Abstract
The circadian clock is an endogenous timer that anticipates and synchronizes biological processes to the environment. Traditional genetic approaches identified the underlying principles and genetic components, but new discoveries have been greatly impeded by the embedded redundancies that confer necessary robustness to the clock architecture. To overcome this, global (omic) techniques have provided a new depth of information about the Arabidopsis clock. Our understanding of the factors, regulation, and mechanistic connectivity between clock genes and with output processes has substantially broadened through genomic (cDNA libraries, yeast one-hybrid, protein binding microarrays, and ChIP-seq), transcriptomic (microarrays, RNA-seq), proteomic (mass spectrometry and chemical libraries), and metabolomic (mass spectrometry) approaches. This evolution in research will undoubtedly enhance our understanding of how the circadian clock optimizes growth and fitness.
Keywords: Arabidopsis, Circadian clock, Genomic, Transcriptomic, Proteomic, Metabolomic
1. Introduction
As the saying goes, timing is everything. Since the sun rises and temperatures crescendo on a daily basis, organisms must react to the repeating and predictable changes of the environment. Most organisms have evolved a self-sustaining endogenous timing mechanism called the circadian clock to anticipate and optimally synchronize biological processes with environmental oscillations. Traditional genetic approaches have been essential for describing the architecture of the circadian clock in organisms such as cyanobacteria, fungi, flies, plants, and mammals. While the precise molecular components may vary, conserved across organisms is a multilayered network of integrated transcription–translation feedback loops that are refined by protein modifications [1,2]. In the model plant Arabidopsis thaliana (Arabidopsis), the circadian clock drives daily rhythms of a broad range of processes (termed outputs) that include hypocotyl growth, leaf movement, stomatal opening, hormone and stress responses, and flowering time. While likely observed even earlier, descriptions of rhythmic behaviours in plants were first recorded millennia ago [3], but only recently have their molecular bases been extensively characterized. With new advances and applications of genome-wide technology, we are realizing that the underlying molecular signatures (from gene expression, protein levels and activity, to metabolite profiles) show distinct differences over the 24-h time period (Fig. 1). By expanding on the pioneering genetic studies of the past, omic approaches provide a more comprehensive snapshot of the simultaneous events contributing to biological processes and overall growth and fitness.
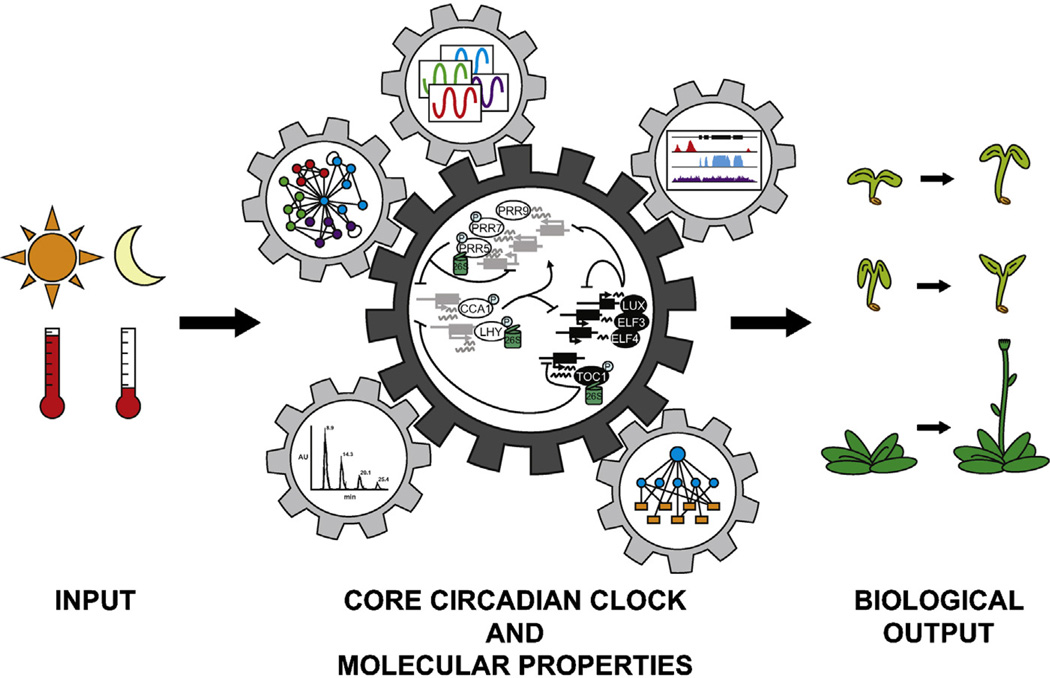
Fig. 1.
Simplified model of the Arabidopsis circadian clock. The core circadian clock is drawn with representative components to illustrate regulatory features including integrated transcription-translation based feed-back loops, alternative splicing, and protein phosphorylation and degradation. The core circadian clock regulates molecular properties including those identified by omic approaches depicted by microarrays and RNA-seq (transcripts), ChIP-seq (protein-DNA interactions), yeast-based screening (protein–protein and protein-DNA interactions), and mass spectrometry (proteins and metabolites). Some molecular rhythms feedback to regulate the core clock while others control overt biological outputs like hypocotyl elongation, leaf movement, and flowering time. Entrainment of the core clock occurs through environmental input pathways including those regulated by light and temperature.
Forward genetic screens were essential for identifying key clock gene components in Arabidopsis. Because Arabidopsis is genetically tractable, short-lived, physically compact, and produces seeds prolifically, thousands of mutagenized individuals can be screened with relative ease. The first circadian clock screen utilized a novel approach with a bioluminescent reporter driven by the circadian-regulated CHLOROPHYLL A-B BINDING PROTEIN 2 (CAB2) gene promoter to uncover mutants with altered circadian rhythms under constant light conditions [4]. This approach identified the evening-expressed clock gene TIMING OF CAB EXPRESSION1 (TOC1; also known as PSEUDORESPONSE REGULATOR1, PRR1), and along with the morning-expressed CIRCADIAN CLOCK-ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) established the first reciprocal feedback loop [5–8]. While bioluminescent reporter screens and the incorporation of other clock-regulated phenotypes (like flowering time and hypocotyl growth) have led to the identification of the majority of the ∼30 clock-associated genes to date [9,10], the discovery of new clock genes has slowed considerably. Independent large-scale forward genetic screens have contributed additional alleles of known clock genes [11–13], suggesting that this particular approach is resulting in diminished returns. Genetic redundancy is also prevalent in the Arabidopsis circadian clock as TOC1 (encoding a DNA-binding transcriptional repressor) and CCA1/ LHY (encoding MYB-like transcription factors) belong to multigene families whose members also have clock activity including PRR9/PRR7/PRR5/PRR3 and RVE8 (REVEILLE8), respectively [14–22]. Similarly, homologs of LOV-domain Kelch-repeat F-box proteins (ZEITLUPE, ZTL; FLAVIN-BINDING KELCH REPEAT F-BOX1, FKF1; and LOV KELCH PROTEIN2, LKP2), MYB-like GARP transcription factors (LUX ARRHYTHMO, LUX; and NOX; also known as PHYTOCLOCK1 and BROTHER OF LUX ARRHTYHMO, BOA respectively), and WD repeat-containing proteins (LIGHT-REGULATED WD1/2, LWD1 and LWD2) are all essential for optimal performance of the circadian clock [23–26]. Of these, only TOC1, ZTL, and LUX were initially recovered in forward genetic clock screens as higher order loss-of-function mutant combinations are needed to detect clock defects in other members. Furthermore, active compensation rather than simple redundancy has been shown to be a property of the mammalian clock network, where knock-down of different clock genes results in the up-regulation of their respective paralogs [27]. This may also be a property of the Arabidopsis clock since TOC1 RNAi plants show up-regulation of PRR9 expression during the evening [28]. As the clock consists of interconnected feedback loops, functional redundancy can also exist between non-related clock factors such as LHY, the TCP transcription factor CCA1-HIKING EXPEDITION (CHE), and PRR9/7/5/1, which all repress CCA1 expression [28–31]. While genetic and functional redundancies (and possibly active compensation) provide robustness essential to maintaining the clock network, this embedded complexity also makes the discovery of additional clock genes and the understanding of underlying network principles inherently challenging.
Since components within the clock structure remain unidentified along with their connections to output processes, alternative initiatives are necessary for more timely gene discovery and characterization. The use of omic technologies is especially suited for clock studies because of the multi-loop feedback architecture, involvement of a large number of genes, time-varying effects, and regulation at multiple levels. A basic definition of omics is the cataloguing of comprehensive sets of biological information from a given sample including genes (genomics), transcripts (transcriptomics), proteins (proteomics), and metabolites (metabolomics). However, the challenge is utilizing this data to gain insight into complex processes such as the circadian clock. Over the last decade, a number of omic-based approaches have been used to identify new clock genes and substantially expand our understanding of clock principles. In this review, we focus on recent contributions that omic approaches have made towards understanding mechanisms underlying the circadian clock in Arabidopsis and speculate on directions of future research.
2. Genomics
One of the most basic questions in circadian clock biology is: what are the genes moderating circadian rhythms? With the sequencing and annotation of the complete Arabidopsis genome [32], we had the first list of parts to begin addressing this question in a comprehensive and curated manner. What the genome revealed is that there is extensive gene duplication relative to non-plant genomes, and distinct genome differences exist when gene categories known to be important to the Arabidopsis clock are examined closely. For example only 8–23% of Arabidopsis proteins involved in transcription have related counterparts in other eukaryotes even though 48–60% of protein synthesis genes are common [32]. Of the ∼2000 transcription factors, Arabidopsis has devoted a larger percent to transcriptional regulators than yeast, Caenorhabditis elegans, and Drosophila [33–35] with almost half belonging to plant-specific families. In addition, F-box domain proteins, which are the substrate specifying module of the canonical SCF-type cullin-RING ligase complex, are over-represented relative to yeast and Drosophila [32,36]. While these observations echo what we have learned from traditional forward genetics, they also foreshadow that reverse genomic approaches may be necessary for dissecting multifaceted processes such as the circadian clock. To test this with three of the largest transcription factor families in Arabidopsis, a reverse genetic screen for altered circadian leaf movement was performed using overexpression lines of MYB, basic helix-loop-helix (bHLH), and bZIP family members (39, 29, and 4 members tested out of 131, 162, and 75 predicted members respectively) [37]. Overexpression of MYB3R2 and bHLH69 altered the phase of a CCA1 promoter-driven LUCIFERASE reporter (CCA1∷LUC) and the expression levels of several clock transcripts. This suggests that these factors influence a clock-regulated output and also modulate clock function itself. It is unknown whether loss-of-function mutants of MYB3R2 and bHLH69 have clock defects, but due to the large number of related family members, they may exhibit weak phenotypes if at all. The molecular mechanisms by which MYB3R2 and bHLH69 affect the clock remain unknown. A different genomic approach was used to directly identify a molecular mechanism regulating CCA1 expression [29]. CCA1 plays a prominent role in the clock as loss-of-function (in combination with LHY loss-of-function) and overexpression mutants exhibit obvious defects in many clock-related phenotypes [5,38,39]. Since most known clock genes exhibit circadian expression, a collection of ∼200 cycling transcription factors was tested in a high throughput yeast one-hybrid screen against fragments of the CCA1 promoter. CHE, belonging to the 23-member TCP transcription factor family, was found to bind the promoter of CCA1 but interestingly not LHY which shows similar expression patterns. Consistent with CHE functioning as a repressor, its overexpression results in down-regulation of CCA1∷LUC expression (which is dependent on the TCP binding site, TBS), and che loss-of-function mutants showed general up-regulation of CCA1∷LUC. Since CHE expression is also regulated by CCA1, another reciprocal feedback loop was established within the clock structure. Interestingly CHE physically interacts with TOC1, and both are associated with the TBS in the CCA1 promoter; however CHE antagonizes the effect of TOC1 on period length suggesting this interaction involves more than simple recruitment to the promoter. The cycling transcription factor library that recovered CHE was also used to identify a family of bHLH transcription factors (FLOWERING BHLHs, FBHs) as regulators of the photoperiodic flowering time gene CONSTANS (CO) [40] supporting that this collection will be valuable for identifying both clock and output regulators that are members of large protein families.
These pilot approaches [29,37] will undoubtedly expand to include all transcription factors, if not all genes, in the Arabidopsis genome. Such systematic surveys are currently limited by the collections of cloned cDNAs available. As transcriptional regulation can precede regulation at the protein and metabolite levels, transcription factors are of special interest to many research fields. In fact, several transcription factor collections have been independently created for Arabidopsis research. The three largest collections to date include up to ∼1600 transcription factors and together include nearly all Arabidopsis transcription factors [41–43]. These were all created in recombinase-compatible vectors that allow for easy transfer to expression vectors such as those used for in planta overexpression or yeast one-hybrid studies. A recent application of these collections, termed the Protoplast TransActivation (PTA) screen, utilized transiently transformed Arabidopsis mesophyll protoplasts to test the capabilities of a high throughput screening protocol for transactivation of promoter∷LUC reporters in a 96-well microtiter plate format [44]. Screening was relatively rapid and reproducible, and more than 700 transcription factors have been transferred to this system for future screening. Protoplasts have been shown to be especially well suited for clock studies since they maintain high amplitude oscillations of clock promoter∷LUC reporters after 6 days under constant light [45]. Interestingly, artificial microRNAs (amiRNAs) can effectively target endogenous clock gene expression in protoplasts and confer transcriptional defects consistent with the respective stable loss-of-function mutants. In fact, an amiRNA that targets all four casein kinase II β-subunits (CKBs) was shown to cause period lengthening, which is opposite to the short period defect previously observed in CKB overexpression transgenic lines [46,47]. The period lengthening in the transient experiment was further confirmed in stable transgenic Arabidopsis lines. Together, these studies indicate that the combination of a complete Arabidopsis transcription factor collection and amenable transient in planta expression systems may be especially useful for reverse genomic studies because they can overcome genetic redundancy issues rapidly. Furthermore, these types of screens will ultimately be enhanced by genome-wide cDNA collections [48], tissue-specific collections [49], and automated screening procedures [50].
Other systematic approaches to characterize the Arabidopsis circadian clock have involved the identification of direct (and ultimately genome-wide) targets of clock-associated transcription factors. Here, complementary in vitro and in vivo approaches have been pursued. A recent in vitro study involved the analysis of LUX, one of the few clock genes that when mutated causes arrhythmicity in clock reporter gene expression and severe defects in hypocotyl growth and flowering time [51,52]. Although LUX was known to transcriptionally regulate CCA1 and LHY and, conversely, be transcriptionally regulated by CCA1 and LHY, placement of LUX within the core clock mechanism was uncertain. Based on its protein sequence, LUX was predicted to possess a GARP DNA-binding domain. As a first step towards identifying target genes, the DNA-binding motif of LUX was determined by an in vitro approach where GST-tagged LUX was expressed and hybridized to a universal protein binding microarray (PBM) consisting of all possible 10 bp DNA sequences [24]. The LUX binding site (LBS) was determined to be GATWCG (where W is A or T). Subsequently, in vivo binding to regions that include the LBS in the PRR9 and LUX promoters was confirmed by chromatin immunoprecipitation followed by PCR (ChIP-PCR). Binding to these regions has functional consequences since up-regulation of PRR9 occurs in the lux loss-of-function mutant and LUX overexpression represses endogenous LUX expression [24,52]. The mechanistic connection between LUX and PRR9 suggests that CCA1 is indirectly activated by LUX through repression of its transcriptional repressor PRR9, illustrating an example of a complex feed-forward connection within the clock [53]. The non-homologous proteins EARLY FLOWERING 3 (ELF3) and EARLY FLOWERING4 (ELF4), which lack domains of known function, also associate with LUX in the Evening Complex at the PRR9 promoter where ELF3 recruitment is dependent on both LUX and its homolog NOX [54–57]. Whether LUX also directly binds the CCA1 promoter (which possesses an LBS motif within its 5′UTR) like NOX [25] is unknown. With the binding motif of LUX clearly established, the genome-wide targets of LUX will be the next obvious step towards understanding how it regulates such a large suite of clock phenotypes.
A powerful in vivo genomics strategy for analyzing the nature of the circadian clock involves the use of ChIP to identify genes directly targeted by transcriptional regulators. However, since ChIP identifies association with large chromatin regions, this method is currently not sensitive enough to determine the precise DNA-binding motif of a given protein. Another consideration is that ChIP involves cross-linking that stabilizes both protein–DNA and protein–protein interactions, so that immunoprecipitated DNA may reflect a pool of direct and indirect associations with DNA. While ChIP-PCR has been used to identify specific in vivo targets for many clock proteins [21,22,24,25,29,30,55,57–69], genome-wide associations using ChIP-followed by high throughput sequencing (ChIP-seq) have only been reported for TOC1 and PRR5 [28,70]. Loss-of-function mutants of PRR5 (in combination with PRR7) and TOC1 exhibit short period defects in gene expression and altered flowering time [7,18]. Both genes are circadian-expressed with a peak near subjective dusk, with PRR5 expression preceding TOC1 (http://diurnal.mocklerlab.org; [71]). TOC1 and PRR5 both possess repressor activity and associate with DNA through their CCT (CONSTANS, CONSTANS-LIKE, and TOC1) domains, but only TOC1 has been shown to directly bind DNA in vitro [28,30,31,70]. ChIP-seq was performed using transgenic lines expressing functional TOC1-and PRR5-GFP translational fusions, and 772 and 1024 potential target genes were reported respectively. Both datasets showed enrichment for morning-phased genes that included the previously identified CCA1 [29,30]. Consistent with morning phase enrichment, genes with G-box/G-box-expanded motifs were prevalent in both datasets. TOC1 and PRR5 both associate with other clock promoters including PRR9, PRR7, and PRR5, but TOC1 alone was reported to associate with the evening-expressed GI (GIGAN-TEA), ELF4, and LUX promoters and only PRR5 associates with the morning-expressed RVE8 promoter (as previously proposed [21]). This is consistent with only the TOC1 dataset showing enrichment for an Evening-Element (EE)-like expanded motif. Interestingly, TOC1 and PRR5 also associate with their own promoters which could support auto-regulation as previously shown for LUX and proposed for both CCA1 and LHY [5,6,24]. Transcriptional consequences of promoter occupation were confirmed for many targets by analyzing lines with altered TOC1 or PRR5 expression [28,30,31,70]. TOC1, PRR9, PRR7, and PRR5 share several target genes supporting their coordinated functions as a wave of inhibitors as proposed initially based on transcription and genetic data [18,28,70,72]. While this genome-wide data is essential for clarifying connectivity between known clock genes, it can also explore new nodes of output regulation from the clock. For example, using a stringent list of 64 PRR5 direct target genes, enrichment was seen for GO annotations associated with “transcription” and included transcription factors that regulate auxin biosynthesis, cotyledon opening, flowering time, hypocotyl elongation, and cold responsiveness [70]. The contribution of PRR5 to these diverse processes remains for future studies.
3. Transcriptomics
Transcriptome studies were the first global experiments to provide hints about how molecular rhythms could translate into rhythmic activities at the whole plant level [73,74]. Time–course studies have generally been performed using whole plants under ambient temperature and constant light with sampling at various intervals over a two-day period. Early time–course studies estimated that 2–16% of the steady state transcriptome is regulated by the circadian clock with peak phases occurring throughout the day [73–76]. However, the integration of multiple datasets and improved methods has increased estimates up to approximately one-third of the expressed genome [77–79]. The combination of phase data and promoter analysis has identified regulatory elements sufficient for circadian expression. These are phased to dusk (Hormone Up at Dawn, HUD, CACATG), early-night (Evening Element, EE, AAAATATCT), and mid-night (Protein Box, PBX, ATGGGCC) [73,77,80]. The Morning Element (ME, AACCAC-GAAAAT), which was identified serendipitously by mutation of the EE, shows peak expression at dawn and is the only other motif sufficient to confer circadian rhythms in reporter assays [81]. While these motifs confer expression peaks throughout the 24-hour period, other motifs are likely required to generate the complete phase diversity observed in circadian expression. Motifs including the G-box, GATA, and others were found to be overrepresented in genes expressed at particular phases but additional studies are required to clarify their role in regulating expression [77,78,82]. Regulatory functions of the ME, G-box, GATA, EE, and PBX motifs appear conserved across plants as transcriptome analysis of rice and poplar also identified enrichment during the same phases of expression [83]. Of note, a tissue-specific circadian time–course has been reported for dark-shielded root tissue where only a subset of the clock genes oscillated and a smaller proportion of expressed genes were rhythmic compared to light-grown shoot tissue [84]. Within the same organ, subsets of cells have also shown differences in clock regulation [85–87] suggesting that future transcriptome studies will need to address the importance of cellular coupling and the control of output regulation across cell types.
A common strategy employed in the circadian clock field is transcript clustering to identify output processes. In parallel, links between the clock and other processes have arisen by comparing independent gene expression data with circadian microarray datasets. While many processes have been associated by transcript data alone, only a subset has been confirmed to be mechanistically linked to the clock. For example, using ChIP-PCR, CCA1 was found to associate with several promoter regions to regulate chlorophyll biosynthesis and starch metabolism, cold sensitivity, and reactive oxygen species (ROS) homeostasis [59,61,64,73,88,89]. In each case, researchers were led to focus on these genes based on information gathered from gene expression microarrays and were able to further show that altered expression of CCA1 affects these biological processes. Together these studies suggest that CCA1 may be a transcriptional hub connecting the clock to output regulation. Reciprocally, temperature and ROS also function as signals that regulate clock and output oscillations; however, the direct mechanisms underlying these arms of regulation remain unknown [16,64,85,90].
Microarray studies have also been used to link several evening-phased clock proteins directly to output genes. PIF4 (PHYTOCHROME-INTERACTING FACTOR4) and PIF5 encode bHLH transcription factors that control the rhythmic growth of hypocotyls under light/dark growth regimes [89]. These genes were selected for characterization from microarray studies by their correlation of expression with hypocotyl elongation. Clock-regulation of PIF4 and PIF5 expression is essential for proper rhythmic growth, and subsequently members of the Evening Complex (LUX, ELF3, and ELF4) were found to directly regulate their expression [54,91].
To identify TOC1-regulated processes, comparisons between microarray datasets from a TOC1 overexpressor and the toc1–2 loss-of-function mutant discovered significant overlap between mis-regulated genes and those associated with ABA signalling [69]. This overlap included the circadian-regulated ABAR gene which encodes a putative ABA-receptor. TOC1 was found to regulate ABA-regulated drought sensitivity by binding the promoter of ABAR in a time-of-day specific manner to repress expression [69]. Mis-regulation of TOC1 affects dehydration tolerance, stomatal aperture and conductance, and water loss (all regulated by ABAR), and genetic analysis confirmed that TOC1 regulation of these responses is dependent on the presence of ABAR. In addition to this, other clock outputs have also been proposed by microarray studies and supported by follow-up biological studies. However in these cases, direct mechanistic connections to the clock have yet to be established. These outputs include auxin responsiveness, nitrogen metabolism, fungal and bacterial defence, glucosinolate metabolism, and insect herbivory [76,92–99].
Circadian time–course array studies have focused on characterizing wildtype or mis-expression lines under a variety of steady state conditions following entrainment. While extremely informative in the general sense, they lack specific insight into the immediate and direct consequences of particular clock proteins. Since the majority of clock proteins regulate transcription and fluctuate daily in abundance, novel insight can be obtained by coupling microarray studies with gene-inducible expression lines such as the established alcohol-inducible CCA1, LHY, and TOC1 lines [100]. For clock studies, inducible lines offer the distinct advantage of precisely controlling the timing and level of transgene expression. This approach was recently taken to identify potential clock and output targets regulated by TOC1 on a genome-wide scale through microarray analysis [31]. Seedlings were induced and collected near dawn and subjective-dawn under light/dark and constant light conditions. Correlating with its own oscillating expression, the genes altered by TOC1 induction (2566 in total) include more genes known to cycle than expected by chance alone. Analysis of the mis-regulated gene set showed that the corresponding promoter regions were enriched for three DNA motifs: the G-box and GA element in up-regulated genes, and the TBS in down-regulated genes. Since the GA element has not been previously associated with clock regulation, it will be of interest to determine how TOC1 affects gene expression through this motif. Although TOC1 was found to bind a short sequence from the CCA1 promoter called the T1ME motif (TGTG) in vitro, it was not recovered in the TOC1 induction dataset suggesting the motif and/or dataset may be incomplete. Output genes associated with TOC1 induction included those involved in metabolism, and biotic and abiotic stress responses. Whether TOC1 regulates these processes directly or indirectly will require additional studies including comparison with genes identified by TOC1 ChIP-seq [28].
The profiling of gene expression through microarrays has made the greatest contribution to Arabidopsis circadian research to date, but recently has been complemented by direct RNA sequencing (RNA-seq). RNA-seq offers greater depth of information for transcripts that are unannotated or indiscernible by microarrays such as newly annotated genes and splice variants. With this approach, alternative splicing was revealed as a new mode of clock gene regulation in Arabidopsis. The first description of alternative splicing identified CCA1 transcripts which retain portions of intron 4 to produce two splice isoforms with premature termination codons [101,102]. Relative to the reference isoform, these transcript variants were substantially increased by high light treatment but decreased in the cold. These splice variants were also detected in poplar and the monocots Brachypodium and rice, indicating that their production is likely conserved across plant species. The functional consequence of CCA1 alternative splicing in the cold was shown more recently with the characterization of a third splice variant (CCA1β) which upon translation retains all domains encoded by the full-length transcript (CCA1 α) except the MYB DNA-binding domain [90]. As the CCA1 α protein forms dimers [103,104] and CCA1β retains its dimerization domain, CCA1β functions as a dominant-negative regulator of CCA1α protein–protein interactions as well as its ability to bind DNA. CCA1β suppresses CCA1α overexpression phenotypes, and causes period-shortening of clock and output genes similar to the cca1 lhy double loss-of-function mutant. As reported for the other CCA1 splice variants, the production of CCA1β is suppressed by the cold, and this suppression is necessary for freezing tolerance since CCA1β overexpression lines are sensitive to freezing [90]. In addition to CCA1, many other clock gene transcripts undergo alternative splicing events [105–110]. Interestingly, the often co-regulated gene pairs CCA1-LHY and PRR9-PRR7 undergo differential alternative splicing in response to temperature changes [106], however, the significance of these observations is unknown.
Little is known about the precise mechanisms that connect alternative splicing and the Arabidopsis circadian clock. Alternative splicing of PRR9 is regulated by PRMT5 (PROTEIN ARGININE METHYL TRANSFERASE5) which is known to transfer methyl groups to histones, components of the transcriptional complex, and spliceosomal proteins in Arabidopsis and other systems [102,105,108]. Mutation of PRMT5 causes an increase in the amount of a PRR9 splice variant that retains intron 3 but decreases the amount of the normal full length PRR9 transcript [105]. In addition, full-length PRR7 transcript is mis-regulated in prmt5 but without alterations in splicing. As overexpression of the PRR9 intron 3 variant had no effect on circadian rhythms, the decrease in both PRR9 and PRR7 transcripts may explain the long period defects of prmt5 mutants. Using tiling microarrays, the effect of PRMT5 mutation was found to extend beyond known clock genes and affect splicing events at a global level [105]. Recently a direct mechanistic link between the clock and splicing was reported when SKIP (a conserved SNW/Ski-interacting protein (SKIP) domain protein) was identified as an interactor of PRR7 and PRR9 pre-mRNAs [109]. SKIP interacts with the spliceosome machinery to regulate alternative splicing. Like prmt5, skip mutants exhibit period lengthening of clock gene expression and outputs, which can again be explained at least in part by a reduction of full length PRR7 and PRR9 transcripts. RNA-seq was used to determine the extent of alternative splicing in the skip mutant, which revealed splicing variation in many clock gene transcripts, as well as other genes involved in a diverse range of functions suggesting that SKIP also regulates general splicing events of mRNAs. Mutation of STIPL1 (SPLICEOSOMAL TIMEKEEPER LOCUS1), encoding a putative RNA binding protein, was also recently shown to affect splicing of several clock transcripts and cause a long period defect [110]. Whether the alternative splice products of any of the known clock genes affects period lengthening in skip or stip1 remains for future studies. The identification of splicing regulators that impinge on clock parameters as well as alternative splice variants of clock transcripts supports that alternative splicing has an essential function within the circadian clock.
4. Proteomics and metabolomics
Relatively few circadian studies in Arabidopsis have utilized proteomic or metabolomic approaches even though both have great potential to reveal new information about the Arabidopsis circadian clock. This can be inferred from mammalian studies where a significant disconnect exists between steady state mRNA and protein levels, and many metabolites form reciprocal feedback loops within the clock [111–114]. In part, the lack of data may reflect the relative technical difficulties associated with obtaining proteomic and metabolomics data.
A variety of approaches have been taken to describe the spectrum of Arabidopsis proteins involved in circadian regulation. The most general is to identify those with rhythmic changes in abundance from bulk tissues collected during a circadian time–course. With this approach, the first characterization of the circadian proteome in a higher plant was obtained using two-dimensional gel electrophoresis followed by mass spectrometry [115]. Fifty-three protein spots from rice shoots showed oscillations in constant darkness. Ten were identified by mass spectrometry, and found to be associated with functions in photosynthesis, central metabolism, amino acid synthesis, and oxidative stress. Since this experimental approach biased towards highly abundant proteins, it may not be surprising that the identified proteins have enzymatic rather than transcriptional or signalling functions. The transcript profile of only one circadian protein (a putative phosphoribulokinase precursor) was measured and found to mirror that of the protein. However, phase comparisons between 84 diurnally-expressed proteins and those reported from microarray studies showed very little correlation [115], suggesting extensive post-transcriptional or post-translational events occur to generate fluctuations in daily protein levels.
Another more targeted proteomic approach was taken to reveal the role of REVEILLE8 (RVE8) within the clock [21]. Using a semi-in vivo technique, plant extracts collected in the afternoon were assayed for proteins that selectively bound immobilized oligonucleotides containing the EE motif. Among the identified transcription factors (which belonged to MYB-like, B3 domain, Basic-leucine zipper, Trihelix, Wrky, and Whirly families), only RVE8 was recovered consistently across replicate experiments. Binding was specific, as RVE8 was not recovered with a mutagenized EE motif. RVE8 expression was found to peak at dawn with protein levels lagging by several hours [21,22]. While RVE8 exhibited similar binding properties as the previously characterized and closely-related CCA1 and RVE1 in yeast one-hybrid and electrophoretic mobility shift assays, genetic analysis of RVE8 loss-of-function and overexpression lines showed distinct differences from its homologs in the misregulation of clock gene expression [21,22]. For instance, unlike the cca1 and rve1 loss-of-function mutants which have a short-period and no effect on clock gene expression respectively, RVE8 mutation causes long period defects [21,22,92,116]. Functions in temperature compensation also differ as RVE8, like LHY, has impacts on period length under high temperatures while CCA1 plays a more dominant role under lower temperatures [21,117]. RVE8 appears to impact PRR5 by binding its promoter, and RVE8 overexpression up-regulates the trough level of PRR5 expression. In turn, PRR5 associates with the RVE8 promoter, and PRR5 mis-expression affects RVE8 transcript levels [21,70] revealing another integral feedback loop within the clock. The loss-of-function phenotypes of RVE8 include subtle sensitivity to light and temperature conditions, consistent with redundancy between its homologs CCA1 and LHY, and supporting that characterization of higher order mutants will likely be informative. RVE8 also regulates TOC1 expression by associating with its promoter and regulating histone H3 acetylation to promote expression [22].
Chemical genetic screens are another strategy to identify regulatory mechanisms of the Arabidopsis circadian clock. In this approach, alterations in biological processes are assayed for by testing chemical effectors that may bind to regulatory proteins and alter their activities, interactions, or stability. This approach may be especially useful in overcoming the genetic redundancy inherent in the circadian clock as chemicals may target a set of related proteins. A natural compound library of 720 chemicals (enriched in steroids, flavonoids, terpenes, limonoids, and coumarins) was screened for perturbations in GI∷LUC rhythms under constant darkness [118]. Prieurianin (Pri) and prieurianin acetate (Pri-Ac) were identified as two related compounds that caused period shortening, dampening, and reduced expression of GI∷LUC and CCA1∷LUC. In addition to affecting circadian expression, both Pri and Pri-Ac caused polarity alterations in different cell types that suggested actin cytoskeletal defects. In fact the known actin inhibitors LatB, CytD, and Jpk shortened the period length of GI∷LUC in the dark in a dose-dependent manner. However LatB treatment under red and blue light conditions caused distinct effects on period length suggesting the photoreceptors regulating the circadian clock are differentially affected by actin impairment. Together these results establish a new link between actin dynamics and the regulation of the circadian clock. Determining the protein target(s) of Pri/Pri-Ac will be the next challenge.
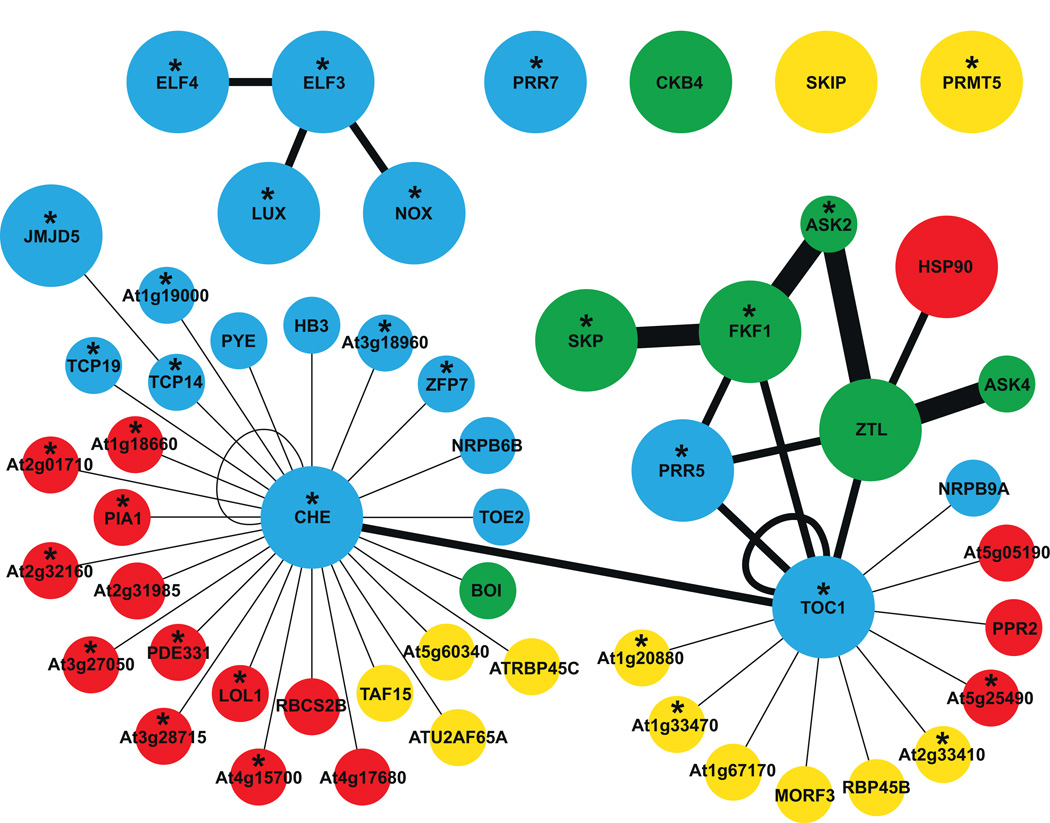
Many protein–protein interactions are essential for circadian clock function. Protein complexes that regulate both clock gene transcription [29,54,56,57,65,103] and protein stability [23,118–120,20,121–124] have been reported. These interactions were identified primarily through detailed genetic and mechanistic studies based on a priori knowledge. To gain an unbiased view of protein–protein interactions relevant to the circadian clock, comprehensive proteomic studies will be required. One approach to generally characterize the binary interactions in the Arabidopsis proteome utilized a high throughput yeast two-hybrid system [48]. In total, ∼8000 proteins were tested in pairwise combinations and the resulting interactome consisted of ∼6200 protein–protein interactions between ∼2700 proteins. This was estimated to represent ∼2% of the interactions occurring in Arabidopsis. Biological relevance for these interactions was supported by enrichment in co-expression and gene ontology (GO) annotations between protein pairs. Interestingly, fourteen clock proteins were included in the screen and are illustrated in Fig. 2. Although most clock proteins did not recover new interactions, CHE and TOC1 each formed communities within the network, with 26 and 10 new interactors, respectively. Interacting proteins have GO annotations associated with transcription, RNA-regulation, and protein modifications (http://www.arabidopsis.org; [125]), and the majority exhibit circadian regulation at the transcript level (http://diurnal.mocklerlab.org; [71]. These features suggest they may have important regulatory functions in the clock network. The depiction shown in Fig. 2 should not be considered a comprehensive clock protein–protein network but rather serves to illustrate the advantages of systematic legacy data and provides examples of protein–protein interactions that should be further characterized by detailed in vivo studies.
Fig. 2.
protein–protein interactions reported for clock proteins by the Arabidopsis Interactome Mapping Consortium (AIMC) [48]. Known clock proteins are denoted by larger circles while others are represented by smaller circles. GO annotations associated with node colours: blue, transcription; yellow, RNA-binding; green, protein modification; and red, other annotations. Edge styles: thin line, yeast two-hybrid interaction from AIMC; medium line, interaction supported by other literature; and thick line, supported by both AIMC yeast two-hybrid interaction and other literature. Asterisks depict proteins that have a circadian-regulated transcript identified by DIURNAL (correlation cutoff value of 0.8 in at least one circadian array; http://diurnal.mocklerlab.org; [71]. Note: individual HSP90 homologs were not included as the specific member(s) involved in clock function have not been reported.
Although much evidence supports the regulation of metabolites by the clock [126], few studies have undertaken a global analysis. The metabolomes of the two arrhythmic clock mutants d975 (the triple mutant for PRR9, PRR7, and PRR5) and CCA1-OX (overexpressor of CCA1) have been compared [127]. While both mutants have similar general morphology, their underlying metabolite profiles are quite distinct. The d975 mutant exhibits a pronounced increase in tricarboxylic acid (TCA) intermediates including citrate and malate, as well as an increase in the secondary metabolite shikimate. Corresponding transcript levels for enzymes involved in the TCA cycle were found to support the changes in metabolite levels in d975. The effects of these metabolites in CCA1-OX were less or not obvious suggesting PRR9/7/5 have a greater impact on these metabolic pathways. In addition transcriptome analysis of d975 suggested the expression of genes involved in carotenoid and ABA biosynthesis is increased which correlated with an increased ABA level and tolerance to ABA-regulated processes including cold, salt and drought [127,128]. Metabolome analysis has also been used to compare clock regulation under constant light with a simultaneous 4°C cold treatment [88]. Under 20°C constant light, 19 metabolites oscillated but only maltose continued to cycle upon transfer to the cold, albeit for a single day. As these analyses focused on compounds including sugars, amino acids and organic acids, it is possible other rhythmic metabolites remain to be linked to the circadian clock. Metabolite profiling has also been used to characterize the effect of clock gene mutation in rice [129]. Mutation of the only GI ortholog in rice (Os-GI) altered the levels of 73 metabolites relative to wildtype under diurnal field conditions. Interestingly, TCA intermediates show increases (aconitate and isocitrate) and decreases (citrate and malate) in abundance which were not supported by transcriptome data collected in parallel. This lack of correlation between transcript and metabolite levels suggests that additional layers of post-transcriptional regulation are involved or that metabolites are indirectly regulated by the clock.
5. Conclusions
The circadian clock displays intricate regulation at multiple levels from transcriptional to post-translational. Traditionally, the principles and molecular components underlying clock regulation were identified on a gene-to-gene basis through classic genetic analyses. Much was learned, but discovery in this vein has slowed considerably. With the proliferation of information from omic technologies including gene expression profiling, yeast-based assays, ChIP-seq, mass spectrometry, and combinations thereof, broader insight has been gained about the mechanisms of clock gene connectivity as well as the control of output processes. Nonetheless, key omic datasets remain uncharacterized. For example, global epigenetic studies over a circadian time–course will be very informative as changes in histone acetylation, methylation, and ubiquitination have been correlated with altered clock gene expression [22,58,59,130–132]. In mouse models, substantial desynchrony has been observed between the nascent and steady state circadian transcriptomes, implying that post-transcriptional regulation is widespread [133–135]; comparable studies in Arabidopsis will determine whether this is conserved across circadian systems. While several RNA-based modes of circadian regulation have been reported, as well as differences in protein translation rates [136–138], their contributions at the global level and mechanistic insights remain unknown. Finally, since protein modifications such as phosphorylation and ubiquitination regulate the activities of key clock proteins [139–141], knowledge about the global circadian changes in protein modifications and the identification of clock protein modifiers will be of great value. Insight into the latter has been gained in mammalian studies using high-throughput RNAi and chemical screens, and the development of omic tools like kinase panels to directly test the phosphorylation of clock proteins will also be useful [142–147]. Key to these and already published omic datasets will be the overlaying of cell-specific information. Ultimately, the integration of all these datasets will provide unprecedented insight into how the circadian clock confers fitness and optimal growth in Arabidopsis.
Acknowledgements
We thank Jose Pruneda-Paz, Joshua M. Gendron, Elsebeth Kolmos, Naden Krogan, and Tsuyoshi Hirota for helpful discussion and critical reading of the manuscript; Naden Krogan for illustrating the figures; and funding by the NIH grants R01 GM50006 and GM67837 to Steve A. Kay.
Contributor Information
Brenda Y. Chow, Email: bchow@ucsd.edu.
Steve A. Kay, Email: stevekay@dornsife.usc.edu.
References
- 1.Harmer SL, Panda S, Kay SA. Molecular bases of circadian rhythms. Annual Review of Cell and Developmental Biology. 2001;17:215–253. doi: 10.1146/annurev.cellbio.17.1.215. [DOI] [PubMed] [Google Scholar]
- 2.Zhang EE, Kay SA. Clocks not winding down: unravelling circadian networks. Nature Reviews Molecular Cell Biology. 2010;11:764–776. doi: 10.1038/nrm2995. [DOI] [PubMed] [Google Scholar]
- 3.McClung CR. Plant circadian rhythms. Plant Cell. 2006;18:792–803. doi: 10.1105/tpc.106.040980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Millar AJ, Carre IA, Strayer CA, Chua NH, Kay SA. Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science. 1995;267:1161–1163. doi: 10.1126/science.7855595. [DOI] [PubMed] [Google Scholar]
- 5.Wang ZY, Tobin EM. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATE 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998;93:1207–1217. doi: 10.1016/s0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- 6.Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carre IA, et al. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell. 1998;93:1219–1229. doi: 10.1016/s0092-8674(00)81465-8. [DOI] [PubMed] [Google Scholar]
- 7.Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Más P, et al. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science. 2000;289:768–771. doi: 10.1126/science.289.5480.768. [DOI] [PubMed] [Google Scholar]
- 8.Alabadi D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- 9.Nakamichi N. Molecular mechanisms underlying the Arabidopsis circadian clock. Plant and Cell Physiology. 2011;52:1709–1718. doi: 10.1093/pcp/pcr118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagel DH, Kay SA. Complexity in the wiring and regulation of plant circadian networks. Current Biology. 2012:22–28. doi: 10.1016/j.cub.2012.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hazen SP, Borevitz JO, Harmon FG, Pruneda-Paz JL, Schultz TF, Yanovsky MJ, et al. Rapid array mapping of circadian clock and developmental mutations in Arabidopsis. Plant Physiology. 2005;138:990–997. doi: 10.1104/pp.105.061408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kevei E, Gyula P, Hall A, Kozma-Bognár L, Kim WY, Eriksson ME, et al. Forward genetic analysis of the circadian clock separates the multiple functions of ZEITLUPE. Plant Physiology. 2006;140:933–945. doi: 10.1104/pp.105.074864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin-Tryon EL, Kreps JA, Harmer SL. GIGANTEA acts in blue light signaling and has biochemically separable roles in circadian clock and flowering time regulation. Plant Physiology. 2007;143:473–486. doi: 10.1104/pp.106.088757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farré EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA. Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Current Biology. 2005;15:47–54. doi: 10.1016/j.cub.2004.12.067. [DOI] [PubMed] [Google Scholar]
- 15.Nakamichi N, Kita M, Ito S, Sato E, Yamashino T, Mizuno T. The Arabidopsis pseudo-response regulators, PRR5 and PRR7, coordinately play essential roles for circadian clock function. Plant and Cell Physiology. 2005;46:609–619. doi: 10.1093/pcp/pci061. [DOI] [PubMed] [Google Scholar]
- 16.Salomé PA, McClung CR. PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell. 2005;17:791–803. doi: 10.1105/tpc.104.029504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujimori T, Sato E, Yamashino T, Mizuno T. PRR5 (PSEUDO-RESPONSE REGULATOR 5) plays antagonistic roles to CCA1 (CIRCADIAN CLOCK-ASSOCIATED 1) in Arabidopsis thaliana . Bioscience, Biotechnology, and Biochemistry. 2005;69:426–430. doi: 10.1271/bbb.69.426. [DOI] [PubMed] [Google Scholar]
- 18.Nakamichi N, Kita M, Ito S, Yamashino T, Mizuno T. PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana . Plant and Cell Physiology. 2005;46:686–698. doi: 10.1093/pcp/pci086. [DOI] [PubMed] [Google Scholar]
- 19.Matsushika A, Murakami M, Ito S, Nakamichi N, Yamashino T, Mizuno T. Characterization of Circadian-associated pseudo-response regulators: I.Comparative studies on a series of transgenic lines misexpressing five distinctive PRR Genes in Arabidopsis thaliana . Bioscience, Biotechnology, and Biochemistry. 2007;71:527–534. doi: 10.1271/bbb.60583. [DOI] [PubMed] [Google Scholar]
- 20.Para A, Farré EM, Imaizumi T, Pruneda-Paz JL, Harmon FG, Kay SA. PRR3 is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock. Plant Cell. 2007;19:3462–3473. doi: 10.1105/tpc.107.054775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rawat R, Takahashi N, Hsu PY, Jones MA, Schwartz J, Salemi MR, et al. REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genetics. 2011;7:e1001350. doi: 10.1371/journal.pgen.1001350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farinas B, Mas P. Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant Journal. 2011;66:318–329. doi: 10.1111/j.1365-313X.2011.04484.x. [DOI] [PubMed] [Google Scholar]
- 23.Baudry A, Ito S, Song YH, Strait AA, Kiba T, Lu S, et al. F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell. 2010;22:606–622. doi: 10.1105/tpc.109.072843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helfer A, Nusinow DA, Chow BY, Gehrke AR, Bulyk ML, Kay SA. LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Current Biology. 2011;21:126–133. doi: 10.1016/j.cub.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai S, Wei X, Pei L, Thompson RL, Liu Y, Heard JE, Ruff TG, Beachy RN. BROTHER OF LUX ARRHYTHMO is a component of the Arabidopsis circadian clock. Plant Cell. 2011;23:961–972. doi: 10.1105/tpc.111.084293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu JF, Wang Y, Wu SH. Two new clock proteins, LWD1 and LWD2, regulate Arabidopsis photoperiodic flowering. Plant Physiology. 2008;148:948–959. doi: 10.1104/pp.108.124917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baggs JE, Price TS, DiTacchio L, Panda S, Fitzgerald GA, Hogenesch JB. Network features of the mammalian circadian clock. PLoS Biology. 2009;7:e52. doi: 10.1371/journal.pbio.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang W, Pérez-García P, Pokhilko A, Millar AJ, Antoshechkin I, Riechmann JL, et al. Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science. 2012;336:75–79. doi: 10.1126/science.1219075. [DOI] [PubMed] [Google Scholar]
- 29.Pruneda-Paz JL, Breton G, Para A, Kay SA. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science. 2009;323:1481–1485. doi: 10.1126/science.1167206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua NH, Sakakibara HP. SEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell. 2010;22:594–605. doi: 10.1105/tpc.109.072892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gendron JM, Pruneda-Paz JL, Doherty CJ, Gross AM, Kang SE, Kay SA. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3167–3172. doi: 10.1073/pnas.1200355109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana . Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 33.Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 34.Guo A, He K, Liu D, Bai S, Gu X, Wei L, et al. DATF: a database of Arabidopsis transcription factors. Bioinformatics. 2005;21:2568–2569. doi: 10.1093/bioinformatics/bti334. [DOI] [PubMed] [Google Scholar]
- 35.Yilmaz A, Mejia-Guerra MK, Kurz K, Liang X, Welch L, Grotewold E. AGRIS: the Arabidopsis gene regulatory information server, an update. Nucleic Acids Research. 2011;39:D1118–D1122. doi: 10.1093/nar/gkq1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD. The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:11519–11524. doi: 10.1073/pnas.162339999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanano S, Stracke R, Jakoby M, Merkle T, Domagalska MA, Weisshaar B, Davis SJ. A systematic survey in Arabidopsis thaliana of transcription factors that modulate circadian parameters. BMC Genomics. 2008;9:182. doi: 10.1186/1471-2164-9-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alabadí D, Yanovsky MJ, Más P, Harmer SL, Kay SA. Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis. Current Biology. 2002;12:757–761. doi: 10.1016/s0960-9822(02)00815-1. [DOI] [PubMed] [Google Scholar]
- 39.Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, Song HR, et al. LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Developmental Cell. 2002;2:629–641. doi: 10.1016/s1534-5807(02)00170-3. [DOI] [PubMed] [Google Scholar]
- 40.Ito S, Song YH, Josephson-Day AR, Miller RJ, Breton G, Olmstead RG, et al. FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:3582–3587. doi: 10.1073/pnas.1118876109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mitsuda N, Ikeda M, Takada S, Takiguchi Y, Kondou Y, Yoshizumi T, et al. Efficient yeast one/two-hybrid screening using a library composed only of transcription factors in Arabidopsis thaliana . Plant and Cell Physiology. 2010;51:2145–2151. doi: 10.1093/pcp/pcq161. [DOI] [PubMed] [Google Scholar]
- 42.Ou B, Yin KQ, Liu SN, Yang Y, Gu T, Wing Hui JM, et al. A high-throughput screening system for Arabidopsis transcription factors and its application to Med25-dependent transcriptional regulation. Molecular Plant. 2011;4:546–555. doi: 10.1093/mp/ssr002. [DOI] [PubMed] [Google Scholar]
- 43.Castrillo G, Turck F, Leveugle M, Lecharny A, Carbonero P, Coupland G, et al. Speeding cis-trans regulation discovery by phylogenomic analyses coupled with screenings of an arrayed library of Arabidopsis transcription factors. PLoS One. 2011;6:e21524. doi: 10.1371/journal.pone.0021524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wehner N, Hartmann L, Ehlert A, Böttner S, Oñate-Sánchez L, Dröge-Laser W. High-throughput protoplast transactivation (PTA) system for the analysis of Arabidopsis transcription factor function. Plant Journal. 2011;68:560–569. doi: 10.1111/j.1365-313X.2011.04704.x. [DOI] [PubMed] [Google Scholar]
- 45.Kim J, Somers DE. Rapid assessment of gene function in the circadian clock using artificial microRNA in Arabidopsis mesophyll protoplasts. Plant Physiology. 2010;154:611–621. doi: 10.1104/pp.110.162271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sugano S, Andronis C, Ong MS, Green RM, Tobin EM. The protein kinase CK2 is involved in regulation of circadian rhythms in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:12362–12366. doi: 10.1073/pnas.96.22.12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perales M, Portolés S, Más P. The proteasome-dependent degradation of CKB4 is regulated by the Arabidopsis biological clock. Plant Journal. 2006;46:849–860. doi: 10.1111/j.1365-313X.2006.02744.x. [DOI] [PubMed] [Google Scholar]
- 48.Arabidopsis Interactome Mapping Consortium. Evidence for network evolution in an Arabidopsis interactome map. Science. 2011;333:601–607. doi: 10.1126/science.1203877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brady SM, Zhang L, Megraw M, Martinez NJ, Jiang E, Yi CS, et al. A stele-enriched gene regulatory network in the Arabidopsis root. Molecular Systems Biology. 2011;7:459. doi: 10.1038/msb.2010.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gaudinier A, Zhang L, Reece-Hoyes JS, Taylor-Teeples M, Pu L, Liu Z, et al. Enhanced Y1H assays for Arabidopsis. Nature Methods. 2011;8:1053–1055. doi: 10.1038/nmeth.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hazen SP, Schultz TF, Pruneda-Paz JL, Borevitz JO, Ecker JR, Kay SA. LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:10387–10392. doi: 10.1073/pnas.0503029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Onai K, Ishiura M. PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes to Cells. 2005;10:963–972. doi: 10.1111/j.1365-2443.2005.00892.x. [DOI] [PubMed] [Google Scholar]
- 53.Carre I, Veflingstad S. Emerging design principles in the Arabidopsis circadian clock. Seminars in Cell and Developmental Biology. 2013 doi: 10.1016/j.semcdb.2013.03.011. current issue. [DOI] [PubMed] [Google Scholar]
- 54.Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, et al. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011;475:398–402. doi: 10.1038/nature10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dixon LE, Knox K, Kozma-Bognar L, Southern MM, Pokhilko A, Millar AJ. Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Current Biology. 2011;21:120–125. doi: 10.1016/j.cub.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chow BY, Helfer A, Nusinow DA, Kay SA. ELF3 recruitment to the PRR9 promoter requires other Evening Complex members in the Arabidopsis circadian clock. Plant Signaling and Behavior. 2012;7:170–173. doi: 10.4161/psb.18766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Herrero E, Kolmos E, Bujdoso N, Yuan Y, Wang M, Berns MC, et al. EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell. 2012;24:428–443. doi: 10.1105/tpc.111.093807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perales M, Más P. A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. Plant Cell. 2007;19:2111–2123. doi: 10.1105/tpc.107.050807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ni Z, Kim ED, Ha M, Lackey E, Liu J, Zhang Y, et al. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature. 2009;457:327–331. doi: 10.1038/nature07523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Portolés S, Más P. The functional interplay between protein kinase CK2 and CCA1 transcriptional activity is essential for clock temperature compensation in Arabidopsis. PLoS Genetics. 2010;6:e1001201. doi: 10.1371/journal.pgen.1001201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dong MA, Farré EM, Thomashow MF. Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7241–7246. doi: 10.1073/pnas.1103741108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu SX, Knowles SM, Webb CJ, Celaya RB, Cha C, Siu JP, et al. The Jumonji C domain-containing protein JMJ30 regulates period length in the Arabidopsis circadian clock. Plant Physiology. 2011;155:906–915. doi: 10.1104/pp.110.167015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lu SX, Webb CJ, Knowles SM, Kim SH, Wang Z, Tobin EM. CCA1 and ELF3 Interact in the control of hypocotyl length and flowering time in Arabidopsis. Plant Physiology. 2012;158:1079–1088. doi: 10.1104/pp.111.189670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lai AG, Doherty CJ, Mueller-Roeber B, Kay SA, Schippers JH, Dijkwel PP. CIRCADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress responses. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:17129–17134. doi: 10.1073/pnas.1209148109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lau OS, Huang X, Charron JB, Lee JH, Li G, Deng XW. Interaction of Arabidopsis DET1 with CCA1 and LHY in mediating transcriptional repression in the plant circadian clock. Molecular Cell. 2011;43:703–712. doi: 10.1016/j.molcel.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sawa M, Nusinow DA, Kay SA, Imaizumi T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science. 2007;318:261–265. doi: 10.1126/science.1146994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science. 2012;336:1045–1049. doi: 10.1126/science.1219644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Y, Wu JF, Nakamichi N, Sakakibara H, Nam HG, Wu SH. LIGHT-REGULATED WD1 and PSEUDO-RESPONSE REGULATOR9 form a positive feedback regulatory loop in the Arabidopsis circadian clock. Plant Cell. 2011;23:486–498. doi: 10.1105/tpc.110.081661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Legnaioli T, Cuevas J, Mas P. TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO Journal. 2009;28:3745–3757. doi: 10.1038/emboj.2009.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nakamichi N, Kiba T, Kamioka M, Suzuki T, Yamashino T, Higashiyama T, et al. Transcriptional repressor PRR5 directly regulates clock-output pathways. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:17123–17128. doi: 10.1073/pnas.1205156109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mockler TC, Michael TP, Priest HD, Shen R, Sullivan CM, Givan SA, et al. The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harbor Symposia on Quantitative Biology. 2007;72:353–363. doi: 10.1101/sqb.2007.72.006. [DOI] [PubMed] [Google Scholar]
- 72.Matsushika A, Makino S, Kojima M, Mizuno T. Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: insight into the plant circadian clock. Plant and Cell Physiology. 2000;41:1002–1012. doi: 10.1093/pcp/pcd043. [DOI] [PubMed] [Google Scholar]
- 73.Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, et al. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 74.Schaffer R, Landgraf J, Accerbi M, Simon V, Larson M, Wisman E. Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell. 2001;13:113–123. doi: 10.1105/tpc.13.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Edwards KD, Anderson PE, Hall A, Salathia NS, Locke JC, Lynn JR, et al. FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell. 2006;18:639–650. doi: 10.1105/tpc.105.038315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Covington MF, Harmer SL. The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biology. 2007;5:e222. doi: 10.1371/journal.pbio.0050222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Michael TP, Mockler TC, Breton G, McEntee C, Byer A, Trout JD, et al. Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genetics. 2008;4:e14. doi: 10.1371/journal.pgen.0040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biology. 2008;9:R130. doi: 10.1186/gb-2008-9-8-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hazen SP, Naef F, Quisel T, Gendron JM, Chen H, Ecker JR, et al. Exploring the transcriptional landscape of plant circadian rhythms using genome tiling arrays. Genome Biology. 2009;10:R17. doi: 10.1186/gb-2009-10-2-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Michael TP, Breton G, Hazen SP, Priest H, Mockler TC, Kay SA, et al. A morning-specific phytohormone gene expression program underlying rhythmic plant growth. PLoS Biology. 2008;6:e225. doi: 10.1371/journal.pbio.0060225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Harmer SL, Kay SA. Positive and negative factors confer phase-specific circadian regulation of transcription in Arabidopsis. Plant Cell. 2005;17:1926–1940. doi: 10.1105/tpc.105.033035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hudson ME, Quail PH. Identification of promoter motifs involved in the network of phytochrome A-regulated gene expression by combined analysis of genomic sequence and microarray data. Plant Physiology. 2003;133:1605–1616. doi: 10.1104/pp.103.030437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Filichkin SA, Breton G, Priest HD, Dharmawardhana P, Jaiswal P, Fox SE, et al. Global profiling of rice and poplar transcriptomes highlights key conserved circadian-controlled pathways and cis-regulatory modules. PLoS One. 2011;6:e16907. doi: 10.1371/journal.pone.0016907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.James AB, Monreal JA, Nimmo GA, Kelly CL, Herzyk P, Jenkins GI, et al. The circadian clock in Arabidopsis roots is a simplified slave version of the clock in shoots. Science. 2008;322:1832–1835. doi: 10.1126/science.1161403. [DOI] [PubMed] [Google Scholar]
- 85.Michael TP, Salome PA, McClung CR. Two Arabidopsis circadian oscillators can be distinguished by differential temperature sensitivity. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:6878–6883. doi: 10.1073/pnas.1131995100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yakir E, Hassidim M, Melamed-Book N, Hilman D, Kron I, Green RM. Cell autonomous and cell-type specific circadian rhythms in Arabidopsis. Plant Journal. 2011;68:520–531. doi: 10.1111/j.1365-313X.2011.04707.x. [DOI] [PubMed] [Google Scholar]
- 87.Wenden B, Toner DL, Hodge SK, Grima R, Millar AJ. Spontaneous spatiotemporal waves of gene expression from biological clocks in the leaf. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6757–6762. doi: 10.1073/pnas.1118814109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Espinoza C, Degenkolbe T, Caldana C, Zuther E, Leisse A, Willmitzer L, et al. Interaction with diurnal and circadian regulation results in dynamic metabolic and transcriptional changes during cold acclimation in Arabidopsis. PLoS One. 2010;5:e14101. doi: 10.1371/journal.pone.0014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Haydon MJ, Hearn TJ, Bell LJ, Hannah MA, Webb AAR. Metabolic regulation of circadian clocks. Seminars in Cell and Developmental Biology. 2013 doi: 10.1016/j.semcdb.2013.03.007. current issue. [DOI] [PubMed] [Google Scholar]
- 90.Seo PJ, Park MJ, Lim MH, Kim SG, Lee M, Baldwin IT, Park CM. A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell. 2012;24:2427–2442. doi: 10.1105/tpc.112.098723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, et al. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448:358–361. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]
- 92.Rawat R, Schwartz J, Jones MA, Sairanen I, Cheng Y, Andersson CR, et al. REVEILLE1, a Myb-like transcription factor, integrates the circadian clock and auxin pathways. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:16883–16888. doi: 10.1073/pnas.0813035106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nozue K, Harmer SL, Maloof JN. Genomic analysis of circadian clock-, light-, and growth-correlated genes reveals PHYTOCHROME-INTERACTING FACTOR5 as a modulator of auxin signaling in Arabidopsis. Plant Physiology. 2011;156:357–372. doi: 10.1104/pp.111.172684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gutiérrez RA, Stokes TL, Thum K, Xu X, Obertello M, Katari MS, et al. Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:4939–4944. doi: 10.1073/pnas.0800211105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang W, Barnaby JY, Tada Y, Li H, Tör M, Caldelari D, et al. Timing of plant immune responses by a central circadian regulator. Nature. 2011;470:110–114. doi: 10.1038/nature09766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bhardwaj V, Meier S, Petersen LN, Ingle RA, Roden LC. Defence responses of Arabidopsis thaliana to infection by Pseudomonas syringae are regulated by the circadian clock. PLoS One. 2011;6:e26968. doi: 10.1371/journal.pone.0026968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kerwin RE, Jimenez-Gomez JM, Fulop D, Harmer SL, Maloof JN, Klieben-stein DJ. Network quantitative trait loci mapping of circadian clock outputs identifies metabolic pathway-to-clock linkages in Arabidopsis. Plant Cell. 2011;23:471–485. doi: 10.1105/tpc.110.082065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Walley JW, Coughlan S, Hudson ME, Covington MF, Kaspi R, Banu G, et al. Mechanical stress induces biotic and abiotic stress responses via a novel ciselement. PLoS Genetics. 2007;3:1800–1812. doi: 10.1371/journal.pgen.0030172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Goodspeed D, Chehab EW, Min-Venditti A, Braam J, Covington MF. Arabidopsis synchronizes jasmonate-mediated defense with insect circadian behavior. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:4674–4677. doi: 10.1073/pnas.1116368109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Knowles SM, Lu SX, Tobin EM. Testing time: can ethanol-induced pulses of proposed oscillator components phase shift rhythms in Arabidopsis? Journal of Biological Rhythms. 2008;23:463–471. doi: 10.1177/0748730408326749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, et al. Genome-wide mapping of alternative splicing in Arabidopsis thaliana . Genome Research. 2010;20:45–58. doi: 10.1101/gr.093302.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Henriques R, Mas P. Chromatin remodeling and alternative splicing: pre- and post-transcriptional regulation of the Arabidopsis circadian clock. Seminars in Cell and Developmental Biology. 2013 doi: 10.1016/j.semcdb.2013.02.009. current issue. [DOI] [PubMed] [Google Scholar]
- 103.Lu SX, Knowles SM, Andronis C, Ong MS, Tobin EM. CIRCADIAN CLOCK ASSO-CIATED1 and LATE ELONGATED HYPOCOTYL function synergistically in the circadian clock of Arabidopsis. Plant Physiology. 2009;150:834–843. doi: 10.1104/pp.108.133272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yakir E, Hilman D, Kron I, Hassidim M, Melamed-Book N, Green RM. Post-translational regulation of CIRCADIAN CLOCK ASSOCIATED1 in the circadian oscillator of Arabidopsis. Plant Physiology. 2009;150:844–857. doi: 10.1104/pp.109.137414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sanchez SE, Petrillo E, Beckwith EJ, Zhang X, Rugnone ML, Hernando CE, et al. A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature. 2010;468:112–116. doi: 10.1038/nature09470. [DOI] [PubMed] [Google Scholar]
- 106.James AB, Syed NH, Bordage S, Marshall J, Nimmo GA, Jenkins GI, et al. Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant Cell. 2012;24:961–981. doi: 10.1105/tpc.111.093948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Filichkin SA, Mockler TC. Unproductive alternative splicing and nonsense mRNAs: a widespread phenomenon among plant circadian clock genes. Biology Direct. 2012;7:20. doi: 10.1186/1745-6150-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hong S, Song HR, Lutz K, Kerstetter RA, Michael TP, McClung CR. Type II protein arginine methyltransferase 5 (PRMT5) is required for circadian period determination in Arabidopsis thaliana . Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21211–21216. doi: 10.1073/pnas.1011987107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang X, Wu F, Xie Q, Wang H, Wang Y, Yue Y, et al. SKIP is a component of the spliceosome linking alternative splicing and the circadian clock in Arabidopsis. Plant Cell. 2012;24:3278–3295. doi: 10.1105/tpc.112.100081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jones MA, Williams BA, McNicol J, Simpson CG, Brown JW, Harmer SL. Mutation of Arabidopsis SPLICEOSOMAL TIMEKEEPER LOCUS1 causes circadian clock defects. Plant Cell. 2012;24:4066–4082. doi: 10.1105/tpc.112.104828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Reddy AB, Karp NA, Maywood ES, Sage EA, Deery M, O’Neill JS, et al. Circadian orchestration of the hepatic proteome. Current Biology. 2006;16:1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 112.Deery MJ, Maywood ES, Chesham JE, Sládek M, Karp NA, Green EW, et al. Proteomic analysis reveals th0e role of synaptic vesicle cycling in sustaining the suprachiasmatic circadian clock. Current Biology. 2009;19:2031–2036. doi: 10.1016/j.cub.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 113.Feng D, Lazar MA. Clocks, metabolism, and the epigenome. Molecular Cell. 2012;47:158–167. doi: 10.1016/j.molcel.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Masri S, Zocchi L, Katada S, Mora E, Sassone-Corsi P. The circadian clock transcriptional complex: metabolic feedback intersects with epigenetic control. Annals of the New York Academy of Sciences. 2012;1264:103–109. doi: 10.1111/j.1749-6632.2012.06649.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hwang H, Cho MH, Hahn BS, Lim H, Kwon YK, Hahn TR, et al. Proteomic identification of rhythmic proteins in rice seedlings. Biochimica et Biophysica Acta. 2011;1814:470–479. doi: 10.1016/j.bbapap.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 116.Green RM, Tobin EM. Loss of the circadian clock-associated protein 1 in Arabidopsis results in altered clock-regulated gene expression. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:4176–4179. doi: 10.1073/pnas.96.7.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gould PD, Locke JC, Larue C, Southern MM, Davis SJ, Hanano S, et al. The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell. 2006;18:1177–1187. doi: 10.1105/tpc.105.039990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tóth R, Gerding-Reimers C, Deeks MJ, Menninger S, Gallegos RM, Tonaco IA, et al. Prieurianin/endosidin 1 is an actin-stabilizing small molecule identified from a chemical genetic screen for circadian clock effectors in Arabidopsis thaliana . Plant Journal. 2012;71:338–352. doi: 10.1111/j.1365-313X.2012.04991.x. [DOI] [PubMed] [Google Scholar]
- 119.Más P, Kim WY, Somers DE, Kay SA. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana . Nature. 2003;426:567–570. doi: 10.1038/nature02163. [DOI] [PubMed] [Google Scholar]
- 120.Kim WY, Fujiwara S, Suh SS, Kim J, Kim Y, Han L, et al. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature. 2007;449:356–360. doi: 10.1038/nature06132. [DOI] [PubMed] [Google Scholar]
- 121.Fujiwara S, Wang L, Han L, Suh SS, Salomé PA, McClung CR, et al. Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. Journal of Biological Chemistry. 2008;283:23073–23083. doi: 10.1074/jbc.M803471200. [DOI] [PubMed] [Google Scholar]
- 122.Yu JW, Rubio V, Lee NY, Bai S, Lee SY, Kim SS, et al. COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Molecular Cell. 2008;32:617–630. doi: 10.1016/j.molcel.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang L, Fujiwara S, Somers DE. PRR5 regulates phosphorylation, nuclear import and subnuclear localization of TOC1 in the Arabidopsis circadian clock. EMBO Journal. 2010;29:1903–1915. doi: 10.1038/emboj.2010.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kim TS, Kim WY, Fujiwara S, Kim J, Cha JY, Park JH, et al. HSP90 functions in the circadian clock through stabilization of the client F-box protein ZEITLUPE. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:16843–16848. doi: 10.1073/pnas.1110406108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, et al. The Arabidopsis information resource (TAIR): improved gene annotation and new tools. Nucleic Acids Research. 2012;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Farré EM, Weise SE. The interactions between the circadian clock and primary metabolism. Current Opinion in Plant Biology. 2012;15:293–300. doi: 10.1016/j.pbi.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 127.Fukushima A, Kusano M, Nakamichi N, Kobayashi M, Hayashi N, Sakakibara H, et al. Impact of clock-associated Arabidopsis pseudo-response regulators in metabolic coordination. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:7251–7256. doi: 10.1073/pnas.0900952106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nakamichi N, Kusano M, Fukushima A, Kita M, Ito S, Yamashino T, et al. Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant and Cell Physiology. 2009 Mar;50(3):447–462. doi: 10.1093/pcp/pcp004. March. [DOI] [PubMed] [Google Scholar]
- 129.Izawa T, Mihara M, Suzuki Y, Gupta M, Itoh H, Nagano AJ, et al. Os-GIGANTEA confers robust diurnal rhythms on the global transcriptome of rice in the field. Plant Cell. 2011;23:1741–1755. doi: 10.1105/tpc.111.083238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Song HR, Noh YS. Rhythmic oscillation of histone acetylation and methylation at the Arabidopsis central clock loci. Molecules and Cells. 2012;34:279–287. doi: 10.1007/s10059-012-0103-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bourbousse C, Ahmed I, Roudier F, Zabulon G, Blondet E, Balzergue S, et al. Histone H2B monoubiquitination facilitates the rapid modulation of gene expression during Arabidopsis photomorphogenesis. PLoS Genetics. 2012;8:e1002825. doi: 10.1371/journal.pgen.1002825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Himanen K, Woloszynska M, Boccardi TM, De Groeve S, Nelissen H, Bruno L, et al. Histone H2B monoubiquitination is required to reach maximal transcript levels of circadian clock genes in Arabidopsis. Plant Journal. 2012;72:249–260. doi: 10.1111/j.1365-313X.2012.05071.x. [DOI] [PubMed] [Google Scholar]
- 133.Menet JS, Rodriguez J, Abruzzi KC, Rosbash M. Nascent-seq reveals novel features of mouse circadian transcriptional regulation. eLife. 2012;1:e00011. doi: 10.7554/eLife.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Doherty CJ, Kay SA. Circadian surprise – it’s not all about transcription. Science. 2012;338:338–340. doi: 10.1126/science.1230008. [DOI] [PubMed] [Google Scholar]
- 135.Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338:349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yakir E, Hilman D, Hassidim M, Green RM. CIRCADIAN CLOCK ASSOCIATED1 transcript stability and the entrainment of the circadian clock in Arabidopsis. Plant Physiology. 2007;145:925–932. doi: 10.1104/pp.107.103812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Staiger D, Green R. RNA-based regulation in the plant circadian clock. Trends in Plant Science. 2011;16:517–523. doi: 10.1016/j.tplants.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 138.Kim JY, Song HR, Taylor BL, Carré IA. Light-regulated translation mediates gated induction of the Arabidopsis clock protein LHY. EMBO Journal. 2003;22:935–944. doi: 10.1093/emboj/cdg075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Más P. Circadian clock function in Arabidopsis thaliana: time beyond transcription. Trends in Cell Biology. 2008;18:273–281. doi: 10.1016/j.tcb.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 140.Kusakina J, Dodd AN. Phosphorylation in the plant circadian system. Trends in Plant Science. 2012;17:575–583. doi: 10.1016/j.tplants.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 141.Ito S, Song YH, Imaizumi T. LOV domain-containing F-box proteins: light-dependent protein degradation modules in Arabidopsis. Molecular Plant. 2012;5:573–582. doi: 10.1093/mp/sss013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Maier B, Wendt S, Vanselow JT, Wallach T, Reischl S, Oehmke S, et al. A large-scale functional RNAi screen reveals a role for CK2 in the mammalian circadian clock. Genes and Development. 2009;23:708–718. doi: 10.1101/gad.512209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Isojima Y, Nakajima M, Ukai H, Fujishima H, Yamada RG, Masumoto KH, et al. CKIε/δ-dependent phosphorylation is a temperature-insensitive, period-determining process in the mammalian circadian clock. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:15744–15749. doi: 10.1073/pnas.0908733106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhang EE, Liu AC, Hirota T, Miraglia LJ, Welch G, Pongsawakul PY, et al. A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell. 2009;139:199–210. doi: 10.1016/j.cell.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hirota T, Lee JW, Lewis WG, Zhang EE, Breton G, Liu X, et al. High-throughput chemical screen identifies a novel potent modulator of cellular circadian rhythms and reveals CKIα as a clock regulatory kinase. PLoS Biology. 2010;8:e1000559. doi: 10.1371/journal.pbio.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chen Z, Yoo SH, Park YS, Kim KH, Wei S, Buhr E, et al. Identification of diverse modulators of central and peripheral circadian clocks by high-throughput chemical screening. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:101–106. doi: 10.1073/pnas.1118034108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, et al. A quantitative analysis of kinase inhibitor selectivity. Nature Biotechnology. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]