Abstract
CD8+ T cell responses to hepatitis C virus (HCV) are important in generating a successful immune response and spontaneously clearing infection. HLA class I presents viral peptides to CD8+ T cells to permit detection of infected cells, and tapasin is an important component of the peptide loading complex for HLA class I. We sought to determine if tapasin polymorphisms affected the outcome of HCV infection.
Patients with resolved or chronic HCV infection were genotyped for the known C/G coding polymorphism in exon 4 of the tapasin gene. In a European, but not a US, Caucasian population, the tapasin G allele was significantly associated with outcome of HCV infection, being found in 82.5% of resolvers versus 71.3% of persistently infected individuals (p=0.02, OR=1.90 95% CI=1.11–3.23). This was more marked at the HLA-B locus at which heterozygosity of both tapasin and HLA-B was protective (p<0.03). Individuals with an HLA-B allele with an aspartate at residue 114 and the tapasin G allele were more likely to spontaneously resolve HCV infection (p<0.00003, OR=3.2 95%CI=1.6–6.6). Additionally individuals with chronic HCV and the combination of an HLA-B allele with an aspartate at residue 114 and the tapasin G allele also had stronger CD8+ T cell responses (p=0.02, OR=2.58, 95% CI-1.05–6.5).
Conclusion
Tapasin alleles contribute to the outcome of HCV infection by synergising with polymorphisms at HLA-B in a population specific manner. This polymorphism may be relevant for peptide vaccination strategies against HCV infection.
Keywords: Hepatitis C virus, HLA, Tapasin, Immunogenetics, CD8+ T cell response
Hepatitis C virus (HCV) is a common chronic viral infection with between 50 and 80% of individuals exposed to HCV become chronically infected. Understanding the immunological determinants of resolution of HCV infection is important in order to develop vaccines and also immunologically based therapeutics. A broad and multi-specific CD8+ T cell response may be important in resolving HCV infection successfully. The strength of this CD8+ response is dependent on a number of factors. These include antigen processing and presentation, an appropriate cytokine microenvironment, and the presence of CD4+ T cell help. In addition to these cellular factors, host genetic factors may also play an important role. Specific HLA class I alleles are associated with the outcome of HCV infection, and certain HLA class I alleles, including HLA-B*27 and B*57, are associated with a strong CD8+ response and hence viral clearance (1–8).
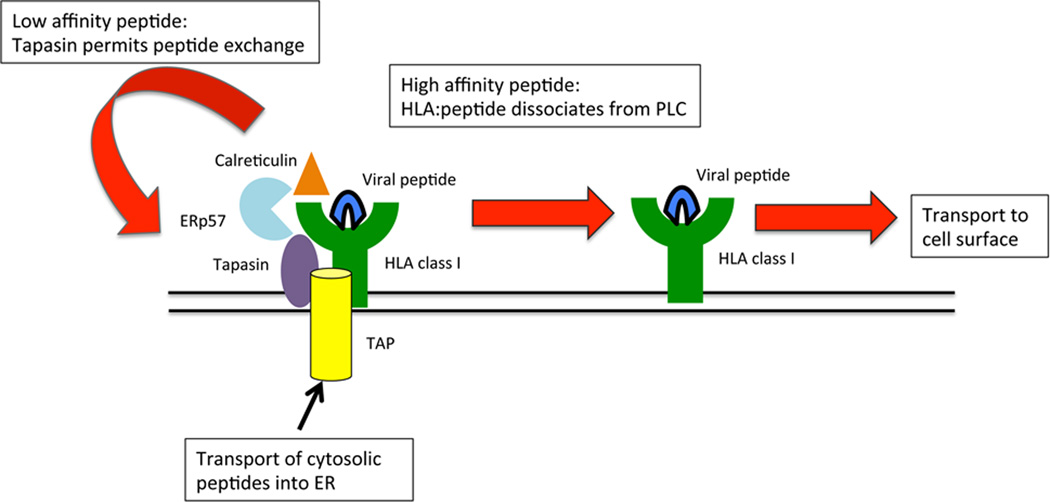
The selection of HCV peptides for presentation is determined by the allelic diversity of HLA and also the supply of HCV peptides to the endoplasmic reticulum. Additionally peptide loading of HLA class I is complex. It involves several proteins including tapasin, TAP, ERp57 and calreticulin that together form a peptide loading complex (PLC) (Figure 1)(9). Overall these proteins act in a collaborative way to properly fold HLA class I with β2-microglobulin and endogenous or virally derived peptides. ERp57 and calreticulin promote folding and disulphide bond formation of HLA class I, whilst tapasin acts as a “bridge” between the components of the PLC (10). In doing so it is thought to promote the binding of high affinity peptides to HLA class I for presentation on the cell surface (11, 12) (Figure 1). In the presence or absence of tapasin the hierarchy of peptides presented by HLA class I can be substantially different (13, 14). However, only certain HLA class I alleles have been shown to be tapasin dependent in biochemical assays. These include HLA-A*3001, HLA-B*2705, HLA-B*3501, HLA-B*4402, HLA-B*5001, HLA-B*5701 (15–17). This dependence is governed by residues 114, 116 and 156 of the HLA class I molecule, in the floor of the peptide binding groove (11, 16, 18, 19). Thus tapasin can have a direct effect on peptide binding to HLA class I, and as the tapasin gene lies at the centromeric end of the MHC, but is beyond a recombination hotspot it is not thought to be in tight linkage with HLA class I (20, 21). The tapasin gene has a number of SNPs (21–24). However of the exonic SNPs only one is non-synonymous and both alleles are commonly found in the Caucasian population (24). This SNP is in exon 4 and codes for a non-conservative arginine to threonine substitution at position 260 of the tapasin protein. The aim of this study was to determine whether polymorphisms in tapasin and HLA class I may interact to etermine the outcome of HCV infection.
Figure 1. Schematic showing the role of tapasin in peptide loading.
Tapasin along with the transporter associated with antigen processing (TAP), ERp57 and calreticulin form the peptide loading complex (PLC), which loads viral peptides onto HLA class I. Tapasin binds to HLA class I and is thought to assist in the dissociation of peptides which bind with low affinity. These dissociated peptides can be replaced by high affinity peptides. When a high affinity peptide binds HLA class I the HLA class I and peptide are released from tapasin and the PLC and egress to the cell surface.
Experimental Procedures
Patients
All patients provided written informed consent and participated in the study in agreement with the relevant local ethics committee’s approval and the declaration of Helsinki.
-
a)
UK cohort 1
These individuals were from the previously described UK cohort of HCV exposed individuals (25), with the addition of 71 chronically infected individuals. Patients were recruited from hepatology clinics at King’s College Hospital, London, Addenbrooke’s Hospital NHS Trust, Cambridge, and Southampton General Hospital, Southampton, UK. There were 120 (37 female, 83 male, mean age 41.5 years) individuals with resolved infection. All had a self-reported Caucasian ethnicity and had at least two negative PCR reactions (HCV COBAS Amplicor (Roche diagnostics, Pleasanton, CA)) at least six months apart. The chronic HCV population consisted of 300 chronically infected individuals (81 female, 219 male, mean age 43.0 years). Two hundred and eighty-six (95.3%) had a self-reported Caucasian ethnicity.
-
b)
UK cohort 2
These consisted of a cohort of 79 (64 male) Caucasian individuals with an extensive history of injection drug use, but repeated negative testing for HCV antibody as previously described (26). This group has been termed HCV exposed seronegative aviraemic. The median duration of injection drug use was 8.62 years (range, 0.3–24) with a median number of injections of 4927 (range, 36– 41,620). Their median age was 28 years. As a control group 79 additional individuals with chronic HCV infection were recruited from the out-patient hepatology clinic at Southampton General Hospital. Fifty-seven were male and 75 (94.9%) were of Caucasian origin. The mean age at diagnosis was 48.4.
-
c)
German Cohort
Forty two patients (17 female, 25 male, mean age 48.6 years) with chronic HCV from the University Hospital Freiburg were included. Thirteen had genotype 1a and 29 genotype 1b infection. All patients had a Caucasian self-reported ethnicity. Sixteen of the patients were positive for HLA-B*18, 17 for HLA-B*35, six for HLA-B*57 and six for HLA-B*58 (three of the patients were positive for two of these HLA alleles). Peripheral blood mononuclear cells (PBMCs) were isolated from patient blood by gradient centrifugation, as previously described (27).
-
c)
US cohort
These individuals were from the previously described USA cohort of HCV exposed individuals limited to Caucasian ethnicity (8). Briefly patients were recruited from the infectious diseases or hepatology clinics at Massachusetts General Hospital with local ethical committee approval. There were 53 (27 female, 26 male, mean age 39.2 years) individuals with resolved infection. Each had at least two negative PCR reactions (HCV COBAS Amplicor (Roche diagnostics, Pleasanton, CA)) at least six months apart; and 196 chronically infected individuals (69 female, 127 male, mean age 41.4 years).
Genotyping
Tapasin genotyping was performed on genomic DNA extracted using a QiaAmp DNA Blood mini kit (Qiagen, Crawley, UK). A 51bp region of the tapasin gene flanking the polymorphism was amplified using the primers 5’-GACCTTCTGGCTGCCTAC-3’ (G allele) and 5’-GACCTTCTGGCTGCCTAG-3’ (C allele) and 5-GCCAGATAGGTGCCCTCCTG-3’ (reverse). PCR reactions were detected using the Quantitect SYBR Green PCR kit (Qiagen) in standard 20ul PCR reactions. Thermal cycling conditions were: 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 58°C for 20 seconds. HLA genotyping was performed by PCR-SSP as previously described (28) or at NCI Frederick (Dr. Mary Carrington) by the PCR-SSOP (sequence-specific oligonucleotide probing) typing protocol and PCR-SBT (sequence based typing) as recommended by the 13th International Histocompatibility Workshop (http://www.ihwg.org).
Generation of peptide-specific T cell lines
Peptides (Table 1) were synthesized by Genaxxon BioScience (Ulm, Germany) and have a purity of at least 70%. Peptide-specific T cell lines were generated as previously described (29). 4 × 106 PBMC were resuspended in 1 mL medium and stimulated with peptide at a final concentration of 10 µg/mL and anti-CD28 (BD PharMingen) at a final concentration of 0.5 µg/mL. On days 3 and 10, 1 mL culture medium and recombinant IL-2 (Hoffmann-La Roche, Basel, Switzerland) at a final concentration of 20 U/mL was added to each well. On day 7, the cultures were restimulated with the corresponding peptide (10 µg/mL) and 106 irradiated autologous feeder cells. After 2 weeks, PBMC were tested for peptide-specific IFN-γ production by intracellular cytokine staining after 5 hours of restimulation with the respective peptide (10 µg/mL) as described (29). Peptides were matched to the HLA-B alleles of the respective patients.
Table 1. CD8+ T cell epitopes analysed.
| HLA allele | protein | position | epitope |
|---|---|---|---|
| B*18 | NS5A | 2143–2151 | DEVSFRVGL(gt 1a)/ DEVTFQVGL(gt 1b) |
| NS3 | 1581–1589 | DNFPYLVAY(gt 1b only) | |
| B*35 | E1 | 234–242 | NASCRCWVAV(gt 1a)/ NSSCRCWVAL(gt 1b) |
| NS3 | 1359–1367 | HPNIEEVAL | |
| NS4A | 1695–1702 | IPDREVLY | |
| NS5A | 2163–2171 | EPEPDVAVL | |
| B*57/B*58 | E2 | 521–529 | RSGAPTYSW |
| E2 | 541–550 | NTRPPLGNWF | |
| E2 | 708–716 | SIASWAIKW | |
| NS3 | 1596–1604 | RAQAPPPSW | |
| NS4 | 1968–1972 | CTTPCSGSW | |
| NS5B | 1801–1809 | LTTSQTLLF | |
| NS5B | 2629–2637 | KSKKTPMGF | |
| NS5B | 2912–2921 | LGVPPLRAWR | |
Statistical analysis
This was performed using SPSS v17.0 and using the Bonferoni correction, where necessary. For the residue analysis n=34, which is the number of different residues at the analysed positions (114,116 and 156) at all three loci.
Results
The tapasin G allele is protective against chronic HCV infection
We typed 120 individuals with resolved and 300 individuals with chronic HCV infection for the G/C SNP in exon 4 of tapasin. Ninety-nine (82.5%) out of 120 individuals with resolved infection versus 214 (71.3%) out of 300 individuals with chronic infection had the G allele, which encodes for the arginine variant (p=0.019, OR=1.90 95% CI=1.11–3.23) (Table 2a). Similar frequencies of individuals with the tapasin C allele had resolved as compared to chronic HCV infection (70.8% vs. 74.3%, p>0.1). Thus the tapasin G allele is associated with the protection against chronic HCV infection in this cohort.
Table 2. Association of tapasin alleles with outcome in the different cohorts.
| a) UK: Spontaneous resolving cohort | |||||
|---|---|---|---|---|---|
| Tapasin allele |
Resolver (n=120) |
Chronic (n=300) |
p value |
OR | 95% CI |
| C | 85 (70.8%) | 223 (74.3%) | 0.47 | 0.84 | 0.52–1.3 |
| G | 99 (82.5%) | 214 (71.3%) | 0.02 | 1.90 | 1.11–3.23 |
| b) USA: Spontaneous resolving cohort | |||
|---|---|---|---|
| Tapasin allele |
Resolver (n=53) |
Chronic (n=196) |
p value |
| C | 38 (71.7%) | 145 (74.0%) | >0.1 |
| G | 43 (81.1%) | 160 (81.7%) | >0.1 |
| c) UK: Exposed uninfected cohort | |||
|---|---|---|---|
|
Tapasin allele |
Exposed Uninfected (n=75) |
Chronic (n=79) |
p value |
| C | 54 (72.0%) | 60 (75.9%) | >0.1 |
| G | 63 (84.0%) | 60 (75.9%) | >0.1 |
Tapasin advantage is related mainly to HLA-B alleles
Tapasin helps to optimise the MHC class I peptide repertoire and some MHC class I alleles are more dependent on tapasin for optimisation of their peptide repertoire than others. We therefore explored as to how this property was manifest in a more in depth immunogenetic analysis. We hypothesized that heterozygosity of MHC class I and also of tapasin could be predicted to increase the number of peptides presented to CD8+ T cells and hence augment the immune response to HCV. We therefore determined the association of heterozygosity for tapasin with the outcome of infection. In this analysis heterozygosity at HLA class I was defined as alleles with different HLA types as determined by four digit typing, which correlates with differences at the single amino acid level. Sixty-four (53.3%) of resolvers and 137 (45.7%) of chronically infected individuals were heterozygous for tapasin (p>0.1). Thus for both spontaneous resolvers and chronically infected individuals the individual gene frequencies were in Hardy-Weinberg equilibrium. However when individual HLA class I loci were considered there was a trend for the combined association of heterozygosity of tapasin with HLA-B heterozygosity with outcome (p=0.028, pc=0.084 (χ2 trend test)), but not for tapasin and heterozygosity at the other loci (Figure 2).
Figure 2. Heterozygosity of tapasin and HLA-B are protective against chronic HCV infection.
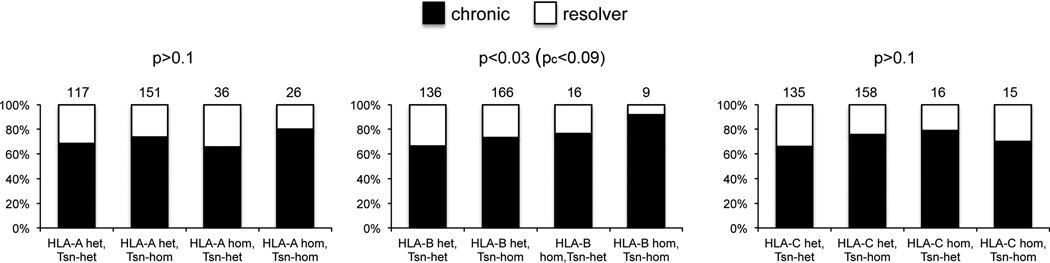
Analysis of the combination of tapasin heterozygosity with HLA heterozygosity at HLA-A, -B and –C. Heterozygosity was determined at the four digit level. Numbers in each group are indicated above the relevant bars. P values were calculated using a Chi-square test for trend and the Bonferoni correction applied (pc).
Although some HLA class I alleles are more tapasin dependent than others, we found only relatively weak associations of outcome and specific HLA class I alleles with the tapasin G and C alleles. These associations include resolution of HCV and the tapasin G allele in individuals who were positive for HLA-A*0101 (p=0.008, OR=3.17, 95% CI 1.4–7.5), B*0702 (p=0.01, OR=4.84, 95% CI 1.3–17.6), and Cw*0701 (p=0.04, OR=2.69, 95% CI 1.07–6.77). Conversely the tapasin C allele was associated with chronic infection in individuals who had the B*0801 allele (p=0.008, OR=0.23, 95% CI 0.08–0.66) and the B*3501 allele (p=0.01, OR=0.16, 95% CI 0.04–0.65).
The protective effect of tapasin G alleles is determined by specific HLA class I residues
Biochemical studies have indicated that specific HLA-class I residues may be relevant for their interaction with tapasin. In particular residues 114, 116 and 156 of the MHC class I heavy chain determine its interaction with tapasin, with the effect of position 114 being dominant over 116 (14–16, 30). We therefore investigated whether these residues determined the association of the tapasin G allele with the outcome of HCV infection. As the three class I loci have distinct sequences we subdivided the analysis by locus, so that in each analysis 2n alleles were considered (Table 3). One hundred and twenty-seven out of 143 (89%) of HLA-B alleles with aspartate at residue 114 (Asp114) were associated with the tapasin G allele in individuals who resolved infection, as compared to 234 out of 330 (71%) of HLA-B alleles from those with chronic infection (p<0.00003 (pc=0.0009), OR=3.26, 95% CI=1.84–5.77) (Table 3). Overall one hundred and five resolvers had at least one HLA-B allele with Asp114, and 93 (89%) of these had a tapasin G allele, as compared to 174 of 246 (71%) individuals with chronic infection (p=0.0002, OR=3.50, 95% CI=1.77–6.93). However the presence of an HLA-B allele with Asp114 per se was not significantly associated with resolution (p>0.1).
Table 3. Association of the Tapasin (Tsn) G allele with specific amino acid residues in the HLA class I heavy chain.
Shown are the number of HLA alleles with each specific residue at positions 114, 116 and 156 in the Tapasin G-positive and -negative groups in individuals that resolved HCV infection or remained chronically infected. Only p values, or Bonferoni corrected p values <0.05 are shown
| a) HLA-A | ||||||||
|---|---|---|---|---|---|---|---|---|
| Residue | N | Resolve Tsn G+ |
Tsn G− | Chronic Tsn G+ |
Tsn G− | p value | OR | 95%CI |
| Glu114 | 11 | 1 | 1 | 7 | 2 | |||
| His114 | 317 | 86 | 16 | 153 | 62 | 0.012 | 2.18 | 1.18–4.01 |
| Gln114 | 99 | 16 | 5 | 57 | 21 | |||
| Arg114 | 399 | 91 | 20 | 203 | 85 | 0.022 | 1.91 | 1.1–3.29 |
| Asp116 | 498 | 107 | 25 | 260 | 106 | 0.028 | 1.75 | 1.07–2.85 |
| His116 | 11 | 1 | 1 | 7 | 2 | |||
| Tyr116 | 317 | 86 | 16 | 153 | 62 | 0.012 | 2.18 | 1.18–4.01 |
| Leu156 | 481 | 114 | 19 | 262 | 86 | 0.014 | 1.97 | 1.14–3.39 |
| Gln156 | 120 | 24 | 7 | 60 | 29 | |||
| Arg156 | 148 | 43 | 8 | 61 | 36 | 0.008 | 3.17 | 1.34–7.5 |
| Try156 | 77 | 13 | 8 | 37 | 19 | |||
| b) HLA-B | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Residue | N | Resolve | Chronic | p value | pc value | OR | 95%CI | ||
| Tsn G+ | Tsn G− | Tsn G+ | Tsn G− | ||||||
| Asp114 | 473 | 127 | 16 | 234 | 96 | <0.00003 | 0.0009 | 3.26 | 1.84–5.77 |
| His114 | 32 | 9 | 4 | 18 | 1 | ||||
| Asn114 | 317 | 62 | 20 | 160 | 75 | ||||
| Asp116 | 185 | 40 | 8 | 102 | 35 | ||||
| Phe116 | 65 | 15 | 5 | 27 | 18 | ||||
| Leu116 | 66 | 9 | 4 | 40 | 13 | ||||
| Ser116 | 200 | 64 | 8 | 93 | 35 | 0.007 | 3.01 | 1.3–6.91 | |
| Try116 | 306 | 70 | 15 | 150 | 71 | 0.011 | 2.21 | 1.18–4.13 | |
| Asp156 | 217 | 46 | 7 | 108 | 56 | 0.003 | 3.41 | 1.45–8.04 | |
| Leu156 | 438 | 100 | 25 | 226 | 87 | ||||
| Arg156 | 110 | 32 | 4 | 51 | 23 | 0.032 | 3.61 | 1.14–11.4 | |
| Try156 | 57 | 20 | 4 | 27 | 6 | ||||
| c) HLA-C | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Residue | N | Resolve | Chronic | p value | pc value | OR | 95%CI | ||
| Tsn G+ | Tsn G− | Tsn G+ | Tsn G− | ||||||
| Asp114 | 583 | 144 | 33 | 286 | 120 | 0.006 | 1.83 | 1.19–2.83 | |
| Asn114 | 231 | 46 | 9 | 124 | 52 | ||||
| Phe116 | 247 | 52 | 10 | 132 | 53 | ||||
| Leu116 | 21 | 1 | 3 | 13 | 4 | ||||
| Ser116 | 424 | 100 | 20 | 202 | 102 | 0.0005 | 0.015 | 2.53 | 1.48–4.32 |
| Try116 | 122 | 37 | 9 | 63 | 13 | ||||
| Asp156 | 15 | 6 | 1 | 7 | 1 | ||||
| Leu156 | 371 | 94 | 23 | 179 | 75 | ||||
| Gln156 | 38 | 6 | 2 | 18 | 12 | ||||
| Arg156 | 258 | 51 | 10 | 139 | 58 | 0.047 | 2.13 | 1.01–4.48 | |
| Try156 | 132 | 33 | 6 | 67 | 26 | ||||
A similar, but weaker, association of the tapasin G allele with Asp114 was seen at HLA-C, as this combination was found in 144 out of 177 alleles (81%) in resolvers but only 286 out of 406 (70%) in chronically infected individuals (p=0.006 (pc>0.05), OR=1.83, 95% CI=1.19–2.83) (Table 3). The only other positive association to remain after Bonferoni correction (n=34) was for serine at residue 116 of an HLA-C allele. One hundred out of 120 HLA-C alleles with Ser116 were associated with tapasin G (83%) in resolvers versus 202 out of 304 (66%) in chronically infected individuals (p=0.0005 (pc=0.015), OR=2.53, 95% CI=1.48–4.32). This association was also shared with HLA-B alleles, in which Ser116 was found in 64 out of 72 (89%) of resolvers versus 93 out of 128 (73%) chronically infected individuals (p=0.007 (pc>0.1), OR=3.01, 95% CI=1.3–6.9). Of note is that neither Asp114 nor Ser116 are present in any of the HLA-A alleles present in our population. Conversely Arg156 is present in alleles at all 3 loci and this was associated with resolution in combination with tapasin G at: HLA-A in 43 out of 51 (84%) versus 61 out of 97 (63%) (p=0.008 (pc>0.1), OR=3.17, 95% CI=1.3–7.5); HLA-B 32 out of 36 (89%) versus 51 out of 74 (69%) (p=0.03 (pc>0.1), OR=3.61, 95% CI=1.1–11.4) and HLA-C 51 out of 61 (84%) versus 139 out of 197 (71%) (p=0.05 (pc>0.1), OR=2.13, 95% CI=1.0–4.5) in resolvers versus chronically infected individuals respectively. Thus we found a consistent trend for this residue to be significantly associated with tapasin G and the outcome of HCV infection.
In order to determine whether the effects we noted were due to the association of specific HLA class I residues with a tapasin G allele or were independent, we performed logistic regression analysis using the most significantly associated residues at each locus (those with p<0.01) and tapasin G as the variables (Tables 4a–c). In these three analyses tapasin G remained significant throughout. However for HLA-A alleles no significant associations remained. For HLA-B alleles Asp114 (p=0.027) and Asp156 (p=0.048) remained significantly associated with resolution in combination with the tapasin G allele, with a trend towards chronicity in the absence of tapasin G (Table 4b). However at HLA-C the strongest effects were for Asp114 with resolution (p=0.002) and Ser116 with chronicity (p=0.006) in the absence of an effect of tapasin, and a weaker protective effect of Ser116 and tapasin G (p=0.011). Thus consistent with our observations on heterozygosity at the MHC, the effect of tapasin is strongest in combination with HLA-B, rather than HLA-A or –C, alleles. This epistatic association is unlikely to be due to linkage between tapasin and specific HLA-B alleles as we found that no specific HLA-B alleles with aspartate at position 114 were significantly (p<0.05) associated with the tapasin G allele (Supplementary Table 1).
Table 4. Logistic regression analysis of tapasin G with specific amino acid residues at the three different HLA class I loci and the outcome of infection.
An OR >1 indicates a positive association with resolution
| a) HLA-A | |||
|---|---|---|---|
| p | OR | 95% CI | |
| Tapasin G | 0.001 | 2.29 | 1.42–3.70 |
| Tapasin G+ Arg 156 | p>0.1 | ||
| Arg 156 | p>0.1 | ||
| b) HLA-B | |||
|---|---|---|---|
| p | OR | 95% CI | |
| Tapasin G | 0.001 | 2.72 | 1.75–5.04 |
| Tapasin G+ Asp114 | 0.027 | 3.00 | 1.13–7.93 |
| Tapasin G+ Asp156 | 0.048 | 2.79 | 1.01–7.68 |
| Asp156 | 0.053 | 0.61 | 0.37–1.01 |
| Asp114 | 0.079 | 0.65 | 0.40–1.05 |
| Tapasin G+ Ser116 | >0.1 | ||
| Ser116 | >0.1 | ||
| c) HLA-C | |||
|---|---|---|---|
| p | OR | 95% CI | |
| Tapasin G | <0.001 | 2.33 | 1.45–3.74 |
| Asp114 | 0.002 | 2.43 | 1.39–4.25 |
| Ser116 | 0.006 | 0.51 | 0.31–0.82 |
| Tapasin G+ Ser116 | 0.011 | 3.48 | 1.32–9.13 |
| Tapasin G+ Asp114 | 0.058 | 0.34 | 0.11–1.04 |
The association of tapasin G with HCV is population specific
To determine if the protective effect of tapasin was universal, we tested a second cohort of Caucasian individuals from the USA. Fifty three individuals had resolved infection and 196 were chronically infected. Similar frequencies of tapasin alleles were found in both groups. The tapasin G allele was found in 43 (81.1%) resolvers versus 160 (81.7%) chronically infected and the tapasin C allele was found in 71.7% of resolvers and 74.0% of chronically infected individuals (all p>0.1) (Table 2b). Thus the effect of tapasin was confined to European Caucasians. Comparison of the different HLA-class I alleles from the two populations revealed slight but significant differences between the HLA-A, -B and –C alleles in both groups, which may be relevant to the lack of association in this cohort (Supplementary Table 2). To confirm that this effect was indeed population specific, we typed a further UK cohort of 75 individuals at high risk of HCV infection due to multiple episodes of intravenous drug-usage but who remained seronegative and aviraemic. These individuals have detectable T cell responses, a protective KIR:HLA-C type and distinct serum cytokine profile, consistent with exposure to HCV infection (26, 31, 32). As a comparator group we typed a further 79 individuals from the UK with chronic HCV infection. The tapasin G allele was present in 63 (84%) individuals in the exposed seronegative avirameic group, similar to that found in the UK spontaneous resolvers, but this was not significantly different from the 60 (76%) with the tapasin G allele in the second UK chronically infected population (Table 2c). However there was an increased frequency of HLA-B Asp114 in combination with tapasin G in the exposed seronegative avirameic individuals as compared to the chronically infected (54.8% versus 40.2%; p<0.04, OR=1.8 95%, CI=1.04–3.14) (Table 5). Furthermore they also had an increased frequency of HLA-B Asp114 in combination with two tapasin G alleles (16.4% versus 6.9%; p<0.04, OR=2.7, 95%, CI=1.05–6.70). Thus tapasin G and HLA-B appear to have a similar protective effect in this cohort.
Table 5. Association of Tapasin (Tsn) alleles with HLA-B alleles with Asp 114 in the exposed seronegative avirameic and second UK chronic populations.
| Exposed Seronegative (2n=104) |
Chronic (2n=102) |
P value | OR (95% CI) | |
|---|---|---|---|---|
| HLA-B Asp114+ Tsn G | 57 (54.8%) | 41 (40.2%) | 0.036 | 1.8 (1.04–3.14) |
| HLA-B Asp114+ Tsn C | 50 (48.1%) | 43 (42.1%) | 0.4 | |
| HLA-B Asp114+ Tsn GG | 17 (16.4%) | 7 (6.9%) | 0.034 | 2.65 (1.05–6.70) |
Tapasin G is associated with stronger interferon-gamma CD8+ T cell responses in chronic HCV infection
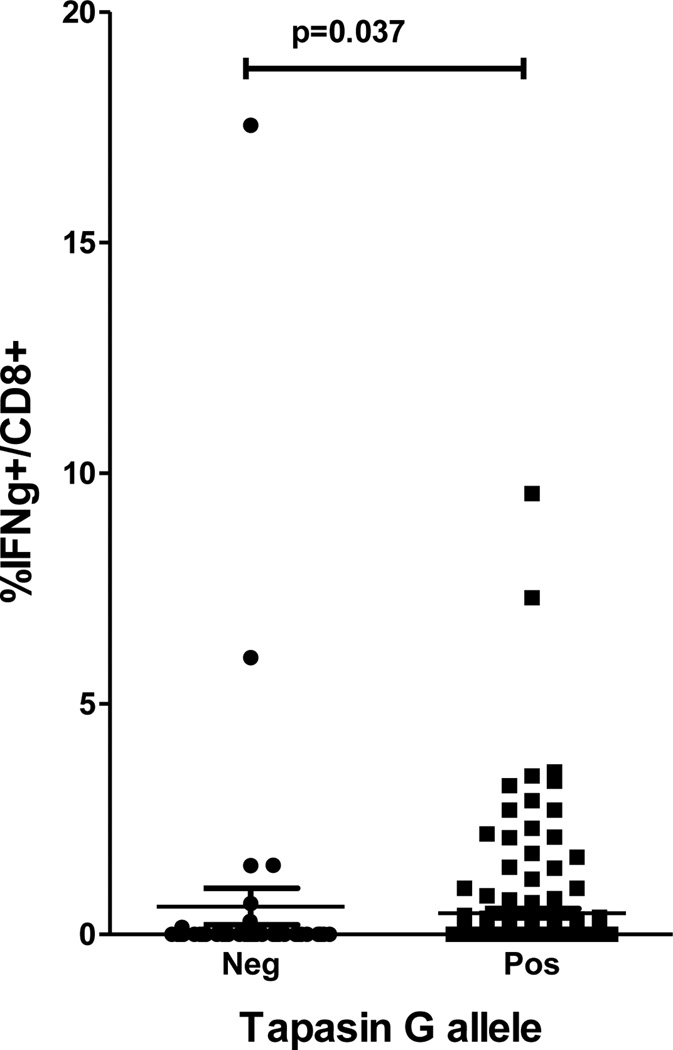
To date the functional role of tapasin polymorphism on CD8+ responses has not been described in viral infection. Tapasin optimizes peptide:MHC class I interactions, likely impacting on priming and induction of CD8+ T cells. In order to determine whether there is an effect of the tapasin polymorphism on anti-viral CD8+ responses, we determined peptide specific CD8+ responses from 42 patients with chronic HCV genotype 1 infection who had HLA-B alleles with aspartate at residue 114, as this was our most significant association with the tapasin polymorphism in the genetic studies. These alleles included B*18, B*35, B*57 and B*58. PBMC were stimulated for two weeks, using specific HLA-B restricted HCV derived peptides (Table 1), matched to the HLA-B allele of the patient and analysed for IFNγ secretion by flow cytometry. In order to avoid confounding effects through HCV genotype mismatches between patients and epitope peptides, we restricted our analysis to patients infected with HCV genotype 1, using only peptides identified from this genotype. One hundred and ninety one–assays using sixteen different peptides were performed in these individuals (145 assays in the 35 patients with the tapasin G allele [median 4, range 1–8] versus 46 assays in the 10 patients without the tapasin G allele [median 3, range 2–8]; p=0.9). In patients with these HLA-B alleles and also a tapasin G allele measurable responses were made in 51 out of 145 (35.1%) assays as compared to 8 out of 46 (17.3%) in patients without a tapasin G allele (p=0.02, OR=2.58, 95% CI=1.05–6.5). Next, the magnitude of the response to these peptides was compared. Individuals carrying a tapasin G allele had a greater overall frequency of CD8+ T cell responses compared to individuals without the tapasin G allele (p=0.037) (Figure 3). Taken together, our data suggest that the tapasin G allele and specific HLA class I alleles may synergize to generate stronger CD8+ T cell responses. As discussed below, future studies performed in acute infection are required to extend our observations in chronically infected patients.
Figure 3. The tapasin G allele is associated with stronger interferon-gamma producing T cell responses in individuals with an HLA-B allele with Asp114.
The results of intracellular cytokine staining for IFN-γ on CD8+ T cells stimulated with various HCV-derived HLA restricted peptides. All patients tested had an HLA-B allele with Asp114 and peptides were matched to the HLA-B allele of the individual. Data are plotted for individuals with and without a tapasin G allele.
Discussion
The association of the G allele with outcome in our cohort is consistent with the role of the CD8+ T cell response in determining the outcome of acute HCV infection. Furthermore the functional data in an unrelated cohort indicates that this polymorphism has a downstream effect on the magnitude of the CD8+ response. Thus, individuals with a protective tapasin allele have greater CD8+ responses than those without. The increased frequency of IFNγ-positive CD8+ T cells following peptide stimulation in these individuals may be related either to a larger pool of memory CD8 T cells or alternatively memory T cells that proliferate more efficiently. It is important to note, however, that our functional CD8+ T cell data have several limitations. First, the strength of HCV-specific CD8+ T cell memory responses may be confounded by viral sequence variations in the respective CD8+ T cell epitopes. We thus analysed the autologous viral sequences in a subset of patients. Importantly, viral sequence mutations occurred at very similar frequencies in patients irrespective of tapasin genotypes (CC: 9/18 epitopes, 50.0%; CG: 12/22, 54.6%; GG: 5/11, 45.45%; p>0.1), indicating that viral sequence variations did not substantial confound our results. Second, the CD8+ T cell response in the chronic phase of infection may not necessarily correlate with the CD8+ T cell response in the acute phase of infection, when the outcome is determined. Longitudinal analyses of patients with acute-persistent infection as well as similar immunodominance of CD8+ T cell responses in acute and chronic infection, however, argue against this notion (8, 29, 33). Third, the rather small number of patients per individual HLA allele may influence our results. Thus, further studies in the acute phase of infection are required to confirm our finding that tapasin polymorphisms and specific HLA class I alleles synergise to generate stronger anti-viral CD8+ responses.
In the tapasin knockout mouse there is an alteration in peptide presentation as compared to the wild-type mouse (34). In these experiments the presence of tapasin favours CD8+ T cell responses to peptides with slow off-rates from MHC class I and its absence is associated with CD8+ responses to peptides with a fast off-rate. How this may be affected by a polymorphism in tapasin is not clear. The R260T polymorphism is present in one of the Ig-like domains of tapasin. In the crystal structure it is in a loop, close to the site of interaction with the oxido-reductase ERp57 which is another key component of the peptide loading complex involved in determining the peptide repertoire expressed on the cell surface (35). Thus the effect of this polymorphism on the interaction with HLA class I is not likely to be a direct effect on tapasin binding to HLA class I. It may be a remote effect on HLA class I binding, or alternatively an indirect effect mediated via the interaction with ERp57.
The finding of an absence of tapasin association in a second unrelated cohort is not unexpected as these are individuals that have a discrete ancestry and are exposed to different viral populations (36, 37). These differences are especially noticeable at the MHC which is characterised by substantial population diversity. In analyses of HLA class I different protective HLA class I alleles are found in European as opposed to USA cohorts (3–5). However despite these population differences, the levels of homozygosity at the MHC were similar between the US and UK populations. For instance at HLA-B 9.2% of the US and 7.1% of the UK were homozygous using four digit HLA typing.
Another source of confounding genetic effects include resolution through different genetic pathways. These include KIR and HLA-C, and also potentially IL-28B. Sub-analysis of the US cohort by KIR ligand (HLA-C) type was not associated with any significant findings (Supplementary Table 3) and IL-28B is thought to be protective in both US and UK Caucasian populations (38). Thus these known protective factors are unlikely to account for our observed population differences. Additionally there is diversity in the prevalent HCV infecting genotypes between the USA and the UK (36, 37), which may also impact on the mechanism of resolution of infection.
Our genetic data correlate well with the biochemistry of tapasin with key residues being 114 and 156 (16). Thus for HLA-B*4402, which has Asp114 and Asp156, the presence of a functional tapasin protein alters the peptide repertoire (39). Thus not only can polymorphisms at these residues impact their interaction with tapasin, but the specific residues which are most significant in our study have been previously identified by biochemical experiments. Overall tapasin is thought to be under purifying selection being well conserved across species with 84% amino acid identity between human and sheep (40). This implies that there is selection pressure to maintain the sequence of tapasin and the coding SNP is likely to be the most relevant due to its potential for functional interaction with HLA or ERp57. Tapasin is thus likely to be important in generating optimal HLA class I peptide repertoires in the anti-viral immune response. Consideration of this polymorphism may be important for the implementation of peptide vaccination strategies for HCV.
Supplementary Material
Acknowledgements
We would like to thank Tim Elliott and Naglaa Shoukry for their scientific insights and the individuals who donated and collected all the samples used in this study.
Financial Support
This study was supported by grants from The Wellcome Trust (SIK), the Deutsche Forschungsgemeinschaft DFG (TH-719/3-1 to RT and NE1567/1-1 to CNH) and The National Institutes of Health, grants U19 AI0663445 to GML and AYK, R01 DA033541 to AYK, and U19 AI082630 to GML.
Abbreviations
- HLA
Human Leukocyte Antigen
- HCV
Hepatitis C virus
References
- 1.Seich Al Basatena NK, Macnamara A, Vine AM, Thio CL, Astemborski J, Usuku K, Osame M, et al. KIR2DL2 enhances protective and detrimental HLA class I-mediated immunity in chronic viral infection. PLoS Pathog. 2011;7:e1002270. doi: 10.1371/journal.ppat.1002270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neumann-Haefelin C, Timm J, Schmidt J, Kersting N, Fitzmaurice K, Oniangue-Ndza C, Kemper MN, et al. Protective effect of human leukocyte antigen B27 in hepatitis C virus infection requires the presence of a genotype-specific immunodominant CD8+ T-cell epitope. Hepatology. 2010;51:54–62. doi: 10.1002/hep.23275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuniholm MH, Kovacs A, Gao X, Xue X, Marti D, Thio CL, Peters MG, et al. Specific human leukocyte antigen class I and II alleles associated with hepatitis C virus viremia. Hepatology. 2010;51:1514–1522. doi: 10.1002/hep.23515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh R, Kaul R, Kaul A, Khan K. A comparative review of HLA associations with hepatitis B and C viral infections across global populations. World J Gastroenterol. 2007;13:1770–1787. doi: 10.3748/wjg.v13.i12.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKiernan SM, Hagan R, Curry M, McDonald GS, Kelly A, Nolan N, Walsh A, et al. Distinct MHC class I and II alleles are associated with hepatitis C viral clearance, originating from a single source. Hepatology. 2004;40:108–114. doi: 10.1002/hep.20261. [DOI] [PubMed] [Google Scholar]
- 6.Fanning LJ, Kenny-Walsh E, Shanahan F. Persistence of hepatitis C virus in a white population: associations with human leukocyte antigen class 1. Hum Immunol. 2004;65:745–751. doi: 10.1016/j.humimm.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 7.Thio CL, Gao X, Goedert JJ, Vlahov D, Nelson KE, Hilgartner MW, O'Brien SJ, et al. HLA-Cw*04 and hepatitis C virus persistence. J Virol. 2002;76:4792–4797. doi: 10.1128/JVI.76.10.4792-4797.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim AY, Kuntzen T, Timm J, Nolan BE, Baca MA, Reyor LL, Berical AC, et al. Spontaneous control of HCV is associated with expression of HLA-B 57 and preservation of targeted epitopes. Gastroenterology. 2011;140:686–696. e681. doi: 10.1053/j.gastro.2010.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bangia N, Cresswell P. Stoichiometric tapasin interactions in the catalysis of major histocompatibility complex class I molecule assembly. Immunology. 2005;114:346–353. doi: 10.1111/j.1365-2567.2005.02103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lehner PJ, Surman MJ, Cresswell P. Soluble tapasin restores MHC class I expression and function in the tapasin-negative cell line .220. Immunity. 1998;8:221–231. doi: 10.1016/s1074-7613(00)80474-4. [DOI] [PubMed] [Google Scholar]
- 11.Williams AP, Peh CA, Purcell AW, McCluskey J, Elliott T. Optimization of the MHC class I peptide cargo is dependent on tapasin. Immunity. 2002;16:509–520. doi: 10.1016/s1074-7613(02)00304-7. [DOI] [PubMed] [Google Scholar]
- 12.Dalchau N, Phillips A, Goldstein LD, Howarth M, Cardelli L, Emmott S, Elliott T, et al. A peptide filtering relation quantifies MHC class I peptide optimization. PLoS Comput Biol. 2011;7:e1002144. doi: 10.1371/journal.pcbi.1002144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howarth M, Williams A, Tolstrup AB, Elliott T. Tapasin enhances MHC class I peptide presentation according to peptide half-life. Proc Natl Acad Sci U S A. 2004;101:11737–11742. doi: 10.1073/pnas.0306294101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thirdborough SM, Roddick JS, Radcliffe JN, Howarth M, Stevenson FK, Elliott T. Tapasin shapes immunodominance hierarchies according to the kinetic stability of peptide-MHC class I complexes. Eur J Immunol. 2008;38:364–369. doi: 10.1002/eji.200737832. [DOI] [PubMed] [Google Scholar]
- 15.Peh CA, Burrows SR, Barnden M, Khanna R, Cresswell P, Moss DJ, McCluskey J. HLA-B27-restricted antigen presentation in the absence of tapasin reveals polymorphism in mechanisms of HLA class I peptide loading. Immunity. 1998;8:531–542. doi: 10.1016/s1074-7613(00)80558-0. [DOI] [PubMed] [Google Scholar]
- 16.Park B, Lee S, Kim E, Ahn K. A single polymorphic residue within the peptide-binding cleft of MHC class I molecules determines spectrum of tapasin dependence. J Immunol. 2003;170:961–968. doi: 10.4049/jimmunol.170.2.961. [DOI] [PubMed] [Google Scholar]
- 17.Belicha-Villanueva A, McEvoy S, Cycon K, Ferrone S, Gollnick SO, Bangia N. Differential contribution of TAP and tapasin to HLA class I antigen expression. Immunology. 2008;124:112–120. doi: 10.1111/j.1365-2567.2007.02746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thammavongsa V, Raghuraman G, Filzen TM, Collins KL, Raghavan M. HLA-B44 polymorphisms at position 116 of the heavy chain influence TAP complex binding via an effect on peptide occupancy. J Immunol. 2006;177:3150–3161. doi: 10.4049/jimmunol.177.5.3150. [DOI] [PubMed] [Google Scholar]
- 19.Turnquist HR, Thomas HJ, Prilliman KR, Lutz CT, Hildebrand WH, Solheim JC. HLA-B polymorphism affects interactions with multiple endoplasmic reticulum proteins. Eur J Immunol. 2000;30:3021–3028. doi: 10.1002/1521-4141(200010)30:10<3021::AID-IMMU3021>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 20.Ahmad T, Neville M, Marshall SE, Armuzzi A, Mulcahy-Hawes K, Crawshaw J, Sato H, et al. Haplotype-specific linkage disequilibrium patterns define the genetic topography of the human MHC. Hum Mol Genet. 2003;12:647–656. [PubMed] [Google Scholar]
- 21.Williams AP, Bevan S, Bunce M, Houlston R, Welsh KI, Elliott T. Identification of novel Tapasin polymorphisms and linkage disequilibrium to MHC class I alleles. Immunogenetics. 2000;52:9–11. doi: 10.1007/s002510000244. [DOI] [PubMed] [Google Scholar]
- 22.Furukawa H, Kashiwase K, Yabe T, Ishikawa Y, Akaza T, Tadokoro K, Tohma S, et al. Polymorphism of TAPASIN and its linkage disequilibria with HLA class II genes in the Japanese population. Tissue Antigens. 1998;52:279–281. doi: 10.1111/j.1399-0039.1998.tb03044.x. [DOI] [PubMed] [Google Scholar]
- 23.Bukulmez H, Fife M, Tsoras M, Thompson SD, Twine NA, Woo P, Olson JM, et al. Tapasin gene polymorphism in systemic onset juvenile rheumatoid arthritis: a family-based case-control study. Arthritis Res Ther. 2005;7:R285–R290. doi: 10.1186/ar1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deshpande A, Wheeler CM, Hunt WC, Peyton CL, White PS, Valdez YE, Nolan JP. Variation in HLA class I antigen-processing genes and susceptibility to human papillomavirus type 16-associated cervical cancer. J Infect Dis. 2008;197:371–381. doi: 10.1086/524300. [DOI] [PubMed] [Google Scholar]
- 25.Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, Cheng J, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 26.Knapp S, Warshow U, Ho KM, Hegazy D, Little AM, Fowell A, Alexander G, et al. A polymorphism in IL28B distinguishes exposed, uninfected individuals from spontaneous resolvers of HCV infection. Gastroenterology. 2011;141:320–325. e321–e322. doi: 10.1053/j.gastro.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thimme R, Oldach D, Chang KM, Steiger C, Ray SC, Chisari FV. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J Exp Med. 2001;194:1395–1406. doi: 10.1084/jem.194.10.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dunn PP, Cox ST, Little AM. Sequencing protocols for detection of HLA class I polymorphism. Methods Mol Biol. 2003;210:191–222. doi: 10.1385/1-59259-291-0:191. [DOI] [PubMed] [Google Scholar]
- 29.Neumann-Haefelin C, McKiernan S, Ward S, Viazov S, Spangenberg HC, Killinger T, Baumert TF, et al. Dominant influence of an HLA-B27 restricted CD8+ T cell response in mediating HCV clearance and evolution. Hepatology. 2006;43:563–572. doi: 10.1002/hep.21049. [DOI] [PubMed] [Google Scholar]
- 30.Williams A, Peh CA, Elliott T. The cell biology of MHC class I antigen presentation. Tissue Antigens. 2002;59:3–17. doi: 10.1034/j.1399-0039.2002.590103.x. [DOI] [PubMed] [Google Scholar]
- 31.Warshow UM, Riva A, Hegazy D, Thurairajah PH, Kaminski ER, Chokshi S, Cramp ME. Cytokine profiles in high risk injection drug users suggests innate as opposed to adaptive immunity in apparent resistance to hepatitis C virus infection. J Viral Hepat. 2012;19:501–508. doi: 10.1111/j.1365-2893.2011.01574.x. [DOI] [PubMed] [Google Scholar]
- 32.Thurairajah PH, Hegazy D, Chokshi S, Shaw S, Demaine A, Kaminski ER, Naoumov NV, et al. Hepatitis C virus (HCV)--specific T cell responses in injection drug users with apparent resistance to HCV infection. J Infect Dis. 2008;198:1749–1755. doi: 10.1086/593337. [DOI] [PubMed] [Google Scholar]
- 33.Cox AL, Mosbruger T, Lauer GM, Pardoll D, Thomas DL, Ray SC. Comprehensive analyses of CD8+ T cell responses during longitudinal study of acute human hepatitis C. Hepatology. 2005;42:104–112. doi: 10.1002/hep.20749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boulanger DS, Oliveira R, Ayers L, Prior SH, James E, Williams AP, Elliott T. Absence of tapasin alters immunodominance against a lymphocytic choriomeningitis virus polytope. J Immunol. 2010;184:73–83. doi: 10.4049/jimmunol.0803489. [DOI] [PubMed] [Google Scholar]
- 35.Dong G, Wearsch PA, Peaper DR, Cresswell P, Reinisch KM. Insights into MHC class I peptide loading from the structure of the tapasin-ERp57 thiol oxidoreductase heterodimer. Immunity. 2009;30:21–32. doi: 10.1016/j.immuni.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delwart E, Slikas E, Stramer SL, Kamel H, Kessler D, Krysztof D, Tobler LH, et al. Genetic diversity of recently acquired and prevalent HIV, hepatitis B virus, and hepatitis C virus infections in US blood donors. J Infect Dis. 2012;205:875–885. doi: 10.1093/infdis/jir862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brant LJ, Ramsay ME, Tweed E, Hale A, Hurrelle M, Klapper P, Ngui SL. Planning for the healthcare burden of hepatitis C infection: Hepatitis C genotypes identified in England, 2002–2007. J Clin Virol. 2010;48:115–119. doi: 10.1016/j.jcv.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 38.Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O'Huigin C, Kidd J, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Badrinath S, Saunders P, Huyton T, Aufderbeck S, Hiller O, Blasczyk R, Bade-Doeding C. Position 156 influences the peptide repertoire and tapasin dependency of human leukocyte antigen B*44 allotypes. Haematologica. 2012;97:98–106. doi: 10.3324/haematol.2011.046037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Subramaniam NS, Morgan EF, Lee CY, Wetherall JD, Groth DM. Polymorphism of sheep MHC Class IIb gene TAPASIN. Vet Immunol Immunopathol. 2010;137:176–180. doi: 10.1016/j.vetimm.2010.04.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.