Abstract
During the last two decades, the studies on ubiquitination in regulating transcription factor NF-κB activation have elucidated the expanding role of ubiquitination in modulating cellular events by non-proteolytic mechanisms, as well as by proteasomal degradation. The significance of ubiquitination has also been recognized in regulating gene transcription, epigenetic modifications, kinase activation, DNA repair and subcellular translocation. This progress has been translated into novel strategies for developing anti-cancer therapeutics, exemplified by the success of the first FDA-approved proteasome inhibitor drug Bortezomib. Here we discuss the current understanding of the ubiquitin-proteasome system and how it is involved in regulating NF-κB signaling pathways in response to a variety of stimuli. We also focus on the recent progress of anti-cancer drug development targeting various steps of ubiquitination process, and the potential of these drugs in cancer treatment as related to their impact on NF-κB activation.
Keywords: Ubiquitin, NF-κB, Cancer, Therapeutics
INTRODUCTION
Ubiquitin-proteasome system (UPS) [1] and Nuclear Factor kappa B (NF-κB) [2] were among the most exciting discoveries during the late 1970s and 1980s, and continue to remain as hotbeds of current research. The finding of protein ubiquitination and subsequent proteasome-dependent degradation shed the first light into how cellular protein turnover is regulated to maintain homeostasis and to cope with cellular stresses [3, 4]. Moreover, following a deeper understanding of the non-proteolytic roles of ubiquitination in mediating signaling transduction, gene transcription, DNA repair and protein translocation [5], the importance of UPS in various human diseases, such as neurodegenerative diseases and cancers, has become well-appreciated [6]. On the other hand, the function of NF-κB has been broadened from its original discovery as a nuclear factor binding to immunoglobin κ light chain gene promoter in B cells to a pivotal player in orchestrating cell proliferation, apoptosis, inflammation, stress response and immunity via regulating expression of a myriad of genes [7]. Moreover, the explosive growth of our knowledge of how ubiquitination controls various NF-κB signaling pathways in the last two decades has shifted the paradigms of both research fields, leading to novel therapeutic strategies for treating cancers, such as multiple myeloma and mantle cell lymphoma, in which dysregulated NF-κB signaling plays a significant role in pathogenesis. The success of Bortezomib (also known as Velcade or PS-341), the first (and the only) proteasome-inhibiting drug approved by FDA, in treating multiple myeloma and relapsed mantle cell lymphoma has inspired increasing enthusiasm in developing novel anti-cancer drugs targeting UPS and ubiquitination–mediated NF-κB signaling [6, 8].
Many comprehensive reviews have been published recently on various aspects related to the ubiquitin system and NF-κB [3, 4, 9–13]. In this review, we first provide an overview of the UPS and the NF-κB transcription factor family, highlighting the recent progress in understanding of the crosstalk between NF-κB activation signaling pathways and various polyubiquitin chain linkages. We then focus on the present state and recent developments in utilizing this understanding to identify novel therapeutic targets, and their potential for therapeutic intervention in cancers.
UBIQUITIN-PROTEASOME SYSTEM AND ITS EXPENDING FUNCTIONS
Ubiquitin is a small 76 amino-acid protein that can be covalently attached to an ε-amino group of the substrate protein’s lysine residues through isopeptide bond linkage [14] (Fig. 1A). It was first purified from bovine thymus [15] and later was found in all eukaryotic cells with a highly conserved protein sequence. Ligation of ubiquitin in the form of polyubiquitin chains to a protein substrate, which consumes ATP, usually leads to its proteasome-dependent degradation. Thus, ubiquitin was also called ATP-dependent proteolysis factor 1 (APF1) [16]. Besides attaching to substrates as a side chain, ubiquitin can also be conjugated to the amino-terminal of substrates linearly which then can be extended into a polyubiquitin chain, leading to protein degradation [17].
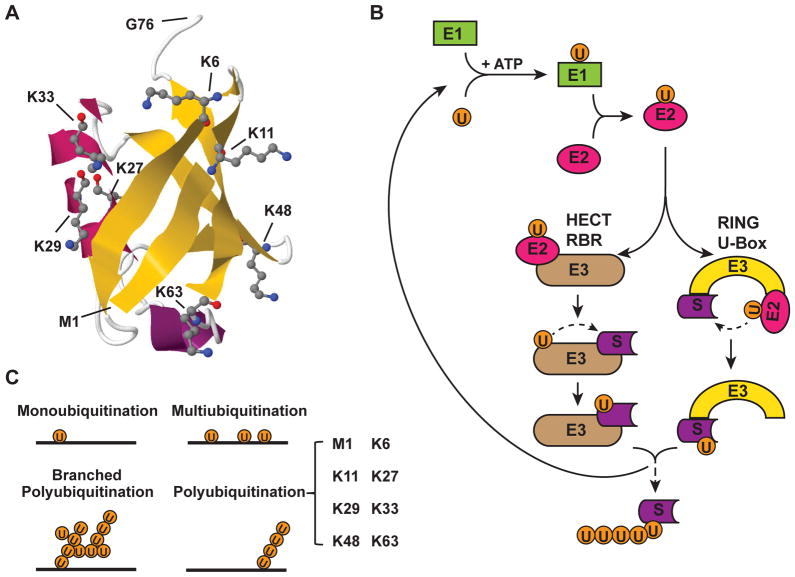
Figure 1. The process of ubiquitination and forms of polyubiquitination.
(A) Structure of a single ubiquitin moiety (Protein Data Bank identifier: 1UBQ) [164] is shown in cartoon style. Secondary structure β-sheet is shown in yellow and α-helix is in magenta. The seven Lys residues and Met1, which can form polyubiquitin linkages with Gly76, are shown in stick-and-ball representation. (B) Ubiquitination is a three-step process which is initiated by activation of ubiquitin. Ubiquitin-activating enzyme (E1) binds to ubiquitin with a high-energy thioester linkage between C-terminus of ubiquitin and active site cysteine residue on the E1, which consumes ATP. Then, the activated ubiquitin moiety is transferred to an ubiquitin-conjugating enzyme (E2) via formation of a second thioester linkage. Finally, with the facilitation of an ubiquitin ligase (E3), ubiquitin is conjugated onto a substrate protein, mostly via an isopeptide bond between the C-terminal Gly76 of the ubiquitin and a internal lysine of the substrate protein. The HECT and RBR family of E3s transfer ubiquitin to substrate via a intermediate step, which involves temporary conjugation of ubiquitin moiety on a internal Cys residue of HECT/RBR E3s. In contrast, the RING and U-box family of E3s modulate direct conjugation of ubiquitin to Lys residue of the substrate from the bound E2. This three-step process can be repeated in multiple cycles, leading to extension of polyubiquitin chain. (C) Depending on the length and number of ubiquitin moiety conjugated on substrate proteins, ubiquitination can be further defined as monoubiquitination (a single ubiquitin moiety conjugated on a single Lys residue of a substrate), multiubiquitination (a single ubiquitin moiety conjugated on multiple Lys residues of a substrate) and polyubiquitination (extended ubiquitin chain forming on a Lys residue of a substrate). Polyubiquitin chain can be linked with homogenous linkages (8 types of linkage as shown in bracket) or with heterogeneous/mixed linkages (also called branched polyubiquitin chain).
Three-Step Process for Ubiquitination
The conjugation of ubiquitin on protein substrates is processed by a three-step cascade mechanism [14] (Fig. 1B). First, ubiquitin is activated by an ubiquitin-activating enzyme (E1) in an ATP-dependent manner which generates a high-energy thioester linkage between the carboxy-terminus of ubiquitin and the E1 enzyme. Then, the activated ubiquitin moiety is transferred by a E2 enzyme (ubiquitin-conjugating enzymes, UBCs), via formation of a second thioester linkage. Finally, with the facilitation of an ubiquitin ligase family of proteins (E3), ubiquitin is conjugated onto a substrate protein, mostly via an isopeptide bond between the C-terminal Gly76 of ubiquitin and an internal lysine of the substrate protein. Less frequently, ubiquitin conjugation can also occured by a peptide linkage between the C-terminus of ubiquitin and the N-terminus of the substrate [18]. In rare occasions, ubiquitin was also found conjugated to internal serine, threonine or cysteine residues of the substrates. The three-step cascade can proceed repeatedly, which leads to further extension of ubiquitin and to form polyubiquitin chains, most commonly via isopeptide linkage between one of seven internal lysines (K6, K11, K27, K29, K33, K48 and K63) of the proximal ubiquitin and the Gly76 of the distal ubiquitin. Recently, a form of linear linkage between Met1 and Gly76 of two ubiquitins was described and referred to as linear, or M1-linked, ubiquitination [19, 20].
In humans, two ubiquitin E1 enzymes, approximately forty E2 enzymes and hundreds of E3 enzymes have been identified. While E1s function universally and E2s often can work with a number of E3s to ubiquitinate different substrates, E3 ligases usually work in a substrate-specific fashion and, along with E2s, determine the linkage specificity of attached polyubiquitin chain [21, 22]. There are several distinct families of E3s, based on specific, commonly shared structural motifs, such as HECT (homologous to the E6-AP C-terminus) domains, RBR (RING between RING) domains, U-box, and RING (really interesting new gene) fingers, which mediate E2 binding and ubiquitination of the substrate. For the HECT and RBR E3s, the ubiquitin moiety is transferred from the E2 to the E3 before its conjugation to the substrate [12, 23]. On the contrary, U-box and RING finger-containing E3s directly transfer the activated ubiquitin from E2 to the E3-associated substrate (Fig. 1B). Many RING-E3 ligases are multi-protein complexes. For example, SCF (Skp1-Cul1-F-box) complex E3 ligases of Cullin–RING ligases (CRL) superfamily, which represents the largest E3 family, consist of multiple subunits, including Cullin (Cul1), S phase kinase-associated protein1 (Skp1), a RING-box protein 1 (RBX1; also known as ROC1), and one of the F-box proteins. Thus far, sixty-nine F-box proteins have been identified in humans, and are classified into three categories based on the additional domains besides the F-box domain, such as WD40 domains (FBXWs), leucine-rich repeats (FBXLs) and other diverse domains (FBXOs) [24]. These subunits of the E3 complex provide multiple targets for pharmacological intervention which we will discuss in the latter section of this review.
Removal of conjugated Ubiquitin
Ubiquitination is a reversible process and the enzymes responsible for removing conjugated ubiquitin from substrates are called deubiquitinating enzymes or deubiquitinases (DUBs). In the human genome, there are 95 DUBs that have been identified based on their activity or protein domain similarity [25]. These DUBs can be divided into five families: ubiquitin carboxy-terminal hydrolase (UCH); ubiquitin specific protease (USP); ovarian tumor domain (OTU); Machado-Joseph disease proteases (MJD); and JAB1/MPN/Mov34 metalloenzyme (JAMM). The former four DUB families belong to cysteine proteases and the fifth is a family of metalloproteases. Different DUBs display distinct affinity and catalytic activity toward specific ubiquitin chain linkages [26]. In an effort to explore the interactome of DUBs in human cells, 774 candidate proteins were identified by association with DUBs, which can be linked to diverse processes, including protein turnover, gene transcription, RNA processing, DNA damage, and endoplasmic reticulum-associated degradation, demonstrating the broad physiological and pathological functions of ubiquitination in cells [27].
Ubiquitination Linkages and Associated Functions
Multiple forms of ubiquitination have been identified and distinct functions have been linked to the different forms of ubiquitination (Fig. 1C). A single ubiquitin moiety can be conjugated to one lysine residue, or to multiple different sites, of the substrate protein, which is termed as monoubiquitination or multiubiquitination, respectively. Diverse functions have been associated with monoubiquitination depending on the ubiquitinated substrate. For example, polycomb complex 1 (PRC1)-dependent histone H2A monoubiquitination results in gene repression or silencing, while monoubiquitination of histone H2B by Bre1 complex promotes transcriptional activation [28]. Monoubiquitination of PCNA affects its affinity toward the replicative polymerases (Polδ and Polε) which are involved in translesion synthesis (TLS) DNA repair [29]. Meanwhile, DNA damage-induced monoubiquitination of Fanconi Anemia (FA) proteins FANCD2 and FANCI by the multi-subunit E3 ubiquitin ligase that is composed of the other 11 members of FA genes directs the chromatin relocation, which is essential for DNA interstrand crosslink repair [30]. Similarly under genotoxic stress condition, low levels of Mdm2 can mediate monoubiquitination of p53 which leads to its nuclear export, thereby inhibiting overactivation of p53 in response to mild DNA damage [31]. Additionally, monoubiquitination of various cell surface receptors serves as a signal for endosomal sorting that targets them for lysosomal degradation [32]. Despite the varied cellular responses upon monoubiquitination, a common theme is that the single ubiquitin moiety conjugated onto a substrate influences the ability of the target protein to interact with other proteins, which carry out the consequent physiological or pathological processes.
The most well-studied ubiquitination form is the K48-linked polyubiquitination. The K48-linked polyubiquitin chain is extended by linkage between G76 of distal ubiquitin and K48 of the proximal ubiquitin. The chain of K48-linked Ub4 adopts a closed structural conformation, which is the minimum signal for efficient proteasome-dependent degradation [33, 34]. The 19S subunit of active proteasomes can recognize polyubiquitinated proteins through directly binding to K48-linked chain and direct the ubiquitinated protein into the barrel shaped 20S subunit where they are cleaved into small peptides. The attached polyubiquitin can be disassembled into free ubiquitins, which can be reused in the UPS system [4].
K11-linked polyubiquitination can also lead to proteasomal degradation, which plays an important role in cell cycle progression [35]. E2s UBE2C (also called UBCH10) along with UBE2S (also known as E2EPF) and E3 anaphase-promoting complex (APC) were found to be the major E2/E3 pairs to catalyze K11 chain formation. Besides its function in regulating cell cycle, K11-linked ubiquitination may also participate in immune signaling independent of its role in promoting proteasomal degradation. Upon TNFα stimulation, cellular inhibitors of apoptosis (cIAP1 and cIAP2) were found to be recruited to TNFR1-associated signaling complexes where they mediated NF-κB activation via promoting K11-linked polyubiquitination of receptor-interacting protein 1 (RIP1) [36].
In contrast to the classical function of K48-ubiquitination in regulating proteasome-dependent protein turnover, the K63-linked polyubiquitination as well as linear polyubiquitination predominantly play a role in regulating cell signaling processes [5, 37]. The studies on the non-proteolytic functions of ubiquitination involved in regulating various NF-κB signaling pathways greatly enhanced our understanding of how UPS system may modulate pathophysiological processes in a variety of diseases which may be exploited as novel drug targets for pharmacological intervention. Therefore, it is not surprising that the crosstalk between UPS and NF-κB signaling has become a hot topic for intensive investigation.
NF-κB FAMILY AND ITS REGULATORY SIGNALING PATHWAYS
NF-κB is a family of transcription factors comprised of five structurally related proteins including RelA/p65, RelB, c-Rel, NF-κB1 (p105, precursor of p50), and NF-κB2 (p100, precursor of p52), which exist in the forms of homo- or heterodimers [38]. RelA/p65, c-Rel and RelB contain transactivation domains, and p65/p50 heterodimer has been studied in depth as the prototypic NF-κB transcription activator. In unstimulated cells, NF-κB resides in the cytoplasm in a latent form by associating with a family of inhibitor proteins called IκBs (Inhibitor of NF-κB), such as IκBα [9, 38]. In response to stimulation, NF-κB is released from associated IκBs and undergoes rapid nuclear translocation, where it can regulate the transcription of its target genes by binding to promoter and/or enhancer regions. In addition to nuclear translocation, the transcriptional activity of NF-κB is also modulated by posttranslational modification, such as p65 phosphorylation, acetylation and methylation [9, 39]. Previous studies demonstrated that NF-κB is involved in the transcriptional regulation of an exceptionally large number of genes (see list at www.nf-kb.org), which participate in a wide range of physiological and pathological processes, such as cell proliferation, innate and adaptive immune responses, inflammation, cell migration, and regulation of apoptosis, among others [9, 40].
A myriad of extracellular stimuli can induce NF-κB activation via two major signaling pathways initiated from membrane-bound receptors, which are called the “canonical” and “non-canonical” pathways [41] (Fig. 2A). The canonical pathway requires IKK (IκB kinase) kinase complex, which is composed of IKKα, IKKβ and IKKγ/NEMO. The IKKβ subunit directly phosphorylates IκBα, leading to its proteasomal degradation through the UPS pathway. NF-κB is then free to translocate into the nucleus and regulate gene transcription. The non-canonical mode of NF-κB activation relies on the NF-κB inducing kinase (NIK)-meditated IKKα homodimer activation, which in turn phosphorylates p100, resulting in proteasome-dependent partial processing of p100 and subsequent p52:RelB dimer nuclear translocation to regulate gene transcription. Genotoxic stress also elicits a retrograde signaling cascade which is initiated from the nucleus to activate cytoplasmic IKK complex, resulting in NF-κB activation [42, 43]. Various forms of ubiquitination have been demonstrated to play essential roles in mediating these NF-κB activation signaling pathways, which we will discuss in detail in following sections (Fig. 2B).
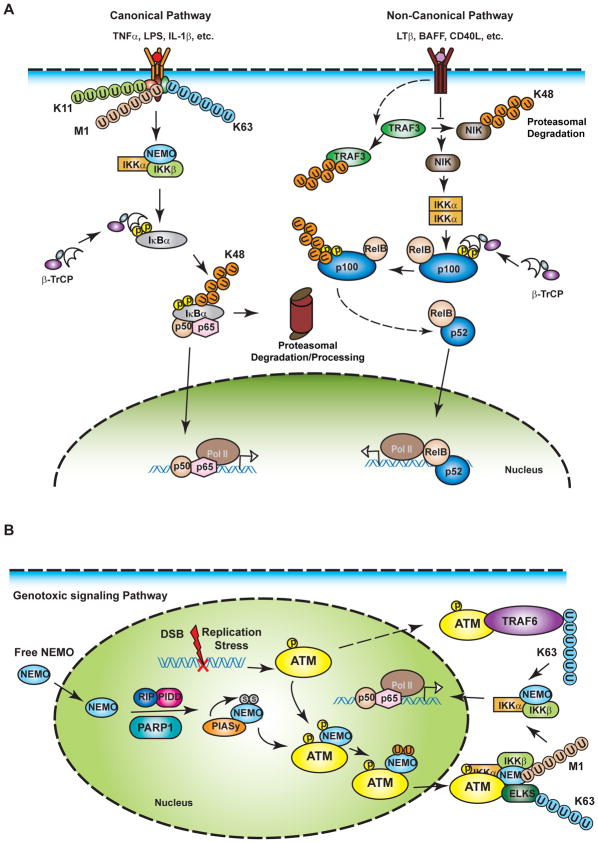
Figure 2. NF-κB activation signaling pathways.
(A) A variety of extracellular stimuli activate NF-κB signaling via the canonical or non-canonical signaling pathway. The canonical pathway is activated in response to engagement of membrane-bound receptors of various cytokines or pathogens, such as TNFα, IL-1β and LPS. One characteristics of the canonical pathway is the activation of NEMO-containing IKK complex which can directly phosphorylate NF-κB-bound IκB proteins, resulting in their β-TrCP-SCF ligase –dependent polyubiquitination and subsequent proteasomal degradation. The activation of IKK is mediated by collaboration of polyubiquitin chains with a variety of linkages, including K63-, K11- and linear/M1-linkages. Degradation of IκBα releases NF-κB which then translocates into the nucleus and regulates gene transcription. The non-canonical NF-κB signaling pathway is activated by a smaller group of stimuli such as lymphotoxin-β and CD40 ligand. The engagement of these stimuli with their receptors leads to ubiquitination and proteasomal degradation of RING ligase TRAF3, and consequently inhibits TRAF3-dependent proteasomal degradation of NIK. The NIK is constitutively active and stabilization of NIK leads to activation of IKKα homodimer, which in turn phosphorylates p100. Phosphorylated p100 then undergoes proteasome-mediated partial processing, yielding the p52-RelB dimer. Subsequently, P52-RelB translocates into the nucleus to regulate gene transcription. (B) Genotoxic agents can induce nuclear DNA damage which initiates a genotoxic NF-κB signaling pathway from the nucleus. DNA damage may induce formation of nuclear PIDDosome and activation of PARP1, which both facilitate E3 PIASy-dependent sumoylation of NEMO. NEMO sumoylation enriches NEMO in the nucleus where it can associate with, and be phosphorylated by, the apical DNA damage responsive kinase ATM. Phosphorylation of NEMO promotes its monoubiquitination, which is required for NEMO-ATM complex export from the nucleus. In the cytoplasm, NEMO-ATM complex further recruits ELKS and facilitates K63-linked and M1-linked polyubiquitination on ELKS and NEMO, respectively, resulting in subsequent activation of IKK and NF-κB. Alternatively, ATM may be independently exported into the cytoplasm, where it associates with TRAF6 and promotes TRAF6 K63-linked polyubiquitination, also leading to activation of IKK and NF-κB.
K48-Linked Polyubiquitination in NF-κB Signaling
The role of UPS in regulating NF-κB signaling was initially based on the finding that proteasome inhibitors effectively blocked NF-κB activation by inhibiting p105 processing into p50 and IκBα degradation upon TNFα treatment [44]. It was soon revealed that IκBα phosphorylation at Ser32 and Ser36, which can be recognized by β-TrCP-containing SCF E3 complex [45], were prerequisite for its polyubiquitination with K48-linked chain, and subsequent proteasomal degradation [46, 47]. The degradation of IκBα unmasks the nuclear localization sequence of NF-κB and permitted its nuclear translocation. Nevertheless, proteasome-dependent processing of p105 can be cotranslational or posttranslational, which may result in complete degradation of p105 or formation of p50. Although both cotranslational and posttranslational processing of p105 requires the proteasome, there is evidence suggesting that K48-linked ubiquitination of p105 may be only involved in the IKK-dependent posttranslational processing of p105, but not in its cotranslational processing [48, 49].
Similar to p105, p100 can also be partially processed by the proteasome to form p52, which is critical for activation of the non-canonical NF-κB signaling pathway in response to stimulation of a subset of the TNFR (tumor necrosis factor receptor) family. Upon treatment with stimuli such as CD40L, BAFF or Lymphotoxin β, proteasomal degradation of NIK is inhibited and stabilized NIK phosphorylates and activates IKKα homodimers. Activated IKKα in turn phosphorylates Ser865 and Ser869 at C-terminus of p100, which leads to β-TrCP-SCF E3-dependent K48-polyubiquitination of p100 and subsequent partial proteasomal processing. Proteasome-mediated partial processing of p100/p105, instead of complete degradation, may depend on a glycine-rich region in both p100 and p105 protein and their binding partners of NF-κB family. The glycine-rich region and/or binding-partner of p100/p105 may serve as “stop” signals for proteasomal processing and their subsequent release in the form of p52/p50 [11].
K48-linked polyubiquitination is also involved in terminating NF-κB signaling by promoting the proteasomal degradation of p65 in the nucleus. COMMD1 (copper metabolism gene MURR1 domain), a subunit of an EC2S multi-subunit E3 ligase complex, has been shown to promote association between p65 and EC2S substrate-binding protein SOCS-1, which promotes the ubiquitination and degradation of p65 [50, 51]. Another E3 ligase PIDLIM2, which contains both PDZ domain and LIM domains, was found to promote both p65 translocation into PML nuclear bodies and its polyubiquitination, resulting in p65 degradation and termination of NF-κB signaling [52]. Both ligases were believed to play important roles in proper control of inflammatory responses by regulating nuclear p65 stability.
Alternatively, nuclear ubiquitination and degradation of p50 may enhance the transcription of a group of NF-κB target genes, leading to hyperactivation of inflammatory responses. B cell leukemia 3 (Bcl3) can bind to the chromatin-bound p50:p50 homodimer in the nucleus, which prevents ubiquitination and the proteasomal degradation of p50 [53]. As p50 does not contain a transactivation domain, p50:p50 homodimer may function as transcription repressor by competing with other transactive NF-κB dimers such as p65:p50 for binding on target gene promoters. A reduction in the abundance of p50:p50 in Bcl3-deficient cells may result in the increased transcription of NF-κB-dependent inflammatory genes.
K63-Linked Polyubiquitination and IKK Activation
The idea that polyubiquitination plays a non-proteolytic role in mediating kinase activation was rather radical when it was first proposed in a study showing that IKK-dependent IκBα phosphorylation relied on ubiquitination [54]. With accumulating evidence from later studies, now the role of K63-linked polyubiquitin in mediating signaling transduction has been well-established. The studies on K63 polyubiquitination in mediating various NF-κB signaling pathways significantly advanced the understanding of the non-proteolytic role of polyubiquitination. We will focus below on several well-characterized NF-κB activation pathways.
TNFR-mediated NF-κB signaling
Tumor necrosis factor-α (TNFα) was originally identified as a cytokine that caused tumor necrosis in a mouse sarcoma model, and whose function was later expanded to regulation of inflammatory responses in a wide variety of autoimmune and inflammatory diseases [55]. Upon TNFα binding, TNFR1 trimerizes, thereby promoting recruitment of the adaptor proteins TRADD (TNF receptor associated protein with a death domain) and RIP1. The formation of TNFR complex can activate pro-survival NF-κB signaling pathway and caspase 8-dependent apoptotic pathway.
In response to TNFα stimulation, TNFR also recruits additional proteins including several RING E3 ligases such as TRAF2 (TNF receptor associated factor), TRAF5, cIAP1, cIAP2, and LUBAC (linear ubiquitin assembly complex). Both TRAF2/5 and cIAP1/2 have been shown to promote K63-linked polyubiquitination of RIP1 [5]. Of note, RING domain of TRAF2 lacks key residues required for binding to E2 Ubc13, which catalyzes K63 polyubiquitin formation, therefore it may be insufficient for promoting RIP1 ubiquitination [56]. However, a recent report suggested that sphingosine-1-phosphate (S1P) binds to TRAF2 which facilitates TRAF2-dependent K63 polyubiquitination of RIP1, likely by assisting the interaction between TRAF2 and Ubc13 [57].
The K63 polyubiquitin chain attached on RIP1 serves as a binding scaffold for further recruitment of TAK1-TAB1-TAB2 complex and IKKα-IKKβ-NEMO complex, resulting in the activation of TAK1 and IKKβ [58–60]. Both TAB2 and NEMO contain ubiquitin binding domains, NZF and UBAN, respectively, which have been shown to bind to K63 chain [60, 61]. Nonetheless, the UBAN domain of NEMO was shown to associate with linear ubiquitin chain, which may also participate in TNFα-induced IKK activation [20] (see discussion below). Besides RIP1, NEMO could be polyubiquitinated with K6-linked and/or linear ubiquitin chain in response to TNFα stimulation, which may be required for efficient IKK activation [62, 63].
Additional evidence supporting the essential role of K63 polyubiquitin chain in mediating IKK activation came from the finding that DUBs, such as CYLD and A20, can inhibit TNFα-induced NF-κB activation through deubiquitinating RIP1 [64]. CYLD has been shown to preferentially disassemble polyubiquitin with a K63-linkage as opposed to a K48-linkage [65]. Mutations and truncations in CYLD C-terminal DUB catalytic domain, which frequently occur in cylindromatosis patients, impair its DUB activity and lead to hyperactivation of NF-κB [66]. A20 is a NF-κB target gene whose induction upon NF-κB activation is considered a critical negative feedback response to avoid overactivation of NF-κB [64]. A20 contains both an OTU DUB domain and multiple ZnF domains which may harbor E3 ligase activity. A20 was proposed to play a dual-function in editing polyubiquitin chains on RIP1 through removing K63-linked ubiquitin chain from RIP1 by the OTU domain and replacing it with K48-linked polyubiquitin chain by the ZnF domains [67]. A recent study suggested that A20 may also interfere with K63 polyubiquitin-mediated TAK1 and IKK activation by binding to NEMO [68]. The ZnF7 of A20 could interact with polyubiquitin chains along with NEMO, which limited the extent of IKK activation by TAK1. Taken together, the negative regulation of NF-κB signaling by CYLD and A20 through disrupting K63 chain and/or ubiquitin-TAK1/IKK kinase complexes underlines an essential role of K63-linked polyubiquitination in mediating NF-κB signaling in response to TNFα stimulation.
Besides the polyubiquitin chains anchored on substrate proteins such as RIP1, the role of unanchored K63-linked polyubiquitin chains, which do not attach on a specific protein, in mediating signaling transduction is an emerging field of studies [69]. Free K63 polyubiquitin chains directly activate TAK1 by binding to the ubiquitin-binding protein TAB2, which leads to sequential activation of TAK1 and IKK. A similar function of unanchored K63 polyubiquitin chains was also found in mediating RIG-I (retinoic acid-inducible gene I) activation in innate immune response [70]. These findings further highlight the significance of K63-linked polyubiquitination in mediating signal transduction, which may involve versatile mechanisms.
It is noteworthy that, although mounting evidence indicates an essential role of K63 polyubiquitin in mediating TNFα-induced NF-κB activation, IKK activation by TNFα treatment was still observed in cells where endogenous ubiquitins were replaced with ubiquitin K63R mutants [71]. It suggested that RIP1 could be ubiquitinated with non-K63-linked chain upon TNFα stimulation, which was sufficient for mediating IKK activation. Indeed, as we will discuss further below, RIP1 was found to be conjugated with linear polyubiquitin as well as K11-linked polyubiquitin chain upon TNFα treatment, which both may facilitate IKK activation [36, 72]. Nonetheless, TRAF6 and Ubc13-dependent K63-linked polyubiquitination is indispensable for IL-1-induced NF-κB activation [71].
IL-1 and Toll-like receptor mediated NF-κB pathways
IL-1 receptor and toll-like receptors (TLRs) share a conserved intracellular domain termed TIR (Toll and IL1 receptor) domain. Upon activation, the TIR domain of these receptors can interact with the adapter proteins TIRAP (Toll/interleukin-1 receptor domain-containing adapter protein) and myeloid differentiation factor 88 (MyD88), which further recruit the IL-1 receptor-associated kinases (IRAK), such as IRAK1, IRAK2, and IRAK4, to the membrane-bound receptor complex. IRAK1 binds to RING E3 ligase TRAF6, along with K63 chain-catalyzing E2 Ubc13-Uev1A, to promote conjugation of K63 polyubiquitination onto IRAK4 and TRAF6, which then facilitate the activation of TAK1 and IKK. Besides anchored polyubiquitin chains, free K63-linked polyubiquitin may also play an important role in mediating TAK1/IKK activation in response to IL-1 treatment [69].
Certain TLRs, such as endosomal TLR4 and TLR3, can also activate IKK/NF-κB via association with additional TIR domain-associated proteins TRAM and TRIF, independent of MyD88 [73]. TRIF recruits both TRAF6 and RIP1 to the proximal receptor signaling complex, where both TRAF6 and RIP1 are thought to be ubiquitinated with K63-linked chains, depending on the E3 ligase activity of TRAF6 and another E3 ligase Pellino-1[74]. The polyubiquitination of TRAF6 and RIP1 then leads to sequential activation of IKK and NF-κB as well as type-I interferon (IFN) production. Another E3 TRAF3 may promote its self polyubiquitination with K63-linked chain, which is critical for the TRIF-induced expression of type I IFNs [75].
T-cell receptor pathway
K63-linked polyubiquitin chain has also been shown to play seminal roles in mediating activation of antigen receptors, such as T-cell receptor (TCR), in immune responses. TCR binds to major histocompatibility complex (MHC)-bound antigenic peptides from pathogens presented by antigen presenting cells, which activates Src/Syc family tyrosine kinases associated with immunoreceptor tyrosine-based activation motif (ITAM) of the TCR. The activation of tyrosine kinases initiates sequential phosphorylation and activation of a kinase cascade including PI3K, PDK1 and PKCθ, which in turn phosphorylates CARMA1, resulting in recruitment of Bcl10 and MALT1 to TCR and assembly of the CARMA1-Bcl10-MALT1 complex [76, 77]. MALT1 contains binding sites for both TRAF2 and TRAF6, which may serve as E3 ligases for assembly of K63-linked polyubiquitin chain [78]. The receptors of K63 chain conjugation have been shown to include TRAF6, NEMO, MALT1 and Bcl10, whose K63 polyubiquitination may facilitate activation of TAK1 and IKK [67, 79].
It is worth noting that K63-linked ubiquitination may also be used as a signal to terminate IKK activation depending on the protein associated with K63 polyubiquitin. A recent report showed that, upon TCR stimulation, K63-linked polyubiquitin conjugated on Bcl10 can also be recognized by UBA domain of p62, which could subsequently direct Bcl10 degradation via autophagy [80]. This finding may serve as a complementary mechanism of Bcl10 degradation upon TCR activation, which can also be mediated by NEDD4 and Itch ligases-dependent polyubiquitination, likely K48-linked, resulting in its lysosomal degradation [81].
DNA damage-induced NF-κB signaling
In contrast to the NF-κB signaling pathways discussed above, NF-κB activation induced by genotoxic agents, such as ionizing radiation (IR) and cytotoxic chemotherapeutic drugs, is initiated in the nucleus [42, 82, 83]. Various forms and linkages of ubiquitination as well as sumoylation, a form of protein modification by ubiquitin-like protein SUMO (small ubiquitin-like modifier), have also been demonstrated to play critical roles in mediating NF-κB activation by DNA damage. In response to genotoxic stress, NEMO can be modified by SUMO in the nucleus, which is facilitated by SUMO E3 ligase PIASy [84, 85]. This sumoylation may also depend on PIDD and PARP1, which may assist in the assembly of nuclear complexes including NEMO and PIASy, resulting in NEMO sumoylation [86, 87]. The sumoylated NEMO then associates with the apical DNA damage responsive kinase ATM which directly phosphorylates NEMO, leading to its subsequent monoubiquitination [88]. The monoubiquitination directs NEMO export from the nucleus, in association with ATM, into the cytoplasm where the NEMO-ATM complex further activates IKK in a manner dependent on K63-linked and linear polyubiquitination. A protein called ELKS was found to be associated with cytoplasmic ATM-NEMO complex which promotes conjugation of K63 polyubiquitin onto ELKS by recruiting E3 ligase XIAP [89]. Alternatively, ATM was also shown to translocate into the cytoplasm in a NEMO-independent fashion [90]. Without NEMO association, cytoplasmic ATM can promote K63 polyubiquitination of TRAF6, likely mediated by itself, upon DNA damage. K63 polyubiquitination of ELKS and TARF6 were shown to recruit TAK1 and IKK complexes, thereby resulting in sequential activation of TAK1, IKK and NF-κB [89, 90].
Linear Ubiquitination in NF-κB Signaling
Linear ubiquitin chain is linked by a peptide-bond between C-terminal G76 of one ubiquitin and N-terminal α-amino group on M1 of another ubiquitin molecule, which renders linear chain in a similar open conformation as K63-linked polyubiquitin chain [91]. To date, the only E3 ligase identified to specifically promote linear ubiquitin chain formation is called LUBAC [19], which is comprised of three subunits, HOIL1 (hemeoxidized IRP2 ubiquitin ligase-1), HOIP (HOIL1-interacting protein) and SHARPIN (shank-associated RH domain interactor) [72, 92, 93].
As K63- and linear linkage adopt similar structural conformations, it is not surprising that LUBAC-mediated linear ubiquitin chain was found to play a mainly non-proteolytic role in mediating signal transduction, such as regulating IKK activation in NF-κB signaling pathways [20, 94], although it was also reported to promote PKC and TRIM25 degradation [95, 96]. Upon TNFα stimulation, LUBAC was found to be recruited to TNFR, which may be facilitated by interaction between the ubiquitin binding domain of HOIP and K63-linked polyubiquitin attached on RIP1 [97]. In the associated complex of TNFR, LUBAC could promote linear ubiquitination of NEMO [63, 97], which may also be recruited to TNFR by binding to K63 chain of RIP1 [58]. Besides NEMO, RIP1 may serve as another acceptor for linear ubiquitin chain attachment [72]. The UBAN domain of NEMO was shown to interact with the linear ubiquitin chain with high affinity [61, 98], suggesting linear ubiquitin chain attached on NEMO or RIP1 in TNFR complex may serve as a binding platform for recruiting additional NEMO/IKK complex, leading to effective activation of IKK. Consistently, deficiency of HOIL1, HOIP or SHARPIN attenuated TNFα-induced NF-κB activation [72, 92, 93]. In addition, LUBAC-mediated NEMO linear ubiquitination was also found to participate in the CD40-induced non-canonical NF-κB pathway as well as genotoxic NF-κB signaling [93, 99]. Depletion of LUBAC or blocking linear ubiquitination of NEMO impaired DNA damage-induced NF-κB activation. Further analyses indicated that both K63 ubiquitination of ELKS and linear ubiquitination of NEMO were required for optimal activation of both TAK1 and IKK upon genotoxic stress [99].
In contrast, LUBAC-mediated polyubiquitination may also negatively regulate NF-κB activation in response to certain stimulation. In RNA virus-infected cells, the RIG-I signaling pathway is activated to elicit anti-viral responses by inducing type-I IFN production as well as NF-κB activation [100]. E3 ligase TRIM25 was shown to induce K63-linked ubiquitination of RIG-I, which is crucial for the antiviral innate immunity mediated by the RIG-I signaling pathway [101]. It was found that LUBAC could induce TRIM25 degradation by promoting its polyubiquitination, as well as inhibiting RIG-I K63 ubiquitination via competing with TRIM25 for RIG-I association, which results in decreased IFN production and potentially NF-κB activation [96]. The nature of LUBAC-dependent polyubiquitination, which leads to proteasomal degradation, is currently unclear. Additional studies will be needed to determine whether linear ubiquitination may also lead to proteasome recognition, or polyubiquitin with isopeptide linkage can be assembled by LUBAC.
Other Type of Ubiquitination in NF-κB Signaling
Accumulating evidence indicates that polyubiquitination with atypical linkages and mono-ubiquitination also plays important roles in NF-κB signaling transduction. As discussed earlier, mono-ubiquitination of NEMO is critical for NEMO nuclear export, which is required for NF-κB activation in response to DNA damage [88]. Another component of the IKK complex, IKKβ, was also found to be monoubiquitinated in response to expression of the Tax protein of human T-cell leukemia virus type 1, which was dependent on IKKβ phosphorylation at its catalytic loop [102]. Monoubiquitination of IKKβ may modulate the phosphorylation status of IKKβ at select C-terminal serines, which could play an important role in controlling IKKβ signaling during chronic inflammation [103]. A recent report showed that nuclear RelA/p65 could be monoubiquitinated. Instead of leading to degradation, monoubiquitination of p65 decreased association with its co-activator CREB-binding protein (CBP), which in turn reduced its transactivity [104].
K11-linked polyubiquitination has been proposed to play both proteolytic and non-proteolytic roles in regulating cellular signaling. The anaphase-promoting complex (APC/C) controls cell-cycle progression by promoting K11-linked ubiquitin chain conjugation on cyclin B1, resulting in its degradation [105]. However, cIAP1 could assemble K11-linked polyubiquitin chain on RIP1 in response to TNFα stimulation, which may facilitate the activation of IKK [36]. It remains to be determined that whether K11-linked ubiquitination also plays a role in mediating protein degradation in NF-κB signaling, and whether other proteins can be conjugated with K11 chain to modulate IKK activation.
Besides the intensively studied K48- and K63-linked ubiquitin chains, a quantitative proteomic analysis revealed endogenous polyubiquitin chains in yeast with linkage of K6, K11, K27, K29, or K33, whose function are likely linked to protein degradation [106]. K33-linked polyubiquitination of TCRζ interferes with its phosphorylation and association with the downstream kinase zeta-chain associated protein kinase 70 kDa (ZAP70) [107], thereby suppressing TCR-initiated downstream signaling events, such as NF-κB activation. K6- and K27-linked ubiquitination of NEMO have also been reported, which facilitate NF-κB activation in response to TNFα treatment or virus infection, respectively [62, 108]. It can be envisioned that more mechanistic insights into different ubiquitin forms and linkages in regulating various NF-κB signaling pathways will be revealed in further investigation.
ANTI-CANCER DRUGS TARGETING UBIQUITINATION AND NF-κB SIGNALING
Both UPS and NF-κB play broad and pivotal roles in regulating physiological and pathophysiological processes in cells. It is not surprising that dysregulation of the UPS and NF-κB signaling pathways are closely linked to various human diseases, such as cancer. Substantial evidence indicates that NF-κB regulates oncogenesis and tumor progression. NF-κB activation (as documented by its nuclear localization) has been observed in a variety of solid tumors, including prostate cancer, breast cancer, melanoma, pancreatic cancer and lung cancer [109, 110]. The tumorigenesis of a number of hematopoietic cancers, such as chronic myelogenous leukemia, multiple myeloma and Hodgkin’s/Non-Hodgkin’s lymphoma, has also been attributed to the aberrant activation of NF-κB [111]. Furthermore, NF-κB-mediated inflammation has been linked to the carcinogenesis and progression of liver and colon cancers [112, 113]. In general, NF-κB is believed to promote cancer development by inhibiting apoptosis. Also, genotoxic chemodrugs and radiation used in cancer treatment have been shown to activate NF-κB [83, 114]. Many cancer cells, either of epithelial or hematopoietic origin, may acquire resistance to anticancer treatments by activating NF-κB [114, 115]. Moreover, the broad functions of NF-κB-regulated genes contribute to all intrinsic hallmarks of cancer [116, 117]. A recent review highlighted a series of stress phenotypes of cancer which may hint at the additional oncogenic activity of NF-κB, given its pivotal role in coordinating cellular stress responses [118].
Many E3 ligases are implicated in either the suppression or the progression of cancer via regulating cell cycle, genomic integrity, hypoxia response, and cancer metastasis [22]. For example, APC/C and SCF ligase complexes control cell cycle progression by inducing degradation of specific substrate during different cell cycle phases. Deficiency (such as FBW7, CDH1 and CDC20) or hyperactivation (such as SKP2 and β-TrCP) of subunits of these E3 complexes is implicated in cancer development [119]. Also, Mdm2, BRCA1 (breast cancer susceptibility gene) and Fanconi anemia (FA) E3s are essential for proper DNA damage responses and efficient DNA repair, and the loss of these genes may cause genomic instability and has been associated with oncogenesis [120–122]. Moreover, a majority of sporadic kidney clear cell cancer patients carry inactivating mutations and/or loss of heterozygosity (LOH) of VHL gene, which is the E3 ligase responsible for the proteasomal degradation of HIF1α (hypoxia inducible factor) [123]. A recent report indicated that loss of FBXO11 protein of SCF E3 complex, which caused stabilization of Bcl6, may contribute to the development of B cell lymphomas [124]. All these studies support the notion that targeting UPS and NF-κB signaling pathways may provide novel avenues of pharmacological intervention for cancer treatment.
Bortezomib and other Proteasome Inhibitors
Bortezomib is the first and still the only proteasome inhibitor approved by the US FDA for use in patients with multiple myeloma and relapsed mantle cell lymphoma. Bortezomib is a peptide boronic acid analog that reversibly binds to the chymotrypsin-like site in the 20S core of the proteasome, resulting in its inhibition [125]. In cell culture models, bortezomib has been shown to induce cell apoptosis in various hematopoietic and solid tumor cell lines. Since numerous proteins are degraded by the proteasome, the tumoricidal activity of bortezomib in different cancer cells probably involves a variety of molecular mechanisms. One of the major mechanisms proposed to be shared in various tumor cells is the inhibition of NF-κB signaling. As proteasome-dependent IκB degradation and p100 processing are the gatekeeping steps of NF-κB activation in response to a large number of stimuli, blocking proteasome activity by bortezomib may effectively inhibit both constitutive and inducible NF-κB activation, favoring increased apoptosis [125, 126]. However, NF-κB inhibition is unlikely the only mechanism for the antitumor capacity of bortezomib, as NF-κB activation in certain multiple myeloma patients are resistant to bortezomib treatment [127], and bortezomib was even shown to activate NF-κB by down-regulating IκBα expression in some multiple myeloma cells [128]. Indeed, inhibition of proteasome by bortezomib may stabilize p53 and cause G2/M cell cycle arrest, which lead to apoptosis [129]. Moreover, bortezomib also induces phosphorylation of c-Jun N-terminal kinase (JNK) and subsequent AP-1activation [130]. AP-1, along with p53 stabilization, may contribute to the upregulation of p21 and p27 upon bortezomib treatment, which leads to apoptosis and cell cycle arrest [129].
Although bortezomib shows improved clinical outcomes when used as a single agent or in combination with radiation or chemotherapies, some patients do not respond to bortezomib treatment and those patients who respond will eventually develop drug resistance. Therefore, additional proteasome inhibitors that act through distinct mechanisms from that of bortezomib are being developed [8] (Table 1). An irreversible proteasome inhibitor, Carfilzomib, is currently being tested in a Phase III trial in relapsed multiple myeloma patients. Carfilzomib induced more potent and selective inhibition of proteasome than bortezomib [131]. Moreover, carfilzomib was shown to be cytotoxic to some bortezomib-resistant cancer cells [131, 132], suggesting its potential for use in patients who do not respond to or acquire resistance to bortezomib. Besides bortezomib and carfilzomib, which both need to be administrated intravenously, orally bioavailable proteasome inhibitors, such as MLN9074, CEP18770 and ONX0912 have been developed [8]. Preclinical studies showed that these proteasome inhibitors increased myeloma cell death by inhibiting chymotrypsin-like activity of the proteasome, reversibly or irreversibly. Their tumoricidal potency is currently being evaluated in clinical trials [6, 8].
Table 1.
Current UPS-targeting anti-cancer drugs in clinical trials. (current as of 07.30.2012 from the National Institutes of Health; www.clinicaltrials.gov)
| Inhibitors | Target | Company | Disease | Stage |
|---|---|---|---|---|
| Bortezomib (Velcade) | Proteasome | Millennium | Multiple myeloma and mantle cell lymphoma | Approved |
| Bortezomib (Velcade) | Proteasome | Millennium | Glioblastoma, Non-small cell lung cancer, | Phase II |
| MLN9708 | Proteasome | Millennium | Multiple myeloma and other cancers | Phase III |
| Carfilzomib (PR-171) | Proteasome | ONYX | Multiple myeloma | Phase III |
| Carfilzomib (PR-171) | Proteasome | ONYX | Ovarian Cancer; Renal Cancer; Non-small Cell Lung Cancer | Phase II |
| Onx 0912 (PR-047) | Proteasome | ONYX | Multiple myeloma and other cancers | Phase II |
| CEP-18770 | Proteasome | Cephalon | Multiple myeloma and other cancers | Phase II |
| NPI-0052 | Proteasome | Nereus | Multiple myeloma and leukemia | Phase I |
| MLN4924 | E1 (Nedd8 activating E1, NAE) | Millennium | Diffuse Large B-cell Lymphoma | Phase II |
| Nutlin (RG7112) | E3 (Mdm2) | Roche | Advanced solid tumors | Phase I |
| JNJ-26854165 | E3 (Mdm2) | Johnson & Johnson | Advanced or refractory solid tumors | Phase I |
| GDC-0152 | E3 (IAPs) | Genentech | Advanced or metastatic malignancies | Phase I |
| GDC-0917 | E3 (IAPs) | Genentech | Refractory Solid Tumors or Lymphoma | Phase I |
| LCL-161 | E3 (IAPs) | Novartis | Breast cancer and other advanced solid tumors | Phase II |
| AT-406 | E3 (IAPs) | Ascenta Therapeutics | Solid tumors and lymphomas, Acute Myelogenous Leukemia | Phase I |
| AEG-40826 (HGS1029) | E3 (IAPs) | Aegera Therapeutics | Advanced Solid Tumors | Phase I |
| TL-32711 | E3 (IAPs) | Tetralogics Pharma | Metastatic solid tumors and Acute Myelogenous Leukemia | Phase II |
Note: only Smac-mimetic inhibitors of E3 IAP were listed. Additional IAP-targeting antagonists undergoing clinical trials, such as AEG 35156 (XIAP, antisense oligos) and YM155 (Survivin inhibitor), were not included in the table.
E1-Targeting Anti-Cancer Drugs
E1-dependent activation of ubiquitin and other ubiquitin-like proteins (Ubls), such as SUMO and NEDD8, are the first step of the ubiquitin/Ubls conjugation cycle. Also, there are only a limited number of E1s, which make them attractive targets for drug development. One successful example is NEDD8 E1 inhibitor MLN4924 [133]. Like ubiquitin, NEDD8 conjugation goes through the E1-E2-E3 three-step process. The most well characterized NEDD8 conjugation substrates are the Cullin family of proteins, which consist of a major component of CRL family of ubiquitin ligases [134, 135]. NEDD8 conjugation on Cullins, which disrupts inhibitory binding by CAND1, enhances activity of the CRL ubiquitin ligases, thereby regulating the turnover of a subset of UPS substrate proteins [136, 137]. MLN4924 is an adenosine sulphamate analogue which is structurally related to AMP and forms a covalent adduct with NEDD8. MLN4924 then stably binds to the nucleotide-binding pocket of the NEDD8 E1, thereby inhibiting further NEDD8 E1 activity and eventual neddylation [138]. Consistently, MLN4924-mediated inhibition of cullin neddylation resulted in stabilization of CRL ubiquitin ligases substrates CDT1, p27 and NRF2, likely via inhibiting ligase activity [133]. MLN4924 treatment induced cell cycle S-phase defects, DNA damage and subsequent apoptosis in several human cancer cell lines, such as HCT-116 cells. Moreover, treatment with MLN4924 also inhibited growth of human tumor xenografts in mice [133]. In acute myeloid leukemia (AML) models, MLN4924 was found to inhibit tumor cell growth both in vitro and in xenograft models, which paralleled the increase of SCF E3 substrates such as IκBα [139]. The increased apoptosis in AML cells exposed to MLN4924 may be due, at least in part, to excessive reactive oxygen species (ROS) generation, resulting from a decrease in NF-κB-dependent expression of antioxidant genes, such as superoxide dismutase 2 (SOD2). Similarly, MLN4924 treatment induced cell death of diffuse large B-cell lymphoma (DLBCL) cells by inhibiting NF-κB activation [140]. Recent reports of tumoricidal effect of MLN4924 in Ewing’s sarcoma cells [141], liver cancer cells [142] and pancreatic cancer cells [143] indicate that MLN4924 may have broad antitumor capacity in hematopoietic and solid tumors. However, it is worth noting that MLN4924 treatment may induce heterozygous mutations in the adenosine triphosphate binding pocket and NEDD8-binding cleft of NEDD8 E1 β-subunit, resulting in resistance to MLN4924 [144]. Therefore, patient selection and the development of next-generation NEDD E1 inhibitors to overcome MLN4924 resistance are needed when considering the clinical application of MLN4924.
Another E1 inhibitor, PYR41, interferes with ubiquitin E1 function by a different mechanism. PYR41 is a pyrazone derivative whose nitrogen dioxide group on the furan ring covalently modifies the ubiquitin E1 active site cysteine [145]. In cells treated with PYR41, cytokine-induced NF-κB activation was inhibited, in part through inhibition of K63-linked ubiquitination of TRAF6 and subsequent IKK activation. NF-κB inhibition was accompanied by stabilization of p53 and the induction of p53-dependent transcription, which induced a pro-apoptotic response, leading to increased cancer cell death [145]. In parallel, a novel small molecule inhibitor, PYZD-4409, was found to induce cell death in malignant cells and primary acute myeloid leukemia cells via inhibiting ubiquitin E1 [146]. In addition, the nitric oxide (NO) prodrug JS-K inhibited ubiquitin E1~ubiquitin thioester formation, which also disrupted E1 function [147]. JS-K treatment increased p53 level and induced cell apoptosis in p53-expressing tumor cells, suggesting its potential application in treating tumors harboring wild-type p53. All these preclinical studies support that inhibiting the E1 enzyme may serve as a promising strategy for developing antitumor drugs; nevertheless, their potential in treating cancer patients remains to be evaluated in clinical trials.
E2-Targeting Anti-Cancer Drugs
There are approximately 40 E2s, which determine ubiquitin/Ubl chain length and linkage topology. Along with E3, E2s control the fate of ubiquitinated substrates by determining whether they are conjugated with mono- or polyubiquitin as well as the specific linkage (e.g. K48 vs K63) [4, 5]. A number of E2s has been found mutated in human cancers such as UBE2Q2 in head and neck squamous cell carcinoma and UBE2T in lung cancer [6]. There are also reports indicating that inhibiting E2 activity may reduce tumor growth. For example, a recent study identified a small molecule compound, NSC697923, which can inhibit the activity of the K63 chain-specific E2 enzyme Ubc13-Uev1A[148]. Treatment with NSC697923 effectively inhibited NF-κB activation in DLBCL cells, likely via inhibiting K63 polyubiquitin-dependent IKK activation. Consequently, significantly increased apoptosis was observed in NSC697923-treated DLBCL cell lines and primary cancer cells from patients. However, to fully take advantage of using E2 inhibitors as anti-cancer drugs, further investigation is needed to improve our understanding of how specific E2s work with varying E3s to determine substrate and ubiquitin chain specificity. Also, more potent and selective E2 inhibitors need to be identified.
E3 Ubiquitin Ligases as Anti-Cancer Drug Targets
General inhibition of the proteasome with Bortezomib blocks the entire UPS-dependent protein degradation, which may lead to unwanted side effects. In contrast, E3 ubiquitin ligases usually regulate ubiquitination of a specific or a small group of substrates, whose inhibition tends to be more targeted. Indeed, MLN4924, which indirectly targets SCF E3 complex, blocks ~20% of all cellular proteins subjected to proteasomal degradation [133]. A number of ubiquitin E3 genes have been implicated as oncogenes or tumor suppressor genes, whose mutation is closely associated with pathogenesis of certain human malignancies [22]. For example, hemizygous frameshift and point mutations of cell cycle-regulating E3 APC/C have been identified in colon cancer cells [149]. Mutation of BRCA1, a E3 ligase involved in DNA repair, is frequently observed in breast cancer and ovarian cancer patients [150]. Moreover, the majority of patients with von Hippel–Lindau familial kidney cancer syndrome harbors inactivated VHL gene, which modulates stability of HIF1α as an E3 ligase [123]. Taken together, this evidence indicates that targeting E3 ubiquitin ligase may serve as an attractive strategy for cancer treatment.
The RING E3 and oncoprotein Mdm2 is a major regulator of p53 that modulates p53 ubiquitination and subsequent proteasomal degradation. Mdm2 usually is amplified in human cancer cells with wild-type p53, suggesting that overexpression of Mdm2 may be involved in inactivating p53 during oncogenesis [151]. The small molecule RITA binds to p53 and blocks its interaction with MDM2 by inducing a conformational change in p53, thereby alleviating it from Mdm2-dependent degradation [152]. Similarly, the small molecule Nutlin-3a binds Mdm2 in its p53-binding domain and thereby disrupts Mdm2–p53 interaction [153]. Treatment with either RITA or Nutlin-3a induced cancer cell apoptosis and inhibited tumor cell growth in vitro and in xenograft mouse models, likely via stabilizing and reactivating p53 in cancer cells. Of note, inhibiting Mdm2 may also stabilize p53 mutant, which still retains the ability to associate with Mdm2, thereby leading to accelerated oncogenesis and increased incidence of metastasis [154]. Therefore, prescreening and selection of patients with wild type p53 is needed when considering using Mdm2 inhibitors as potential therapeutics.
Another category of E3-targeting anti-cancer compounds called Smac (second mitochondria-derived activator of caspase, also known as Diablo) mimetics. Smac is a mitochondrial protein, which is capable of inhibiting IAP family of proteins and promoting apoptosis [155]. The IAP proteins can directly bind to caspases, such as caspase9, and/or interfere with the assembly of pro-apoptotic protein signaling complexes, resulting in the inactivation of caspases [156]. Smac mimetics can mimic the action of Smac and inhibit the function of IAPs, which in turn potentiate apoptosis by intrinsic and extrinsic inducers [157]. Besides directly inhibiting caspase activation, IAPs, such as cIAP1/2 and XIAP, have been shown to function as RING E3 ligases that regulate NF-κB activation in response to various stimuli, including TNFα and genotoxic agents [42, 79]. Smac mimetics has been reported to induce cell death primarily by promoting autoubiquitination and degradation of cIAPs [158–160]. Degradation of cIAP1 promoted RIP1 recruitment to TNFR which enhanced NF-κB activation via the canonical NF-κB pathway. In parallel, cIAP1/2 degradation also stabilized NIK due to decreased NIK ubiquitination, which lead to activation of the non-canonical NF-κB signaling pathway. Escalated NF-κB activation could enhance TNFα expression which in turn functions in autocrine/paracrine fashion to induce cell death. Meanwhile, degradation of cIAP1/2 also promoted formation of RIP1-FADD-caspase8 signaling complex, which could activate downstream apoptotic caspase cascade. The increased TNFα expression combined with enhanced RIP1-FADD-caspase8 complex formation resulted in aggravated cell death upon Smac treatment [155].
How Smac mimetics induce cIAP1/2 degradation is not entirely clear. Recent studies indicate that the BIR domains of cIAP1 could inhibit the RING finger dimerization, which is required for cIAP1 ligase activity. The binding of Smac mimetics to BIR domain may induce conformational change of cIAP1 which could relieve this inhibition, thereby promoting cIAP1 dimerization. The dimerization of cIAP1 RING domain could lead to ligase activation and promote its self-ubiquitination and proteasomal degradation [161]. Another RING E3, TRAF2, may also participate in cIAP1 degradation, and both TRAF2 and cIAP1 are indispensible for cIAP2 degradation. Taken together, these studies support that inhibiting E3 ligase IAPs with Smac mimetic drugs holds promise as a potential therapeutic approach to treat cancer.
CONCLUSIONS AND PERSPECTIVES
Our understanding of the crosstalk between UPS and NF-κB activation has changed dramatically in the last two decades. We now know that ubiquitination regulates NF-κB at different levels of various signaling cascades, and exerts distinct functions depending on the protein substrates being ubiquitinated as well as the types of linkage forming polyubiquitin chains. K48-linked polyubiquitination may activate or inhibit NF-κB by inducing proteasomal degradation of inhibitors (e.g. IκBα in the canonical pathway) or activators (e.g. NIK in the non-canonical pathway) of NF-κB signaling. The collaborative and/or compensative roles of nonproteolytic ubiquitin chains with K63-, K11- and linear linkages render efficient, resilient and fine-tuned NF-κB signaling cascades in response to various stimuli. On the other hand, NF-κB activation can induce up-regulation of genes involved in ubiquitination cascade, such as ubiquitin ligase cIAPs and DUB A20, which will serve as a feedback regulation of NF-κB signaling via modulating ubiquitination process. The success of the proteasome inhibitor bortezomib in treating multiple myeloma and mantle cell lymphoma, in which aberrant NF-κB activation is believed to drive tumor initiation and progression, further supports the promise of developing anti-cancer drugs that target UPS and NF-κB signaling.
However, there are still many questions that need to be addressed, before we can fully take advantage of ubiquitin-dependent regulation of various NF-κB signaling pathways in developing the next generation of anti-cancer drugs. For example, various non-proteolytic polyubiquitination (K63-, K11- and linear linkages) could be induced by TNFα stimulation as well as by other stimuli. All of these different linkages appeared to be important for prompt and effective NF-κB activation. How are these distinct ubiquitin chain conformations, facilitated by different or same ligase, coordinated in response to stimulation? Is there a hierarchy between these different linkages? Are there temporal and/or spatial orders for assembly of these linkages? Ubiquitination is a dynamic process which is controlled by the balance between E1-E2-E3 ubiquitin assembling machinery and DUB-dependent deubiquitination. A proteomic analysis in human cells revealed that there is significant difference between the abundance of polyubiquitin chains with various linkages [162], suggesting that certain types of ubiquitin chain are probably localized and/or transient. The molecular mechanisms by which those rare chain linkages, such as K6, K27, K33 and linear, are enriched in NF-κB signaling complexes and how their turnover is regulated remains unknown. Both K11 and linear ubiquitin chains can exercise proteolytic or non-proteolytic functions, depending on the substrates being ubiquitinated. It will be interesting to uncover the mechanism determining the final outcome of these ubiqutinations, and their influence on NF-κB activation. Moreover, recent studies have demonstrated a conserved and critical function of unanchored K63-linked polyubiquitin chains in mediating various signaling pathways [79]. It remains to be determined how these free K63 chains involved in NF-κB signaling are initiated, whether they are cleaved from an anchored substrate or assemble independently, and how they interplay with anchored polyubiquitin chains in mediating TAK1/IKK activation. Besides, we should bear in mind that NF-κB has important function in regulating physiological processes, such as innate and adaptive immunity, when considering therapeutic strategies involving inhibiting NF-κB. A more pathway-selective and ideally cell-type specific inhibition of NF-κB will be a desired approach for future cancer treatment [163]. Therefore, further investigation of how ubiquitination controls substrate behavior and how the ubiquitin signal is translated into downstream cellular events may lead to more selective, targeted and efficient anti-cancer drugs targeting NF-κB by modulating ubiquitination.
Acknowledgments
We apologize to investigators whose important contributions and original articles were not cited in this review due to space limitations. We thank Drs. L. Pfeffer, R. Laribee and members of the Wu laboratory for critical reading of the manuscript. The research in the author’s laboratory is support by grant CA149251 from NIH.
References
- 1.Hershko A, Ciechanover A, Rose IA. Resolution of the ATP-dependent proteolytic system from reticulocytes: a component that interacts with ATP. Proc Natl Acad Sci U S A. 1979;76:3107–10. doi: 10.1073/pnas.76.7.3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sen R, Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986;46:705–16. doi: 10.1016/0092-8674(86)90346-6. [DOI] [PubMed] [Google Scholar]
- 3.Pickart CM, Cohen RE. Proteasomes and their kin: proteases in the machine age. Nat Rev Mol Cell Biol. 2004;5:177–87. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- 4.Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol. 2005;6:79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- 5.Chen ZJ, Sun LJ. Nonproteolytic Functions of Ubiquitin in Cell Signaling. Mol Cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 6.Bedford L, Lowe J, Dick LR, Mayer RJ, Brownell JE. Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets. Nat Rev Drug Discov. 2011;10:29–46. doi: 10.1038/nrd3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayden MS, Ghosh S. NF-κB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 2012;26:203–234. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruschak AM, Slassi M, Kay LE, Schimmer AD. Novel proteasome inhibitors to overcome bortezomib resistance. J Natl Cancer Inst. 2011;103:1007–17. doi: 10.1093/jnci/djr160. [DOI] [PubMed] [Google Scholar]
- 9.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–62. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 10.Chen ZJ. Ubiquitin signalling in the NF-kappaB pathway. Nat Cell Biol. 2005;7:758–65. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skaug B, Jiang X, Chen ZJ. The Role of Ubiquitin in NF-kB Regulatory Pathways. Annu Rev Biochem. 2009;78:769–796. doi: 10.1146/annurev.biochem.78.070907.102750. [DOI] [PubMed] [Google Scholar]
- 12.Kulathu Y, Komander D. Atypical ubiquitylation — the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat Rev Mol Cell Biol. 2012;13:508–523. doi: 10.1038/nrm3394. [DOI] [PubMed] [Google Scholar]
- 13.Iwai K. Diverse ubiquitin signaling in NF-κB activation. Trends cell biol. 2012;22:355–364. doi: 10.1016/j.tcb.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 14.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–79. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 15.Goldstein G, Scheid M, Hammerling U, Schlesinger DH, Niall HD, Boyse EA. Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells. Proc Natl Acad Sci U S A. 1975;72:11–15. doi: 10.1073/pnas.72.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciechanover A, Hod Y, Hershko A. A heat-stable polypeptide component of an ATP-dependent proteolytic system from reticulocytes. Biochem Biophys Res Commun. 1978;81:1100–5. doi: 10.1016/0006-291x(78)91249-4. [DOI] [PubMed] [Google Scholar]
- 17.Bloom J, Amador V, Bartolini F, DeMartino G, Pagano M. Proteasome-mediated degradation of p21 via N-terminal ubiquitinylation. Cell. 2003;115:71–82. doi: 10.1016/s0092-8674(03)00755-4. [DOI] [PubMed] [Google Scholar]
- 18.Ciechanover A, Ben-Saadon R. N-terminal ubiquitination: more protein substrates join in. Trends in cell biology. 2004;14:103–6. doi: 10.1016/j.tcb.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 19.Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, Sano S, Tokunaga F, Tanaka K, Iwai K. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 2006;25:4877–87. doi: 10.1038/sj.emboj.7601360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iwai K, Tokunaga F. Linear polyubiquitination: a new regulator of NF-[kappa]B activation. EMBO Rep. 2009;10:706–713. doi: 10.1038/embor.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu YC. Ubiquitin ligases and the immune response. Annu Rev Immuno. 2004;22:81–127. doi: 10.1146/annurev.immunol.22.012703.104813. [DOI] [PubMed] [Google Scholar]
- 22.Lipkowitz S, Weissman AM. RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat Rev Cancer. 2011;11:629–43. doi: 10.1038/nrc3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eisenhaber B, Chumak N, Eisenhaber F, Hauser M-T. The ring between ring fingers (RBR) protein family. Genome Biol. 2007;8:209. doi: 10.1186/gb-2007-8-3-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and [beta]-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nijman SMB, Luna-Vargas MPA, Velds A, Brummelkamp TR, Dirac AMG, Sixma TK, Bernards R. A Genomic and Functional Inventory of Deubiquitinating Enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 26.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 27.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the Human Deubiquitinating Enzyme Interaction Landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammond-Martel I, Yu H, Affar EB. Roles of ubiquitin signaling in transcription regulation. Cell Signal. 2012;24:410–421. doi: 10.1016/j.cellsig.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 29.Moldovan G-L, Pfander B, Jentsch S. PCNA, the Maestro of the Replication Fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 30.Bergink S, Jentsch S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 2009;458:461–467. doi: 10.1038/nature07963. [DOI] [PubMed] [Google Scholar]
- 31.Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. Mono-versus polyubiquitination: differential control of p53 fate by Mdm2. Science. 2003;302:1972–5. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- 32.Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- 33.Varadan R, Walker O, Pickart C, Fushman D. Structural Properties of Polyubiquitin Chains in Solution. J Mol Biol. 2002;324:637–647. doi: 10.1016/s0022-2836(02)01198-1. [DOI] [PubMed] [Google Scholar]
- 34.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Behrends C, Harper JW. Constructing and decoding unconventional ubiquitin chains. Nat Struct Mol Biol. 2011;18:520–528. doi: 10.1038/nsmb.2066. [DOI] [PubMed] [Google Scholar]
- 36.Dynek JN, Goncharov T, Dueber EC, Fedorova AV, Izrael-Tomasevic A, Phu L, Helgason E, Fairbrother WJ, Deshayes K, Kirkpatrick DS, Vucic D. c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J. 2010;29:4198–4209. doi: 10.1038/emboj.2010.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–60. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 39.Perkins ND, Gilmore TD. Good cop, bad cop: the different faces of NF-kappaB. Cell Death Differ. 2006;13:759–772. doi: 10.1038/sj.cdd.4401838. [DOI] [PubMed] [Google Scholar]
- 40.Vallabhapurapu S, Karin M. Regulation and Function of NF-kappaB Transcription Factors in the Immune System. Annu Rev Immuno. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 41.Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 42.McCool KW, Miyamoto S. DNA damage-dependent NF-kappaB activation: NEMO turns nuclear signaling inside out. Immunol Rev. 2012;246:311–26. doi: 10.1111/j.1600-065X.2012.01101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu ZH, Miyamoto S. Many faces of NF-kappaB signaling induced by genotoxic stress. J Mol Med. 2007;85:1187–202. doi: 10.1007/s00109-007-0227-9. [DOI] [PubMed] [Google Scholar]
- 44.Palombella VJ, Rando OJ, Goldberg AL, Maniatis T. The ubiquitinproteasome pathway is required for processing the NF-κB1 precursor protein and the activation of NF-κB. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 45.Yaron A, Hatzubai A, Davis M, Lavon I, Amit S, Manning AM, Andersen JS, Mann M, Mercurio F, Ben-Neriah Y. Identification of the receptor component of the IkappaBalpha-ubiquitin ligase. Nature. 1998;396:590–4. doi: 10.1038/25159. [DOI] [PubMed] [Google Scholar]
- 46.Alkalay I, Yaron A, Hatzubai A, Orian A, Ciechanover A, Ben-Neriah Y. Stimulation-dependent I kappa B alpha phosphorylation marks the NF-kappa B inhibitor for degradation via the ubiquitin-proteasome pathway. Proceedings of the National Academy of Sciences. 1995;92:10599–10603. doi: 10.1073/pnas.92.23.10599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Z, Hagler J, Palombella VJ, Melandri F, Scherer D, Ballard D, Maniatis T. Signal-induced site-specific phosphorylation targets I kappa B alpha to the ubiquitin-proteasome pathway. Genes Dev. 1995;9:1586–1597. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- 48.Cohen S, Achbert-Weiner H, Ciechanover A. Dual effects of IkappaB kinase beta-mediated phosphorylation on p105 Fate: SCF(beta-TrCP)-dependent degradation and SCF(beta-TrCP)-independent processing. Mol Cell Biol. 2004;24:475–86. doi: 10.1128/MCB.24.1.475-486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moorthy AK, Savinova OV, Ho JQ, Wang VY, Vu D, Ghosh G. The 20S proteasome processes NF-kappaB1 p105 into p50 in a translation-independent manner. EMBO J. 2006;25:1945–56. doi: 10.1038/sj.emboj.7601081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maine GN, Mao X, Komarck CM, Burstein E. COMMD1 promotes the ubiquitination of NF-[kappa]B subunits through a cullin-containing ubiquitin ligase. EMBO J. 2007;26:436–447. doi: 10.1038/sj.emboj.7601489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ryo A, Suizu F, Yoshida Y, Perrem K, Liou Y-C, Wulf G, Rottapel R, Yamaoka S, Lu KP. Regulation of NF-κB Signaling by Pin1-Dependent Prolyl Isomerization and Ubiquitin-Mediated Proteolysis of p65/RelA. Mol Cell. 2003;12:1413–1426. doi: 10.1016/s1097-2765(03)00490-8. [DOI] [PubMed] [Google Scholar]
- 52.Tanaka T, Grusby MJ, Kaisho T. PDLIM2-mediated termination of transcription factor NF-[kappa]B activation by intranuclear sequestration and degradation of the p65 subunit. Nat Immunol. 2007;8:584–591. doi: 10.1038/ni1464. [DOI] [PubMed] [Google Scholar]
- 53.Carmody RJ, Ruan Q, Palmer S, Hilliard B, Chen YH. Science. 2007. Negative Regulation of Toll-Like Receptor Signaling by NF-{kappa}B p50 Ubiquitination Blockade; pp. 675–678. [DOI] [PubMed] [Google Scholar]
- 54.Chen ZJ, Parent L, Maniatis T. Site-specific phosphorylation of IkappaBalpha by a novel ubiquitination-dependent protein kinase activity. Cell. 1996;84:853–62. doi: 10.1016/s0092-8674(00)81064-8. [DOI] [PubMed] [Google Scholar]
- 55.Chen G, Goeddel DV. TNF-R1 signaling: a beautiful pathway. Science. 2002;296:1634–5. doi: 10.1126/science.1071924. [DOI] [PubMed] [Google Scholar]
- 56.Yin Q, Lamothe B, Darnay BG, Wu H. Structural Basis for the Lack of E2 Interaction in the RING Domain of TRAF2. Biochemistry. 2009;48:10558–10567. doi: 10.1021/bi901462e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alvarez SE, Harikumar KB, Hait NC, Allegood J, Strub GM, Kim EY, Maceyka M, Jiang H, Luo C, Kordula T, Milstien S, Spiegel S. Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature. 2010;465:1084–1088. doi: 10.1038/nature09128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ea C-K, Deng L, Xia Z-P, Pineda G, Chen ZJ. Activation of IKK by TNF[alpha] Requires Site-Specific Ubiquitination of RIP1 and Polyubiquitin Binding by NEMO. Mol Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 59.Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation. Nat Cell Biol. 2006;8:398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- 60.Kanayama A, Seth RB, Sun L, Ea CK, Hong M, Shaito A, Chiu YH, Deng L, Chen ZJ. TAB2 and TAB3 activate the NF-kappaB pathway through binding to polyubiquitin chains. Mol Cell. 2004;15:535–48. doi: 10.1016/j.molcel.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 61.Lo YC, Lin SC, Rospigliosi CC, Conze DB, Wu CJ, Ashwell JD, Eliezer D, Wu H. Structural basis for recognition of diubiquitins by NEMO. Mol Cell. 2009;33:602–15. doi: 10.1016/j.molcel.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tang ED, Wang C-Y, Xiong Y, Guan K-L. A Role for NF-kB Essential Modifier/IkB Kinase-gamma (NEMO/IKKgamma) Ubiquitination in the Activation of the IkB Kinase Complex by Tumor Necrosis Factor-a. J Biol Chem. 2003;278:37297–37305. doi: 10.1074/jbc.M303389200. [DOI] [PubMed] [Google Scholar]
- 63.Tokunaga F, Sakata S, Saeki Y, Satomi Y, Kirisako T, Kamei K, Nakagawa T, Kato M, Murata S, Yamaoka S, Yamamoto M, Akira S, Takao T, Tanaka K, Iwai K. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat Cell Biol. 2009;11:123–32. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- 64.Harhaj EW, Dixit VM. Regulation of NF-kappaB by deubiquitinases. Immunol Rev. 2012;246:107–24. doi: 10.1111/j.1600-065X.2012.01100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Komander D, Lord CJ, Scheel H, Swift S, Hofmann K, Ashworth A, Barford D. The Structure of the CYLD USP Domain Explains Its Specificity for Lys63-Linked Polyubiquitin and Reveals a B Box Module. Mol Cell. 2008;29:451–464. doi: 10.1016/j.molcel.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 66.Kovalenko A, Chable-Bessia C, Cantarella G, Israel A, Wallach D, Courtois G. The tumour suppressor CYLD negatively regulates NF-[kappa]B signalling by deubiquitination. Nature. 2003;424:801–805. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- 67.Wertz IE, O’Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, Ma A, Koonin EV, Dixit VM. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–9. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 68.Skaug B, Chen J, Du F, He J, Ma A, Chen Zhijian J. Direct, Noncatalytic Mechanism of IKK Inhibition by A20. Mol Cell. 2011;44:559–571. doi: 10.1016/j.molcel.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xia ZP, Sun L, Chen X, Pineda G, Jiang X, Adhikari A, Zeng W, Chen ZJ. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–9. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, Xu M, Chen ZJ. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–30. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu M, Skaug B, Zeng W, Chen ZJ. A ubiquitin replacement strategy in human cells reveals distinct mechanisms of IKK activation by TNFalpha and IL-1beta. Mol Cell. 2009;36:302–14. doi: 10.1016/j.molcel.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gerlach B, Cordier SM, Schmukle AC, Emmerich CH, Rieser E, Haas TL, Webb AI, Rickard JA, Anderton H, Wong WWL, Nachbur U, Gangoda L, Warnken U, Purcell AW, Silke J, Walczak H. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature. 2011;471:591–596. doi: 10.1038/nature09816. [DOI] [PubMed] [Google Scholar]
- 73.Kawai T, Akira S. Toll-like Receptors and Their Crosstalk with Other Innate Receptors in Infection and Immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 74.Chang M, Jin W, Sun S-C. Peli1 facilitates TRIF-dependent Toll-like receptor signaling and proinflammatory cytokine production. Nat Immunol. 2009;10:1089–1095. doi: 10.1038/ni.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tseng P-H, Matsuzawa A, Zhang W, Mino T, Vignali DAA, Karin M. Different modes of ubiquitination of the adaptor TRAF3 selectively activate the expression of type I interferons and proinflammatory cytokines. Nat Immunol. 2010;11:70–75. doi: 10.1038/ni.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee K-Y, D’Acquisto F, Hayden MS, Shim J-H, Ghosh S. PDK1 Nucleates T Cell Receptor-Induced Signaling Complex for NF-κB Activation. Science. 2005;308:114–118. doi: 10.1126/science.1107107. [DOI] [PubMed] [Google Scholar]
- 77.Wang D, You Y, Case SM, McAllister-Lucas LM, Wang L, DiStefano PS, Nunez G, Bertin J, Lin X. A requirement for CARMA1 in TCR-induced NF-kappa B activation. Nature immunology. 2002;3:830–5. doi: 10.1038/ni824. [DOI] [PubMed] [Google Scholar]
- 78.Sun L, Deng L, Ea CK, Xia ZP, Chen ZJ. The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol Cell. 2004;14:289–301. doi: 10.1016/s1097-2765(04)00236-9. [DOI] [PubMed] [Google Scholar]
- 79.Chen ZJ. Ubiquitination in signaling to and activation of IKK. Immunol Rev. 2012;246:95–106. doi: 10.1111/j.1600-065X.2012.01108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Paul S, Kashyap Anuj K, Jia W, He Y-W, Schaefer Brian C. Selective Autophagy of the Adaptor Protein Bcl10 Modulates T Cell Receptor Activation of NF-κB. Immunity. 2012;36:947–958. doi: 10.1016/j.immuni.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Scharschmidt E, Wegener E, Heissmeyer V, Rao A, Krappmann D. Degradation of Bcl10 Induced by T-Cell Activation Negatively Regulates NF-κB Signaling. Mol Cell Biol. 2004;24:3860–3873. doi: 10.1128/MCB.24.9.3860-3873.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hadian K, Krappmann D. Signals from the Nucleus: Activation of NF-{kappa}B by Cytosolic ATM in the DNA Damage Response. Sci Signal. 2011;4:pe2. doi: 10.1126/scisignal.2001712. [DOI] [PubMed] [Google Scholar]
- 83.Miyamoto S. Nuclear initiated NF-[kappa]B signaling: NEMO and ATM take center stage. Cell Res. 2011;21:116–130. doi: 10.1038/cr.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang TT, Wuerzberger-Davis SM, Wu ZH, Miyamoto S. Sequential modification of NEMO/IKKgamma by SUMO-1 and ubiquitin mediates NF-kappaB activation by genotoxic stress. Cell. 2003;115:565–76. doi: 10.1016/s0092-8674(03)00895-x. [DOI] [PubMed] [Google Scholar]
- 85.Mabb AM, Wuerzberger-Davis SM, Miyamoto S. PIASy mediates NEMO sumoylation and NF-kappaB activation in response to genotoxic stress. Nat Cell Biol. 2006;8:986–93. doi: 10.1038/ncb1458. [DOI] [PubMed] [Google Scholar]
- 86.Janssens S, Tinel A, Lippens S, Tschopp J. PIDD mediates NF-kappaB activation in response to DNA damage. Cell. 2005;123:1079–92. doi: 10.1016/j.cell.2005.09.036. [DOI] [PubMed] [Google Scholar]
- 87.Stilmann M, Hinz M, Arslan SC, Zimmer A, Schreiber V, Scheidereit C. A Nuclear Poly(ADP-Ribose)-Dependent Signalosome Confers DNA Damage-Induced IkB Kinase Activation. Mol Cell. 2009;36:365–378. doi: 10.1016/j.molcel.2009.09.032. [DOI] [PubMed] [Google Scholar]
- 88.Wu ZH, Shi Y, Tibbetts RS, Miyamoto S. Molecular linkage between the kinase ATM and NF-kappaB signaling in response to genotoxic stimuli. Science. 2006;311:1141–6. doi: 10.1126/science.1121513. [DOI] [PubMed] [Google Scholar]
- 89.Wu ZH, Wong ET, Shi Y, Niu J, Chen Z, Miyamoto S, Tergaonkar V. ATM- and NEMO-Dependent ELKS Ubiquitination Coordinates TAK1-Mediated IKK Activation in response to Genotoxic Stress. Mol Cell. 2010;40:75–86. doi: 10.1016/j.molcel.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hinz M, Stilmann M, Arslan Sc, Khanna KK, Dittmar G, Scheidereit C. A Cytoplasmic ATM-TRAF6-cIAP1 Module Links Nuclear DNA Damage Signaling to Ubiquitin-Mediated NF-kappaB Activation. Mol Cell. 2010;40:63–74. doi: 10.1016/j.molcel.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 91.Komander D, Reyes-Turcu F, Licchesi JDF, Odenwaelder P, Wilkinson KD, Barford D. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 2009;10:466–473. doi: 10.1038/embor.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ikeda F, Deribe YL, Skanland SS, Stieglitz B, Grabbe C, Franz-Wachtel M, van Wijk SJL, Goswami P, Nagy V, Terzic J, Tokunaga F, Androulidaki A, Nakagawa T, Pasparakis M, Iwai K, Sundberg JP, Schaefer L, Rittinger K, Macek B, Dikic I. SHARPIN forms a linear ubiquitin ligase complex regulating NF-[kgr]B activity and apoptosis. Nature. 2011;471:637–641. doi: 10.1038/nature09814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tokunaga F, Nakagawa T, Nakahara M, Saeki Y, Taniguchi M, Sakata S-i, Tanaka K, Nakano H, Iwai K. SHARPIN is a component of the NF-kB-activating linear ubiquitin chain assembly complex. Nature. 2011;471:633–636. doi: 10.1038/nature09815. [DOI] [PubMed] [Google Scholar]
- 94.Schmukle AC, Walczak H. No one can whistle a symphony alone - how different ubiquitin linkages cooperate to orchestrate NF-kappaB activity. J Cell Sci. 2012;125:549–59. doi: 10.1242/jcs.091793. [DOI] [PubMed] [Google Scholar]
- 95.Nakamura M, Tokunaga F, Sakata S-i, Iwai K. Mutual regulation of conventional protein kinase C and a ubiquitin ligase complex. Biochem Biophys Res Commun. 2006;351:340–347. doi: 10.1016/j.bbrc.2006.09.163. [DOI] [PubMed] [Google Scholar]
- 96.Inn K-S, Gack MU, Tokunaga F, Shi M, Wong L-Y, Iwai K, Jung JU. Linear Ubiquitin Assembly Complex Negatively Regulates RIG-I- and TRIM25-Mediated Type I Interferon Induction. Mol Cell. 2011;41:354–365. doi: 10.1016/j.molcel.2010.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Haas TL, Emmerich CH, Gerlach B, Schmukle AC, Cordier SM, Rieser E, Feltham R, Vince J, Warnken U, Wenger T, Koschny R, Komander D, Silke J, Walczak H. Recruitment of the Linear Ubiquitin Chain Assembly Complex Stabilizes the TNF-R1 Signaling Complex and is Required for TNF-Mediated Gene Induction. Mol Cell. 2009;36:831–844. doi: 10.1016/j.molcel.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 98.Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D, Randow F, Wakatsuki S, Dikic I. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136:1098–109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 99.Niu J, Shi Y, Iwai K, Wu Z-H. LUBAC regulates NF-[kappa]B activation upon genotoxic stress by promoting linear ubiquitination of NEMO. EMBO J. 2011;30:3741–3753. doi: 10.1038/emboj.2011.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yan N, Chen ZJ. Intrinsic antiviral immunity. Nat Immunol. 2012;13:214–222. doi: 10.1038/ni.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gack MU, Shin YC, Joo C-H, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 102.Carter RS, Pennington KN, Ungurait BJ, Arrate P, Ballard DW. Signal-induced Ubiquitination of IκB Kinase-β. J Biol Chem. 2003;278:48903–48906. doi: 10.1074/jbc.M310686200. [DOI] [PubMed] [Google Scholar]
- 103.Carter RS, Pennington KN, Arrate P, Oltz EM, Ballard DW. Site-specific Monoubiquitination of IκB Kinase IKKβ Regulates Its Phosphorylation and Persistent Activation. J Biol Chem. 2005;280:43272–43279. doi: 10.1074/jbc.M508656200. [DOI] [PubMed] [Google Scholar]
- 104.Hochrainer K, Racchumi G, Zhang S, Iadecola C, Anrather J. Monoubiquitination of nuclear RelA negatively regulates NF-kappaB activity independent of proteasomal degradation. Cell Mol Life Sci. 2012;69:2057–73. doi: 10.1007/s00018-011-0912-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jin L, Williamson A, Banerjee S, Philipp I, Rape M. Mechanism of Ubiquitin-Chain Formation by the Human Anaphase-Promoting Complex. Cell. 2008;133:653–665. doi: 10.1016/j.cell.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. Quantitative Proteomics Reveals the Function of Unconventional Ubiquitin Chains in Proteasomal Degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Huang H, Jeon M-s, Liao L, Yang C, Elly C, Yates JR, III, Liu Y-C. K33-Linked Polyubiquitination of T Cell Receptor-ζ Regulates Proteolysis-Independent T Cell Signaling. Immunity. 2010;33:60–70. doi: 10.1016/j.immuni.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Arimoto K-i, Funami K, Saeki Y, Tanaka K, Okawa K, Takeuchi O, Akira S, Murakami Y, Shimotohno K. Polyubiquitin conjugation to NEMO by triparite motif protein 23 (TRIM23) is critical in antiviral defense. Proc Natl Acad Sci U S A. 2010;107:15856–15861. doi: 10.1073/pnas.1004621107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kim HJ, Hawke N, Baldwin AS. NF-kappaB and IKK as therapeutic targets in cancer. Cell Death Differ. 2006;13:738–47. doi: 10.1038/sj.cdd.4401877. [DOI] [PubMed] [Google Scholar]
- 110.Perkins ND. The diverse and complex roles of NF-κB subunits in cancer. Nat Rev Cancer. 2012;12:121–132. doi: 10.1038/nrc3204. [DOI] [PubMed] [Google Scholar]
- 111.Braun T, Carvalho G, Fabre C, Grosjean J, Fenaux P, Kroemer G. Targeting NF-kappaB in hematologic malignancies. Cell Death Differ. 2006;13:748–58. doi: 10.1038/sj.cdd.4401874. [DOI] [PubMed] [Google Scholar]
- 112.Karin M, Greten FR. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5:749–59. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 113.Karin M, Lawrence T, Nizet V. Innate Immunity Gone Awry: Linking Microbial Infections to Chronic Inflammation and Cancer. Cell. 2006;124:823–835. doi: 10.1016/j.cell.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 114.Baldwin AS. Control of oncogenesis and cancer therapy resistance by the transcription factor NF-kappaB. J Clin Invest. 2001;107:241–6. doi: 10.1172/JCI11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nakanishi C, Toi M. Nuclear factor-kappaB inhibitors as sensitizers to anticancer drugs. Nat Rev Cancer. 2005;5:297–309. doi: 10.1038/nrc1588. [DOI] [PubMed] [Google Scholar]
- 116.Hanahan D, Weinberg Robert A. Hallmarks of Cancer: The Next Generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 117.Baud V, Karin M. Is NF-[kappa]B a good target for cancer therapy? Hopes and pitfalls. Nat Rev Drug Discov. 2009;8:33–40. doi: 10.1038/nrd2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Luo J, Solimini NL, Elledge SJ. Principles of Cancer Therapy: Oncogene and Non-oncogene Addiction. Cell. 2009;136:823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Skaar JR, Pagano M. Control of cell growth by the SCF and APC/C ubiquitin ligases. Curr Opin Cell Biol. 2009;21:816–24. doi: 10.1016/j.ceb.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Huen MS, Sy SM, Chen J. BRCA1 and its toolbox for the maintenance of genome integrity. Nat Rev Mol Cell Biol. 2010;11:138–48. doi: 10.1038/nrm2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Moldovan GL, D’Andrea AD. How the fanconi anemia pathway guards the genome. Annu Rev Genet. 2009;43:223–49. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lee JT, Gu W. The multiple levels of regulation by p53 ubiquitination. Cell Death Differ. 2010;17:86–92. doi: 10.1038/cdd.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gnarra JR, Tory K, Weng Y, Schmidt L, Wei MH, Li H, Latif F, Liu S, Chen F, Duh FM, et al. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat genet. 1994;7:85–90. doi: 10.1038/ng0594-85. [DOI] [PubMed] [Google Scholar]
- 124.Duan S, Cermak L, Pagan JK, Rossi M, Martinengo C, di Celle PF, Chapuy B, Shipp M, Chiarle R, Pagano M. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature. 2012;481:90–93. doi: 10.1038/nature10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Adams J. The development of proteasome inhibitors as anticancer drugs. Cancer Cell. 2004;5:417–421. doi: 10.1016/s1535-6108(04)00120-5. [DOI] [PubMed] [Google Scholar]
- 126.Richardson PG, Mitsiades C, Hideshima T, Anderson KC. Bortezomib: proteasome inhibition as an effective anticancer therapy. Annu Rev Med. 2006;57:33–47. doi: 10.1146/annurev.med.57.042905.122625. [DOI] [PubMed] [Google Scholar]
- 127.Markovina S, Callander NS, O’Connor SL, Kim J, Werndli JE, Raschko M, Leith CP, Kahl BS, Kim K, Miyamoto S. Bortezomib-Resistant Nuclear Factor-κB Activity in Multiple Myeloma Cells. Mol Cancer Res. 2008;6:1356–1364. doi: 10.1158/1541-7786.MCR-08-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hideshima T, Ikeda H, Chauhan D, Okawa Y, Raje N, Podar K, Mitsiades C, Munshi NC, Richardson PG, Carrasco RD, Anderson KC. Bortezomib induces canonical nuclear factor-κB activation in multiple myeloma cells. Blood. 2009;114:1046–1052. doi: 10.1182/blood-2009-01-199604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hideshima T, Mitsiades C, Akiyama M, Hayashi T, Chauhan D, Richardson P, Schlossman R, Podar K, Munshi NC, Mitsiades N, Anderson KC. Molecular mechanisms mediating antimyeloma activity of proteasome inhibitor PS-341. Blood. 2003;101:1530–4. doi: 10.1182/blood-2002-08-2543. [DOI] [PubMed] [Google Scholar]
- 130.Yang Y, Ikezoe T, Saito T, Kobayashi M, Koeffler HP, Taguchi H. Proteasome inhibitor PS-341 induces growth arrest and apoptosis of non-small cell lung cancer cells via the JNK/c-Jun/AP-1 signaling. Cancer sci. 2004;95:176–80. doi: 10.1111/j.1349-7006.2004.tb03200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Demo SD, Kirk CJ, Aujay MA, Buchholz TJ, Dajee M, Ho MN, Jiang J, Laidig GJ, Lewis ER, Parlati F, Shenk KD, Smyth MS, Sun CM, Vallone MK, Woo TM, Molineaux CJ, Bennett MK. Antitumor activity of PR-171, a novel irreversible inhibitor of the proteasome. Cancer Res. 2007;67:6383–91. doi: 10.1158/0008-5472.CAN-06-4086. [DOI] [PubMed] [Google Scholar]
- 132.Kuhn DJ, Chen Q, Voorhees PM, Strader JS, Shenk KD, Sun CM, Demo SD, Bennett MK, van Leeuwen FW, Chanan-Khan AA, Orlowski RZ. Potent activity of carfilzomib, a novel, irreversible inhibitor of the ubiquitin-proteasome pathway, against preclinical models of multiple myeloma. Blood. 2007;110:3281–90. doi: 10.1182/blood-2007-01-065888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, Cullis CA, Doucette A, Garnsey JJ, Gaulin JL, Gershman RE, Lublinsky AR, McDonald A, Mizutani H, Narayanan U, Olhava EJ, Peluso S, Rezaei M, Sintchak MD, Talreja T, Thomas MP, Traore T, Vyskocil S, Weatherhead GS, Yu J, Zhang J, Dick LR, Claiborne CF, Rolfe M, Bolen JB, Langston SP. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 134.Watson IR, Irwin MS, Ohh M. NEDD8 Pathways in Cancer, Sine Quibus Non. Cancer Cell. 2011;19:168–176. doi: 10.1016/j.ccr.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 135.Schulman BA, Wade Harper J. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural Insights into NEDD8 Activation of Cullin-RING Ligases: Conformational Control of Conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yamoah K, Oashi T, Sarikas A, Gazdoiu S, Osman R, Pan Z-Q. Autoinhibitory regulation of SCF-mediated ubiquitination by human cullin 1’s C-terminal tail. Proc Natl Acad Sci U S A. 2008;105:12230–12235. doi: 10.1073/pnas.0806155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Brownell JE, Sintchak MD, Gavin JM, Liao H, Bruzzese FJ, Bump NJ, Soucy TA, Milhollen MA, Yang X, Burkhardt AL, Ma J, Loke HK, Lingaraj T, Wu D, Hamman KB, Spelman JJ, Cullis CA, Langston SP, Vyskocil S, Sells TB, Mallender WD, Visiers I, Li P, Claiborne CF, Rolfe M, Bolen JB, Dick LR. Substrate-Assisted Inhibition of Ubiquitin-like Protein-Activating Enzymes: The NEDD8 E1 Inhibitor MLN4924 Forms a NEDD8-AMP Mimetic In Situ. Mol Cell. 2010;37:102–111. doi: 10.1016/j.molcel.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 139.Swords RT, Kelly KR, Smith PG, Garnsey JJ, Mahalingam D, Medina E, Oberheu K, Padmanabhan S, O’Dwyer M, Nawrocki ST, Giles FJ, Carew JS. Inhibition of NEDD8-activating enzyme: A novel approach for the treatment of acute myeloid leukemia. Blood. 2010;115:3796–3800. doi: 10.1182/blood-2009-11-254862. [DOI] [PubMed] [Google Scholar]
- 140.Milhollen MA, Traore T, Adams-Duffy J, Thomas MP, Berger AJ, Dang L, Dick LR, Garnsey JJ, Koenig E, Langston SP, Manfredi M, Narayanan U, Rolfe M, Staudt LM, Soucy TA, Yu J, Zhang J, Bolen JB, Smith PG. MLN4924, a NEDD8-activating enzyme inhibitor, is active in diffuse large B-cell lymphoma models: rationale for treatment of NF-κB–dependent lymphoma. Blood. 2010;116:1515–1523. doi: 10.1182/blood-2010-03-272567. [DOI] [PubMed] [Google Scholar]
- 141.Mackintosh C, Garcia-Dominguez DJ, Ordonez JL, Ginel-Picardo A, Smith PG, Sacristan MP, de Alava E. WEE1 accumulation and deregulation of S-phase proteins mediate MLN4924 potent inhibitory effect on Ewing sarcoma cells. Oncogene. 2012 doi: 10.1038/onc.2012.153. [DOI] [PubMed] [Google Scholar]
- 142.Luo Z, Yu G, Lee HW, Li L, Wang L, Yang D, Pan Y, Ding C, Qian J, Wu L, Chu Y, Yi J, Wang X, Sun Y, Jeong LS, Liu J, Jia L. The Nedd8-Activating Enzyme Inhibitor MLN4924 Induces Autophagy and Apoptosis to Suppress Liver Cancer Cell Growth. Cancer Res. 2012;72:3360–3371. doi: 10.1158/0008-5472.CAN-12-0388. [DOI] [PubMed] [Google Scholar]
- 143.Wei D, Li H, Yu J, Sebolt JT, Zhao L, Lawrence TS, Smith PG, Morgan MA, Sun Y. Radiosensitization of Human Pancreatic Cancer Cells by MLN4924, an Investigational NEDD8-Activating Enzyme Inhibitor. Cancer Res. 2012;72:282–293. doi: 10.1158/0008-5472.CAN-11-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Milhollen Michael A, Thomas Michael P, Narayanan U, Traore T, Riceberg J, Amidon Benjamin S, Bence Neil F, Bolen Joseph B, Brownell J, Dick Lawrence R, Loke H-K, McDonald Alice A, Ma J, Manfredi Mark G, Sells Todd B, Sintchak Mike D, Yang X, Xu Q, Koenig Erik M, Gavin James M, Smith Peter G. Treatment-Emergent Mutations in NAEβ Confer Resistance to the NEDD8-Activating Enzyme Inhibitor MLN4924. Cancer Cell. 2012;21:388–401. doi: 10.1016/j.ccr.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 145.Yang Y, Kitagaki J, Dai R-M, Tsai YC, Lorick KL, Ludwig RL, Pierre SA, Jensen JP, Davydov IV, Oberoi P, Li C-CH, Kenten JH, Beutler JA, Vousden KH, Weissman AM. Inhibitors of Ubiquitin-Activating Enzyme (E1), a New Class of Potential Cancer Therapeutics. Cancer Res. 2007;67:9472–9481. doi: 10.1158/0008-5472.CAN-07-0568. [DOI] [PubMed] [Google Scholar]
- 146.Xu GW, Ali M, Wood TE, Wong D, Maclean N, Wang X, Gronda M, Skrtic M, Li X, Hurren R, Mao X, Venkatesan M, Zavareh RB, Ketela T, Reed JC, Rose D, Moffat J, Batey RA, Dhe-Paganon S, Schimmer AD. The ubiquitin-activating enzyme E1 as a therapeutic target for the treatment of leukemia and multiple myeloma. Blood. 2010;115:2251–2259. doi: 10.1182/blood-2009-07-231191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kitagaki J, Yang Y, Saavedra JE, Colburn NH, Keefer LK, Perantoni AO. Nitric oxide prodrug JS-K inhibits ubiquitin E1 and kills tumor cells retaining wild-type p53. Oncogene. 2008;28:619–624. doi: 10.1038/onc.2008.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Pulvino M, Liang Y, Oleksyn D, DeRan M, Van Pelt E, Shapiro J, Sanz I, Chen L, Zhao J. Inhibition of proliferation and survival of diffuse large B-cell lymphoma cells by a small-molecule inhibitor of the ubiquitin-conjugating enzyme Ubc13-Uev1A. Blood. 2012 doi: 10.1182/blood-2012-02-406074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang Q, Moyret-Lalle C, Couzon F, Surbiguet-Clippe C, Saurin J-C, Lorca T, Navarro C, Puisieux A. Alterations of anaphase-promoting complex genes in human colon cancer cells. Oncogene. 2003;22:1486–1490. doi: 10.1038/sj.onc.1206224. [DOI] [PubMed] [Google Scholar]
- 150.Roy R, Chun J, Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer. 2012;12:68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Marine JC, Lozano G. Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Differ. 2009;17:93–102. doi: 10.1038/cdd.2009.68. [DOI] [PubMed] [Google Scholar]
- 152.Issaeva N, Bozko P, Enge M, Protopopova M, Verhoef LGGC, Masucci M, Pramanik A, Selivanova G. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med. 2004;10:1321–1328. doi: 10.1038/nm1146. [DOI] [PubMed] [Google Scholar]
- 153.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA. In Vivo Activation of the p53 Pathway by Small-Molecule Antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 154.Terzian T, Suh Y-A, Iwakuma T, Post SM, Neumann M, Lang GA, Van Pelt CS, Lozano G. The inherent instability of mutant p53 is alleviated by Mdm2 or p16INK4a loss. Genes Dev. 2008;22:1337–1344. doi: 10.1101/gad.1662908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wu H, Tschopp J, Lin S-C. Smac Mimetics and TNFα: A Dangerous Liaison? Cell. 2007;131:655–658. doi: 10.1016/j.cell.2007.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Fulda S, Vucic D. Targeting IAP proteins for therapeutic intervention in cancer. Nat Rev Drug Discov. 2012;11:109–124. doi: 10.1038/nrd3627. [DOI] [PubMed] [Google Scholar]
- 157.Li L, Thomas RM, Suzuki H, De Brabander JK, Wang X, Harran PG. A Small Molecule Smac Mimic Potentiates TRAIL- and TNFα-Mediated Cell Death. Science. 2004;305:1471–1474. doi: 10.1126/science.1098231. [DOI] [PubMed] [Google Scholar]
- 158.Vince JE, Wong WW-L, Khan N, Feltham R, Chau D, Ahmed AU, Benetatos CA, Chunduru SK, Condon SM, McKinlay M, Brink R, Leverkus M, Tergaonkar V, Schneider P, Callus BA, Koentgen F, Vaux DL, Silke J. IAP Antagonists Target cIAP1 to Induce TNFα-Dependent Apoptosis. Cell. 2007;131:682–693. doi: 10.1016/j.cell.2007.10.037. [DOI] [PubMed] [Google Scholar]
- 159.Varfolomeev E, Blankenship JW, Wayson SM, Fedorova AV, Kayagaki N, Garg P, Zobel K, Dynek JN, Elliott LO, Wallweber HJA, Flygare JA, Fairbrother WJ, Deshayes K, Dixit VM, Vucic D. IAP Antagonists Induce Autoubiquitination of c-IAPs, NF-κB Activation, and TNFα-Dependent Apoptosis. Cell. 2007;131:669–681. doi: 10.1016/j.cell.2007.10.030. [DOI] [PubMed] [Google Scholar]
- 160.Petersen SL, Wang L, Yalcin-Chin A, Li L, Peyton M, Minna J, Harran P, Wang X. Autocrine TNFα Signaling Renders Human Cancer Cells Susceptible to Smac-Mimetic-Induced Apoptosis. Cancer Cell. 2007;12:445–456. doi: 10.1016/j.ccr.2007.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Feltham R, Bettjeman B, Budhidarmo R, Mace PD, Shirley S, Condon SM, Chunduru SK, McKinlay MA, Vaux DL, Silke J, Day CL. Smac Mimetics Activate the E3 Ligase Activity of cIAP1 Protein by Promoting RING Domain Dimerization. J Biol Chem. 2011;286:17015–17028. doi: 10.1074/jbc.M111.222919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Dammer EB, Na CH, Xu P, Seyfried NT, Duong DM, Cheng D, Gearing M, Rees H, Lah JJ, Levey AI, Rush J, Peng J. Polyubiquitin Linkage Profiles in Three Models of Proteolytic Stress Suggest the Etiology of Alzheimer Disease. J Biol Chem. 2011;286:10457–10465. doi: 10.1074/jbc.M110.149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Enzler T, Sano Y, Choo M-K, Cottam HB, Karin M, Tsao H, Park JM. Cell-Selective Inhibition of NF-κB Signaling Improves Therapeutic Index in a Melanoma Chemotherapy Model. Cancer Discovery. 2011;1:496–507. doi: 10.1158/2159-8290.CD-11-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Vijay-Kumar S, Bugg CE, Cook WJ. Structure of ubiquitin refined at 1. 8Å resolution. J Mol Biol. 1987;194:531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]