SUMMARY
Carriers of BRCA1 germline mutations are predisposed to breast and ovarian cancers. Accumulated evidence shows that BRCA1 is quickly recruited to DNA lesions and plays an important role in the DNA damage response. However, the mechanism by which BRCA1 is recruited to DNA damage sites remains elusive. BRCA1 forms a Ring-domain heterodimer with BARD1, a major partner of BRCA1 that contains tandem BRCT motifs. Here, we identify the BRCTs of BARD1 as a poly(ADP-ribose) (PAR)-binding module. The binding of the BARD1 BRCTs to PAR targets the BRCA1/BARD1 heterodimer to DNA damage sites. Thus, our study uncovers a PAR-dependent mechanism of rapid recruitment of BRCA1/BARD1 to DNA damage sites.
INTRODUCTION
Cells encounter numerous environmental and internal hazards that induce various kinds of DNA damage. To cope with these threats, cells have developed a DNA damage response system to sense and repair DNA lesions. Loss of this DNA damage response lead to the accumulation of DNA lesions, triggers genomic instability, and ultimately promotes tumorigenesis. Thus, many DNA damage response proteins are important tumor suppressors.
BRCA1 is a breast and ovarian cancer suppressor (Miki et al., 1994). Germline BRCA1 mutation carriers are predisposed to breast and ovarian cancers (King et al., 2003; Rahman and Stratton, 1998; Turner et al., 2004; Venkitaraman, 2002). Accumulated evidence suggests that BRCA1 plays important roles in several biological events during the DNA damage response including cell cycle checkpoint activation and repair of DNA double strand breaks (DSBs) (Huen et al., 2010; Roy et al., 2011; Scully et al., 1997; Scully and Livingston, 2000). As a result, tumor cells bearing BRCA1 mutations are hypersensitive to DSBs-inducing agents, such as ionizing radiation (IR) (Abbott et al., 1999; Shen et al., 1998). In addition to DSB repair, cells have other pathways to repair different types of DNA lesions, such as DNA single-strand breaks (SSBs). It has been shown that poly(ADP-ribose) polymerases (PARPs) play important roles in SSB repair (Burkle, 2001; Fisher et al., 2007; Okano et al., 2003). Interestingly, suppression of PARPs by inhibitors can specifically kill breast cancer cells bearing BRCA1 mutations. It has been hypothesized that cells lacking both PARP-dependent SSB repair and BRCA1-dependent DSB repair are inviable (Farmer et al., 2005; Fong et al., 2009; Rouleau et al., 2010). Following DNA damage, PARP1, one of the major PARPs in the DNA damage response, quickly relocates to DNA damage sites and catalyzes protein PARylation (Kim et al., 2005). Although the function of this PARylation is not clear, some evidence suggests that it can function as a docking signal to recruit other DNA damage response factors to DNA lesions (Masson et al., 1998; Okano et al., 2003; Ruscetti et al., 1998). Interestingly, recent structural analyses indicate that PARP1 also recognizes DSBs (Ali et al., 2012), although the function of PARP1 in DSB repair is unknown.
Like PARP1, BRCA1 is also quickly recruited to DNA damage sites (Scully et al., 1997). The molecular mechanism by which BRCA1 is recruited to DNA damage sites remains elusive. Two important DNA damage response factors, γH2AX and MDC1, have been shown to facilitate the recruitment of BRCA1 to DNA damage sites (Harper and Elledge, 2007). However, BRCA1 can still be transiently recruited to DNA damage sites in the absence of γH2AX, although it cannot be stably retained at DNA damage sites (Celeste et al., 2003), suggesting that H2AX provides the platform to stabilize BRCA1 at DNA damage sites instead of directly recruiting it. Recently, it has been shown that a DNA damage-induced protein ubiquitination pathway governs the relocation of BRCA1 to sites of DNA damage via the RAP80 complex) (Kim et al., 2007; Sobhian et al., 2007; Wang et al., 2007). However, deletion of RAP80 does not completely abolish the IR-induced foci formation of BRCA1(Hu et al., 2011), suggesting that alternative mechanism of recruitment of BRCA1 to DNA damage sites exists. Since the DNA damage-induced protein ubiquitination pathway is γH2AX-dependent (Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007), it is likely that protein ubiquitination at DNA damage sites, like γH2AX, only stabilizes BRCA1 foci instead of acting in the initial recruitment of BRCA1. Thus, in this study, we examined the molecular mechanism by which BRCA1 is recruited to the sites of DNA damage.
RESULTS
BARD1 mediates the rapid recruitment of BRCA1 to DNA damage sites
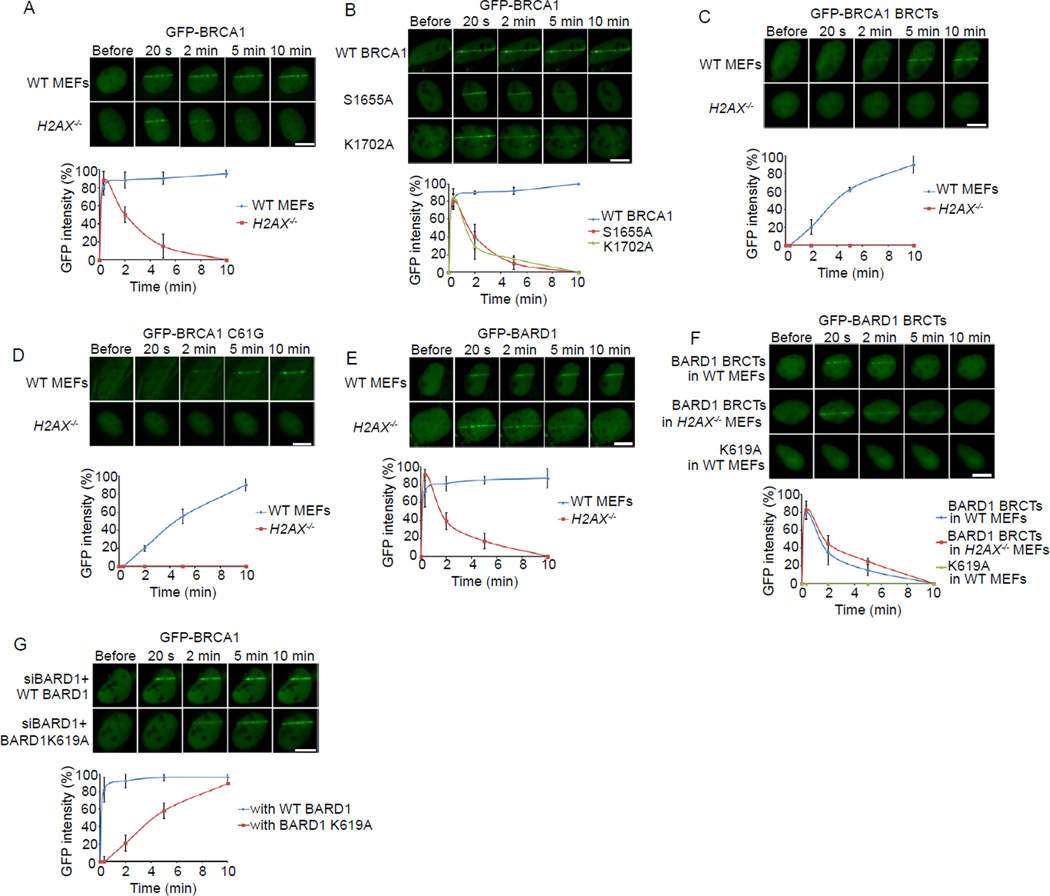
To search for the molecular mechanism by which BRCA1 is recruited to DNA damage sites, we measured the kinetics of BRCA1’s relocation to sites of DNA damage. Using laser microirradiation and live cell imaging, we found that BRCA1 was rapidly recruited to sites of DNA damage within 20 seconds following irradiation in mouse embryonic fibroblasts (MEFs) (Figure 1A). In H2AX−/− MEFs, BRCA1 was still recruited to DNA damage sites within 20 seconds. However, the majority of the BRCA1 dissociated from the DNA damage sites within 5 minutes following laser microirradiation (Figure 1A). To rule out the possibility that the relocation kinetics fluctuated based on transfection efficiency in each experiment, we transfected the H2AX−/− MEFs with a high (0.6 µg/1.5 cm glass-bottomed dish ) and a low (0.2 µg/1.5 cm glass-bottomed dish) concentration of GFP-BARD1 BRCTs plasmid, then investigated the relocation kinetics of BARD1 BRCTs following laser microirradiation. As shown in Figure S1A, different protein expression levels did not affect the relocation kinetics. Thus, these results are in agreement with a previous report that H2AX is important for BRCA1 retention but not for its initial recruitment to DNA damage sites (Celeste et al., 2003).
Figure 1. The recruitments of BRCA1 and BARD1 to DNA damage sites.
(A) The relocation kinetics of BRCA1 to DNA damage sites. GFP-BRCA1 was expressed in WT or H2AX−/− MEFs. The relocation kinetics was monitored in a time course following laser microirradiation (the same for below). (B) The relocation kinetics of the S1655A or K1702A mutants of BRCA1 to DNA damage sites. GFP-WT BRCA1, S1655A or K1702A mutants were expressed in U2OS cells. (C) The relocation kinetics of the BRCA1 BRCTs to DNA damage sites. The GFP-BRCA1 BRCTs was expressed in WT or H2AX−/− MEFs. (D) The relocation kinetics of the C61G mutant of BRCA1 to DNA damage sites. The GFP-BRCA1 C61G mutant was expressed in WT or H2AX−/− MEFs. (E) The relocation kinetics of BARD1 to DNA damage sites. GFP-BARD1 was expressed in WT or H2AX−/− MEFs. (F) The relocation kinetics of the BARD1 BRCTs and the K619A mutant to DNA damage sites. GFP-WT BARD1 BRCTs or the K619A mutant was expressed in WT or H2AX−/− MEFs. (G) The effect of the BARD1 K619A mutant on the recruitment of BRCA1 to DNA damage sites. U2OS cells stably expressing siRNA-resistant WT BARD1 or the K619A mutant were transfected with BARD1 siRNA to deplete endogenous BARD1. GFP-BRCA1 was expressed in these stable cell lines. GFP fluorescence at the laser line was converted into a numerical value (relative fluorescence intensity) using Axiovision software (version 4.5). Normalized fluorescent curves from 20 cells from three independent experiments were averaged. The error bars represent the standard deviation. Scale bar = 10 µm. See also Figure S1.
BRCA1 contains a C-terminal BRCT domain and an N-terminal Ring domain. We next examined the importance of these two domains to the recruitment of BRCA1. It is well known that the tandem BRCA1 BRCT motifs recognize phosphoserine (pSer) motifs and are involved in targeting BRCA1 to DNA damage sites (Manke et al., 2003; Yu et al., 2003). Either S1655A or K1702A mutation in the BRCA1 BRCTs abolishes pSer-binding (Botuyan et al., 2004; Clapperton et al., 2004; Williams et al., 2004). Unexpectedly, both mutated forms of BRCA1 were still recruited to DNA damage sites within 20 seconds after laser microirradiation. However, neither could be stably accumulated at DNA damage sites, and both dissociated from the DNA damage sites within 5 minutes (Figure 1B). These results suggest that the BRCT domains of BRCA1 are required for the retention of BRCA1 at DNA damage sites but not for the initial, rapid recruitment of BRCA1 to sites of DNA damage. To confirm our conclusion, we expressed the recombinant BRCT domain of BRCA1 in wild-type (WT) MEFs. The BRCA1 BRCTs slowly accumulated at DNA damage sites and could be visualized there within ~ 10 minutes following laser microirradiation (Figure 1C). Moreover, the BRCA1 BRCTs failed to be recruited to DNA damage sites in the absence of H2AX (Figure 1C), suggesting that H2AX is required for the BRCT-dependent BRCA1 retention at DNA damage sites.
Since the BRCA1 BRCT motifs do not directly target BRCA1 to DNA damage sites during the early DNA damage response, we searched for other possible factors that might facilitate the relocation of BRCA1. Beside the C-terminal BRCTs, BRCA1 contains an N-terminal Ring domain (Koonin et al., 1996). It has been reported that the Ring domain of BRCA1 is important for IR-induced BRCA1 foci formation (Au and Henderson, 2005). To explore the role of the Ring domain in the recruitment of BRCA1 to sites of DNA damage, we generated the C61G mutation in BRCA1 that abolishes the structure of the Ring domain. Interestingly, BRCA1 bearing the C61G mutation failed to be rapidly recruited to DNA damage sites following laser microirradiation treatment, but retained its ability to accumulate at later times (Figure 1D). These observations suggest that the Ring domain of BRCA1 is required for its early recruitment to DNA damage sites. Moreover, the slow accumulation of the C61G mutant was abolished in H2AX−/− MEFs (Figure 1D), further suggesting that the H2AX-dependent pathway maintains the stability of BRCA1 at DNA damage sites.
The Ring domain of BRCA1 associates with the Ring domain of BARD1, which forms a Ring domain heterodimer and functions as an E3 ubiquitin ligase (Hashizume et al., 2001; Meza et al., 1999; Wu et al., 1996). However, this Ring domain heterodimer per se could not relocate to DNA damage sites (Au and Henderson, 2005), suggesting that BARD1, the Ring domain partner of BRCA1, may target the heterodimer to DNA damage sites. Like BRCA1, BARD1 quickly relocates to DNA damage sites regardless of the status of H2AX, and H2AX is also required for the retention of BARD1 at DNA damage sites (Figure 1E).
Similar to BRCA1, BARD1 has tandem BRCT motifs at its C-terminus. However, unlike the BRCA1 BRCTs, the isolated BARD1 BRCTs relocated to DNA damage sites within 20 seconds after laser microirradiation, but dissociated within 5 minutes. Moreover, the relocation of the BARD1 BRCTs to DNA damage sites was independent of the status of H2AX (Figure 1F). These results suggest that the BARD1 BRCTs might target BRCA1 to DNA damage sites during the early DNA damage response. Structural analysis of the BARD1 BRCTs suggests that, similar to the pSer-binding pocket of the BRCA1 BRCTs, the tandem BARD1 BRCTs fold together and form a binding pocket with K619 as a key residue (Birrane et al., 2007). Thus, we generated a K619A mutant of the BARD1 BRCTs. This mutant form failed to relocate to DNA damage sites, suggesting that this potential binding pocket is important for the rapid relocation of BARD1 to DNA damage sites (Figure 1F).
Next, we asked whether K619 of BARD1 is also important for the recruitment of BRCA1 to DNA damage sites. U2OS cells stably expressing either siRNA-resistant WT BARD1 or siRNA-resistant K619A mutant were treated with BARD1 siRNA to deplete endogenous BARD1 (Figure S1B and S1C). When cells were treated with laser microirradiation, BRCA1 quickly relocated to DNA damage sites in the presence of WT BARD1 but not the K619A mutant, suggesting that the BARD1 BRCTs are critical for targeting BRCA1 to DNA lesions during early DNA damage response (Figure 1G). However, the BARD1 BRCTs mutation did not impair the slow accumulation of BRCA1 to DNA damage sites (Figure 1G). We also generated the L44P mutation in the BARD1 Ring domain, which abolishes BRCA1/BARD1 heterodimer formation (Morris et al., 2002). Like the C61G mutation in the BRCA1 Ring domain, the BARD1 L44P mutant could not facilitate the early recruitment of BRCA1 to DNA damage site (Figure S1D). Taken together, these results suggest that the early recruitment of BRCA1 to DNA damage sites is mediated by the BARD1 BRCTs.
The BARD1 BRCTs bind PAR in vitro and in vivo
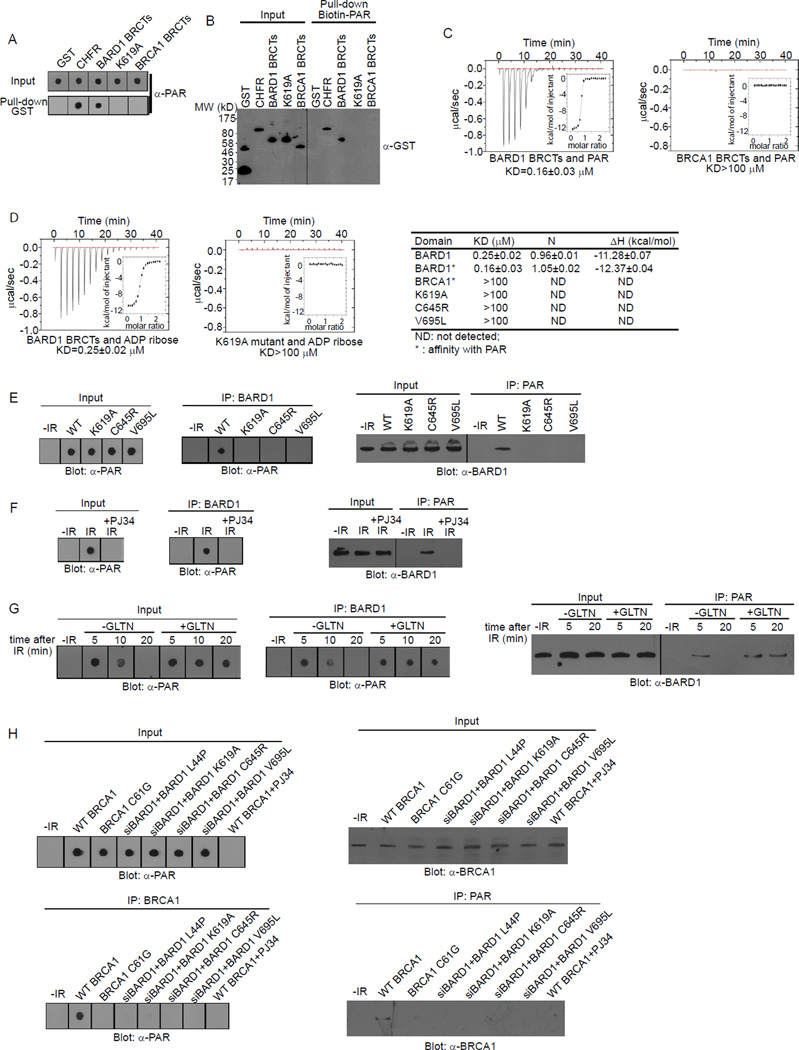
Next we sought the binding partner of the BARD1 BRCTs. Since the relocation kinetics of the BARD1 BRCTs to DNA damage sites is very similar to that of PAR at DNA damage sites (Gibson and Kraus, 2012; Kim et al., 2005), we hypothesized that the BARD1 BRCTs may recognize PAR. We synthesized and purified PAR from an established in vitro assay (Fahrer et al., 2007; Kiehlbauch et al., 1993) and generated recombinant BARD1 BRCTs and the K619A mutant. The WT BARD1 BRCTs but not the K619A mutant could directly co-immunoprecipitate PAR (Figure 2A). Moreover, a reciprocal pull down further confirmed the direct interaction between the BARD1 BRCTs and PAR (Figure 2B). Next, we measured the affinity between the BARD1 BRCTs and PAR using Isothermal Titration Calorimetry (ITC) (Figure 2C). The KD was ~ 0.16 mM, which is very similar to the affinity between the BRCA1 BRCTs and pSer peptide, and the affinity between PAR and its other binding partners (Karras et al., 2005; Yu et al., 2003). Importantly, unlike the BARD1 BRCTs, the BRCA1 BRCTs did not interact with PAR (Figure 2A–C).
Figure 2. The BARD1 BRCTs directly bind PAR.
(A) The interaction between GST (negative control), GST-CHFR (positive control), GST-BARD1 BRCTs, GST-BARD1 BRCTs K619A mutant, or GST-BRCA1 BRCTs and PAR was examined by dot blot using anti-PAR antibody. PAR was blotted and shown as the input. (B) The interaction between the recombinant proteins in (A) and biotin-PAR was examined by the reciprocal pull-down assay with anti-GST antibody. Recombinant proteins were blotted and shown as the input. (C) The affinity between GST-BARD1 BRCTs or GST-BRCA1 BRCTs and PAR was measured by ITC. Titration of PAR was injected into a solution containing the purified protein. The inset shows the fit of the data to an equilibrium binding isotherm. The fit provides an equilibrium dissociation constant (KD) for the binding of PAR to the protein. (D) The affinity between GST-BARD1 BRCTs or GST-BARD1 BRCTs K619A mutant and ADP-ribose was measured by ITC (left). Affinities between the BRCA1 BRCTs, BARD1 BRCTs, or BARD1 BRCTs mutants and ADP ribose or PAR are summarized in the table (right). (E) The in vivo interaction between BARD1 or the mutants and PAR was measured by co-IP and reciprocal co-IP. (F) The in vivo interaction between BARD1 and PAR with or without the treatment of PJ34 was measured by co-IP and reciprocal co-IP. (G) The in vivo interaction between BARD1 and PAR with or without the treatment of GLTN was measured by co-IP and reciprocal co-IP. (H) The in vivo interaction between WT BRCA1, BRCA1 C61G, or WT BRCA1 with the indicated BARD1 mutants and PAR was measured by co-IP and reciprocal co-IP. Whole cell lysates were blotted and shown as the input (E)-(H). See also Figure S2.
Since PAR is a branched polymer of ADP-ribose, we could not precisely determine the structure of PAR synthesized both in vitro and in vivo. Thus, the affinity between the BARD1 BRCTs and PAR might not have been measured accurately by ITC. To clarify this, we measured the affinity between the BARD1 BRCTs and ADP-ribose, the basic unit of PAR. This KD was ~ 0.25 µM, which is again similar to the affinity between other PAR-binding domains and ADP-ribose (Karras et al., 2005). To further examine the interaction between the BARD1 BRCTs and PAR, we performed a competition experiment using excess ADP-ribose in the pull down and reciprocal pull down assays. In these assays, we used 30 ~ 50 mer PAR. At a 100:1 molar ratio between free ADP-ribose and PAR, ADP-ribose could significantly suppress the interaction between the BARD1 BRCTs and PAR (Figure S2A). The results support our interpretation that the BARD1 BRCTs bind each ADP-ribose in PAR. Moreover, we could not detect any interaction between the K619A mutant and ADP-ribose, suggesting that K619 mutation abolishes the BARD1 ADP-ribose binding pocket (Figure 2D).
A previous study suggests that the BARD1 BRCT may selectively recognize the pSDDE motif (Rodriguez et al., 2003). However, the affinity between the XXXXpSDDE peptide (“X” stands for random amino acid) and the BARD1 BRCTs was much weaker than that between PAR and the BARD1 BRCTs (Figure S2B). Accordingly, the pSer peptide could not compete away the interaction between PAR and the BARD1 BRCTs in the same manner as ADP-ribose (Figure S2C).
Germline mutations in the BARD1 BRCTs have been identified in familial breast cancer patients (Ishitobi et al., 2003; Sauer and Andrulis, 2005; Thai et al., 1998). We randomly picked two of these mutations, C645R and V695L, for further study. The BARD1 BRCTs bearing either mutation failed to bind PAR (Figure 2D and Figure S2D).
Next, we asked whether BARD1 could interact with PAR in vivo. Proteins with long poly(ADP-ribose) chains (>100 ADP-ribose) cannot easily migrate into the SDS-PAGE because of the size and phosphate moieties in PAR (Figure S2E). Thus, we used dot blot to examine PAR binding in vivo, which is a better approach to recover the long PAR chains during the analysis (Affar et al., 1998; Fiorillo et al., 2005; Vilchez Larrea et al., 2011). Following DNA damage, PAR is quickly synthesized at the DNA damage sites (D'Amours et al., 1999; Kim et al., 2005). Thus, without DNA damage, we could not detect PAR in vivo (Figure 2E). Following IR treatment, we could not only detect PAR, but also found that PAR interacted with BARD1 using co-immunoprecipitation (co-IP) assays. The results were further confirmed by reciprocal co-IP (Figure 2E). Moreover, BARD1 itself was not PARylated (Figure S2F), and the K619A, C645R and V695L mutants of BARD1 could not interact with PAR (Figure 2E), consistent with the in vitro analysis. Next, we treated cells with PJ34, a potent PARP inhibitor, to suppress PAR synthesis at DNA damage sites. With PJ34 treatment, BARD1 could no longer interact with PAR after IR treatment (Figure 2F).
Immediately following DNA damage, the PAR that is synthesized in a few seconds by PARPs is hydrolyzed quickly by PAR glycohydrolase (PARG) (D'Amours et al., 1999; Kim et al., 2005). Without any treatment, the BARD1/PAR complex is diminished within 20 minutes following IR. However, when we pretreated cells with Gallotannin (GLTN), a cell-permeable PARG inhibitor that suppresses PAR degradation in vivo (Fathers et al., 2012; Ying et al., 2001), DNA damage-induced PAR remained elevated. Thus, we could detect the BARD1/PAR complex in a significantly prolonged period following DNA damage (Figure 2G).
Since BRCA1 forms a stable complex with BARD1, we asked whether BRCA1 also associates with PAR in vivo. As shown in Figure 2H, WT BRCA1 did associate with PAR in vivo following IR treatment. However, the C61G mutation that disrupts the interaction with BARD1 also abolished the PAR interaction. Moreover, cells depleted of endogenous BARD1 by siRNA and reconstituted with the L44P, K619A, C645R or V659L BARD1 mutants lacked the interaction between BRCA1 and PAR (Figure 2H). Taken together, these results demonstrate that the BARD1 BRCTs interact with PAR both in vitro and in vivo and mediate an association of BRCA1 with PAR in vivo.
PARP inhibition suppresses the early recruitment of the BRCA1/BARD1 complex to DNA lesions
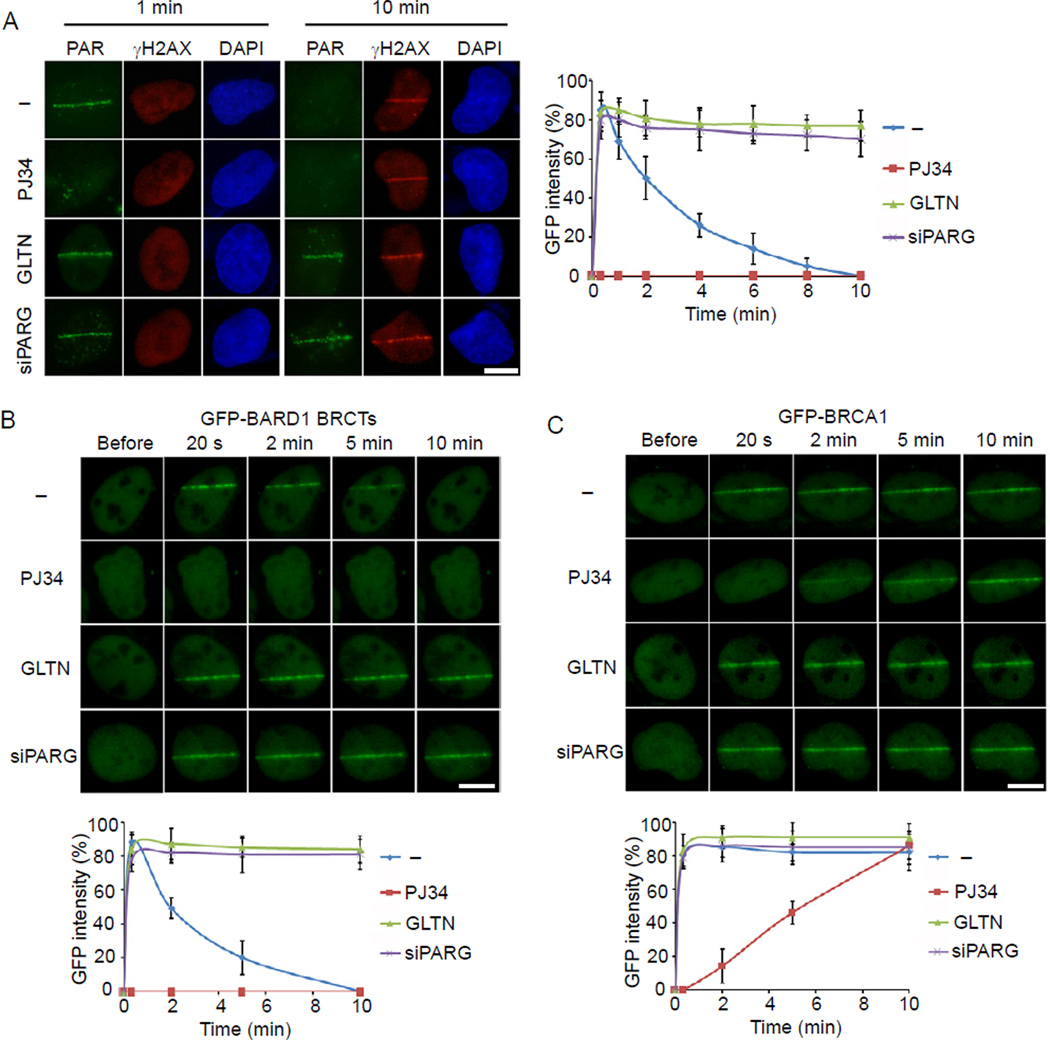
Accumulated evidence shows that cells bearing BRCA mutations are hypersensitive to PARP inhibitors (Bryant et al., 2005; Farmer et al., 2005; Rouleau et al., 2010). Since the BARD1 BRCTs bind PAR, we examined the effects of PARP inhibitor treatment on the recruitment of BRCA1 to DNA damage sites. As expected, PAR was quickly synthesized at DNA damage sites, appearing within less than a minute, and almost degraded within 10 minutes following laser microirradiation (Figure 3A). With PJ34 treatment to suppress PARP activity, we could not detect PAR at DNA damage sites (Figure 3A; Figure S3A). However, PJ34 treatment did not affect the phosphorylation of H2AX at DNA damage sites (Figure 3A; Figure S3B), suggesting that laser microirradiation still induced DNA damage when cells were treated with PJ34. With GLTN treatment or PARG knockdown (Figure S1B) to suppress PARG activity, PAR could be detected at DNA damage sites for a prolonged period (Figure 3A).
Figure 3. The effect of PARP inhibitor on the recruitment of the BRCA1/BARD1 heterodimer to DNA lesions during early DNA damage response.
(A) Representative staining of PAR and γH2AX in cells pretreated by PJ34, GLTN, or PARG knockdown at 1 min or 10 min after laser microirradiation (left). Cells pretreated by PJ34, GLTN, or PARG knockdown were fixed at the indicated time points after laser microirradiation, and the kinetics of PAR staining was examined and summarized in the panel graph (right). (B) The effects of PJ34, GLTN and PARG knockdown on the recruitment of BARD1 BRCTs to DNA damage sites. GFP-BARD1 BRCTs was expressed in U2OS cells pretreated by PJ34, GLTN or PARG knockdown. The relocation of GFP-BARD1 BRCTs was monitored in a time course following laser microirradiation. (C) The effects of PJ34, GLTN and PARG knockdown on the recruitment of BRCA1 to DNA damage sites. GFP-BRCA1 was expressed in U2OS cells pretreated by PJ34, GLTN or PARG knockdown. The relocation of BRCA1 to DNA damage sites was monitored in a time course following laser microirradiation. GFP fluorescence at the laser line was converted into a numerical value (relative fluorescence intensity) using Axiovision software (version 4.5). Normalized fluorescent curves from 20 cells from three independent experiments were averaged. The error bars represent the standard deviation. Scale bar = 10 µm. See also Figure S3.
We next examined the relocation kinetics of the BARD1 BRCTs to DNA damage sites. Similar to the kinetics of PAR at DNA damage sites, PJ34 treatment abolished the recruitment of the BARD1 BRCTs to DNA damage sites (Figure 3B; Figure S3A), whereas GLTN treatment or PARG knockdown significantly prolonged the retention (Figure 3B). A similar relocation kinetics of BRCA1 to DNA damage sites was also observed with these treatments (Figure 3C). The fast relocation of the BARD1 BRCTs and BRCA1 to DNA damage sites was also suppressed by the distinct PARP inhibitors olaparib and ABT-888 (Figure S3C and S3D). We further confirmed the results using Parp1−/− MEFs (Figure S3E), in which the early recruitment of BARD1, the BARD1 BRCTs and BRCA1 were all impaired. Although a small amount of BARD1 and BARD1 BRCTs was still recruited to DNA lesions during the early DNA damage response in Parp1−/− MEFs, it is likely due to the small amount PAR synthesized at DNA lesions in the Parp1−/− cells, since PARP1 synthesizes most but not all of the PAR in response to DNA damage (Kim et al., 2005; Schreiber et al., 2006). Collectively, these data suggest that BRCA1 is recruited by PAR during the early DNA damage response.
BRCA1/BARD1 is quickly recruited to an I-SceI-induced DSB
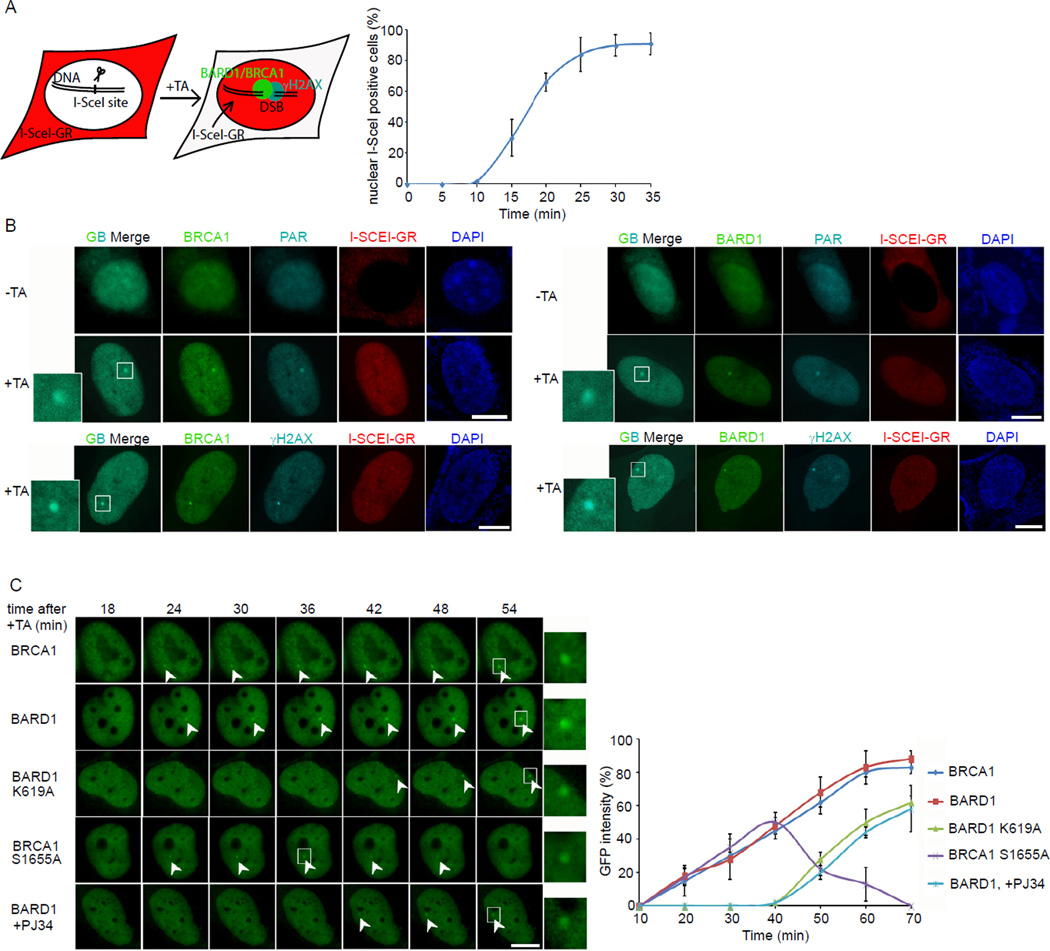
Laser microirradiation not only induces DSBs, but might also generate SSBs. Since BRCA1 mainly participates in HR repair for DSBs, we wondered whether PAR mediates the recruitment of BRCA1 to DSBs. We adopted an inducible I-SceI system to generate a single DSB in vivo (Soutoglou et al., 2007). I-SceI was fused with glucocorticoid receptor (GR) and RFP. Through triamcinolone acetonide (TA) induction, I-SceI translocated into the nucleus between 10 to 20 minutes and its location was monitored by RFP fluorescence (Figure 4A). Twenty minutes following TA induction, we observed a single BRCA1 and BARD1 focus, which colocalized with a PAR focus (Figure 4B). The focus of BRCA1 or BARD1 also colocalized with γH2AX, the surrogate marker of DSB presence. These results suggest that PAR participates in the DSB-induced DNA damage response.
Figure 4. The recruitment of BRCA1/BARD1 complex to DSB.
(A) Schematics of the inducible I-SceI system. TA treatment induces the translocation of RFP-I-SceI-GR fusion protein (red) from the cytoplasm to the nucleus (left). A time course shows the translocation kinetics of RFP-I-SceI-GR from the cytoplasm to the nucleus after TA addition (right). (B) The localization of BRCA1 or BARD1 and PAR before and after TA induction. The DSB (focus) was also marked by γH2AX. Magnified boxes denote the co-localization of BRCA1 or BARD1 with PAR or γH2AX at the DSB. (C) Real time images of the recruitments of GFP-BRCA1, GFP-BARD1 and their GFP-mutants in the inducible I-SceI system. Magnified boxes denote the GFP-fusion proteins at focus. The error bars represent the standard deviation. Scale bar = 10 µm.
Using the I-SceI system, we found that both BRCA1 and BARD1 relocated to a DSB very quickly. However, the K619A mutant of BARD1 that disrupts the interaction with PAR could not relocate to the DSB during early DNA damage response. The S1655A mutant of BRCA1 that associates with BARD1 but disrupts the interaction with its phosphoprotein binding partners was recruited to but not stabilized at the DSB. Moreover, PJ34 pretreatment abolished the rapid recruitment of BARD1 (Figure 4C). These results are consistent with the relocation kinetics seen with laser microirradiation. It further confirms that the interaction between PAR and BARD1 mediates the early recruitment of the BRCA1/BARD1 complex to DNA damage sites, and that the intact BRCA1 BRCTs are critical for the retention (or the slow accumulation) of the BRCA1/BARD1 complex at DNA damage sites.
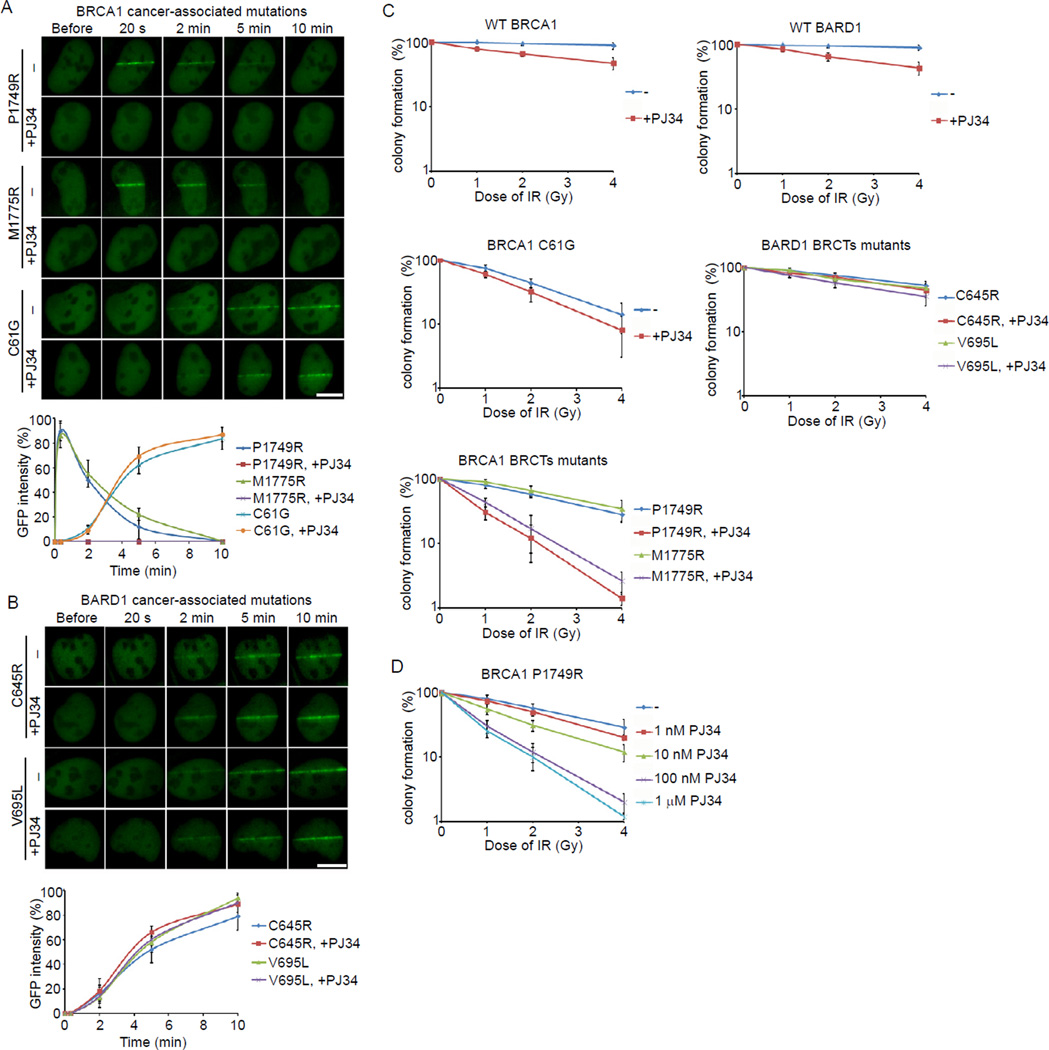
Efficacies of PARP inhibition on cancer-associated BRCA1 and BARD1 mutants
Since PAR targets the BRCA1/BARD1 complex to DNA damage sites, we examined the effects of PARP inhibition on the recruitments of cancer-associated BRCA1 and BARD1 mutants to DNA damage sites. We first selected several cancer-associated BRCA1 and BARD1 mutants and categorized them into three groups: the BRCA1 BRCT mutants (P1749R and M1775R), the BRCA1 Ring domain mutant (C61G) and the BARD1 BRCT mutants (C645R and V695L). As shown in Figure 5A and B, without PJ34 treatment, only the BRCA1 BRCT mutants could rapidly relocate to DNA damage sites. Both the BRCA1 Ring domain mutant and the BARD1 BRCT mutants failed to quickly relocate to DNA damage sites. The BRCA1 Ring domain mutant abolishes the interaction with BARD1, thus this mutant could only slowly accumulate at DNA damage sites through the intact BRCA1 BRCTs. The BARD1 BRCT mutants abolished the interaction with PAR. Thus, these BARD1 BRCT mutants could only slowly accumulate at DNA damage sites since they associate with WT BRCA1 and the interaction between the BRCA1 BRCTs and pSer motifs is intact. Although the BRCA1 BRCT mutants could quickly relocate to DNA damage sites, they could not stably exist there since these mutations abolish the interaction with the pSer-motifs. With PJ34 treatment, which suppresses PAR synthesis at DNA damage sites, the relocation of the BRCA1 BRCT mutants to DNA damage sites was abolished. However, PJ34 treatment affected neither the slow accumulation of the BRCA1 Ring domain mutant nor the BARD1 BRCT mutants.
Figure 5. Efficacy of PARP inhibitor on cancer-associated BRCA1 and BARD1 mutants.
(A) The effect of PJ34 on the recruitment of the P1749R and M1775R mutants of BRCA1 to DNA damage sites. GFP-BRCA1 mutants were expressed in U2OS cells with or without the treatment of PJ34. The relocation of BRCA1 mutants to DNA damage sites was monitored in a time course following laser microirradiation. (B) The effect of PJ34 on the recruitment of the C645R and V695L mutants of BARD1 to DNA damage sites. GFP-BARD1 mutants were expressed in U2OS cells with or without the treatment of PJ34. The relocation of BARD1 mutants to DNA damage sites was monitored in a time course following laser microirradiation. Scale bar = 10 µm. (C) The sensitivities of cells bearing cancer-associated BRCA1 or BARD1 mutants to low dose of IR in the presence or absence of PJ34. (D) The effect of different doses of PJ34 on the cells bearing the P1749R mutant treated by IR. The error bars represent the standard deviation.
We then explored the sensitivities of cells with the different BRCA1 and BARD1 mutations to PJ34 during DNA damage. The siRNA-resistant cDNA of these mutants was generated. U2OS cells stably expressing these constructs were transfected with siRNA to deplete endogenous BRCA1 or BARD1 (Figure S1B and S1C). Cells were treated with or without PJ34 followed by a low dose of ionizing radiation (IR). We found that the recruitment to DNA damage sites correlated well with the sensitivity of these cancer-associated BRCA1 and BARD1 mutants to PARP inhibition. Since PARP inhibitor treatment only suppresses the quick recruitment of the BRCA1 BRCT mutants to DNA damage sites but not the slow accumulation of the BRCA1 Ring domain mutant and the BARD1 BRCT mutants at DNA damage sites, only cells expressing the BRCA1 BRCT mutants were hypersensitive to PARP inhibitor treatment and IR (Figure 5C). Moreover, we treated cells bearing the P1749R mutation with different doses of PJ34. Only a higher dose of PJ34 that suppressed the relocation of BRCA1 to DNA damage sites could sensitize the cells to IR treatment (Figure 5D). Taken together, these results suggest that PARP inhibition sensitizes cells with BRCA1 BRCT mutant to IR treatment.
DISCUSSION
In this study, we identified the BARD1 BRCTs as a PAR-binding motif. DNA damage induces massive PAR synthesis in a very short period of time at DNA lesions (D'Amours et al., 1999; Kim et al., 2005). Previous results suggest that protein PARylation is involved in SSB repair. However, recent structural analysis of PARP1, one of the major PAR polymerases in the DNA damage response, shows that it can recognize DSBs (Ali et al., 2012; Langelier et al., 2012). Our results show that DSBs also induce protein PARylation. Protein PARylation functions as a signal to recruit DNA damage repair proteins like the BRCA1/BARD1 complex to repair DSBs. Suppression of protein PARylation in turn impairs BRCA1/BARD1 recruitment. Our findings also explain the molecular mechanism by which BRCA1/BARD1 is rapidly recruited to DNA damage sites even in the H2AX-deficient cells. Following DSB formation, the BARD1 BRCTs first recognize PAR at DNA lesions, which mediates the rapid recruitment of BRCA1. The retention of BRCA1 is mediated by the BRCA1 BRCTs and is H2AX dependent (Figure S3F). Since SSBs also induce PAR synthesis, it is likely that the BRCA1/BARD1 complex is recruited to SSBs. However, without DNA damage-induced H2AX phosphorylation, the BRCA1/BARD1 complex would be quickly released from DNA damage sites following rapid PAR degradation. Thus, H2AX phosphorylation is still a key factor in facilitating BRCA1 function in DNA damage response.
In our study, the BARD1 BRCTs bind ADP-ribose, the basic unit of PAR, but not phospho-proteins. Although previous peptide library screening showed that the BARD1 BRCT domain preferentially recognizes the pSDDE motif (Rodriguez et al., 2003), the protein binding partner of the BARD1 BRCTs has not yet been identified. Of note, there are two phosphate groups in one ADP-ribose. The phosphate group of Ser and negative charged residues following the pSer in the pSDDE motif might mimic the negatively charged phosphate groups in ADP-ribose, potentially explaining why pSDDE was identified in library screening. Future structural analysis of the BARD1 BRCTs/ADP-ribose complex should reveal the molecular details of the interaction.
Since the BARD1 BRCTs bind ADP-ribose, free ADP-ribose competed with the BARD1-PAR interaction in vitro. Such competition could not occur in vivo because high level of free ADP-ribose in vivo is toxic to cells (Dunn et al., 1999; Hassa et al., 2006). The intracellular concentration of ADP-ribose in mammals is maintained below 100 µM (Gasser and Guse, 2005) and is tightly controlled by specific ADP-ribose hydrolases/pyrophosphatases, which act as protective factors that limit free ADP-ribose accumulation and protein glycation (D'Amours et al., 1999; Fernandez et al., 1996; Hassa et al., 2006; Miro et al., 1989; Ribeiro et al., 1995; Ribeiro et al., 2001). The concentration of NAD+ in undamaged mammalian cells is around 400–500 µM, with 65–75 % of NAD+ utilized to synthesize PAR in response to DNA damage (D'Amours et al., 1999; Hassa et al., 2006). Thus, free ADP-ribose cannot reach a sufficient concentration to compete away PAR in vivo.
The recruitment of the BRCA1/BARD1 complex to DNA damage sites by PAR is important for cells during the DNA damage response. This process ensures that cells which lose certain phosphorylation-dependent pathways could still repair DNA lesions. It might increase cell viability when cells bear germline or somatic mutations of BRCA1. In particular, the most frequent cancer-associated BRCA1 mutations are hypomorphic mutations that lose the C-terminal BRCT domain, which is required to bind to functional partners with pSer motifs. Cells with mutations that abolish the interaction between the BRCA1 BRCTs and pSer motifs could be hypersensitive to PARP inhibitor treatment in part because, in the absence of PAR synthesis, the BRCA1/BARD1 complex could neither quickly relocate to nor slowly accumulate at DNA damage sites. This “double hit” might induce tumor cell lethality. This model also provides an explanation for a recent observation that disrupting the interaction between the BRCA1 BRCTs and phosphoproteins enhances the cytotoxic effect of PARP inhibitors in cancer cells (Langelier et al., 2012).
Notably, PARP inhibitors also kill BRCA1-null cells for which the double-hit model described above is irrelevant (Drost et al., 2011). It has been shown that impairment of base excision repair by PARP inhibitors aggravates the DNA damage repair deficiency in BRCA1-deficient cells and promotes synthetic lethality (Kummar et al., 2012; Rios and Puhalla, 2011). Moreover, PARP inhibitors can trap PARP1 at DNA damage sites (Murai et al., 2012), blocking normal DNA repair. Together with our data, these observations indicate that there are multiple mechanisms by which PARP inhibition can kill breast cancer cells.
EXPERIMENTAL PROCESURES
All other experimental procedures can be found in the Supplemental Information.
Generation and purification of PAR
PAR (or biotin labeled PAR) was synthesized and purified in vitro according to the previous work (Fahrer et al., 2007) with some modifications. Briefly, PAR was synthesized in a 15 ml incubation mixture comprising 100mM Tris-HCl pH 7.8, 10mM MgCl2, 1mM NAD+, 10mM DTT, 60 mg/ml histone H1, 60 mg/ml histone type IIa, 50 mg/ml octameric oligonucleotide GGAATTCC and 150nM human PARP1. The reaction was stopped after 60 min by addition of 20 ml ice-cold 20 % TCA. Following precipitation the pellet was washed with ice-cold 99.8 % ethanol. Polymer was detached using 0.5 M KOH/50 mM EDTA and was purified by phenol-chloroform extraction and isopropanol precipitation. For the ITC assay, PAR was diluted to the indicated concentrations by the buffer containing 10 mM Na2HPO4 (pH 7.5), 100 mM NaCl.
GST fusion protein expression and Dot blot
GST fusion proteins were expressed in Escherichia coli or using the Bac-to-Bac Baculovirus expression system (Invitrogen, for CHFR) and purified under standard procedures. Purified GST fusion proteins (10 pmol) were conjugated to the Glutathione beads and incubated with PAR (100 pmol, calculated as the ADP-ribose unit) for 2 hours at 4°C; For the competition assays, 30 ~ 50 mer PAR was fractionated by anion exchange HPLC protocol as described previously (Fahrer et al., 2007; Kiehlbauch et al., 1993), and used in the experiments. GST-BARD1 BRCTs (10 pmol) were conjugated to the Glutathione beads and incubated with PAR (100 pmol, calculated as the ADP-ribose unit) plus 0.1, 1, or 10 nmol ADP-ribose or pSer peptide respectively for 2 hours at 4°C. The beads were washed with NETN-100 buffer (0.5 % Nonidet P-40, 2 mM EDTA, 50 mM Tris-HCl pH 8.0, 100 mM NaCl) four times. GST-fusion proteins were eluted from beads by glutathione and spotted onto a nitrocellulose membrane. The membrane was blocked with TBST buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.05 % Tween 20) supplemented with 5 % milk, extensively washed with TBST. After drying in the air, the membrane was examined by anti-PAR antibody.
Pull-down assay
Purified GST fusion proteins (1 pmol) were incubated with biotin labeled PAR (5 pmol) and streptavidin beads for 2 hours at 4 °C; For the competition assays, 30 ~ 50 mer biotin-PAR was fractionated by anion exchange HPLC protocol and used in the experiments. GST-BARD1 BRCTs (1 pmol) were incubated with biotin- PAR (5 pmol) and streptavidin beads plus 5, 50, or 500 pmol ADR-ribose or pSer peptide respectively for 2 hours at 4°C. After washing with NETN-100 buffer four times, the samples were boiled in the SDS-sample buffer. The eluates were analyzed by Western blot with anti-GST antibody.
Isothermal Titration Calorimetry (ITC)
ITC was carried out at 16 °C with an ITC 200 Microcalorimeter (GE Healthcare). Proteins were dialyzed extensively into the buffer containing 10 mM Na2HPO4 (pH 7.5), 100 mM NaCl at the final concentrations of 20 ~ 60 µM. Ligands (PAR, ADP-ribose, or pSer peptide) in the injection syringe were also diluted by the same buffer at the final concentration of 150 ~ 750 µM (the concentration of PAR was calculated as the ADP-ribose unit). A typical titration consisted of 19 consecutive 2-µl injections ligands following a pre-injection of 0.4 µl into the protein solution at time intervals of 120 s while stirring at 1000 rpm. Binding isotherms were integrated and analyzed using the software Origin 7.0 (OriginLab, USA) provided by the manufacturer.
Laser microirradiation and live cell imaging
U2OS cells or MEFs transfected with the indicated plasmids were plated on glass-bottomed culture dishes (Mat Tek Corporation). Laser microirradiation was performed using an IX 71 microscope (Olympus) coupled with the MicroPoint Laser Illumination and Ablation System (Photonic Instruments, Inc.). A 337.1 nm laser diode (3.4 mW) transmits through a specific Dye Cell and then yields 365 nm wavelength laser beam that is focused through ×60 UPlanSApo/1.35 oil objective to yield a spot size of 0.5–1 µm. The time of cell exposure to the laser beam was around 3.5 ns. The pulse energy is 170 µJ at 10 Hz. Images were taken by the same microscope with the CellSens software (Olympus). GFP fluorescence at the laser line was converted into a numerical value (relative fluorescence intensity) using Axiovision software (version 4.5). Normalized fluorescent curves from 20 cells from three independent experiments were averaged. The error bars represent the standard deviation.
Inducible Double-Strand Break system
U2OS cells stably expressing RFP-I-SceI-GR were used in this system. The synthetic glucocorticoid ligand triamcinolone acetonide (TA, Sigma) was added (0.1 µM) to induce the translocation of RFP-I-SceI-GR from cytoplasm into nucleus (Zeitlin et al., 2009). Images were taken using the same microscope of laser microirradiation with the CellSens software (Olympus).
Ionizing radiation treatment and colony formation assay
Cells were cultured and irradiated 16 h later with a 137Cs source at a dose of 10 Gy. After irradiation, cells were lysed at the indicated time points for immunoprecipitation or Western blot. For colony formation assay, cells were split into 6-well plates and then treated by various doses of IR with or without PJ34. After a 7-day culture, the viable cells were fixed and stained with crystal violet. The number of colonies (more than 50 cells for each colony) was calculated.
Supplementary Material
SIGNIFICANCE.
Familial breast cancers are often derived from germline mutations of BRCA1,with mutation carriers having a ~90% lifetime risk of developing breast cancer. PARP inhibitors selectively kill BRCA1-deficient cells and several PARP inhibitors are currently in breast cancer clinical trials. However, the mechanism underlying the sensitivity of tumor cells bearing BRCA1 mutations to PARP inhibition is not clear. Here, we found that PARP inhibition directly suppresses the fast recruitment of the BRCA1/BARD1 heterodimer to DNA damage sites and impairs DNA repair. These findings suggest a mechanism by which PARP inhibitors specifically kill breast cancer cells bearing BRCA1 mutations.
Highlights.
The BARD1 BRCT domain targets BRCA1 to DNA lesions during early DNA damage response
The BARD1 BRCT domain is a PAR-binding domain
PARP inhibitors suppress the fast recruitment of BRCA1 to DNA lesions
PARP inhibition sensitizes the cells bearing mutations in the BRCA1 BRCT domain to IR
ACKNOWLEDGEMENTS
We thank Dr. Ming Lei for technical support, Drs. Mats Ljungman, Thomas Wilson, Jennifer Keller and Henry Kuang for editing and proofreading of the manuscript. This work was supported by the National Institute of Health (CA132755 and CA130899 to X.Y.). X.Y. is a recipient of the Era of Hope Scholar Award from the Department of Defense.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abbott DW, Thompson ME, Robinson-Benion C, Tomlinson G, Jensen RA, Holt JT. BRCA1 expression restores radiation resistance in BRCA1-defective cancer cells through enhancement of transcription-coupled DNA repair. J Biol Chem. 1999;274:18808–18812. doi: 10.1074/jbc.274.26.18808. [DOI] [PubMed] [Google Scholar]
- Affar EB, Duriez PJ, Shah RG, Sallmann FR, Bourassa S, Kupper JH, Burkle A, Poirier GG. Immunodot blot method for the detection of poly(ADP-ribose) synthesized in vitro and in vivo. Anal Biochem. 1998;259:280–283. doi: 10.1006/abio.1998.2664. [DOI] [PubMed] [Google Scholar]
- Ali AA, Timinszky G, Arribas-Bosacoma R, Kozlowski M, Hassa PO, Hassler M, Ladurner AG, Pearl LH, Oliver AW. The zinc-finger domains of PARP1 cooperate to recognize DNA strand breaks. Nat Struct Mol Biol. 2012;19:685–692. doi: 10.1038/nsmb.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au WW, Henderson BR. The BRCA1 RING and BRCT domains cooperate in targeting BRCA1 to ionizing radiation-induced nuclear foci. J Biol Chem. 2005;280:6993–7001. doi: 10.1074/jbc.M408879200. [DOI] [PubMed] [Google Scholar]
- Birrane G, Varma AK, Soni A, Ladias JA. Crystal structure of the BARD1 BRCT domains. Biochemistry. 2007;46:7706–7712. doi: 10.1021/bi700323t. [DOI] [PubMed] [Google Scholar]
- Botuyan MV, Nomine Y, Yu X, Juranic N, Macura S, Chen J, Mer G. Structural basis of BACH1 phosphopeptide recognition by BRCA1 tandem BRCT domains. Structure. 2004;12:1137–1146. doi: 10.1016/j.str.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- Burkle A. Physiology and pathophysiology of poly(ADP-ribosyl)ation. Bioessays. 2001;23:795–806. doi: 10.1002/bies.1115. [DOI] [PubMed] [Google Scholar]
- Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol. 2003;5:675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- Clapperton JA, Manke IA, Lowery DM, Ho T, Haire LF, Yaffe MB, Smerdon SJ. Structure and mechanism of BRCA1 BRCT domain recognition of phosphorylated BACH1 with implications for cancer. Nat Struct Mol Biol. 2004;11:512–518. doi: 10.1038/nsmb775. [DOI] [PubMed] [Google Scholar]
- D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342(Pt 2):249–268. [PMC free article] [PubMed] [Google Scholar]
- Drost R, Bouwman P, Rottenberg S, Boon U, Schut E, Klarenbeek S, Klijn C, van der Heijden I, van der Gulden H, Wientjens E, et al. BRCA1 RING function is essential for tumor suppression but dispensable for therapy resistance. Cancer Cell. 2011;20:797–809. doi: 10.1016/j.ccr.2011.11.014. [DOI] [PubMed] [Google Scholar]
- Dunn CA, O'Handley SF, Frick DN, Bessman MJ. Studies on the ADP-ribose pyrophosphatase subfamily of the nudix hydrolases and tentative identification of trgB, a gene associated with tellurite resistance. J Biol Chem. 1999;274:32318–32324. doi: 10.1074/jbc.274.45.32318. [DOI] [PubMed] [Google Scholar]
- Fahrer J, Kranaster R, Altmeyer M, Marx A, Burkle A. Quantitative analysis of the binding affinity of poly(ADP-ribose) to specific binding proteins as a function of chain length. Nucleic Acids Res. 2007;35:e143. doi: 10.1093/nar/gkm944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- Fathers C, Drayton RM, Solovieva S, Bryant HE. Inhibition of poly(ADP-ribose) glycohydrolase (PARG) specifically kills BRCA2-deficient tumor cells. Cell Cycle. 2012;11:990–997. doi: 10.4161/cc.11.5.19482. [DOI] [PubMed] [Google Scholar]
- Fernandez A, Ribeiro JM, Costas MJ, Pinto RM, Canales J, Cameselle JC. Specific ADP-ribose pyrophosphatase from Artemia cysts and rat liver: effects of nitroprusside, fluoride and ionic strength. Biochim Biophys Acta. 1996;1290:121–127. [PubMed] [Google Scholar]
- Fiorillo C, Nediani C, Ponziani V, Giannini L, Celli A, Nassi N, Formigli L, Perna AM, Nassi P. Cardiac volume overload rapidly induces oxidative stress-mediated myocyte apoptosis and hypertrophy. Biochim Biophys Acta. 2005;1741:173–182. doi: 10.1016/j.bbadis.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Fisher AE, Hochegger H, Takeda S, Caldecott KW. Poly(ADP-ribose) polymerase 1 accelerates single-strand break repair in concert with poly(ADP-ribose) glycohydrolase. Mol Cell Biol. 2007;27:5597–5605. doi: 10.1128/MCB.02248-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O'Connor MJ, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- Gasser A, Guse AH. Determination of intracellular concentrations of the TRPM2 agonist ADP-ribose by reversed-phase HPLC. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;821:181–187. doi: 10.1016/j.jchromb.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol. 2012;13:411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ. The DNA damage response: ten years after. Molecular cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Hashizume R, Fukuda M, Maeda I, Nishikawa H, Oyake D, Yabuki Y, Ogata H, Ohta T. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J Biol Chem. 2001;276:14537–14540. doi: 10.1074/jbc.C000881200. [DOI] [PubMed] [Google Scholar]
- Hassa PO, Haenni SS, Elser M, Hottiger MO. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol Mol Biol Rev. 2006;70:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Scully R, Sobhian B, Xie A, Shestakova E, Livingston DM. RAP80-directed tuning of BRCA1 homologous recombination function at ionizing radiation-induced nuclear foci. Genes Dev. 2011;25:685–700. doi: 10.1101/gad.2011011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Sy SM, Chen J. BRCA1 and its toolbox for the maintenance of genome integrity. Nat Rev Mol Cell Biol. 2010;11:138–148. doi: 10.1038/nrm2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitobi M, Miyoshi Y, Hasegawa S, Egawa C, Tamaki Y, Monden M, Noguchi S. Mutational analysis of BARD1 in familial breast cancer patients in Japan. Cancer Lett. 2003;200:1–7. doi: 10.1016/s0304-3835(03)00387-2. [DOI] [PubMed] [Google Scholar]
- Karras GI, Kustatscher G, Buhecha HR, Allen MD, Pugieux C, Sait F, Bycroft M, Ladurner AG. The macro domain is an ADP-ribose binding module. EMBO J. 2005;24:1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehlbauch CC, Aboul-Ela N, Jacobson EL, Ringer DP, Jacobson MK. High resolution fractionation and characterization of ADP-ribose polymers. Anal Biochem. 1993;208:26–34. doi: 10.1006/abio.1993.1004. [DOI] [PubMed] [Google Scholar]
- Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007;316:1202–1205. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: 'PAR-laying' NAD+ into a nuclear signal. Genes Dev. 2005;19:1951–1967. doi: 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]
- King MC, Marks JH, Mandell JB. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302:643–646. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Altschul SF, Bork P. BRCA1 protein products … Functional motifs. Nat Genet. 1996;13:266–268. doi: 10.1038/ng0796-266. [DOI] [PubMed] [Google Scholar]
- Kummar S, Chen A, Parchment RE, Kinders RJ, Ji J, Tomaszewski JE, Doroshow JH. Advances in using PARP inhibitors to treat cancer. BMC Med. 2012;10:25. doi: 10.1186/1741-7015-10-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier MF, Planck JL, Roy S, Pascal JM. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science. 2012;336:728–732. doi: 10.1126/science.1216338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, de Murcia G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol Cell Biol. 1998;18:3563–3571. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meza JE, Brzovic PS, King MC, Klevit RE. Mapping the functional domains of BRCA1. Interaction of the ring finger domains of BRCA1 and BARD1. J Biol Chem. 1999;274:5659–5665. doi: 10.1074/jbc.274.9.5659. [DOI] [PubMed] [Google Scholar]
- Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- Miro A, Costas MJ, Garcia-Diaz M, Hernandez MT, Cameselle JC. A specific, low Km ADP-ribose pyrophosphatase from rat liver. FEBS Lett. 1989;244:123–126. doi: 10.1016/0014-5793(89)81176-7. [DOI] [PubMed] [Google Scholar]
- Morris JR, Keep NH, Solomon E. Identification of residues required for the interaction of BARD1 with BRCA1. J Biol Chem. 2002;277:9382–9386. doi: 10.1074/jbc.M109249200. [DOI] [PubMed] [Google Scholar]
- Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, Ji J, Takeda S, Pommier Y. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 2012;72:5588–5599. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano S, Lan L, Caldecott KW, Mori T, Yasui A. Spatial and temporal cellular responses to single-strand breaks in human cells. Mol Cell Biol. 2003;23:3974–3981. doi: 10.1128/MCB.23.11.3974-3981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman N, Stratton MR. The genetics of breast cancer susceptibility. Annu Rev Genet. 1998;32:95–121. doi: 10.1146/annurev.genet.32.1.95. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Cameselle JC, Fernandez A, Canales J, Pinto RM, Costas MJ. Inhibition and ADP-ribose pyrophosphatase-I by nitric-oxide-generating systems: a mechanism linking nitric oxide to processes dependent on free ADP-ribose. Biochem Biophys Res Commun. 1995;213:1075–1081. doi: 10.1006/bbrc.1995.2237. [DOI] [PubMed] [Google Scholar]
- Ribeiro JM, Carloto A, Costas MJ, Cameselle JC. Human placenta hydrolases active on free ADP-ribose: an ADP-sugar pyrophosphatase and a specific ADP-ribose pyrophosphatase. Biochim Biophys Acta. 2001;1526:86–94. doi: 10.1016/s0304-4165(01)00113-1. [DOI] [PubMed] [Google Scholar]
- Rios J, Puhalla S. PARP inhibitors in breast cancer: BRCA and beyond. Oncology (Williston Park) 2011;25:1014–1025. [PubMed] [Google Scholar]
- Rodriguez M, Yu X, Chen J, Songyang Z. Phosphopeptide binding specificities of BRCA1 COOH-terminal (BRCT) domains. J Biol Chem. 2003;278:52914–52918. doi: 10.1074/jbc.C300407200. [DOI] [PubMed] [Google Scholar]
- Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R, Chun J, Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer. 2011;12:68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruscetti T, Lehnert BE, Halbrook J, Le Trong H, Hoekstra MF, Chen DJ, Peterson SR. Stimulation of the DNA-dependent protein kinase by poly(ADP-ribose) polymerase. J Biol Chem. 1998;273:14461–14467. doi: 10.1074/jbc.273.23.14461. [DOI] [PubMed] [Google Scholar]
- Sauer MK, Andrulis IL. Identification and characterization of missense alterations in the BRCA1 associated RING domain (BARD1) gene in breast and ovarian cancer. J Med Genet. 2005;42:633–638. doi: 10.1136/jmg.2004.030049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- Scully R, Chen J, Ochs RL, Keegan K, Hoekstra M, Feunteun J, Livingston DM. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell. 1997;90:425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- Scully R, Livingston DM. In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature. 2000;408:429–432. doi: 10.1038/35044000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen SX, Weaver Z, Xu X, Li C, Weinstein M, Chen L, Guan XY, Ried T, Deng CX. A targeted disruption of the murine Brca1 gene causes gamma-irradiation hypersensitivity and genetic instability. Oncogene. 1998;17:3115–3124. doi: 10.1038/sj.onc.1202243. [DOI] [PubMed] [Google Scholar]
- Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E, Dorn JF, Sengupta K, Jasin M, Nussenzweig A, Ried T, Danuser G, Misteli T. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol. 2007;9:675–682. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai TH, Du F, Tsan JT, Jin Y, Phung A, Spillman MA, Massa HF, Muller CY, Ashfaq R, Mathis JM, et al. Mutations in the BRCA1-associated RING domain (BARD1) gene in primary breast, ovarian and uterine cancers. Hum Mol Genet. 1998;7:195–202. doi: 10.1093/hmg/7.2.195. [DOI] [PubMed] [Google Scholar]
- Turner N, Tutt A, Ashworth A. Hallmarks of 'BRCAness' in sporadic cancers. Nat Rev Cancer. 2004;4:814–819. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- Vilchez Larrea SC, Alonso GD, Schlesinger M, Torres HN, Flawia MM, Fernandez Villamil SH. Poly(ADP-ribose) polymerase plays a differential role in DNA damage-response and cell death pathways in Trypanosoma cruzi. Int J Parasitol. 2011;41:405–416. doi: 10.1016/j.ijpara.2010.11.008. [DOI] [PubMed] [Google Scholar]
- Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Lee MS, Hau DD, Glover JN. Structural basis of phosphopeptide recognition by the BRCT domain of BRCA1. Nat Struct Mol Biol. 2004;11:519–525. doi: 10.1038/nsmb776. [DOI] [PubMed] [Google Scholar]
- Wu LC, Wang ZW, Tsan JT, Spillman MA, Phung A, Xu XL, Yang MC, Hwang LY, Bowcock AM, Baer R. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat Genet. 1996;14:430–440. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- Ying W, Sevigny MB, Chen Y, Swanson RA. Poly(ADP-ribose) glycohydrolase mediates oxidative and excitotoxic neuronal death. Proc Natl Acad Sci U S A. 2001;98:12227–12232. doi: 10.1073/pnas.211202598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302:639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- Zeitlin SG, Baker NM, Chapados BR, Soutoglou E, Wang JY, Berns MW, Cleveland DW. Double-strand DNA breaks recruit the centromeric histone CENP-A. Proc Natl Acad Sci U S A. 2009;106:15762–15767. doi: 10.1073/pnas.0908233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.