently in clinical development (Figure 1 and Table 1) [67]. GSK2118436 is a BRAF inhibitor that showed promising results in an early clinical trial [68]. Interestingly, regression of brain metastasis after treatment with GSK2118436 has been observed in several patients. A Phase II clinical trial is now ongoing to test its effect in melanoma patients with BRAF mutation (NCT01266967).
CRAF may also be an effective target for melanoma therapy [69,70], particularly in BRAFWT cells, because CRAF seems to be the key mediator of MEK activation in NRAS-mutated melanomas [71]. PRIs may be more relevant for NRAS-mutated melanomas and non-V600E BRAF mutants, which tend to activate MEK through CRAF signaling [71,72].
MEK inhibitor
MEK is the major downstream molecule of oncogenic BRAF. An early study found that melanoma cells with BRAF mutations tend to be more sensitive to MEK inhibitors than those with NRAS mutations [73]. Therefore, BRAF mutational status may also predict sensitivity to MEK inhibitors in the clinic.
MEK inhibitors reduce proliferation, colony formation and invasiveness of BRAFV600E mutant human melanoma cells in vitro and tumor growth in vivo [74]. Several MEK inhibitors have been investigated in clinical trials in which patients with advanced melanoma were treated (Figure 1). PD0325901 was evaluated in a Phase I trial: 2 of 27 patients had an objective response and another 5 patients showed disease stabilization [67]. However, dose-limiting side effects such as diarrhea and rash precluded the high amount of target inhibition required to adequately suppress the MAPK pathway in tumor cells. Because MEK inhibitors inhibit MAPK pathway signaling in normal cells as well as tumor cells, it may not be possible to achieve sufficient target effects in tumors owing to normal tissue toxicity at the drug concentrations required. Phase II trials of PD0325901 in non-small-cell lung cancer were suspended because of limited activity and intolerable side effects such as visual disturbances [75]. In a recent Phase I trial, another MEK inhibitor, AZD6244, showed only moderate effects in a very small subgroup of patients with metastatic melanoma harboring BRAFV600E mutations [76]. However, in the follow-up phase II trial with AZD6244, 12% of patients whose tumors harbored BRAFV600E showed significant tumor regression, although the regression was not complete [67]. This limited response may be due to insufficient target inhibition or failure to induce cell death. In vitro studies have also demonstrated that BRAF/MEK inhibitors lead to mainly cytostatic effects in BRAFV600E-mutated melanoma cells [56,73,77] and therefore AZD6244 may not be adequate as a single agent in melanoma treatment. GSK1120212 is an allosteric MEK inhibitor [78] that showed promising antitumor activity in a Phase I clinical trial [79] and is now being evaluated in a Phase III trial (NCT01245062). Finally, the lethal toxin anthrax, which selectively degrades and inactivates MEK1 and MEK2 [80], is also being tested in melanoma clinical trials [53].
Concluding remarks
It is clear that single-agent approaches in melanoma are not capable of achieving a cure, a finding that is not surprising given the genetic complexity of melanomas and the concomitant activation of multiple signaling pathways. The experience with BRAF inhibitors has demonstrated that melanoma often resurrects itself, even after the main growth signals are abrogated. Therefore, simultaneous targeting of several pathways is likely to result in better outcomes. The redundancy within the multiple signaling pathways activated in melanoma, such as PTEN loss with consequent AKT activation, raises the possibility of combining MAPK and AKT pathway inhibitors in new formulations. Studies have shown that PI3K and MEK inhibitors synergize to reduce growth and survival of melanoma cells in 3D cell culture systems [81,82] and thus larger signaling networks may need to be considered. Moreover, melanomas with BRAFV600E often have other genetic disruptions in molecules such as cyclin D1, CDK2, CDK4, MITF and AKT3 [67], which suggests that additional inhibitor combinations may enhance efficacy.
Melanomas are genetically heterogeneous, and the use of personalized cancer therapy has already been demonstrated in this cancer. To maximize success, future targeted therapy may need to be tested in patients for whom the relevant combination of genetic aberrations in the tumors have been predetermined.
Acknowledgments
We want to thank the National Institutes of Health (K24 CA149202 to H.T.), the Melanoma Research Alliance, the American Skin Association and the generous donors to the Massachusetts General Hospital Millennium Melanoma Fund for their support during the writing of this review.
References
- 1.Berwick M, et al. Melanoma epidemiology and public health. Dermatol. Clin. 2009;27:205–214. doi: 10.1016/j.det.2008.12.002. viii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R, et al. Cancer statistics, 2011: The impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J. Clin. 2011;61:212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 3.Tsao H, et al. Management of cutaneous melanoma. N. Engl. J. Med. 2004;351:998–1012. doi: 10.1056/NEJMra041245. [DOI] [PubMed] [Google Scholar]
- 4.Agarwala SS. Current systemic therapy for metastatic melanoma. Expert Rev. Anticancer Ther. 2009;9:587–595. doi: 10.1586/era.09.25. [DOI] [PubMed] [Google Scholar]
- 5.Fecher LA, Flaherty KT. Where are we with adjuvant therapy of stage III and IV melanoma in 2009? J. Natl. Compr. Cancer Netw. 2009;7:295–304. doi: 10.6004/jnccn.2009.0022. [DOI] [PubMed] [Google Scholar]
- 6.Tarhini AA, Agarwala SS. Cutaneous melanoma: available therapy for metastatic disease. Dermatol. Ther. 2006;19:19–25. doi: 10.1111/j.1529-8019.2005.00052.x. [DOI] [PubMed] [Google Scholar]
- 7.Kirkwood JM, et al. A pooled analysis of eastern cooperative oncology group and intergroup trials of adjuvant high-dose interferon for melanoma. Clin. Cancer Res. 2004;10:1670–1677. doi: 10.1158/1078-0432.ccr-1103-3. [DOI] [PubMed] [Google Scholar]
- 8.Agarwala SS, Kirkwood JM. Interferons in melanoma. Curr. Opin. Oncol. 1996;8:167–174. doi: 10.1097/00001622-199603000-00015. [DOI] [PubMed] [Google Scholar]
- 9.Atkins MB, et al. High-dose recombinant interleukin 2 therapy for patients with metastatic melanoma: analysis of 270 patients treated between 1985 and 1993. J. Clin. Oncol. 1999;17:2105–2116. doi: 10.1200/JCO.1999.17.7.2105. [DOI] [PubMed] [Google Scholar]
- 10.Atkins MB, et al. High-dose recombinant interleukin-2 therapy in patients with metastatic melanoma: long-term survival update. Cancer J. Sci. Am. 2000;6(Suppl. 1):S11–S14. [PubMed] [Google Scholar]
- 11.Ji Z, et al. Molecular therapeutic approaches to melanoma. Mol. Aspects Med. 2010;31:194–204. doi: 10.1016/j.mam.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Wagner JD, et al. Current therapy of cutaneous melanoma. Plast. Reconstr. Surg. 2000;105:1774–1799. doi: 10.1097/00006534-200004050-00028. quiz 1800-1771. [DOI] [PubMed] [Google Scholar]
- 13.Mays SR, Nelson BR. Current therapy of cutaneous melanoma. Cutis. 1999;63:293–298. [PubMed] [Google Scholar]
- 14.Chapman PB, et al. Phase III multicenter randomized trial of the Dartmouth regimen versus dacarbazine in patients with metastatic melanoma. J. Clin. Oncol. 1999;17:2745–2751. doi: 10.1200/JCO.1999.17.9.2745. [DOI] [PubMed] [Google Scholar]
- 15.Chin L, et al. Malignant melanoma genetics and therapeutics in the genomic era. Genes Dev. 2006;20:2149–2182. doi: 10.1101/gad.1437206. (PMID:16912270) [DOI] [PubMed] [Google Scholar]
- 16.Puzanov I, et al. Biological challenges of BRAF inhibitor therapy. Mol. Oncol. 2011;5:116–123. doi: 10.1016/j.molonc.2011.01.005. (PMID:21393075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hocker TL, et al. Melanoma genetics and therapeutic approaches in the 21st century: moving from the benchside to the bedside. J. Invest. Dermatol. 2008;128:2575–2595. doi: 10.1038/jid.2008.226. [DOI] [PubMed] [Google Scholar]
- 18.Natali PG, et al. Progression of human cutaneous melanoma is associated with loss of expression of c-kit proto-oncogene receptor. Int. J. Cancer. 1992;52:197–201. doi: 10.1002/ijc.2910520207. [DOI] [PubMed] [Google Scholar]
- 19.Lassam N, Bickford S. Loss of c-kit expression in cultured melanoma cells. Oncogene. 1992;7:51–56. [PubMed] [Google Scholar]
- 20.Curtin JA, et al. Somatic activation of KIT in distinct subtypes of melanoma. J. Clin. Oncol. 2006;24:4340–346. doi: 10.1200/JCO.2006.06.2984. [DOI] [PubMed] [Google Scholar]
- 21.Ashida A, et al. Pathological activation of KIT in metastatic tumors of acral and mucosal melanomas. Int. J. Cancer. 2009;124:862–868. doi: 10.1002/ijc.24048. [DOI] [PubMed] [Google Scholar]
- 22.Rivera RS, et al. C-kit protein expression correlated with activating mutations in KIT gene in oral mucosal melanoma. Virchows Arch. 2008;452:27–32. doi: 10.1007/s00428-007-0524-2. [DOI] [PubMed] [Google Scholar]
- 23.Antonescu CR, et al. L576P KIT mutation in anal melanomas correlates with KIT protein expression and is sensitive to specific kinase inhibition. Int. J. Cancer. 2007;121:257–264. doi: 10.1002/ijc.22681. [DOI] [PubMed] [Google Scholar]
- 24.Smalley KS, et al. Identification of a novel subgroup of melanomas with KIT/cyclin-dependent kinase-4 overexpression. Cancer Res. 2008;68:5743–5752. doi: 10.1158/0008-5472.CAN-08-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lutzky J, et al. Dose-dependent, complete response to imatinib of a metastatic mucosal melanoma with a K642E KIT mutation. Pigment Cell Melanoma Res. 2008;21:492–493. doi: 10.1111/j.1755-148X.2008.00475.x. [DOI] [PubMed] [Google Scholar]
- 26.Hodi FS, et al. Major response to imatinib mesylate in KIT-mutated melanoma. J. Clin. Oncol. 2008;26:2046–2051. doi: 10.1200/JCO.2007.14.0707. [DOI] [PubMed] [Google Scholar]
- 27.Jiang X, et al. Imatinib targeting of KIT-mutant oncoprotein in melanoma. Clin. Cancer Res. 2008;14:7726–7732. doi: 10.1158/1078-0432.CCR-08-1144. [DOI] [PubMed] [Google Scholar]
- 28.Smalley KS, et al. c-KIT signaling as the driving oncogenic event in sub-groups of melanomas. Histol. Histopathol. 2009;24:643–650. [Google Scholar]
- 29.Hofmann UB, et al. Overexpression of the KIT/SCF in uveal melanoma does not translate into clinical efficacy of imatinib mesylate. Clin. Cancer Res. 2009;15:324–329. doi: 10.1158/1078-0432.CCR-08-2243. [DOI] [PubMed] [Google Scholar]
- 30.Carvajal RD, et al. KIT as a therapeutic target in metastatic melanoma. J. Am. Med. Assoc. 2011;305:2327–2334. doi: 10.1001/jama.2011.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo J, et al. Phase II, open-label, single-arm trial of imatinib mesylate in patients with metastatic melanoma harboring c-Kit mutation or amplification. J. Clin. Oncol. 2011;29:2904–2909. doi: 10.1200/JCO.2010.33.9275. [DOI] [PubMed] [Google Scholar]
- 32.Paul C, et al. Masitinib for the treatment of systemic and cutaneous mastocytosis with handicap: a Phase 2a study. Am. J. Hematol. 2010;85:921–925. doi: 10.1002/ajh.21894. [DOI] [PubMed] [Google Scholar]
- 33.Prickett TD, et al. Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in ERBB4. Nat. Genet. 2009;41:1127–1132. doi: 10.1038/ng.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eskandarpour M, et al. Suppression of oncogenic NRAS by RNA interference induces apoptosis of human melanoma cells. Int. J. Cancer. 2005;115:65–73. doi: 10.1002/ijc.20873. [DOI] [PubMed] [Google Scholar]
- 35.James GL, et al. Benzodiazepine peptidomimetics: potent inhibitors of Ras farnesylation in animal cells. Science. 1993;260:1937–1942. doi: 10.1126/science.8316834. [DOI] [PubMed] [Google Scholar]
- 36.Sun J, et al. Ras CAAX peptidomimetic FTI 276 selectively blocks tumor growth in nude mice of a human lung carcinoma with K-Ras mutation and p53 deletion. Cancer Res. 1995;55:4243–4247. [PubMed] [Google Scholar]
- 37.Caponigro F, et al. Farnesyl transferase inhibitors in clinical development. Expert Opin. Investig. Drugs. 2003;12:943–954. doi: 10.1517/13543784.12.6.943. [DOI] [PubMed] [Google Scholar]
- 38.Smalley KS, Eisen TG. Farnesyl transferase inhibitor SCH66336 is cytostatic, pro-apoptotic and enhances chemosensitivity to cisplatin in melanoma cells. Int. J. Cancer. 2003;105:165–175. doi: 10.1002/ijc.11064. [DOI] [PubMed] [Google Scholar]
- 39.Weisz B, et al. A new functional Ras antagonist inhibits human pancreatic tumor growth in nude mice. Oncogene. 1999;18:2579–2588. doi: 10.1038/sj.onc.1202602. [DOI] [PubMed] [Google Scholar]
- 40.Aharonson Z, et al. Stringent structural requirements for anti-Ras activity of S-prenyl analogues. Biochim. Biophys Acta. 1998;1406:40–50. doi: 10.1016/s0925-4439(97)00077-x. [DOI] [PubMed] [Google Scholar]
- 41.Jansen B, et al. Novel Ras antagonist blocks human melanoma growth. Proc. Natl. Acad. Sci. U.S.A. 1999;96:14019–14024. doi: 10.1073/pnas.96.24.14019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smalley KS, Eisen TG. Farnesyl thiosalicylic acid inhibits the growth of melanoma cells through a combination of cytostatic and pro-apoptotic effects. Int. J. Cancer. 2002;98:514–522. doi: 10.1002/ijc.10213. [DOI] [PubMed] [Google Scholar]
- 43.Johnson ML, et al. A Phase II trial of salirasib in patients with stage IIIB/IV lung adenocarcinoma enriched for KRAS mutations. J. Clin. Oncol. 2009;27(15 Suppl.):8012. [Google Scholar]
- 44.Laheru D, et al. Integrated development of S-trans,trans-farnesylthiosalicyclic acid (FTS, salirasib) in advanced pancreatic cancer. J. Clin. Oncol. 2009;27(15 Suppl.):4529. [Google Scholar]
- 45.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 46.Wilhelm SM, et al. BAY 43-9006 exhibits broad spectrum oral antitumor activity and targets the RAF/MEK/ERK pathway and receptor tyrosine kinases involved in tumor progression and angiogenesis. Cancer Res. 2004;64:7099–7109. doi: 10.1158/0008-5472.CAN-04-1443. [DOI] [PubMed] [Google Scholar]
- 47.Amaravadi R, et al. Updated results of a randomized Phase II study comparing two schedules of temozolomide in combination with sorafenib in patients with advanced melanoma. J. Clin. Oncol. 2007;25(18 Suppl.):8527. [Google Scholar]
- 48.Flaherty KT, et al. Phase I/II trial of BAY 43-9006, carboplatin (C) and paclitaxel (P) demonstrates preliminary antitumor activity in the expansion cohort of patients with metastatic melanoma. J. Clin. Oncol. 2004;22(14 Suppl.):7507. [Google Scholar]
- 49.Flaherty KT. Chemotherapy and targeted therapy combinations in advanced melanoma. Clin. Cancer Res. 2006;12:2366s–2370s. doi: 10.1158/1078-0432.CCR-05-2505. [DOI] [PubMed] [Google Scholar]
- 50.Hauschild A, et al. Results of a Phase III, randomized, placebo-controlled study of sorafenib in combination with carboplatin and paclitaxel as second-line treatment in patients with unresectable stage III or stage IV melanoma. J. Clin. Oncol. 2009;27:2823–2830. doi: 10.1200/JCO.2007.15.7636. [DOI] [PubMed] [Google Scholar]
- 51.Flaherty KT, et al. Phase I/II, pharmacokinetic and pharmacodynamic trial of BAY 43-9006 alone in patients with metastatic melanoma. J. Clin. Oncol. 2005;23(16 Suppl.):3037. [Google Scholar]
- 52.Eisen T, et al. Sorafenib in advanced melanoma: a Phase II randomised discontinuation trial analysis. Br. J. Cancer. 2006;95:581–586. doi: 10.1038/sj.bjc.6603291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gray-Schopfer V, et al. Melanoma biology and new targeted therapy. Nature. 2007;445:851–857. doi: 10.1038/nature05661. [DOI] [PubMed] [Google Scholar]
- 54.Adnane L, et al. Sorafenib (BAY 43-9006, Nexavar), a dual-action inhibitor that targets RAF/MEK/ERK pathway in tumor cells and tyrosine kinases VEGFR/PDGFR in tumor vasculature. Methods Enzymol. 2006;407:597–612. doi: 10.1016/S0076-6879(05)07047-3. [DOI] [PubMed] [Google Scholar]
- 55.Whittaker S, et al. Gatekeeper mutations mediate resistance to BRAF-targeted therapies. Sci. Transl. Med. 2010;2 doi: 10.1126/scitranslmed.3000758. 35ra41. [DOI] [PubMed] [Google Scholar]
- 56.Tsai J, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc. Natl. Acad. Sci. U.S.A. 2008;105:3041–3046. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Poulikakos PI, et al. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–30. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bollag G, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Flaherty K, et al. Phase I study of PLX4032: proof of concept for V600E BRAF mutation as a therapeutic target in human cancer. J. Clin. Oncol. 9000;27(15 Suppl.) [Google Scholar]
- 60.Flaherty KT, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl. J. Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chapman PB, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl. J. Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heidorn SJ, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140:209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nazarian R, et al. Melanomas acquire resistance to B-RAFV600E inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johannessen CM, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wagle N, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J. Clin. Oncol. 2011;29:3085–3096. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Villanueva J, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smalley KS, Flaherty KT. Integrating BRAF/MEK inhibitors into combination therapy for melanoma. Br. J. Cancer. 2009;100:431–35. doi: 10.1038/sj.bjc.6604891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kefford RA, et al. Phase I/II study of GSK2118436, a selective inhibitor of oncogenic mutant BRAF kinase, in patients with metastatic melanoma and other solid tumors. J. Clin. Oncol. 2010;28(15 Suppl.):8503. [Google Scholar]
- 69.Garnett MJ, et al. Wild-type and mutant B-RAF activate C-RAF through distinct mechanisms involving heterodimerization. Mol. Cell. 2005;20:963–969. doi: 10.1016/j.molcel.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 70.Gray-Schopfer VC, et al. The role of B-RAF in melanoma. Cancer Metastasis Rev. 2005;24:165–183. doi: 10.1007/s10555-005-5865-1. [DOI] [PubMed] [Google Scholar]
- 71.Dumaz N, et al. In melanoma, RAS mutations are accompanied by switching signaling from BRAF to CRAF and disrupted cyclic AMP signaling. Cancer Res. 2006;66:9483–9491. doi: 10.1158/0008-5472.CAN-05-4227. [DOI] [PubMed] [Google Scholar]
- 72.Smalley KS, et al. CRAF inhibition induces apoptosis in melanoma cells with non-V600E BRAF mutations. Oncogene. 2009;28:85–94. doi: 10.1038/onc.2008.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Solit DB, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439:358–362. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Collisson EA, et al. Treatment of metastatic melanoma with an orally available inhibitor of the Ras-Raf-MAPK cascade. Cancer Res. 2003;63:5669–5673. [PubMed] [Google Scholar]
- 75.Haura EB, et al. A phase II study of PD-0325901, an oral MEK inhibitor, in previously treated patients with advanced non-small cell lung cancer. Clin. Cancer Res. 2010;16:2450–2457. doi: 10.1158/1078-0432.CCR-09-1920. [DOI] [PubMed] [Google Scholar]
- 76.Adjei AA, et al. Phase I pharmacokinetic and pharmacodynamic study of the oral, small-molecule mitogen-activated protein kinase kinase 1/2 inhibitor AZD6244 (ARRY-142886) in patients with advanced cancers. J. Clin. Oncol. 2008;26:2139–2146. doi: 10.1200/JCO.2007.14.4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Haass NK, et al. The mitogen-activated protein/extracellular signal-regulated kinase kinase inhibitor AZD6244 (ARRY-142886) induces growth arrest in melanoma cells and tumor regression when combined with docetaxel. Clin. Cancer Res. 2008;14:230–239. doi: 10.1158/1078-0432.CCR-07-1440. [DOI] [PubMed] [Google Scholar]
- 78.Gilmartin AG, et al. GSK1120212 (JTP-74057) is an inhibitor of MEK activity and activation with favorable pharmacokinetic properties for sustained in vivo pathway inhibition. Clin. Cancer Res. 2011;17:989–1000. doi: 10.1158/1078-0432.CCR-10-2200. [DOI] [PubMed] [Google Scholar]
- 79.Infante JR, et al. Safety and efficacy results from the first-inhuman study of the oral MEK 1/2 inhibitor GSK1120212. J. Clin. Oncol. 2010;28(15 Suppl.):2503. [Google Scholar]
- 80.Koo HM, et al. Apoptosis and melanogenesis in human melanoma cells induced by anthrax lethal factor inactivation of mitogen-activated protein kinase kinase. Proc. Natl. Acad. Sci. U.S.A. 2002;99:3052–3057. doi: 10.1073/pnas.052707699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meier F, et al. Combined targeting of MAPK and AKT signalling pathways is a promising strategy for melanoma treatment. Br. J. Dermatol. 2007;156:1204–1213. doi: 10.1111/j.1365-2133.2007.07821.x. [DOI] [PubMed] [Google Scholar]
- 82.Smalley KS, et al. Multiple signaling pathways must be targeted to overcome drug resistance in cell lines derived from melanoma metastases. Mol. Cancer Ther. 2006;5:1136–1144. doi: 10.1158/1535-7163.MCT-06-0084. [DOI] [PubMed] [Google Scholar]
- 83.McDermott U, et al. Genomics and the continuum of cancer care. N Engl. J. Med. 2011;364:340–350. doi: 10.1056/NEJMra0907178. [DOI] [PubMed] [Google Scholar]
- 84.Rix U, et al. Chemical proteomic profiles of the BCR-ABL inhibitors imatinib, nilotinib, and dasatinib reveal novel kinase and nonkinase targets. Blood. 2007;110:4055–4063. doi: 10.1182/blood-2007-07-102061. [DOI] [PubMed] [Google Scholar]
- 85.dos Santos SC, Sa-Correia I. Genome-wide identification of genes required for yeast growth under imatinib stress: vacuolar H+-ATPase function is an important target of this anticancer drug. Omics. 2009;13:185–198. doi: 10.1089/omi.2008.0086. [DOI] [PubMed] [Google Scholar]
- 86.Capdeville R, et al. Glivec (STI571, imatinib), a rationally developed, targeted anticancer drug. Nat. Rev. Drug Discov. 2002;1:493–502. doi: 10.1038/nrd839. [DOI] [PubMed] [Google Scholar]
- 87.Atkins M, et al. Sunitinib maleate. Nat. Rev. Drug Discov. 2006;5:279–280. doi: 10.1038/nrd2012. [DOI] [PubMed] [Google Scholar]
- 88.Jarkowski A, Sweeney RP. Nilotinib: a new tyrosine kinase inhibitor for the treatment of chronic myelogenous leukemia. Pharmacotherapy. 2008;28:1374–1382. doi: 10.1592/phco.28.11.1374. [DOI] [PubMed] [Google Scholar]
- 89.Weisberg E, et al. Characterization of AMN107, a selective inhibitor of native and mutant Bcr-Abl. Cancer Cell. 2005;7:129–141. doi: 10.1016/j.ccr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 90.Wei G, et al. First-line treatment for chronic myeloid leukemia: dasatinib, nilotinib, or imatinib. J. Hematol. Oncol. 2010;3:47. doi: 10.1186/1756-8722-3-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Braun T, Fenaux P. Farnesyltransferase inhibitors and their potential role in therapy for myelodysplastic syndromes and acute myeloid leukaemia. Br. J. Haematol. 2008;141:576–586. doi: 10.1111/j.1365-2141.2008.07099.x. [DOI] [PubMed] [Google Scholar]
- 92.Charette N, et al. Salirasib inhibits the growth of hepatocarcinoma cell lines in vitro and tumor growth in vivo through ras and mTOR inhibition. Mol. Cancer. 2010;9:256. doi: 10.1186/1476-4598-9-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Board RE, et al. Detection of BRAF mutations in the tumour and serum of patients enrolled in the AZD6244 (ARRY-142886) advanced melanoma phase II study. Br. J. Cancer. 2009;101:1724–1730. doi: 10.1038/sj.bjc.6605371. [DOI] [PMC free article] [PubMed] [Google Scholar]