Abstract
Aims: Cervical Cancer (CC) is one of the most important health problems in women. It frequently presents genetic changes at chromosome region 3q21. This region contains the Cellular Retinol Binding Protein 1 gene (CRBP1) which has been implicated as an important element in the development of other types of cancer. The main goal of the present work was to determine the molecular alterations of CRBP1 and its relationship to CC. Methods: To determine the molecular alterations of CRBP1 gene in CC; twenty-six CC and twenty-six healthy cervix samples were evaluated for: 1) Copy number gain by real-time PCR analysis, 2) expression levels by an immunohistochemistry assay on tissue microarray, and 3) the methylation status of the CRBP1 promoter region. Results: The increase in CRBP1 copy number was observed in 10 out of the 26 CC samples analyzed, while healthy cervices samples showed no changes in the copy number. In addition, there was a lack of expression of the CRBP1 gene in an important number of the CC samples (17/26), and the CRBP1 gene promoter was methylated in 15/26 of the CC samples. Interestingly, there was a significant association between the lack of expression of the CRBP1 gene and its methylation status. Conclusions: The data indicates that, both activating and inactivating changes in the CRBP1 gene could be significant events in the development and progression of CC, and the lack of expression of the CRBP1 protein could be related with to the development of CC. We believe that there is enough evidence to consider to CRBP1 gene as a tumor suppressor gene for CC.
Keywords: Cervical cancer, CRBP1, expression, copy number, suppressor gene
Introduction
Cervical cancer (CC) is an important public health problem worldwide, including Mexico [1]. It is widely known that persistent Human Papillomavirus (HPV) infection is the main risk factor for CC development [2]; however, HPV infection is not sufficient for malignant transformation involving molecular alterations in cellular genes [3,4]. Several studies have investigated genomic alterations in CC, involving the loss of tumor suppressor genes (TSG) and the gain of oncogenes [4]. Specifically, TSG are related to protecting the genome from mutagenic events, impeding deregulated progression through the cell cycle, inducing apoptosis in altered cells that escape normal cell cycle control, etc. [5]. But different alterations such as deletions, mutations and hypermethylation can lead to suppression of these genes [6,7].
Molecular cytogenetic studies demonstrated frequent aberrations in CC [8-14], showing that the gain of DNA in the chromosome 3q region comprises one of the most common genetic alterations. We previously showed that the most common gain in CC corresponds to the cytogenetic region 3q21-q22, where Cellular Retinol Binding Protein 1 gene (CRBP1) is located [15], which codifies a protein involved in retinoid metabolism [16]. Retinoid compounds are analogues of vitamin A, and play an important role in development and homeostasis through their regulatory effects on cell differentiation, proliferation, and apoptosis [17,18]. Thus, CRBP1 is essential for vitamin A homeostasis; maintaining normal liver retinol storage and aiding in esterification [19]. Retinol is much more active when complexed with CRBP1 [19]. CRBP1 downregulation has been associated with the malignant phenotype in breast, and ovarian cancer in 30% of cases, while for nasopharyngeal cases 80% was observed [20-24]. However, the possible role of altered CRBP1 in human carcinogenesis has not yet been established. Thus, we decided to investigate the possible role of alterations such as DNA copy number changes, expression, and methylation in the promoter region of CC samples.
Methods
Biological samples
Twenty-six CC samples were collected from patients who attended the Colposcopy Service at Hospital General of Mexico, S.S., Mexico City. The local Ethics Committees of Hospital General de Mexico, Ministry of Health (SSa) and the Mexican Institute of Social Security (IMSS) approved the described procedures, and all samples were taken after informed consent from the patients.
The biopsies were divided into three sections: the central part was used for genomic DNA extraction using the Wizard Genomic kit (Promega, Madison, Wi, USA), and both extremes were fixed with 70% ethanol overnight and paraffin embedded. Hematoxylin and Eosin (H & E)-stained sections were analyzed to confirm the presence of at least 80% tumor cells in each sample. All CC samples were classified as squamous cervical carcinoma. Normal cervix samples (n = 26) were collected from patients who attended the colposcopy clinic for routine gynecological inspection. In this case, twenty-six women consented to participate as control subjects in the work. All CC samples were HPV positive and normal cervix samples were HPV negative (data not shown).
Cervical cancer cell lines
HPV18 positive CC cell lines: HeLa, RoVa. HPV16 positive CC cell lines: SiHa. Cell lines were maintained in minimal essential medium containing Earle’s salts and L-glutamic acid (Cellgro; Mediatech, Herdon, VA, USA), and supplemented with non-essential amino acids, sodium pyruvate, and 10% fetal bovine serum. All the cell lines were grown until 70% confluence. RoVa cell line has been previously reported [25].
DNA copy number by quantitative Real-time PCR
In order to determine the CRBP1 gene copy number, normal cervices, CC cell lines, and CC clinical samples were analyzed using relative quantitation real-time PCR. The reactions were designed using TaqMan® Genotyping Master Mix, No. 4371355 (Applied Biosystem, USA). The PCR amplification was performed in an ABI PRIMS 7500 from Applied Biosystem (Applied Biosystem) with 100 ng of DNA. Amplification conditions were as follow: 95°C 10 min, following 40 cycles of 15 sec at 95°C, 1 min at 60°C. CRBP1 Hs01437985_cn probe, and Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) Hs00894322_cn probe were used as reference; the relative genomic copy number was calculated using the comparative Ct methods [26].
Immunodetection of CRBP1
A tissue microarray (TMA) was constructed as follow including the 26 CC cases. Core samples were taken using 0.6 mm2 blunt-tip needles and placed on the recipient microarray block using a Tissue Microarrayer (Chemicon Co., MA, USA). Sections (4 μm) were cut and placed on coated slides.
The immunostaining was performed using a streptavidin-biotin complex peroxidase method (Dako, Glostrup, Denmark). TMA slides were deparaffinized with xylene followed by ethanol and rehydrated in water. Briefly, after dewaxing the tissue section, endogenous peroxide activity was inhibited with freshly prepared 0.5% H2O2 in distilled water for 20 minutes. Next, the sections were processed in a 600-W microwave oven, at maximum power, three times for 5 minutes in citrate buffer (pH 6.0). Incubation with the monoclonal mouse anti-CRBP1 antibody (ab24090 Abcam) was performed overnight at 4°C, at 1:100 dilution in 1% bovine serum albumin in phosphate buffered saline (PBS). All incubations were performed in a humidified chamber. Sections were developed with a peroxidase substrate solution (0.05% 3,3-diaminobenzidine tetrahydrochloride, 0.01% H2O2 in PBS), counterstained with hematoxylin, dehydrated, and mounted. Appropriate positive control was used for the reaction (human liver tissue) and human heart tissue as negative control. The assessment of CRBP1 expression was performed by light microscope at 40X original magnification. The immunostaining was evaluated for positive or negative staining.
HeLa, RoVa and SiHa cells were cultured onto sterile glass coverslips in twelve-well plates at an approximate density of 0.5 x 105 cells/well in DMEM (Gibco) containing 10% FBS (Gibco), 100 U/ml penicillin, and 100 mg/ml streptomycin (Gibco). The cells were incubated at 37°C with 5% CO2 (Gibco) overnight, fixed with 3% paraformaldehyde, made permeable with 0.2% Triton X-100, and incubated with the monoclonal mouse anti-CRBP1 antibody overnight at 4°C, at 1:50 dilution. After three washes with 1X PBS, cells were incubated with anti-mouse Alexa Fluor 488 secondary antibody (Molecular Probes) at a dilution of 1:500. Cells were washed in 1X PBS and mounted on slides using Vectashield (Vector Laboratories, Inc. Burlin-game). Confocal images were taken with a Leica TCS SP5 confocal microscope.
Bisulfite treatment and CRBP1 promoter methylation status
DNA methylation patterns in the CpG island of CRBP1 were determined by methylation-specific PCR (MSP) as demonstrated in previous reports [27]. Bisulfite treatment of DNA converted un-methylated cytosines to uracil, but the methylated bases remaining as cytosines. Two μl of bisulfite-modified DNA was added to produce a final volume of 25 μl PCR mix containing 1X PCR buffer (16.6 mM ammonium sulphate, 67 mM Tris pH 8.8, 6.7 mM MgCl2, and 10 mM 2-mercaptoethanol), dNTPs (each at 1.25 mM), 1 U Hot Start Taq DNA polymerase (Qiagen, Hilden, Germany), and primers (25 nM each reaction). The CRBP1 primer sequences used were previously described by Esteller et al. [28] MSP was carried out using the following conditions: one cycle at 95°C for 1 min, followed by 35 cycles of 1 min at 95°C, 1 min at 62°C, and 1 min at 72°C. CRBP1 DNA promoters of the MCF-7 cell line (un-methylated) and HeLa cells (methylated) were used as controls. The PCR products were directly loaded on to a 2% agarose gel, stained with ethidium bromide, and visualized under a UV transilluminator.
Statistical analysis
All comparisons for significance were performed by means of the X2 exact test. All p values represent two-tailed tests and were considered significant at 0.001. The statistical analysis was performed using the SPSS v15 statistical software.
Results
For CRBP1 copy number analysis, purified DNA from CC and healthy tissues was subjected to real-time PCR assays. As expected, no variation in CRBP1 gene copy number was observed in healthy cervix samples (data not shown). In contrast, 38% of CC samples showed a gain in copy number (2-20 copies) (Figure 1). In the case of CC cell lines, only SiHa showed a high copy number of the CRBP1 gene, the rest presented a similar pattern to normal cervix samples (data not shown).
Figure 1.
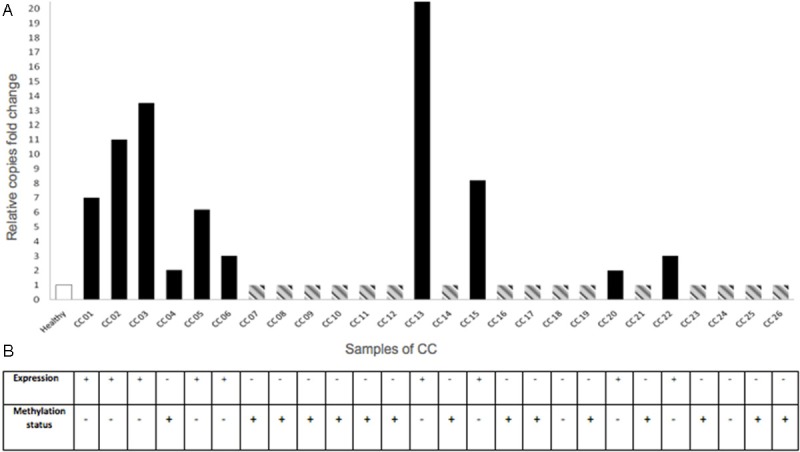
Molecular events for CRBP1 gene in cervical epithelium samples. A: In order to know the gain of copy number of the CRBP1 gene, DNA of healthy cervix and CC samples, were subjected to real time PCR with specific Taqman probes. White bar (healthy cervix samples) represents the mean of the normal cervices (n = 26) without extra copies of CRBP1 gene. Black bars show CC samples with gain of copy number (2-20X); while gray dotted line bars are showing CC samples that do not change in the copies number. Values above the cut-off line (as 1), being assigned as increased gene copy number compared with normal cervical epithelium. CRBP1 Hs01437985_cn probe, and Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) Hs00894322_cn probe were used as reference; the relative genomic copy number was calculated using the comparative Ct methods [26]. In X-axis represents cervical samples, Y-axis relative copies fold change of CRBP1 gene. B: CRBP1 expression was observed as positive immunostaining result on tissue microarray as mentioned in Methods section The DNAs used for gain of copy number (panel A) were also used for the methylation assay. Methylation result represents the methylation of the CRBP1 promoter. In this case, each healthy or CC sample, correspond to each column for CRBP1 expression and methylation status. Interestingly, in most of the cases, there was an association between the lack of expression of the CRBP1 gene and its methylation status.
In order to establish a possible relationship between gene amplification and CRBP1 gene expression, cervical cancer tissues were subjected to an immunohistochemistry assay on tissue microarray. Figure 2A depicts representative normal cervical epithelium and CC samples for CRBP1 immunostaining. Normal cervical tissues showed positive immunostaining in the cytoplasm of cells from the cellular basal layer. Interestingly for CC samples, the lack of CRBP1 expression was observed in most of the cases (≈ 66%), while the remaining samples showed positive immunostaining in the transformed cells with variations in signal intensity. A significant association was obtained between CRBP1 expression and copy number gain (p = 0.001; see Table 1). Furthermore, a significant association was also observed between early sexual activity early in life and degrees of differentiation (Table 2).
Figure 2.
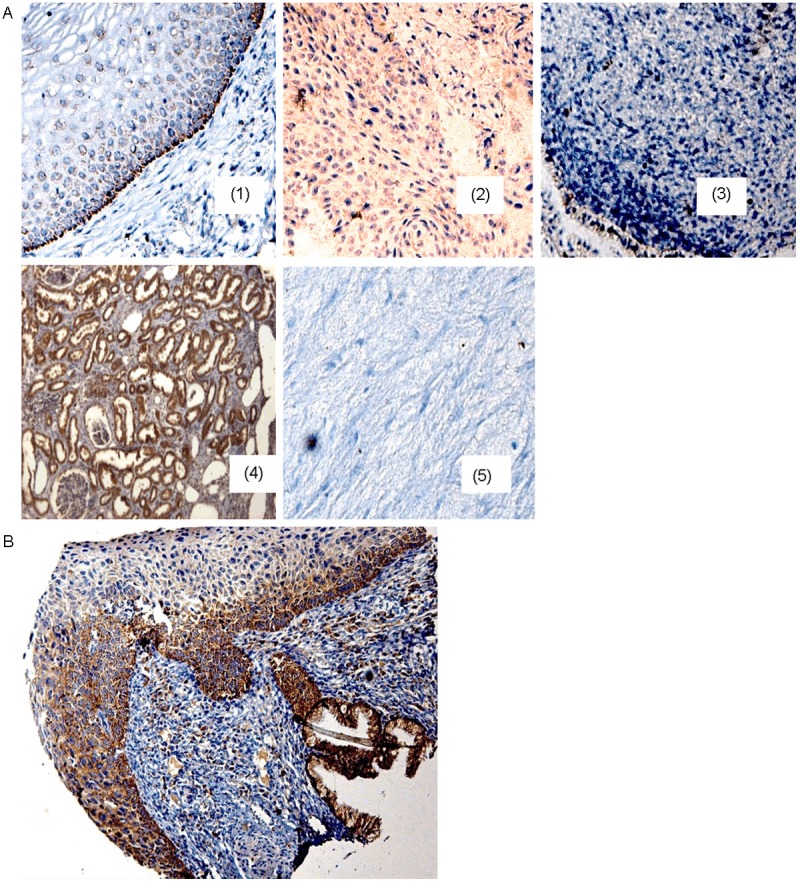
CRBP1 immunodetection in the uterine cervix samples. A: (1) Cytoplasmic CRBP1 expression is present in cells of the basal layer of normal cervical epithelium (healthy tissue); (2) the immunodetection in the transformed cells of a cervical cancer (CC03) tissue harboring gain of CRBP1 gene. (3) CC samples without gain CRBP1 gene showing negative immunostaining (CC16 sample). A kidney tissue section (4) was used as positive control, while a heart tissue section for negative control (5). B: cervical progression spectrum. The tissue section shows a brownish reaction (positive reaction) in the basal cell layer of the “normal” region, in the high-grade lesion, and also in the invasive region. All tissue sections were hematoxylin counterstained, 200X original amplification.
Table 1.
Association between CRBP1 gene gain copy number and its expression in cervical cancer samples
| CRBP1 gene copy number | CRBP1 immunodetection | P value* | |
|---|---|---|---|
|
| |||
| (+) | (-) | ||
| 2 | 0 | 16 | 0.001 |
| >2 | 9 | 1 | 0.001 |
X2-test.
Table 2.
Correlation between CRBP1 expression and clinic pathological variables in cervical cancer
| Clinical variable | P value |
|---|---|
| Age (< 50) | 0.211 |
| Clinical state (II/II, III/IV) | 0.768 |
| Histological differentiation (moderate) | 0.034* |
| HPV (+) | 0.192 |
| Pregnancies (> 2) | 0.086 |
| Onset of sexual activity (< 18 years) | 0.034* |
| Family history of cancer (Yes) | 0.899 |
| Age at menarche (< 12) | 0.946 |
Represents statistical significance.
To establish whether CRBP1 expression could be related as an early event in cervical carcinogenesis, a tissue section harboring the cervical lesions spectrum was subjected to immunohistochemistry assay. The result showed that CRBP1 expression was present since the precursor lesion (Figure 2B), indicating that the upregulation of CRBP1 expression plays a role in cancer progression.
In the case of CC cell lines, all (HeLa, SiHa and RoVa) were positive for CRBP1 expression (Figure 3).
Figure 3.
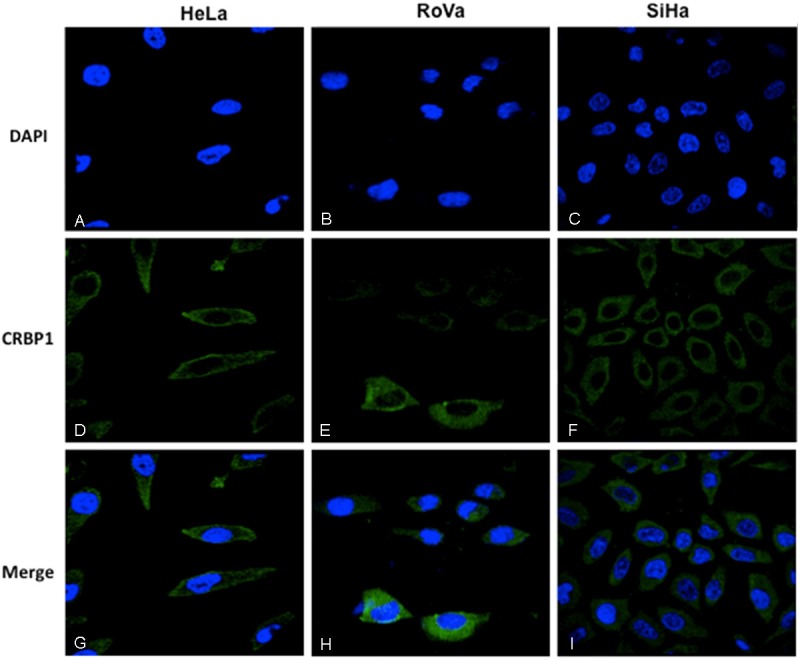
Immunolocalization of CRBP1 by immunofluorescence in cervical cells. Nuclei were Dapi stained in blue color (A-C). The immunodetection of CRBP1 was observed in green color (D-F). Cytoplasmic immunodetection of CRBP1 in the merge imaging (G-I). 100X original amplification.
Tissue specimens corresponding to those analyzed by immunohistochemistry and gene copy number were then used for methylation by specific PCR analysis. It was noted that CRBP1 promoter CpG islands were found methylated in 15/26 CC samples, while 11/26 malignant samples were observed to be un-methylated (Figure 4). So, a direct relationship was observed between the lacks of expression of CRBP1 and specimen methylation status (Figure 1).
Figure 4.
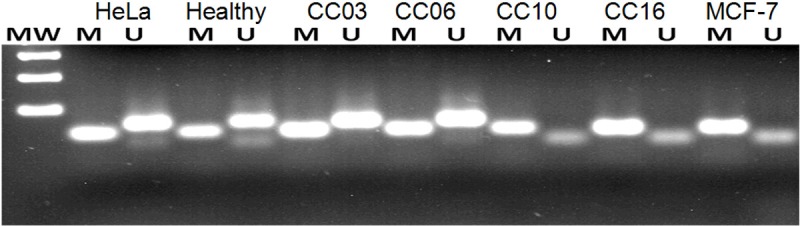
Methylation promoter of CRBP1 gene in cervical cancer samples. Example of CRBP1 gene promoter methylation analysis. Lanes: Healthy cervix sample, CC03 and CC06 samples with un-methylated status; lanes CC 10 and CC 16 with methylated status; HeLa cells as un-methylated control (109 bp), or MCF-7 cells as methylated control (99 bp). MW: molecular weight marker of 100 bp.
Discussion
To date, CC is the second most common cancer in females of the Mexican population. Despite the wide acceptance of a screening program for CC, in Mexico more than 12,000 women are diagnosed with CC annually, and unfortunately, this is accompanied by a 50% mortality rate. Thus, there are clearly deficiencies in the current system of screening. Due to high number of false negatives or positives and also the lack of molecular markers, there is a need for supplementary cervical screening techniques [29].
One interesting issue on CC comprises the genetic changes that could correlate with the disease. Several reports on this topic have been described, indicating that the gain of the chromosome 3q could be considered as one of the most prevalent chromosomal alterations in this tumor type [8].
It is noteworthy that the chromosomal imbalances profile between CC and larynx cancer is quite similar (Peralta R., manuscript in preparation), with the 3q arm being the most common DNA gain. We have hypothesized that the CRBP1 gene could also present common alterations in CC. Very recently, we have published about the multiple molecular alterations involved in the CRBP1 gene in larynx cancer [23].
It was previously reported, employing DNA microarray technology on CC cells that the CRBP1 gene presented a gain in copy number [17]. In the present work, we demonstrate and confirm that the majority of patients do not present changes in copy number of CRBP1 gene and only one third of patients with CC presented amplification in copy number of CRBP1 gene. In that scenario, the data suggests that of a subgroup of epithelial tumors of the cervix harboring the amplification of 3q21 region, the CRBP1 gene could represent an important and common clue for the carcinogenesis process.
CRBP1 gene alterations in cancer research are being explored. It is known that CRBP1 protein is expressed in several human tissues. In contrast, a lack of expression is present in human cancer types such as ovary, prostate, larynx and breast cancer [21,23,30,31]. Our present data shows that CRBP1 protein is only expressed in the basal cell layer of normal cervical tissue, supporting previous results found in stratified epithelial tissues [23]. With regard to CRBP1 expression in CC, we show two tumor types; one CRBP1 positive and CRBP1 negative. Interestingly, a predominant lack of expression of CRBP1 is showed in CC, supporting previous reports [22]. This finding could represent an important molecular event in carcinogenesis. Then, we decided to investigate whether CRBP1 gene amplification could be related to protein expression in CC. We observed a relationship between gene amplification and protein expression. This result supports previous results [23], and could suggest that the gene amplification could be related to its expression.
In order to determine the probable mechanism of CRBP1 expression, the cervical samples were then subjected to methylation assay. Most of the samples showed methylated sequences. Interestingly the analysis showed a correlation between methylation status and lack of expression. This data could support the idea that CRBP1 gene of lack expression is consistently related with methylation of its promoter, independently of the gene amplification event. Similar results could be extended for prostate, breast, and ovarian human cancer types [20,21,27,31].
According to the result for immunodetection of CRBP1 in cervical cancer progression in which the HG-SIL is positive, this could suggest that un-methylated gene promoter precedes (the transcription and translation events) invasive lesion. In this case, we hypothesize that those HG-SIL CRBP1 positive, correspond to samples that will progress to CRBP1 positive invasive lesions as an early step (a landmark) in cervical progression. In contrast, in CRBP1 invasive samples an aberrant methylation event also occurs in the invasive tumors, inactivating the CRBP1 gene. To address the clinical implications of these molecular alterations for CRBP1 gene in CC, we have already initiated the proper clinical and follow-up protocols.
It is classically known that TSGs have been described to acquire loss of function by mutations or deletions leading to their inability to impede malignant transformation [5]. Alternatively, the methylation epigenetic event represents a distinct mechanism of tumor suppressor gene inactivation. Aberrant gene promoter methylation is associated with gene silencing and is functionally equivalent to a deleted gene [5]. Based on this scenario where the CRBP1 lack expression could be due to aberrant gene promoter methylation, we suggest the role of CRBP1 as potential TSG in CC and probably for some other human tumors.
On the other hand, it is widely known that persistence of HPV infection in women who to develop dysplasia or carcinoma [32], and the long latency of the transition from HG-SIL to carcinoma strongly suggest that cellular factors in addition to HPV infection are required for the malignant transformation of epithelial cells [33]. The present work clearly demonstrates that the alterations in CRBP1 are not influenced by HPV.
Conclusions
The current data demonstrates several molecular alterations of the CRBP1 gene in cervical tissues. CRBP1 un-methylated status and its expression are essential molecular mechanisms involved in the basal cell layer of healthy cervical epithelium, while CRBP1 methylated status and its lack of expression could be one of the most prevalent event in CC; this fact suggests that CRBP1 gene could act as a tumor suppressor gene associated with cervical cancer.
Acknowledgements
This work was partially supported by grants 69719 and 87244 from Fondos Sectoriales CONACYT-Mexico. During this work MMR, HA, VV, PR, BA, LP were recipients of scholarships from CONACYT Mexico. This work was submitted in partial fulfillment of the requirements for the D.Sc. degree for MM. at Doctorado en Biomedicina Molecular, Centro de Investigación y de Estudios Avanzados, IPN, Mexico, and for and D.Sc. degree HA. at Doctorado en Ciencias Biomédicas, Universidad Nacional Autónoma de Mexico. We want to mention the special critical review of the manuscript to Miss Grace O´Malley from M.I.T. at Cambridge, MA, during her Summer Training Course. FIS-IMSS G12/1153. To Dr Alberto Monroy (IMMS) to provide te ROVA cells.
Disclosure of conflict of interest
All other authors declared no conflict of interest.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Wani K, Huilgol N, Hongyo T, Shah K, Chatterjee N, Nair CK, Nomura T. Genetic alterations in the coding region of the bak gene in uterine cervical carcinoma. Br J Cancer. 2003;88:1584–1586. doi: 10.1038/sj.bjc.6600944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitra A, Murty V, Li R, Pratap M, Luthra UK, Chaganti RS. Allelotype analysis of cervical carcinoma. Cancer Res. 1994;54:4481–4487. [PubMed] [Google Scholar]
- 4.Kloth J, Oosting J, Wezel T, Szuhai K, Knijnenburg J, Gorter A, Kenter GG, Fleuren GJ, Jordanova ES. Combined array-comparative genomic hybridization and single-nucleotide polymorphism-loss of heterozygosity analysis reveals complex genetic alterations in cervical cancer. BMC Genomics. 2007;8:1–13. doi: 10.1186/1471-2164-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayslip J, Montero A. Tumor suppressor gene methylation in follicular lymphoma: a comprehensive review. Mol Cancer. 2006;5:44. doi: 10.1186/1476-4598-5-44. 1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonazzi VF, Irwin D, Hayward NK. Identification of candidate tumor suppressor genes inactivated by promoter methylation in melanoma. Genes Chromosomes Cancer. 2009;48:10–21. doi: 10.1002/gcc.20615. [DOI] [PubMed] [Google Scholar]
- 7.Rothhammer T, Bosserhoff AK. Epigenetic events in malignant melanoma. Pigment Cell Res. 2007;20:92–111. doi: 10.1111/j.1600-0749.2007.00367.x. [DOI] [PubMed] [Google Scholar]
- 8.Heselmeyer K, Schrock E, du Manoir S, Blegen H, Shah K, Steinbeck R, Auer G, Ried T. Gain of chromosome 3q defines the transition from severe dysplasia to invasive carcinoma of the uterine cervix. Proc Natl Acad Sci U S A. 1996;93:479–484. doi: 10.1073/pnas.93.1.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ried T, Heselmeyer-Haddad K, Blegen H, Schröck E, Auer G. Genomic changes defining genesis, progression and malignancy potential in human solid tumors: a phenotype/genotype correlation. Genes Chromosomes Cancer. 1999;25:195–204. doi: 10.1002/(sici)1098-2264(199907)25:3<195::aid-gcc1>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 10.Dellas A, Torhorst J, Jiang F, Proffitt J, Schultheiss E, Holzgreve W, Sauter G, Mihatsch MJ, Moch H. Prognostic value of genomic alterations in invasive cervical squamous cell carcinoma of clinical Stage IB detected by comparative genomic hybridization. Cancer Res. 1999;59:3475–3479. [PubMed] [Google Scholar]
- 11.Kirchhoff M, Rose H, Petersen B, Maahr J, Gerdes T, Lundsteen C, Bryndorf T, Kryger-Baggesen N, Christensen L, Engelholm SA, Philip J. Comparative genomic hybridization reveals a recurrent pattern of chromosomal aberrations in severe dysplasia/carcinoma in situ of the cervix and in advanced-stage cervical carcinoma. Genes Chromosomes Cancer. 1999;24:144–150. doi: 10.1002/(sici)1098-2264(199902)24:2<144::aid-gcc7>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 12.Allen D, White D, Hutchins A, Scurry JP, Tabrizi SN, Garland SM, Armes JE. Progressive genetic aberrations detected by comparative genomic hybridization in squamous cell cervical cancer. Br J Cancer. 2000;83:1659–1663. doi: 10.1054/bjoc.2000.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oh EK, Kim YW, Kim IW, Liu HB, Lee KH, Chun HJ, Park DC, Oh EJ, Lee AW, Bae SM, Ahn WS. Differential DNA copy number aberrations in the progression of cervical lesions to invasive cervical carcinoma. Int J Oncol. 2012;41:2038–46. doi: 10.3892/ijo.2012.1644. [DOI] [PubMed] [Google Scholar]
- 14.Rao PH, Arias-Pulido H, Lu XY, Harris CP, Vargas H, Zhang FF, Narayan G, Schneider A, Terry MB, Murty VV. Chromosomal amplifications, 3q gain and deletions of 2q33-q37 are the frequent genetic changes in cervical carcinoma. BMC Cancer. 2004;4:5. doi: 10.1186/1471-2407-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hidalgo A, Baudis M, Petersen I, Arreola H, Piña P, Vázquez-Ortiz G, Hernández D, González J, Lazos M, López R, Pérez C, García J, Vázquez K, Alatorre B, Salcedo M. Microarray comparative genomic hybridization detection of chromosomal imbalances in uterine cervix carcinoma. BMC Cancer. 2005;5:77. doi: 10.1186/1471-2407-5-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuppumbatti Y, Rexer S, Nakajo K, Nakaya K, Mira-y-Lopez R. CRBP suppresses breast cancer cell survival and anchorage-independent growth. Oncogene. 2001;20:7413–7419. doi: 10.1038/sj.onc.1204749. [DOI] [PubMed] [Google Scholar]
- 17.Ross S, McCaffery P, Drager U, De Luca LM. Retinoids in embryonal development. Physiol Rev. 2001;80:1021–1054. doi: 10.1152/physrev.2000.80.3.1021. [DOI] [PubMed] [Google Scholar]
- 18.Altucci L, Gronemeyer H. Nuclear receptors in cell life and death. Trends Endocrinol Metab. 2001;12:460–468. doi: 10.1016/s1043-2760(01)00502-1. [DOI] [PubMed] [Google Scholar]
- 19.Ghyselinck B, Bavik C, Sapin V, Mark M, Bonnier D, Hindelang C, Dierich A, Nilsson CB, Håkansson H, Sauvant P, Azaïs-Braesco V, Frasson M, Picaud S, Chambon P. Cellular retinol-binding protein I is essential for vitamin A homeostasis. EMBO J. 1999;18:4903–4914. doi: 10.1093/emboj/18.18.4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuppumbatti Y, Bleiweiss I, Mandeli J, Waxman S, Mira-Y-Lopez R. Cellular retinol-binding protein expression and breast cancer. J Natl Cancer Inst. 2000;92:475–480. doi: 10.1093/jnci/92.6.475. [DOI] [PubMed] [Google Scholar]
- 21.Cvetkovic D, Williams S, Hamilton T. Loss of cellular retinol binding protein 1 gene expression in microdissected human ovarian cancer. Clin Cancer Res. 2003;9:1013–1020. [PubMed] [Google Scholar]
- 22.Kwong J, Lo K, Chow L, To KF, Choy KW, Chan FL, Mok SC, Huang DP. Epigenetic silencing of cellular retinol-binding proteins in nasopharyngeal carcinoma. Neoplasia. 2005;7:67–74. doi: 10.1593/neo.04370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peralta R, Baudis M, Vazquez G, Juárez S, Ortiz R, Decanini H, Hernandez D, Gallegos F, Valdivia A, Piña P, Salcedo M. Increased expression of cellular retinol- binding protein 1 in laryngeal squamous cell carcinoma. J Cancer Res Clin Oncol. 2010;136:931–938. doi: 10.1007/s00432-009-0735-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peralta R, Valdivia A, Alvarado-Cabrero I, Gallegos F, Apresa T, Hernández D, Mendoza M, Romero P, Paniagua L, Ibáñez M, Cabrera L, Salcedo M. Correlation between expression of cellular retinol-binding protein 1 and its methylation status in larynx cancer. J Clin Pathol. 2011;65:46–50. doi: 10.1136/jclinpath-2011-200304. [DOI] [PubMed] [Google Scholar]
- 25.Caceres-Cortes J, Alvarado J, Waga K, Rangel-Corona R, Monroy-Garcia A, Rocha-Zavaleta L, Urdiales-Ramos J, Weiss-Steider B, Haman A, Hugo P, Brousseau R, Hoang T. Implication of tyrosine kinase receptor and steel factor in cell density-dependent growth in cervical cancers and leukemias. Cancer Res. 2001;61:6281–6289. [PubMed] [Google Scholar]
- 26.Livak K, Schmittgen T. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) Methods. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Herman J, Graff J, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esteller M, Guo M, Moreno V, Peinado MA, Capella G, Galm O, Baylin SB, Herman JG. Hypermethylation-associated inactivation of the cellular retinol-binding protein 1 gene in human cancer. Cancer Res. 2002;62:5902–5905. [PubMed] [Google Scholar]
- 29.Valdivia A, Peralta R, Matute-González M, García Cebada JM, Casasola I, Jiménez-Medrano C, Aguado-Pérez R, Villegas V, González-Bonilla C, Manuel-Apolinar L, Ibáñez M, Salcedo M. Co-expression of metalloproteinases 11 and 12 in cervical scrapes cells from cervical precursor lesions. Int J Clin Exp Pathol. 2011;4:674–82. [PMC free article] [PubMed] [Google Scholar]
- 30.Farias E, Ong D, Ghyselinck N, Nakajo S, Kuppumbatti YS, Mira y Lopez R. Cellular retinol-binding protein I, a regulator of breast epithelial retinoic acid receptor activity, cell differentiation, and tumorigenicity. J Natl Cancer Inst. 2005;97:21–29. doi: 10.1093/jnci/dji004. [DOI] [PubMed] [Google Scholar]
- 31.Jeronimo C, Henrique R, Oliveira J, Lobo F, Pais I, Teixeira MR, Lopes C. Aberrant cellular retinol binding protein 1 (CRBP1) gene expression and promoter methylation in prostate cancer. J Clin Pathol. 2004;57:872–876. doi: 10.1136/jcp.2003.014555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hildesheim A, Schiffman M, Gravitt P, Glass AG, Greer CE, Zhang T, Scott DR, Rush BB, Lawler P, Sherman ME, et al. Persistence of type-specific human papillomavirus infection among cytologically normal women. J Infect Dis. 1994;169:235–240. doi: 10.1093/infdis/169.2.235. [DOI] [PubMed] [Google Scholar]
- 33.zur Hausen H. Disrupted dichotomous intracellular control of human papillomavirus infection in cancer of the cervix. Lancet. 1994;343:955–957. doi: 10.1016/s0140-6736(94)90070-1. [DOI] [PubMed] [Google Scholar]


