Abstract
Dietary restriction (DR) extends lifespan in a wide range of animal models. A major obstacle to understanding how DR modulates lifespan and aging-related dysfunction is the multiplicity of physiological and molecular changes associated with DR. Unraveling their importance to the longevity effect of DR remains a major challenge. In this Perspective, we review the marked genetic variation in the response to DR of multiple recombinant inbred (RI) mouse strains. We illustrate how this genetic variation can be exploited to probe the mechanisms mediating lifespan extension by DR, as well as uncover its limits as an intervention. RI strains exhibit marked variation in their lifespan as well as physiological responses to DR. Quantitative genetic and statistical tools can use this phenotypic variation to probe the importance of physiological and molecular changes that have been hypothesized to play roles in DR-mediated lifespan extension.
Keywords: Dietary Restriction, Lifespan, Recombinant Inbred, Mouse, Genetics
Introduction
Aging is a complex, multi-factorial process that likely results from many biological processes. The predominant genetic approach has been to target specific genes using transgenic approaches to overexpress or nullify specific genes in order to identify genes that can influence aging (Liang et al., 2003). The dramatic success of this approach is evidence of its merit. However, modulation of traits such as aging are undoubtedly multigenic and the single-gene approach inherent in most transgenic studies may not adequately address the goal to identify the multifactorial causality underlying aging. Indeed, single-gene-modulated models of extended longevity are likely the outcome of pleiotropic actions across tissues and over time, themselves dependent on multigenic interactions. Natural genetic variation has been recognized as a powerful means to probe mechanisms of prolongevity for several decades (Johnson and Wood, 1982). Such studies, when undertaken in mammals, have used the genetic variation associated with multiple laboratory mouse strains to probe the biological basis for strain-differences in lifespan and other aging traits (Yuan et al., 2011) (http://phenome.jax.org). Until recently, however, there have been no systematic studies of strain variation in the response to dietary restriction (DR), one of the most robust lifespan-extending interventions in a variety of taxa (Weindruch and Walford, 1988). Examining genetic variation of particular aging phenotypes, such as longevity, across genetically distinct strains that have withstood the test of laboratory survival provides an alternative approach to the single-gene approaches to probing mechanisms of aging. This inherently polygenic approach to the study of aging can confirm, modify and even provide novel insights into the genetic, and ultimately, physiological and molecular underpinnings of longevity. In other words, we can identify the array of genes that modulate and are thus responsible for longevity and other aging traits by screening for phenotypic variation across a species (i.e. forward genetics). Such an approach may also uncover genetic variation in the mechanisms of longevity, i.e., that genetic context may determine efficacy of a given anti-aging intervention, be it nutritional, pharmacologic, or genetic.
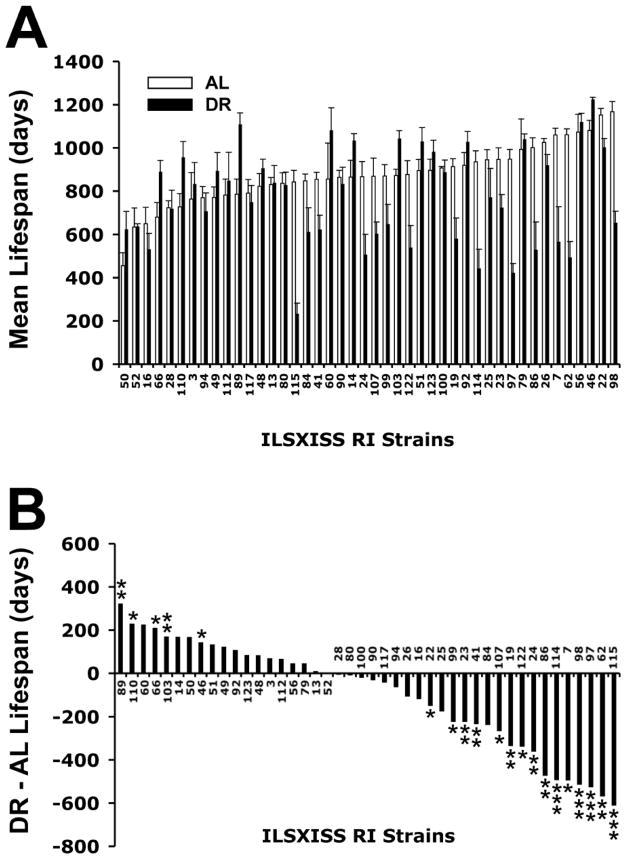
As a first step to study the longevity effects of DR in a wide variety of mouse genetic backgrounds, we took advantage of one of the largest groups of recombinant inbred (RI) strains of mice currently available, ILSXISS (Williams et al., 2004), now maintained at the Jackson Laboratory. We tested the hypothesis that lifespan response to DR would exhibit genetic variation across these genotypes. As shown in Figure 1, a major finding was that, contrary to an extensive literature, at least as many strains showed lifespan shortening by DR as showed lifespan lengthening (Liao et al., 2010a; Rikke et al., 2010). Although the negative effects of DR especially have garnered critical commentary (Mattson, 2010), the profound genetic variation observed in these studies is consistent with recent comparative and meta-analytic studies showing that life extension by DR is genetically dependent and variable in a variety of taxa (Nakagawa et al., 2012; Swindell, 2012). Also, a recent study of DR in a large number yeast mutants strikingly mirrors our result, showing a continuum of responses from life-shortening to –lengthening (Johnson et al., 2012; Schleit et al., 2012). Variation in the effect of DR on longevity has recently extended to the long-lived rhesus monkeys. One study provided evidence that DR extends lifespan (Colman et al., 2009) while a more recent study reported that DR had no effect on animals’ survival (Mattison et al., 2012). In principle, these results should not be surprising given the knowledge that intra-species genetic variation is a fundamental tenet of biology. They also raise the likelihood that, although DR and its presumptive pharmacologic mimetics are attractive as a means to modulate aging in humans, the benefits are likely to vary between individuals. Indeed, DR and its mimetics, as indicated by aforementioned studies, could even be deleterious for some. In this Perspective, we discuss these and other issues arising from our DR studies in ILSXISS RI mice and also discuss avenues of aging research needed to follow up on these studies.
Figure 1.
Strain variation in mean lifespan of ILSXISS recombinant inbred (RI) mice fed ad libitum (AL) or dietary restriction (DR; 60% AL) diets. Lifespans were obtained from 10 AL and 10 DR mice from each strain (data are combined for 5 males and 5 females in each dietary group). Mean lifespans in panel A are ranked in ascending order according the AL means. Panel B illustrates the deviation (positive and negative) of the mean DR lifespan from the mean AL lifespan for the same strains, ranked from the strain with the greatest increase in lifespan under DR to the strain with the greatest decrease. Error bars represent SEM. * p<0.05; ** p<0.01, *** p<0.001 by t-test (no experiment-wise Bonferroni correction). Data redrawn from Liao et al., 2010a.
How representative are the ILSXISS RI strains as a model for examining genetic variation in the response to DR?
The unexpected finding that DR shortened lifespan in as many strains as the number showing lengthened lifespan is a departure from the literature (Liao et al., 2010). An obvious concern is that the ILSXISS RI strains may not be representative of either non-laboratory mice or even the commonly used laboratory strains that are considered good models for human genetic variation. Indeed, it was argued that the lifespan shortening effects of DR in these strains is a specific and thus somewhat trivial consequence of “fierce competition” among cagemates for limited food—a result of the mice being housed multiply (Mattson, 2010). Mattson argued that the results are therefore confounded by lifespan-shortening effects of competition stress counteracting and, in some cases, overwhelming lifespan lengthening effects of DR. In response, we published body weight and observational studies providing no evidence of a dominance hierarchy (Liao et al., 2010a). The strongest evidence against competition stress or any aspect of multiple housing as a factor was that both lifespan shortening and lengthening effects of DR were also observed when strains were housed singly (Liao et al., 2010b). It should also be noted that the variation of lifespan responses to DR as well as lifespan shortening by DR was replicated in two separate cohorts of ILSXISS RI strains housed at two different sites: one in San Antonio, TX and the other at Boulder, CO (Liao et al., 2010a; Rikke et al., 2010). Moreover, the strain variation in lifespan observed under ad libitum (AL) feeding was similar in range to that reported for the classic inbred murine models that have been used for genetic studies for nearly a century (Yuan et al., 2009). Indeed, the longest lived ILSXISS strains have mean lifespans that not only exceed those of the classic inbred strains (Yuan et al., 2009), but match the lifespans of the longest-lived mouse models reported in the literature, including the dwarf mice (Bartke, 2008; Bartke et al., 2001). Because the intent is to use strain variation to identify the genes and underlying processes mediating DR longevity, variation in lifespan is an absolute necessity. Thus, even if the strains are unrepresentative in terms of the response of other mouse strains or of humans to DR, their profound genetic variation in response to DR remains a tool for understanding biological mechanism. Indeed, it is arguable that the model is more informative biologically than a model in which the variable response to DR was only in the life extending spectrum. A spectrum of response from positive to negative provides the basis for identifying both the genes and mechanisms they specify that give rise to lifespan extension and those that give rise to lifespan shortening. Understanding both life extension and life shortening is informative from the point of understanding the mechanisms that enhance or limit the anti-aging effects of DR. The results of this research have already identified such: namely, factors related to the response of fat to DR (Liao et al., 2011), rate of hair regrowth and recovery of fertility upon return to AL feeding (Rikke et al., 2010) appear to play a role in both the beneficial and deleterious effects of DR. It is the magnitude of these response that distinguishes the negative from the positive responders. For example, the strains with the greatest reduction of adiposity were at greatest risk for lifespan shortening and, conversely, those with the least or even no reduction in adiposity were more likely to exhibit lifespan extension by DR (Liao et al., 2011) Determining the generality of the lifespan shortening effect of DR and the profound variation overall in response to DR requires similar studies in other genetically diverse populations such as the BXD RI lines (Lang et al., 2010) and the conventional inbred strains from the Jackson Laboratory (Yuan et al., 2009). Determining the effects of DR in F1 crosses of the ILSXISS RI’s or other groups of inbred strains is particularly important -- to determine whether the lifespan shortening effects are consequences of homozygosity of recessive alleles. There have been too few studies of DR in wild-derived (Harper et al.,2006) and F1 hybrids (Turturro et al., 1999) to draw any conclusions. Our study will also be informed by studies at different degrees of DR (e.g., 30%, 20%, 10% reductions from AL intake). If, for example, studies in other models or at other degrees of DR reveal qualitatively similar variation of lifespan modulation by DR, the notion that DR is not universally life extending and/or can be life shortening will be strengthened. Further study is needed to establish more clearly both the genetic/physiologic and environmental the limits of DR as a life-extending mechanism. Whether or not the results from the ILSXISS studies are matched using intercrosses or other allellically heterogeneous populations, the genetic variation of this model provides an useful tool for probing genetic, physiological and biochemical factors underlying DR effects on aging.
The influence of genetic background in lifespan response to DR
Mice are a powerful tool for research on the genetics of mammalian aging (Yuan et al., 2011). During the past two decades, genetic manipulation, utilizing spontaneous mutations or transgenic mice, has demonstrated that single genes can have marked effects on lifespan (Liang et al., 2003). However, most of these studies have been conducted in only one genetic background. The impact on longevity of manipulating a single gene is likely to vary depending on the genetic background on which it is expressed. The few studies that have examined a life-extending genetic manipulation on more than one background have revealed the importance of genetic background on the outcome. In at least one study (Garcia et al., 2008) that, is seldom cited, genetic background reversed the outcome of DR -- from life-extending to life-shortening—a result that parallels our findings. DR extended lifespan of Propdf Ames dwarf mice (Bartke et al., 2001), but shortened lifespan of Propdf mice on the C57BL/6 background (Garcia et al., 2008). It is also noteworthy that lifespan increases induced by the Propdf allele under ad libitum feeding also show marked variation, and some of this variation correlates with the different backgrounds on which it has been placed (Bartke et al., 2001; Brown-Borg et al., 1996; Garcia et al., 2008). Remarkably, few studies showing lifespan extension in transgenic or knockout models have been conducted in more than one strain, raising the question of the extent to which the identified genes are robust in extending lifespan across disparate genotypes.
Using multiple mouse strains can mitigate against strain-specific idiosyncrasies that can complicate interpretation of data from a single genetic background. Clearly, when using a rodent model to test the mechanisms underlying the DR effects, or the effects of any genetic or pharmacologic intervention that affects lifespan, the choices of species and strain, as well as sex, can have a large effect on the outcome. Had the first DR studies been done in strains or a sex that exhibit minimal benefit from life extension by DR (such as DBA/2 males, and many of the RI strains in Liao et al. 2010a), this very informative line of research pioneered by the work of McCay and colleagues (1935) might not have been pursued. Studies using RI mouse strains highlight the marked genetic differences that exist among strains of mice, and raise the question of how robust are observations limited to the strains that are commonly used to make transgenic and knockout mice.
Is there a species-specific limit to lifespan extension in mice?
Of note, as shown in Figure 1A, the maximum average lifespan achieved by any RI strain under DR did not exceed that achieved by the longest lived strains under AL (Liao et al., 2010a). Also, there was no overlap between the strains with longest lifespans under DR and those with longest lifespans under AL feeding. Moreover, a cursory survey of the literature on murine longevity indicates that no mouse strain, whether spontaneously mutated, genetically modified, or pharmacologically treated, has been reported to have an average lifespan exceeding that observed in these RI strains (Bartke, 2008; Bartke et al., 2001; Harrison et al., 2009; Liang et al., 2003). If upheld by further studies, this observation has several implications. First, it suggests there is a ceiling to lifespan extension in mice that is limited by the allelic variation intrinsic to the species. Second, if there are a limited number of pathways by which lifespan can be extended, the lifespan extending biochemical pathways modulated by DR (e.g., insulin/IGF-1, mTOR, Sirtuins; see following discussion) may already be maximally modulated in terms of their longevity potential in strains that are long lived under AL conditions. One reason that some of the longest-lived strains showed lifespan shortening under DR might be that further activation (or suppression if the pathway extends life by being downregulated) becomes deleterious and hence lifespan shortening. These intriguing hypotheses are amenable to experimental testing. For example, one experiment would be to evaluate the aforementioned anti-aging pathways in the longest-lived strains under AL feeding and in the longest-lived strains (which are distinct from the AL strains) under DR. One hypothesis would be that among the longest-lived strains under either AL or DR feeding, insulin/IGF-1 signaling will be similarly reduced, and/or mTOR pathways will be similarly reduced, etc. Likewise, among the shortest-lived strains under AL and DR conditions, respectively, those life-prolonging pathways will be similarly attenuated in relation to their activation/suppression in the longest-lived models. This is only one possible outcome of such a study, but it exemplifies a new and potentially useful direction in aging research that emanates from the multi-strain RI approach to aging research.
Phenotypic strain variation across RI-strains provides an unbiased evaluation of mechanisms hypothesized to mediate lifespan modulation by DR
DR, because of its robust lifespan extending effect, has been a widely used tool for elucidating the mechanisms of aging, using both invertebrate and rodent models (McCay et al., 1935; Weindruch and Walford, 1988). A major obstacle to elucidating the molecular mechanisms of DR is the vast array of physiological, cellular, and biochemical changes induced by DR that are potentially causal (Masoro, 2005; Speakman and Mitchell, 2011). By using multiple RI strains, we can test the role of potential anti-aging DR-associated traits by exploiting the marked strain variation in those traits and determining whether that variation corresponds to the strain variation in the effect o DR on lifespan. This strain variation enables determination of the genetic factors mediating and hence playing a role in lifespan modulation by DR. If any plausible trait indeed involves in lifespan modulation, the relationship between traits and lifespan should be correlated in multiple strains. As an example, we tested the hypothesis that fat reduction under DR is important for life extension in ILSXISS RI strains. We found that the reduction of adiposity, which exhibited marked genetic variation, was inversely correlated to the extension of lifespan by DR (Liao et al., 2011). Thus, strains with the least reduction in fat were more likely to show life extension, and those with the greatest reduction were more likely to have shortened lifespan under DR. This result counters the notion that reduction of fat mass, presumably by reducing deleterious metabolic effects associated with obesity, contributes to the life-extension effect of DR (Barzilai and Gabriely, 2001; Barzilai and Gupta, 1999). The result suggests instead that factors associated with maintaining adiposity are important for survival and life extension under DR. The informative results from the adiposity studies are evidence of the value of this model for dissecting the importance of other potential traits implicated in DR’s anti-aging actions (Masoro, 2005), which presumably also exhibit genetic variation across RI strains. High-throughput phenotyping experiments of the effect of DR on these strains, including RNA and protein expression profiling, are obvious examples of ways to capitalize on the power of this approach to provide genetic and mechanistic insight to the longevity and healthspan modulating effects of DR.
Although the general effect of life extension by DR has been demonstrated in multiple inbred, laboratory rodents (Weindruch and Walford, 1988), the numbers of strains examined have been small and thus do not represent the various genotypes present and the various responses of DR on lifespan that would be expected in humans. Using a small number of animal strains without cognizance of the impact of strain variation on outcome or failing to consider the differential sensitivity to each stimulus may bias or mislead the interpretation of experimental findings. Correlation of plausible traits between two strains offers extremely limited evidence of an association, let alone causality. Too often such differences have been over-interpreted. For example, in a recent study using only two inbred strains, the conclusion was drawn that only relatively obese mice, as represented by the C57BL/6J strain benefited from DR. The leaner strain was DBA/2J, which did not show life extension in that study (Sohal et al., 2009). The results were interpreted to indicate that life extension by DR only occurs in genotypes with a positive energy imbalance (i.e., gaining weight) under AL feeding. In the report, body weight under AL feeding increased from 4 to 22 mo of age in the C57BL/6J mice, whereas body weight did not increase significantly in DBA/2J mice during that period (Forster et al., 2003). If this conclusion were correct, one would expect a positive correlation between body weight change with age under AL feeding and lifespan under DR in multiple mouse strains. However, we found no correlation between AL body weight change from 3 to 22 mo and DR lifespan in ILSXISSS RI strains (males: r = 0.11, P = 0.50; females: r = −0.11, P = 0.50; data is derived from the study in Liao et al. 2011(Liao et al., 2011)). This result illustrates the fallacy of drawing conclusions from 2-strain comparisons, yet such comparisons are often made in the study of DR.
The other advantage of using a panel of RI mice is the potential to identify chromosomal regions that modulate the traits under study—in this case, those that modulate the response to DR. These regions can be identified by quantitative trait loci (QTL) mapping, which is the first step to identify candidate genes that underlie quantitative traits. We have found significant correlations between lifespan and several traits that have been hypothesized to mediate the response to DR in RI mice. These correlations at the least suggest that the correlated variables have a genetically shared basis. For example, we identified correlations and shared QTLs between the lifespan and fertility responses to DR (Rikke et al., 2010) as well as lifespan and the fuel efficiency response to DR (Rikke et al., 2010). Congenics of the two allelic variants at this “fuel efficiency” locus confirmed its role in the weight-loss response to DR—evidence that with greater power, QTL mapping using RI strains will uncover informative loci. These studies have also found that some responses to DR are not correlated and hence are uncoupled. For example, body temperature and BW responses to DR are uncoupled in the ILSXISS RI strains (Rikke and Johnson, 2007). Identifying which responses are coupled and which are not can lead to a more comprehensive understanding of the genetically specified factors involved in the DR response.
There is growing evidence that many life-extending pathways identified by genetic manipulation in other animal models may also be exploited by DR: these include reduced insulin/IGF-1 signaling (Bartke et al., 2007), reduced mTOR signaling (Fontana et al., 2010), and up-regulation of Sirtuin-mediated pathways (Haigis and Guarente, 2006; Masoro, 2004; Picard and Guarente, 2005). Definitive experiments establishing linkage between these pathways and the DR response are lacking. RI strains can be used to test for correlation and genetic coregulation of these hypothesized causal pathway and lifespan modulation by DR. For example, if reduced mTOR signaling plays a role in life extension under DR, the magnitiude of reduction in mTOR signaling (e.g., mTOR activity or downstream effectors) should be directly correlated with lifespan extension under DR. Moreover, some genetic loci regulating the two DR responses (mTOR and longevity) should be shared and identifiable. In this way, the use of the RI model approach can be used as an important tool for probing the mechanisms whereby DR modulates lifespan.
In summary, life span and other physiologic responses to DR are genetically specified traits that exhibit marked variation across a panel of RI strains, and are likely to do so in other genetically diverse animal models. Our studies have revealed the power of the genetic variation found in RI mouse strains to identify the relative importance of hypothesized traits and mechanisms in the actions of DR. Although further study using varying DR protocols and other genetic models is needed to establish the generality of the lifespan shortening effect by DR, the large number of RI strains found to respond to DR with shortened lifespan is sobering and raises an important caveat to the widely held assumption that DR is almost universally effective in extending longevity.
Highlights.
We review variable lifespan response of recombinant inbred mice to diet restriction (DR).
We discuss significance of shortened lifespan responses of some strains to DR.
We show how this genetic variation can be used to probe mechanisms of DR.
We show how this genetic variation can reveal limits of anti-aging interventions.
Acknowledgments
We thank Vivian Diaz, Matt Battaglia, Christine Martin, Colin Larson, Galen Miller, Kristina Williams, and John Yerg for data collection and animal husbandry. We thank Phyllis Carosone-Link for managing the data base and assisting with the statistical analyses. We also thank the many undergraduate students who assisted with data collection and animal care. We apologize to colleagues in this field that we could not cite all of the important papers due to strict limitations on space and numbers of references.
Funding
This project was supported by grants from the National Institute on Aging (1 RO1 AG024354), the Glenn Foundation, and the Ellison Medical Foundation..
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bartke A. Impact of reduced insulin-like growth factor-1/insulin signaling on aging in mammals: novel findings. Aging Cell. 2008;7:285–290. doi: 10.1111/j.1474-9726.2008.00387.x. [DOI] [PubMed] [Google Scholar]
- Bartke A, Masternak MM, Al-Regaiey KA, Bonkowski MS. Effects of dietary restriction on the expression of insulin-signaling-related genes in long-lived mutant mice. Interdiscip. 2007;35:69–82. doi: 10.1159/000096556. [DOI] [PubMed] [Google Scholar]
- Bartke A, Wright JC, Mattison JA, Ingram DK, Miller RA, Roth GS. Extending the lifespan of long-lived mice. Nature. 2001;414:412. doi: 10.1038/35106646. [DOI] [PubMed] [Google Scholar]
- Barzilai N, Gabriely I. The role of fat depletion in the biological benefits of caloric restriction. J Nutr. 2001;131:903S–906S. doi: 10.1093/jn/131.3.903S. [DOI] [PubMed] [Google Scholar]
- Barzilai N, Gupta G. Revisiting the role of fat mass in the life extension induced by caloric restriction. J Gerontol A Biol Sci Med Sci. 1999;54:B89–96. doi: 10.1093/gerona/54.3.b89. discussion B97–88. [DOI] [PubMed] [Google Scholar]
- Brown-Borg HM, Borg KE, Meliska CJ, Bartke A. Dwarf mice and the ageing process. Nature. 1996;384:33. doi: 10.1038/384033a0. [DOI] [PubMed] [Google Scholar]
- Colman RJ, Anderson RM, Johnson SC, Kastman EK, Kosmatka KJ, Beasley TM, Allison DB, Cruzen C, Simmons HA, Kemnitz JW, et al. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325:201–204. doi: 10.1126/science.1173635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L, Partridge L, Longo VD. Extending healthy life span--from yeast to humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster MJ, Morris P, Sohal RS. Genotype and age influence the effect of caloric intake on mortality in mice. Faseb J. 2003;17:690–692. doi: 10.1096/fj.02-0533fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia AM, Busuttil RA, Calder RB, Dolle MET, Diaz V, McMahan CA, Bartke A, Nelson J, Reddick R, Vijg J. Effect of Ames dwarfism and caloric restriction on spontaneous DNA mutation frequency in different mouse tissues. Mech Ageing Dev. 2008;129:528–533. doi: 10.1016/j.mad.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis MC, Guarente LP. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- Harper JM, Leathers CW, Austad SN. Does caloric restriction extend life in wild mice? Aging Cell. 2006;5:441–449. doi: 10.1111/j.1474-9726.2006.00236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TE, Wood WB. Genetic analysis of life-span in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1982;79:6603–6607. doi: 10.1073/pnas.79.21.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S, Schleit J, Kaeberlein M, Bennett C. Molecular mechanisms underlying genotype-dependent responses to dietary restriction. 65th Annual Meeting of the Gerontological Society of America; 2012. p. 3. Abstract. [Google Scholar]
- Lang DH, Gerhard GS, Griffith JW, Vogler GP, Vandenbergh DJ, Blizard DA, Stout JT, Lakoski JM, McClearn GE. Quantitative trait loci (QTL) analysis of longevity in C57BL/6J by DBA/2J (BXD) recombinant inbred mice. Aging Clin. 2010;22:8–19. doi: 10.1007/BF03324809. [DOI] [PubMed] [Google Scholar]
- Liang H, Masoro EJ, Nelson JF, Strong R, McMahan CA, Richardson A. Genetic mouse models of extended lifespan. Exp Gerontol. 2003;38:1353–1364. doi: 10.1016/j.exger.2003.10.019. [DOI] [PubMed] [Google Scholar]
- Liao CY, Rikke BA, Johnson TE, Diaz V, Nelson JF. Genetic variation in the murine lifespan response to dietary restriction: from life extension to life shortening. Aging Cell. 2010a;9:92–95. doi: 10.1111/j.1474-9726.2009.00533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao C-Y, Rikke BA, Johnson TE, Diaz V, Nelson JF. No evidence that competition for food underlies lifespan shortening by dietary restriction in multiply housed mice. Aging Cell. 2010b;9:450–452. [Google Scholar]
- Liao CY, Rikke BA, Johnson TE, Gelfond JA, Diaz V, Nelson JF. Fat maintenance is a predictor of the murine lifespan response to dietary restriction. Aging Cell. 2011;10:629–639. doi: 10.1111/j.1474-9726.2011.00702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoro EJ. Role of sirtuin proteins in life extension by caloric restriction. Mech Ageing Dev. 2004;125:591–594. doi: 10.1016/j.mad.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Masoro EJ. Overview of caloric restriction and ageing. Mech Ageing Dev. 2005;126:913–922. doi: 10.1016/j.mad.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Mattison JA, Roth GS, Beasley TM, Tilmont EM, Handy AM, Herbert RL, Longo DL, Allison DB, Young JE, Bryant M, et al. Impact of caloric restriction on health and survival in rhesus monkeys from the NIA study. Nature. 2012 doi: 10.1038/nature11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP. Genes and behavior interact to determine mortality in mice when food is scarce and competition fierce. Aging Cell. 2010;9:448–449. doi: 10.1111/j.1474-9726.2010.00561.x. discussion 450–442. [DOI] [PubMed] [Google Scholar]
- McCay CM, Crowell MF, Maynard LA. The effect of retarded growth upon the length of life and upon the ultimate body size. J Nutr. 1935;10:63–79. [PubMed] [Google Scholar]
- Nakagawa S, Lagisz M, Hector KL, Spencer HG. Comparative and meta-analytic insights into life extension via dietary restriction. Aging Cell. 2012;11:401–409. doi: 10.1111/j.1474-9726.2012.00798.x. [DOI] [PubMed] [Google Scholar]
- Picard F, Guarente L. Molecular links between aging and adipose tissue. Int J Obes (Lond) 2005;29(Suppl 1):S36–39. doi: 10.1038/sj.ijo.0802912. [DOI] [PubMed] [Google Scholar]
- Rikke BA, Johnson TE. Physiological genetics of dietary restriction: uncoupling the body temperature and body weight responses. Am J Physiol Regul Integr Comp Physiol. 2007;293:R1522–1527. doi: 10.1152/ajpregu.00215.2007. [DOI] [PubMed] [Google Scholar]
- Rikke BA, Liao CY, McQueen MB, Nelson JF, Johnson TE. Genetic dissection of dietary restriction in mice supports the metabolic efficiency model of life extension. Exp Gerontol. 2010;45:691–701. doi: 10.1016/j.exger.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikke BA, Yerg JE, 3rd, Battaglia ME, Nagy TR, Allison DB, Johnson TE. Quantitative trait Loci specifying the response of body temperature to dietary restriction. J Gerontol A Biol Sci Med Sci. 2004;59:118–125. doi: 10.1093/gerona/59.2.b118. [DOI] [PubMed] [Google Scholar]
- Schleit J, Wasko BM, Kaeberlein M. Yeast as a model to understand the interaction between genotype and the response to calorie restriction. FEBS Let. 2012;586:2868–2873. doi: 10.1016/j.febslet.2012.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal RS, Ferguson M, Sohal BH, Forster MJ. Life span extension in mice by food restriction depends on an energy imbalance. J Nutr. 2009;139:533–539. doi: 10.3945/jn.108.100313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speakman JR, Mitchell SE. Caloric restriction. Mol Aspects Med. 2011;32:159–221. doi: 10.1016/j.mam.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Swindell WR. Dietary restriction in rats and mice: A meta-analysis and review of the evidence for genotype-dependent effects on lifespan. Ageing Res Rev. 2012;11:254–270. doi: 10.1016/j.arr.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turturro A, Witt WW, Lewis S, Hass BS, Lipman RD, Hart RW. Growth curves and survival characteristics of the animals used in the Biomarkers of Aging Program. J Gerontol A Biol Sci Med Sci. 1999;54:B492–501. doi: 10.1093/gerona/54.11.b492. [DOI] [PubMed] [Google Scholar]
- Weindruch R, Walford R. The Retardation of Aging and Disease by Dietary Restriction. Springfield, Ill: Charles C Thomas Publisher; 1988. [Google Scholar]
- Williams RW, Bennett B, Lu L, Gu J, DeFries JC, Carosone-Link PJ, Rikke BA, Belknap JK, Johnson TE. Genetic structure of the LXS panel of recombinant inbred mouse strains: a powerful resource for complex trait analysis. Mamm Genome. 2004;15:637–647. doi: 10.1007/s00335-004-2380-6. [DOI] [PubMed] [Google Scholar]
- Yuan R, Peters LL, Paigen B. Mice as a mammalian model for research on the genetics of aging. Ilar J. 2011;52:4–15. doi: 10.1093/ilar.52.1.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan R, Tsaih SW, Petkova SB, de Evsikova CM, Xing S, Marion MA, Bogue MA, Mills KD, Peters LL, Bult CJ, et al. Aging in inbred strains of mice: study design and interim report on median lifespans and circulating IGF1 levels. Aging Cell. 2009;8:277–287. doi: 10.1111/j.1474-9726.2009.00478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]