Abstract
Small interfering RNA (siRNA) is an emerging class of therapeutics, working by regulating the expression of a specific gene involved in disease progression. Despite the promises, effective transport of siRNA with minimal side effects remains a challenge. In this study, a non-viral nanoparticle gene carrier has been developed and its efficiency for siRNA delivery and transfection has been validated at both in vitro and in vivo levels. Such a nanocarrier, abbreviated as Alkyl-PEI2k-IO, was constructed with a core of iron oxide (IO) and a shell of alkylated PEI2000 (Alkyl-PEI2k). It was found to be able to bind with siRNA, resulting in well-dispersed nanoparticles with a controlled clustering structure and narrow size distribution. Electrophoresis studies showed that the Alkyl-PEI2k-IOs could retard siRNA completely at N/P ratios above 10, protect siRNA from enzymatic degradation in serum and release complexed siRNA efficiently in the presence of polyanionic heparin. The knockdown efficiency of the siRNA loaded nanocarriers was assessed with 4T1 cells stably expressing luciferase (fluc-4T1) and further, with a fluc-4T1 xenograft model. Significant downregulation of luciferase was observed, and unlike the high molecular weight analogs, the Alkyl-PEI2k coated IOs showed a good biocompatibility. In conclusion, Alkyl-PEI2k-IOs demonstrate highly efficient delivery of siRNA and an innocuous toxic profile, making it a potential carrier for gene therapy.
Keywords: Biomaterials, Superparamagnetic nanoparticles, Polytehyleneimine, Small interfering RNA
1. Introduction
Gene therapy shows great potential to treat many forms of human diseases by replacing, altering, or supplementing a gene that is absent or abnormal.[1] Small interfering RNA (siRNA), known as silencing RNA or short interfering RNA, plays a variety of roles in biology[2] and its discovery has led to a surge in interest in harnessing RNA interference for biomedical research and drug development.[3] To date, siRNA has been experimentally introduced as a nucleic acid-based medicine and is expected to be developed into a promising novel strategy to down-regulate the targeted oncogenes for cancer therapy.[4]
However, there has been only modest success in the development of siRNA-based cancer therapy, mainly due to the lack of safe, broadly applicable delivery methods.[5] Viral delivery is the more conventional approach because viral vectors have been proven to have high transfection efficiency. However, human clinical trials have demonstrated substantial obstacles to their use, such as insertional mutagenesis and immunological problems.[6] Due to the inherent problems associated with viral vectors, there has been increased interest in harnessing non-viral vectors, such as polymers and nanoparticles[7] for delivery of siRNA, with the emphasis on high efficiency, stability, and biocompatibility.
Iron oxide nanoparticles (IOs) have shown remarkable potentials in biomedical research, due to their innate biocompatibility, magnetic properties, scalability, high surface area-to-volume ratio, and easily adaptable surface for bioagent attachment.[8] Indeed, IOs have been functionalized with a range of therapeutics, antibodies, peptides, nucleic acid, and other assorted biological agents [9]. As for rationally designing IOs-based gene delivery vectors, surface modification is central, which determines the loading and release of the therapeutic gene [7i–k, 9c]. A common strategy is to convert the nanoparticle surface a polycation layer, onto which the highly negatively charged therapeutic genes can be electrostatically attracted and tethered. This is mostly achieved by introducing one layer of cationic polymers [10] onto nanoparticle surface and various cationic polymers-coated IOs have been reported as non-viral gene delivery vehicles [11].
Polyethylenimine (PEI) is an excellent transfection reagent which is able to bind to DNA via electrostatic interactions, protect the DNA from degradation, and enter cells through rapid endocytosis.[12] In addition, PEI is also able to delay acidification and fusion of intracellular organelles through a “proton sponge effect”, which causes osmotic swelling of endosomes and subsequent release of the trapped materials into the cytosol.[12b, 13] The transfection efficiency and toxicity of PEI correlate strongly with its molecular weight. Low molecular weight PEI exhibits much lower transfection efficiency as well as cytotoxicity compared to its high molecular weight analogs.[12a] However, it has been shown that cross-linked low molecular weight PEI or low molecular weight PEI coated IOs/gold nanoparticles could enhance gene delivery efficacy without compromising its low cytotoxicity.[14] In our previously study, we chose low molecular weight amphiphilic Alkyl-PEI2k to form stable nanocomplexes with IOs nanoparticles (Alkyl-PEI2k-IOs).[15] Different from many other systems involving high molecular polycations, Alkyl-PEI2k can hold multiple IOs nanoparticles with a controlled clustering structure, leading to much higher T2 relaxivities than single IOs. Additionally, Alkyl-PEI2k-IOs have shown good stability, cell labeling efficiency and little cytotoxicity.
We propose and validate in this study that this controlled clustering nanocomposite has a great potential as non-viral vectors for gene delivery. We found, through agarose gel electrophoresis, toxicity and magnetic resonance imaging (MRI), that Alkyl-PEI2k-IOs were capable of stably binding, protecting, and delivering siRNA for gene silence while maintaining magnetic properties and high biocompatibility. We further validated, with success, their use as gene vector in vivo in a 4T1 xenograft tumor mouse model. Through these systematic studies, we demonstrated that Alkyl-PEI2k-IOs possess low cytotoxicity and high siRNA transfer efficiency, promising them as a safe and efficient avenue for gene therapy.
2. Results and discussion
Alkyl-PEI2k-IOs were prepared following our previous protocol.[15–16] It is worth noting that Alkyl-PEI2k-IOs showed high T2 relaxivity with a unique controlled clustering structure (Fig. S1A). Both Transmission Electron Microscopy (TEM) and Dynamic Light Scattering (DLS) analyses (Fig. S1B,C) showed that the Alkyl-PEI2k-IOs were monodispersed without obvious aggregation. In fact, such nanoparticles were found to remain stable in water for over 6 months at room temperature. Zeta potential results showed that Alkyl-PEI2k-IOs were positively charged, with a potential around +40 mV (Fig. S1D). This was attributable to the presence of multiple amino groups from the alkylated-PEI2k.
The Alkyl-PEI2k-IO/siRNA complexes with different N/P ratios (i.e. N/P ratio from 5 to 50) were prepared by adding an appropriate amount of the Alkyl-PEI2k-IOs into firefly Luciferase siRNA (lucsiRNA) in PBS under agitation. The N/P ratios were calculated based on PEI nitrogen per nucleic acid phosphate (1 μg of siRNA is 3 nmol of phosphate and 0.9 μg PEI contains 10 nmol of amine nitrogen).[17] The formed Alkyl-PEI2k-IOs/siRNA complexes (Fig. 1) in pure water showed an average hydrodynamic diameter of about 100 nm with relatively narrow distribution (Fig. 2A). As expected, the zeta potentials of the complexes decreased from +40.8 mV to −2.64 mV when increasing the initial siRNA concentration (i.e. N/P ratio from 50 to 5) (Fig. 2B), suggesting successful loading of siRNA onto the nanoparticle surface. Compact nanoparticles are highly desirable for efficient siRNA delivery, since the biophysical characteristics play an important role in the eventual transfection efficiency. To determine the structural integrity of the Alkyl-PEI2k-IOs during the siRNA binding process, we performed TEM studies on the complexes. We found that the Alkyl-PEI2k-IO/siRNA complex inherited clustering feature of the parent nanostructures (Fig. 2C).[15] The morphology of the Alkyl-PEI2k-IO/siRNA complexes was further visually characterized by Atomic Force Microscopy (AFM) (Fig. 2D). It can be seen that the Alkyl-PEI2k-IO/siRNA complexes were homogeneous, compact and spherical, and the diameter of the complexes observed by AFM was in good agreement with that from the DLS measurement. These findings demonstrated the rigidity of the Alkyl-PEI2k-IOs throughout the siRNA loading process.
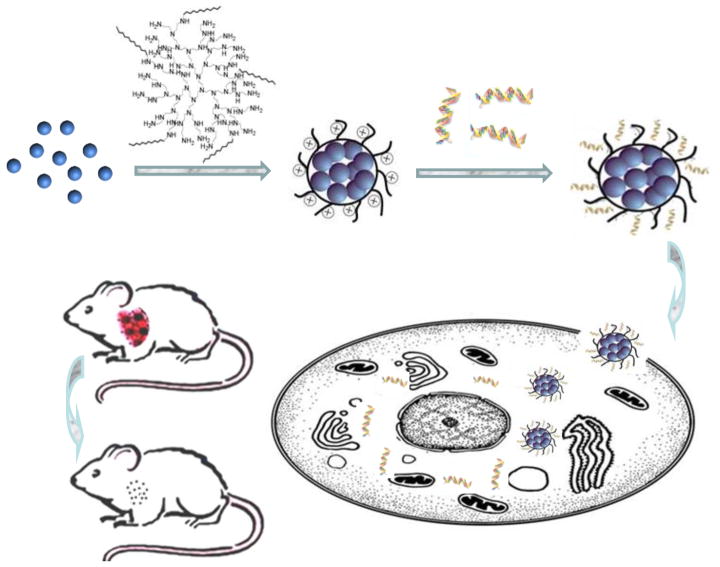
Figure 1.
Schematic illustration of the preparation of Alkyl-PEI2k-IO/siRNA complexes and their intracellular uptake.
Figure 2.
Physical characterization of Alkyl-PEI2k-IO/siRNA complexes. (A) Average diameter and (B) Zeta potential of Alkyl-PEI2k-IO/siRNA complexes at various N/P ratios; (C) TEM and (D) AFM images of Alkyl-PEI2k-IOs/siRNA with an N/P ratio of 20.
To assess the siRNA binding ability, Alkyl-PEI2k-IOs were mixed with lucsiRNA and analyzed by agarose gel electrophoresis. The results of the retardation assay were well matched with the zeta potential values of all complexes and the retardation efficiency increased when decreasing the initial siRNA concentration (Figure 3A). When the N/P ratios were higher than 10, the migration of siRNA was completely blocked, indicating successful siRNA binding via electrostatic interactions. On the contrary, in a heparin decomplexation assay, naked siRNA was found released from Alkyl-PEI2k-IO/siRNA complexes when heparin was added, presumably due to the latter’s stronger association with the particle surface (Fig. 3B). A qualified gene delivery carrier should protect the cargos from enzymatic degradation during the transportation. To assess such a qualification, we subjected Alkyl-PEI2k-IOs/siRNA to incubation with 50% fetal bovine serum (FBS) at 37 °C and studied the genes’ stability. Alkyl-PEI2k-IOs/siRNA complexes were obviously more stable than naked siRNA in the presence of serum. As shown in Figure 3C, D, siRNA alone was digested thoroughly within 12 h, but when complexed with Alkyl-PEI2k-IOs, it could survive beyond 24 h.
Figure 3.
Agarose gel electrophoresis of Alkyl-PEI2k-IO/siRNA complexes: (A) Electrophoretic retardation analysis of siRNA binding with Alkyl-PEI2k-IOs/siRNA samples; (B) Release of siRNA with the addition of heparin at various concentrations; (C) Serum stability of siRNA when complexed with Alkyl-PEI2k-IOs at an N/P ratio of 20. The study was performed in 50% serum solution for a predetermined incubation time.
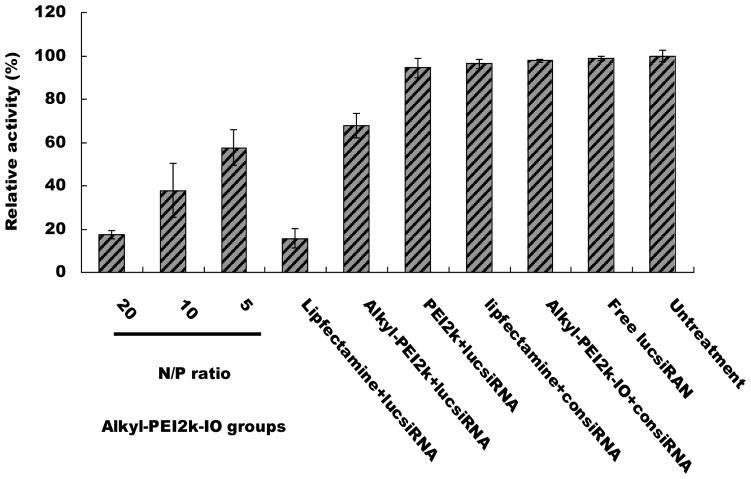
To evaluate the potential application of Alkyl-PEI2k-IOs for siRNA delivery, in vitro transfection experiments were conducted with 4T1-fluc cells. Gene-silencing efficiency of the lucsiRNA complexes was quantitatively determined by measuring the bioluminescence imaging (BLI) signal intensity of 4T1-fluc cells. The BLI signal intensity of the control (untreated) 4T1-fluc cells was set to 100% luciferase expression. 4T1-fluc cells treated with negative control siRNA (consiRNA) or free lucsiRNA did not show any reduced gene expression (Fig. 4). Similar to other reports,[12a, 18] the low-molecular-weight PEI2k is an inferior transfection agent, showing a slightly reduced gene expression (94%) at an N/P ratio of 20. Alkyl-PEI2k showed a slightly enhanced silencing effect compared to PEI2k. It is believed that the partial alkylation of the primary amine on PEI2k helped genetic material condensation and strengthen the interaction of the complexes with cell membranes.[14c, 19] The presence of a cationic surface could also promote nanoparticle interaction and binding with the endosomal membrane, and as a result of that, induced membrane destablisation and cytosolic relocalisation.[10] Thus, we hypothesize that the Alkyl-PEI2k coating on SPIOs facilitated not only siRNA condensation but also its escape from the endosome to the cytoplasm. As expected, Alkyl-PEI2k-IO/lucsiRNA complexes showed remarkably high gene silencing efficacy (Fig. 4). Interestingly, the Alkyl-PEI2k-IO/lucsiRNA complexes lead to much higher transfection rates than Alkyl-PEI2k. At an N/P ratio of 20, the gene-suppression extent of fluc for Alkyl-PEI2k-IO/lucsiRNA complexes was comparable to that of Lipofectamine™ 2000, one of the most widely used transfection reagents. The improved gene-silencing effects of Alkyl-PEI2k-IO/lucsiRNA complexes could be due to an increased intracellular, as reported by Mok et al.[20], physically stable siRNA polyelectrolyte complexes with a size around 100 nm are more readily internalized by cells through the endocytic pathway.
Figure 4.
Inhibition of fluc gene expression by Alkyl-PEI2k-IOs/siRNA (siRNA = 6 pm) at various N/P ratios. The study was performed with fluc-4T1 cells in serum-deficient condition.
To further assess the capacity of the nanovector for siRNA loading and subsequent cellular internalization, we carried out confocal laser scanning microscopy (CLSM) imaging with FITC-labeled siRNA. Shown in Figure 5A, green fluorescence was exclusively observed in the cytoplasm of 4T1 cells treated with Alkyl-PEI2k-IO/siRNA complexes. Cellular TEM also revealed that the nanocomplexes were confined in the membrane-bound intracellular space and not found in the cell nuclei (Fig. 5B). Such a feature indicated that the Alkyl-PEI2k-IOs played a role in steering the intracellular distribution of siRNA, as it is well known that unmodified PEI/siRNA is localized not only in the cytoplasm but also in the nuclei.[7f, 21] It is possible that such efficient unpacking of complexes and subsequent siRNA release from Alkyl-PEI2k-IOs into cytoplasm contributed to an enhanced gene silencing efficiency when compared with Alky-PEI2k/siRNA complexes.
Figure 5.
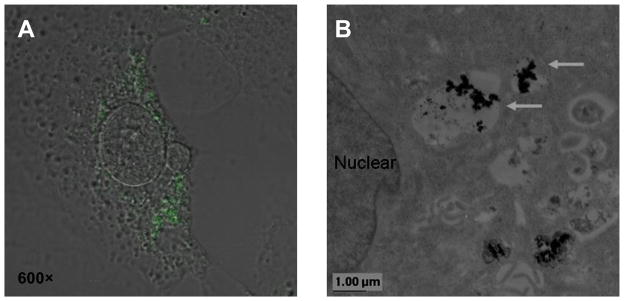
Cellular uptake of 4T1-fluc cells. (A) Confocal microscopic images of 4T1-fluc cells after treatment with Alkyl-PEI2k-IO/siRNA complexes. The internalized siRNA appeared green in the fluorescence images (Magnification 600×). (B) Electron microscopic image of 4T1-fluc cells transfected with Alkyl-PEI2k-IO/siRNA complexes (arrows). Scale bar = 0.6 μm.
The ideal way to track the delivered genes in a therapy is to use a specific imaging probe. With functional contrast agent such as IOs, molecular MRI provides a promising tool.[7k, 22] To investigate the potential MRI contrast enhancement associated with the uptake of Alky-PEI2k-IO/siRNA complexes, the transfected cells were collected and imaged using MRI. At a 7T magnetic field, the Alky-PEI2k-IO/siRNA complexes transfected cells exhibited obviously decreased signal intensities on T2 weighted images. As shown in Figure 6, the internalized nanoparticles shortened the spin-spin relaxation time by dephasing the spins of neighboring water protons, resulting in hypointensities on T2 weighted images. Higher N/P ratios were associated with decreased signal intensities, which is not surprising since a high N/P ratio indicates a more positive zeta potential and a higher internalization rate.
Figure 6.
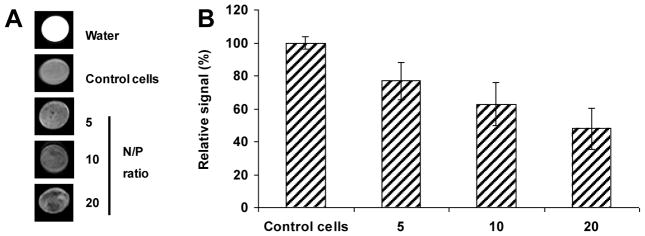
(A) T2-weighted magnetic resonance images of agarose gel phantoms containing the fluc-4T1 cells treated with Alkyl-PEI2k-IO complexes; (B) Negative correlation between the T2 contrast and the N/P ratios of the Alkyl-PEI2k-IO/siRNA complexes (p < 0.05).
To rule out the possibility that the decreased gene expression was caused by nonspecific cytotoxicity, we performed MTT assay with the Alkyl-PEI2k-IOs/siRNA complexes at six different concentrations, ranging from 3.125 to 25 μg Fe/mL. The viability of 4T1-fluc cells was expressed as a fraction of viable cells and normalized to that of untreated cells (blank control). The MTT results indicate that the Alkyl-PEI2k-IO/siRNA complexes had little cytotoxicity even at the highest concentration (25 μg Fe/mL), which exceeds by 5 times the concentration used in our cell transfection experiments (Fig. 7). It also appears that the cytotoxicity of Alkyl-PEI2k-IOs was slightly improved when complexed with siRNA. Thus, the Alkyl-PEI2k-IO/siRNA complexes have no obvious cytotoxicity on 4T1-fluc cells within the transfection concentrations, confirming that the observed gene-silencing was a pure consequence of RNAi effect.
Figure 7.
Cytotoxic effect of Alkyl-PEI2k-IO/siRNA complexes on 4T1-fluc cells. There was no significant difference in viability between untransfected and Alkyl-PEI2k-IO/siRNA complexes transfected cells.
The new vector was investigated on a 4T1-fluc xenograft model to evaluate its potential of delivering siRNA. As intratumoral injection is a routine method for local gene delivery that can improve interstitial transport of alien genes in tumor tissues, four groups of Balb/C mice (n = 3/group) bearing subcutaneous 4T1-fluc tumors were intratumorally injected with Alkyl-PEI2k-IO/lucsiRNA complexes, Alkyl-PEI2k-IOs, naked lucsiRNA or PBS, respectively. The complexes or control materials (50 μL) were administered by several injections into the tumors. After each injection, the needle was left in the tumor for about 30 s to prevent leakage of the injected materials. The fluc gene-silencing effect was measured by in vivo BLI. Figure 8 showed in vivo optical images of fluc-expressing tumors after intratumoral injection of Alkyl-PEI2k-IO/siRNA complexes. The relative level of fluc expression was significantly reduced for the Alkyl-PEI2k-IO/lucsiRNA complexes, but was not observed in the other control groups. To determine whether the intratumoral injection of complexes had inhibited tumor growth in vivo, tumors from each group were harvested and weighed after the last optical imaging experiment. As shown in Figure S2A, no significant difference in tumor weight was observed between the Alkyl-PEI2k-IO/lucsiRNA complex group and the other three control groups. In addition, no loss in body weight was found in the Alkyl-PEI2k-IOs/siRNA group. In tumors injected with Alkyl-PEI2k-IO/siRNA complexes, a large number of IOs were found at the injection sites under TEM (Fig. S2B). Taken together, down-regulation of fluc was not due to the toxicity of the nanosystem but rather the RNAi effect.
Figure 8.
Gene-silencing effect by Alkyl-PEI2k-IOs/siRNA complexes in vivo. (A) In vivo optical images and (B) quantitative analysis of fluc-expressing tumors after intratumoral injection of Alkyl-PEI2k-IOs/siRNA complexes.
Overall, Alkyl-PEI2k-IOs meet several criteria for becoming a successful siRNA delivery system while maintaining the salient features pertinent to IOs. While continued studies on investigating the mechanisms behind Alkyl-PEI2k-IO enhanced gene delivery are being explored, the nanosystem can be extended to include other biologically relevant components by tethering onto the amine groups on Alkyl-PEI2k-IOs surface. For example, Alkyl-PEI2k-IOs can be easily modified to incorporate functionalities such as charge shielding brushes (e.g. PEG chains) for long blood circulation times, biovectors (e.g. RGD) for tumor targeted delivery, or chemotherapeutic agents (e.g. doxorubicin) that can work in conjunction with therapeutic genes to achieve more comprehensive therapeutic results. Apart from this, other imaging nanoprobes, such as quantum dots, can be co-loaded as part of the clustering structures, and the resulted conjugates are of multimodal imaging capabilities.
3. Conclusions
Alkyl-PEI2k-IOs were successfully synthesized using Alkyl-PEI2k as the phase transfer materials, resulting in well-dispersed nanoparticles with a uniform structure and narrow size distribution. Alkyl-PEI2k-IOs possess many outstanding features in favor of siRNA delivery, including high siRNA binding capability, protecting siRNA from enzymatic degradation and releasing complexed siRNA in the presence of polyanionic heparin. When loaded with fluc siRNA, Alkyl-PEI2k-IOs induced enhanced down regulation of luciferase in fluc-4T1 cells without exhibiting cellular toxicity. Meanwhile, the transfected cells displayed strong signal contrast compared to untreated cells on T2 weighted imaging. Furthermore, the Alkyl-PEI2k-IOs/siRNA complexes show the remarkably high gene silencing efficacy on a 4T1-fluc tumor xenograft model with good biocompatibility. Altogether, Alkyl-PEI2k-IOs may open up exciting new horizons for gene delivery and nanomedicine.
4. Experimental Section
Nanovector Synthesis
Alkyl-PEI2k-IOs were synthesized following a previously published protocol.[15–16] Briefly, branched PEI2k (Alfa Aesar) was reacted with 1-iodododecane (Aldrich) in ethanol. The purified product was obtained as a gummy solid after lyophilization and was confirmed by 1H NMR (CDCl3). For IO synthesis, Fe(acac)3 (2 mmol) was mixed with 1,2-hexadecanediol (10 mmol), oleic acid (6 mmol), and oleylamine (6 mmol) in benzyl ether (20 mL), and the mixture was heated up to reflux under nitrogen (~300 °C) for 1 h. The product was washed three times with ethanol and was resuspended and kept in hexane. Before surface modification, the nanocrystals were dried under argon and redispersed in a chloroform solution of alkylated PEI2k. The solution was slowly added into water with sonication to form Alkyl-PEI2k-IOs nanoparticles.
The Alkyl-PEI2k-IO/siRNA complexes with different N/P ratios (i.e. N/P ratio from 5 to 50) were prepared by adding an appropriate amount of the Alkyl-PEI2k-IOs into firefly luciferase siRNA (Ambion) in PBS under agitation. The resulting mixture was incubated at room temperature for 20 min and was characterized by dynamic light scattering (DLS), zeta-potential, atomic force microscopy (AFM) and transmission electron microscopy (TEM).
Agarose Gel Electrophoresis of Alkyl-PEI2k-IOs/siRNA Complexes
Alkyl-PEI2k-IOs/siRNA complexes (N/P ratios 5:1; 10:1; 20:1) were analyzed by 2% agarose gel electrophoresis.[23] The gels were prepared with 2% agarose in Tris-acetate-EDTA buffer containing 0.5 μg/mL ethidium bromide. As for gel retardation assay, samples were incubated at room temperature for 20 min, after which an appropriate amount of DNA loading buffer was added to each sample. Gel electrophoresis was carried out at 100 V for 15 min and the gel was subsequently imaged using a LAS-3000 gel documentation system (Fujifilm Life Science, Japan).
For heparin decomplexation assay, the Alkyl-PEI2k-IOs were first complexed with siRNA (N/P ratios 20:1) at room temperature for 20 min. Then, various amounts of heparin (heparin/siRNA weight ratio 0; 2; 10; 25 and 100) were added and the mixtures were further incubated for 15 min. The samples were loaded on a 2% agarose gel and subjected to electrophoresis as described above.
For serum stability assay, after adding serum at a final 50% concentration, samples (N/P ratios 20:1) were incubated for predetermined periods at 37°C. The resulting samples were treated with heparin (10 μg) and then loaded on a 2% agarose gel. The released siRNA fraction was imaged as described above.
Cell Culture
The murine 4T1 breast cancer cells were purchased from the American Type Culture Collection (ATCC). The cells were maintained in RPMI 1640 medium (Invitrogen) supplemented with 10% FBS at 37°C with 5% CO2. For establishing the 4T1 cell lines stably expressing luciferase gene (4T1-fluc), transfection was done with pcDNA 3.1 cytomegalovirus-firefly luciferase (fluc) DNA and Lipofectamine 2000 (Invitrogen) using 80% confluent 4T1 cells.[24] The cells were incubated for 48 h before the culture medium was changed. Selection was made by adding selective medium containing 1 mg/mL G418 antibiotic (Mediatech, Inc.) every 2–3 days. Two weeks later, the cells were analyzed and subcloned in 24-well plates. When reaching 80% confluence, the cells in the plate were imaged using a Xenogen IVIS-100 system (Caliper Life Sciences) after addition of the substrate D-luciferin (20 μL per well of 3 mg/mL stock). The 4T1-fluc clone with the highest fluc activity was selected for further studies. No significant difference between 4T1 and 4T1-fluc cells was observed in terms of proliferation or tumorigenicity. The cells were used when they reached 80% confluence.
In Vitro Transfection of Alkyl-PEI2k-IOs/siRNA Complexes
Transfection complexes were prepared as follows: siRNA (6 pm per well) and appropriate amount of the Alkyl-PEI2k-IOs were both diluted to 25 μL with fresh medium and were mixed at room temperature for 5 min; after that, the solution was incubated at room temperature for another 20 min. In the meantime, the cells in 96-well plate were washed with PBS and the medium was replaced with fresh medium (50 μL, each well). The transfection complexes were then added to the wells and the cells were incubated at 37 °C, 5% CO2, 95–100% humidity for the indicated time period. Cells for positive controls were transfected with Lipofectamine using the recommended protocol from the manufacturer (Invitrogen). The luciferase expression of fluc-4T1 cells was visualized 48 h post-transfection as described above.
Cellular Uptake Study
Confocal laser scanning microscopy (CLSM) was used to assess the intracellular trafficking of siRNA. 4T1-fluc cells (2×105) were seeded on coverslips in a 6-well tissue culture plate and transfected 24 h later with Alkyl-PEI2k-IO complexes of FITC-labeled siRNA for 3 h at 37 °C. Cells were washed and fixed with 4% formaldehyde and the slides were mounted and observed with a Zeiss LSM510 CLSM imaging system (Carl Zeiss, Germany) with an upright confocal microscope.[25]
Electron microscopy was performed using a Philips CM-10 transmission electron microscope (Eindhoven, Netherlands).[15] After 3 h of incubation with Alkyl-PEI2k-IO/siRNA complexes, the cells were fixed in 2.5% buffered glutaraldehyde for 1 h at 4°C, followed by 1% osmium tetroxide for 2 h. The cells were then dehydrated in ascending concentrations of ethanol, immersed in propylene-oxide, and embedded in resin. After that, the samples were cut into ultrathin sections and then examined.
MRI Study of Transfected Cells
After transfection for 3 h, cells were washed three times with PBS, harvested, and processed for MR imaging as described below. Cells (106) were dispersed in a final concentration of 5% gelatin phantom (100 μL) crosslinked with glutaraldehyde inside microcentrifuge tubes.[16] T2 relaxation measurements were performed on a 7T Bruker magnet (Bruker, Germany) with a 5 cm volume coil and using spin-echo imaging a sequence. Images were acquired using a repetition time (TR) of 4200 ms and an echo time (TE) of 36.0 ms. The spatial resolution parameters were as follows: acquisition matrix of 256×128, field-of-view of 30×30 mm, section thickness of 1 mm and two averages. The T2 weighted imaging was generated by NIH Image J (Bethesda, MD) and the signal intensity of transfected cells was displayed as percentages relative to untreated cells.
Cytotoxicity
The cytotoxicity of Alkyl-PEI2k-IO/siRNA complexes was evaluated using the standard MTT assay protocol.[15] Briefly, 4T1-fluc cells were seeded onto 96-well plates at a density of 1×104 cells/well and incubated for 24 h. siRNA was complexed with the Alkyl-PEI2k-IOs at predetermined N/P ratios in PBS buffer and incubated for 20 min before use. Alkyl-PEI2k-IO/siRNA complexes were incubated with the cells for 3 h in 100 μL of serum-free RPMI 1640 medium followed by 48 h in 200 μL of RPMI 1640 medium containing 10% FBS. The medium was replaced with 200 μL fresh media including 20 μL 5 mg/mL MTT solution, and the incubation proceeded for another 4 h. The media was then removed, and 150 μL dimethyl sulfoxide was added into each well to dissolve the internalized purple formazan crystals. An aliquot of 100 μL was taken from each well and transferred into a fresh 96-well plate. The absorption at 570 nm was measured using a microplate reader. The absorption from the control cells, which were not subjected to transfection, was set as 100% cell viability.
In Vivo Studies
All animal experiments were conducted in accordance with an NIH approved protocol. Flank xenograft tumors models were prepared by subcutaneous injection of 106 4T1-fluc cells suspended in serum free medium into female athymic nude mice (18 – 20 g). When the diameter of tumors reached about 6 mm, 250 pmoles of lucsiRNA complexed with Alkyl-PEI2k-IOs at an N/P ratio of 20 were intratumorally injected every other day for 3 times. The control groups were injected with an equal volume of PBS, free siRNA or IOs. The luciferase expression was visualized and quantified by a Xenogen IVIS-100 system. In brief, mice were anesthetized with 2% isoflurane in O2 and received i.p. injection of D-luciferin solution in PBS at a dose of 150 mg/kg. Serial images were acquired between 5 to 20 min after D-luciferin administration. The bioluminescence signal intensity (BLI) was quantified as total photon counts from the region of interest after subtraction of background luminescence. The animals were sacrificed at the end of the longitudinal imaging experiment and the tumors were removed and weighed immediately. The tumor tissue samples were fixed with 0.1 M cacodylate buffer and 1% osmium tetroxide. After being stained using 1% uranyl acetate and fully dehydrated, the samples were imaged using TEM.[15]
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Program (IRP) of the National Institutes of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH), and the International Cooperative Program of National Science Foundation of China (NSFC) (81028009). Dr. Jin Xie was partially supported by the Pathway to Independence (K99/R00) Fellowship. The work also was supported by Program for New Century Excellent Talents in University (NCET-06-0781), Distinguished Young Scholars Project of Sichuan Province and National Natural Science Foundation of China (20974065, 50603015, and 50830107).
Footnotes
Supporting Information is available on the WWW under http://www.small-journal.com or from the author.
Contributor Information
Dr. Gang Liu, Laboratory of Molecular Imaging and Nanomedicine (LOMIN), National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH), Bethesda, MD 20892 (USA). National Engineering Research Center for Biomaterials, Sichuan University, Chengdu, 610064 (China). Sichuan Key Laboratory of Medical Imaging, Department of Radiology, Affiliated Hospital of North Sichuan Medical College, Nanchong 637007 (China).
Dr. Jin Xie, Laboratory of Molecular Imaging and Nanomedicine (LOMIN), National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH), Bethesda, MD 20892 (USA).
Dr. Fan Zhang, Laboratory of Molecular Imaging and Nanomedicine (LOMIN), National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH), Bethesda, MD 20892 (USA)
Dr. Zhi-Yong Wang, National Engineering Research Center for Biomaterials, Sichuan University, Chengdu, 610064 (China)
Dr. Kui Luo, National Engineering Research Center for Biomaterials, Sichuan University, Chengdu, 610064 (China)
Dr. Lei Zhu, Laboratory of Molecular Imaging and Nanomedicine (LOMIN), National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH), Bethesda, MD 20892 (USA)
Dr. Qi-Meng Quan, Laboratory of Molecular Imaging and Nanomedicine (LOMIN), National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH), Bethesda, MD 20892 (USA)
Dr. Gang Niu, Laboratory of Molecular Imaging and Nanomedicine (LOMIN), National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH), Bethesda, MD 20892 (USA)
Dr. Seulki Lee, Laboratory of Molecular Imaging and Nanomedicine (LOMIN), National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH), Bethesda, MD 20892 (USA)
Prof. Hua Ai, Email: huaai@scu.edu.cn, National Engineering Research Center for Biomaterials, Sichuan University, Chengdu, 610064 (China). Department of Radiology, West China Hospital, Sichuan University, Chengdu, 610041 (China).
Dr. Xiaoyuan Chen, Email: shawn.chen@nih.gov, Laboratory of Molecular Imaging and Nanomedicine (LOMIN), National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH), Bethesda, MD 20892 (USA).
References
- 1.a) Friedmann T, Roblin R. Science. 1972;175:949. doi: 10.1126/science.175.4025.949. [DOI] [PubMed] [Google Scholar]; b) Cavazzana-Calvo M, Thrasher A, Mavilio F. Nature. 2004;427:779. doi: 10.1038/427779a. [DOI] [PubMed] [Google Scholar]; c) Ott MG, Schmidt M, Schwarzwaelder K, Stein S, Siler U, Koehl U, Glimm H, Kuhlcke K, Schilz A, Kunkel H, Naundorf S, Brinkmann A, Deichmann A, Fischer M, Ball C, Pilz I, Dunbar C, Du Y, Jenkins NA, Copeland NG, Luthi U, Hassan M, Thrasher AJ, Hoelzer D, von Kalle C, Seger R, Grez M. Nat Med. 2006;12:401. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- 2.a) Hamilton AJ, Baulcombe DC. Science. 1999;286:950. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]; b) Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature. 2001;411:494. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 3.a) Hannon GJ, Rossi JJ. Nature. 2004;431:371. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]; b) de Fougerolles A, Vornlocher HP, Maraganore J, Lieberman J. Nat Rev Drug Discov. 2007;6:443. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Kim VN, Han J, Siomi MC. Nat Rev Mol Cell Biol. 2009;10:126. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 4.a) Iorns E, Lord CJ, Turner N, Ashworth A. Nat Rev Drug Discov. 2007;6:556. doi: 10.1038/nrd2355. [DOI] [PubMed] [Google Scholar]; b) Ashihara E, Kawata E, Maekawa T. Curr Drug Targets. 2010;11:345. doi: 10.2174/138945010790711897. [DOI] [PubMed] [Google Scholar]; c) Phalon C, Rao DD, Nemunaitis J. Expert Rev Mol Med. 2010;12:e26. doi: 10.1017/S1462399410001584. [DOI] [PubMed] [Google Scholar]
- 5.a) Gao K, Huang L. Mol Pharm. 2009;6:651. doi: 10.1021/mp800134q. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Devi GR. Cancer Gene Ther. 2006;13:819. doi: 10.1038/sj.cgt.7700931. [DOI] [PubMed] [Google Scholar]
- 6.a) Woods NB, Bottero V, Schmidt M, von Kalle C, Verma IM. Nature. 2006;440:1123. doi: 10.1038/4401123a. [DOI] [PubMed] [Google Scholar]; b) Edelstein ML, Abedi MR, Wixon J. J Gene Med. 2007;9:833. doi: 10.1002/jgm.1100. [DOI] [PubMed] [Google Scholar]
- 7.a) Geusens B, Lambert J, De Smedt SC, Buyens K, Sanders NN, Van Gele M. J Control Release. 2009;133:214. doi: 10.1016/j.jconrel.2008.10.003. [DOI] [PubMed] [Google Scholar]; b) Green JJ, Langer R, Anderson DG. Acc Chem Res. 2008;41:749. doi: 10.1021/ar7002336. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Malek A, Czubayko F, Aigner A. J Drug Target. 2008;16:124. doi: 10.1080/10611860701849058. [DOI] [PubMed] [Google Scholar]; d) Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA, Yen Y, Heidel JD, Ribas A. Nature. 2010;464:1067. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Meng H, Liong M, Xia T, Li Z, Ji Z, Zink JI, Nel AE. ACS Nano. 2010;4:4539. doi: 10.1021/nn100690m. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Song WJ, Du JZ, Sun TM, Zhang PZ, Wang J. Small. 2010;6:239. doi: 10.1002/smll.200901513. [DOI] [PubMed] [Google Scholar]; g) Guo S, Huang Y, Jiang Q, Sun Y, Deng L, Liang Z, Du Q, Xing J, Zhao Y, Wang PC, Dong A, Liang XJ. ACS Nano. 2010;4:5505. doi: 10.1021/nn101638u. [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Mok H, Veiseh O, Fang C, Kievit FM, Wang FY, Park JO, Zhang M. Mol Pharm. 2010;7:1930. doi: 10.1021/mp100221h. [DOI] [PMC free article] [PubMed] [Google Scholar]; i) Veiseh O, Kievit FM, Fang C, Mu N, Jana S, Leung MC, Mok H, Ellenbogen RG, Park JO, Zhang M. Biomaterials. 2010;31:8032. doi: 10.1016/j.biomaterials.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]; j) Taratula O, Garbuzenko O, Savla R, Wang YA, He H, Minko T. Curr Drug Deliv. 2011;8:59. doi: 10.2174/156720111793663642. [DOI] [PubMed] [Google Scholar]; k) Liu G, Swierczewska M, Lee S, Chen X. Nano today. 2010;5:524. doi: 10.1016/j.nantod.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Xie J, Huang J, Li X, Sun S, Chen X. Curr Med Chem. 2009;16:1278. doi: 10.2174/092986709787846604. [DOI] [PubMed] [Google Scholar]; b) Liu G, Yang H, Zhang XM, Shao Y, Jiang H. Contrast Media Mol Imaging. 2010;5:53. doi: 10.1002/cmmi.362. [DOI] [PubMed] [Google Scholar]; c) Xie J, Wang J, Niu G, Huang J, Chen K, Li X, Chen X. Chem Commun (Camb) 2010;46:433. doi: 10.1039/b917195a. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Xie J, Chen K, Huang J, Lee S, Wang J, Gao J, Li X, Chen X. Biomaterials. 2010;31:3016. doi: 10.1016/j.biomaterials.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]; e)M. M. Yallapu, S. F. Othman, E. T. Curtis, B. K. Gupta, M. Jaggi, S. C. Chauhan, Biomaterials 2010; f) Yallapu MM, Othman SF, Curtis ET, Gupta BK, Jaggi M, Chauhan SC. Biomaterials. 2011;32:1890. doi: 10.1016/j.biomaterials.2010.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Gao J, Gu H, Xu B. Acc Chem Res. 2009;42:1097. doi: 10.1021/ar9000026. [DOI] [PubMed] [Google Scholar]; b)H. Ai, Adv Drug Deliv Rev 2011, accepted; c) Hao R, Xing R, Xu Z, Hou Y, Gao S, Sun S. Adv Mater. 2010;22:2729. doi: 10.1002/adma.201000260. [DOI] [PubMed] [Google Scholar]
- 10.Leroueil PR, Hong S, Mecke A, Baker JR, Jr, Orr BG, Banaszak Holl MM. Acc Chem Res. 2007;40:335. doi: 10.1021/ar600012y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a) Scherer F, Anton M, Schillinger U, Henke J, Bergemann C, Kruger A, Gansbacher B, Plank C. Gene Ther. 2002;9:102. doi: 10.1038/sj.gt.3301624. [DOI] [PubMed] [Google Scholar]; b) Arsianti M, Lim M, Marquis CP, Amal R. Langmuir. 2010;26:7314. doi: 10.1021/la9041919. [DOI] [PubMed] [Google Scholar]; c) Cheong SJ, Lee CM, Kim SL, Jeong HJ, Kim EM, Park EH, Kim DW, Lim ST, Sohn MH. Int J Pharm. 2009;372:169. doi: 10.1016/j.ijpharm.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 12.a) Godbey WT, Wu KK, Mikos AG. J Control Release. 1999;60:149. doi: 10.1016/s0168-3659(99)00090-5. [DOI] [PubMed] [Google Scholar]; b) Demeneix B, Behr JP. Adv Genet. 2005;53:217. [PubMed] [Google Scholar]
- 13.Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. Proc Natl Acad Sci U S A. 1995;92:7297. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.a) Huang H, Yu H, Tang G, Wang Q, Li J. Biomaterials. 2010;31:1830. doi: 10.1016/j.biomaterials.2009.11.012. [DOI] [PubMed] [Google Scholar]; b) Kievit FM, Veiseh O, Bhattarai N, Fang C, Gunn JW, Lee D, Ellenbogen RG, Olson JM, Zhang M. Adv Funct Mater. 2009;19:2244. doi: 10.1002/adfm.200801844. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Thomas M, Klibanov AM. Proc Natl Acad Sci U S A. 2003;100:9138. doi: 10.1073/pnas.1233634100. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Hu C, Peng Q, Chen F, Zhong Z, Zhuo R. Bioconjug Chem. 2010;21:836. doi: 10.1021/bc900374d. [DOI] [PubMed] [Google Scholar]
- 15.Liu G, Wang Z, Lu J, Xia C, Gao F, Gong Q, Song B, Zhao X, Shuai X, Chen X, Ai H, Gu Z. Biomaterials. 2011;32:528. doi: 10.1016/j.biomaterials.2010.08.099. [DOI] [PubMed] [Google Scholar]
- 16.Wang ZY, Liu G, Sun JY, Wu B, Gong Q, Song B, Ai H, Gu Z. J Nanosci Nanotechnol. 2009;9:378. doi: 10.1166/jnn.2009.j033. [DOI] [PubMed] [Google Scholar]
- 17.Werth S, Urban-Klein B, Dai L, Hobel S, Grzelinski M, Bakowsky U, Czubayko F, Aigner A. J Control Release. 2006;112:257. doi: 10.1016/j.jconrel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 18.Gebhart CL, Kabanov AV. J Control Release. 2001;73:401. doi: 10.1016/s0168-3659(01)00357-1. [DOI] [PubMed] [Google Scholar]
- 19.Thomas M, Klibanov AM. Proc Natl Acad Sci U S A. 2002;99:14640. doi: 10.1073/pnas.192581499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mok H, Park TG. Biopolymers. 2008;89:881. doi: 10.1002/bip.21032. [DOI] [PubMed] [Google Scholar]
- 21.a) Shim MS, Kwon YJ. Bioconjug Chem. 2009;20:488. doi: 10.1021/bc800436v. [DOI] [PubMed] [Google Scholar]; b) Zintchenko A, Philipp A, Dehshahri A, Wagner E. Bioconjug Chem. 2008;19:1448. doi: 10.1021/bc800065f. [DOI] [PubMed] [Google Scholar]
- 22.Medarova Z, Pham W, Farrar C, Petkova V, Moore A. Nat Med. 2007;13:372. doi: 10.1038/nm1486. [DOI] [PubMed] [Google Scholar]
- 23.Mok H, Lee SH, Park JW, Park TG. Nat Mater. 2010;9:272. doi: 10.1038/nmat2626. [DOI] [PubMed] [Google Scholar]
- 24.Cao Q, Cai W, Niu G, He L, Chen X. Clin Cancer Res. 2008;14:6137. doi: 10.1158/1078-0432.CCR-08-0049. [DOI] [PubMed] [Google Scholar]
- 25.Liu G, Tian J, Liu C, Ai H, Gu Z, Gou J, Mo X. J Mater Res. 2009;24:1317. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.