Abstract
Found in inflammatory zone (FIZZ1), also known as hypoxia-induced mitogenic factor (HIMF), is a secreted protein formed by 111 amino acid residues. FIZZ1 is mainly located in alveolar epithelial cells, white adipose tissue and the heart. This study aimed to explore the effects of FIZZ1 on the angiogenic ability of cultured rat aortic endothelial cells (RAECs) and the potential mechanism. The RAECs were cultured in the extracellular matrix (ECM) supplemented with 10% fetal bovine serum (FBS). Matrigel assay was used to detect the angiogenic ability of the RAECs and Agilent Rat Microarray containing 41,000 genes/ESTs was used to screen the differentially expressed genes of the RAECs after they were treated with FIZZ1 (5 x 10-9~2 x 10-8 mol/L). The results were verified using RT-PCR method. We found that FIZZ1 markedly enhanced the angiogenic ability of RAECs (22.6 ± 2.94 vs. 19.7 ± 2.57, P < 0.01; 28.5 ± 3.32 vs. 19.7 ± 2.57, P < 0.01; 36.9 ± 5.01 vs. 19.7 ± 2.57, P < 0.01) in a dose-dependent manner (5 x 10-9~2 x 10-8 mol/L). 440 genes (Gng8, Atg9a, Gdf6, etc.) were found to be up-regulated and 497 genes (Hbb-b1, Camk1g, etc.) down-regulated in the experimental group. Changes in Gng8 and Atg9a were revealed by RT-PCR. FIZZ1 could enhance angiogenesis of RAECs by up-regulating Gng8 and Atg9a.
Keywords: FIZZ1/RELMα, RAECs, angiogenesis, gene expression profiling, gene chip
Introduction
Bronchoalveolar lavage fluid from mice with experimentally induced allergic pulmonary inflammation contains a novel 9.4 kDa cysteine-rich secreted protein, which was FIZZ1 formed by 111 amino acid residues [1-6]. Its structure can be separated to 3 areas: N-Signal sequence, unfixed middle part, and unique C-ribbon formed by highly conservative Cysteine repeat motif (1CXl1 2CX8 3CX4CX3 5CXl0 6CX7CX8CX9 9C10C). It was also called RELMα because the function area was consistent with that of other members of Resistin [7]. The Resistin family included 4 members: Relm-α/Fizz1, Relm-β/Fizz2, Resistin/Fizz3, and Relm-γ/Fizz4 [8].
Resistin-like molecules or those found in inflammatory zone (FIZZ) are a family of cysteine-rich secreted small proteins. They played an important role in the cell differentiation implicated in the pathophysiology of chronic diseases such as obesity, type 2 diabetes and helminth infections [9-14]. FIZZ1 was high expressive under the conditions of hypoxia, inflammation, peripheral blood mononuclear cells and macrophage activation [3]. Another recent study using FIZZ1 knockout (KO) mice demonstrated that the differential regulation of FIZZ1 expression is likely determined by the relative expression levels of IL-4, IL-13, and their corresponding receptors, which are differentially expressed by divergent cells [4]. In addition, FIZZ1 has been shown to stimulate myofibroblast differentiation [10,11] and enhance the production of collagen type I and spinal muscular atrophy (SMA) in lung fibroblast cell lines [15,16].
Angiogenesis included a cascade of intricately regulated processes occurring in growing tissues. For example, conditions of hypoxia have turned on the production of angiogenic growth factors, such as the families of vascular endothelial growth factors (VEGFs) and fibroblast growth factors (FGFs). This can be sensed by preexisting endothelial cells (ECs) in capillaries and subsequently proteases are produced to dissolve the basement membrane and extracellular matrix. Therefore, ECs migrate to the direction of the stimulus, subsequently proliferate and form new vascular sprouts [17].
FIZZ1 has been shown to induce VEGF production by murine epithelial cells [18] as well as proliferation of EC, which correlating with angiogenesis in a murine model of asthma [19]. It has also been shown to induce the expression of VCAM-1 by ECs [20] which supports eosinophil trafficking by interacting with α4β1 on the cell surface of human eosinophils [21]. Since VEGF has the capacity of promoting the generation of vessel, we hypothesize that FIZZ1 may have the same capacity. In addition, it was also shown that reduction in FIZZ1 expression may contribute to the decreased angiogenesis [22]. To validate our hypothesis, we cultivate the SVARECs of rat by reorganizing the FIZZ1, and explore the effect of FIZZ1 on the angiogenic ability of cultured aortic rat aortic endothelial cells (RAECs) and the relevant potential mechanism.
Materials and methods
Cell culture
Male Wistar rats (the Department of Experimental Animals of Shandong University, Jinan, China) between 80~100 g were anesthetized with pentobarbital (30-40 mg/kg body weight, intraperitoneally). The aorta was isolated and cut into approximately 2-mm rings. 15-20 rings were placed on the bottom of 100 mm dishes and cultured in complete ECM at pH 7.45 without disturbance for 60 h. The rings were then discarded and the cells were maintained as regular endothelial cell culture. The RAECs were confirmed as endothelial in origin by using DiI-Ac-LDL direct labeling and indirect immunofluorescence microscopy using anti-rat CD31 antibody staining. The cells were maintained in ECM, supplemented with 10% fetal bovine serum (FBS, Gibco BRL), 100 U/ml penicillin (Invitrogen) and 100 μg/ml treptomycin (Invitrogen) and incubated at 37°C, 95% air and 5% CO2. These cells were also used for up to 6-7 passages.
In vitro capillary tube formation assay in Matrigel
Tube formation assay was performed as described by Nagata, Mogi, and Walsh [24]. Twenty-four-well culture plates (Costar, Corning Incorporated) were coated with growth factor-reduced Matrigel (BD Biosciences) in a total volume of 500 μl/well and allowed to solidify for 30 min at 37°C. RAECs were trypsinized, neutralized with trypsin neutralizing solution, and resuspended in medium ECM at a density of 2 x 105/ml, and 300 μl of this cell suspension was added into each well. The wells were divided into four groups: control group, single-dose FIZZ1 group (5 x 10-9 mol/L), double-dose FIZZ1 group (10-8 mol/L) and 4-time-dose FIZZ1 group (2 x 10-8 mol/L). FIZZ1 were added to the appropriate well and the cells were incubated at 37°C for 12 h. Tube formation was observed under an inverted microscope (Olympus Corporation, Japan, x200), and photos were taken. Five visual fields were randomly selected from each well to count the tubes, and the average value was taken for statistical analysis.
RNA extraction
Extraction and purification of total RNA: RAECs were trypsinized and resuspended in medium ECM (supplemented with 10% FBS) at a density of 4 x 105/ml, and 2 ml of this cell suspension was added to a six-well culture plate, and then to the FBS-free medium after the cells were incubated at 37°C for 24 h. The same amount of 5 x 10-9 mol/L FIZZ1 or saline was added to the six-well plate, separately. Total RNA was extracted from cells to which FIZZ1 or saline was added.
RNA labeling and array hybridization
Sample labeling and array hybridization were performed according to the Agilent One-Color Microarray-Based Gene Expression Analysis protocol (Agilent Technology). Briefly, 1 μg of total RNA from each sample was linearly amplified and labeled with Cy3-dCTP. The labeled cRNAs were purified by RNAeasy Mini Kit (Qiagen). The concentration and specific activity of the labeled cRNAs (pmol Cy3/μg cRNA) were measured by NanoDrop ND-1000. 1 μg of each labeled cRNA was fragmented by adding 11 μl 10 x blocking agent and 2.2 μl of 25 x fragmentation buffer, and then heated at 60°C for 30 min. Finally 55 μl 2 x GE hybridization buffer was added to dilute the labeled cRNA. 100 μl of hybridization solution was dispensed into the gasket slide and assembled to the gene expression microarray slide. The slides were incubated for 17 h at 65°C in an Agilent hybridization oven. The hybridized arrays were washed, fixed and scanned using the Agilent DNA Microarray Scanner (part number G2505B).
Real-time quantitative PCR analysis
To verify gene changes identified by microarray analysis, we used quantitative real-time PCR (RT-PCR) as previously described. Atg9a and Gng8 were randomly selected from the differentially expressed genes screened. The techniques used were identical to those previously described. Using TRIzol (Invitrogen, USA), we extracted total cellular RNA from the experimental (5 x 10-9 mol/L FIZZ1) and control groups. RNA samples were reverse-transcribed using the ReverTraAce® qPCR RT Kit (Toyobo, Osaka, Japan) according to the manufacturer’s protocol. RT-PCR was performed with 10 μl SYBR® Green Real-time PCR Master Mix (Toyobo, Osaka, Japan) in a 20 μl reaction mix containing cDNAs (2 μl), 6 μl nuclease-free water, 1 μl each (forward and reverse) primer. Primer sequences were shown in Table 1. mplification was performed in a thermal cycler (ABI PRISM® 7700; Applied Biosystems, Foster City, CA) under the following conditions: initial heating at 95°C for 60 s, followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 15 s, and then final melting curve analysis.
Table 1.
Primer sequences
| Gene symbol | Genbank ID | Primer sequences | Product size, bp |
|---|---|---|---|
| Gng8 | NM_139185 | 5’-CCAAGATCGCTGAGGCCCGC | 168 |
| 3’-CGCGGAAGGGATTCTCGGCG | |||
| Atg9a | NM_001014218 | 5’-CCCACGGGCCCTGGAGATCA | 105 |
| 3’-CCGTGCTGGCGAACGTCCAT | |||
| Rat-actin | 5’-AGACCTTCAAGACCCCAG | 136 | |
| 3’-CACGATTTCCCTCTCAGC |
Statistical analysis
The results from tube formation assay and RT-PCR were presented as Means ± SD. The treatment effects were analyzed using SPSS 15.0. Data were analyzed by analysis of variance (ANOVA), and the difference between the groups was analyzed by t-test. P values < 0.05 were considered to be statistically significant.
Agilent Feature Extraction software (version 10.7.3.1) was used to analyze the acquired array images. Median normalization and subsequent data processing were performed using the GeneSpring GX v11.5.1 software package (Agilent Technologies). After median normalization of the raw data, genes that at least 2 out of 2 samples have flags in Detected (“All Targets Value”) were chosen for further data analysis. Differentially expressed genes were identified through fold change filtering. Hierarchical Clustering was performed using the Agilent GeneSpring GX software (version 11.5.1). Gene Ontology (GO) analysis and Pathway analysis were performed in the standard enrichment computation method.
Results
FIZZ1 enhanced the angiogenic ability of RAECs
In a dose-dependent manner(5 x 10-9~2 x 10-8 mol/L), FIZZ1 markedly enhanced the angiogenic ability of RAECs (Figure 1). There was statistically significant difference between the experimental and the normal control groups. (P < 0.05) (Table 2).
Figure 1.
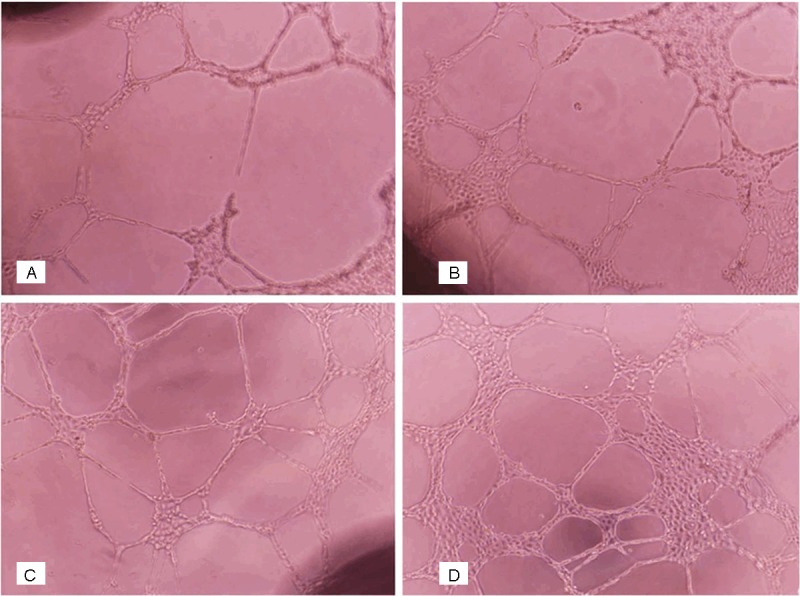
A: Control group. B: 2 x 10-8 mol/L FIZZ1 group. C: 10-8 mol/L FIZZ1 group. D: 5 x 10-9 mol/L FIZZ1 group. In vitro capillary tube formation assay in Matrigel for each group (x200). Five visual fields were randomly selected from each well to count the tubes, and the average value was taken for statistical analysis. The average tubes in control groups, 5 x 10-9 mol/L FIZZ1 groups, 10-8 mol/L FIZZ1 groups, 2 x 10-8 mol/L FIZZ1 groups were 19.7, 22.6, 28.5 and 36.9, respectively. There were statistically significant difference between the experimental and the normal control groups. (22.6 ± 2.94 vs. 19.7 ± 2.57, P < 0.01; 28.5 ± 3.32 vs. 19.7 ± 2.57, P < 0.01; 36.9 ± 5.01 vs. 19.7 ± 2.57, P < 0.01).
Table 2.
The number of the tubes in each group (x̅ ± S, n = 4)
| Group | Number of the tubes (one/HP) |
|---|---|
| Control | 19.7 ± 2.57 |
| 5 × 10-9 mol/L FIZZ1 | 22.6 ± 2.94* |
| 10-8 mol/L FIZZ1 | 28.5 ± 3.32* |
| 2 × 10-8 mol/L FIZZ1 | 36.9 ± 5.01* |
P < 0.05 (compared with the control group).
Global gene expression profiles
In order to evaluate the difference of gene expression in the control and experimental groups, we conducted microarray analysis using the Agilent DNA Microarray Scanner (part number G2505B), and found significant differences (Figure 2). Median normalization and subsequent data processing were performed using the Gene Spring GX v11.5.1 software package (Agilent Technologies). After median normalization of the raw data, genes that at least 2 out of 2 samples have flags in Detected (“All Targets Value”) were chosen for further data analysis. We identified 937 differentially expressed transcripts in FIZZ1 through fold change filtering. Among the identified genes, 497 transcripts were down-regulated and 440 up-regulated in FIZZ1 (Tables 3 and 4).
Figure 2.
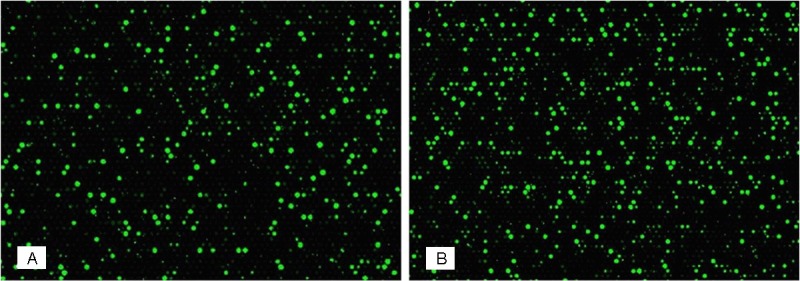
The hybridized arrays were scanned using the Agilent DNA Microarray Scanner (part number G2505B) and significant differences were found. A was the hybridized array from the control group; B was the hybridized array from the experimental group.
Table 3.
Genes representing upregulated expression in FIZZ1 cultures
| GenBank accession | Gene symbol | Fold change (Treat/Control) | Description |
|---|---|---|---|
| NM_001107354 | Hist1h2an | 5.7152147 | Histone cluster 1, H2an (Hist1h2an) |
| NM_001135046 | Arpp-21 | 5.677911 | Cyclic AMP-regulated phosphoprotein (Arpp-21) |
| NM_001002813 | Ctsql2 | 5.3080893 | Cathepsin Q-like 2 (Ctsql2) |
| NM_031343 | Slc6a2 | 4.9812746 | Solute carrier family 6 (neurotransmitter transporter) |
| NM_001014779 | Pcdhb8 | 4.9663377 | Protocadherin beta 8 (Pcdhb8) |
| NM_032070 | Hmga2 | 4.871653 | High mobility group AT-hook 2 (Hmga2) |
| NM_001007235 | Itpr1 | 4.7131433 | Inositol 1, 4, 5-triphosphate receptor |
| NM_001014218 | Atg9a | 4.6005354 | ATG9 autophagy related 9 homolog A (Atg9a) |
| NM_139185 | Gng8 | 4.3666644 | Guanine nucleotide binding protein (G protein) |
| NM_198049 | Slc10a6 | 4.2526517 | Solute carrier family 10 member 6 (Slc10a6) |
Table 4.
Genes representing downregulated expression in FIZZ1 cultures
| Genbank accession | Gene symbol | Fold change (Treat/Control) | Description |
|---|---|---|---|
| BF290343 | -14.0022 | UNKNOWN | BF290343 EST454934 Rat Gene Index |
| NM_001014059 | -11.245 | RGD1304952 | Similar to RIKEN cDNA C530028O21 gene (RGD1304952) |
| NM_198776 | -8.75434 | Hbb-b1 | Beta-glo (MGC72973) |
| NM_182842 | -7.93352 | Camk1g | Calcium/calmodulin-dependent protein kinase IG (Camk1g) |
| NM_013088 | -7.78243 | Ptpn11 | Protein tyrosine phosphatase, non-receptor type 11 (Ptpn11), transcript variant 2 |
| NM_001013853 | -7.64786 | LOC287167 | Globin, alpha (LOC287167) |
| NM_001000020 | -7.22534 | Olr1454 | Olfactory receptor 1454 (Olr1454) |
| NM_001025002 | -7.21841 | LOC310926 | Hypothetical protein LOC310926 (LOC310926) |
| XM_001059821 | -7.17914 | Elf4 | PREDICTED: E74-like factor 4 (ets domain transcription factor) (Elf4) |
| M83679 | -7.09958 | Rab15 | Sprague-Dawley (clone LRB9) RAB15 Mrna |
mRNA analysis of control and FIZZ1 group
To monitor the transcriptional regulation of FIZZ1 proteins, we performed real-time quantitative PCR analysis using primers designed based on the GenBank sequences of Rat-actin, Gng8 and Atg9a. Rat-actin was used as an internal RNA loading control. Our data showed significant difference in the up-regulation of Gng8 and Atg9a between the control and FIZZ1 groups (Figure 3).
Figure 3.
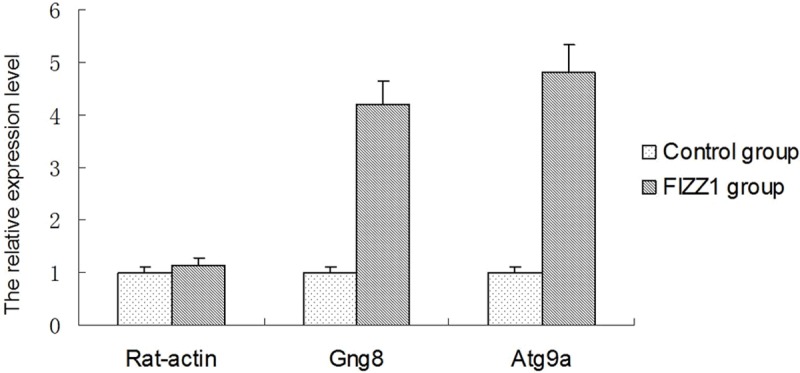
mRNA expressions of Rat-actin, Gng8 and Atg9a in two groups. Total RNA was extracted from RAECs and RAECs with FIZZ1 at the end of 12 h after drug administration. Results are expressed as fold relative to mRNA levels in RAECs after FIZZ1 treated with control group.*P < 0.05, compared to respective control.
Discussion
It is reported that FIZZ1 possesses functions such as stimulating the generation and movement of pulmonary artery smooth muscle cells, contracting vessels and stimulating myofibroblastic differentiation [23,24], and it plays a significant role in pathogenic mechanism of many diseases such as pulmonary fibrosis, asthma, hypoxia, silicosis, and chronic obstructive pulmonary disease (COPD). However, whether it can promote the generation of endothelial cells has not been reported. Tong et al. [15,17] showed that FIZZ1 had the functions of stimulating VCAM-1 and VEGF, which can stimulate vessel generation and affect the receptors on endothelial cells, activate P13K and phosphorylate Akt which was key element in the signaling pathway of angiogenesis. They reported that FIZZ1 can further induce VEGF expression through PI3K-AKt-NF-κB signaling pathway, PI3K-depressor (LY294002) can only partially restrain the proliferation of pulmonary vascular smooth muscle cells and the migration of FIZZ1. In addition, Yamaji et al [25] found that RELMα can promote the generation of pulmonary microvascular endothelial cells, while anti-VEGF can only partly inhibit angiogenesis. Therefore, multiple ways to stimulate the generation of vessels may exist, it is possible that FIZZ1 could stimulate the aortic endothelial cells of new blood vessels and the mechanism is complicated and diversified.
The results of this study showed that FIZZ1 can obviously promote the formation of RAECs newborn tube. After RAECs are treated with the reorganized FIZZl at the concentration of 5 x 10-9~2 x 10-8 mol/L, FIZZ1 increased the angiogenic ability of RAECs obviously. It means that FIZZ1 can significantly promote the vessel generation of RAECs.
To further explore the mechanism underlying the stimulation of vessel generation by FIZZ1, microarray analysis and RT-PCR were used to analyze the control and experimental groups. The results showed that RAECs of the experimental group are quite different from those of the control group in number after the expression of FIZZ1. The analysis result of Gene Spring GX v11.5.1 showed that over 440 genes (Gng8, Atg9a, Gdf6, etc.) existed. Atg9a was known as a transmembrane protein and the molecule of N and C-terminal transmembrane 6 participated in signal transduction of the membrane outside [26]. Gng8 participated in the multi-cell signal transduction through the cell membrane G-protein phosphorylation and dephosphorylation [27]. Gng8 and Atg9a may be associated with the ability of the FIZZ1 to promote RAECs in vitro angiogenesis.
To sum up, our research explored the mechanism and the ability of FIZZ1 to stimulate SVAREC vessel generation. It was found that FIZZ1 can promote the vessel generation ability of RAECs through the up-regulated expression of Gng8, Atg9a and other genes besides P13K/Akt pathway. Whether the increased expression of Gng8, Atg9a and other genes can promote angiogenesis ability of specific signal transduction pathways in RAECs cultued with FIZZ1 still needs further research.
Acknowledgements
This work was funded by a grant from Natural Science Foundation of Shandong Province (No. ZR2010HM202) and Science Foundation of Guizhou Province (No. SY(2010)3081).
Disclosure of conflict of interest
None.
References
- 1.Holcomb IN, Kabakoff RC, Chan B, Baker TW, Gurney A, Henzel W, Nelson C, Lowman HB, Wright BD, Skelton NJ, Frantz GD, Tumas DB, Peale FV Jr, Shelton DL, Hebert CC. FIZZ1, a novel cysteine-rich secreted protein associated with pulmonary inflammation, defines a new gene family. EMBO J. 2000;19:4046–4055. doi: 10.1093/emboj/19.15.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Libby P, Ridker PM, Maseri A. Inflammation and atherosclerosis. Circulation. 2002;105:1135–1143. doi: 10.1161/hc0902.104353. [DOI] [PubMed] [Google Scholar]
- 3.Steppan CM, Brown EJ, Wright CM, Bhat S, Banerjee RR, Dai CY, Enders GH, Silberg DG, Wen X, Wu GD, Lazar MA. A family of tissue-specific resistin-like molecules. Proc Natl Acad Sci U S A. 2001;98:502–506. doi: 10.1073/pnas.98.2.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munitz A, Cole ET, Karo-Atar D, Finkelman FD, Rothenberg ME. Resistin-like molecule-alpha regulates IL-13-induced chemokine production but not allergen-induced airway responses. Am J Respir Cell Mol Biol. 2012;46:703–713. doi: 10.1165/rcmb.2011-0391OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung MJ, Liu T, Ullenbruch M, Phan SH. Antiapoptotic effect of found in inflammatory zone (FIZZ)1 on mouse lung fibroblasts. J Pathol. 2007;212:180–187. doi: 10.1002/path.2161. [DOI] [PubMed] [Google Scholar]
- 6.Liu T, Dhanasekaran SM, Jin H, Hu B, Tomlins SA, Chinnaiyan AM, Phan SH. FIZZ1 stimulation of myofibroblast differentiation. Am J Pathol. 2004;164:1315–1326. doi: 10.1016/S0002-9440(10)63218-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raes G, De Baetselier P, Noel W, Beschin A, Brombacher F, Hassanzadeh Gh G. Differential expression of FIZZ1 and Ym1 in alternatively versus classically activated macrophages. J Leukoc Biol. 2002;71:597–602. [PubMed] [Google Scholar]
- 8.Madala SK, Edukulla R, Davis KR, Schmidt S, Davidson C, Kitzmiller JA, Hardie WD, Korfhagen TR. Resistin-like molecule alpha1 (Fizz1) recruits lung dendritic cells without causing pulmonary fibrosis. Respir Res. 2012;13:51. doi: 10.1186/1465-9921-13-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pesce JT, Ramalingam TR, Wilson MS, Mentink-Kane MM, Thompson RW, Cheever AW, Urban JF Jr, Wynn TA. Retnla (relmalpha/fizz1) suppresses helminth-induced Th2-type immunity. PLoS Pathog. 2009;5:e1000393. doi: 10.1371/journal.ppat.1000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamaji-Kegan K, Su Q, Angelini DJ, Myers AC, Cheadle C, Johns RA. Hypoxia-induced mitogenic factor (HIMF/FIZZ1/RELMalpha) increases lung inflammation and activates pulmonary microvascular endothelial cells via an IL-4-dependent mechanism. J Immunol. 2010;185:5539–5548. doi: 10.4049/jimmunol.0904021. [DOI] [PubMed] [Google Scholar]
- 11.Nair MG, Du Y, Perrigoue JG, Zaph C, Taylor JJ, Goldschmidt M, Swain GP, Yancopoulos GD, Valenzuela DM, Murphy A, Karow M, Stevens S, Pearce EJ, Artis D. Alternatively activated macrophage-derived RELM-{alpha} is a negative regulator of type 2 inflammation in the lung. J Exp Med. 2009;206:937–952. doi: 10.1084/jem.20082048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu T, Hu B, Choi YY, Chung M, Ullenbruch M, Yu H, Lowe JB, Phan SH. Notch1 signaling in FIZZ1 induction of myofibroblast differentiation. Am J Pathol. 2009;174:1745–1755. doi: 10.2353/ajpath.2009.080618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herbert DR, Yang JQ, Hogan SP, Groschwitz K, Khodoun M, Munitz A, Orekov T, Perkins C, Wang Q, Brombacher F, Urban JF Jr, Rothenberg ME, Finkelman FD. Intestinal epithelial cell secretion of RELM-beta protects against gastrointestinal worm infection. J Exp Med. 2009;206:2947–2957. doi: 10.1084/jem.20091268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munitz A, Seidu L, Cole ET, Ahrens R, Hogan SP, Rothenberg ME. Resistin-like molecule alpha decreases glucose tolerance during intestinal inflammation. J Immunol. 2009;182:2357–2363. doi: 10.4049/jimmunol.0803130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong L, Wang SJ, Camoretti-Mercado B, Li HJ, Chen M, Bi WX. FIZZ1 plays a crucial role in early stage airway remodeling of OVA-induced asthma. J Asthma. 2008;45:648–653. doi: 10.1080/02770900802126941. [DOI] [PubMed] [Google Scholar]
- 16.Teng X, Li D, Champion HC, Johns RA. FIZZ1/RELMalpha, a novel hypoxia-induced mitogenic factor in lung with vasoconstrictive and angiogenic properties. Circ Res. 2003;92:1065–1067. doi: 10.1161/01.RES.0000073999.07698.33. [DOI] [PubMed] [Google Scholar]
- 17.Mulder WJ, Griffioen AW. Imaging of angiogenesis. Angiogenesis. 2010;13:71–74. doi: 10.1007/s10456-010-9178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tong Q, Zheng L, Lin L, Li B, Wang D, Huang C, Li D. VEGF is upregulated by hypoxia-induced mitogenic factor via the PI-3K/Akt-NF-kappaB signaling pathway. Respir Res. 2006;7:37. doi: 10.1186/1465-9921-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun Y, Wang J, Li H, Han X. Found in inflammatory zone 1 induces angiogenesis in murine models of asthma. Lung. 2008;186:375–380. doi: 10.1007/s00408-008-9099-1. [DOI] [PubMed] [Google Scholar]
- 20.Tong Q, Zheng L, Lin L, Li B, Wang D, Li D. Hypoxia-induced mitogenic factor promotes vascular adhesion molecule-1 expression via the PI-3K/Akt-NF-kappaB signaling pathway. Am J Respir Cell Mol Biol. 2006;35:444–456. doi: 10.1165/rcmb.2005-0424OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rao SP, Wang Z, Zuberi RI, Sikora L, Bahaie NS, Zuraw BL, Liu FT, Sriramarao P. Galectin-3 functions as an adhesion molecule to support eosinophil rolling and adhesion under conditions of flow. J Immunol. 2007;179:7800–7807. doi: 10.4049/jimmunol.179.11.7800. [DOI] [PubMed] [Google Scholar]
- 22.Ge XN, Bahaie NS, Kang BN, Hosseinkhani MR, Ha SG, Frenzel EM, Liu FT, Rao SP, Sriramarao P. Allergen-induced airway remodeling is impaired in galectin-3-deficient mice. J Immunol. 2010;185:1205–1214. doi: 10.4049/jimmunol.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagata D, Mogi M, Walsh K. AMP-activated protein kinase (AMPK) signaling in endothelial cells is essential for angiogenesis in response to hypoxic stress. J Biol Chem. 2003;278:31000–31006. doi: 10.1074/jbc.M300643200. [DOI] [PubMed] [Google Scholar]
- 24.Karlsson A, Follin P, Leffler H, Dahlgren C. Galectin-3 activates the NADPH-oxidase in exudated but not peripheral blood neutrophils. Blood. 1998;91:3430–3438. [PubMed] [Google Scholar]
- 25.Yamaji-Kegan K, Su Q, Angelini DJ, Champion HC, Johns RA. Hypoxia-induced mitogenic factor has proangiogenic and proinflammatory effects in the lung via VEGF and VEGF receptor-2. Am J Physiol Lung Cell Mol Physiol. 2006;291:L1159–1168. doi: 10.1152/ajplung.00168.2006. [DOI] [PubMed] [Google Scholar]
- 26.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryba NJ, Tirindelli R. A novel GTP-binding protein gamma-subunit, G gamma 8, is expressed during neurogenesis in the olfactory and vomeronasal neuroepithelia. J Biol Chem. 1995;270:6757–6767. doi: 10.1074/jbc.270.12.6757. [DOI] [PubMed] [Google Scholar]


