SUMMARY
Prion proteins undergo self-sustaining conforma-tional conversions that heritably alter their activities. Many of these proteins operate at pivotal positions in determining how genotype is translated into pheno-type. But the breadth of prion influences on biology and their evolutionary significance are just beginning to be explored. We report that a prion formed by the Mot3 transcription factor, [MOT3+], governs the acquisition of facultative multicellularity in the budding yeast Saccharomyces cerevisiae. The traits governed by [MOT3+] involved both gains and losses of Mot3 regulatory activity. [MOT3+]-dependent expression of FLO11, a major determinant of cell-cell adhesion, produced diverse lineage-specific multicellular phenotypes in response to nutrient deprivation. The prions themselves were induced by ethanol and eliminated by hypoxia—conditions that occur sequentially in the natural respiro-fermen-tative cycles of yeast populations. These data demonstrate that prions can act as environmentally responsive molecular determinants of multicellularity and contribute to the natural morphological diversity of budding yeast.
INTRODUCTION
The evolution of multicellularity is among the most notable transitions in the history of life (Grosberg and Strathmann, 2007). Despite its reputation as a simple, “unicellular” eukaryote, the budding yeast Saccharomyces cerevisiae has proven to be a powerful model for this transition (Koschwanez et al., 2011; Ratcliff et al., 2012). It frequently and reversibly abandons a solitary lifestyle in favor of diverse multicellular growth forms (reviewed in Brückner and Mösch, 2012). In doing so, individual yeast cells cooperate to protect themselves from the environment, from other organisms, and from starvation.
The cell surface adhesins that drive multicellularity are encoded by some of the most heavily regulated and yet rapidly evolving genes in the yeast genome (Hahn et al., 2005; Brückner and Mösch, 2012). Repeated selection for new adhesin pheno-types might support mechanisms that expedite their genetic diversification, including intragenic tandem repeats, gene multiplicity, and a subtelomeric location, all of which produce high recombination rates that drive the frequent appearance of new functional variants (Brückner and Mösch, 2012).
In opposition to the environmental pressures for diversification, multicellularity itself necessitates conformity: individual cells must act concertedly and cooperatively to maintain the integrity and therefore adaptive benefits of multicellular structures. This dichotomy may have favored the evolution of switch-like regulation in adhesin expression, resulting in binary or “dimorphic” transitions. Such switches are both genetic and epigenetic in nature. Mutations occur frequently in trans-acting regulators of adhesins (Halme et al., 2004), whereas position-dependent genomic silencing (Halme et al., 2004), cis-interfering noncoding RNAs (Bumgarner et al., 2012), and stochastic associations with low-abundance transcription factors (Octavio et al., 2009) can each act as epigenetic toggle switches of adhesin expression. Self-perpetuating switches in the folding of certain proteins, known as prions, might also play a role (Patel et al., 2009; Halfmann et al., 2012). Among these very different molecular mechanisms, the fact that prions are based on protein folding might give them an intrinsic capacity to transduce environmental changes into dominant and highly stable phenotypic changes.
Prion-forming proteins can exist in profoundly different structural states, at least one of which is a self-templating (prion) state. Proteins in the prion conformation interact with nonprion conformers of the same protein, converting them to prion conformers. The biochemically characterized prions are insoluble amyloid-like fibrils that cause heritable changes in the cellular distribution of the protein. Prions create phenotypes by diverse means—in many cases by sequestering proteins away from their normal cellular activities, but in others, endowing them with new activities (Derkatch et al., 2001; Rogoza et al., 2010; Suzuki et al., 2012). Notably, the self-templating nature of prion conformations allows such phenotypes to be immediately and robustly heritable (Serio et al., 2000; Satpute-Krishnan and Serio, 2005). This has generated a vigorous discussion about the potential significance of prions in yeast evolution (Pigliucci, 2008; Wickner et al., 2011; Koonin, 2012).
Yeast cells normally switch between prion and nonprion states at low frequencies, but protein folding is extraordinarily sensitive to environmental stress (Chiti and Dobson, 2006). Therefore, when cells are not well suited to their environment, i.e., when they are stressed, prion switching may accelerate (Tyedmers et al., 2008). The net result is that a small fraction of stressed cells explores alternative phenotypes. Often, newly arising prions are detrimental (McGlinchey et al., 2011). On occasion, however, the phenotypes revealed by prions are adaptive, enabling the prion-containing lineages to survive at times when they otherwise might perish (True and Lindquist, 2000; Halfmann and Lindquist, 2010). Theoretical work supports the concept that prion switching may constitute a sophisticated form of “bet-hedging” (Griswold and Masel, 2009; Lancaster et al., 2010). Despite the plausible functions of prion-based switches in gene regulation, the topic remains intensely controversial (Liebman and Chernoff, 2012). Definitive evidence for prion functionality, including mechanisms that would link specific prion-protein conformational switches to environmental changes that are explicitly relevant to their functions, is wanting.
Here, we report that prion formation by the Mot3 transcriptional repressor regulates the acquisition of multicellular growth forms in budding yeast. The formation, elimination, and phenotypic manifestation of Mot3 prions each respond to specific environmental conditions. The effects of Mot3 prions are further determined by heritable variation between different yeast isolates. Mot3 prion switching is thus a molecular mechanism that couples natural environmental changes to heritable changes in gene expression. It might also provide a new route to the evolution of cooperative multicellular behaviors.
RESULTS
A Convergence of Prions at the Multicellularity Determinant, FLO11
The functions of known yeast prions are heavily biased for gene regulation (Halfmann and Lindquist, 2010). To identify regulatory functions that might be associated with prion switching, we compared the published regulons of transcription factors that we and others have shown to be capable of forming prions (Alberti et al., 2009; Patel et al., 2009; Rogoza et al., 2010). The regulons of all three experimentally verified prion-forming transcription factors (Cyc8, Mot3, and Sfp1), as well as Gln3, a prion-like transcription factor that is itself the major regulatory target of the Ure2 prion (Wickner, 1994; Kulkarni et al., 2001), overlapped at only two genes: FLO11, encoding a cell wall-anchored glycoprotein; and HXT2, encoding a high-affinity glucose transporter (Figure 1A). Both genes are naturally induced by stress and by nutrient deprivation (Ozcan and Johnston, 1999; Türkel, 1999; Brückner and Mösch, 2012). The probability that any given ORF would be regulated by all four of these transcription factors, by chance, is 1.21 × 10−5 (Extended Experimental Procedures available online). Notably, FLO11 is also regulated by the chromatin-remodeling factor Swi1 (Barrales et al., 2012), yet another type of transcriptional regulator that is capable of forming a prion (Du et al., 2008). Other transcription factors are likely to be prions. Therefore, to reduce bias, we also undertook a complementary computational analysis to determine if the 32 transcription factors annotated to regulate FLO11 are significantly enriched for the presence of prion-like domains, including those that have not yet been discovered to act as prions. We used a modified hidden Markov model algorithm (Alberti et al., 2009) to predict the prion propensities for all yeast transcription factors. We then asked whether the observed distribution of this propensity score for the regulators of FLO11 was significantly different from random samples of 32 transcription factors using a bootstrap approach (Extended Experimental Procedures). We found that regulators of FLO11 had a higher enrichment for prion propensity than expected by chance (p = 0.017).
Figure 1. Prions Converge on the Principal Determinant of Yeast Multicellularity FLO11.
(A) Venn diagram demonstrating the overlap in the regulons of known prion-forming yeast transcription factors. Only two genes, FLO11 and HXT2, are regulated by all four.
(B) A genetic reporter for [MOT3+]. Mot3 represses transcription of DAN1 under normal growth conditions. When the DAN1 ORF is replaced with URA3, laboratory yeast strains can become prototrophic for uracil by prion-driven inactivation of Mot3.
(C) Cells harboring the reporter for [MOT3+] were transformed with either empty vector (E.V.) or a galactose-inducible vector encoding Mot3. Following 24 hr of growth in galactose, 5-fold serial dilutions of cells were plated to media lacking uracil (– Uracil) to select for those that contained stably inactivated Mot3. Cells plated to uracil-containing media (+ Uracil), at the highest dilution, are also shown. Transient overexpression of Mot3 strongly induces conversion to a Ura+ phenotype.
(D) qRT-PCR using mRNA isolated from isogenic [MOT3+] and [mot3−] colonies reveals that [MOT3+] cells have increased FLO11 expression. Values are normalized to ACT1. Error bars represent SD from triplicate colonies.
Why might the FLO11 regulatory interactome be inundated with prion regulators? Three aspects of its biology suggest an answer. First, as the principal determinant of multicellularity; Flo11 enables single cells to differentiate into multicellular structures, including cell clumps, chains, and biofilms (Brückner and Mösch, 2012). The success of any multigenerational developmental program of this nature necessitates a stable molecular commitment (which prions provide) rather than the short-term responses of conventional regulatory networks. Second, FLO11 expression switches between expression states at multiple frequencies, ranging from approximately once every three cell divisions to once in a thousand cell divisions (Kuthan et al., 2003; Halme et al., 2004; Octavio et al., 2009). This is relevant because prion-based switches have also been found to occur at multiple frequencies, from 10−7 to 10−2 per cell division (Liebman and Chernoff, 2012). Finally, FLO11 is induced by, and is thought to protect against, environmental stresses (Brückner and Mösch, 2012). Prions are intrinsically sensitive to changes in protein homeostasis (Tyedmers et al., 2008) and may, therefore, be able to act as “sensors” of environmental stress. These considerations led us to test whether the gain and loss of prion states by prion-forming transcription factors might induce facultative multicellular transitions in response to environmental adversities.
Isolation of [MOT3+] Colonies
Given the well-known instability of FLO11 expression, and the multitude of factors responsible for regulating it, we reasoned that simply correlating FLO11 expression dynamics with individual prions would not provide a sufficiently rigorous test of our hypothesis. Instead, we employed a reverse approach, in which we isolated stable prion states on the basis of a completely orthogonal phenotype and then queried their effects on FLO11. To this end, and to facilitate follow-up experiments, we focused on Mot3, a transcription factor with a relatively discrete set of gene targets and well-characterized biology. The consensus binding motif for Mot3 (nucleotide sequence HAGGYA) occurs 16 times upstream of FLO11 (Table S1), far more frequently than expected by chance (cumulative binomial probability of 0.007). Mot3 normally represses FLO11 (Carter et al., 2007) and other genes involved in remodeling the yeast cell surface (Table S2; annotation clusters 1, 2, and 6 designate cell wall-and cell membrane-localized gene products; cluster 4 designates gene products involved with membrane biosynthesis). Oxygen depletion alleviates this repression (Sertil et al., 2003; Lai et al., 2006).
To provide a facile orthogonal measure of Mot3 prion switching, we placed a URA3 gene under the control of a Mot3-regulated promoter (DAN1), in a strain that also carries a ura3 deletion (Alberti et al., 2009). Mot3 normally represses this promoter. When Mot3 switches to the prion state, however, it should sequester the protein away from the genome, derepressing URA3 and allowing cells to grow without uracil (Figure 1B). In keeping with prion nomenclature, this state is designated [MOT3+] (Alberti et al., 2009), with capital letters denoting dominance in genetic crosses and brackets denoting cytoplasmic inheritance. Because most laboratory yeast have recessive null mutations in FLO8, a master regulator of FLO11 (Liu et al., 1996), we mated the Mot3-reporter strain with a ura3 FLO8 strain (Σ1278b) to produce a hybrid diploid competent for FLO11 expression. This strain spontaneously acquired a Ura+ phenotype at a frequency of ~10−4 to 10−3, depending on the stringency of selection (Figure S1).
One defining feature of prion biology is that overexpression of the prion protein increases the frequency at which the prion appears in a population of prion minus cells (Wickner, 1994; Alberti et al., 2009). A second is that once the protein has acquired the prion conformation, overexpression is no longer necessary to maintain that state. To induce [MOT3+], we therefore transformed cells with a galactose-inducible plasmid encoding Mot3, or with an empty vector, and grew the cells in galactose media for 24 hr. We then plated the cells to glucose media lacking uracil to restore endogenous Mot3 expression levels. This selected for cells in which Mot3 was inactivated in a heritable way. Indeed, the transient overexpression greatly increased the appearance of Ura+ colonies (Figure 1C). Such colonies retained their phenotype after repeated passaging.
Another characteristic common to prions formed by amyloidogenic proteins is a requirement for Hsp104. This protein-remodeling factor fragments prion amyloid fibers and enables prion templates to be inherited by daughter cells. To test if the Ura+ phenotype was due to a prion switch, we passaged cells on media containing 3 mM guanidine hydrochloride (GdHCl), a selective inhibitor of Hsp104 (Ferreira et al., 2001). We then passaged them to media lacking GdHCl before testing for the continued inheritance of the Ura+ phenotype. As was previously demonstrated in a different genetic background by Alberti et al. (2009), this treatment restored Ura+ cells to the original, ura− phenotype (Figure S1B). To ensure that this was not due to an off-target effect of GdHCl, we used a genetic approach—transiently expressing a dominant-negative variant of Hsp104 (Chernoff et al., 1995), Hsp104DN. This treatment also restored the ura− phenotype. Overexpression of WT Hsp104 did not (Figures S1C and S1D). Thus, the inheritance of the Ura+ phenotype requires the continuous activity of Hsp104.
As noted above, the self-templating structure for most prions is an amyloid fiber. Most amyloids are extraordinarily resistant to solubilization by detergents, and this property can be used to distinguish amyloids from other noncovalent protein complexes (Alberti et al., 2009). To verify that the prion responsible for the Ura+ phenotype is indeed formed by Mot3, we used semidenaturing detergent-agarose gel electrophoresis (SDD-AGE). SDD-AGE resolves amyloids from nonamyloid aggregates and soluble proteins based on size and insolubility in SDS. By probing SDD-AGE blots for the naturally occurring hexa-histidine motif of Mot3, we found that SDS-resistant aggregates of Mot3 occurred in Ura+ cells, but not in ura− cells (Figure S1B). We conclude that the Ura+ cells are [MOT3+].
Having isolated [MOT3+] by virtue of the transcriptional derepression of the DAN1 locus, we next asked if FLO11 was simultaneously affected. We prepared mRNA from isogenic [MOT3+] and [mot3−] cells and used quantitative RT-PCR (qRT-PCR) to evaluate FLO11 expression. Indeed, [MOT3+] increased FLO11 transcripts by ~10-fold (Figure 1D).
[MOT3+] Governs the Acquisition of Multicellular Phenotypes
FLO11 expression mediates the development of diverse multicellular phenotypes in response to specific environmental signals. Under standard nutrient-rich laboratory growth conditions, [MOT3+] conferred only modest FLO11 -related phenotypes (data not shown). We therefore explored the synergistic effects of [MOT3+] with environmental conditions that naturally induce FLO11. The best characterized of these is nitrogen starvation. When challenged with limiting or poor nitrogen sources, cells elongate, bud in a unipolar fashion, and remain attached after cell division (Gimeno et al., 1992). The resulting chains of cells extend well beyond the original colony boundaries and can even invade the underlying growth substratum.
To explore the effects of [MOT3+] on invasive growth, we plated cells on media in which the only source of nitrogen is a poor one, proline. After 5 days of growth, noninvasive cells were dislodged by rinsing the plates vigorously with running water. Prion minus cells, [mot3−], were easily washed away. In contrast, [MOT3+] colonies acquired invasive filaments that could not be dislodged (Figure 2A).
Figure 2. [MOT3+] Enables Facultative Multicellular Growth.
(A) Schematics on the left depict multicellular phenotypes (dark cells) that are induced by each of the common growth conditions indicated. On plates containing only proline as a nitrogen source, [MOT3+] cells formed invasive filaments that penetrated the agar surface (top; invasive growth remains after dislodging surface cells under running water). On plates containing only ethanol as a carbon source, [MOT3+] cells formed complex colony morphologies (middle). The schematic for colony morphology is based on Váchová et al. (2011). When grown to saturation in liquid media, [MOT3+] cells exhibited a decreased tendency to flocculate (bottom). Each of these behaviors of [MOT3+] cells was eliminated by treating the cells transiently with GdHCl, an inhibitor of the prion-partitioning factor Hsp104.
(B) [MOT3+] cells form elaborate, biofilm-like structures when grown on semisolid ethanol media.
Scale bars, 1 cm. See also Figure S2.
In addition to invasive growth in response to nitrogen starvation, Flo11 can induce complex colony architectures in response to starvation for fermentable carbon sources (Granek and Magwene, 2010). Emerging evidence suggests that such colonies are, in fact, rudimentary biofilms that protect cells from stress and enable cells to cooperate metabolically (Váchová et al., 2011). To investigate, we plated [MOT3+] and [mot3−] cells to media containing a variety of different carbon sources. [mot3−] cells formed smooth, simple colonies regardless of carbon source. [MOT3+] colonies were indistinguishable from those of [mot3−] on glucose and galactose, which are fermentable carbon sources. But on glycerol and ethanol, which are nonfer-mentable, [MOT3+] cells formed elaborate colonies with prominent ridges and invaginations (Figure 2A; data not shown).
Another stimulus of Flo11-dependent colony differentiation is growth on a semisolid substratum (Reynolds and Fink, 2001). When grown on semisolid media, with ethanol as a carbon source, [MOT3+] cells differentiated into an elaborate compound structure consisting of well-developed microcolonies embedded in a transparent gelatinous matrix (Figure 2B). Notably, the matrix was impenetrable to isogenic yeast cells applied to the exterior of the colony (Figure S2). In striking contrast, [mot3−] colonies again failed to differentiate, instead producing a simple smooth colony lacking a prominent gelatinous exterior.
Finally, we examined a quite different multicellular behavior. Some yeast strains clump together, or flocculate, toward the end of fermentation in liquid media. Unlike phenotypes typically driven by Flo11, which result from cell-cell interactions between mother cells and their daughters, flocculation results from “horizontal” association between cells. This behavior is industrially desirable but notoriously unstable; flocculation competence is frequently and stochastically gained and lost (Verstrepen et al., 2003). We asked if [MOT3+] can also contribute to flocculation phenotypes by allowing cells to grow to saturation in rich liquid media. [MOT3+] cultures became moderately flocculent 1 day before [mot3−] cultures. The final extent of flocculation, however, was greater in [mot3−] cultures (Figure 2A). The flocculation phenotype, in both [MOT3+] and [mot3−] cells, was driven by Flo1, a cell-wall-anchored adhesin similar to Flo11 (Supplemental Information). Like FLO11, the FLO1 promoter contains numerous binding motifs for Mot3.
[MOT3+] Prion Phenotypes Result from Both Losses and Gains of Mot3 Function
Most prion phenotypes mimic loss-of-function mutations in the genes that encode them. To determine whether [MOT3+] phenotypes are due to simple inactivation of Mot3, we employed two genetic approaches. First, we asked whether the prion’s phenotypes could be complemented by supplying Mot3’s normal cellular activity in trans. To this end, we constructed a variant of Mot3 with a deletion in the prion-forming region of the protein, known as the PrD (Alberti et al., 2009). This variant, ΔPrD, was immune to prion-mediated inactivation. When expressed from a plasmid in [MOT3+] cells, it maintained solubility and fully reversed the Ura+ phenotype (Figure 3). It also reversed the complex colony morphology and the hypoflocculent phenotypes (Figure 4A). Thus, a loss of Mot3’s normal cellular activity is necessary for the prion phenotypes. Importantly, when the plasmid expressing ΔPrD was lost, [MOT3+] phenotypes returned, demonstrating that ΔPrD simply masked the prion phenotypes while allowing the full-length protein to maintain the prion state.
Figure 3. The N-Terminal Region Is Required for Prion-Mediated Inactivation of Mot3.
(A) A schematic of full-length Mot3 and two functionally distinct truncation mutants. Regions of high predicted prion-forming propensity are indicated in red. The C-terminal nonprion-like region of Mot3 contains two C2H2 zinc fingers, indicated in blue, which are involved in DNA binding. The prion-determining region, “PrD,” as initially defined by Alberti et al. (2009) encompasses the N-terminal 295 residues of the protein. A poly-asparagine (poly-N) tract stretches from residues 143–157. In the present study, we construct a variant, “ΔPrD,” that lacks much of the PrD including the poly-N tract. An endogenous hexa-histidine motif is indicated in green.
(B) Ectopic expression of ΔPrD (↑ΔPrD) from a constitutive promoter (TEF1) suppresses the Ura+ phenotype of [MOT3+]. In contrast, [MOT3+] cells ectopically expressing full-length Mot3 (↑WT) remain Ura+. This is because WT Mot3, but not ΔPrD, accumulates as SDS-resistant aggregates, as visualized by SDD-AGE and immunoblotting against the endogenous hexa-histidine motif. Amyloids from endogenous Mot3 are too low abundance to be detected in this exposure. An SDS-PAGE blot of lysates boiled in 2% LDS demonstrates comparable expression of the Mot3 variants.
Figure 4. Prion-Mediated Inactivation of Mot3 Is Required but Is Not Sufficient to Convey the Full Spectrum of [MOT3+] Phenotypes.
(A) Ectopic expression of ΔPrD (↑ΔPrD) from a constitutive promoter (TEF1) suppresses the complex colony morphology, invasion, and hypo-flocculation phenotypes of [MOT3+] cells. In contrast, [MOT3+] cells that ectopically express full-length Mot3 (↑WT) retain [MOT3+] phenotypes.
(B) Genetic deletion of MOT3 confers some phenotypes that mimic those of [MOT3+], including biofilm formation and invasion. Δmot3 cells do not flocculate when grown to saturation in liquid media.
(C) Genetic deletion of MOT3 is not sufficient to fully derepress the DAN1 promoter (PDAN1-URA3). Unlike [MOT3+] cells (which are Ura+), Δmot3 cells partition dynamically between fully repressed (ura−) and derepressed (Ura+) states.
(D) Ectopic expression of WT Mot3 stabilizes the derepressed (Ura+) state, whereas the non-aggregating variant of Mot3, ΔPrD, restores the fully repressed (ura−) state. Expression of PrD or empty vector has no effect. Duplicate transformants are shown, and serial dilutions were made onto the same plate. Bottom view is a western blot comparing expression levels of the Mot3 variants.
(E) In a hypothetical model for the gain of function of Mot3 prion conformers, Mot3 interacts with another protein that corepresses transcription from the DAN1 promoter. In the [MOT3+] state, Mot3 prion particles sequester this protein, resulting in the full derepression of the DAN1 promoter. In cells altogether lacking Mot3, this protein can still bind to and partially repress the DAN1 promoter.
Scale bars, 1 cm.
Next, we asked if genetic deletion of MOT3 conferred the same spectrum of phenotypes as [MOT3+]. To a limited extent, it did: Δmot3 cells formed ruffled colonies on ethanol media and invasive filaments on nitrogen-limited media (Figure 4B). However, other phenotypes were not fully recapitulated. Whereas [MOT3+] prions altered both the kinetics and magnitude of flocculation, Δmot3 cells were entirely nonflocculent. Most surprisingly, deletion of MOT3 did not fully derepress the URA3 reporter for Mot3 activity. Instead, derepression was only partially penetrant: Δmot3 cells were predominantly ura− but produced abundant papilla with unstable Ura+ phenotypes (Figure 4C).
What might explain the difference between [MOT3+] and Δmot3 phenotypes? Taking advantage of the facile readout of the URA3 reporter, we investigated potential gains of function by the prion. We first tested if ectopically expressed prion particles could stabilize a Ura+ phenotype in Δmot3 cells. To do so, we transformed cells with a plasmid that overexpressed full-length Mot3 from the strong TEF1 promoter. These cells became Ura+ at an increased frequency (Figure 4D). The phenotype was not maintained when the plasmid was lost, indicating that it required continuous expression of Mot3 (data not shown). To verify that the phenotype was due to PrD-mediated aggregation, and not some other effect of overexpressed Mot3, we repeated the experiment with plasmids encoding either the ΔPrD variant of Mot3 or the highly aggregation-prone PrD alone (Alberti et al., 2009). As expected, expression of ΔPrD suppressed the formation of Ura+ colonies. To our surprise, however, expression of the PrD had no effect (Figure 4D). These results suggest that the Mot3 prion templates are themselves insufficient to stabilize the Ura+ phenotype of Δmot3 cells. Instead, the stable phenotype requires the prion-mediated sequestration of a region of the Mot3 protein that is outside the prion-forming region. It seems likely that the gain of function of [MOT3+] results from the cosequestration of one or more transcriptional corepressors that interact with this region of Mot3, as illustrated in Figure 4E.
A Typical Environmental Condition, Ethanol Stress, Induces [MOT3+]
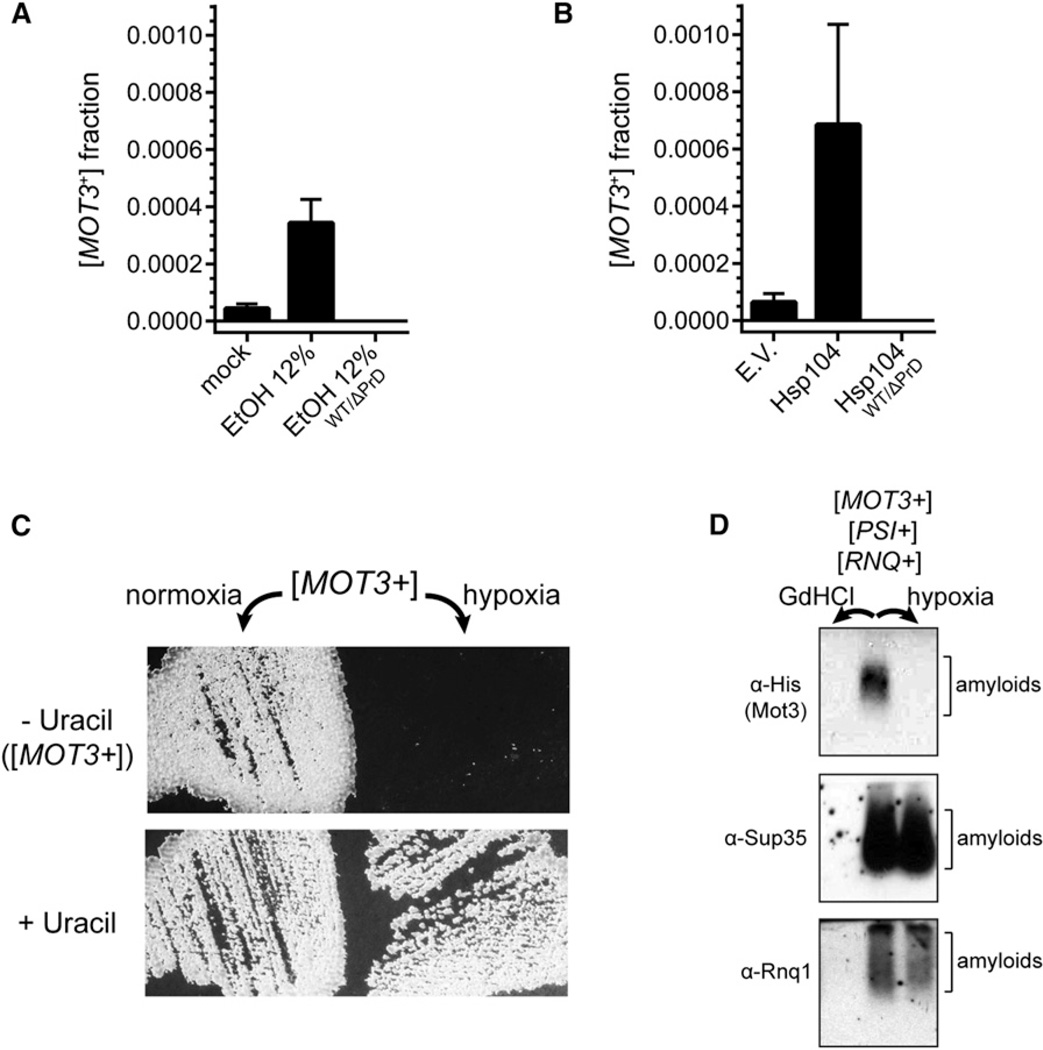
Multicellular behaviors are commonly induced by environmental stresses. At least one prion, formed by the translation termination factor, Sup35, also shares this property (Tyedmers et al., 2008). To determine if [MOT3+] can be induced by stress, we exposed [mot3−] cells to a variety of chemical stressors. Following 6 hr of treatment, cells were plated to uracil-deficient media to select for those that had acquired [MOT3+]. Most of the treatments did not change the frequency of [MOT3+] (Figure S3A; data not shown). Only one stressor, ethanol, had an effect. Treating cells for 6 hr with a concentration of ethanol that is commonly reached in wine fermentations (12%) increased the frequency of Ura+ colonies by ~10-fold (Figures 5A and S3). Cells bearing a ΔPrD MOT3 gene were unable to form Ura+ colonies in response to ethanol. Importantly, the treatment was not overtly toxic to cells, indicating that ethanol did not merely select for pre-existing [MOT3+] cells.
Figure 5. Environmental Conditions Govern Mot3 Prion Switching.
(A) The spontaneous switching of Mot3 to its prion state is increased by ethanol stress. Diploid [mot3−] cells, or as a negative control, WT/ΔPrD heterozygotes, were incubated for 6 hr in media alone (mock), or media containing 12% ethanol (EtOH), prior to plating to media lacking uracil. Ura+ colonies were counted after 4 days and normalized by the number of total colony-forming units. Data represent mean ± SD from three independent experiments.
(B) [mot3−] cells were transformed with either empty vector or a plasmid expressing Hsp104 from a strong constitutive promoter (GPD). Transformants were inoculated to rich media overnight, and then the fraction of [MOT3+] cells was determined as in (A).
(C) Passaging [MOT3+] cells under hypoxic conditions reverts them to [mot3−].
(D) The effect of hypoxia is specific to [MOT3+]. A strain harboring prion states of three different proteins ([MOT3+], [PSI+], and [RNQ+]) was treated transiently with either GdHCl or hypoxia. Amyloids representing each prion state were detected by SDD-AGE. GdHCl, which targets the prion-partitioning factor Hsp104, eliminated all three prions, whereas hypoxia specifically eliminated [MOT3+]. See also Figure S3.
How does ethanol stimulate Mot3 prion conversion? It did not simply increase Mot3 protein levels (Figure S3B). Ethanol stress causes protein misfolding and strongly induces the protein-remodeling factor Hsp104 (Figure S3B; Sanchez et al., 1992). Hsp104 has previously been found to stimulate prion conversion by other prion proteins (Shorter and Lindquist, 2006; Kryndushkin et al., 2011). To determine if Hsp104 also stimulates [MOT3+], we transformed [mot3−] cells with a plasmid expressing Hsp104 from a strong constitutive promoter. This produced a greater than 10-fold increase in the frequency of Ura+ colonies (Figure 5B). Overexpressed Hsp104 did not induce Ura+ colonies in cells bearing a ΔPrD MOT3 gene. These experiments suggest that ethanol stress accelerates the formation of [MOT3+] by perturbing protein homeostasis.
Another Typical Environmental Condition, Hypoxia, Eliminates [MOT3+]
A strategy used by yeast cells to inhibit competition from other organisms is to rapidly ferment glucose to ethanol. The subsequent respiration of that ethanol can produce an anoxic environment by the end of the respiro-fermentative cycle. Hypoxia represses MOT3 transcription (Figure S3C; Sertil et al., 2003). To test if this is sufficient to eliminate [MOT3+] prions and reset cells back to the [mot3−] state, we passaged [MOT3+] cells under hypoxic (<1% O2) conditions. After 5 days, cells were repassaged on fresh nonselective media and incubated under normal atmospheric conditions. Colonies were then replica-plated to uracil-deficient media to determine the presence or absence of [MOT3+]. The hypoxic treatment proved remarkably effective: [MOT3+] cultures reverted uniformly back to [mot3−] (Figure 5C). Cells that were passaged in parallel under normoxic conditions remained [MOT3+]. Notably, all [MOT3+]-associated phenotypes, including the ethanol- and proline-dependent multicellular growth forms, were also eliminated. Thus, by way of a [MOT3+] to [mot3−] prion switch, transient oxygen depletion induced specific and heritable changes to respiration-associated phenotypes.
To determine if the effect of hypoxia is specific to [MOT3+], we created a single yeast strain harboring the prion states of three different proteins: Mot3, Sup35, and Rnq1. We then subjected cells of this strain to hypoxia followed by SDD-AGE to detect the continued presence of amyloids of each prion protein. Mot3 amyloids were eliminated. Sup35 and Rnq1 amyloids remained (Figure 5D).
Hypoxia did not simply counterselect [MOT3+] cells (Figures S3D and S3E). To verify that the unique response of [MOT3+] to hypoxia results from the transcriptional repression of MOT3, we supplemented [MOT3+] cells with Mot3 expressed from a heterologous promoter (SUP35) that is not regulated by hypoxia. When these [MOT3+] cells were passaged under hypoxia, they retained the prion state (Figure S3F). Cells that contained empty vector did not, demonstrating that repression of MOT3 under hypoxia is required for prion elimination. This natural transcriptional regulation of MOT3 ensures that [MOT3+] prions will be eliminated from cells as they reach the end of the respiro-fermentative cycle.
[MOT3+] Produces Different Multicellular Behaviors in Different Genetic Backgrounds
Natural yeast isolates exhibit a rich diversity of multicellular behaviors and fermentative characteristics (Casalone et al., 2005; Liti et al., 2009; Granek and Magwene, 2010). The profound effects that [MOT3+] exerted on FLO11 -dependent phenotypes in the laboratory strain suggested that it might also contribute to the acquisition of new phenotypes in the wild.
To analyze Mot3 prion formation in nonlaboratory yeast, we integrated a dominant drug-resistance reporter for Mot3 activity (Figure 6A) into genetically and ecologically diverse, yet experimentally tractable, natural isolates of S. cerevisiae (Liti et al., 2009). We then overexpressed the Mot3 prion-determining region and selected [MOT3+] derivatives on drug-containing media. Of 32 strains tested (Table S3), 28 (Figure S4A) readily acquired the resistance phenotype indicative of the prion. We further focused on ten divergent strains (Figure 6B) that retained strong resistance phenotypes even after loss of the inducing plasmid (as expected for cells harboring the prion). When examined by SDD-AGE, they contained Mot3 amyloids (Figure S4C). Hypoxia eliminated the phenotype and eliminated the amyloids (Figures S4B and S4C). These results demonstrate that diverse genetic backgrounds have the capacity to acquire [MOT3+] and to naturally regulate its inheritance through a hypoxic growth phase.
Figure 6. [MOT3+] Phenotypes Are Dictated by Genetic Variation.
(A) A genetic reporter for [MOT3+] in prototrophic strains. A gene encoding resistance to the antibiotic nourseothricin (NAT) was placed under control of the DAN1 promoter, such that yeast carrying the reporter are NAT resistant only when they are [MOT3+].
(B) The reporter was integrated into genetically and ecologically diverse strains of S. cerevisiae. Each node on the phylogenetic tree (adapted from Liti et al., 2009) represents a different yeast strain that could be induced to form a NAT-resistant phenotype by overexpressed Mot3 PrD. Labels denote strains that were evaluated for [MOT3+]-dependent multicellular phenotypes. Green labels indicate that [MOT3+] produced multiple robust multicellular phenotypes, yellow indicates more limited effects of [MOT3+], and red indicates no observable effect. The two strains labeled in blue exhibited [MOT3+]-dependent changes in flocculation.
(C) [MOT3+] produces a different array of multicellular phenotypes in each strain, as demonstrated by invasive behaviors and colony morphologies in four divergent strains. ferm., fermenting.
(D) [MOT3+]-dependent morphological differences between L-1374 and UWOPS83-787.3 are reproduced by deleting MOT3. [MOT3+]-dependent multicellularity in L-1374 requires FLO11.
(E) Demonstrative qRT-PCR analyses of the expression of Mot3-regulated genes for isogenic [MOT3+], [mot3−], and Δmot3 cells in L-1374 and UWOPS83-787.3. The direct effect of Δmot3 on FLO11 was not determined. Error bars represent SD from separate experiments.
Scale bars, 1 cm.
See also Figure S4 and Tables S4 and S5.
We then compared the Flo11-dependent phenotypes of [MOT3+] isolates and their [mot3−] counterparts. These differed widely between genetic backgrounds (Figures 6C, 6D, and S4D–S4H; Table S4). Two strains, the grape/wine isolates L-1374 and Y-55, exhibited strong increases in multiple multicellular behaviors. In the absence of the prion, they were noninvasive and formed smooth colonies under all growth conditions. But when they contained [MOT3+], they became invasive on proline media and acquired complex colony morphologies on ethanol media. The colony morphologies were also manifest, to a lesser extent, on glucose media.
[MOT3+] had specific multicellular effects in other strains. In the fermenting fruit juice isolate DBVPG6040, [MOT3+] conferred a strong invasive phenotype but had no effect on colony morphology. In contrast, the tecc (honey wine) isolate DBVPG1853 had altered colony morphologies in the [MOT3+] state but acquired invasive filaments independently of prion status. Only one of the ten strains investigated, the soil isolate DBVPG1373, had no apparent multicellular effect of [MOT3+].
There are two general mechanisms by which [MOT3+] might produce different phenotypes in different genetic backgrounds. First, the physical properties of the prion (including the nature of the prion fold and its efficiency of propagation) may themselves be influenced by the genetic background, which in turn would influence how it interacts with Mot3-regulated loci. Second, [MOT3+] may regulate (directly or indirectly) genes that contain polymorphisms that are phenotypically silent in the absence of the prion. To distinguish between these possibilities, we deleted MOT3 from two genetically divergent strains, L-1374 and UWOPS83–787.3, that had very different phenotypic manifestations of [MOT3+]. If the causative genetic differences acted exclusively by influencing the prion itself, we would expect the deletion to have the same phenotypic effect in both backgrounds. If, conversely, the causative genetic differences are themselves subject to prion regulation, the genetic ablation of Mot3 activity would be expected to produce different effects in each background. As shown in Figure 6D, the latter scenario proved true: MOT3 deletions produced different effects in the two strains and, in both, phenocopied [MOT3+]. Altogether, these data demonstrate that [MOT3+] alters the expression of standing genetic variation in S. cerevisiae, resulting in combinations of multicellular phenotypes that differ between lineages.
To verify the role of FLO11 in [MOT3+]-dependent multicellularity, we deleted FLO11 from a [MOT3+] isolate of L-1374. The resulting cells lost the multicellular phenotypes of [MOT3+], despite retaining the prion itself (Figure 6D; data not shown). The number and arrangement of Mot3 binding motifs in the FLO11 promoter did not differ significantly between strains and, therefore, are unlikely to account for [MOT3+]-dependent phenotypic differences (Table S1). Instead, we discovered that the FLO11-coding region is itself highly polymorphic, with different strains harboring different repeat length variants (Figure S4I). The number of tandem repeats within cell surface adhesins correlates with their activity (Verstrepen et al., 2005), suggesting that prion formation by Mot3 could enable the expression of functional differences between FLO11 alleles. Differences in the activities of the other numerous regulators of FLO11 are quite likely to further synergize with [MOT3+] in diversifying multicellular behaviors.
To gain insight into the extent of [MOT3+]-induced transcriptional changes, we analyzed the expression of other Mot3-regulated genes in both L-1374 and UWOPS83–787.3. Using qRT-PCR, we found that [MOT3+] derepressed loci throughout the genomes of both strains (Figure 6E; Table S5). Two important trends emerged. First, the effects of genetic background were quite specific. Although multiple transcripts exhibited quantitatively different responses between strains, only FLO11 differed qualitatively: it was derepressed by [MOT3+] in L-1374 but was unaffected in UWOPS83–787.3. Second, the relative effect of [MOT3+] and Δmot3 differed among individual genes. DAN1, for example, was derepressed much more strongly by the prion than by the deletion, whereas the reverse was true for ANB1. Therefore, the gain of function of [MOT3+] appears to impact a specific subset of the Mot3 regulon. These findings illustrate that, despite genetic divergence between the strains leading to distinct multicellular effects of [MOT3+], the unique transcrip-tional response of the prion is nevertheless shared between them.
DISCUSSION
The evolutionary constraints on multicellularity are stringent (Grosberg and Strathmann, 2007). Multicellularity involves mechanisms for cell differentiation, cooperation, and the control of defector cells that might promote their own growth at the expense of the integrity of the multicellular structure. We have demonstrated that a self-perpetuating conformational switch in a protein provides a mechanism to drive multicellular growth in a “unicellular” organism.
The Mot3 transcription factor lies at the helm of a Flo11-dependent developmental program. The successful deployment of that program necessitates an epigenetic commitment that spans multiple generations. The self-perpetuating nature of prion propagation ensures that commitment. Prion conversion by Mot3 activates the multicellularity program and stably perpetuates that state to subsequent generations. By virtue of their common descent and shared prion inheritance, all cells in the lineage remain physically and metabolically connected. The complex heritable phenotypes that emerge from this cooperation would be difficult to achieve by a group of cells each acting independently.
Yeast multicellularity is deeply intertwined with carbon metabolism. The multicellularity determinants FLO11 and FLO1 are coregulated with glucose, sucrose, and starch metabolic programs (Verstrepen and Klis, 2006). Mot3 appears to be an important regulator of carbon metabolism: its targets are most heavily enriched for sugar transporters, sterol biosynthetic enzymes, and alcohol dehydrogenases (Table S2). Mathematical models suggest that multicellularity may have evolved in response to competition between carbon metabolism strategies (Pfeiffer et al., 2001), a hypothesis supported by correlations between dimorphic transitions and glucose depletion (Verstrepen and Klis, 2006). Thus, Mot3 establishes a paradigm that links prions with carbon metabolism and facultative multicellularity. Importantly, there may be many such prions. Two major regulators of carbon metabolism and multicellularity, Cyc8 and Swi1, were recently found to form prions (Du et al., 2008; Patel et al., 2009). Another prion, which involves the transmembrane proton pump, Pma1, also governs carbon source utilization (Brown and Lindquist, 2009).
Prion Formation as a Potential Function of Mot3
Mot3 is a key player in the cellular response to oxygen availability. When oxygen is limiting, Mot3 levels rapidly decrease, resulting in the derepression of its genetic targets (Sertil et al., 2003; Lai et al., 2006). Paradoxically, however, hypoxia does not produce the same multicellular responses as [MOT3+]. In fact, oxygen is strictly required for Flo11-dependent colony morphologies and filamentation (Wright et al., 1993; Zupan and Raspor, 2010). It seems that prion conversion, therefore, enables phenotypic manifestations of the Mot3 regulon that may otherwise be inaccessible.
How might Mot3 regulation, via prion formation, be advantageous during aerobic growth? [MOT3+] effectively primed cells to respond to limitations in either glucose or nitrogen. Under those conditions, yeast cells fulfill most of their carbon and nitrogen needs by oxidative metabolism. Ethanol is the predominant source of carbon in the postdiauxic phase of wine fermentations, and proline is by far the most abundant source of nitrogen in grapes (Ough and Stashak, 1974). Each of these nutrients can only be metabolized in the presence of oxygen (Ingledew et al., 1987). Indeed, the strongest [MOT3+]-dependent responses occurred when cells were forced to grow with either ethanol as a carbon source or proline as a nitrogen source. Physiological responses to these nutrients would be counterproductive during anaerobic growth. The natural regulation of MOT3 by hypoxia parsimoniously overrides the carbon- and nitrogen-limitation signals by eliminating [MOT3+].
Why should cells employ a prion—a semistochastic switch — to regulate fundamental metabolic pathways? Prion switching divides large populations into mosaics of distinct subpopulations, and recent work reveals that cell-to-cell heterogeneity in metabolic processes can be maintained through cooperative interactions that benefit the population as a whole (Beardmore et al., 2011; MacLean, 2008). For a population of [mot3−] and [MOT3+] cells, the whole may be greater than the sum of its parts.
Environmentally Regulated Inheritance
Our data are consistent with a function for [MOT3+] during postdiauxic growth. Two environmental signals that delimit the beginning and end of that period, ethanol and hypoxia, inversely regulated prion switching by Mot3 (Figure 7). These signals naturally happen in sequence: as ethanol concentrations reach a peak, yeast switch from fermentative to respirative metabolism. The oxidative respiration of ethanol and remaining sugars then depletes molecular oxygen from the local environment (Bauer and Pretorius, 2000). The ensuing hypoxia reverts cells back to the [mot3−] state. Accordingly, Mot3 prion switching may constitute an explicit mechanism that contributes to the recently discovered (Mitchell et al., 2009) capacity of yeast to adaptively “anticipate” the environmental changes that occur sequentially in the course of wine production. With the exception of phenomena that involve the uptake of foreign nucleic acids (Koonin, 2012), the elimination of [MOT3+] by hypoxia is, to our knowledge, the only example of a stably inherited phenotypic change that is induced en masse by a specific environmental change.
Figure 7. Model for a Function for Mot3 Prion Switching in the Respiro-Fermentative Cycle of Wine Yeasts.
Yeast cells begin the cycle upon inoculation to glucose-rich grape must. Glucose is fermented to ethanol during the exponential growth phase. The ensuing ethanol stress triggers prion conversion to [MOT3+], which, in conjunction with glucose exhaustion, drives some of the cells into a multicellular growth program that protects them from stress and/or increases metabolic efficiency. As the population respires ethanol and remaining sugars, oxygen levels decline resulting in the accelerated reappearance of [mot3−] cells within [MOT3+] subpopulations.
Complexity of Multicellular Phenotypes
[MOT3+] differs in two key respects from other mechanisms that underlie rapid morphological switching in yeast. First, unlike other genetic and epigenetic elements, [MOT3+] is structurally autonomous from nucleic acids, enabling it to act in a dominant and promoter-extrinsic manner to alter gene expression throughout the genome. Second, the prion is not simply an “on/off switch” for Mot3. Instead, it represses the protein’s normal activity while endowing it with new activities. Both effects are required for the full spectrum of [MOT3+] phenotypes. One simple explanation for the gain of function of [MOT3+] is that the prion particles sequester other transcriptional regulators that interact with Mot3. Indeed, Mot3 functions within a regulatory framework rife with other transcription factors whose sequences are particularly well suited for prion-like interactions. To what extent these proteins influence each other’s aggregation and how this, in turn, contributes to the complexity and plasticity of [MOT3+] phenotypes are exciting subjects for exploration.
In sum, we have demonstrated that prions formed by a yeast transcription factor enable the differentiation of yeast cells into facultative multicellular structures. The formation, inheritance, and phenotypic manifestation of the prions were dictated by natural environmental signals and by an abundance of heritable genetic variation between yeast isolates. These relationships explicate some of the natural diversity and plasticity of multicellular phenotypes and promise to further our understanding of the molecular mechanisms that enable the evolution of social behaviors.
EXPERIMENTAL PROCEDURES
Computational Analyses
Genetic targets of transcription factors were obtained from YEASTRACT (Teixeira et al., 2006) and from Wong and Struhl for the Cyc8-Tup1 complex (Wong and Struhl, 2011). Prion propensities were predicted using a previously developed algorithm (Alberti et al., 2009) with parameters updated according to experimentally confirmed prion hits. An independent algorithm (Toombs et al., 2012) was also used, with comparable results. Functional annotations were determined using the functional clustering analysis in DAVID release 6.7 (Huang et al., 2009) with the highest classification stringency. Mot3 binding motifs in the 3 kb regulatory region 5′ upstream of the FLO11 gene (Rupp et al., 1999) were identified in all strains according to a frequency matrix for the Mot3 motif (Bryne et al., 2008).
Yeast Genetics and Molecular Biology
Standard cloning procedures were used (Alberti et al., 2009). Oligos and plasmid sequences are available upon request. Chromosomal integrations were achieved using PCR-based mutagenesis and the delitto perfetto method (Goldstein and McCusker, 1999; Storici and Resnick, 2006). Yeast were transformed with a standard lithium-acetate protocol as described by Gietz et al. (1992). Yeast strains are listed in Table S3.
Yeast Media and Phenotypic Analyses
Standard yeast media and growth conditions were used. Where appropriate, glucose was replaced with 2% ethanol or glycerol, and ammonium sulfate was replaced with 0.05% proline. Semisolid media contained 0.3% agar. For agar invasion analyses, colonies were allowed to grow for 5 days at 25°C. Plates were photographed before and after the removal of noninvasive cells by dislodging surface cells under running water. Flocculation was assayed after growth to saturation in YPD containing 20% glucose.
Hypoxia Treatments
Cells were point inoculated to YPD plates supplemented with Tween 80 and ergosterol (Hongay et al., 2002) and placed in a sealed bag containing a BD GasPak EZ Anaerobe Pouch System sachet. After 5 days at 30°C, cells were retrieved from the outer perimeter of colonies to a fresh YPD plate and allowed to form colonies under standard laboratory conditions prior to subsequent analyses.
SDD-AGE
SDD-AGE was performed as described (Halfmann et al., 2012).
qRT-PCR
Total RNA was extracted using MasterPure Yeast RNA Purification Kit (Epicenter) as instructed by the manufacturer. cDNAs were generated using oligo(dT)20 and the Superscript III First-Strand Synthesis System (Invitrogen). qPCR was carried out using the DyNAmo HS SYBR Green qPCR Kit (Thermo Fisher Scientific) on the HT 7900 Real-Time PCR System (Applied Bio-systems). RQ values were normalized against those for ACT1 or TFC1. Primers are listed in Table S6.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported primarily by the NIH Director’s Early Independence Award Program, grant number DP5-OD009152-01 (to R.H.) and by the Sara and Frank McKnight Fellowship (R.H.). S.L. is a Howard Hughes Medical Institute (HHMI) investigator. This work was supported by grants from the G. Harold and Leila Y. Mathers Foundation and HHMI (to S.L.).We thank Cintia Hongay for fruitful early discussions about Mot3, Oliver King for assistance with the prion prediction algorithm, and Sunil Laxman and members of the S.L. lab for critical reading of the manuscript. We thank Gerry Fink and Fred Winston for yeast strains Σ1278b and FY2609, respectively, and Mikko Taipale for unpublished plasmids.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, four figures, and six tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2013.02.026.
REFERENCES
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrales RR, Korber P, Jimenez J, Ibeas JI. Chromatin modulation at the FLO11 promoter of Saccharomyces cerevisiae by HDAC and Swi/ Snf complexes. Genetics. 2012;191:791–803. doi: 10.1534/genetics.112.140301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer FF, Pretorius IS. Yeast stress response and fermentation efficiency: how to survive the making of wine – a review. S. Afr. J. Enol. 2000;21:27–51. [Google Scholar]
- Beardmore RE, Gudelj I, Lipson DA, Hurst LD. Metabolic trade-offs and the maintenance of the fittest and the flattest. Nature. 2011;472:342–346. doi: 10.1038/nature09905. [DOI] [PubMed] [Google Scholar]
- Brown JC, Lindquist S. A heritable switch in carbon source utilization driven by an unusual yeast prion. Genes Dev. 2009;23:2320–2332. doi: 10.1101/gad.1839109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brückner S, Mösch HU. Choosing the right lifestyle: adhesion and development in Saccharomyces cerevisiae . FEMS Microbiol. Rev. 2012;36:25–58. doi: 10.1111/j.1574-6976.2011.00275.x. [DOI] [PubMed] [Google Scholar]
- Bryne JC, Valen E, Tang MH, Marstrand T, Winther O, da Piedade I, Krogh A, Lenhard B, Sandelin A. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 2008;36:D102–D106. doi: 10.1093/nar/gkm955. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumgarner SL, Neuert G, Voight BF, Symbor-Nagrabska A, Grisafi P, van Oudenaarden A, Fink GR. Single-cell analysis reveals that noncoding RNAs contribute to clonal heterogeneity by modulating transcription factor recruitment. Mol. Cell. 2012;45:470–482. doi: 10.1016/j.molcel.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter GW, Prinz S, Neou C, Shelby JP, Marzolf B, Thorsson V, Galitski T. Prediction of phenotype and gene expression for combinations of mutations. Mol. Syst. Biol. 2007;3:96. doi: 10.1038/msb4100137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casalone E, Barberio C, Cappellini L, Polsinelli M. Characterization of Saccharomyces cerevisiae natural populations for pseudohyphal growth and colony morphology. Res. Microbiol. 2005;156:191–200. doi: 10.1016/j.resmic.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN(+)] Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- Du Z, Park KW, Yu H, Fan Q, Li L. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae . Nat. Genet. 2008;40:460–465. doi: 10.1038/ng.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira PC, Ness F, Edwards SR, Cox BS, Tuite MF. The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol. Microbiol. 2001;40:1357–1369. doi: 10.1046/j.1365-2958.2001.02478.x. [DOI] [PubMed] [Google Scholar]
- Gietz D, St Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR. Unipolar cell divisions in the yeast S cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae . Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Granek JA, Magwene PM. Environmental and genetic determinants of colony morphology in yeast. PLoS Genet. 2010;6:e1000823. doi: 10.1371/journal.pgen.1000823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold CK, Masel J. Complex adaptations can drive the evolution of the capacitor [PSI], even with realistic rates of yeast sex. PLoS Genet. 2009;5:e1000517. doi: 10.1371/journal.pgen.1000517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosberg RK, Strathmann RR. The evolution of multicellularity: a minor major transition? Annu. Rev. Ecol. Evol. Syst. 2007;38:621–654. [Google Scholar]
- Hahn MW, De Bie T, Stajich JE, Nguyen C, Cristianini N. Estimating the tempo and mode of gene family evolution from comparative genomic data. Genome Res. 2005;15:1153–1160. doi: 10.1101/gr.3567505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R, Lindquist S. Epigenetics in the extreme: prions and the inheritance of environmentally acquired traits. Science. 2010;330:629–632. doi: 10.1126/science.1191081. [DOI] [PubMed] [Google Scholar]
- Halfmann R, Jarosz DF, Jones SK, Chang A, Lancaster AK, Lind-quist S. Prions area commonmechanism for phenotypic inheritancein wild yeasts. Nature. 2012;482:363–368. doi: 10.1038/nature10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halme A, Bumgarner S, Styles C, Fink GR. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell. 2004;116:405–415. doi: 10.1016/s0092-8674(04)00118-7. [DOI] [PubMed] [Google Scholar]
- Hongay C, Jia N, Bard M, Winston F. Mot3 is a transcriptional repressor of ergosterol biosynthetic genes and is required for normal vacuolar function in Saccharomyces cerevisiae . EMBO J. 2002;21:4114–4124. doi: 10.1093/emboj/cdf415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Sherman BT, Lempicki RA. Systematic and integra-tive analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Ingledew WM, Magnus CA, Sosulski FW. Influence of oxygen on proline utilization during the wine fermentation. Am. J. Enol. Vitic. 1987;38:246–248. [Google Scholar]
- Koonin EV. Does the central dogma still stand? Biol. Direct. 2012;7:27. doi: 10.1186/1745-6150-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koschwanez JH, Foster KR, Murray AW. Sucrose utilization in budding yeast as a model for the origin of undifferentiated multicellularity. PLoS Biol. 2011;9:e1001122. doi: 10.1371/journal.pbio.1001122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin DS, Engel A, Edskes H, Wickner RB. Molecular chaperone Hsp104 can promote yeast prion generation. Genetics. 2011;188:339–348. doi: 10.1534/genetics.111.127779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni AA, Abul-Hamd AT, Rai R, El Berry H, Cooper TG. Gln3p nuclear localization and interaction with Ure2p in Saccharomyces cere-visiae . J. Biol. Chem. 2001;276:32136–32144. doi: 10.1074/jbc.M104580200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuthan M, Devaux F, Janderová B, Slaninová I, Jacq C, Palková Z. Domestication of wild Saccharomyces cerevisiae is accompanied by changes in gene expression and colony morphology. Mol. Microbiol. 2003;47:745–754. doi: 10.1046/j.1365-2958.2003.03332.x. [DOI] [PubMed] [Google Scholar]
- Lai LC, Kosorukoff AL, Burke PV, Kwast KE. Metabolic-state-dependent remodeling of the transcriptome in response to anoxia and subsequent reoxygenation in Saccharomyces cerevisiae . Eukaryot. Cell. 2006;5:1468–1489. doi: 10.1128/EC.00107-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster AK, Bardill JP, True HL, Masel J. The spontaneous appearance rate of the yeast prion [PSI+] and its implications for the evolution of the evolvability properties of the [PSI+] system. Genetics. 2010;184:393–400. doi: 10.1534/genetics.109.110213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebman SW, Chernoff YO. Prionsinyeast. Genetics. 2012;191:1041–1072. doi: 10.1534/genetics.111.137760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Styles CA, Fink GR. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics. 1996;144:967–978. doi: 10.1093/genetics/144.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean RC. The tragedy of the commons in microbial populations: insights from theoretical, comparative and experimental studies. Heredity (Edinb) 2008;100:471–477. [PubMed] [Google Scholar]
- McGlinchey RP, Kryndushkin D, Wickner RB. Suicidal [PSI+] is a lethal yeast prion. Proc. Natl. Acad. Sci. USA. 2011;108:5337–5341. doi: 10.1073/pnas.1102762108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A, Romano GH, Groisman B, Yona A, Dekel E, Kupiec M, Dahan O, Pilpel Y. Adaptive prediction of environmental changes by microorganisms. Nature. 2009;460:220–224. doi: 10.1038/nature08112. [DOI] [PubMed] [Google Scholar]
- Octavio LM, Gedeon K, Maheshri N. Epigenetic and conventional regulation is distributed among activators of FLO11 allowing tuning of population-level heterogeneity in its expression. PLoS Genet. 2009;5:e1000673. doi: 10.1371/journal.pgen.1000673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ough CS, Stashak RM. Further studies on proline concentration in grapes and wines. Am. J. Enol. Vitic. 1974;25:7–12. [Google Scholar]
- Ozcan S, Johnston M. Function and regulation of yeast hexose transporters. Microbiol. Mol. Biol. Rev. 1999;63:554–569. doi: 10.1128/mmbr.63.3.554-569.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel BK, Gavin-Smyth J, Liebman SW. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat. Cell Biol. 2009;11:344–349. doi: 10.1038/ncb1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer T, Schuster S, Bonhoeffer S. Cooperation and competition in the evolution of ATP-producing pathways. Science. 2001;292:504–507. doi: 10.1126/science.1058079. [DOI] [PubMed] [Google Scholar]
- Pigliucci M. Is evolvability evolvable? Nat. Rev.Genet. 2008;9:75–82. doi: 10.1038/nrg2278. [DOI] [PubMed] [Google Scholar]
- Ratcliff WC, Denison RF, Borrello M, Travisano M. Experimental evolution of multicellularity. Proc. Natl. Acad. Sci. USA. 2012;109:1595–1600. doi: 10.1073/pnas.1115323109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds TB, Fink GR. Bakers’ yeast, a model for fungal biofilm formation. Science. 2001;291:878–881. doi: 10.1126/science.291.5505.878. [DOI] [PubMed] [Google Scholar]
- Rogoza T, Goginashvili A, Rodionova S, Ivanov M, Viktorovskaya O, Ru-bel A, Volkov K, Mironova L. Non-Mendelian determinant [ISP+] in yeast is a nuclear-residing prion form of the global transcriptional regulator Sfp1. Proc. Natl. Acad. Sci. USA. 2010;107:10573–10577. doi: 10.1073/pnas.1005949107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp S, Summers E, Lo HJ, Madhani H, Fink G. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 1999;18:1257–1269. doi: 10.1093/emboj/18.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y, Taulien J, Borkovich KA, Lindquist S. Hsp104 is required for tolerance to many forms of stress. EMBO J. 1992;11:2357–2364. doi: 10.1002/j.1460-2075.1992.tb05295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpute-Krishnan P, Serio TR. Prion protein remodelling confers an immediate phenotypic switch. Nature. 2005;437:262–265. doi: 10.1038/nature03981. [DOI] [PubMed] [Google Scholar]
- Serio TR, Cashikar AG, Kowal AS, Sawicki GJ, Moslehi JJ, Serpell L, Arnsdorf MF, Lindquist SL. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- Sertil O, Kapoor R, Cohen BD, Abramova N, Lowry CV. Synergistic repression of anaerobic genes by Mot3 and Rox1 in Saccharomyces cerevisiae . Nucleic Acids Res. 2003;31:5831–5837. doi: 10.1093/nar/gkg792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol. Cell. 2006;23:425–438. doi: 10.1016/j.molcel.2006.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storici F, Resnick MA. The delitto perfetto approach to in vivo site-directed mutagenesis and chromosome rearrangements with synthetic oligonucleotides in yeast. Methods Enzymol. 2006;409:329–345. doi: 10.1016/S0076-6879(05)09019-1. [DOI] [PubMed] [Google Scholar]
- Suzuki G, Shimazu N, Tanaka M. A yeast prion, Mod5, promotes acquired drug resistance and cell survival under environmental stress. Science. 2012;336:355–359. doi: 10.1126/science.1219491. [DOI] [PubMed] [Google Scholar]
- Teixeira MC, Monteiro P, Jain P, Tenreiro S, Fernandes AR, Mira NP, Alenquer M, Freitas AT, Oliveira AL, Sá-Correia I. The YEASTRACT database: a tool for the analysis of transcription regulatory associations in Saccharomyces cerevisiae . Nucleic Acids Res. 2006;34(Database issue):D446–D451. doi: 10.1093/nar/gkj013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toombs JA, Petri M, Paul KR, Kan GY, Ben-Hur A, Ross ED. De novo design of synthetic prion domains. Proc. Natl. Acad. Sci. USA. 2012;109:6519–6524. doi: 10.1073/pnas.1119366109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- Türkel S. Hyperosmotic stress represses the transcription of HXT2 and HXT4 genes in Saccharomyces cerevisiae . Folia Microbiol. (Praha) 1999;44:372–376. doi: 10.1007/BF02903707. [DOI] [PubMed] [Google Scholar]
- Tyedmers J, Madariaga ML, Lindquist S. Prion switching in response to environmental stress. PLoS Biol. 2008;6:e294. doi: 10.1371/journal.pbio.0060294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Váchová L, Stovícek V, Hlavácek O, Chernyavskiy O, Stěpánek L, Kubí-nová L, Palková Z. Flo11p, drug efflux pumps, and the extracellular matrix cooperatetoform biofilm yeast colonies. J. Cell Biol. 2011;194:679–687. doi: 10.1083/jcb.201103129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstrepen KJ, Klis FM. Flocculation, adhesion and biofilm formation yeasts. Mol. Microbiol. 2006;60:5–15. doi: 10.1111/j.1365-2958.2006.05072.x. [DOI] [PubMed] [Google Scholar]
- Verstrepen KJ, Derdelinckx G, Verachtert H, Delvaux FR. Yeast flocculation: what brewers should know. Appl. Microbiol. Biotechnol. 2003;61:197–205. doi: 10.1007/s00253-002-1200-8. [DOI] [PubMed] [Google Scholar]
- Verstrepen KJ, Jansen A, Lewitter F, Fink GR. Intragenic tandem repeats generate functional variability. Nat. Genet. 2005;37:986–990. doi: 10.1038/ng1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae . Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- Wickner RB, Edskes HK, Bateman D, Kelly AC, Gorkovskiy A. The yeast prions [PSI+] and [URE3] are molecular degenerative diseases. Prion. 2011;5:258–262. doi: 10.4161/pri.5.4.17748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KH, Struhl K. The Cyc8-Tup1 complex inhibits transcription primarily bymasking the activation domain of the recruiting protein. Genes Dev. 2011;25:2525–2539. doi: 10.1101/gad.179275.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright RM, Repine T, Repine JE. Reversible pseudohyphal growth in haploid Saccharomyces cerevisiae is an aerobic process. Curr. Genet. 1993;23:388–391. doi: 10.1007/BF00312623. [DOI] [PubMed] [Google Scholar]
- Zupan J, Raspor P. Invasive growth of Saccharomyces cerevisiae depends on environmental triggers: a quantitative model. Yeast. 2010;27:217–228. doi: 10.1002/yea.1746. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.