Abstract
There can be substantial negative consequences for insects colonizing a resource in the presence of competitors. We hypothesized that bacteria, associated with an oviposition resource and the insect eggs deposited on that resource, serve as a mechanism regulating subsequent insect attraction, colonization, and potentially succession of insect species. We isolated and identified bacterial species associated with insects associated with vertebrate carrion and used these bacteria to measure their influence on the oviposition preference of adult black soldier flies which utilizes animal carcasses and is an important species in waste management and forensics. We also ascertained that utilizing a mixture of bacteria, rather than a single species, differentially influenced behavioral responses of the flies, as did bacterial concentration and the species of fly from which the bacteria originated. These studies provide insight into interkingdom interactions commonly occurring during decomposition, but not commonly studied.
Interactions between microbes and multicellular organisms are often challenging to characterize. No other place is this more apparent than in systems where there is competition for ephemeral resources. Janzen1 proposed that single-celled organisms on decomposing materials, such as seed, fruits and even carrion, function as more than simple nutrient recyclers. They are in fact members of the complex community competing for these resources, and through evolutionary time, have developed strategies for reducing competition with prokaryote and eukaryote consumers. However, it took 30 years before Janzen's concept was validated when Burkepile et al.2 reported that fish carrion contaminated with fewer bacteria were attractive to scavengers for a much longer period of time, and to a wider array of scavengers, than those with uninhibited bacterial fauna. These results indicated bacterial activity reduced competition with scavengers for the resource2. However, this effect did not apply to all competitors as some scavengers were actually more successful on the bacteria laden resource2.
Microbes have long been recognized for their functional importance in driving colonization of a resource by arthropods. Holdaway3 and Seddon4 proposed that ammonia produced by bacterial putrefaction on sheep stimulated oviposition by blow flies (Diptera: Calliphoridae). In comparison, gravid mosquitoes, Aedes aegypti (L.) (Diptera: Culicidae), must locate water sources that exhibit the appropriate environmental conditions for the development of their offspring5. A primary factor regulating attraction and colonization of these sites by female mosquitos is the associated microbial flora6, where it is the specific combination of 14 bacterial species responsible for the attraction5. Gravid house flies, Musca domestica L. (Diptera: Muscidae) evaluate volatiles produced by microbes on conspecific eggs to ensure synchronous larval development which allows for aggregative feeding and reduced likelihood of cannibalism7. Bacteria associated with these eggs also provided initial food resources8 and protection from pathogenic fungi on carrion9.
Recent advances in technology have expanded the tools available for the study of microbial ecology. Limitations existed in the ability to recover most community bacteria via conventional culture-based methods, thus providing gross under-estimates of microbial diversity in nature. Some anaerobic or microoxic bacteria require specific nutrients, or interactions with other organisms to grow and reproduce; factors that have made it problematic to replicate appropriate culture conditions in the laboratory. More recently, non-culture based techniques, such as molecular identification of bacteria via 16s rDNA, are being used to describe a more comprehensive bacterial community structure in different habitats and natural settings10. Consequently, more studies are using metagenomic methods to investigate microbial interactions with higher trophic levels11,12.
These tools can be directly applied to sustainable waste management and forensics. An organism that bridges these disciplines is the black soldier fly, Hermetia illucens L. (Diptera: Stratiomyidae). Adult black soldier flies lay eggs in a host of decomposing materials ranging from animal wastes13 to carrion14,15. Colonization of animal waste by black soldier flies often results in the exclusion of competing species, such as the house fly16. Past researchers speculated that this exclusion was due to the reduction in Escherichia coli, one of the primary food substrates of house flies located in the waste, by black soldier fly larvae17. The elimination or reduction of E. coli and other bacteria also is important to food safety to decrease transmission of pathogens to animals. In terms of forensics, this fly species frequently colonizes human remains and can be used to estimate a minimum postmortem interval18,19,20.
The black soldier fly is not the only species in these environments and often compete with a number of other insect species. The lesser mealworm, Alphitobius diaperinus (Panzer) (Coleoptera: Tenebrionidae) commonly occurs with the black soldier fly on decomposing matter in poultry operations. For decomposing carcasses, other insects such as the secondary screwworm, Cochliomyia macellaria (Fabr.) (Diptera: Calliphoridae) and the hairy maggot fly, Chrysomya rufifacies (Macquart) (Diptera: Calliphoridae), also are known to occur on the same resources as the black soldier fly. The occurrence of multiple species at the same ephemeral resource creates an environment for competition for resources, where direct and indirect interactions between insect species and microbes likely influence colonization patterns and local community structure; however there is little known about these interkingdom interactions.
Using bacteria isolated from black soldier fly food and identified through metagenomic 16S rDNA analyses, we ask if these bacteria attracted gravid females? We also determined if bacteria associated with black soldier fly eggs attract conspecific adult females. If there was increased attraction, we attempted to determine if it was associated with a single bacteria species or a more complex community. We hypothesized that bacteria found at a site used by the black soldier fly for egg deposition could influence insect attraction, colonization, and potentially succession. We demonstrated that bacteria from various life stages and species of insects significantly influenced oviposition preference by gravid black soldier fly females. Ultimately, such information could lead to the development of approaches to disrupt and manipulate the microbiota to produce communities engineered to repel pest insects, pathogen vectors, or attract beneficial insects.
Results
Conspecific eggs and substrate preference
Ephemeral resources played a role in attracting gravid black soldier fly females to oviposition sites (Table 2). The most significant (t = 3.36, df = 4, P = 0.028) increase in egg deposition in the presence of conspecific eggs occurred when no media was present. There was also an increase in oviposition in the presence of conspecific eggs when sterile substrate was present (t = 3.82, df = 4, P = 0.019). The number of eggs deposited between the different substrate treatments was similar (F = 1.45, df = 2, 6, P = 0.307) when conspecific eggs were already present. However, in the absence of conspecific eggs, there was less oviposition (F = 5.98, df = 2, 6, P = 0.037), primarily between the no substrate and non-sterile substrate treatments (Tukey's q = 4.66, P < 0.05).
Table 1. Sanger sequencing results of 16S rDNA of isolated bacterial strains. Lists insect species from which initial bacterial isolate was obtained (source) and GenBank (http://www.ncbi.nlm.nih.gov/genbank/) accession numbers of newly submitted sequences.
| Source | Accession Number | Length* | RDP ID† | Bootstrap support (%) ‡ |
|---|---|---|---|---|
| Alphitobius diaperinus | JQ979481 | 733 | Acinetobacter sp. | 100 |
| Chrysomya rufifacies | JQ979484 | 737 | Klebsiella sp. | 100 |
| JQ979485 | 725 | Morganella sp. | 100 | |
| JQ979486 | 732 | Proteus sp. | 100 | |
| JQ979480 | 677 | Providencia sp. | 100 | |
| Cochliomyia macellaria | JQ979483 | 723 | Hafnia sp. | 100 |
| JQ979477 | 600 | Ignatzschineria sp. | 100 | |
| JQ979478 | 656 | Ignatzschineria sp. | 100 | |
| JQ979479 | 662 | Ignatzschineria sp. | 100 | |
| Hermetia illucens | JQ979475 | 688 | Bacillus sp. | 100 |
| JQ979469 | 682 | Cellulomonas sp | 100 | |
| JQ979474 | 615 | Empedobacter sp. | 71 | |
| JQ979482 | 663 | Enterobacter sp. | 97 | |
| JQ979470 | 718 | Gordonia sp. | 100 | |
| JQ979476 | 691 | Kurthia sp. | 99 | |
| JQ979471 | 666 | Microbacterium sp. | 100 | |
| JQ979472 | 664 | Micrococcus sp. | 100 | |
| JQ979473 | 666 | Micrococcus sp. | 100 |
*Number of base pairs of the consensus sequence represented by a minimum of 2× sequence coverage.
†Taxonomic identification made for that sequence by the Ribosomal Database Project naive Bayesian classifier.
‡Bootstrap percentage support for that identification.
Table 2. Percent ± SE (n = 9 experiments) black soldier fly, Hermetia illucens, egg deposition in oviposition sites with and without conspecific eggs nested within different substrate treatments.
| Substrate Treatments | |||
|---|---|---|---|
| No Substrate | Non-sterile Substrate | Sterile Substrate† | |
| Eggs | 79.3 ± 11.5a,c | 52.2 ± 10.7a,c | 60.1 ± 8.3a,c |
| No Eggs | 9.5 ± 6.0b,c | 36.6 ± 9.6a,d | 28.9 ± 5.1b,d |
†Diet sterilized by autoclaving.
a–bSample groups (Eggs and No Eggs) with the same letter (a or b) are not significantly different (P < 0.05) as compared within Substrate Treatment.
c–dSubstrate treatments (No substrate, non-sterile substrate and Sterile substrate) with the same letter (c or d) are not significantly different (P < 0.05) as compared within sample group (Eggs or No Eggs).
Sterility preference
There was a difference in egg deposition among sterility preference treatments (F = 28.39, df = 4, 12, P < 0.0001). Sterilization to remove microbes from eggs (T1) reduced oviposition preference by gravid females compared to the non-sterile (T2) (Tukey's q = 6.47, P < 0.05) and H2O rinsed (C1) egg (Tukey's q = 5.20, P < 0.05) treatments (Table 3). No eggs (C2) had the lowest level of attraction accounting for 7.4% of the eggs deposited. In contrast H2O rinsed (C1) (Tukey's q = 10.38, P < 0.05) or the Non-Sterile egg (T1) (Tukey's q = 11.65, P < 0.05) treatments had the highest levels of attraction, 34.8 and 39.2 respectively.
Table 3. Percent ± SE* black soldier fly, Hermetia illucens, egg deposition in oviposition sites inoculated with differing egg treatments using conspecific eggs.
| Sterile† | Non-Sterile | H2O Rinse‡ | No Eggs | |
|---|---|---|---|---|
| (T1) | (T2) | (C1) | (C2) | |
| Eggs Deposited1 | 18.6 ± 1.6a | 39.2 ± 2.4b | 34.8 ± 2.8b | 7.4 ± 2.9c |
1Treatments with the same letter are not significantly different (P < 0.05: Tukey's Post-hoc Pairwise comparisons).
*Twelve replicate experiments; †Eggs sterilized with Sporgon®; ‡Eggs rinsed once with sterile water.
Bacterial species preference
Black soldier fly preference for bacterial isolates was tested by oviposition responses (Table 4). All bacterial isolates tested were identified by Sanger sequencing using 16S rDNA sequence. Alignment of two universal bacterial primer sets allowed for the creation of a minimum 2× coverage consensus sequence of approximately 600 to 750 bp. Summary of the accession numbers, length of the sequences, and Ribosomal Database Project Naïve Baysian classifier identification can be found in Table 1. Of the isolates tested, Ignatzschineria sp. 2 and 3 from C. macellaria eggs, Providencia sp. from Ch. rufifacies 3rd instar and Acintobacter sp. from Alphitobius diaperinus stimulated a significant response from the black soldier fly.
Table 4. Percent ± SE* oviposition response of black soldier flies, Hermetia illucens, to concentration curve of identified bacteria isolated from various fly sources and life stages and the lesser mealworm, Alphitobius diaperinus.
| Concentrations (cfu/ml) | Statistics | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Source | Stage | Bacteria | Gram | 0 | 104 | 106 | 108 | ANOVA | Kruskal-Wallis |
| Percent Oviposition ± SE | |||||||||
| CM | egg | Hafnia sp. | − | 22.0 ± 5.5 | 31.9 ± 5.2 | 25.6 ± 4.7 | 20.4 ± 2.2 | P = 0.361 | P = 0.320 |
| Ignatzschineria sp. 1 | − | 17.4 ± 2.6 | 29.3 ± 2.1 | 29.9 ± 3.2 | 23.5 ± 6.7 | P = 0.180 | P = 0.223 | ||
| Ignatzschineria sp. 2 | − | 42.9 ± 0.9a | 24.0 ± 0.9b | 13.8 ± 1.7c | 19.3 ± 1.9b,c | P = 0.001 | P = 0.02 | ||
| Ignatzschineria sp. 3 | − | 49.5 ± 5.8a | 22.4 ± 5.1b | 12.3 ± 3.0b | 15.7 ± 1.0b | P = 0.0009 | P = 0.044 | ||
| CR | 3rd | Klebsiella sp. | − | 22.6 ± 6.4 | 28.1 ± 3.4 | 25.1 ± 3.3 | 24.2 ± 3.4 | P = 0.841 | P = 0.764 |
| instar | Morganella sp. | − | 21.9 ± 5.4 | 28.7 ± 3.8 | 29.7 ± 4.4 | 19.6 ± 5.4 | P = 0.491 | P = 0.618 | |
| Proteus sp. | − | 25.0 ± 2.8 | 20.8 ± 3.4 | 30.6 ± 2.0 | 23.6 ± 1.0 | P = 0.103 | P = 0.218 | ||
| Providencia sp. | − | 17.2 ± 0.6a | 19.1 ± 3.7a | 35.6 ± 3.7b | 28.1 ± 2.0a,b | P = 0.006 | P = 0.044 | ||
| Staphylococcus sp. | − | 23.6 ± 8.6 | 18.5 ± 7.4 | 22.8 ± 3.0 | 35.1 ± 13.1 | P = 0.607 | P = 0.715 | ||
| HI | egg | Bacillus sp. | + | 48.7 ± 7.9 | 10.3 ± 4.1 | 18.8 ± 3.5 | 22.3 ± 7.6 | P = 0.011 | P = 0.063 |
| Empedobacter sp. | − | 19.3 ± 2.8 | 36.1 ± 6.3 | 23.6 ± 3.1 | 20.9 ± 2.9 | P = 0.068 | P = 0.113 | ||
| Enterobacter sp. | − | 19.2 ± 0.7 | 25.0 ± 4.0 | 25.5 ± 1.0 | 30.3 ± 5.4 | P = 0.065 | P = 0.123 | ||
| Kurthia sp. | + | 24.3 ± 4.3 | 25.6 ± 8.0 | 19.3 ± 1.1 | 30.1 ± 3.7 | P = 0.489 | P = 0.392 | ||
| Mixture | 21.2 ± 9.4a | 26.6 ± 8.2a,c | 23.0 ± 6.5b,c | 29.2 ± 7.7a,c | P = 0.0004 | P = 0.02 | |||
| Constituents: | |||||||||
| Cellulomonas sp. | + | 12.0 ± 1.0 | 30.0 ± 19.5 | 16.1 ± 0.5 | 41.9 ± 20.0 | P = 0.495 | P = 0.321 | ||
| Gordonia sp. | + | 17.6 ± 4.6a | 31.3 ± 3.3b | 22.9 ± 1.1a,b | 28.3 ± 1.3a,b | P = 0.0004 | P = 0.02 | ||
| Microbacterium sp. | + | 17.9 ± 4.0 | 35.1 ± 8.3 | 25.0 ± 1.8 | 22.0 ± 3.3 | P = 0.172 | P = 0.340 | ||
| Micrococcus sp. | + | 19.1 ± 5.0 | 29.4 ± 1.8 | 24.8 ± 3.1 | 26.7 ± 2.2 | P = 0.396 | P = 0.418 | ||
| AD | adult | Acinetobacter sp. | − | 36.2 ± 0.7a | 23.2 ± 4.0a | 21.3 ± 1.0a | 19.3 ± 5.4b | P = 0.0009 | P = 0.044 |
*Twelve replicate experiments. AD = Alphitobius diaperinus (Lesser Mealworm); HI = Hermetia illucens (Black soldier fly); CM = Cochliomyia macellaria (secondary screwworm); CR = Chrysomya rufifacies (hairy maggot blow fly).
a–cSample groups with the same letter are not significantly different (P < 0.05) as compared across concentrations.
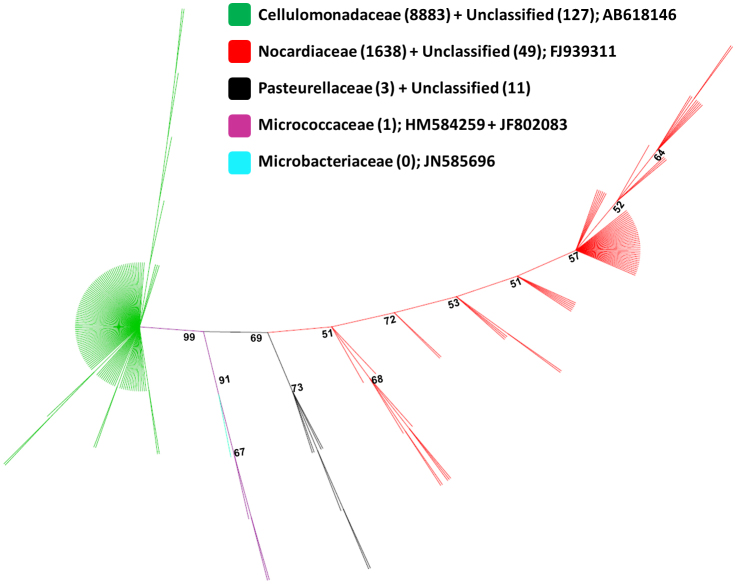
In one instance, a mixture of gram positive bacteria was obtained from black soldier fly eggs (Mixture), which preferentially grew collectively and proved challenging to separate into individual isolates by culture methods. When this mix was tested for olfactory response as a collective it produced a significant oviposition response. Therefore, pyrosequencing was performed to determine its constituents. Out of total 10712 sequences, ranging in length from 250 bp to 518 bp (average length = 387 bp), 10702 (99.90%) were classified into order Actinomycetales (99.88%) and Pasteurelles (0.03%). Similarly, 10525 (98.25%) sequences were classified into 4 families Cellulomonadaceae (82.93%), Nocardiaceae (15.29%), Pasteurelleceae (0.03%), and Micrococcaceae (0.01%). At the genus level only 1623 (15.15%) of the sequence could be classified with ≥80% bootstrap support (default setting); Gordonia (99.51%), Cellulomonas (0.04%), Gallibacterium (0.03%) and Micrococcus (0.01%). Reducing the bootstrap cutoff to 50%, resulted in Cellulomonas identified as the most abundant genera (63.29%) followed by Gordonia (36%). In the Neighbor-joining (NJ) tree, all families (except Microbacteriaceae) were well supported monophyletic group (Fig. 1). Sequences that were unclassified at the family level using RDP classifier were clustered with the classified families with strong bootstrap supports (Fig. 1). Ultimately, four bacterial genera were isolated from the mix by subculturing using phenotypic characteristics and identified using capillary sequencing of the 16s rDNA as Microbacterium, Cellulomonas, Gordonia and two phenotypically different Micrococcus. These identifications complemented pyrosequencing results for the Mixture; therefore these isolates were tested individually for olfactory response. Only the Gordonia isolate, from the mixture, produced a significant response by the black soldier fly.
Figure 1. Unrooted neighbor-joining tree of 16S rRNA gene sequences associated with the bacterial mixture isolated from black soldier fly eggs.
Values in the parentheses indicate total number of sequences obtained from pyrosequencing and assigned to a particular family or unknown group. GenBank accession numbers after the semicolon indicate those sequences that were downloaded from GenBank based on the best blast match with Sanger sequences. Sequences from both methods were used for construction of NJ tree. Numbers on the node indicate bootstrap values (bootstrap values for some terminal nodes are not shown). Branches are colored to indicate sequence assignment at family level.
Discussion
Ephemeral resources, such as carrion and plant material, represent valuable nutrients for a variety of species. The occurrence of carrion in an ecosystem is unpredictable and tends to degrade relatively fast. Further, competition between vertebrate scavengers and decomposers such as arthropods and microbes is intense21. Some arthropod species, such as the black soldier fly, have evolved to detect and locate these resources at the time of, or soon after, their demise14. Detection is predominately due to olfactory cues produced by these resources22,23,24 or conspecific offspring25.
We cultured aerobic bacteria from four insect species and isolated and characterized bacteria from 15 genera to test in this study. One of the bacteria was identified as Empedobacter sp. [JQ979474]; however Kämpfer, et al.26 suggest that the identification of accession #EU276091.1 may be Wautersiella falsenii. The Ribosomal Database Project naïve Baysian classifier identification of this isolate resulted in only a 71% bootstrap support, suggesting that identification to the genus level is tenuous. Three Ignatzschineria sp. were isolated from C. macellaria and sequence analysis determined only minor mismatches or gaps between the three sequences. Ignatzschineria sp. 1 [JQ979477] and 2 [JQ979478] aligned from the 4th to the 661st position with a single mismatch at the 53rd position. Ignatzschineria sp. 3 [JQ979479] align beginning at the 61st position, with a gap at the 169th and 171st position. This suggests that these isolates are all closely related. Similarity was also very high between the two Micrococcus isolates obtained from black soldier flies, Y [JQ979472] and W [JQ979473], with a single mismatch at the 626th position.
Our study found that bacteria isolated from conspecifics on decomposing materials attract gravid black soldier flies, presumably by emission of volatiles. The same has been determined for other species including blow flies27,28. Bovine blood inoculated with bacteria isolated from wounds infested with Cochliomyia hominivorax (Coquerel) (Diptera: Calliphoridae) released volatiles attracting intraspecific adults24. House fly eggs are coated with microbes that release volatiles7. Concentrations thresholds of these volatiles dictate attraction and repulsion of conspecifics for oviposition behavior7. Colonization attempts when volatiles were above a threshold resulted in reduced survivorship of deposited eggs, while the opposite was determined for cohorts deposited with eggs emitting volatile concentrations below threshold levels7.
Oviposition responses of black soldier flies to bacteria isolated from competing arthropods were mixed. Significantly more eggs were laid in sites without the bacteria when black soldier flies were given a choice between sites with and without specific bacteria from C. macellaria (Ignatzschineria sp.) and A. diaperinus (Acinetobacter sp.), indicating repellency. This response by the flies is not unexpected as the species compete for the same resources. However in one instance, the black soldier flies responded to a Providencia sp. isolated from C. rufifacies. This unexpected response to bacteria from another species could be due to C. rufifacies being a newly introduced species to North America and consequently H. illucens had limited prior exposure to it. Secondly, C. rufifacies utilizes disparate resources, as they are predators on other blow fly species; therefore there could be less selection to avoid resources with C. rufifacies due to lack of competition.
A common approach when examining microbe-arthropod interactions is to isolate a single bacteria species and determine its impact on the arthropod of interest24. Such an approach is known to be limiting in terms of deciphering the true biological relevance of bacterial interactions with arthropod, as the behavior of the bacteria in isolation can be quite different than in the community mixtures typically encountered in the environment. We investigated the response of black soldier flies to a bacterial mixture isolated from conspecific eggs, as well as to several of its constituent species. We determined that the level of response was higher to the mixture than to any individual species. At a 50% bootstrap cutoff, a large component of the mixture were identified as genus Cellulomonas; a soil inhabiting Actinobacteria which have the ability to hydrolyze cellulose. The degradation of this major carbohydrate synthesized by plants represents an important part of the carbon cycle and this bacterium participates in the reduction of this biomass within the biosphere29,30. However, it was one of the lesser constituent species, Gordonia sp., which induced a significantly higher level of oviposition indicating its importance to the mixture and possibly to the ecology of the fly. Many Gordoniae can degrade xenobiotics, environmental pollutants, and other natural polymers; however, some are opportunistic pathogens31.
It is not surprising to have these bacteria present in such high numbers on black soldier fly eggs as these flies are known to colonize and develop efficiently on decomposing plant material13,32. On animal tissue, their development is greatly retarded with larvae needing an extra two weeks to complete development33. Such a response could be due to a lack of the carriage of appropriate bacteria to facilitate degradation of the different food source thus reducing required nutrient absorption. A potential solution could be taking a probiotic approach when using black soldier flies to reduce wastes other than plant materials. Inoculating a resource with the necessary bacteria prior to introducing black soldier flies could enhance their ability to recycle associated nutrients. Such an approach has been demonstrated in the past by inoculating poultry manure with Bacillus subtilis strains isolated from black soldier fly larvae, which enhanced larval weight by 30% and reduced development time up to 10%34.
Community level approaches to understanding microbial-insect interactions are integral for deciphering the “natural” mechanisms of ecosystem function. Researchers are now beginning to examine the interactions between bacterial communities, arthropods5,35,36 and hosts37. This should provide insight into these ecological interactions as it allows the bacteria to respond in a natural setting and results in a more accurate reflection of responses by arthropods. For example, oviposition response of the mosquitoes, A. aegypti and Aedes albopictus (Skuse) (Diptera: Culicidae) is influenced by the bacterial diversity and abundance associated decomposing leaf litter in artificial oviposition sites5. However, even in that study, a true appreciation of the bacterial diversity cannot be made due to limitations in techniques (i.e., polymerase chain reaction-denaturing gradient gel electrophoresis (DGGE)) utilized5. Unlike the Ponnusamy et al.5 study, we were able to use 454 sequencing to gain a greater appreciation of the bacterial diversity present in the microbial mixture isolated from black soldier fly eggs.
We were not able to conduct an in depth study of bacteria diversity associated with black soldier fly eggs due to financial limitations associated with 454 sequencing. However, with the trend towards reduced costs associated with high-throughput sequencing, collaborations between organismal and molecular ecologists are much more practical. This should allow future studies of insect-microbe interactions to explore a more full community level approach with microbes. In fact, future studies conducted with single microbe species, while valuable, should be explained within the context of the bacterial community as related to behavioral ecology.
Methods
Source of insects
Black soldier flies used in this study were obtained from a colony housed in a cage (1.8 m3 and 1.5 mm mesh screen) maintained year round in a greenhouse, outside the Forensic Laboratory for Investigative Entomological Sciences (FLIES) Facility located at Texas A&M University in College Station, TX, USA. The colony was established in the spring of 2009 from the eggs of a laboratory colony initiated at the Coastal Plain Experiment Station, University of Georgia, Tifton, GA, USA, which originated from material collected at a poultry facility in Bacon Co., GA, USA in 1998. Lesser mealworm colonies maintained at the Southern Plains Agricultural Research Center (SPARC, College station, TX, USA) were started from specimens isolated from a poultry farm located in Wake County, NC, USA. The SPARC colony has been in production since 200438. Secondary screwworm and hairy maggot fly colonies were initiated from material collected from carrion located in the College Station, TX, USA vicinity during the summers of 2008 and 2009.
Maintenance of the colony
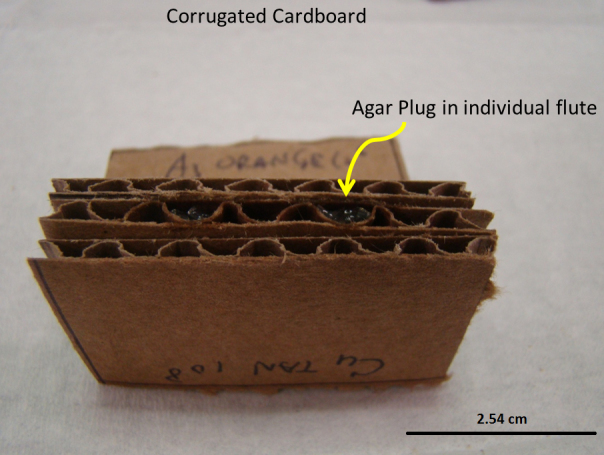
Black soldier flies were reared according to the methods of Sheppard et al.39. Eggs were collected in a three layer, 3 × 5 cm corrugated cardboard block, held together with Elmer's® white glue with 3 × 4 mm flutes used as an oviposition substrate. Blocks were taped to the sides of a 22 × 22 × 10 cm square pan 5 cm above the oviposition substrate (moist-to-wet Gainesville diet)40 with the flutes perpendicular to the substrate. Collected eggs were labeled according to their date of oviposition to keep track of cohorts of the same generation. Briefly, eggs were held in 10 (L) × 10 (W) × 8 (D) cm plastic tubs at 27°C with ambient humidity until eclosion. The neonatal larvae were given approximately 200 g of Gainesville diet (5:3:2 hand mixture of wheat bran, alfalfa and corn meal, respectively, Producers Cooperative Association, Bryan, TX, USA)13. After 48 h, a fresh diet was added and the larvae were transferred into 40 (L) × 15 (W) × 10 (D) cm plastic pans as needed. The top of the pan was covered once per day with 2 cm of fresh diet (or as needed). Larvae were divided into new 50 (L) × 35 (W) × 12 (D) cm pans after approximately two weeks to maintain a density of approximately 2500 larvae per pan. When dispersing larvae accounted for 50% of the population in each pan, feeding was stopped and the remaining moist day-old food was allowed to dry to serve as a pupation substrate. Each pan of pupae was covered with white polyester organza fabric. Emerged adults were released into 1.5 m3 experiment cages held in the green house under natural light.
Lesser mealworm colonies were maintained using methods described in Crippen et al.38. Insects were housed in 15 (L) × 15 (W) × 30 (D) cm cages at 30°C on a 8:16 L:D cycle. Colonies were provided 1000 ml wheat bran (Morrison Milling Co., Denton, TX, USA) and 30 ml of fishmeal (Omega Protein, Inc., Hammond, LA, USA) as food. Deionized water was provided via a 36 cm3 sponge in each cage along with a 0.5-cm-thick slice of apple which was provided twice per week. Secondary screwworm and hairy maggot flies were maintained in separate cages using methods by Boatright and Tomberlin41. Adult flies of each species were housed in 30 cm3 Bioquip cages (Bioquip Products, Rancho Dominguez, CA, USA) in a 136LLVL Percival (Percival Scientific, Perry, IA, USA) growth chamber at ~27°C with 14:10 (L:D). Adult flies were provided a 50:50 sugar:powdered milk mixture ad libitum. Larvae were fed beef liver in 1.1 L styrene mosquito-breeding containers (Bioquip Products, Rancho Dominguez, CA, USA) held in the growth chamber previously described.
Bacterial characterization by culture methods
Samples of approximately 0.2 g eggs, aged 24 hours, or ten 3rd instar larvae were placed into a beaker with 2 ml DEPC dH2O and incubated at room temperature for 5 min, briefly shaken once per minute. The resulting supernatant was spread by 10-fold serial dilution onto Brain Heart Infusion (BHI) plates (Biolink Scientific, Austin, TX). Three replicates of each were incubated for 3 days at 26, 30 and 37°C, respectively. Individual colonies were selected phenotypically and sub-cultured for isolation onto trypic soy agar +5% sheep's blood plates (TSAB, BVA Scientific, San Antonio, TX, USA). The isolated bacteria were characterized by Analytical Profile Index (API) biotyping system (bioMerieux, Hazlewood, MO) and by 16S ribosomal DNA (16S rDNA) sequencing.
DNA extraction, PCR amplification and sequencing
DNA template for Sanger sequencing was prepared by mixing 10 μl of an isolated colony in DEPC H2O and incubating at 100°C for 15 min followed by a brief centrifugation. The DNA for 454-pyrosequencing was prepared by modification of the protocol previously described in Zheng et al.42. Briefly, polymerase chain reaction (PCR) was performed from extracted DNA for the amplification of 16S rDNA using two independent sets of primers (short read primers (300 bp) forward: 5′- ACT TAA CCC AAC ATC TCA CGA, and reverse: 5′- AGG ATT AGA TAC CCT GGT AGT43, and long read primers (750 bp) forward: 5′- ACT CCT ACG GGA GGC AGC AG44, and reverse: 5′- AGG ATT AGA TAC CCT GGT AGT (Integrated DNA Technologies, Coralville, IA, USA) and PCR Master Mix (2×) (Fermentas, Glen Burnie, MD, USA). Since we were working with a wide variety of bacteria species, two different primer sets were used in two independent PCR's to assure good coverage and identification of a diversity of bacterial species. Thermal cycling for the short read primers consisted of an initial denaturation at 95°C for five minutes, then 30 cycles of denaturation at 94°C for one minute, annealing at 55°C for one minute, and extension at 72°C for one minute, and then a final extension at 72°C for 10 minutes. Thermal cycling for the long read primers consisted of an initial denaturation at 95°C for five minutes, then 40 cycles at denaturation at 94°C for 30 seconds, annealing at 50°C for one minute, and extension at 72°C for two minutes, and a final extension at 72°C for 10 minutes. The PCR products were purified using ExoSAP-IT (USB Corp., Cleveland, OH, USA) according to manufacturer's protocol and sequenced in both directions using standard ABI BigDye-terminator Cycle Sequencing (Applied Biosystems Inc., Carlsbad, CA, USA) protocols.
Sequences were edited for quality using 4Peaks (Mekentosj, Amsterdam, Netherlands)45 and aligned using MEGA version 546 and a consensus sequence of a minimum 2× coverage was generated. The consensus sequences were submitted to GenBank, (see Table 1 for accession numbers). Consensus sequences were identified at genus level using Naïve Bayesian rRNA classifier version 2.247 as implemented in Ribosomal Database Project (RDP) (http://rdp.cme.msu.edu/classifier/classifier.jsp) (accessed on October 4 2011) and at the species level using ≥97% sequence similarity cut-off in “blastn” algorithm of GenBank (NCBI, http://www.ncbi.nlm.nih.gov). When multiple species yielded identical scores, the sample was identified only to the genus level. Gordonia PCR products did not produce sequences sufficient to make a strong identification from the two universal bacterial primers, so a specific primer pair48 was used to confirm the identification of accession #JQ979470 (see Table 1).
DNA extraction and pyrosequencing
One aliquot of 10 μl of the mixed bacterial culture was added in 500 μl Tris-EDTA (pH = 8), 50 μl 10% SDS, 3 μl proteinase K (20 mg/ml), 1.5 μl of lysozyme (50 mg/ml) and then incubated for 1 h with shaking (900 rpm) at 56°C in a water bath. After incubation, 100 μl NaCl (5 M) and 80 μl CTAB extraction solution (Cat# C2190, TEKNOVA) were added and samples were thoroughly mixed and incubated at 65°C for 10 minutes. Sequential extraction in a 1× volume was performed using phenol (pH = 8.0), phenol/chloroform/isoamyl alcohol (25:24:1), and chloroform/isoamyl alcohol (24:1) by centrifugation at 6000 × g for six minutes. The DNA was precipitated in 0.7 volume of isopropanol, washed twice in 70% ethanol, dissolved in nuclease free water, and quantified by spectrophotometry. Extracted DNA sent to Research and Testing Laboratory (http://www.researchandtesting.com/) for 16S rDNA 454-pyrosequecning using universal bacterial primer pair 28F (5′- GAGTTTGATCNTGGCTCAG) and 519R (5′- GTNTTACNGCGGCKGCTG) by bacterial tag-encoded FLX-Titanium pyrosequencing (bTEFAP) method11,49 in Genome Sequencer FLX System (Roche, Nutley, NJ, USA). All FLX related procedures were performed following Genome Sequencer FLX System manufacturers instructions (Roche, Nutley, NJ, USA).
Pyrosequencing data analysis
Hierarchical classification of 10712 16S rDNA sequences were carried out according to the Bergey's bacterial taxonomy50 using Naïve Bayesian rRNA classifier version 2.247 as implemented in Ribosomal Database Project (RDP) (http://rdp.cme.msu.edu/classifier/classifier.jsp) (accessed on October 04, 2011). Only sequences having ≥80% bootstrap support were considered classified at a particular hierarchical level.
To reduce computation load during phylogenetic analysis, almost identical reads (sequences with ≥98% similarity) (10505 sequences) were filtered using default parameters in cdhit-45451. Total 212 sequences (207 from pyrosequencing after cdhit-454 and 5 from Sanger sequencing) were aligned based on 16S rRNA secondary structure in Infernal aligner52,53, as implemented in the RDP under tool Aligner (http://rdp.cme.msu.edu/) (accessed on October 12, 2011). Because 16S ribosomal sequences obtained using pyrosequencing did not overlap with those obtained using Sanger sequencing in the multiple sequence alignment, we replaced Sanger sequences with those that were best match in NCBI BLAST search (AB618146, FJ939311, JN585696, HM584259, JF802083), but were longer in length. These sequences were realigned based on 16S rRNA secondary structure in Infernal aligner52,53, as implemented in the RDP under tool Aligner (http://rdp.cme.msu.edu/) (accessed on October 12, 2011). Hypervariable ambiguous regions were manually deleted from the multiple sequence alignment in MEGA546. Evolutionary distances from 377 bp of aligned sequences were calculated by neighbor joining (NJ) method with the Kimura two-parameter correction54 for 1000 bootstrap replications in PAUP* v.4.0b1055. Calculated evolutionary distances were used for construction of unrooted NJ tree in PAUP* v.4.0b1055. All trees were edited using Archaeoptryx version 0.957 beta56.
Experimental design for oviposition preference testing
Pans of pupae and newly emerged adults from the same generation were collected from the colony and released into three 1.5 m3 mesh cages over 5 consecutive days, providing a population of 5 cohorts of flies from the same generation, totaling approximately 2500 adults. On the fifth day, oviposition experiments began, having allotted 3 d for mating and 2 additional days for ovary development as described by Tomberlin and Sheppard57. Clear plastic tubs (22 (L) × 22 (W) × 10 (D) cm) sterilized with 80% ethanol were used as treatment containers. Corrugated cardboard blocks were sterilized under UV light for 30 minutes, treatments were applied (eggs, bacteria or nothing) and the blocks were taped one per side to the treatment containers. Oviposition preference was determined by measuring the mass of eggs oviposited into each flute of 5.0 (L) × 5.0 (W) × 0.2 (D) cm, triple layered, corrugated cardboard blocks. A female will only deposit one egg clutch and then will die57. Therefore, eggs deposit only represents clutches from individual females. Three replicates of each experiment were conducted for each of the three preference tests using approximately 2000 flies per cage with a 50:50 ♀:♂ ratio.
Conspecific eggs and substrate preference
Two substrate diets were tested as long distance oviposition attractant: sterile, moist Gainesville diet and non-sterile, moist Gainesville diet. The non-sterile substrate consisted of 250 g of fresh Gainesville diet moistened with 700 ml dH2O and the sterile substrate consisted of 250 g of fresh Gainesville diet which was autoclaved, then moistened with 700 ml of autoclaved dH2O. The control consisted of an identical container with no diet substrate. The short distance oviposition attractant tested was the presence or absence of black soldier fly eggs which was tested with each of the two diets. Cardboard blocks were seeded with approximately 0.04 g of conspecific 1-d-old black soldier fly eggs divided evenly between two cardboard flutes. Of the four cardboard blocks, two containing eggs and two, taped on opposite sides, containing no eggs.
Oviposition preference site was tested for 60 minutes following the first active oviposition observed (t = 0). Three replicates in each cage with one cohort of flies were run consecutively in one day, rotating the position of each treatment and control within the cages. These replicates were run daily over 3 consecutive days, rotating treatments and controls between cages, totaling 9 replicates (N = 9). Oviposited eggs were dissected out of the cardboard flutes and weighed. The percent oviposition per treatment per substrate was determined gravimetrically.
Sterility preference
Approximately 0.04 g of 1-d-old black soldier fly eggs were used for each of two treatments, Sporgon® sterilized 1-day-old eggs (T1) and non-sterile 1-day-old eggs (T2), and two controls, non-sterile 1-day-old eggs washed with sterile water (C1) and no eggs (C2). The T1 eggs were sterilized by placement into 1 ml of Sporgon® (Decon Laboratories, Inc., King of Prussia, PA) for one minute, with occasional gentle shaking, then removed from the solution and allowed to stand at room temperature for two minutes, followed by a rinse with two 1 ml aliquots of sterile dH2O and air dried for 30 minutes. T2 non-sterilized eggs were not treated with Sporgon® prior to use. C1 control eggs were rinsed twice with 1 ml sterile dH2O and air dried for 30 minutes prior to use. Using sterile forceps, the eggs were then evenly divided between two cardboard flutes of UV sterilized cardboard blocks.
For this experiment, four cardboard blocks (T1, T2, C1 and C2) were taped 3 cm above the Gainesville diet substrate. Oviposition preference was tested for 60 minutes after t = 0. Three replicates were run simultaneously in three cages. During each day, treatments were replaced in the cages at four consecutive times. With each replacement, treatments were rotated within and between cages for a total of twelve replicates per day. Eggs deposited were dissected out of the cardboard flutes and weighed. Percent oviposition per treatment per substrate was determined.
Bacterial preference
An aliquot of 8 μl of 18 bacteria strains and one bacterial mixture, at 104, 106, or 108 colony-forming units (cfu)/ml or a PBS (control) was added to the top of two plugs of BHI agar (approximately 0.5 g) sterilely inserted in two flutes each of a UV sterilized cardboard block (Fig. 2). One bacterial concentration was used per treatment block. Three treatment blocks (one treatment bacteria at each of three different concentrations) and one control block (consisting of sterile agar) were taped, one block per side, on a clear 22 (W) × 22 (L) × 10 (D) cm container, 3 cm above the moist-to-wet Gainesville diet to test oviposition site preference (Fig. 3). These replicates were run over three consecutive days at the same time each day, in four replicate cages (N = 12). In each cage the treatment faced a different direction (N, S, E, or W); which was rotated within each cage, each day. During the experiment the naturally lit green-house was kept at a constant temperature of 26°C. Eggs were removed from the cardboard and weighed as previously described. Percent oviposition per treatment was determined.
Figure 2. Olfactory test block showing 2 × 5 cm triple layered corrugated cardboard block into which a 0.5 g sterile agar plug could be added to the flute.
An aliquot of 8 μl bacteria in PBS at the desired concentration could then be added onto the top of plug.
Figure 3. Olfactory test showing triple layered, corrugated cardboard block, taped one per side in 22 × 22 × 10 cm container, at 3 cm above the diet.

This design was used to present bacteria (in the flutes) to black soldier flies and test oviposition site preference. In this view 108 cfu/ml of the bacterial mix is in the left block and control (agar only) is in the right block.
Statistical analyses
Oviposition differences for conspecific eggs and sterile/non-sterile substrates were analyzed using a student's t-test while the effect of egg sterilization was tested using one-way ANOVA Tukey's post-tests. Both analyses were performed after arc-sine square root transformation of percentages.
Oviposition preference for different bacterial strains was analyzed using both parametric (one-way ANOVA with Tukey's post-tests) and non-parametric (Kruskal-Wallis with Dunn's post-tests) approaches to allow for appropriate statistical interpretation. This was done to balance interpretation and reduce the risk of Type II error. We preferred the more powerful one-way ANOVA where the assumptions of normality and homogeneity of variances were violated even after arc-sine square root transformation of percentages; however, we also used the Kruskal-Wallis rank test that is less powerful but does not have data structure assumptions. Significance was set at P < 0.05.
Author Contributions
L.Z., L.H., T.L.C. and J.K.T. conducted the behavioral research. B.S. and M.L.P. conducted the molecular research and associated analyses. S.D. conducted the sequencing. M.E.B. conducted the statistical analyses on the behavioral data. T.L.C., J.K.T., A.M.T., Z.Y., S.L.V. and T.K.W. provided financial support. All authors assisted with drafting the manuscript for submission.
Acknowledgments
J.K.T. and A.M.T. would like to thank Agrilife research for providing funds to conduct this research. L.Z. and Z.Y. were funded through State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University. Additional funds for T.L.C., M.E.B., B.S., J.K.T. and A.M.T. were provided by the National Institute of Justice, Office of Justice Programs, U.S. Department of Justice through Grant 2010-DN-BX-K243. Sequencing services by S.D. were also funded by 2010-DN-BX-K243. Funds for L.H. and S.L.V. were provided by the National Sciences and Engineering Research Council of Canada, Strategic Project Grant #357002-07 and University of Windsor. T.K.W. is the T Michael O'Connor Endowed Professor at Texas A&M University. Points of view in this document are those of the authors and do not necessarily represent the official position or policies of the U.S. Department of Justice. Mention of trade names, companies, or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement of the products by the U.S. Department of Agriculture.
References
- Janzen D. H. Why fruits rot, seeds mold, and meat spoils. Am. Nat. 111, 691–713 (1977). [Google Scholar]
- Burkepile D. E. et al. Chemically mediated competition between microbes and animals: microbes as consumers in food webs. Ecology 87, 2821–2831 (2006). [DOI] [PubMed] [Google Scholar]
- Holdaway F. G. Field population and natural control of Lucilia sericata. Nature 126, 648–649 (1930). [Google Scholar]
- Seddon H. R. Conditions which predispose sheep to blowfly attack. Agricultural Gazette, New South Wales 42, 581–594 (1931). [Google Scholar]
- Ponnusamy L., Wesson D. M., Arellano C., Schal C. & Apperson C. S. Species composition of bacterial communities influences attraction of mosquitoes to experimental plant infusions. Microbial Ecol. 59, 158–173 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnusamy L. et al. Identification of bacteria and bacteria-associated chemical cues that mediate oviposition site preferences by Aedes aegypti. Proc. Nat. Acad. Sci. 105, 9262–9267 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam K. et al. Proliferating bacterial symbionts on house fly eggs affect oviposition behaviour of adult flies. Ani. Behav. 74, 81–92 (2007). [Google Scholar]
- Lam K., Geisreiter C. & Gries G. Ovipositing female house flies provision offspring larvae with bacterial food. Entomol. Exp. Appl. 133, 292–295 (2009). [Google Scholar]
- Lam K., Thu K., Tsang M., Moore M. & Gries G. Bacteria on housefly eggs, Musca domestica, suppress fungal growth in chicken manure through nutrient depletion or antifungal metabolites. Naturwissenschaften 9, 1127–1132 (2009). [DOI] [PubMed] [Google Scholar]
- Dowd S. E., Sun Y. & Secor P. R. Survey of bacterial diversity in chronic wounds using Pyrosequencing, DGGE, and full ribosome shotgun sequencing. Bmc Microbiol 8, 43 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd S. E., Sun Y. & Secor P. R. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol. 8, 125 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomberlin J. K., Sheppard D. C. & Joyce J. A. Selected life-history traits of black soldier flies (Diptera: Stratiomyidae) reared on three artificial diets. Ann. Entomol. Soc. Am. 95, 379–386 (2002). [Google Scholar]
- Tomberlin J. K., Sheppard D. C. & Joyce J. A. Black soldier fly (Diptera: Stratiomyidae) colonization of pig carrion in south Georgia. J. Forensic Sci. 50, 152–153 (2005). [PubMed] [Google Scholar]
- Lord W. D., Goff M. L., Adkins T. R. & Haskell N. H. The black soldier fly Hermetia illucens (Diptera: Stratiomyidae) as a potential measure of human postmortem interval: observations and case histories. J. Forensic Sci. 39, 215–222 (1994). [PubMed] [Google Scholar]
- Bradley S. W. & Sheppard D. C. House fly oviposition inhibition by larvae of Hermetia illucens, the black soldier fly. J. Chemical Ecol. 10, 853–859 (1984). [DOI] [PubMed] [Google Scholar]
- Liu Q., Tomberlin J. K., Brady J. A., Sanford M. R. & Yu Z. Black soldier fly (Diptera: Stratiomyidae) larvae reduce Escherichia coli in dairy manure. Environ. Entomol. 37, 1525–1530 (2008). [DOI] [PubMed] [Google Scholar]
- Heo C. C. et al. Study of insect succession and rate of decomposition on a partially burned pig carcass in an oil palm plantation in Malaysia. Trop. Biomed. 25, 202–208 (2008). [PubMed] [Google Scholar]
- Pujol-Luz J. R., Francez P. A., Ururahy-Rodrigues A. & Constantino R. The black soldier-fly, Hermetia illucens (Diptera, Stratiomyidae), used to estimate the postmortem interval in a case in Amapa State, Brazil. J. Forensic Sci. 53, 476–478 (2008). [DOI] [PubMed] [Google Scholar]
- Sánchez A. M., Magaña C., Soloña M. & Rojo S. First record of Hermetia illucens (Diptera: Stratiomyidae) on human corpses in Iberian Peninsula. Forensic Sci. Int. (2010). [DOI] [PubMed] [Google Scholar]
- DeVault T. L., Rhodes J. O. E. & Shivik J. A. Scavenging by vertebrates: behavioral, ecological, and evolutionary perspectives on an important energy transfer pathway in terrestrial ecosystems. Oikos 102, 225–234 (2003). [Google Scholar]
- Watts J. E., Merritt G. C. & Goodrich B. S. The ovipositional response of the Australian sheep blowfly, Lucilia cuprina, to fleece-rot odours. Aust. Vet. J. 57, 45045–45044 (1981). [DOI] [PubMed] [Google Scholar]
- Schröder R. & Hilker M. The relevance of background odor in resource location by insects: A behavioral approach. BioScience 58, 308–316 (2008). [Google Scholar]
- Chaudhury M. F., Skoda S. R., Sagel A. & Welch J. B. Volatiles emitted from eight wound-isolated bacteria differentially attract gravid screwworms (Diptera: Calliphoridae) to oviposit. J. Med. Entomol. 47, 349–354 (2010). [DOI] [PubMed] [Google Scholar]
- Hilker M. & Meiners T. Chemoecology of Insect Eggs adn Egg Deposition. (Blackwell Publishing, 2002). [Google Scholar]
- Kämpfer P. et al. Description of Wautersiella falsenii gen. nov., sp. nov., to accommodate clinical isolates phenotypically resembling members of the genera Chryseobacterium and Empedobacter. International J. Syst. Evol. Microbiol. 56, 2323–2329 (2006). [DOI] [PubMed] [Google Scholar]
- Ashworth J. R. & Wall R. Response of the sheep blowflies Lucilia sericata and L. cuprina to odour and the development of semiochemical baits. Med. Vet. Entomol. 8, 303–309 (1994). [DOI] [PubMed] [Google Scholar]
- Vass A. A. et al. Decomposition chemistry of human remains: a new methodology for determining the postmortem interval. J. Forensic Sci. 47, 542–553 (2002). [PubMed] [Google Scholar]
- Béguin P. & Aubert J.-P. The biological degradation of cellulose. FEMS Microbiol. Rev. 13, 25–58 (1994). [DOI] [PubMed] [Google Scholar]
- Stackebrandt E., Schumann P. & Prauser H. in The Prokaryotes (eds Martin Dworkin et al.) 983–1001 (Springer, 2006). [Google Scholar]
- Arenskötter M., Bröker D. & Steinbüchel A. Biology of the metabolically diverse genus Gordonia. App. Environ. Microbiol. 70, 3195–3204 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomberlin J. K., Adler P. H. & Myers H. M. Development of the black soldier fly (Diptera: Stratiomyidae) in relation to temperature. Environ. Entomol. 38, 930–934 (2009). [DOI] [PubMed] [Google Scholar]
- St. Hilaire S. et al. Fish offal recycling by the black soldier fly produces a foodstuff high in omega-3 fatty acids. J. World Aqua. Soc. 38, 309–313 (2007). [Google Scholar]
- Yu G. et al. Inoculating poultry manure with companion bacteria influences growth and development of black soldier fly (Diptera: Stratiomyidae) larvae. Environ. Entomol. 40, 30–35 (2011). [DOI] [PubMed] [Google Scholar]
- Verhulst N. O., Takken W., Dicke M., Schraa G. & Smallegange R. C. Chemical ecology of interactions between human skin microbiota and mosquitoes. FEMS Microbiol. Ecol. 35, 1–9 (2010). [DOI] [PubMed] [Google Scholar]
- Verhulst N. O. et al. Composition of human skin microbiota affects attractiveness to malaria mosquitoes. PLoS ONE 6, e28991 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis T. S., Boundy-Mills K. L. & Landolt P. J. Volatile emissions from an epiphytic fungus are semiochemicals for eusocial wasps. Microbial Ecology 64, 1056–1063 (2012). [DOI] [PubMed] [Google Scholar]
- Crippen T. L., Sheffield C. L., Esquivel S. V., Droleskey R. E. & Esquivel J. F. The acquisition and internalization of Salmonella by the lesser mealworm, Alphitobius diaperinus (Coleoptera: Tenebrionidae). Vector Borne Zoo. Dis. 9, 65–72 (2009). [DOI] [PubMed] [Google Scholar]
- Sheppard D. C., Tomberlin J. K., Joyce J. A., Kiser B. & Sumner S. M. Rearing methods for the black soldier fly (Diptera: Stratiomyidae). J. Med. Entomol. 39, 695–698 (2002). [DOI] [PubMed] [Google Scholar]
- Hogsette J. A. New diets for production of house flies and stable flies (Diptera: Muscidae) in the laboratory. J. Economic Entomol. 85, 2291–2294 (1985). [DOI] [PubMed] [Google Scholar]
- Boatright S. A. & Tomberlin J. K. Effects of temperature and tissue type on the development of Cochliomyia macellaria (Diptera: Calliphoridae). J. Med. Entomol. 47, 917–923 (2010). [DOI] [PubMed] [Google Scholar]
- Zheng L. et al. Bacterial diversity from successive life stages of black soldier fly (Diptera: Stratiomyidae) using 16S rDNA pyrosequencing. J. Med. Entomol. 50, 647–658 (2013). [DOI] [PubMed] [Google Scholar]
- Campbell P. W. et al. Detection of Pseudomonas (Burkholderia) cepacia using PCR. Ped. Pulmonol. 20, 44–49 (1995). [DOI] [PubMed] [Google Scholar]
- Moreno L. The application of amplicon length heterogeneity PCR (LH-PCR) for monitoring the dynamics of soils microbial communitities associated with cadaver decomposition. J. Microbiol. Meth. 84, 388–393 (2011). [DOI] [PubMed] [Google Scholar]
- Griekspoor A. & Groothuis T. 4peaks: a program that helps molecular biologists to visualize and edit their DNA sequence files <http://mekentosj.com/4peaks/> (2005). [Google Scholar]
- Tamura K. et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Garrity G., Tiedje J. & Cole J. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen F. T. & Young C. C. Rapid detection and identification of the metabolically diverse genus Gordonia by 16S rRNA-gene-targeted genus-specific primers. FEMS Microbiol. Lett. 250, 221–227 (2005). [DOI] [PubMed] [Google Scholar]
- Handl S., Dowd S. E., Garcia-Mazcorro J. F., Steiner J. M. & Suchodolski J. S. Massive parallel 16S rRNA gene pyrosequencing reveals highly diverse fecal bacterial and fungal communities in healthy dogs and cats. FEMS Microbiol. Ecol. 76, 301–310 (2011). [DOI] [PubMed] [Google Scholar]
- Garrity G., Bell J. & Lilburn T. Bergey's Manual of Systematic Bacteriology. 2nd edn, 161 (Springer-Verlag, 2005). [Google Scholar]
- Niu B., Fu L., Sun S. & Li W. Artificial and natural duplicates in pyrosequencing reads of metagenomic data. BMC Bioinformatics 11, 187 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki E. & Eddy S. Query-dependent banding (QDB) for faster RNA similarity searches. PLOS Comp. Biol. 3, 0540–0554 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki E., Kolbe D. & Eddy S. Infernal 1.0: inference of RNA alignments. Bioinformatics 25, 1335–1337 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N. & Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987). [DOI] [PubMed] [Google Scholar]
- PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods) v. Version 4 (Sinauer Associates, Sunderland, Massachusetts 2003).
- Han M. & Zmasek C. PhyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinformatics, 1–6 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomberlin J. K. & Sheppard D. C. Factors influencing mating and oviposition of black soldier flies (Diptera: Stratiomyidae) in a colony. J. Entomol. Sci. 37, 345–352 (2002). [Google Scholar]