Abstract
The mechanistic target of rapamycin (mTOR) pathway integrates diverse environmental inputs, including immune signals and metabolic cues, to direct T cell fate decisions1. Activation of mTOR, comprised of mTORC1 and mTORC2 complexes, delivers an obligatory signal for proper activation and differentiation of effector CD4+ T cells2,3, whereas in the regulatory T cell (Treg) compartment, the Akt-mTOR axis is widely acknowledged as a crucial negative regulator of Treg de novo differentiation4–8 and population expansion9. However, whether mTOR signaling affects the homeostasis and function of Tregs remains largely unexplored. Here we show that mTORC1 signaling is a pivotal positive determinant of Treg function. Tregs have elevated steady-state mTORC1 activity compared to naïve T cells. Signals via T cell receptor (TCR) and IL-2 provide major inputs for mTORC1 activation, which in turn programs suppressive function of Tregs. Disruption of mTORC1 through Treg-specific deletion of the essential component Raptor leads to a profound loss of Treg suppressive activity in vivo and development of a fatal early-onset inflammatory disorder. Mechanistically, Raptor/mTORC1 signaling in Tregs promotes cholesterol/lipid metabolism, with the mevalonate pathway particularly important for coordinating Treg proliferation and upregulation of suppressive molecules CTLA-4 and ICOS to establish Treg functional competency. In contrast, mTORC1 does not directly impact the expression of Foxp3 or anti- and pro-inflammatory cytokines in Tregs, suggesting a non-conventional mechanism for Treg functional regulation. Lastly, we provide evidence that mTORC1 maintains Treg function partly through inhibiting the mTORC2 pathway. Our results demonstrate that mTORC1 acts as a fundamental ‘rheostat’ in Tregs to link immunological signals from TCR and IL-2 to lipogenic pathways and functional fitness, and highlight a central role of metabolic programming of Treg suppressive activity in immune homeostasis and tolerance.
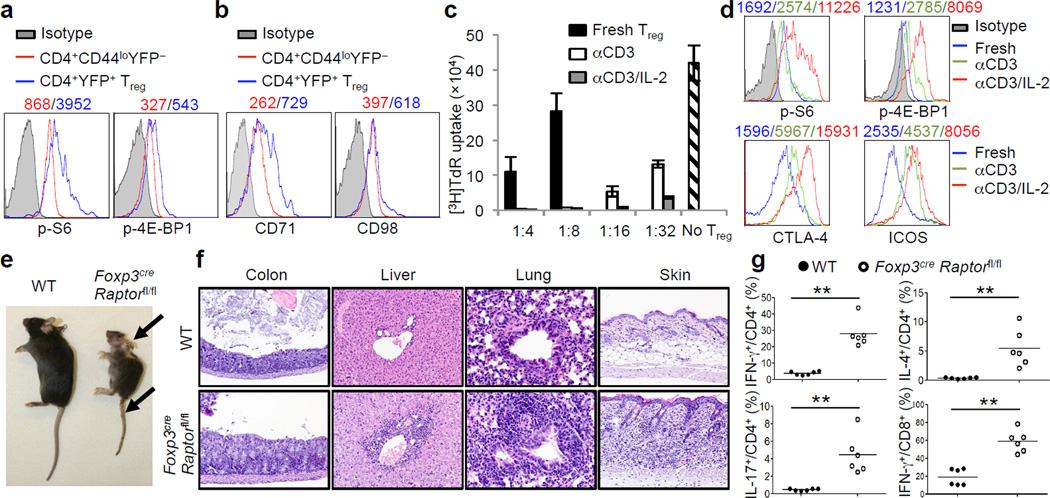
The evolutionarily conserved mTOR signaling couples cell growth and proliferation to nutrient availability and metabolic cues10. To investigate the function of mTORC1 in naturally occurring Tregs, we compared mTORC1 activity between Tregs and naïve T cells at steady state. Tregs had elevated phosphorylation of S6 and 4E-BP1, two major mTORC1 downstream targets (Fig. 1a and Supplementary Fig. 1a–c), whereas STAT5 phosphorylation was similar between these cells (Supplementary Fig. 1d). This finding is consistent with a recent study describing elevated S6 phosphorylation in Tregs compared to non-Tregs11. Tregs also contained higher abundance of CD71 (the transferrin receptor) and to a lesser extent, CD98 (a subunit of L-amino acid transporter), key nutrient receptors that depend upon mTORC1 activity for expression12, (Fig. 1b). Previous studies have demonstrated a requirement of mTORC1 for mitochondrial metabolism13, the dysregulation of which could affect homeostasis of memory T cells14 and hematopoietic stem cells15. Tregs had reduced mitochondrial membrane potential, whereas their mitochondrial mass and reactive oxygen species (ROS) production were largely comparable to naïve cells (Supplementary Fig. 1e). Thus, Tregs exhibit distinct regulation of mTORC1 activity and metabolism under steady state.
Figure 1.
mTORC1 signaling is constitutively active in Tregs and its disruption results in a fatal early-onset inflammatory disorder. a, Comparison of phosphorylation of S6 and 4E-BP1 between naïve T cells (CD4+CD44loFoxp3−) and Tregs (CD4+Foxp3+; from Foxp3YFP-Cre mice). b, Comparison of CD71 and CD98 expression between naïve T cells and Tregs. c, Tregs from C57BL/6J mice were activated with anti-CD3 or anti-CD3 and IL-2 for 3 days, washed and then used for in vitro suppression assays at multiple Treg vs Teff ratios; freshly isolated Tregs were included for comparison (shown only for 1:4 and 1:8 ratios). Error bars represent s.d. (n=3). d, Comparison of phosphorylation of S6 and 4E-BP1 and expression of CTLA-4 and ICOS between freshly isolated and pre-activated Tregs. e, Images of 42-day-old wild-type (WT) and Foxp3creRaptorfl/fl mice. Arrows indicate the scaly tail and ulceration of the body. f, Hematoxylin and eosin staining of colon, lung, liver and skin from 21-day-old wild-type and Foxp3creRaptorfl/fl mice. g, IFN-γ, IL-17 and IL-4 production in CD4+ and IFN-γ production in CD8+ cells from wild-type and Foxp3creRaptorfl/fl mice. P values are determined by Mann Whitney test. Mean fluorescent intensity (MFI) is presented above the plots of the relevant staining (a, b, d). Results represent 5 (a, g), 3 (b) and 2 (c-f) independent experiments.
One of the hallmarks of Tregs is that they are anergic in vitro upon TCR stimulation alone16, but are highly proliferative in vivo17,18. Whereas reversal of anergy has been suggested to impair Treg function16, Tregs stimulated by TCR together with IL-2, or under lymphopenic conditions, possess enhanced suppressive activity18,19. Indeed, prior stimulation of Tregs with anti-CD3 upregulated their suppressive activity, and this was further enhanced by concomitant treatment with IL-2 (Fig. 1c). Such functional enhancements were associated with increased mTORC1 activity and expression of CTLA-4 and ICOS, important effector molecules mediating Treg function20,21 (Fig. 1d). Compared with IL-2, CD28-mediated co-stimulation had more modest enhancing effects on these events (Supplementary Fig. 1f,g). Thus, TCR and IL-2 represent predominant signals at promoting mTORC1 activity and suppressive function of Tregs.
Although these results indicate that mTORC1 is positively correlated with Treg suppressive activity, a number of recent studies have revealed a negative role of mTORC1 in Tregs, including suppression of Treg differentiation2,4–8 and population expansion9. To establish the physiological relevance of mTORC1 signaling in Tregs, we deleted Raptor, an obligatory component of mTORC1 complex10, in Tregs by crossing Raptorfl/fl mice with Foxp3YFP-Cre mice22 (designated Foxp3creRaptorfl/fl). Raptor expression and S6 and 4E-BP1 phosphorylation were abolished in Raptor-deficient Tregs, but were normal in Foxp3− populations (Supplementary Fig. 2a–c). Additionally, a slight increase of phosphorylation of Akt Ser473 was observed in mutant Tregs, whereas phosphorylation of Akt Thr308 and Erk in Tregs was not affected (Supplementary Fig. 2d). Thus, deletion of Raptor specifically abrogated mTORC1 activity.
Surprisingly, Foxp3creRaptorfl/fl mice developed profound inflammatory diseases, indicated by a reduced body size and hunched posture, crusting of ears, eyelids and tail, and skin ulceration particularly on the head and upper back (Fig. 1e). Foxp3creRaptorfl/fl mice showed extensive lymphadenopathy (Supplementary Fig. 3a) and infiltrations of lymphocytes and myeloid cells in colon mucosa, lung, liver sinusoids, skin (Fig. 1f), among other organs, before they died at a young age (Supplementary Fig. 3b). Foxp3creRaptorfl/fl mice had increased CD8+ cell percentage and altered CD4+/CD8+ ratio, highly elevated memory/effector phenotype (CD44hiCD62Llo) T cells, and expansion of CD11b+ granulocytes and macrophages (Supplementary Fig. 3c–e). Further, T cells from Foxp3creRaptorfl/fl mice showed increased IFN-γ, IL-4 and IL-17 producing CD4+ cells and IFN-γ producing CD8+ cells (Fig. 1g). These phenotypes are reminiscent of those observed in mice carrying scurfy mutation23, indicating a loss of Treg function.
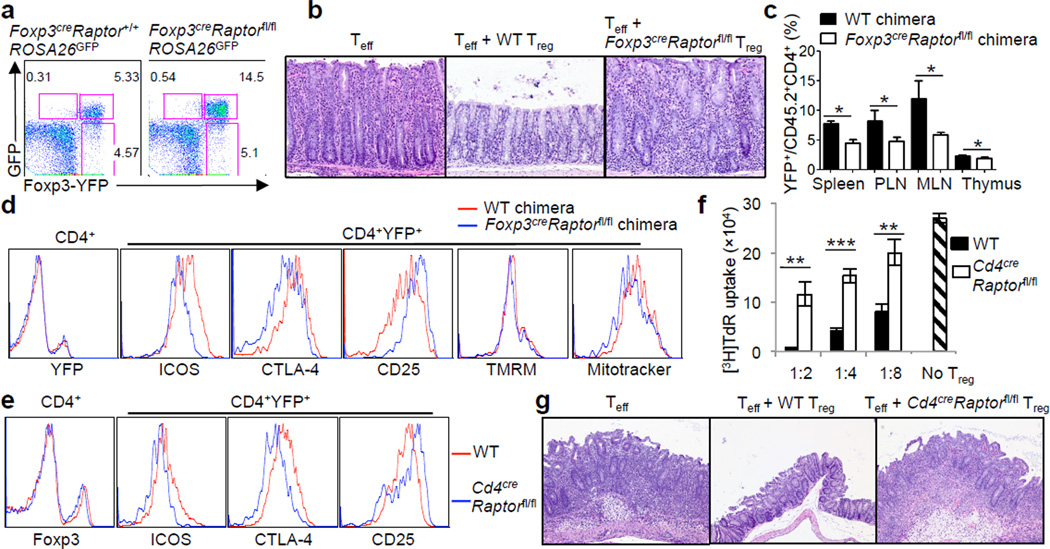
Despite severe autoimmune diseases, Foxp3creRaptorfl/fl mice had increased Foxp3+ Tregs (Supplementary Fig. 4a). Lineage tracing experiments indicated that the increase of Tregs was mainly ascribed to a more abundant population of canonical Tregs, as indicated by concomitant expression of YFP-Foxp3 and Cre activity (Fig. 2a). Raptor-deficient Tregs contained normal Foxp3 expression and either increased or normal expression of many Treg signature molecules examined, whereas CD62L expression was reduced (Supplementary Fig. 4b). Thus, in settings of immune activation, Raptor-deficient Tregs acquired activated phenotypes. Additional analysis revealed highly reduced mitochondrial membrane potential and mass and ROS production in mutant Tregs, whereas in vitro suppressive activity appeared largely undisturbed (Supplementary Fig. 4b,c). To test function of Raptor-deficient Tregs in vivo, we used a model of colitis induced by the transfer of Teff (CD4+CD45RBhiCD25−) cells into Rag1−/− hosts, which could be prevented by cotransfer with wild-type Tregs. However, cotransfer with Raptor-deficient Tregs failed to inhibit colitis (Fig. 2b), or the expansion and IFN-γ production of Teff cells (Supplementary Fig. 4d). Therefore, Raptor is required for suppressive function of Tregs in vivo.
Figure 2.
Raptor deletion abrogates Treg suppressive activity. a, Foxp3-YFP and GFP expression in CD4+ T cells from 25-day-old Foxp3creRaptor+/+ROSA26GFP and Foxp3creRaptorfl/flROSA26GFP mice. b, Representative colon histology in Rag1−/− mice given Teff cells alone or in combination with wild-type or Foxp3creRaptorfl/fl Tregs. c, Percentages of Tregs in mixed chimeras reconstituted with bone marrow cells from CD45.1+ mice and wild-type or Foxp3creRaptorfl/fl mice (n=3). d, Expression of Treg signature molecules and mitochondrial membrane potential (tetramethylrhodamine methyl ester, TMRM) and mass (Mitotracker) in the reconstituted chimeras. e, Expression of Treg signature molecules in 6-week-old wild-type and Cd4creRaptorfl/fl mice. f, In vitro suppression assays mediated by Tregs from wild-type and Cd4creRaptorfl/fl mice. Error bars represent s.d. (n=3). g, Representative colon histology in Rag1−/− mice given Teff cells alone or in combination with wild-type or Cd4creRaptorfl/fl Tregs. P values are determined by Mann Whitney test (c) or t-test (f). Results represent 3 (a, d, f), 2 (b, c, g) and 5 (e) independent experiments.
We speculated that the increased number and heightened activation phenotypes of Tregs in Foxp3creRaptorfl/fl mice could be due to a compensatory response to the ongoing inflammation24. To test this, we generated mixed bone marrow chimeras by reconstituting sub-lethally irradiated Rag1−/− mice with bone marrow cells from CD45.1+ mice mixed with those from either wild-type or Foxp3creRaptorfl/fl mice. Raptor-deficient Tregs were under-represented in the chimeras (Fig. 2c), with impaired expression of CTLA-4 and ICOS but largely normal CD25 levels and mitochondrial parameters (Fig. 2d). These findings were recapitulated in 10-day-old Foxp3creRaptorfl/fl mice that exhibited minimal immune activation (Supplementary Fig. 4e). Thus, in a disease-free environment, loss of Raptor in Tregs renders them a competitive disadvantage and compromises expression of CTLA-4 and ICOS. In contrast, the pronounced mitochondrial defects of Tregs from Foxp3creRaptorfl/fl mice (Supplementary Fig. 4b) were not intrinsic due to Raptor deficiency, but were likely a compensatory response, as observed in hematopoietic stem cells lacking the metabolic sensor LKB115.
To facilitate mechanistic studies of Raptor-deficient Tregs without the influence of ongoing inflammation, we generated Cd4-Cre;Raptorfl/fl (Cd4creRaptorfl/fl) mice, in which Raptor was deleted in all αβ T cells. Cd4creRaptorfl/fl mice showed a small reduction of Treg numbers in peripheral lymph nodes but not the spleen (Supplementary Fig. 5a), and immunoblots confirmed the loss of S6 and 4E-BP1 phosphorylation in these Tregs (Supplementary Fig. 5b). Expression of CTLA-4, ICOS and CD71 was impaired in Tregs from Cd4creRaptorfl/fl mice, whereas the levels of Foxp3, CD25 and other markers were normal (Fig. 2e and Supplementary Fig. 5c). In vitro assays revealed loss of suppressive activity of Tregs from Cd4creRaptorfl/fl mice (Fig. 2f). These Tregs also exhibited severely impaired in vivo suppressive activity, because they failed to suppress colitis or IFN-γ production mediated by Teff cells (Fig. 2g and Supplementary Fig. 5d). Analysis of mixed chimeras comprised of CD45.1+ and Cd4creRaptorfl/fl cells confirmed that Raptor-deficient Tregs were reduced in percentage with impaired CTLA-4 and ICOS expression (Supplementary Fig. 5e). Thus, Tregs deficient in mTORC1 have an intrinsic defect in suppressive activity. Nonetheless, Cd4creRaptorfl/fl mice exhibited normal health status, which was likely due to impaired activation of conventional T cells (to be described elsewhere). We found a similar requirement of Raptor in Treg suppressive activity following Cre-ERT2-mediated acute deletion in adult mice (Supplementary Fig. 6).
Cytokines produced by Tregs represent an important immunosuppressive mechanism25. The anti-inflammatory cytokines TGF-β1 and IL-10 were expressed normally in Raptor-deficient Tregs (Supplementary Fig. 7a,b). Although Tregs from Foxp3creRaptorfl/fl mice expressed increased IFN-γ that may compromise Treg function26, Tregs from Cd4creRaptorfl/fl did not (data not shown), thereby excluding such phenotypes as cell-intrinsic. Moreover, ablation of IFN-γ did not ameliorate the immune activation phenotypes of Foxp3creRaptorfl/fl mice (Supplementary Fig. 7c). Thus, the severe autoimmune phenotypes of Foxp3creRaptorfl/fl mice likely occur independently of cytokine dysregulation.
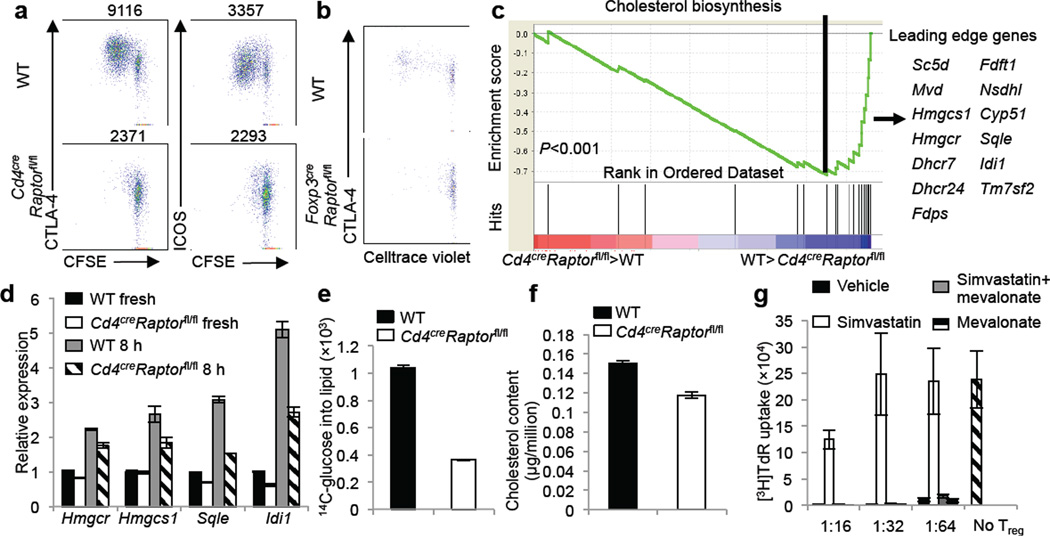
We therefore focused on the effects of Raptor on Treg surface effector molecules especially CTLA-4 and ICOS, which were highly reduced in Tregs from multiple Raptor-deficient genetic models in an intrinsic manner (Fig, 2d,e and Supplementary Fig. 4e,5e). Moreover, CTLA-4 and to a lesser extent, ICOS, were further increased upon in vitro stimulation, and this upregulation correlated with Treg proliferation. Such upregulation was blunted in Tregs from Cd4creRaptorfl/fl mice, associated with impaired proliferation (Fig. 3a and Supplementary Fig. 8a). Moreover, when wild-type Tregs were transferred into unmanipulated CD45.1+ mice, a proportion of them divided and upregulated CTLA-4 (Fig. 3b). However, Tregs from Cd4creRaptorfl/fl or Foxp3creRaptorfl/fl mice failed to proliferate or upregulate CTLA-4 upon transfer into CD45.1+ mice (Fig. 3b and Supplementary Fig. 8b). Markedly diminished homeostatic proliferation and CTLA-4 upregulation were also observed when Raptor-deficient Tregs were transferred into Rag1−/− mice (Supplementary Fig. 8c). Altogether, loss of Raptor inhibits Treg proliferation in vitro and in vivo. Furthermore, the proliferative defect was associated specifically with impaired CTLA-4 and ICOS expression, whereas apoptosis or other activation-induced events such as upregulation of GATA-3 and CD103 were independent of Raptor (data not shown). Thus, mTORC1 is important for coordinating Treg proliferation and CTLA-4 and ICOS expression that may collectively contribute to Treg suppressive function.
Figure 3.
Raptor coordinates Treg proliferation and effector molecule expression by orchestrating cholesterol/lipid biosynthetic metabolism, especially the mevalonate pathway. a, CFSE-labeled Tregs from wild-type and CD4creRaptorfl/fl mice were stimulated with anti-CD3, anti-CD28 and IL-2 for 3 days, followed by analysis for CFSE dilution and CTLA-4 and ICOS expression (MFI presented above the plots). b, Celltrace violet-labeled Tregs from wild-type and Foxp3creRaptorfl/fl mice were transferred into unmanipulated CD45.1+, and Celltrace violet dilution and CTLA-4 expression were analyzed 7 days later. c, Gene-set enrichment analysis reveals the under-representation of cholesterol biosynthesis genes in Raptor-deficient Tregs. The lower part of the plot shows the distribution of the genes in the cholesterol biosynthesis signature gene set (‘Hits’) against the ranked list of genes, and the list on the right shows the top hit genes. d, Real-time PCR quantification of lipogenic gene expression in wild-type and Cd4creRaptorfl/fl freshly isolated Tregs or those stimulated with anti-CD3, anti-CD28 and IL-2 for 8 hours. e, De novo lipogenesis of Tregs from wild-type and Cd4creRaptorfl/fl mice after 24 hours of anti-CD3, anti-CD28 and IL-2 stimulation, with the final 4 hours labeled with 14C-glucose. f, Total cholesterol level was measured using lysates of activated Tregs from wild-type and Cd4creRaptorfl/fl mice. g, In vitro suppression assays mediated by Tregs previously activated by anti-CD3, anti-CD28 and IL-2 for 3 days in the presence of vehicle, simvastatin (2 μM), simvastatin and mevalonate (100 μM) or mevalonate alone. Error bars represent s.d. (n=3). Results represent 2 independent experiments except for c (n=4 from one experiment).
To explore Raptor/mTORC1-dependent molecular mechanisms, we used functional genomics and compared gene expression profiles of Tregs from Cd4creRaptorfl/fl and wild-type mice. Gene-set enrichment analysis revealed that cholesterol biosynthesis was the most significantly altered pathway in a Raptor-dependent manner: Expression of many genes in this pathway was downregulated after Raptor deletion (Fig. 3c and Supplementary Fig. 9a), including those encoding 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR), squalene epoxidase (SQLE) and isopentenyl-diphosphate delta isomerase 1 (IDI-1), which were verified by real-time PCR analysis (Fig. 3d). Further, upon TCR stimulation, Raptor-deficient Tregs failed to efficiently synthesize lipids from glucose (Fig. 3e), and had reduced cellular cholesterol (Fig. 3f). These results establish a role of mTORC1 in promoting cholesterol/lipid biosynthesis. Given the interconnected relationship between the biosynthetic and bioenergetic programs27, we determined the role of mTORC1 in Treg bioenergetics by measuring extracellular acidification rate (ECAR) and oxygen consumption rate (OCR), which denoted glycolysis and mitochondrial respiration, respectively. Raptor-deficient Tregs showed decreased ECAR and OCR (Supplementary Fig. 9b,c), indicating a requirement for mTORC1 in bioenergetics. We therefore determined de novo lipid synthesis rate by measuring the incorporation of 14C-acetate into cellular lipids, thereby bypassing the requirement of mTORC1 in glycolysis or mitochondrial activity. Indeed, Raptor deficiency diminished synthesis of lipids from 14C-acetate in Tregs (Supplementary Fig. 9d). These results demonstrated a specific role of mTORC1 in orchestrating the lipogenic program.
To test functional significance of lipid metabolism, we activated Tregs in the presence of 25-hydroxycholesterol, and this general lipid synthesis inhibitor potently blocked Treg suppressive activity (Supplementary Fig. 10a). Direct inhibition of HMGCR, the rate-limiting enzyme in the synthesis of cholesterol and isoprenoid lipids, by simvastatin also impaired Treg suppressive activity. Importantly, simvastatin-induced inhibition was completely reversed by the simultaneous addition of mevalonate, the metabolite downstream of HMGCR (Fig. 3g). Similar effects were observed after treatments with atorvastatin and lovastatin (Supplementary Fig. 10b). The inhibitory effects of these agents on Treg function were associated with impaired Treg proliferation and effector molecule upregulation (Supplementary Fig. 10c), in a mevalonate-dependent manner (Supplementary Fig. 10d). Further, proliferation and CTLA-4 and ICOS upregulation in Tregs transferred into congenic CD45.1+ mice were diminished by statin treatment in vivo (Supplementary Fig. 11). Altogether, Raptor/mTORC1 signaling promotes the lipogenic program, with the mevalonate pathway particularly important for mediating Treg proliferation, CTLA-4 and ICOS upregulation, and functional fitness.
Moreover, lipid metabolism-dependent regulation of CTLA-4 and ICOS is also operative in conventional T cells, because naïve T cells treated with inhibitors of this pathway or deficient in Raptor failed to effectively upregulate these molecules following TCR stimulation (Supplementary Fig. 12). We speculate this constitutes a feedback mechanism to account for the immunosuppressive activity of activated non-Treg cells previously described28,29, a notion that awaits further investigation.
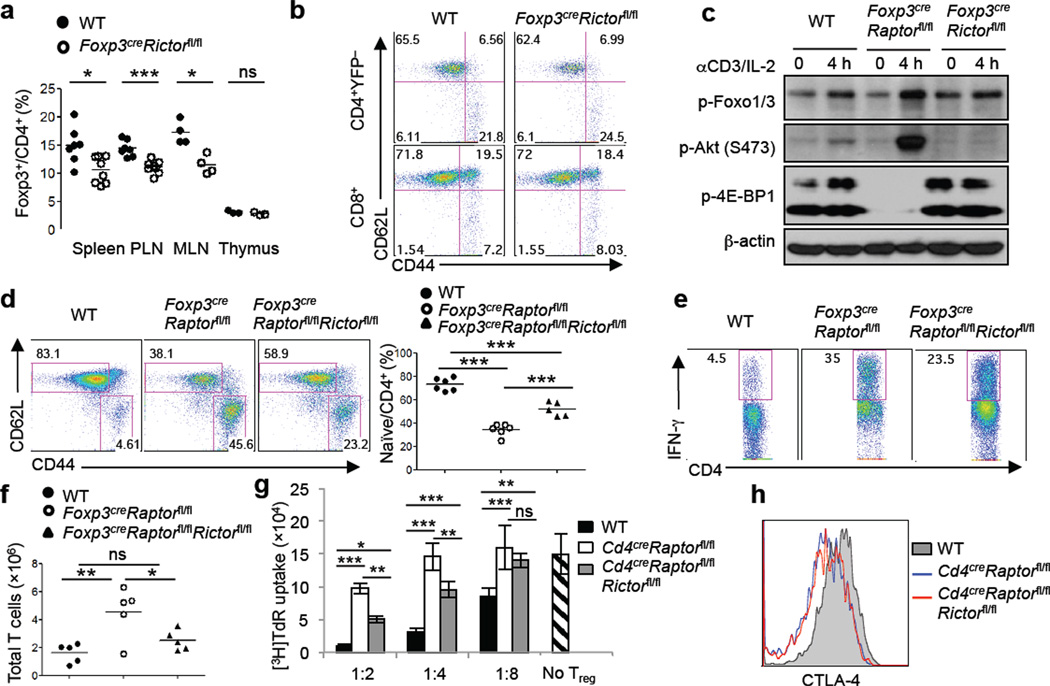
Crosstalk between mTORC1 and mTORC2 signaling pathways has been reported10, although molecular details and functional significance are unclear. Notably, mTORC2 contributes to Akt activation that in turn signals to Foxo126, and acts in synergy with mTORC1 to negatively control generation of induced Tregs2. We therefore examined the role of mTORC2 and its interaction with mTORC1 in naturally occurring Treg cells. Treg-specific deletion of Rictor, the defining component of mTORC2, resulted in a small reduction of Tregs in the periphery (Fig. 4a). However, loss of Rictor in Tregs did not perturb T cell homeostasis, cellularity or cytokine expression at steady state (Fig. 4b and Supplementary Fig. 13a,b), or the health status of the mice. Also, Rictor-deficient Tregs had normal expression of Treg signature molecules, mitochondrial activity and in vitro suppressive activity (Supplementary Fig. 13c,d). Thus, in contrast to a crucial requirement of mTORC1 in programming Treg activity, mTORC2 likely contributes to Treg maintenance but is dispensable for Treg function.
Figure 4.
Deletion of Rictor does not alter Treg function but partially rescues inflammation in Foxp3creRaptorfl/fl mice. a, Treg percentage in 2–3 month old wild-type and Foxp3creRictorfl/fl mice. b, Expression of CD62L and CD44 on splenic T cells from wild-type and Foxp3creRictorfl/fl mice. c, Tregs were activated by anti-CD3 and IL-2 for 4 hours followed by immunoblots. d, Expression of CD62L and CD44 expression on splenic CD4+ T cells from 3–4 week old wild-type, Foxp3creRaptorfl/fl and Foxp3creRaptorfl/flRictorfl/fl mice. Right, percentage of CD62LhiCD44lo naïve CD4+ cells in the spleen. e, IFN-γ production in splenic CD4+ T cells from wild-type, Foxp3creRaptorfl/fl and Foxp3creRaptorfl/flRictorfl/fl mice. f, Numbers of total TCRβ+ cells in peripheral lymph nodes from wild-type, Foxp3creRaptorfl/fl and Foxp3creRaptorfl/flRictorfl/fl mice. g, In vitro suppression assays mediated by Tregs from 3–4 week old wild-type, CD4creRaptorfl/fl and CD4creRaptorfl/flRictorfl/fl mice. Error bars represent s.d. (n=3). h, CTLA-4 expression in Tregs from the spleen of wild-type, CD4creRaptorfl/fl and CD4creRaptorfl/flRictorfl/fl mice. P values are determined by Mann Whitney test (a) and ANOVA (d, f, g). Results represent 3 (a-c, e-g) and 2 (d, h) independent experiments.
To examine the crosstalk between the two mTOR complexes, we analyzed mTORC2 activity in Raptor-deficient Tregs. Phosphorylation of well-established mTORC2 targets, Akt Ser473 and Foxo1/3, was elevated in Raptor-deficient Tregs after anti-CD3/IL-2 (Fig. 4c) or anti-CD3/CD28 stimulation (Supplementary Fig. 14), consistent with the observation in freshly isolated Raptor-deficient Tregs (Supplementary Fig. 2d). In contrast, Akt Ser473 phosphorylation was nearly abolished after Rictor deletion (Fig. 4c and Supplementary Fig. 14a). Thus, mTORC1 institutes a negative feedback on mTORC2 in Tregs. To determine the contribution of mTORC2 activity to Foxp3creRaptorfl/fl phenotypes, we generated Foxp3creRaptorfl/flRictorfl/fl mice. Compared with T cells in Foxp3creRaptorfl/fl mice, those in Foxp3creRaptorfl/flRictorfl/fl mice contained an increased percentage of naïve phenotypes (Fig. 4d) and showed modestly reduced IFN-γ production (Fig. 4e). Also, the lymphadenopathy phenotype observed in Foxp3creRaptorfl/fl mice was ameliorated in Foxp3creRaptorfl/flRictorfl/fl mice (Fig. 4f). To test the intrinsic suppressive activity of Raptor/Rictor-deficient Tregs, we generated Cd4creRaptorfl/flRictorfl/fl mice that eliminated both mTORC1 and mTORC2 activities (Supplementary Fig. 15a,b). In vitro suppression assay revealed an intermediate phenotype of Raptor/Rictor-deficient Tregs compared with Raptor-deficient and wild-type cells (Fig. 4g). However, impaired CTLA-4 expression and TCR-induced proliferation upon Raptor deletion were not restored by the concomitant loss of Rictor (Fig. 4h and Supplementary Fig. 15c). Further, Foxp3creRaptorfl/flRictorfl/fl mice still succumbed to an inflammatory disorder, albeit with a small extension of lifespan and less inflammation as compared with Foxp3creRaptorfl/fl mice (Supplementary Fig. 16). Therefore, Raptor/mTORC1 signaling promotes Treg function in part through inhibiting mTORC2, although the predominant effect of mTORC1 is to coordinate metabolic programs and Treg suppressive molecules.
Much emphasis has been placed on the transcriptional mechanisms that orchestrate Treg suppressive activity, but how immunological signals are sensed and integrated by Tregs for their functional activation remains obscure. Here we describe that mTORC1-dependent metabolic programing is a central mechanism to couple immune signals including TCR and IL-2 and Treg suppressive function. Raptor/mTORC1 promotes the lipogenic program, with the mevalonate pathway particularly important for coordinating Treg proliferation and optimal induction of effector molecules CTLA-4 and ICOS (Supplementary Fig. 17). Thus, mTORC1-dependent lipid metabolism provides a novel link between two crucial Treg regulators, IL-2 and CTLA-4, the loss of which causes fulminant autoimmune diseases21,30. An additional mechanism is via mTORC1-dependent inhibition of mTORC2, although the modest rescue effect of Rictor deletion in Foxp3creRaptorfl/fl mice suggests a relatively minor contribution of this feedback pathway. Notably, mTORC1-dependent metabolic programming operates both under steady state and immune stimulation, and this provides important mechanistic insight into the apparently conflicting observations between Tregs’ in vitro anergy and in vivo antigen-primed state16–18. We propose that Tregs adopt the evolutionarily ancient mTORC1 signaling to link immunological inputs to metabolic activity and functional fitness, therefore implicating mTORC1 as a fundamental rheostat to program Treg suppressive activity via a non-conventional mechanism.
METHODS
Mice
C57BL/6J, CD45.1+, Raptorfl, Rag1−/−, Ifng−/−, and ROSA26GFP (a loxP site-flanked STOP cassette followed by the GFP-encoding sequence was inserted into the ROSA26 locus) mice were purchased from the Jackson Laboratory. Rictorfl/fl, Cd4-Cre and ROSA26-Cre-ERT2 mice have been described previously31. Foxp3YFP-Cre mice were a generous gift from Dr. Alexander Rudensky22. Foxp3creRaptorfl/fl mice were used at 3–4 weeks old unless otherwise noted, with the age and gender-matched wild-type mice containing the Foxp3cre allele as controls. Other mice were used at 8–10 weeks old unless otherwise noted. Bone marrow chimeras were generated by transferring 1 × 107 T cell-depleted bone marrow cells into sublethally irradiated (5 Gy) Rag1−/− mice, followed by reconstitution for at least 2 months. For treatment with tamoxifen, mice were injected intraperitoneally with tamoxifen (1 mg per mouse) in corn oil daily for 4 consecutive days and then analyzed 7 days after the last injection. All mice were kept in specific pathogen free facility in Animal Resource Center at St. Jude Children’s Research Hospital. Animal protocols were approved by the Institutional Animal Care and Use Committee of St. Jude Children’s Research Hospital.
Flow cytometry
For analysis of surface markers, cells were stained in PBS containing 2% (wt/vol) fetal bovine serum (FBS) with antibodies from eBioscience, unless otherwise noted. Foxp3 staining was performed per manufacturer’s instruction (eBioscience). Intracellular staining of CTLA-4 was performed together with Foxp3 using anti-CTLA-4 antibody (UC10-4F10-11; BD Biosciences). BrdU and caspase-3 staining was performed per manufacturer’s instruction (BD Biosciences). For detection of phosphorylated signaling proteins, lymphocytes were rested in complete medium for 1 hour. They were fixed with Phosflow Lyse/Fix buffer, followed by permeabilization with Phosflow Perm buffer III (BD Biosciences) and staining with antibodies to S6 phosphorylated at Ser235 and Ser236 (D57.2.2E; Cell Signaling Technology), 4E-BP1 phosphorylated at Thr37 and Thr46 (236B4; Cell Signaling Technology), Akt phosphorylated at Ser473 (M89-61; BD Biosciences) and Thr308 (J1-223.371; BD Biosciences), and Erk phosphorylated at Thr202 and Tyr204 (20A; BD Biosciences). For staining mitochondria, lymphocytes were incubated for 30 minutes at 37°C with 10 nM MitoTracker Deep Red (Life Technologies) or 20 nM TMRM (tetramethylrhodamine, methyl ester; ImmunoChemistry Technologies) after staining surface markers. ROS were measured by incubation with 10 μM CM-H2DCFDA (5-(and-6)-chloromethyl-2’,7’-dichlorodihydrofluorescein diacetate, acetyl ester; Life Technologies) or 2.5 μM CellROX Deep Red (Life Technologies) after staining surface markers. Flow cytometry data were acquired on LSRII or LSR Fortessa (BD Biosciences) and analyzed using Flowjo software (Tree Star).
Cell purification and culture
Lymphocytes were isolated from lymphoid organs (spleen and peripheral lymph nodes that included inguinal, auxiliary and cervical lymph nodes) and naïve and Treg cells were sorted on a MoFlow (Beckman-Coulter) or Reflection (i-Cyt). Sorted naïve (CD4+CD62L+CD44loCD25−) and Treg cells were used for in vitro culture in Click’s medium (plus β-mercaptoethanol) supplemented with 10% (vol/vol) FBS and 1% (vol/vol) penicillin-streptomycin. Tregs were activated with plate-coated anti-CD3 (5 μg/ml; 2C11; Bio X Cell), anti-CD28 (5 μg/ml; 37.51; Bio X Cell) and human IL-2 (200 U/ml), in the presence or absence of simvastatin (2 μM; EMD Millipore), or 25-Hydroxycholesterol (0.5 μg/ml; Sigma). Thymidine incorporation assays were performed by adding 0.2 × 105 T cells and 1 × 105 irradiated splenocytes together with soluble anti-CD3 (0.2 μg/ml) in the presence or absence of human IL-2 (100 U/ml) for 72 hours, followed by pulsing with [3H]-thymidine at 1 µCi per well for the last 12–16 hours of culture. For labeling with CFSE and Celltrace violet (both from Life Technologies), T cells were incubated in Click’s medium with 5% FBS and 4 μM of either dye at 37°C for 25 minutes, followed by extensive washes. In vitro Treg suppression assays were performed as described7. Briefly, freshly sorted Teff (CD4+CD45RBhiCD25−; 5 × 104) and Treg cells (at different ratios with Teff) were cultured in 96-well plates along with anti-CD3 antibody and irradiated splenocytes for 72 hours, followed by Thymidine incorporation assays. For Treg suppression assays using previously activated Tregs, Tregs were sorted from C57BL/6J mice and activated with anti-CD3, anti-CD28 and IL-2 for 3 days in the absence or presence of simvastatin (2 μM), 25-hydroxycholesterol (0.5 μg/ml), 22(R)-hydroxycholesterol (5 μM; Sigma), atorvastatin (5 μM; EMD Millipore), lovastatin (5 μM; EMD Millipore), mevalonate (100 μM; Sigma), or certain combinations as specified; vehicle control did not have measurable effects (not shown). Viable Tregs were purified using Ficoll and incubated with freshly isolated Teff cells for additional analysis as described above.
Colitis model
A total of 4 × 105 Teff cells (CD4+CD45RBhiCD25−) from CD45.1+ mice were mixed with 2 × 105 wild-type or Raptor-deficient Tregs (from Foxp3creRaptorfl/fl or Cd4creRaptorfl/fl mice), and were transferred intraperitoneally into Rag1−/− mice. Mice were assessed for clinical signs of colitis weekly and were analyzed 8–10 weeks after transfer. Colons were fixed in 10% (vol/vol) neutral buffered formalin, and sectioned at 4 μm thickness. Colon pathology was assigned scores by an experienced pathologist (P. V.) as described7. Lymphocytes were isolated from spleen and mesenteric lymph nodes and analyzed by flow cytometry.
De novo lipogenesis, cholesterol measurement and bioenergetics assays
Tregs were activated with anti-CD3, anti-CD28 plus IL-2 for 20 hours, and [1-14C]-acetic acid (Perkin Elmer; 4 μCi/ml) was added for additional 4 hours of culture. For labeling with [U-14C]-glucose (American radiolabeled chemicals), Tregs activated for 20 hours were washed with PBS and incubated in glucose-free medium with dialyzed FBS and [U-14C]-glucose (4 μCi/ml) for additional 4 hours of culture. Following incubation, cells were collected, washed twice with PBS and lysed in 0.5% Triton X-100. Lipids were extracted by the addition of chloroform and methanol mixture (2:1 vol/vol) with vortexing, followed by the addition of water with vortexing. After centrifugation, the lipid-containing phase (at the bottom) was separated and 14C incorporation was measured using a Beckman LS6500 scintillation counter. Results are normalized to cell number. Cellular cholesterol level was measured using Amplex Red assay (Life Technologies). Briefly, cells were washed with PBS and lysed in the Amplex Red reaction buffer. After 15 minute of incubation, cell lysates were centrifuged at maximum speed for 5 minutes. Fifty μl of supernatants were pipetted into a 96-well tissue culture plate and a 50 ul aliquot of Amplex Red working solution was added to each well. The plate was incubated for 120 minutes at 37°C, protected from light. Fluorescence was subsequently measured on a fluorescence microplate reader (Molecular Devices). A cholesterol standard curve was determined for each plate using a cholesterol reference standard. The bioenergetic activities of the ECAR and OCR pathways were measured using the Seahorse XF24-3 extracellular flux analyzer per manufacturer’s instructions (Seahorse Bioscience).
Gene expression profiling and gene-set enrichment analysis
RNA samples from freshly isolated Tregs from wild-type and Cd4creRaptorfl/fl mice were analyzed using the Affymetrix HT_MG-430_PM GeneTitan peg array and expression signals were summarized using the RMA algorithm (Affymetrix Expression Console v1.1). Gene-set enrichment analysis within canonical pathways was performed as described32.
RNA and immunoblot analysis
Real-time PCR analysis was done as described with primers and probe sets from Applied Biosystems7, or using Power SYBR Green Master Mix from Life Technologies. Immunoblots were performed and quantified as described previously31, using the following antibodies: p-S6, p-Akt Ser473, p-4E-BP1, p-Foxo1/3, Raptor (all from Cell Signaling Technology) and β-actin (Sigma).
Statistical analysis
P values were calculated with Student’s t test, Mann Whitney test, or ANOVA (GraphPad Prism) as specified in figure legends, with proper post-test analysis performed. Statistical analysis of mouse survival and respective P values were determined using the Logrank test. *, P < 0.05; **, P <0.01; ***, P < 0.001; ns, not significant.
Supplementary Material
Acknowledgements
The authors acknowledge A. Rudensky for Foxp3YFP-Cre mice, D. Green for help with metabolic assays, N. Brydon for animal colony management, Y. Wang for editing of the manuscript, and St. Jude Immunology FACS core facility for cell sorting. This work was supported by US National Institutes of Health (K01 AR053573, R21 AI094089 and R01 NS064599), the Lupus Research Institute, and the American Lebanese Syrian Associated Charities (all to H.C.).
Footnotes
Author Contributions. H.Z. designed and performed experiments, and wrote the manuscript; K.Y. contributed to cellular experiments; C.C. contributed to survival curves and technical support; G.N. performed bioinformatic analyses; P.V. performed histological analysis; H.C designed experiments, contributed to writing the manuscript, and provided overall direction.
The authors declare no competing financial interests.
REFERENCES
- 1.Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol. 2012;12:325–338. doi: 10.1038/nri3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Delgoffe GM, et al. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol. 2011;12:295–303. doi: 10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee K, et al. Mammalian target of rapamycin protein complex 2 regulates differentiation of Th1 and Th2 cell subsets via distinct signaling pathways. Immunity. 2010;32:743–753. doi: 10.1016/j.immuni.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delgoffe GM, et al. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 2009;30:832–844. doi: 10.1016/j.immuni.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haxhinasto S, Mathis D, Benoist C. The AKT-mTOR axis regulates de novo differentiation of CD4+Foxp3+ cells. J Exp Med. 2008;205:565–574. doi: 10.1084/jem.20071477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sauer S, et al. T cell receptor signaling controls Foxp3 expression via PI3K, Akt, and mTOR. Proc Natl Acad Sci U S A. 2008;105:7797–7802. doi: 10.1073/pnas.0800928105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu G, et al. The receptor S1P1 overrides regulatory T cell-mediated immune suppression through Akt-mTOR. Nat Immunol. 2009;10:769–777. doi: 10.1038/ni.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu G, Yang K, Burns S, Shrestha S, Chi H. The S1P(1)-mTOR axis directs the reciprocal differentiation of T(H)1 and T(reg) cells. Nat Immunol. 2010;11:1047–1056. doi: 10.1038/ni.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Battaglia M, Stabilini A, Roncarolo MG. Rapamycin selectively expands CD4+CD25+FoxP3+ regulatory T cells. Blood. 2005;105:4743–4748. doi: 10.1182/blood-2004-10-3932. [DOI] [PubMed] [Google Scholar]
- 10.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Procaccini C, et al. An oscillatory switch in mTOR kinase activity sets regulatory T cell responsiveness. Immunity. 2010;33:929–941. doi: 10.1016/j.immuni.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelly AP, et al. Notch-induced T cell development requires phosphoinositidedependent kinase 1. The EMBO Journal. 2007;26:3441–3450. doi: 10.1038/sj.emboj.7601761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cunningham JT, et al. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- 14.van der Windt GJ, et al. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity. 2012;36:68–78. doi: 10.1016/j.immuni.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gurumurthy S, et al. The Lkb1 metabolic sensor maintains haematopoietic stem cell survival. Nature. 2010;468:659–663. doi: 10.1038/nature09572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi T, et al. Immunologic self-tolerance maintained by CD25+CD4+ naturally anergic and suppressive T cells: induction of autoimmune disease by breaking their anergic/suppressive state. Int Immunol. 1998;10:1969–1980. doi: 10.1093/intimm/10.12.1969. [DOI] [PubMed] [Google Scholar]
- 17.Fisson S, et al. Continuous activation of autoreactive CD4+ CD25+ regulatory T cells in the steady state. J Exp Med. 2003;198:737–746. doi: 10.1084/jem.20030686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gavin MA, Clarke SR, Negrou E, Gallegos A, Rudensky A. Homeostasis and anergy of CD4(+)CD25(+) suppressor T cells in vivo. Nat Immunol. 2002;3:33–41. doi: 10.1038/ni743. [DOI] [PubMed] [Google Scholar]
- 19.Thornton AM, Donovan EE, Piccirillo CA, Shevach EM. Cutting edge: IL-2 is critically required for the in vitro activation of CD4+CD25+ T cell suppressor function. J Immunol. 2004;172:6519–6523. doi: 10.4049/jimmunol.172.11.6519. [DOI] [PubMed] [Google Scholar]
- 20.Herman AE, Freeman GJ, Mathis D, Benoist C. CD4+CD25+ T regulatory cells dependent on ICOS promote regulation of effector cells in the prediabetic lesion. J Exp Med. 2004;199:1479–1489. doi: 10.1084/jem.20040179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wing K, et al. CTLA-4 control over Foxp3+ regulatory T cell function. Science. 2008;322:271–275. doi: 10.1126/science.1160062. [DOI] [PubMed] [Google Scholar]
- 22.Rubtsov YP, et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 23.Kanangat S, et al. Disease in the scurfy (sf) mouse is associated with overexpression of cytokine genes. Eur J Immunol. 1996;26:161–165. doi: 10.1002/eji.1830260125. [DOI] [PubMed] [Google Scholar]
- 24.Chaudhry A, et al. Interleukin-10 signaling in regulatory T cells is required for suppression of Th17 cell-mediated inflammation. Immunity. 2011;34:566–578. doi: 10.1016/j.immuni.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol. 2012;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ouyang W, et al. Novel Foxo1-dependent transcriptional programs control T(reg) cell function. Nature. 2012;.491:554–559. doi: 10.1038/nature11581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang R, Green DR. Metabolic checkpoints in activated T cells. Nat Immunol. 2012;13:907–915. doi: 10.1038/ni.2386. [DOI] [PubMed] [Google Scholar]
- 28.Tai X, et al. Basis of CTLA-4 function in regulatory and conventional CD4(+) T cells. Blood. 2012;119:5155–5163. doi: 10.1182/blood-2011-11-388918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corse E, Allison JP. Cutting edge: CTLA-4 on effector T cells inhibits in trans. J Immunol. 2012;189:1123–1127. doi: 10.4049/jimmunol.1200695. [DOI] [PubMed] [Google Scholar]
- 30.Sadlack B, et al. Ulcerative colitis-like disease in mice with a disrupted interleukin-2 gene. Cell. 1993;75:253–261. doi: 10.1016/0092-8674(93)80067-o. [DOI] [PubMed] [Google Scholar]
- 31.Yang K, Neale G, Green DR, He W, Chi H. The tumor suppressor Tsc1 enforces quiescence of naive T cells to promote immune homeostasis and function. Nat Immunol. 2011;12:888–897. doi: 10.1038/ni.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.