SUMMARY
The trypanosomes cause two neglected tropical diseases, Chagas disease in the Americas and African trypanosomiasis in sub-Saharan Africa. Over recent years a raft of molecular tools have been developed enabling the genetic dissection of many aspects of trypanosome biology, including the mechanisms underlying resistance to some of the current clinical and veterinary drugs. This has led to the identification and characterization of key resistance determinants, including transporters for the anti-Trypanosoma brucei drugs, melarsoprol, pentamidine and eflornithine, and the activator of nifurtimox-benznidazole, the anti-Trypanosoma cruzi drugs. More recently, advances in sequencing technology, combined with the development of RNA interference libraries in the clinically relevant bloodstream form of T. brucei have led to an exponential increase in the number of proteins known to interact either directly or indirectly with the anti-trypanosomal drugs. In this review, we discuss these findings and the technological developments that are set to further revolutionise our understanding of drug-trypanosome interactions. The new knowledge gained should inform the development of novel interventions against the devastating diseases caused by these parasites.
Key words: Trypanosoma brucei, Trypanosoma cruzi, drug resistance, RNA interference
INTRODUCTION
The trypanosomatids belong to the order Kinetoplastida and include the parasitic protozoa Trypanosoma and Leishmania (the latter are considered elsewhere in this special issue of Parasitology). The trypanosomes represent a significant threat to human and animal health, primarily impacting populations in sub-Saharan Africa and Latin America. There is a limited set of drugs available to treat the diseases caused by these parasites (Wilkinson and Kelly, 2009; Simarro et al. 2012b), few new drugs in development and evidence that drug resistance among both animal and human-infective trypanosomes is a significant problem (Geerts et al. 2001; Barrett et al. 2011). Therefore, understanding the mechanisms by which parasites can and have become resistant to the available drugs is of paramount importance. This review presents the approaches used to dissect the mechanisms of drug resistance in trypanosomes, and how recently developed high throughput techniques are contributing to this process and identifying factors responsible for drug efficacy.
African trypanosomiasis
The African trypanosomes, transmitted by tsetse flies, are extracellular parasites responsible for human African trypanosomiasis (HAT) (Brun et al. 2010). They have a broad host range, also causing disease in other mammals. Infections with Trypanosoma congolense, T. vivax and T. brucei in wild ungulates (‘hoofed mammal’) result in mild symptoms, while domestic livestock suffer a progressive wasting disease known as Nagana, making stock farming challenging in sub-Saharan Africa (Steverding, 2008). HAT is caused by T. brucei gambiense and T. brucei rhodesiense, with the former causing at least 95% of reported cases. Both forms of HAT progress through two stages. In stage one, the parasites spread through the haemo-lymphatic system from the site of the tsetse bite. In stage two, parasites cross the blood brain barrier and establish an infection in the central nervous system (CNS), typically causing death if the patient remains untreated (Brun et al. 2010). However, a recent report demonstrated parasite clearance and declining serological response in some individuals even in the absence of treatment, though the prevalence of this apparent trypanotolerance in the human population is unknown (Jamonneau et al. 2012). The latest estimates indicate that more than 69 million people in sub-Saharan Africa live in known HAT-endemic areas, and of these, more than five million are at high risk of contracting HAT (Simarro et al. 2012a). Increased case surveillance and improved access to anti-HAT drugs over recent years has led to a significant drop in disease incidence (Brun et al. 2010). Indeed, since 2009 fewer than 10 000 new cases per annum have been reported, with less than 7000 cases reported in 2011 (Simarro et al. 2012a), though this is likely to be a substantial underestimate due in part to misdiagnosis and limited access to health-care in some areas (Odiit et al. 2005; Matemba et al. 2010; Mumba et al. 2011).
The necessity for highly co-ordinated efforts to control HAT means disease incidence is vulnerable to the impact of civil conflict and the subsequent breakdown of health-care infrastructure and control programmes; disease incidence reached a historical low in the 1960s, but the cessation of surveillance activities and the outbreak of post-independence conflicts saw a resurgence in HAT (Berrang Ford, 2007), peaking in the late 1990s (Brun et al. 2010). Recent findings suggest that the current decline in HAT incidence has already led to a concomitant decrease in control efforts in some areas, which could lead to disease resurgence (Ruiz-Postigo et al. 2012). The downward trend is also vulnerable to the development and spread of parasite resistance to the available drugs.
Chagas disease
Trypanosoma cruzi is the causative agent of Chagas disease, the most important parasitic infection in the Americas. Blood-sucking triatomine bugs are the main vectors of transmission, although ingestion of contaminated food and drink, organ transplantation, blood transfusion and the congenital route also result in a significant number of cases. More than 10 million people in Latin America are estimated to be infected with T. cruzi, resulting in 10–20 000 deaths annually (Moncayo and Silveira, 2009). As a result of migration, the disease is also becoming a global public health issue. For example, in the USA there are thought to be 300 000 individuals with undiagnosed Chagas disease (Bern and Montgomery, 2009). In Europe, there have been 4000 confirmed cases in the last 10 years, and more than 80 000 people are estimated to be infected (WHO, 2009).
There are three distinct phases to Chagas disease; acute, indeterminate and chronic. The ‘acute’ stage, which occurs 1–3 weeks after infection, is often asymptomatic. However in children, the outcome can be more serious, with death in 5% of diagnosed cases, mainly due to myocarditis or meningoencephalitis. In most instances, parasitaemia is suppressed following the development of a cellular immune response, although this does not give rise to sterile immunity. Individuals in this ‘indeterminate’ stage remain a source of infection throughout their lives. In about 30% of cases, the disease progresses from the ‘indeterminate’ to the ‘chronic’ stage, sometimes decades after the primary infection (Kirchhoff, 2011), leading to clinical outcomes such as cardiomyopathy, alimentary tract pathology (typically, megacolon and megaoesophagus) and/or damage to the peripheral nervous system. In several regions of South America, Chagasic heart disease is a common cause of sudden cardiac failure. Co-infections with HIV can lead to activation of chronic Chagas disease, often with atypical clinical manifestations, including CNS involvement (Diazgranados et al. 2009). Chagas disease presents significant challenges in terms of diagnosis, vector control and treatment, and the situation is further complicated by the development of resistance to the available drugs.
CHEMOTHERAPY
It is unlikely that a vaccine will be developed against any human trypanosome in the near future. In the case of HAT, this is primarily due to antigenic variation of the surface coat, enabling T. brucei to evade the immune response until the host succumbs (Horn and McCulloch, 2010). T. cruzi vaccine development shows some promise (Cazorla et al. 2009), but it has been notoriously challenging to develop any anti-protozoal vaccine. Therefore, public health measures have focused on the insect vectors, making a significant contribution to the control of Chagas disease and HAT (Hashimoto and Schofield, 2012; Welburn and Maudlin, 2012). Despite this, there have been concerns about the sustainability of the programmes. Hence, though limited by issues of specificity, toxicity and developing parasite resistance, chemotherapy is fundamental to the control of African trypanosomiasis and Chagas disease.
African trypanosomiasis
There are five drugs currently in use for the treatment of HAT; their application is dependent upon disease stage and the identity of the infecting subspecies (Table 1). Stage one T. b. gambiense and T. b. rhodesiense HAT are treated with pentamidine or suramin, respectively; both drugs have been in constant use for decades – suramin was developed in 1916 and pentamidine in 1937 (Steverding, 2008). Since 1990 and until recently, eflornithine has been used as a monotherapy to treat stage two T. b. gambiense HAT. It is now recommended to be used as part of NECT (nifurtimox-eflornithine combination therapy), which has equivalent therapeutic outcomes to eflornithine monotherapy, but allows for reduced dosing resulting in greater patient compliance (Priotto et al. 2009).
Table 1.
Current drugs – their application, and resistance mechanisms identified by low throughput analysis of laboratory-derived and clinical resistant isolates
| Drug | Stage/Trypanosome | Resistance determinant(s) laboratory | Clinical |
|---|---|---|---|
| Berenil | T. congolense, T. vivax, T. b. brucei |
|
AT1 |
| Isometamidium |
|
nd | |
| Homidium |
|
nd | |
| Suramin | Stage 1 T. b. rhodesiense |
|
ndc |
| Pentamidine | Stage 1 T. b. gambiense |
|
ndd |
| Melarsoprol | Stage 2 T. brucei spp. |
|
AT1e |
| Eflornithine | Stage 2 T. b. gambiense |
|
ndf |
| Nifurtimox | T. cruzi, stage 2 T. b. gambiense |
|
NTR |
| Benznidazole | T. cruzi |
|
NTR |
Notes
Not determined.
Following selection, a suramin-resistant phenotype was expressed in bloodstream but not insect stage parasites (Scott et al. 1996).
Treatment failures reported in west Africa in the 1950s (Pepin and Milord, 1994).
Pentamidine resistant T. brucei can be generated in the laboratory, but they have not been reported in the field (Barrett et al. 2011).
Mutation or loss of AT1 renders T. brucei less sensitive to melarsoprol, however not all resistant clinical isolates have modified this locus (Matovu et al. 2001).
Melarsoprol, which has been in use since 1949, is the only drug effective against both forms of HAT during stage two disease, though its use can lead to a devastating reactive encephalitis in 5–10% of cases. This is thought to be due to massive release of parasite antigens and a subsequent autoimmune reaction (Pepin and Milord, 1994). Until the introduction of eflornithine and NECT for the treatment of T. b. gambiense HAT, melarsoprol was the only drug effective against either late stage HAT. Since these alternative treatments have become available, melarsoprol use has declined, being used in only 12% of reported cases in 2010 (Simarro et al. 2012b). However, melarsoprol is still the only available treatment for late stage T. b. rhodesiense HAT. All the licensed anti-HAT drugs require administration in hospital and come with significant side effects (Barrett and Croft, 2012). There may soon be new chemotherapeutic options, with fexnidazole, an orally active nitroimidazole, and the benzoxaborole, SCYX-7158, about to enter phase II and phase I clinical trials, respectively (Jacobs et al. 2011; Barrett and Croft, 2012). In addition, novel diamidines are showing early promise in animal models of HAT (Thuita et al. 2012).
As with HAT, the range of drugs for the treatment of Nagana is limited and resistant parasites are widespread. There are three drugs currently used to treat Nagana, all of which have been in use for more than 50 years: homidium bromide and the related isometamidium chloride, and diminazene aceturate (berenil). The widespread and often unregulated use of these drugs, both as treatments and in prophylaxis, has inevitably led to the development of drug resistance (Geerts et al. 2001). This is a particularly acute problem in highly endemic areas with very high drug use (Delespaux and de Koning, 2007).
Chagas disease
For more than 40 years, the nitroheterocyclic agents, nifurtimox and benznidazole, have been the front line drugs for the treatment of Chagas disease (Wilkinson and Kelly, 2009). However, these drugs are far from optimal. They are effective against infections in the acute phase, but their usefulness in preventing or alleviating symptoms in the chronic stage remains controversial (Marin-Neto et al. 2008; Urbina, 2010). Both drugs have been reported to be carcinogenic and display a wide range of side effects which include CNS toxicity, leukopenia, muscle weakness and severe dermatitis (Castro et al. 2006; Wilkinson and Kelly, 2009). This, coupled with treatment regimes that extend over several months, frequently results in a failure to complete the therapeutic schedule. T. cruzi strains refractory to treatment are encountered throughout South America (Castro et al. 2006), though the extent to which this reflects acquired resistance or natural variation in sensitivity is unknown.
The large number of infected individuals throughout Latin America and beyond makes the development of new therapies to treat Chagas disease a research priority. A major question is whether drugs targeted at the parasite are the best strategy for treating the chronic stage, given the numerous reports that autoimmune responses could be a significant determinant of disease pathology. However, recent studies using murine models have shown that the continued presence of the parasite is both necessary and essential for development of cardiac disease (Tarleton et al. 1997; Tarleton, 2003; Garcia et al. 2005). Consistent with this, antiparasitic drugs can block chronic cardiomyopathy, and give rise to a stable protective T cell memory (Bustamante et al. 2008). Taken together, these reports suggest that chemotherapeutic intervention is appropriate, even against the chronic stage of the disease.
The research community is taking a twin-track approach to improving therapy against Chagas disease. Firstly, as outlined below, there has been a drive to identify resistance mechanisms so that treatment regimes based on current drugs can be optimized. Secondly, the development of new drugs is being widely pursued (Wilkinson and Kelly, 2009; Le Loup et al. 2011). Promising drug targets currently under investigation include enzymes of the ergosterol biosynthetic pathway (Buckner et al. 2003; Suryadevara et al. 2009; Urbina, 2009) and the cathepsin L-like cysteine protease, cruzipain, with several inhibitors at various stages of development (McKerrow et al. 2009; Beaulieu et al. 2010). Two anti-fungal triazole compounds, posoconazole and E1224 (inhibitors of lanosterol 14 α-demethylase, CYP51), are presently undergoing clinical trials (Barrett and Croft, 2012).
Are treatment failures associated with drug resistance?
Treatment failures have been reported for all of the currently available HAT monotherapies, though in the case of pentamidine these are rare (Barrett et al. 2011). Difficulties encountered in the staging of HAT infections and the subsequent administration of chemotherapy, mean that it is often difficult to ascribe these failures to the presence of bona fide drug-resistant parasites. However, data derived from experiments on drug-resistant laboratory strains of T. brucei enabled genotypic analysis of parasites from relapse patients in Uganda, indicating that at least some melarsoprol treatment failures are due to the development of resistance (Matovu et al. 2001). Until the 1950s suramin was used to treat T. b. gambiense HAT, since when its use has been restricted to treating T. b. rhodesiense HAT, in part due to a large number of reported treatment failures in West Africa. More recently, there have been a number of anecdotal reports of eflornithine monotherapy treatment failures, though it is not known if these were due to the presence of eflornithine-resistant parasites (Barrett et al. 2011). NECT has only been in use since 2009, however its use against T. b. gambiense HAT is now widespread, with 59% of all second stage cases treated with this combination therapy in 2010 (Simarro et al. 2012b), and there are already reports of small numbers of relapses (Franco et al. 2012). It is not known whether these were due to the presence of drug-resistant trypanosomes, although this is a possibility, given the ease with which parasites resistant to either drug can be selected in the laboratory (see below).
Understanding how trypanosomes can become resistant to the available drugs enables the development of diagnostic tools for the identification of genuinely resistant parasites in the field. Indeed, genetic assays designed to identify the presence of known determinants of drug resistance have revealed high prevalence of resistance to berenil amongst T. congolense isolated from cattle and game animals in endemic areas of Ethiopia and Southern Africa (Chitanga et al. 2011; Moti et al. 2012).
Distinguishing between treatment failure due to drug resistance or for other reasons is even more challenging in the case of Chagas disease. The toxicity of the available drugs in combination with the need for prolonged treatment regimes has an inevitable impact on patient compliance, providing a selective environment for the development of drug resistant parasites (see above).
IDENTIFYING RESISTANCE MECHANISMS – LOW THROUGHPUT APPROACHES
Trypanosomes resistant to each of the licensed HAT and Chagas disease monotherapies have been derived by in vitro or in vivo selection in the laboratory. With the exception of suramin resistance (Scott et al. 1996), it has been possible to define the underlying mechanism in each case (Table 1). Biochemical assays, inspired by insights into the chemistry of the compound under investigation, have been used to define the character of the resistance mechanism. Using various genetic approaches it has been possible to identify the gene products responsible for some of the observed resistance phenotypes. This broad approach was used to identify the genes encoding the trypanosome P2 adenosine transporter, AT1 (Carter and Fairlamb, 1993; Maser et al. 1999), and the amino acid transporter, AAT6 (Vincent et al. 2010), as key determinants of melarsoprol and eflornithine uptake, respectively, in T. brucei. Candidate-based reverse genetic approaches have also identified potential drug resistance determinants, including the melarsoprol-trypanothione (Mel-T) transporter, multidrug resistance protein A (MRPA) (Shahi et al. 2002), and two nucleobase transporters related to AT1, NT11·1 and NT12·1, capable of taking up pentamidine (Ortiz et al. 2009). A similar methodology identified the trypanosome nitroreductase (NTR), as the activator of the pro-drugs nifurtimox and benznidazole in T. cruzi (Wilkinson et al. 2008). These studies were facilitated by the sequencing of the trypanosome genomes (Berriman et al. 2005; El-Sayed et al. 2005), the development of reverse genetic tools, such as stable transfection, inducible protein expression and RNA interference (RNAi) systems (Lee and Van der Ploeg, 1990; Wirtz and Clayton, 1995; Wirtz et al. 1999; LaCount et al. 2000; Wang et al. 2000; Alsford et al. 2005b; Taylor and Kelly, 2006; Alsford and Horn, 2008), and early forward genetic tools, such as trypanosome cDNA and genomic DNA libraries (Maser et al. 1999; Shahi et al. 2002).
Characterizing melarsoprol uptake and efflux
Melarsoprol is a melaminophenyl arsenical developed in the 1940s (Friedheim, 1949) and thought to act via the formation of Mel-T, a toxic adduct with the trypanosome-specific thiol, trypanothione (Fairlamb et al. 1989; Alsford et al. 2012). Melarsoprol contains a melamine ring, a nitrogen-rich heterocycle with similarities to several natural metabolites, an observation that led to the ‘melamine receptor’ hypothesis (Barrett and Fairlamb, 1999). Assessing the ability of more than 100 nitrogen-containing biochemicals to inhibit cell lysis by melarsoprol revealed adenine and adenosine as potent inhibitors, and subsequent biochemical characterization identified the P2 adenosine transporter as a mediator of melarsoprol uptake (Carter and Fairlamb, 1993). This work was done prior to the sequencing of the T. brucei genome, at a time when the identification of the proteins responsible for a particular phenotype could be a laborious process. Mäser and colleagues came up with a creative solution. Saccharomyces cerevisiae is normally incapable of taking up adenosine, so they reasoned that expression of the trypanosome transporter in yeast defective for adenosine biogenesis would enable their survival. The mutant yeast was transformed with a T. brucei cDNA library and grown on media supplemented with adenosine, and cells expressing the trypanosome P2 purine transporter, AT1, were able to grow (Maser et al. 1999).
AT1 was subsequently found to be mutated or absent from a number of melarsoprol-resistant strains (Maser et al. 1999; Matovu et al. 2001; Stewart et al. 2010). However, loss of AT1 function was only seen in some isolates from melarsoprol-treated relapse patients (Matovu et al. 2001). It was subsequently found that AT1 gene deletion confers only a two-fold decrease in melarsoprol sensitivity (Matovu et al. 2003), suggesting that an additional or alternative factor must be driving high level resistance. Contemporary work on drug resistance in Leishmania tarentolae identified the ABC transporter, LtMRPA, as contributing to resistance to trivalent antimony via efflux of the metal-trypanothione conjugate (Legare et al. 2001). It was proposed that T. brucei might be able to remove Mel-T using a similar transporter. The T. brucei orthologue of LtMRPA was isolated by screening a genomic DNA library with a known T. brucei ABC transporter sequence, and its over-expression led to a ten-fold increase in melarsoprol EC50 in vitro (Shahi et al. 2002). However, MRPA over-expression has not been found in melarsoprol-resistant clinical isolates (Alibu et al. 2006), so its contribution to treatment failures in the field remains equivocal.
Soon after the identification of the role of AT1 in the uptake of melarsoprol, it was shown that this transporter also contributed to the uptake of pentamidine and other diamidines, such as the anti-nagana drug, berenil (Barrett et al. 1995; Carter et al. 1995; de Koning et al. 2004). Two other uptake mechanisms distinct from AT1 have been biochemically characterized, the high and low affinity pentamidine transporters (or HAPT and LAPT) (de Koning 2001, 2008; Bridges et al. 2007). The two AT1-related nucleobase transporters, NT11·1 and NT12·1, have been shown to be capable of pentamidine uptake in T. brucei, though neither seems to correspond to HAPT or LAPT (Ortiz et al. 2009). Therefore, pentamidine may be able to access the T. brucei interior via multiple routes, possibly explaining why treatment failures with this drug are rare. Those that do occur may arise from factors other than changes in pentamidine uptake by the infecting parasite.
Identifying the eflornithine transporter
Eflornithine, an analogue of ornithine, blocks spermidine synthesis and the formation of trypanothione through the inhibition of ornithine decarboxylase (ODC) (Bacchi et al. 1983), which normally catalyses the conversion of ornithine to putrescine. It is effective against T. b. gambiense, but T. b. rhodesiense is naturally tolerant to eflornithine as a result of several factors; these include reduced drug uptake, increased putrescine uptake or higher ODC turnover, depending on the isolate analysed (Bacchi et al. 1993; Iten et al. 1997). The existence of T. b. gambiense resistant to eflornithine has yet to be confirmed in the field, in spite of recent anecdotal reports of monotherapy treatment failures (Barrett et al. 2011). It should be noted that eflornithine use has only become widespread since the introduction of NECT (Priotto et al. 2009; Simarro et al. 2012b). Indeed, eflornithine-resistant trypanosomes can be generated easily in the laboratory, and cells exhibiting significantly reduced drug uptake were derived before the drug was widely used (Phillips and Wang, 1987). However, it was not until 2010 that the resistance mechanism was defined (Vincent et al. 2010). By this time, significant advances had been made in the field of metabolomics, enabling the quantification of multiple metabolites through time in treated and untreated cells (Creek et al. 2012). This led to the observation that, while the levels of spermidine and the intermediates of its synthesis were no different in resistant and sensitive cell lines, eflornithine accumulation was markedly reduced in the resistant line (Vincent et al. 2010). A systematic analysis of the amino acid transporters in the eflornithine-resistant line revealed that the AAT6 transporter had been lost from these cells. Its key role in eflornithine uptake was confirmed using RNAi knockdown (Vincent et al. 2010).
The nifurtimox-benznidazole activator
The genetic tools applicable to T. cruzi include a wide range of episomal expression vectors and robust methodology that allows targeted gene deletion (Taylor et al. 2011). However, T. cruzi reverse genetics procedures are generally less flexible than those of T. brucei, mainly due to the length of time required for the isolation of transformed parasites (∼4 weeks as opposed to 5 days) and the lack of RNAi-based systems. T. cruzi is unable to generate an RNAi response since genes encoding the Argonaute and DICER-like proteins are absent (Lye et al. 2010). Evidence suggests they were lost after divergence of the two trypanosome species. Despite these limitations, the available tools have been experimentally crucial in increasing our understanding of T. cruzi infection biology and the mechanisms of drug activity and resistance.
Nifurtimox and benznidazole are nitroheterocyclic compounds in which a nitro-group is linked to a furan and an imidazole, respectively (Fig. 1). Both are pro-drugs, but the specific NTRs required for their activation and the nature of the resulting toxic metabolites had remained unresolved, despite being a major focus of research for more than 40 years. With nifurtimox, early experiments had suggested that trypanocidal activity could arise from the generation of toxic oxygen metabolites, mediated by one-electron reduction of the drug by type II NTR activity (Docampo, 1990; Viode et al. 1999). In an aerobic environment, this results in a ‘futile cycling’ process in which NADPH is consumed, superoxide radicals produced and nifurtimox regenerated. However, parasites engineered to have an enhanced oxidative defence capacity were no more resistant to nifurtimox than wild type T. cruzi, suggesting that this process has limited in vivo significance (Taylor et al. 2011).
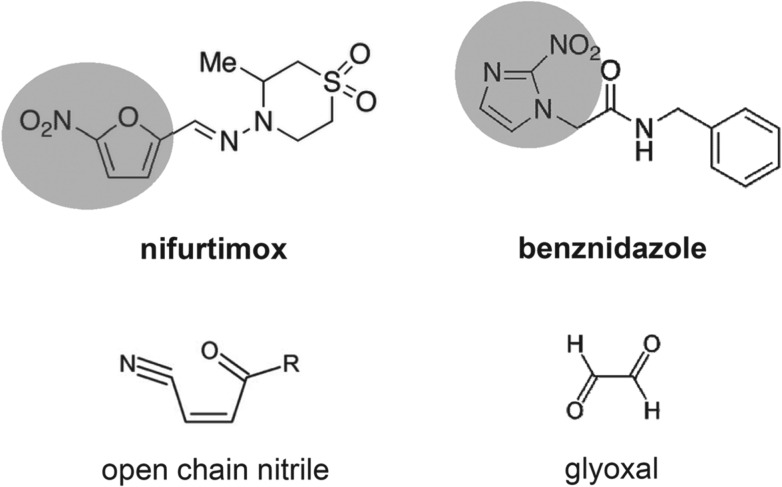
Fig. 1.
Structures of the nitroheterocyclic drugs used to treat T. cruzi infections and their toxic metabolites. The highlighted regions of nifurtimox and benznidazole correspond to the 5-nitrofuran and the 2-nitroimidazole groups, respectively. The structures of the toxic unsaturated open chain nitrile and glyoxal metabolites (Hall et al. 2011; Hall and Wilkinson, 2012) are shown below the corresponding drugs.
In bacteria, resistance to nitrofuran pro-drugs results from mutations in the flavin-dependent oxidoreductases of the type I NTR family which catalyse the O2-insensitive NAD(P)H-dependent two-electron reduction of the nitro group (Whiteway et al. 1998; Parkinson et al. 2000). This results in a hydroxylamine product, and ultimately the generation of nitrenium ions, which can cause lesions in chromosomal DNA and damage to other biological molecules (McCalla et al. 1971; Streeter and Hoener, 1988). A type I NTR-like protein was identified in the T. cruzi genome (Wilkinson et al. 2008). Biochemical analysis demonstrated that TcNTR could reduce nifurtimox, benznidazole and other nitroheterocyclic drugs in an NADH-dependent manner. TcNTR-mediated nifurtimox reduction leads to the production of an unsaturated open-chain nitrile (Fig. 1), the metabolite responsible for the trypanocidal activity (Hall et al. 2011). With benznidazole, reductive metabolism leads to the formation of glyoxal (Fig. 1), which has a range of cytotoxic properties (Hall and Wilkinson, 2012). Deletion of one copy of TcNTR results in 2–5 fold resistance against a range of nitroheterocyclics, presumably because of the reduced rate of drug metabolism (Wilkinson et al. 2008). The observation that heterozygotes do not display reduced infectivity, suggests a possible route for the development of acquired cross-resistance to this class of drugs. Further experimentation demonstrated that TcNTR null mutants have a much reduced capacity to infect mammalian cells and to divide as amastigotes, indicating an upper limit to clinically relevant resistance by this mechanism (Wilkinson et al. 2008; Mejia et al. 2012).
T. cruzi cross-resistant to nitroheterocyclic drugs are readily selectable in the laboratory. This can result from the loss of one copy of the entire chromosome containing the TcNTR gene, a mechanism that does not result in deleterious phenotypic consequences (Wilkinson et al. 2008; Mejia et al. 2012). Other routes to acquired resistance include point mutations in TcNTR that lead to inactivation of the protein product by disruption of FMN-binding capacity. The ease with which this can occur has been demonstrated in a report that three distinct TcNTR inactivating mutations arose independently within a single population undergoing benznidazole selection (Mejia et al. 2012). Although resistance linked to TcNTR is now well characterized, there is also evidence that other mechanisms can act on drug efficacy. For example, resistance to nifurtimox and benznidazole can occur independently, suggesting a non-involvement of TcNTR in these cases (Filardi and Brener, 1987). Similarly, a recent survey has found a 10-fold variation in benznidazole sensitivity in parasite isolates from a variety of biological and geographical backgrounds, which is not associated with differences in the sequence of the TcNTR genes (Mejia et al. 2012), although an association with the level of expression cannot be excluded. Identifying other possible mechanisms of resistance must be regarded as a priority for Chagas disease researchers.
T. brucei resistance to nitroheterocyclic pro-drugs, including nifurtimox and the clinical trial candidate fexnidazole, is also easily selected in the laboratory and is not associated with reduced virulence (Sokolova et al. 2010). Sensitivity to these drugs is also associated with activation by NTR in T. brucei (Wilkinson et al. 2008) and in Leishmania (Wyllie et al. 2012).
INCREASING THROUGHPUT – GENETIC SCREENS
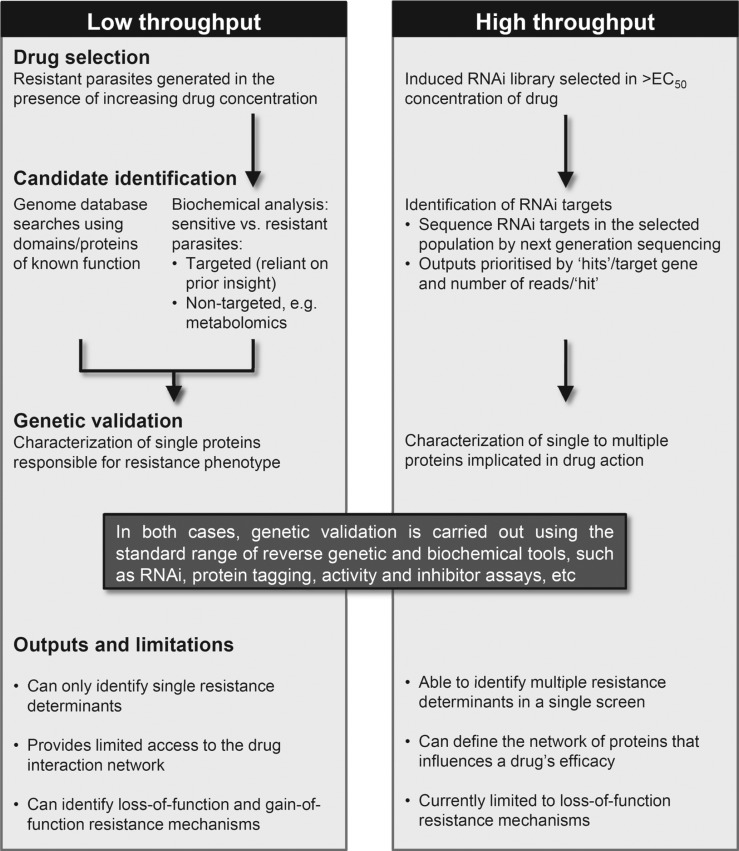
The ‘low throughput’ approaches described above have identified several resistance mechanisms, some of which have subsequently been found to be clinically important (Table 1). However, they are often dependent upon long-term selection of resistant parasites and prior knowledge enabling the identification of suitable candidate proteins. Also, they provide little access to the wider network of factors that influence drug efficacy. These limitations can be overcome by the development of high throughput systems that enable the contribution of every gene product to be simultaneously assessed (see Fig. 2 for a comparison of low and high throughput approaches). New high throughput approaches, as well as improved tools for low throughput analysis, have led to a significant growth in our understanding of drug-trypanosome interactions over recent years (Fig. 3A). This transition has accompanied the generation, release and exploitation of genome sequence data (Berriman et al. 2005; El-Sayed et al. 2005; Ivens et al. 2005). During this period, researchers have developed novel approaches to exploiting these data with impressive effect and these developments have already had a tremendous impact on our understanding of drug resistance in trypanosomes. Indeed, new genetic approaches have recently led to the identification of many genes associated with drug action and resistance phenotypes.
Fig. 2.
A comparison of low and high throughput approaches to understanding drug resistance in trypanosomes.
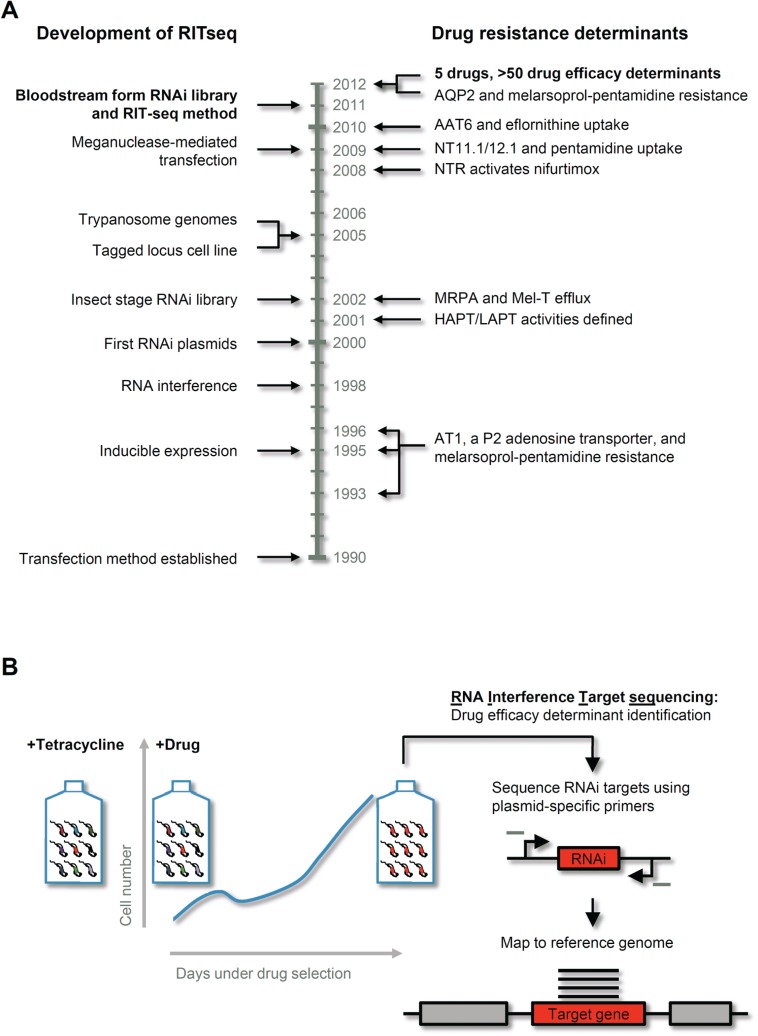
Fig. 3.
Developments in T. brucei molecular genetics and advances in the understanding of drug resistance in trypanosomes since 1990. (A) The development of the bloodstream form T. brucei RNAi library and the RIT-seq methodology was dependent upon a number of earlier methodological advances. Although concurrent progress was made in dissecting drug resistance mechanisms in trypanosomes, the establishment of a high throughput approach rapidly led to a significant increase in our understanding of both potential resistance mechanisms and the networks of proteins that influence drug efficacy. See text for abbreviations and references. (B) Drug selection of the tetracycline induced RNAi library can rapidly generate a resistant population. The RNAi targets, whose expression confers drug resistance, are sequenced using RNAi plasmid-specific primers (grey bars) on a next generation sequencing platform, such as Illumina, and mapped to the reference genome to identify candidate drug efficacy determinants.
Prior to sequencing of the trypanosome genomes, the power of the classical forward genetic screen was demonstrated when the gene encoding ODC was recovered by complementation of putrescine auxotrophy in odc mutant T. brucei (Sommer et al. 1996). This feasibility study suggested that cells exhibiting a phenotype of interest could be isolated from a mutagenized population and the mutagenized gene(s) could be recovered by complementation. Despite the promise of identifying novel genes that control important phenotypes, this form of forward genetic screening has not been widely used. In terms of drug resistance (and many other phenotype screens), this can be partly explained by the difficulty associated with complementation of the drug-resistance phenotype; gain-of-function would increase drug sensitivity, which cannot typically be selected for in the context of a complex population. Another challenge for mutagenesis screens is the typically low frequency of loss-of-heterozygocity in trypanosomes, since many phenotypes are likely to be mild when one allele remains intact in these diploid cells.
Transposon mutagenesis in trypanosomatids (Gueiros-Filho and Beverley, 1997) raised the possibility of loss-of-function and ‘signature-tagged mutagenesis’ in a single step. This approach was validated using a transposon mutagenesis screen for lectin (concanavalin A) resistance in T. brucei, which lead to the identification of ALG12, a gene involved in N-linked oligosaccharide synthesis (Leal et al. 2004). However, as above, the frequency of loss-of-heterozygosity may also represent a limiting factor for these screens.
RNA interference
Potent and specific genetic interference by double-stranded RNA (dsRNA), or RNA interference (RNAi), was first reported in the nematode, Caenorhabditis elegans (Fire et al. 1998). In the same year, dsRNA was shown to induce mRNA degradation in T. brucei (Ngo et al. 1998) and RNAi has subsequently had a huge impact on genetic studies in African trypanosomes. The genes encoding the RNAi machinery were found to be degenerate or absent in the T. cruzi and Leishmania major genomes, but L. braziliensis retains the machinery for RNAi, indicating that RNAi can be used as a genetic tool in some Leishmania species (Lye et al. 2010).
Several technical innovations and developments have helped to facilitate the wider use of RNAi technology for functional genomics in trypanosomes (Fig. 3A). One of the early innovations was the development of dsRNA inducible expression vectors with head-to-head or opposing promoters (LaCount et al. 2000; Wang et al. 2000); tools for inducible expression were developed previously (Wirtz and Clayton, 1995; Wirtz et al. 1999). This allowed the rapid assembly of multiple RNAi-targeting vectors, and several hundred T. brucei genes have now been analysed individually using such vectors, including the full complement of RNA polymerase II transcribed genes on chromosome 1 (Subramaniam et al. 2006).
RNA interference libraries
A particularly important innovation, made possible by the string of technical advances that went before (Fig. 3A), was the development of the T. brucei RNAi library. In fact, the first RNAi library screen in T. brucei was the first RNAi screen carried out in any organism (Morris et al. 2002). In this case, concanavalin-A resistance was linked to changes in glycolysis that affected cell-surface protein glycosylation status in procyclic, insect stage cells. This RNAi library has also subsequently been used to select for tubercidin-resistant cells, revealing hexose transporter knockdown and inhibition of glycolysis by this drug (Drew et al. 2003). The library has also been used to generate large numbers of individual RNAi strains which have been screened for a variety of phenotypes; see Zhao et al. (2008) for example. The potential for further drug-resistance screening was clearly demonstrated at this point in time, but there were some outstanding technical issues. One particularly prominent challenge was to transfer RNAi library screening from the insect-stage of T. brucei to the developmentally distinct and pathogenic bloodstream-form parasite.
One difficulty, which makes comparison among some T. brucei RNAi strains challenging, and may also be responsible for a number of ‘false negative’ or even ‘false positive’ assignments in terms of RNAi-associated phenotypes, is a phenomenon known as position effect. This comes about because dsRNA expression vectors are typically integrated at one of several related chromosomal loci, which support reproducible, but highly variable expression levels. One solution to this problem was to modify a single, expression-validated integration site (a non-transcribed ribosomal spacer) such that all expression vectors are specifically targeted to that site by homologous recombination (Alsford et al. 2005b). Another challenge was low transfection efficiency in bloodstream-form T. brucei, the stage where drug-resistance studies would typically be more informative. This hurdle was overcome by priming a chromosomal site for homologous recombination by meganuclease cleavage (Glover and Horn, 2009), or by using an improved transfection protocol and pooling many transformed populations (Schumann Burkard et al. 2011). Both approaches can now be used to generate genome-scale RNAi libraries, with the former approach also minimising the position effects described above. Thus, it is now possible to assemble bloodstream-form T. brucei RNAi libraries with approximately 10× genome coverage.
Proof-of-principle, in terms of screening for drug action and resistance mechanisms relevant to the bloodstream life-cycle stage, emerged in 2011. Drug transporters, AT1 and AAT6, and a drug activator, NTR, identified previously by other means (see above), were rapidly identified in RNAi library screens for melarsoprol, eflornithine, and nifurtimox-benznidazole resistance, respectively (Baker et al. 2011; Schumann Burkard et al. 2011). Drugs were no longer taken up by these cells or no longer activated once inside, due to the loss-of-function defects associated with inducible RNAi. Another challenge, in terms of whole-genome screening, was then to develop outputs that report phenotypes associated with multiple genes. An early feasibility study showed that it would be possible to report relative representations of large numbers of RNAi clones within complex populations, using a slot-blot or microarray read-out format, for example (Alsford et al. 2005a). However, DNA sequencing technology has now become the method of choice for generating complex genetic read-outs and this has been applied to genome-scale libraries using an approach called RNA interference target sequencing, or RIT-seq (Alsford et al. 2011). RIT-seq analysis, following RNAi screens for eflornithine, nifurtimox, melarsoprol, pentamidine and suramin resistance (Fig. 3B), revealed more than fifty new genes linked to the uptake or action of these drugs (Alsford et al. 2012), including aquaglyceroporin 2, the loss of which leads to melarsoprol-pentamidine cross-resistance (Baker et al. 2012). This approach can also be used to gain insights into the uptake and intracellular transit of drugs used against related parasites, such as T. cruzi and Leishmania, which while similar to T. brucei are genetically less tractable (Kolev et al. 2011). Indeed, the RNAi screen using nifurtimox implicated several factors, in addition to NTR (Alsford et al. 2012). More recently, initial RNAi screens using the anti-leishmanials, sodium stibogluconate, amphotericin-B, miltefosine and paromomycin, have identified several proteins already known to interact with these drugs, as well as a number of additional candidate transporters (Alsford, unpublished results).
FUTURE PROSPECTS
We have advanced from single gene analysis to high-throughput, genome-scale screens in a relatively short time. These developments, driven by the availability of genome sequence data, should facilitate further progress. For example, a genome-scale gain-of-function screening approach could now be developed for trypanosomatids. This would involve the generation of an over-expression library, which could be amenable to the same phenotype screens applied to the RNAi loss-of-function libraries, including drug-resistance screens. In T. cruzi, episomal expression (Kelly et al. 1992) and cosmid vectors (Kelly et al. 1994) offer an approach to generating such libraries. The availability of both loss-of-function and gain-of-function libraries for parallel screening could provide further insight into drug-resistance mechanisms, revealing, for example, efflux channels and even the drug-targets themselves. This approach could be applied to new drugs as they advance through development, providing useful information to facilitate the development of new and more effective compounds and combination therapies.
FINANCIAL SUPPORT
Work in the authors’ laboratories is supported by the Wellcome Trust (S. A., Institutional Strategic Support Fellowship; J. M. K., grant number 092573; D. H., grant number 093010/Z/10/Z) and the Medical Research Council (S. A., grant number MR/K011987/1; D. H., grant number MR/K000500/1). N. B. was also supported by a Bloomsbury Colleges doctoral studentship.
REFERENCES
- Alibu V. P., Richter C., Voncken F., Marti G., Shahi S., Renggli C. K., Seebeck T., Brun R. and Clayton C. (2006). The role of Trypanosoma brucei MRPA in melarsoprol susceptibility. Molecular and Biochemical Parasitology 146, 38–44. 10.1016/j.molbiopara.2005.10.016 [DOI] [PubMed] [Google Scholar]
- Alsford S., Eckert S., Baker N., Glover L., Sanchez-Flores A., Leung K. F., Turner D. J., Field M. C., Berriman M. and Horn D. (2012). High-throughput decoding of antitrypanosomal drug efficacy and resistance. Nature 482, 232–236. 10.1038/nature10771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsford S., Glover L. and Horn D. (2005a). Multiplex analysis of RNA interference defects in Trypanosoma brucei. Molecular and Biochemical Parasitology 139, 129–132. 10.1016/j.molbiopara.2004.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsford S. and Horn D. (2008). Single-locus targeting constructs for reliable regulated RNAi and transgene expression in Trypanosoma brucei. Molecular and Biochemical Parasitology 161, 76–79. 10.1016/j.molbiopara.2008.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsford S., Kawahara T., Glover L. and Horn D. (2005b). Tagging a T. brucei RRNA locus improves stable transfection efficiency and circumvents inducible expression position effects. Molecular and Biochemical Parasitology 144, 142–148. 10.1016/j.molbiopara.2005.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsford S., Turner D. J., Obado S. O., Sanchez-Flores A., Glover L., Berriman M., Hertz-Fowler C. and Horn D. (2011). High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Research 21, 915–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacchi C. J., Garofalo J., Mockenhaupt D., Mccann P. P., Diekema K. A., Pegg A. E., Nathan H. C., Mullaney E. A., Chunosoff L., Sjoerdsma A. and Hutner S. H. (1983). In vivo effects of alpha-DL-difluoromethylornithine on the metabolism and morphology of Trypanosoma brucei brucei. Molecular and Biochemical Parasitology 7, 209–225 [DOI] [PubMed] [Google Scholar]
- Bacchi C. J., Garofalo J., Ciminelli M., Rattendi D., Goldberg B., McCann P. P. and Yarlett N. (1993). Resistance to DL-alpha-difluoromethylornithine by clinical isolates of Trypanosoma brucei rhodesiense. Role of S-adenosylmethionine. Biochemical Pharmacology 46, 471–481 [DOI] [PubMed] [Google Scholar]
- Baker N., Alsford S. and Horn D. (2011). Genome-wide RNAi screens in African trypanosomes identify the nifurtimox activator NTR and the eflornithine transporter AAT6. Molecular and Biochemical Parasitology 176, 55–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker N., Glover L., Munday J. C., Aguinaga Andres D., Barrett M. P., De Koning H. P. and Horn D. (2012). Aquaglyceroporin 2 controls susceptibility to melarsoprol and pentamidine in African trypanosomes. Proceedings of the National Academy of Sciences, USA 109, 10996–11001. 10.1073/pnas.1202885109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett M. P. and Croft S. L. (2012). Management of trypanosomiasis and leishmaniasis. British Medical Bulletin 104, 175–196. 10.1093/bmb/lds031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett M. P. and Fairlamb A. H. (1999). The biochemical basis of arsenical-diamidine crossresistance in African trypanosomes. Parasitology Today 15, 136–140 [DOI] [PubMed] [Google Scholar]
- Barrett M. P., Vincent I. M., Burchmore R. J., Kazibwe A. J. and Matovu E. (2011). Drug resistance in human African trypanosomiasis. Future Microbiology 6, 1037–1047. 10.2217/fmb.11.88 [DOI] [PubMed] [Google Scholar]
- Barrett M. P., Zhang Z. Q., Denise H., Giroud C. and Baltz T. (1995). A diamidine-resistant Trypanosoma equiperdum clone contains a P2 purine transporter with reduced substrate affinity. Molecular and Biochemical Parasitology 73, 223–229 [DOI] [PubMed] [Google Scholar]
- Beaulieu C., Isabel E., Fortier A., Masse F., Mellon C., Methot N., Ndao M., Nicoll-Griffith D., Lee D., Park H. and Black W. C. (2010). Identification of potent and reversible cruzipain inhibitors for the treatment of Chagas disease. Bioorganic and Medicinal Chemistry Letters 20, 7444–7449. 10.1016/j.bmcl.2010.10.015 [DOI] [PubMed] [Google Scholar]
- Bern C. and Montgomery S. P. (2009). An estimate of the burden of Chagas disease in the United States. Clinical Infectious Diseases 49, e52–e54. 10.1086/605091 [DOI] [PubMed] [Google Scholar]
- Berrang Ford L. (2007). Civil conflict and sleeping sickness in Africa in general and Uganda in particular. Conflict and Health 1, 6. 10.1186/1752-1505-1-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriman M., Ghedin E., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D. C., Lennard N. J., Caler E., Hamlin N. E., Haas B., Bohme U., Hannick L., Aslett M. A., Shallom J., Marcello L., Hou L., Wickstead B., Alsmark U. C., Arrowsmith C., Atkin R. J., Barron A. J., Bringaud F., Brooks K., Carrington M., Cherevach I., Chillingworth T. J., Churcher C., Clark L. N., Corton C. H., Cronin A., Davies R. M., Doggett J., Djikeng A., Feldblyum T., Field M. C., Fraser A., Goodhead I., Hance Z., Harper D., Harris B. R., Hauser H., Hostetler J., Ivens A., Jagels K., Johnson D., Johnson J., Jones K., Kerhornou A. X., Koo H., Larke N., Landfear S., Larkin C., Leech V., Line A., Lord A., Macleod A., Mooney P. J., Moule S., Martin D. M., Morgan G. W., Mungall K., Norbertczak H., Ormond D., Pai G., Peacock C. S., Peterson J., Quail M. A., Rabbinowitsch E., Rajandream M. A., Reitter C., Salzberg S. L., Sanders M., Schobel S., Sharp S., Simmonds M., Simpson A. J., Tallon L., Turner C. M., Tait A., Tivey A. R., Van Aken S., Walker D., Wanless D., Wang S., White B., White O., Whitehead S., Woodward J., Wortman J., Adams M. D., Embley T. M., Gull K., Ullu E., Barry J. D., Fairlamb A. H., Opperdoes F., Barrell B. G., Donelson J. E., Hall N., Fraser C. M., Melville S. E. and El-Sayed N. M. (2005). The genome of the African trypanosome Trypanosoma brucei. Science 309, 416–422 [DOI] [PubMed] [Google Scholar]
- Bridges D. J., Gould M. K., Nerima B., Maser P., Burchmore R. J. and De Koning H. P. (2007). Loss of the high-affinity pentamidine transporter is responsible for high levels of cross-resistance between arsenical and diamidine drugs in African trypanosomes. Molecular Pharmacology 71, 1098–1108. 10.1124/mol.106.031351 [DOI] [PubMed] [Google Scholar]
- Brun R., Blum J., Chappuis F. and Burri C. (2010). Human African trypanosomiasis. Lancet 375, 148–159. 10.1016/S0140-6736(09)60829-1 [DOI] [PubMed] [Google Scholar]
- Buckner F., Yokoyama K., Lockman J., Aikenhead K., Ohkanda J., Sadilek M., Sebti S., Van Voorhis W., Hamilton A. and Gelb M. H. (2003). A class of sterol 14-demethylase inhibitors as anti-Trypanosoma cruzi agents. Proceedings of the National Academy of Sciences, USA 100, 15149–15153. 10.1073/pnas.2535442100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante J. M., Bixby L. M. and Tarleton R. L. (2008). Drug-induced cure drives conversion to a stable and protective CD8+ T central memory response in chronic Chagas disease. Nature Medicine 14, 542–550. 10.1038/nm1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter N. S., Berger B. J. and Fairlamb A. H. (1995). Uptake of diamidine drugs by the P2 nucleoside transporter in melarsen-sensitive and -resistant Trypanosoma brucei brucei. Journal of Biological Chemistry 270, 28153–28157 [DOI] [PubMed] [Google Scholar]
- Carter N. S. and Fairlamb A. H. (1993). Arsenical-resistant trypanosomes lack an unusual adenosine transporter. Nature 361, 173–176. 10.1038/361173a0 [DOI] [PubMed] [Google Scholar]
- Castro J. A., De Mecca M. M. and Bartel L. C. (2006). Toxic side effects of drugs used to treat Chagas’ disease (American trypanosomiasis). Human and Experimental Toxicology 25, 471–479 [DOI] [PubMed] [Google Scholar]
- Cazorla S. I., Frank F. M. and Malchiodi E. L. (2009). Vaccination approaches against Trypanosoma cruzi infection. Expert Review of Vaccines 8, 921–935. 10.1586/erv.09.45 [DOI] [PubMed] [Google Scholar]
- Chitanga S., Marcotty T., Namangala B., Van Den Bossche P., Van Den Abbeele J. and Delespaux V. (2011). High prevalence of drug resistance in animal trypanosomes without a history of drug exposure. PLoS Science Neglected Tropical Diseases 5, e1454. 10.1371/journal.pntd.0001454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creek D. J., Anderson J., McConville M. J. and Barrett M. P. (2012). Metabolomic analysis of trypanosomatid protozoa. Molecular and Biochemical Parasitology 181, 73–84. 10.1016/j.molbiopara.2011.10.003 [DOI] [PubMed] [Google Scholar]
- De Koning H. P. (2001). Uptake of pentamidine in Trypanosoma brucei brucei is mediated by three distinct transporters: implications for cross-resistance with arsenicals. Molecular Pharmacology 59, 586–592 [DOI] [PubMed] [Google Scholar]
- De Koning H. P. (2008). Ever-increasing complexities of diamidine and arsenical crossresistance in African trypanosomes. Trends in Parasitology 24, 345–349. 10.1016/j.pt.2008.04.006 [DOI] [PubMed] [Google Scholar]
- De Koning H. P., Anderson L. F., Stewart M., Burchmore R. J., Wallace L. J. and Barrett M. P. (2004). The trypanocide diminazene aceturate is accumulated predominantly through the TbAT1 purine transporter: additional insights on diamidine resistance in African trypanosomes. Antimicrobial Agents and Chemotherapy 48, 1515–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delespaux V. and De Koning H. P. (2007). Drugs and drug resistance in African trypanosomiasis. Drug Resistance Updates 10, 30–50. 10.1016/j.drup.2007.02.004 [DOI] [PubMed] [Google Scholar]
- Diazgranados C. A., Saavedra-Trujillo C. H., Mantilla M., Valderrama S. L., Alquichire C. and Franco-Paredes C. (2009). Chagasic encephalitis in HIV patients: common presentation of an evolving epidemiological and clinical association. Lancet Infectious Diseases 9, 324–330. 10.1016/S1473-3099(09)70088-X [DOI] [PubMed] [Google Scholar]
- Docampo R. (1990). Sensitivity of parasites to free radical damage by antiparasitic drugs. Chemico-Biological Interactions 73, 1–27 [DOI] [PubMed] [Google Scholar]
- Drew M. E., Morris J. C., Wang Z., Wells L., Sanchez M., Landfear S. M. and Englund P. T. (2003). The adenosine analog tubercidin inhibits glycolysis in Trypanosoma brucei as revealed by an RNA interference library. Journal of Biological Chemistry 278, 46596–46600. 10.1074/jbc.M309320200 [DOI] [PubMed] [Google Scholar]
- El-Sayed N. M., Myler P. J., Bartholomeu D. C., Nilsson D., Aggarwal G., Tran A. N., Ghedin E., Worthey E. A., Delcher A. L., Blandin G., Westenberger S. J., Caler E., Cerqueira G. C., Branche C., Haas B., Anupama A., Arner E., Aslund L., Attipoe P., Bontempi E., Bringaud F., Burton P., Cadag E., Campbell D. A., Carrington M., Crabtree J., Darban H., Da Silveira J. F., De Jong P., Edwards K., Englund P. T., Fazelina G., Feldblyum T., Ferella M., Frasch A. C., Gull K., Horn D., Hou L., Huang Y., Kindlund E., Klingbeil M., Kluge S., Koo H., Lacerda D., Levin M. J., Lorenzi H., Louie T., Machado C. R., Mcculloch R., Mckenna A., Mizuno Y., Mottram J. C., Nelson S., Ochaya S., Osoegawa K., Pai G., Parsons M., Pentony M., Pettersson U., Pop M., Ramirez J. L., Rinta J., Robertson L., Salzberg S. L., Sanchez D. O., Seyler A., Sharma R., Shetty J., Simpson A. J., Sisk E., Tammi M. T., Tarleton R., Teixeira S., Van Aken S., Vogt C., Ward P. N., Wickstead B., Wortman J., White O., Fraser C. M., Stuart K. D. and Andersson B. (2005). The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science 309, 409–415. 10.1126/science.1112631 [DOI] [PubMed] [Google Scholar]
- Fairlamb A. H., Henderson G. B. and Cerami A. (1989). Trypanothione is the primary target for arsenical drugs against African trypanosomes. Proceedings of the National Academy of Sciences, USA 86, 2607–2611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filardi L. S. and Brener Z. (1987). Susceptibility and natural resistance of Trypanosoma cruzi strains to drugs used clinically in Chagas disease. Transactions of the Royal Society of Tropical Medicine and Hygiene 81, 755–759 [DOI] [PubMed] [Google Scholar]
- Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E. and Mello C. C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. 10.1038/35888 [DOI] [PubMed] [Google Scholar]
- Franco J. R., Simarro P. P., Diarra A., Ruiz-Postigo J. A., Samo M. and Jannin J. G. (2012). Monitoring the use of nifurtimox-eflornithine combination therapy (NECT) in the treatment of second stage gambiense human African trypanosomiasis. Research and Reports in Tropical Medicine 3, 93–101. 10.2147/RRTM.S34399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedheim E. A. (1949). Mel B in the treatment of human trypanosomiasis. American Journal of Tropical Medicine and Hygiene 29, 173–180 [DOI] [PubMed] [Google Scholar]
- Garcia S., Ramos C. O., Senra J. F., Vilas-Boas F., Rodrigues M. M., Campos-De-Carvalho A. C., Ribeiro-Dos-Santos R. and Soares M. B. (2005). Treatment with benznidazole during the chronic phase of experimental Chagas’ disease decreases cardiac alterations. Antimicrobial Agents and Chemotherapy 49, 1521–1528. 10.1128/AAC.49.4.1521-1528.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geerts S., Holmes P. H., Eisler M. C. and Diall O. (2001). African bovine trypanosomiasis: the problem of drug resistance. Trends in Parasitology 17, 25–28 [DOI] [PubMed] [Google Scholar]
- Glover L. and Horn D. (2009). Site-specific DNA double-strand breaks greatly increase stable transformation efficiency in Trypanosoma brucei. Molecular and Biochemical Parasitology 166, 194–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueiros-Filho F. J. and Beverley S. M. (1997). Trans-kingdom transposition of the Drosophila element mariner within the protozoan Leishmania. Science 276, 1716–1719 [DOI] [PubMed] [Google Scholar]
- Hall B. S., Bot C. and Wilkinson S. R. (2011). Nifurtimox activation by trypanosomal type I nitroreductases generates cytotoxic nitrile metabolites. Journal of Biological Chemistry 286, 13088–13095. 10.1074/jbc.M111.230847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. S. and Wilkinson S. R. (2012). Activation of benznidazole by trypanosomal type I nitroreductases results in glyoxal formation. Antimicrobial Agents and Chemotherapy 56, 115–123. 10.1128/AAC.05135-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K. and Schofield C. J. (2012). Elimination of Rhodnius prolixus in Central America. Parasites and Vectors 5, 45. 10.1186/1756-3305-5-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn D. and Mcculloch R. (2010). Molecular mechanisms underlying the control of antigenic variation in African trypanosomes. Current Opinion in Microbiology 13, 700–705. 10.1016/j.mib.2010.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iten M., Mett H., Evans A., Enyaru J. C., Brun R. and Kaminsky R. (1997). Alterations in ornithine decarboxylase characteristics account for tolerance of Trypanosoma brucei rhodesiense to D,L-alpha-difluoromethylornithine. Antimicrobial Agents and Chemotherapy 41, 1922–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivens A. C., Peacock C. S., Worthey E. A., Murphy L., Aggarwal G., Berriman M., Sisk E., Rajandream M. A., Adlem E., Aert R., Anupama A., Apostolou Z., Attipoe P., Bason N., Bauser C., Beck A., Beverley S. M., Bianchettin G., Borzym K., Bothe G., Bruschi C. V., Collins M., Cadag E., Ciarloni L., Clayton C., Coulson R. M., Cronin A., Cruz A. K., Davies R. M., De Gaudenzi J., Dobson D. E., Duesterhoeft A., Fazelina G., Fosker N., Frasch A. C., Fraser A., Fuchs M., Gabel C., Goble A., Goffeau A., Harris D., Hertz-Fowler C., Hilbert H., Horn D., Huang Y., Klages S., Knights A., Kube M., Larke N., Litvin L., Lord A., Louie T., Marra M., Masuy D., Matthews K., Michaeli S., Mottram J. C., Muller-Auer S., Munden H., Nelson S., Norbertczak H., Oliver K., O'neil S., Pentony M., Pohl T. M., Price C., Purnelle B., Quail M. A., Rabbinowitsch E., Reinhardt R., Rieger M., Rinta J., Robben J., Robertson L., Ruiz J. C., Rutter S., Saunders D., Schafer M., Schein J., Schwartz D. C., Seeger K., Seyler A., Sharp S., Shin H., Sivam D., Squares R., Squares S., Tosato V., Vogt C., Volckaert G., Wambutt R., Warren T., Wedler H., Woodward J., Zhou S., Zimmermann W., Smith D. F., Blackwell J. M., Stuart K. D., Barrell B. and Myler P. J. (2005). The genome of the kinetoplastid parasite, Leishmania major. Science 309, 436–442. 10.1126/science.1112680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs R. T., Nare B., Wring S. A., Orr M. D., Chen D., Sligar J. M., Jenks M. X., Noe R. A., Bowling T. S., Mercer L. T., Rewerts C., Gaukel E., Owens J., Parham R., Randolph R., Beaudet B., Bacchi C. J., Yarlett N., Plattner J. J., Freund Y., Ding C., Akama T., Zhang Y. K., Brun R., Kaiser M., Scandale I. and Don R. (2011). SCYX-7158, an orally-active benzoxaborole for the treatment of stage 2 human African trypanosomiasis. PLoS Neglected Tropical Diseases 5, e1151. 10.1371/journal.pntd.0001151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamonneau V., Ilboudo H., Kabore J., Kaba D., Koffi M., Solano P., Garcia A., Courtin D., Laveissiere C., Lingue K., Buscher P. and Bucheton B. (2012). Untreated human infections by Trypanosoma brucei gambiense are not 100% fatal. PLoS Neglected Tropical Diseases 6, e1691. 10.1371/journal.pntd.0001691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J. M., Das P. and Tomas A. M. (1994). An approach to functional complementation by introduction of large DNA fragments into Trypanosoma cruzi and Leishmania donovani using a cosmid shuttle vector. Molecular and Biochemical Parasitology 65, 51–62 [DOI] [PubMed] [Google Scholar]
- Kelly J. M., Ward H. M., Miles M. A. and Kendall G. (1992). A shuttle vector which facilitates the expression of transfected genes in Trypanosoma cruzi and Leishmania. Nucleic Acids Research 20, 3963–3969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhoff L. V. (2011). Epidemiology of American trypanosomiasis (Chagas disease). Advances in Parasitology 75, 1–8. 10.1016/B978-0-12-385863-4.00001-0 [DOI] [PubMed] [Google Scholar]
- Kolev N. G., Tschudi C. and Ullu E. (2011). RNA interference in protozoan parasites: achievements and challenges. Eukaryotic Cell 10, 1156–1163. 10.1128/EC.05114-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacount D. J., Bruse S., Hill K. L. and Donelson J. E. (2000). Double-stranded RNA interference in Trypanosoma brucei using head-to-head promoters. Molecular and Biochemical Parasitology 111, 67–76 [DOI] [PubMed] [Google Scholar]
- Le Loup G., Pialoux G. and Lescure F. X. (2011). Update in treatment of Chagas disease. Current Opinion in Infectious Diseases 24, 428–434. 10.1097/QCO.0b013e32834a667f [DOI] [PubMed] [Google Scholar]
- Leal S., Acosta-Serrano A., Morris J. and Cross G. A. (2004). Transposon mutagenesis of Trypanosoma brucei identifies glycosylation mutants resistant to concanavalin A. Journal of Biological Chemistry 279, 28979–28988. 10.1074/jbc.M403479200 [DOI] [PubMed] [Google Scholar]
- Lee M. G. and Van Der Ploeg L. H. (1990). Homologous recombination and stable transfection in the parasitic protozoan Trypanosoma brucei. Science 250, 1583–1587 [DOI] [PubMed] [Google Scholar]
- Legare D., Richard D., Mukhopadhyay R., Stierhof Y. D., Rosen B. P., Haimeur A., Papadopoulou B. and Ouellette M. (2001). The Leishmania ATP-binding cassette protein PGPA is an intracellular metal-thiol transporter ATPase. Journal of Biological Chemistry 276, 26301–26307. 10.1074/jbc.M102351200 [DOI] [PubMed] [Google Scholar]
- Lye L. F., Owens K., Shi H., Murta S. M., Vieira A. C., Turco S. J., Tschudi C., Ullu E. and Beverley S. M. (2010). Retention and loss of RNA interference pathways in trypanosomatid protozoans. PLoS Pathogens 6, e1001161. 10.1371/journal.ppat.1001161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Neto J. A., Rassi A. Jr., Morillo C. A., Avezum A., Connolly S. J., Sosa-Estani S., Rosas F. and Yusuf S. (2008). Rationale and design of a randomized placebo-controlled trial assessing the effects of etiologic treatment in Chagas’ cardiomyopathy: the BENznidazole Evaluation For Interrupting Trypanosomiasis (BENEFIT). American Heart Journal 156, 37–43. 10.1016/j.ahj.2008.04.001 [DOI] [PubMed] [Google Scholar]
- Maser P., Sutterlin C., Kralli A. and Kaminsky R. (1999). A nucleoside transporter from Trypanosoma brucei involved in drug resistance. Science 285, 242–244 [DOI] [PubMed] [Google Scholar]
- Matemba L. E., Fevre E. M., Kibona S. N., Picozzi K., Cleaveland S., Shaw A. P. and Welburn S. C. (2010). Quantifying the burden of rhodesiense sleeping sickness in Urambo District, Tanzania. PLoS Neglected Tropical Diseases 4, e868. 10.1371/journal.pntd.0000868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matovu E., Geiser F., Schneider V., Maser P., Enyaru J. C., Kaminsky R., Gallati S. and Seebeck T. (2001). Genetic variants of the TbAT1 adenosine transporter from African trypanosomes in relapse infections following melarsoprol therapy. Molecular and Biochemical Parasitology 117, 73–81 [DOI] [PubMed] [Google Scholar]
- Matovu E., Stewart M. L., Geiser F., Brun R., Maser P., Wallace L. J., Burchmore R. J., Enyaru J. C., Barrett M. P., Kaminsky R., Seebeck T. and De Koning H. P. (2003). Mechanisms of arsenical and diamidine uptake and resistance in Trypanosoma brucei. Eukaryotic Cell 2, 1003–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCalla D. R., Reuvers A. and Kaiser C. (1971). Breakage of bacterial DNA by nitrofuran derivatives. Cancer Research 31, 2184–2188 [PubMed] [Google Scholar]
- McKerrow J. H., Doyle P. S., Engel J. C., Podust L. M., Robertson S. A., Ferreira R., Saxton T., Arkin M., Kerr I. D., Brinen L. S. and Craik C. S. (2009). Two approaches to discovering and developing new drugs for Chagas disease. Memórias do Instituto Oswaldo Cruz 104(Suppl. 1), 263–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejia A. M., Hall B. S., Taylor M. C., Gomez-Palacio A., Wilkinson S. R., Triana-Chavez O. and Kelly J. M. (2012). Benznidazole-resistance in Trypanosoma cruzi is a readily acquired trait that can arise independently in a single population. Journal of Infectious Diseases 206, 220–228. 10.1093/infdis/jis331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncayo A. and Silveira A. C. (2009). Current epidemiological trends for Chagas disease in Latin America and future challenges in epidemiology, surveillance and health policy. Memórias do Instituto Oswaldo Cruz 104(Suppl. 1), 17–30 [DOI] [PubMed] [Google Scholar]
- Morris J. C., Wang Z., Drew M. E. and Englund P. T. (2002). Glycolysis modulates trypanosome glycoprotein expression as revealed by an RNAi library. EMBO Journal 21, 4429–4438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moti Y., Fikru R., Van Den Abbeele J., Buscher P., Van Den Bossche P., Duchateau L. and Delespaux V. (2012). Ghibe river basin in Ethiopia: present situation of trypanocidal drug resistance in Trypanosoma congolense using tests in mice and PCR-RFLP. Veterinary Parasitology 189, 197–203. 10.1016/j.vetpar.2012.04.022 [DOI] [PubMed] [Google Scholar]
- Mumba D., Bohorquez E., Messina J., Kande V., Taylor S. M., Tshefu A. K., Muwonga J., Kashamuka M. M., Emch M., Tidwell R., Buscher P. and Meshnick S. R. (2011). Prevalence of human African trypanosomiasis in the Democratic Republic of the Congo. PLoS Neglected Tropical Diseases 5, e1246. 10.1371/journal.pntd.0001246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo H., Tschudi C., Gull K. and Ullu E. (1998). Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proceedings of the National Academy of Sciences, USA 95, 14687–14692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odiit M., Coleman P. G., Liu W. C., Mcdermott J. J., Fevre E. M., Welburn S. C. and Woolhouse M. E. (2005). Quantifying the level of under-detection of Trypanosoma brucei rhodesiense sleeping sickness cases. Tropical Medicine and International Health 10, 840–849. 10.1111/j.1365-3156.2005.01470.x [DOI] [PubMed] [Google Scholar]
- Ortiz D., Sanchez M. A., Quecke P. and Landfear S. M. (2009). Two novel nucleobase/pentamidine transporters from Trypanosoma brucei. Molecular and Biochemical Parasitology 163, 67–76. 10.1016/j.molbiopara.2008.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson G. N., Skelly J. V. and Neidle S. (2000). Crystal structure of FMN-dependent nitroreductase from Escherichia coli B: a prodrug-activating enzyme. Journal of Medicinal Chemistry 43, 3624–3631 [DOI] [PubMed] [Google Scholar]
- Pepin J. and Milord F. (1994). The treatment of human African trypanosomiasis. Advances in Parasitology 33, 1–47 [DOI] [PubMed] [Google Scholar]
- Phillips M. A. and Wang C. C. (1987). A Trypanosoma brucei mutant resistant to alpha-difluoromethylornithine. Molecular and Biochemical Parasitology 22, 9–17 [DOI] [PubMed] [Google Scholar]
- Priotto G., Kasparian S., Mutombo W., Ngouama D., Ghorashian S., Arnold U., Ghabri S., Baudin E., Buard V., Kazadi-Kyanza S., Ilunga M., Mutangala W., Pohlig G., Schmid C., Karunakara U., Torreele E. and Kande V. (2009). Nifurtimox-eflornithine combination therapy for second-stage African Trypanosoma brucei gambiense trypanosomiasis: a multicentre, randomised, phase III, non-inferiority trial. Lancet 374, 56–64. 10.1016/S0140-6736(09)61117-X [DOI] [PubMed] [Google Scholar]
- Ruiz-Postigo J. A., Franco J. R., Lado M. and Simarro P. P. (2012). Human African trypanosomiasis in South Sudan: how can we prevent a new epidemic? PLoS Neglected Tropical Diseases 6, e1541. 10.1371/journal.pntd.0001541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann Burkard G., Jutzi P. and Roditi I. (2011). Genome-wide RNAi screens in bloodstream form trypanosomes identify drug transporters. Molecular and Biochemical Parasitology 175, 91–94 [DOI] [PubMed] [Google Scholar]
- Scott A. G., Tait A. and Turner C. M. (1996). Characterisation of cloned lines of Trypanosoma brucei expressing stable resistance to MelCy and suramin. Acta Tropica 60, 251–262 [DOI] [PubMed] [Google Scholar]
- Shahi S. K., Krauth-Siegel R. L. and Clayton C. E. (2002). Overexpression of the putative thiol conjugate transporter TbMRPA causes melarsoprol resistance in Trypanosoma brucei. Molecular Microbiology 43, 1129–1138 [DOI] [PubMed] [Google Scholar]
- Simarro P. P., Cecchi G., Franco J. R., Paone M., Fevre E. M., Diarra A., Postigo J. A., Mattioli R. C. and Jannin J. G. (2012a). Risk for human African trypanosomiasis, Central Africa, 2000–2009. Emerging Infectious Diseases 17, 2322–2324. 10.3201/eid1712.110921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simarro P. P., Franco J., Diarra A., Postigo J. A. and Jannin J. (2012b). Update on field use of the available drugs for the chemotherapy of human African trypanosomiasis. Parasitology 139, 842–846. 10.1017/S0031182012000169 [DOI] [PubMed] [Google Scholar]
- Sokolova A. Y., Wyllie S., Patterson S., Oza S. L., Read K. D. and Fairlamb A. H. (2010). Cross-resistance to nitro drugs and implications for treatment of human African trypanosomiasis. Antimicrobial Agents and Chemotherapy 54, 2893–2900. 10.1128/AAC.00332-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer J. M., Hua S., Li F., Gottesdiener K. M. and Wang C. C. (1996). Cloning by functional complementation in Trypanosoma brucei. Molecular and Biochemical Parasitology 76, 83–89 [DOI] [PubMed] [Google Scholar]
- Steverding D. (2008). The history of African trypanosomiasis. Parasites and Vectors 1, 3. 10.1186/1756-3305-1-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. L., Burchmore R. J., Clucas C., Hertz-Fowler C., Brooks K., Tait A., MacLeod A., Turner C. M., De Koning H. P., Wong P. E. and Barrett M. P. (2010). Multiple genetic mechanisms lead to loss of functional TbAT1 expression in drug-resistant trypanosomes. Eukaryotic Cell 9, 336–343. 10.1128/EC.00200-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streeter A. J. and Hoener B. A. (1988). Evidence for the involvement of a nitrenium ion in the covalent binding of nitrofurazone to DNA. Pharmaceutical Research 5, 434–436 [DOI] [PubMed] [Google Scholar]
- Subramaniam C., Veazey P., Redmond S., Hayes-Sinclair J., Chambers E., Carrington M., Gull K., Matthews K., Horn D. and Field M. C. (2006). Chromosome-wide analysis of gene function by RNA interference in the African trypanosome. Eukaryotic Cell 5, 1539–1549. 10.1128/EC.00141-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryadevara P. K., Olepu S., Lockman J. W., Ohkanda J., Karimi M., Verlinde C. L., Kraus J. M., Schoepe J., Van Voorhis W. C., Hamilton A. D., Buckner F. S. and Gelb M. H. (2009). Structurally simple inhibitors of lanosterol 14alpha-demethylase are efficacious in a rodent model of acute Chagas disease. Journal of Medicinal Chemistry 52, 3703–3715. 10.1021/jm900030h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarleton R. L. (2003). Chagas disease: a role for autoimmunity? Trends in Parasitology 19, 447–451 [DOI] [PubMed] [Google Scholar]
- Tarleton R. L., Zhang L. and Downs M. O. (1997). ‘Autoimmune rejection’ of neonatal heart transplants in experimental Chagas disease is a parasite-specific response to infected host tissue. Proceedings of the National Academy of Sciences, USA 94, 3932–3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M. C. and Kelly J. M. (2006). pTcINDEX: a stable tetracycline-regulated expression vector for Trypanosoma cruzi. BMC Biotechnology 6, 32. 10.1186/1472-6750-6-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M. C., Huang H. and Kelly J. M. (2011). Genetic techniques in Trypanosoma cruzi. Advances in Parasitology 75, 231–250. 10.1016/B978-0-12-385863-4.00011-3 [DOI] [PubMed] [Google Scholar]
- Thuita J. K., Wang M. Z., Kagira J. M., Denton C. L., Paine M. F., Mdachi R. E., Murilla G. A., Ching S., Boykin D. W., Tidwell R. R., Hall J. E. and Brun R. (2012). Pharmacology of DB844, an orally active aza analogue of pafuramidine, in a monkey model of second stage human African trypanosomiasis. PLoS Neglected and Tropical Diseases 6, e1734. 10.1371/journal.pntd.0001734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbina J. A. (2009). Ergosterol biosynthesis and drug development for Chagas disease. Memórias do Instituto Oswaldo Cruz 104(Suppl. 1), 311–318 [DOI] [PubMed] [Google Scholar]
- Urbina J. A. (2010). Specific chemotherapy of Chagas disease: relevance, current limitations and new approaches. Acta Tropica 115, 55–68. 10.1016/j.actatropica.2009.10.023 [DOI] [PubMed] [Google Scholar]
- Vincent I. M., Creek D., Watson D. G., Kamleh M. A., Woods D. J., Wong P. E., Burchmore R. J. and Barrett M. P. (2010). A molecular mechanism for eflornithine resistance in African trypanosomes. PLoS Pathogens 6, e1001204. 10.1371/journal.ppat.1001204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viode C., Bettache N., Cenas N., Krauth-Siegel R. L., Chauviere G., Bakalara N. and Perie J. (1999). Enzymatic reduction studies of nitroheterocycles. Biochemical Pharmacology 57, 549–557 [DOI] [PubMed] [Google Scholar]
- Wang Z., Morris J. C., Drew M. E. and Englund P. T. (2000). Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. Journal of Biological Chemistry 275, 40174–40179. 10.1074/jbc.M008405200 [DOI] [PubMed] [Google Scholar]
- Welburn S. C. and Maudlin I. (2012). Priorities for the elimination of sleeping sickness. Advances in Parasitology 79, 299–337. 10.1016/B978-0-12-398457-9.00004-4 [DOI] [PubMed] [Google Scholar]
- Whiteway J., Koziarz P., Veall J., Sandhu N., Kumar P., Hoecher B. and Lambert I. B. (1998). Oxygen-insensitive nitroreductases: analysis of the roles of nfsA and nfsB in development of resistance to 5-nitrofuran derivatives in Escherichia coli. Journal of Bacteriology 180, 5529–5539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2009). Statement – Chagas disease in Europe. World Health Organisation, Geneva, Switzerland. http://www.who.int/neglected_diseases/integrated_media_chagas_statement/en/index.html
- Wilkinson S. R. and Kelly J. M. (2009). Trypanocidal drugs: mechanisms, resistance and new targets. Expert Reviews in Molecular Medicine 11, e31. 10.1017/S1462399409001252 [DOI] [PubMed] [Google Scholar]
- Wilkinson S. R., Taylor M. C., Horn D., Kelly J. M. and Cheeseman I. (2008). A mechanism for cross-resistance to nifurtimox and benznidazole in trypanosomes. Proceedings of the National Academy of Sciences, USA 105, 5022–5027. 10.1073/pnas.0711014105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz E. and Clayton C. (1995). Inducible gene expression in trypanosomes mediated by a prokaryotic repressor. Science 268, 1179–1183 [DOI] [PubMed] [Google Scholar]
- Wirtz E., Leal S., Ochatt C. and Cross G. A. (1999). A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Molecular and Biochemical Parasitology 99, 89–101 [DOI] [PubMed] [Google Scholar]
- Wyllie S., Patterson S., Stojanovski L., Simeons F. R., Norval S., Kime R., Read K. D. and Fairlamb A. H. (2012). The anti-trypanosome drug fexinidazole shows potential for treating visceral leishmaniasis. Science Translational Medicine 4, 119re111. 10.1126/scitranslmed.3003326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Lindsay M. E., Roy Chowdhury A., Robinson D. R. and Englund P. T. (2008). p166, a link between the trypanosome mitochondrial DNA and flagellum, mediates genome segregation. EMBO Journal 27, 143–154. 10.1038/sj.emboj.7601956 [DOI] [PMC free article] [PubMed] [Google Scholar]