Abstract
Introduction
We performed a whole-transcriptome correlation analysis, followed by the pathway enrichment and testing of innate immune response pathways analyses to evaluate the hypothesis that transcriptional activity can predict cortical gray matter thickness (GMT) variability during normal cerebral aging
Methods
Transcriptome and GMT data were availabe for 379 individuals (age range=28–85) community-dwelling members of large extended Mexican-American families. Collection of transcriptome data preceded that of neuroimaging data by 17 years. Genome-wide gene transcriptome data consisted of 20,413 heritable lymphocytes-based transcripts. GMT measurements were performed from high-resolution (isotropic 800µm) T1-weighted MRI. Transcriptome-wide and pathway enrichment analysis was used to classify genes correlated with GMT. Transcripts for sixty genes from seven innate immune pathways were tested as specific predictors of GMT variability.
Results
Transcripts for eight genes (IGFBP3, LRRN3, CRIP2, SCD, IDS, TCF4, GATA3, HN1) passed the transcriptome-wide significance threshold. Four orthogonal factors extracted from this set predicted 31.9% of the variability in the whole-brain and between 23.4 and 35% of regional GMT measurements. Pathway enrichment analysis identified six functional categories including cellular proliferation, aggregation, differentiation, viral infection, and metabolism. The integrin signaling pathway was significantly (p<10−6) enriched with GMT. Finally, three innate immune pathways (complement signaling, toll-receptors and scavenger and immunoglobulins) were significantly associated with GMT.
Conclusion
Expression activity for the genes that regulate cellular proliferation, adhesion, differentiation and inflammation can explain a significant proportion of individual variability in cortical GMT. Our findings suggest that normal cerebral aging is the product of a progressive decline in regenerative capacity and increased neuroinflammation.
Introduction
The aging process is characterized by a decline in regenerative capacity, reduced repair potential, increased susceptibility to disease and eventual loss of function (Lazarov et al., 2011; Villeda et al., 2011). Cerebral aging is associated with a precipitous decline in adult stem cell capacity beyond what is necessary to maintain the integrity of cerebral tissue (Sharpless and DePinho, 2007; Shook et al., 2011; van Praag et al., 2005) and with activation of the inflammatory pathways that prime the brain for neurodegenerative cascades (Cribbs et al., 2011; Cunningham et al., 2005; Holmes et al., 2009). Cerebral aging trajectories can vary greatly among individuals. Healthy cerebral aging is characterized by the lack of cerebral atrophy and retention of normal cognitive function, with some decline in regenerative capacity but no increase in neuroinflammation (Salthouse, 2009). Neurodegenerative aging is characterized by an abrupt decline in cognition and is associated with increased neuroinflammation, microglial activation, and accumulation of neuroinflammatory proteins and beta-amyloid plaques (Capell et al., 2007; Dodge et al., 2011; Kirkpatrick et al., 2008; Royall et al., 2011; Salthouse, 2009; Schillerstrom et al., 2008; Wolkowitz et al., 2011). Neuroinflammation was assumed to be minimal in healthy aging; yet more recent observations suggest that activation of innate immune pathways may still occur even in healthy cerebral aging (Berchtold et al., 2008; Cribbs et al., 2011). Familial history explains a very large proportion (40–80%) of the variance in individual trajectories in many imaging-based phenotypes of in cerebral aging, including cortical gray matter thickness (GMT) and others (Chiang et al., 2011; DeStefano et al., 2006; Kent et al., 2012; Kochunov et al., 2010a; Kochunov et al., 2009a; Turner et al., 2005; Winkler et al., 2010). However, the search for specific genotypes leading to aging-related disorders had so far identified candidate genes that explain only a small proportion (1–2%) of the total risk (Biffi et al., 2011; Chouliaras et al., 2010). The risk of neurodegenerative aging is likely to be modulated by both genotype and environment, and genome-wide association analyses generally cannot account for their interaction (Chouliaras et al., 2010; Kamboh et al., 2011; Kent et al., 2012; Tanzi, 2012; Weinstein et al., 2011). In contrast, transcriptional profiling analyses that measure the expression of genes are sensitive to both genotype and environment and therefore may offer the potential to clarify the pathophysiology of cerebral aging. Hence, we proposed to examine how the changes in transcriptional profiles associated with regenerative and immune functions influence cerebral aging. We performed a correlation analysis between gene expression data and measurements of cortical GMT, collected seventeen years apart. We hypothesized that transcriptional profiling can be used to identify specific genes and pathways associated with decline in cortical GMT during this time period. Second, we hypothesized that markers of activated immune system, reported from a brain-tissue microarray study (Cribbs et al., 2011), will predict decline in GMT. Both hypotheses were tested in a large number of well-characterized, community-dwelling, cognitively-normal aging Mexican-Americans from large extended families (Mitchell et al., 1996).
Analysis of transcriptional profiling is a promising method for identifications of risk factors for measurements of brain aging, such as reduced GMT (Bihaqi et al., 2011; Cribbs et al., 2011; Maloney et al., 2011; Sequeira et al., 2012; Wood et al., 2012). We chose the GMT because it is sensitive to aging-related decline in cortical integrity (Kochunov et al., 2008; Kochunov et al., 2007; Leritz et al., 2011; Salat et al., 2009). GMT is calculated as a symmetric distance from the outer cortical surface to the inner cortical gray-white-matter interface (Aganj et al., 2009; Fischl and Dale, 2000; Lerch and Evans, 2005). Cortical GMT undergoes a significant reduction during aging, which is related to decline in neuronal density and inter-layer myelin contents (Jelsing et al., 2005; Lerch and Evans, 2005; Selemon et al., 1995; Sowell ER et al., 2003). During normal aging, the average thickness of the gray matter mantle is reported to decrease from an average thickness of approximately 2.5mm during the 3rd-to-4th decades of life to less than 2mm during the 8-to-9th decades (Kochunov et al., 2011b; Magnotta VA et al., 1999; Raz N et al., 1997). Importantly, the rate of GMT decline is correlated with progression of neuropsychological deficits in aging and psychiatric disorders (Carless et al., 2011; Eskildsen et al., 2012; Kochunov et al., 2009b; Narr et al., 2007; Turner et al., 2012) and is a sensitive diagnostic factor for neurodegenerative aging (Li et al., 2012; Risacher et al., 2009; Thompson PM et al., 2003; Thompson et al., 2004). The whole-brain and regional GM measurements are influenced by additive genetic factors and demonstrate high heritability (40–60%) (Panizzon et al., 2011; Rijsdijsk et al., 2011; Turner et al., 2012; Winkler et al., 2010).
We performed these analyses in a well-studied, community dwelling, normally aging population of randomly selected Mexican American families. Genome-wide gene expression measures were collected seventeen years prior to neuroimaging, hence providing a prospective element to cortical outcomes. Prior analyses in this population established significant heritability for both the transcriptional measurements (Goring et al., 2007; Moses et al., 2007) and the GM thickness (Winkler et al., 2010). Here, we tested the next logical hypothesis: that there should be a significant degree of shared variance between gene expression and cortical integrity, and that transcriptional profiling data can be used to identify pathways that control cerebral maintenance and repair. Additionally, we tested the hypothesis that activation of innate immune response will be associated with reduced cortical integrity as suggested by a microarray expression analysis in postmortem brain samples (Cribbs et al., 2011). Innate immune response is the first line of defense in response to pathogens and tissue damage. A microarray analysis of human brain tissue demonstrated a strong aging-related upregulation in innate immune pathways including: complement signaling, toll-like receptors, inflammasome, chemokine ligands and receptors, scavenger and immunoglobulin receptors and major histocompatibility I and II pathways (Cribbs et al., 2011). The transcripts for the gene sets identified by that analysis were used to test the hypothesis of activated immune system involvement in contributing to reduced cortical integrity in normal aging.
Methods
Subjects
The data are derived from 379 (40.6% male) active participants in the Getetics of Brain Structure and Function (GOBS) study (Glahn et al., 2012), for whom gene expression and brain imaging data were available. These individuals are of Mexican American descent from large extended families selected randomly from the community. Subjects ranged from 28 to 85 years of age (47.1± 12.7 years) at the time of imaging and were members of 47 families (9.3±8.3 individuals/family; range 2–38). Subjects’ demographic and health information is detailed in Table 1. All subjects provided written informed consent on forms approved by the Institutional Review Board of the University of Texas Health Science Center at San Antonio (UTHSCSA) and Yale University.
Table 1.
Subjects’ demographic and health information including obesity (body mass index>30), the number of subjects with hypercholesterolemia (total cholesterol levels > 200 mg/dl) and hyperlipidemia (blood lipids>150 mg/dL), the number of subjects diagnosed with coronary heart disorder, diabetes, hypertension, the number of subjects taking antihypertensive medications, the number of subjects with alcohol dependence (AD) and the number of subject with a lifetime major depression (MD) episode.
| Age | N subjects (M/F) | Nobese | Nhyper- cholesterole mia |
Nhyper- lipidemia |
Nheart disorder |
Ndiabetes | Nhypertension | Nanti- hypertensive meds |
Nad | Nmd |
|---|---|---|---|---|---|---|---|---|---|---|
| 28–30 | 19 (9/10) | 10 | 1 | 0 | 0 | 0 | 1 | 0 | 2 | 3 |
| 31–40 | 117 (56/61) | 32 | 8 | 3 | 0 | 3 | 18 | 3 | 8 | 18 |
| 41–50 | 92 (30/62) | 32 | 22 | 6 | 2 | 19 | 16 | 8 | 7 | 27 |
| 51–60 | 68 (22/46) | 26 | 23 | 4 | 4 | 24 | 15 | 10 | 2 | 24 |
| 61–70 | 54 (22/32) | 16 | 27 | 10 | 7 | 32 | 18 | 10 | 2 | 9 |
| 71–85 | 19 (8/11) | 6 | 10 | 3 | 3 | 12 | 7 | 5 | 0 | 5 |
Magnetic resonance imaging and data processing
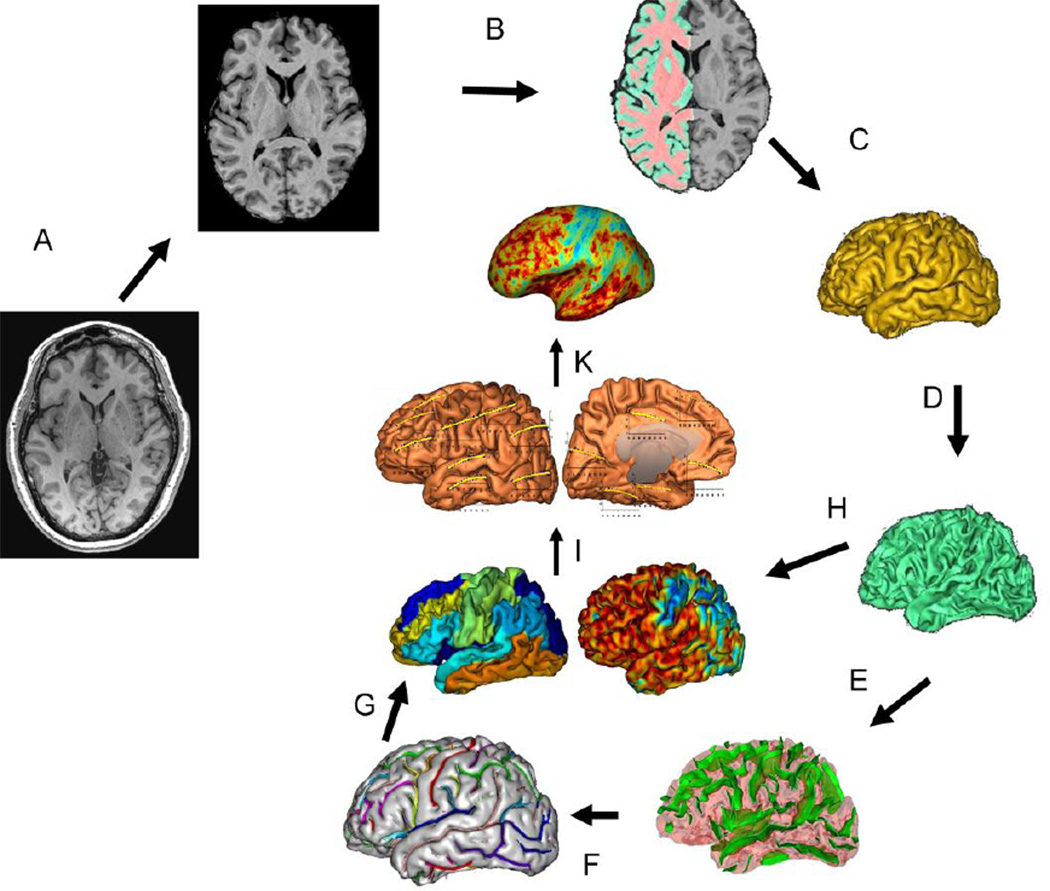
Imaging data were collected using a Siemens 3 Tesla Trio scanner located at the Research Imaging Institute, UTHSCSA. The details of the imaging protocol specifically optimized for measurements of GMT and data collection procedures are described elsewhere (Kochunov et al., 2011b). In short, the protocol for measurements of GMT was designed to collect data capable of resolving the cortical ribbon across the cerebral cortex using an isotropic spatial resolution of 800µm (voxel volume=0.5mm3). T1-weighted contrast was achieved using a magnetization prepared sequence with an adiabatic inversion contrast-forming pulse (scan parameters: TE/TR/TI=3.04/2100/785 ms, flip angle=11 degrees). A retrospective motion correction technique (Kochunov et al., 2006) was used to reduce artifacts related to subject motion. Whole-brain average and regional measurements of cortical thickness were performed using a freely available BrainVisa software (Kochunov et al., 2011d) (Figure 1) (http://www.nitrc.org/projects/brainvisa_ext/). This tool uses a “normal-average” algorithm that has been reported to be a good compromise between accuracy and performance (Lerch and Evans, 2005). The GMT is measured as the Euclidean distance from an inner mesh vertex to the outer mesh along the direction normal to the inner mesh polygon. The measurement is repeated, inward, along the direction normal the outer mesh and the two distances are averaged (Kochunov et al., 2008). The consistency of the distance measurements is ensured by verifying that the line connecting the two surfaces does not intersect other polygons, inner or outer, along the way. GMT measurements were averaged for individual cortical areas across hemispheres; the whole-brain regional GMT was calculated as average values for fourteen major cortical areas and vertex-wise as described elsewhere (Kochunov et al., 2012; Kochunov et al., 2011b).
Figure 1.
T1-w image processing pipelines. A T1-w image is skull-stripped, globally spatially normalized, and RF-inhomogeneity corrected (A). Next, cerebral hemispheres and cerebellum and identified and tissue classified (B); cortical surfaces for GM and WM are calculated (C;D) and homotopic erosion operation and crevasse detector are used to reconstruct sulcal surface as the medial surface of the two opposing gyral banks (E). Sulcal identification pipeline uses a congregation of 500 artificial neural network-based pattern classifiers to identify (F) sulcal landmarks and to perform gyral segmentation of the cortex (G). GM thickness (GTM) was calculate as the distance between the pial (C) and GM/WM interface surfaces (H). Analyses are performed by measuring GMT for fourteen cortical areas (I) and vertex-wise (K).
Gene expression measurements
Transcriptome-wide gene expression data consisted of 20,413 individual, significantly heritable transcript measurements (Goring et al., 2007). Transcriptome-wide gene expression data consisted of 20,413 individual, significantly heritable transcript measurements generated by Göring and colleagues (Goring et al., 2007). Gene expression measures were derived from untransformed leukocytes collected seventeen years prior to imaging and stored in liquid nitrogen until technology for transcriptional profiling became readily available (Goring et al., 2007). The details of synthesis, amplification, purification and gene expression measurement protocols are described elsewhere (Goring et al., 2007). In short, lymphocytes were obtained from blood samples collected in the morning after an overnight fast. Lymphocytes were isolated from a 10-ml sample using Histopaque (Sigma Chemical Co.), following the suggested protocol of the manufacturer. The isolated and washed lymphocytes were frozen and transferred to liquid nitrogen tanks to prevent RNA degradation during long term storage. Total RNA was isolated from lymphocyte samples using a modified procedure of the QIAGEN RNeasy 96 protocol for isolating total RNA from animal cells using spin technology (QIAGEN, Inc.). Its integrity was verified and anti-sense RNA was synthesized, amplified and purified. Hybridization of anti-sense RNA was carried out using Illumina’s BeadChip 6×2 protocol. Total isolated RNA was hybridized to Illumina Sentrix Human Whole Genome (WG-6) Series 1 BeadChips as previously described (Moses et al., 2007). Processing of the sample was performed in a single batch, thereby eliminating the batch effect. Expression levels were z-normalized to make the expression phenotypes comparable among individuals and across transcripts (Goring et al., 2007; Moses et al., 2007). Finally, each transcript’s residual expression scores were normalized by using an inverse Gaussian transformation across individuals to ensure normality (Goring et al., 2007; Moses et al., 2007). The final dataset consisted of 20,413 transcripts that passed quality control and assurance (Goring et al., 2007).
Data analyses
Variance component, mixed-model analysis methods, as implemented in SOLAR version 6.61 (Almasy et al., 1997) and optimized for binary imaging data (http://www.nitrc.org/projects/se_linux/) were used to calculate the degree of shared variability between gene expression measurements and indexes of cerebral integrity.
Step 1. Whole-transcriptome correlation analysis
Bivariate correlation analysis was used to calculate the magnitude and significance of phenotypic correlation coefficient between GM thickness and the 20,413 individual heritable transcripts. A transcriptome-wide Bonferroni threshold (p=0.05/20413=2.44•10− 6) was set for the significance of the correlation. Age and sex were included as covariates in all analyses. The health screening data shown in Table 1 were not used as covariates due to the loss of statistical power to detect genetic relationships caused by the overlap in risk factors (Kochunov et al., 2010b; Kochunov et al., 2011c).
Step 2. Factor analysis
Factor analysis was performed on transcript data that passed the statistical threshold for multiple comparisons to produce the smallest network. The purpose of this step was to reduce the number of correlated variables and to produce non-collinear estimates of transcriptional activity. The results of the factor analysis were used to estimate the proportion of variance explained by the transcriptional activity and to test its regional specificity. Factor analysis used principal components analysis (PCA) to extract linear composites of correlated variables with eigenvalues greater than one. PCA yielded eigenvalues describing the amount of variance among variables explained by a factor. A varimax rotation was then used to remove co-linearity, for example to orthogonalize individual eigenvectors. Results of the factor analysis yielded factor loadings (correlations between a variable and a factor) and factor scores (transcript’s standardized score on each factor).
Step 3. Regression analysis
A two-stage regression was performed to probe the multivariate effects of transcriptional factors on variability in the global and regional GMT. The GMT values were used as dependent variable, transcript data was inserted at the first step and age was inserted at the second step as predictor variables. This yielded the degree of variance described at each entry step and whether the change was significant. It also produced standardized significance estimates of the linear associations between GMT (the dependent) and transcriptional factors (the predictors). The threshold for statistical significance for this analysis was set at p≤1.62•10−5, which corresponded to a 1% FDR value for the correlation between GMT and transcriptome data.
Step 4. Pathway analysis
Genes whose expression levels were correlated with GMT at 10% (this corresponded to p≤2•10−3) false discovery rate (FDR) (Benjamini and Hochberg, 1995), as suggested by others (Lee et al., 2011; Purcell et al., 2009) were analyzed using Ingenuity Pathway Analysis (www.ingenuity.com) by mapping them to objects within the Ingenuity Knowledge Base. IPA uses literature-curated data to identify over-representation of functional classes and canonical pathways within the imported set of genes. The right-tailed Fisher’s exact test was used to calculate a p-value determining the probability that each biological function, canonical pathway or disease assigned to that dataset was due to chance alone; by comparing the number of user-imported genes in a given function or pathway relative to the expected number of occurrences based on the reference set. For these analyses the entire set of 20,413 detected transcripts was used as the reference set. The genes of interest were overlaid onto a global molecular network developed from literature reported connectivity recorded in the Knowledge Base, allowing the generation of gene networks; graphical representation of the molecular relationships between genes/gene products.
Step 5. Innate-Immune pathway analysis
Expression levels for sixty genes in seven innate immune response pathways that showed a significant aging-related change in brain tissue(Cribbs et al., 2011) were used as predictors of GM variability (Table S1, see supplement). The details of the genes and pathways selection are described in the original study (Cribbs et al., 2011). This analysis was performed using the Affymetrix Human Genome Hg-U133 chip, precluding an exact match with our transcriptional probes; therefore the match was made by selecting transcripts within the same genes from the Illumina array. The gene sets that were included in the analysis are shown in Table S1. The complement signaling pathway was represented by fourteen genes that modulate antibody response. The toll-like receptors (TLR) pathway was represented by twelve genes that regulate the activation of microglia and perivascular macrophages. The inflammasome and the scavenger and immunoglobulin pathways were represented by genes (six and eight respectively) that regulate initiation of inflammation. The chemokine receptor pathway was represented by six genes that recruit immune cells to the site of inflammation. The major histocompatibility II/I pathways were represented by nine and seven genes respectively that that mediate interaction between leukocytes and other cells. Two sets of analyses were performed; first, we calculated correlation coefficients between individual transcripts and whole-brain average GM thickness and second, we calculated the degree of variability explained by each of the pathways in the whole-brain and three regional GMT measurements for the superior, post-central and fusiform gyri. The regional measurements were chosen to match the location of brain tissue where the original microarray study was performed (Cribbs et al., 2011).
Results
The transcriptome-wide correlation analysis identified eight transcripts that satisfied the threshold (p≤2.44•10−6) for significant correlation with whole-brain GM thickness (Table 2). The most significant was a negative correlation with the insulin-like growth factor-binding protein 3 (IGFBP3), followed by positive correlation with the transcript for the leucine-rich repeat neuronal protein 3 (LRRN). The factor analysis of the eight transcripts produced four orthogonal factors (eigenvalues>1) that captured 71% of the variability (Tables 3). Factor 1 loaded equally on the transacting T-cell-specific transcription factor (GATA3) and cysteine-rich intestinal protein (CRIP) gene transcripts and explained 23.8% of variability. Factor 2, loaded the iduronate-2-sulfatase (IDS) gene transcript, explained 17.9% of variability. Factors 3 and 4 loaded on the stearoyl-CoA desaturase (SCD) and the transcription factor 4 (TCF4) gene transcripts and explained 16.5% and 13.2% of the variability in the eight transcripts, respectively (Table 3).
Table 2.
Results of the whole-transcriptom correlation analysis (r (p)) for three global measurements of cerebral integrity. Bolded values indicate correlations coefficients that satisfied correction for whole-transcriptome multiple comparison (p<2.5•10−6 ).
| Transcript | Gene | Whole-brain GM thickness |
|---|---|---|
| GI_19923110-S | IGFBP3 | −.35 (1.1•10−10) |
| GI_37059785-S | LRRN3 | .35 (1.4•10−10) |
| GI_31542322-S | CRIP2 | −.32 (8.7•10−10) |
| GI_31982900-S | SCD | .31 (1.8•10−9) |
| GI_5360215-I | IDS | −.28 (8.6•10−8) |
| GI_4507398-S | TCF4 | .29 (1.5•10−7) |
| GI_4503928-S | GATA3 | −.26 (5.8•10−7) |
| GI_7705876-S | HN1 | −.27 (8.4•10−7) |
Table 3.
Results of the factor analysis for eight transcripts. Principal components extraction was followed by varimax rotation to produce four orthogonal factors.
| Transcript | Gene | GATA/CRIP- factor |
IDS- factor |
SCD- factor |
TCF4- factor |
|---|---|---|---|---|---|
| GI_4503928-S | GATA3 | 0.45 | 0.06 | 0.19 | −0.04 |
| GI_31542322-S | CRIP2 | 0.45 | −0.28 | −0.22 | 0.13 |
| GI_19923110-S | IGFBP3 | 0.36 | 0.17 | 0.03 | 0.11 |
| GI_37059785-S | LRRN3 | 0.13 | −0.53 | 0.16 | −0.10 |
| GI_4507398-S | TCF4 | 0.11 | 0.02 | 0.05 | 0.94 |
| GI_5360215-I | IDS | 0.10 | 0.59 | 0.20 | −0.05 |
| GI_31982900-S | SCD | 0.02 | 0.13 | 0.60 | 0.23 |
| GI_7705876-S | HN1 | −0.01 | −0.02 | −0.57 | 0.15 |
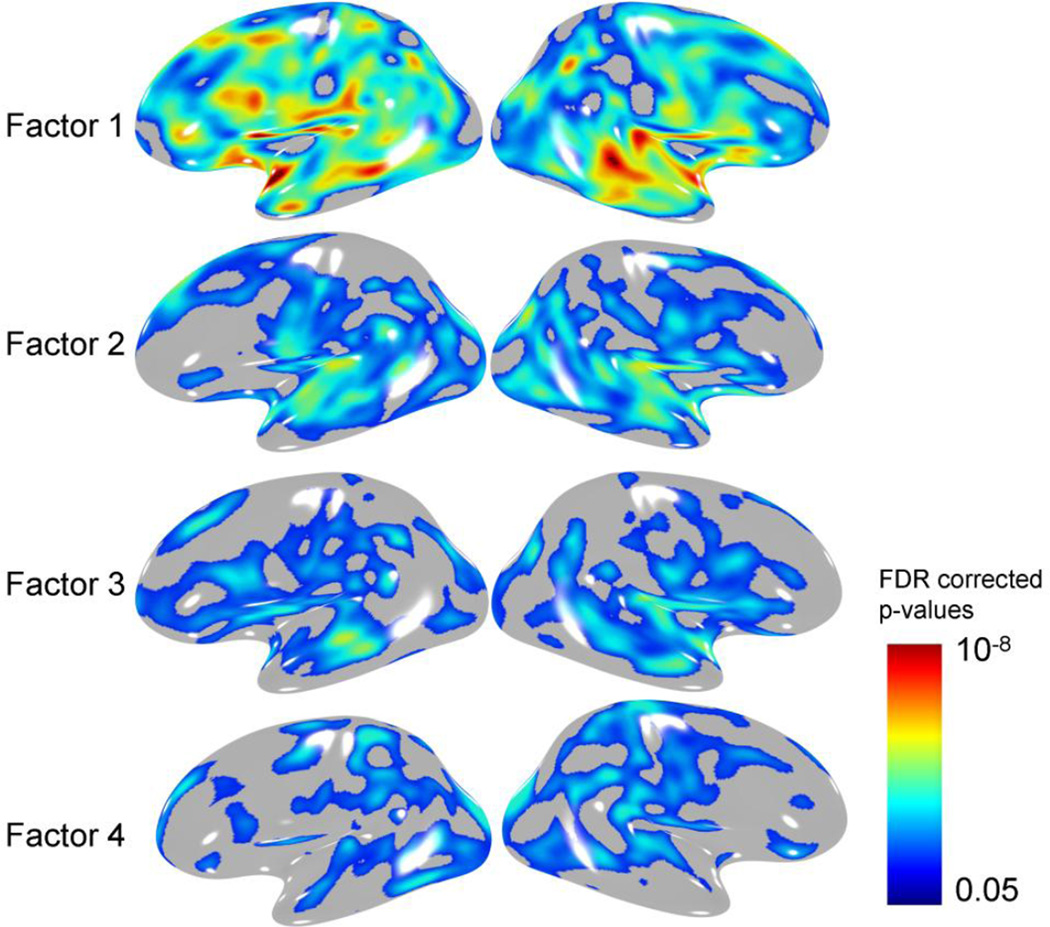
Regression analysis where the four orthogonal factors were used as predictors at the first step, explained 31.9% of the variability in the whole-brain average GM thickness and between 23.4 and 35% of regional GM thickness measurements (Table 4). These factors fully accounted for variability captured by age as its insertion at the second step did not significantly add to the model. Whole-brain GMT measurement showed the strongest association (t=−8.94, p<1•10−18) with the GATA/CRIP-Factor. This factor also explained the largest proportion of variability for eight out of fourteen regional GM measurements, followed by the SCD-factor, which explained the largest proportion of variability in four regional measurements. The TCF4-factor explained the smallest proportional of variability and was only nominally significant (1.6·10−5<p≤.05) in association with the GMT measurements (Table 4). The results of the vertex-wise GMT analysis (Figure 2) closely mirrored this pattern.
Table 4.
Results (t-score (probability) ) of the two stage regression analysis performed for gray matter thickness (GMT) measurements. Bolded values are significant at p<1.6 10−5
| GMT traits | Primary function |
GATA/CRIP- factor (t-score, (p)) |
IDS-factor (t- score, (p)) |
SCD-factor (t- score, (p)) |
TCF4-factor (t-score, (p)) |
%Variance Explained (p) |
|---|---|---|---|---|---|---|
| Whole-brain | −8.94 (<1•10−18) | −5.76 (1.8•10−8) | 7.62 (1.4•10−13) | 4.4 (1.4•10−5) | 31.9% (<1•10−18) | |
| Sup. Frontal gyrus | Associative | −8.21 (<1•10−15) | −6.6 (2.2•10−10) | 7.6 (2.6•10−13) | 4.0 (1.1•10−5) | 34.4% (<1•10−18) |
| Interm. Frontal gyrus | Associative | −8.0 (1.8•10−14) | −6.0 (6.0•10−9) | 7.8 (6.0•10−14) | 4.4 (1.5•10−5) | 35.0 (<1•10-18) |
| Inf. Frontal Gyrus | Associative + Motor |
−7.7 (1.3•10−13) | −5.3 (2.4•10−7) | 7.5 (2.6•10−13) | 5.69 (3.8•10−4) | 31.7 (<1•10−18) |
| Precentral gyrus | Motor | −7.3 (1.8•10−12) | −6.0 (4.7•10−9) | 6.8 (5.7•10−11) | 4.0 (7.5•10−5) | 29.3 (<1•10−18) |
| Postcentral gyrus | Sensory | −7.2 (4.0•10−12) | −7.6 (5.4•10−7) | 5.78 (1.6•10−8) | 4.3 (2.5•10−5) | 27.7 (<1•10−18) |
| Sup. Parietal Lobule | Associative | −6.3 (1.1•10−9) | −5.5 (7.5•10−8) | 5.8 (1.7•10−8) | 3.7 (2.8•10−4) | 24.6 (<1•10−18) |
| Inf. Parital Lobule | Associative | −7.1 (6.7•10−12) | −5.8 (1.6•10−8) | 6.6 (1.7•10−10) | 3.2 (1.5•10−4) | 27.9 (<1•10−18) |
| Sup. Temporal gyrus | Sensory + Associative |
−7.8 (4.8•10−14) | −6.1 (3.6•10−9) | 6.5 (2.2•10−10) | 3.9 (1•10−4) | 30.0 (<1•10−18) |
| Inter+Inf Temporal gyri | Associative | −6.3 (7.3•10−13) | −5.2 (2.8•10−7) | 5.9 (6.4•10−9) | 3.81 (1.5•10−4) | 24.9 (<1•1018) |
| Cingulate gyrus | Associative | −8.3 (2.2•10−15) | −5.8 (1.8•10−8) | 8.2 (4.7•10−15) | 3.1 (1.9•10−3) | 34.1 (<1•10−18) |
| Medial Occipital area | Visual | −7.1 (6.7•10−12) | −5.8 (1.6•10−8) | 6.6 (1.6•10−10) | 3.83 (1.5•10−4) | 27.9 (<1•10−18) |
| Lateral Occipital area | Visual | −6.2 (1.7•10−9) | −5.3 (1.7•10−7) | 5.9 (9.5•10−10) | 3.31 (1.3•10−4) | 23.4 (<1•10−18) |
| OrbitoFrontal gyrus | Associate+ Sensory |
−7.1 (6.3•10−12) | −4.6 (4.8•10−6) | 7.4 (8.8•10−13) | 3.2 (1.3•10−4) | 28.8 (<1•10−18) |
| Fusiform gyrus | Associative + Memory |
−5.9 (7.1•10−9) | −3.4 (8.0•10−4) | 6.4 (4.9•10−10) | 2.4 (0.02) | 26.4 (<1•10−18) |
Figure 2.
The pattern of significant (FDR-corrected p-values of 0.05) correlation between four transcriptional factors and regional GMT measurements.
IP A analysis of whole-brain average GMT was performed using the 474 gene transcript significant at the 10% FDR threshold (Table S2, see supplement). GMT correlated transcripts showed significant (p≤1.6×10−5) enrichment for seven functional categories (Table 5); including proliferation and aggregation of cells, quantity of blood cells, cell differentiation, viral infection, free radical scavenging and vascular diseases (Table 5). Transcripts for five of the genes significant at the Bonferroni level were enriched in the cell proliferation category (CRIP2, GATA3, IGFBP3, LNRN3 and TCF4; Table 5). Analysis of canonical pathway enrichment identified a strong signal for the integrin signaling pathway (p=3.2·10−6), which included seventeen significant genes primarily negatively correlated with GM thickness (Figure S1).
Table 5.
Functional categories for the gene significantly associated with GMT that were identified by the Inginuity pathways analysis.
| Function Annotation |
Enrichment (p-value) |
Effect on function (z score) |
Number of genes |
Genes correlated with GMT at a 10% FDR |
|---|---|---|---|---|
| Proliferation of cells |
1.3·10−8 | −1.88 | 143 |
ABCC3, ACRBP, ADORA2A, ALDH1A1, ALOX12, ATP2A2, BAD, BATF, BCL2L1 BMP6, CAMK2N1, CAPN1, CAPNS1, CBX7, CCDC92, CCL5, CD163, CD2, CD320 CD38, CD99, CDCA7L, CDKN2D, CELA1, CLECL1, CLU, CNR2, CRIP1, CRIP2 CRLF3, CTBP2, CTSL1, CXCL5, E2F2, E2F5, EFEMP1, EGF, EIF2AK1, ENTPD1 FABP5, FBLN2, FDXR, FGF18, FHL1, FHL2, FLNA, FXYD1, GAPT, GATA3 GDPD5, GFI1, GLCE, GRAP2, HRAS, IFNGR1, IGFBP3, IL10RA, IL12RB1 IL13RA1, IL2RA, IL32, IL6ST, ILK, INPP4B, IRF1, ITGA2B, ITGB1, ITGB3 KIDINS220, LCN2, LMNA, LRRN3, LTBP4, MCM5, MEF2C, MEIS1, MGAT4B MORC3, MPL, MSI2, MVP, MYL9, MYLK, NFIB, NRCAM, NREP, NRIP1, OPTN PAK1IP1, PARVB, PCBP4, PDIA5, PDZK1IP1, PF4, PFKP, PIM1, PLCD1, PNPT1 POLD4, PPBP, PPP1CA, PRKCSH, PRKD3, PTGS1, PURA, RALGDS, RAPGEF1 RARRES3, RHOC, RORA, SELP, SERPINF1, SERTAD2, SH2D2A, SLC9A3R1 SMOX, SNCA, SPARC, SPTAN1, STUB1, TAGLN2, TAL1, TAX1BP3, TCF4, TFAM TFPI, TIMP1, TJP3, TNFRSF17, TNFRSF21, TNFRSF4, TRADD, TRPV2, TSPAN3 TXNDC5, VASH1, VAV1, VWF, WBP2, WDR12, XBP1, XPC, XRCC5 |
| Aggregation of cells |
2.1·10−8 | −3.35 | 26 |
ADORA2A, ALOX12, CAPN1, CCL5, CD2, CD38, CLEC1B, CLU, EGF, ENTPD1 ESAM, FERMT3, GP1BA, GP5, GP6, ITGA2B, ITGB1, ITGB3, MPL, SELP, SEPT5, SERPINF1, TIMP1, TREML1, VAV1, VWF |
| Quantity of blood cells |
5.8·10−8 | −0.12 | 58 |
ABCC10, ADORA2A, AHSP, BAD, BATF, BCL11A, BCL2L1, CAPNS1, CCL5, CD38 CFH, CLEC1B, CNR2, E2F2, EIF2AK1, ENTPD1, EPB49, FABP5, FERMT3, FLNA GAPT, GATA3, GFI1, GP1BA, GPR68, GRAP2, HBA1/HBA2, HBB, HRAS, IFNGR1 IGJ, IL10RA, IL12RB1, IL13RA1, IL2RA, IL6ST, IRF1, ITGA2B, ITGB3, LCN2, MAF MEIS1, MPL, PF4, PIM1, PURA, SELP, SNCA, TAL1, TCF4, TIMP1, TNFRSF17 TNFRSF21, TNFRSF4, TREML1, VAV1, XBP1, XRCC5 |
| Differentiation of cells |
7.9·10−8 | −3.21 | 90 |
ADORA2A, AHSP, ALOX12, BAD, BATF, BCL11A, BCL2L1, BMP6, CA2, CAPNS1 CCDC92, CCL5, CD2, CD38, CDKN2D, CELA1, CLU, CNR2, CRIP2, CTBP2, E2F2 EGF, FABP5, FGF18, FHL2, GATA3, GDPD5, GFI1, GP1BA, GPR68, GRAP2, HBB, HDAC9, HEMGN, HRAS, IFITM2, IFNGR1, IGFBP3, IL10RA, IL12RB1, IL2RA, IL32 IL6ST, ILK, INPP4B, IRF1, ITGB1, ITGB3, ITM2C, L3MBTL1, LCN2, LMNA, LTBP4 MAF, MEF2C, MEIS1, MME, MPL, NDST1, NFIB, NREP, PACSIN1, PBXIP1, PF4 PIM1, PLCD1, PPP1CA, PRKD3, PTGS1, RAB23, RALGDS, RARRES3, RDH10 RORA, SCAND1, SCD, SELP, SERPINF1, SLA2, SNCA, SPARC, TAL1, TCF4 TFAM, TIMP1, TMBIM1, TNFRSF4, VAV1, XBP1, XRCC5 |
| Viral Infection | 2.1·10−7 | −1.56 | 82 |
ABCE1, ABLIM3, ALOX12, APOL1, ARPC3, ATP6V1A, BCL11A, BCL2L1, CA2 CACNA2D3, CCL5, CD38, CLU, CTBP2, CTSL1, CXCL5, DDX10, DLGAP4, E2F2 EGF, EXOSC5, FERMT3, FGD6, FLNA, HEMGN, HIST2H2BE, HRAS, IFITM2 IFNGR1, IGJ, IL10RA, IL12RB1, IL2RA, IL32, IRF1, ITGA2B, ITGB1, ITGB3 KATNB1, KBTBD11, LCN2, LMNA, LRRN3, MARCH2, MED16, MGLL, MPL, OPTN PARVB, PF4, POLR2I, PPBP, PTGS1, PURA, RAPGEF1, RARRES3, RHOC RNF214, SBF2, SLC9A3R1, SNRPA, SNX10, SPARC, SPATS2L, SPTAN1 TAGLN2, TAL1, TCFL5, TECR, TIMP1, TNFRSF4, TRADD, TRPV2, TUBA4A TUBB4B, UBE2E2, VAV1, VPS4A, VWF, XBP1, ZNF594, ZSCAN16 |
| Metabolism of reactive oxygen species |
4.1·10−7 | −2.23 | 30 |
AHSP, BCL2L1, CCL5, CFH, CTTN, EGF, ENTPD1, FAAH, FDXR, GZMH HBA1/HBA2, HBB, HRAS, IL32, ILK, ITGB1, ITGB3, MYLK, PIM1, PNPT1, PTGS1 RALGDS, RAPGEF1, RRAS, SELP, SERPINF1, SMOX, SNCA, TFAM, XBP1 |
The predictive power for the transcripts of genes involved in the regulation of the innate immune response was analyzed in two ways. First, we performed a correlation analysis for whole-brain GM thickness (Table 2S). The correlation coefficients for two transcripts, the clusterin gene (CLU) from the signaling pathway and the Fc fragment of IgG binding protein (FCGBP) gene from the scavenging pathway passed the threshold of statistical significance (p=8·10−4) after correcting for multiple (N=60) comparisons. The correlation with the CLU transcript was negative (r=−0.23; p=3.1•10−5), while the correlation with the FCGBP transcript was positive (r=0.22; p=5.2•10−5). Ten more transcripts (C1QA, C1QB, C4A, TLR2, TLR4, TLR8, CASP1, CD163, HLA-DPA1, HLA-DRA) showed nominally significant correlations (8·10−4<p≤.05). Regression analyses for the whole-brain and regional GM thickness were significant for the complement signaling and scavenger and immunoglobulins pathways (Table 6). In addition, the toll-receptors pathway demonstrated significant association with GM thickness of the fusiform gyrus.
Table 6.
Results of the regression analysis for the whole-brain and three regional GMT measurement and seven innate-immune response pathways. Bolded values are significant at p<1.610−5
| Innate Inflammation Pathways |
Whole-brain, % variance explained (p) |
Superior Frontal Gyrus% variance explained (p) |
Post-Central Gyrus, % variance explained (p) |
Fusiform gyrus, % variance explained (p) |
|---|---|---|---|---|
| Complement Signaling |
10.1 (1.8•10 −9) | 11.7 (8.1•10 −11) | 7.26 (4.2•10−7) | 5.71 (7.9•10 −6) |
| Toll Receptors | 4.10 (1.6•10 −4) | 3.51 (5.6•10−4) | 4.2 (1.710 −4) | 5.91 (5.2•10 −6) |
| Inflammatosome | 0.01 (0.6) | 0.00 (0.8) | 0.00 (0.7) | 2.12 (0.02) |
| Chemochimes | 0.15 (0.6) | 0.09 (0.7) | 0.02 (0.9) | 0.11 (0.6) |
| Scavenger and Immunoglobulins |
8.15 (8.510 −8) | 9.53 (5.5•10−9) | 7.6 (1.1•107) | 5.27 (2.4•10−6) |
| MHI | 4.2 (1.710 −4) | 3.61 (4.1•10 −4) | 3.15 (0.001) | 1.91 (0.01) |
| MHII | 2.31 (0.004) | 2.46 (0.003) | 1.67 (0.01) | 1.93 (0.01) |
Discussion
This is the first study, to our knowledge, to examine the effects of gene expression on the variability of cortical GMT during normal aging. Our results demonstrated that expression activity for the genes that regulate cellular proliferation, adhesion, differentiation and inflammation can explain a large (20–30%) proportion of individual variability in cortical GMT. Overall, our findings agree with conclusions derived from animal literature that define cerebral aging as the product of a progressive downregulation in the adult neurogenesis and upregulation in neuro-inflammatory pathways (Cribbs et al., 2011; Lazarov et al., 2011; Villeda et al., 2011). Specifically, the top four functional categories that predicted GMT decline (cellular proliferation, aggregation and differentiation, metabolism and the quantity of blood cells) are reported to regulate adult stem cell neurogenesis in mice (van Praag et al., 2005; Villeda et al., 2011). The same functional categories were also implicated in development of Alzheimer’s disease (Silva et al., 2011). This age-related downregulation of adult cellular proliferation pathways is a natural strategy to prevent unregulated cell growth. Uncontrolled upregulation of cellular proliferation pathways is observed in neoplasm (Beausejour and Campisi, 2006; Fumagalli and d'Adda di Fagagna, 2009) and in fact, the integrin-signaling pathway, that showed the strongest association with GMT, is upregulated in invasive glial tumors (Kobayashi et al., 2012; Tchaicha et al., 2011). Finally, the analysis of seven innate immune regulation pathways supports the hypothesis that neuroinflammation plays a role in cerebral decline even during normal aging. Five out of seven immune activation pathways predicted GMT variability at p<0.05 level and the pattern of association suggested that upregulation of these pathways corresponded to lower GMT values.
The eight genes (IGFBP3, LRRN3, CRIP2, SCD, IDS, TCF4, GATA3, HN1) whose expressions were significantly correlated with GMT at the transcriptome-wide level have relevant functions that include regulation of neural development, angiogenesis, transcriptional activity and metabolism. The network constructed from these transcripts using the factor analysis explained a large proportion of the global (31.9%) and regional (23.4–35%) of variability in GMT. GMT was negatively correlated with the expression levels of five genes (IGFBP3, CRIP2, IDS, GATA3 and HN1). IGFBP3, GATA3 and HN1 play a regulatory role in proliferation and differentiation of neural progenitor cells during development and in response to brain injury, and their expression is known to be upregulated with age and downregulated in tumors (Achim et al., 2012; Ajo et al., 2003; Goto et al., 2012; Hong et al., 2008; Ishii et al., 1996; Kalluri and Dempsey, 2011; Kizil et al., 2011; Tang et al., 1997). CRIP2 is upregulated during angiogenesis, including development of cerebral vasculature (Kihara et al., 2011; Wei et al., 2011). IDS gene codes an enzyme that is essential for normal metabolism of large sugars in CNS (Calias et al., 2012) and its haploinsufficiency is associated with abnormal brain development and cognitive impairment (Muenzer et al., 2012). Upregulation of IDS expression was recently reported as a transcriptome-wide significant finding in Alzheimer’s disease patients (Lunnon et al., 2011). There were three significant genes (LRRN3, SCD and TCF4) whose expression was positively correlated with GMT. LRRN3 codes for a leucine-rich repeat neuronal protein 3 that organizes synaptic connections and therefore is highly expressed in the cerebral cortex and its expression level declines with age (Hong et al., 2008; Tang et al., 1997). It confers susceptibility to autism, presumably because of its role in cortical development (Hutcheson et al., 2004). SCD codes for Stearoyl-CoA desaturase-1; an enzyme involved in production of essential phospholipids, including myelin, and its expression also declines with age (Castro et al., 2011; Cermenati et al., 2011; Hong et al., 2008; Lengi and Corl, 2007). TCF4 encodes a transcriptional factor that is highly expressed in the brain and its haploinsufficiency causes mental retardation and severe structural brain abnormalities (Van Balkom et al., 2011).
Increasing the threshold for statistical significance to 10% FDR threshold (p=2×10−3) yielded 474 transcripts including these for four genes (SELP, RORA, CLU, CCL5) that were previously identified as candidates for brain integrity using genome-wide-association and quantitative-trait loci (QTL) analyses of cerebral integrity traits by this and other groups. Specifically, the Selectin-P gene (SELP) was identified, by this and other groups as a potential candidate gene that can pleiotropically influence blood-pressure and the cerebral integrity (Kochunov et al., 2010b; Kochunov et al., 2012; Melville et al., 2012). Likewise, the Retinotopic Acid Receptor (RAR)-Related Orphan Receptor Alpha (RORA) was previously identified as a possible candidate gene responsible for a significant bivariate QTL at 15q23 for GMT and fractional anisotropy of water diffusion measurements (Kochunov et al., 2011a). A common polymorphism in the CLU gene (rs11136000) is involved in activation of neuroinflammatory cascades and confers a 1.16 greater risk of developing late-onset Alzheimer’s dementia (Kamboh et al., 2012), as initially identified and replicated in two genome wide association studies of Alzheimer’s disease patients (Harold et al., 2009; Lambert et al., 2009). Its polymorphic variability was shown to be associated with higher rate of cognitive decline(Rodriguez-Rodriguez et al., 2012), lower WM integrity (Braskie et al., 2011) and higher brain atrophy and faster cognitive decline (Thambisetty et al., 2012b). Plasma levels of plasma clusterin, the protein product of CLU transcription was shown to play a role in amyloid clearance and accumulation (DeMattos et al., 2004) and is associated to rates of brain atrophy in elderly individuals (Thambisetty et al., 2012a; Thambisetty et al., 2010). Similarly, functional polymorphisms of the Chemokine (C-C motif) ligand 5 (CCL5) gene were shown to influence the course of neuroinflammation in multiple sclerosis and Gaucher's disease (van Veen et al., 2007; Vitner et al., 2012).
Further analysis of the association between individual transcripts was performed by studying the functional relationship between GMT and the networks of genes. The network analyses can clarify the relationships between traits and transcripts because it accounts for epistatic, gene-to-gene, regulations in functional networks and pathways. The smallest network was constructed based on the relationships between GMT and transcripts by performing a factor analysis of the eight transcripts that passed transcriptome-wide correction and did not account for potential epistatic and gene-to-gene regulations. Three out of four factors showed a significant (p≤1.6•10−5) association with the whole-brain average and regional GMT measurements. The highest association was observed for the factor that loaded equally on the CRIP2 and GATA3 genes both of which have regulatory function that involves cell proliferation and differentiations. This factor predicted the highest degree of variance in the multimodal functional areas including frontal and superior parietal lobes and the cingulate gyrus. SCD and IDS-factors showed the second and third highest association with GMT suggesting that reduction in phospholipids and myelin production and large sugar metabolism enzymes also have stronger effect on the multimodal cortical areas.
A broader set of networks was obtained by performing pathway enrichment analyses. Cell-proliferation and aggregation were two functional categories that most significantly associated (p=1.3·10−8 and 2.1·10−8, respectively) with GMT. The direction of the correlation suggests that downregulation in both categories was associated with lower GMT values. The pathway analysis identified significant (p= 3.2×10−6) over-representation of GMT correlated transcripts in the integrin signaling pathway (Figure S1). The seventeen significant genes involved in this pathway include one alpha integrin (IGRA2B) and two beta integrins (IGTB1 and ITGB3), as well as two components of the calpain complex (CAPN1 and CAPNS1) and two members of the actin related protein complex (ARPC3 and ARPC5L). The integrin signaling pathway is involved in cell-cell and cell-extracellular matrix (ECM) interactions; allowing rapid response to changes in environment (Berman et al., 2003; Danen and Yamada, 2001). While integrins are generally associated with cell proliferation, movement and attachment, integrin signalling plays an important role in modulating neuronal survival by protecting against oxidative stress and apoptosis (Gary and Mattson, 2001). Interactions between integrins and the ECM play a major role in neurogenesis such as adult neurogenesis, axonal outgrowth and path finding(Myers et al., 2011). The primarily negative direction of the correlation between GMT and the expression of genes in this pathway suggested that a downregulation of this important signaling pathway that responds to environmental stress and facilitates remodeling and apoptosis co-occurred with a decline in cerebral integrity in our data.
Neuroinflammation plays a pivotal role in both the development and progression of Alzheimer’s dementia (AD) (McGeer et al., 2006; Rogers, 2008). Innate immune pathways may also play a role in normal cerebral aging (Cribbs et al., 2011). Upregulation of innate immune pathways causes the release of proinflammatory factors, activation of inflammasome and recruitment of immune cell. This can be neuroprotective in response to exogenous pathogens and brain trauma (Rivest, 2009; Wyss-Coray, 2006). However, chronic activation of immune pathways by the endogenous amyloid-beta peptide is thought to be the chief contributing factor to neurodegeneration in AD (Perry et al., 2007; Rivest, 2009). Our findings support the hypothesis that innate immune activation play a role in normal aging. We observed a highly significant association between GMT and the cascade of three pathways implicated in AD neurodegeneration: complement signaling, the Toll Receptors pathways and the scavenger and immunoglobulins. The complement signaling pathway showed the strongest association with GMT (p=1.8·10−9). This pathway has long been in the center of attention, being investigated as a potential culprit for the neurotoxicity of amyloid-beta peptide in AD (Rogers et al., 1992). This association replicates the findings that AD-like neurodegenerative processes may be present to a some degree in subjects who undergo normal aging (Cribbs et al., 2011). In particular, CLU inhibits complement system activation and may potentially prevent the recognition of fibrillar structures by the immune system (Nuutinen et al., 2009; Sleegers et al., 2010).
Conclusions and limitation
Our findings in a cohort of normally aging, community-dwelling, Mexican Americans demonstrated the utility of lymphocyte-based expression level measurements to predict individual variability in GMT. The cross-sectional nature of these data and the long gap between transcriptional and imaging data is suboptimal and there are limits to the interpretations that can be made about longitudinal processes from cross-section data. For instance, individual differences in cerebral integrity associated with regenerative and inflammatory pathways may have also arisen from development rather than aging. However, our observation that subjects with downregulated regenerative pathways demonstrated lower GMT values agrees with findings from longitudinal animal studies on normal aging (Sharpless and DePinho, 2007; Shook et al., 2011; van Praag et al., 2005). Moreover, expression measurements performed on human brain and lymphocytes demonstrate that transcriptome activity for several of the genes identified in our analysis, including IGFBP3, LRRN3 and SCD shows significant decline with age, supporting the hypothesis of the age-related downregulation in the regenerative potential (Hong et al., 2008). Further, longitudinal studies that include cerebral integrity measurements in conjunction with transcriptome profiling, such as those proposed by the Alzheimer's Disease Neuroimaging Initiative (Walhovd et al., 2010), will be necessary to replicate our findings and help identify the genetic factors contributing to cerebral aging.
This research was conducted in Mexican Americans, a population with significant Native American admixture. If relatively rare variants are involved in the determination of quantitative variability, we may expect considerable population differences across populations (Blangero et al., 2003). Our analytical methods explicitly considered and exploited the relatedness among participants. Estimates of mean effect parameters are unbiased even in the presence of relatedness but their error covariance structure is biased low. Our approach which explicitly models biological relatedness among individuals eliminates this bias and leads to valid hypothesis testing that exhibits appropriate asymptotic behavior. Our data are made available on collaborative basis through the imaging genetics consortium, known as the Enhancing Neuro Imaging Genetic through Meta Analysis (ENIGMA) group (http://enigma.loni.ucla.edu/). Our group has already contributed the imaging and genetic data to the ENIGMA study of hippocampal and intracranial volumes (Stein et al., 2011) and to ENIGMA-DTI study (Jahanshad et al., 2013). In addition, both imaging and genetics data will be made available through the NIMH Genetics Repository.
Supplementary Material
Acknowledgements
This research was supported by National Institute of Biomedical. Imaging and Bioengineering (EB015611 and EB006395) grants to P.K., the National Heart Lung and Blood Institute (P01HL045522) to J.B., and the National Institute of Mental Health (R37MH059490 and R01MH078111) to J.B. and (R01MH0708143 and R01MH083824) to D.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest, Authors have no conflicts of interest to disclose.
References
- Achim K, Peltopuro P, Lahti L, Li J, Salminen M, Partanen J. Distinct.developmental origins and regulatory mechanisms for GABAergic neurons associated with dopaminergic nuclei in the ventral mesodiencephalic region. Development. 2012;139:2360–2370. doi: 10.1242/dev.076380. [DOI] [PubMed] [Google Scholar]
- Aganj I, Sapiro G, Parikshak N, Madsen SK, Thompson PM. Measurement of cortical thickness from MRI by minimum line integrals on soft-classified tissue. Hum Brain Mapp. 2009;30:3188–3199. doi: 10.1002/hbm.20740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajo R, Cacicedo L, Navarro C, Sanchez-Franco F. Growth hormone action on proliferation and differentiation of cerebral cortical cells from fetal rat. Endocrinology. 2003;144:1086–1097. doi: 10.1210/en.2002-220667. [DOI] [PubMed] [Google Scholar]
- Almasy L, Dyer TD, Blangero J. Bivariate quantitative trait linkage analysis: pleiotropy versus co-incident linkages. Genet Epidemiol. 1997;14:953–958. doi: 10.1002/(SICI)1098-2272(1997)14:6<953::AID-GEPI65>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Beausejour CM, Campisi J. Ageing: balancing regeneration and cancer. Nature. 2006;443:404–405. doi: 10.1038/nature05221. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
- Berchtold NC, Cribbs DH, Coleman PD, Rogers J, Head E, Kim R, Beach T, Miller C, Troncoso J, Trojanowski JQ, Zielke HR, Cotman CW. Gene expression changes in the course of normal brain aging are sexually dimorphic. Proc Natl Acad Sci U S A. 2008;105:15605–15610. doi: 10.1073/pnas.0806883105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman AE, Kozlova NI, Morozevich GE. Integrins: structure and signaling. Biochemistry (Mosc) 2003;68:1284–1299. doi: 10.1023/b:biry.0000011649.03634.74. [DOI] [PubMed] [Google Scholar]
- Biffi A, Anderson CD, Desikan RS, Sabuncu M, Cortellini L, Schmansky N, Salat D, Rosand J. Genetic variation and neuroimaging measures in Alzheimer disease. Arch Neurol. 2011;67:677–685. doi: 10.1001/archneurol.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bihaqi SW, Schumacher A, Maloney B, Lahiri DK, Zawia NH. Do epigenetic pathways initiate late onset Alzheimer disease (LOAD): towards a new paradigm. Curr Alzheimer Res. 2011;9:574–588. doi: 10.2174/156720512800617982. [DOI] [PubMed] [Google Scholar]
- Blangero J, Williams JT, Almasy L. Novel family-based approaches to genetic risk in thrombosis. J Thromb Haemost. 2003;1:1391–1397. doi: 10.1046/j.1538-7836.2003.00310.x. [DOI] [PubMed] [Google Scholar]
- Braskie MN, Jahanshad N, Stein JL, Barysheva M, McMahon KL, de Zubicaray GI, Martin NG, Wright MJ, Ringman JM, Toga AW, Thompson PM. Common Alzheimer's disease risk variant within the CLU gene affects white matter microstructure in young adults. J Neurosci. 2011;31:6764–6770. doi: 10.1523/JNEUROSCI.5794-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calias P, Papisov M, Pan J, Savioli N, Belov V, Huang Y, Lotterhand J, Alessandrini M, Liu N, Fischman AJ, Powell JL, Heartlein MW. CNS penetration of intrathecal-lumbar idursulfase in the monkey, dog and mouse: implications for neurological outcomes of lysosomal storage disorder. PLoS One. 2012;7:e30341. doi: 10.1371/journal.pone.0030341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capell BC, Collins FS, Nabel EG. Mechanisms of cardiovascular disease in accelerated aging syndromes. Circ Res. 2007;101:13–26. doi: 10.1161/CIRCRESAHA.107.153692. [DOI] [PubMed] [Google Scholar]
- Carless MA, Glahn DC, Johnson MP, Curran JE, Bozaoglu K, Dyer TD, Winkler AM, Cole SA, Almasy L, MacCluer JW, Duggirala R, Moses EK, Goring HH, Blangero J. Impact of DISC1 variation on neuroanatomical and neurocognitive phenotypes. Molecular psychiatry. 2011;16:1096–1104. doi: 10.1038/mp.2011.37. 1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro LF, Wilson JM, Goncalves O, Galante-Oliveira S, Rocha E, Cunha I. The evolutionary history of the stearoyl-CoA desaturase gene family in vertebrates. BMC Evol Biol. 2011;11:132. doi: 10.1186/1471-2148-11-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermenati G, Abbiati F, Cermenati S, Brioschi E, Volonterio A, Cavaletti G, Saez E, De Fabiani E, Crestani M, Garcia-Segura LM, Melcangi RC, Caruso D, Mitro N. Diabetes-induced myelin abnormalities are associated with an altered lipid pattern: protective effects of LXR activation. J Lipid Res. 2011;53:300–310. doi: 10.1194/jlr.M021188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang MC, McMahon KL, de Zubicaray GI, Martin NG, Hickie I, Toga AW, Wright MJ, Thompson PM. Genetics of white matter development: a DTI study of 705 twins and their siblings aged 12 to 29. Neuroimage. 2011;54:2308–2317. doi: 10.1016/j.neuroimage.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouliaras L, Rutten BP, Kenis G, Peerbooms O, Visser PJ, Verhey F, van Os J, Steinbusch HW, van den Hove DL. Epigenetic regulation in the pathophysiology of Alzheimer's disease. Prog Neurobiol. 2010;90:498–510. doi: 10.1016/j.pneurobio.2010.01.002. [DOI] [PubMed] [Google Scholar]
- Cribbs DH, Berchtold NC, Perreau V, Coleman PD, Rogers J, Tenner AJ, Cotman CW. Extensive innate immune gene activation accompanies brain aging, increasing vulnerability to cognitive decline and neurodegeneration: a microarray study. J Neuroinflammation. 2011;9:179. doi: 10.1186/1742-2094-9-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham C, Wilcockson DC, Campion S, Lunnon K, Perry VH. Central and systemic endotoxin challenges exacerbate the local inflammatory response and increase neuronal death during chronic neurodegeneration. J Neurosci. 2005;25:9275–9284. doi: 10.1523/JNEUROSCI.2614-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danen EH, Yamada KM. Fibronectin, integrins, and growth control. J Cell Physiol. 2001;189:1–13. doi: 10.1002/jcp.1137. [DOI] [PubMed] [Google Scholar]
- DeMattos RB, Cirrito JR, Parsadanian M, May PC, O’Dell MA, Taylor JW, Harmony JA, Aronow BJ, Bales KR, Paul SM, Holtzman DM. ApoE and clusterin cooperatively suppress Abeta levels and deposition: evidence that ApoE regulates extracellular Abeta metabolism in vivo. Neuron. 2004;41:193–202. doi: 10.1016/s0896-6273(03)00850-x. [DOI] [PubMed] [Google Scholar]
- DeStefano AL, Atwood LD, Massaro JM, Heard-Costa N, Beiser A, Au R, Wolf PA, DeCarli C. Genome-wide scan for white matter hyperintensity: the Framingham Heart Study. Stroke. 2006;37:77–81. doi: 10.1161/01.STR.0000196987.68770.b3. [DOI] [PubMed] [Google Scholar]
- Dodge HH, Wang CN, Chang CC, Ganguli M. Terminal decline and practice effects in older adults without dementia: the MoVIES project. Neurology. 2011;77:722–730. doi: 10.1212/WNL.0b013e31822b0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskildsen SF, Coupe P, Garcia-Lorenzo D, Fonov V, Pruessner JC, Collins DL. Prediction of Alzheimer's disease in subjects with mild cognitive impairment from the ADNI cohort using patterns of cortical thinning. Neuroimage. 2012 doi: 10.1016/j.neuroimage.2012.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci U S A. 2000;97:11050–11055. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli M, d'Adda di Fagagna F. SASPense and DDRama in cancer and ageing. Nat Cell Biol. 2009;11:921–923. doi: 10.1038/ncb0809-921. [DOI] [PubMed] [Google Scholar]
- Gary DS, Mattson MP. Integrin signaling via the PI3-kinase-Akt pathway increases neuronal resistance to glutamate-induced apoptosis. J Neurochem. 2001;76:1485–1496. doi: 10.1046/j.1471-4159.2001.00173.x. [DOI] [PubMed] [Google Scholar]
- Goring HH, Curran JE, Johnson MP, Dyer TD, Charlesworth J, Cole SA, Jowett JB, Abraham LJ, Rainwater DL, Comuzzie AG, Mahaney MC, Almasy L, MacCluer JW, Kissebah AH, Collier GR, Moses EK, Blangero J. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- Goto T, Tokunaga F, Hisatomi O. Hematological- and Neurological-Expressed Sequence 1 Gene Products in Progenitor Cells during Newt Retinal Development. Stem Cells Int. 2012;2012:436042. doi: 10.1155/2012/436042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schurmann B, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Hull M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O’Donovan M, Owen MJ, Williams J. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes C, Cunningham C, Zotova E, Woolford J, Dean C, Kerr S, Culliford D, Perry VH. Systemic inflammation and disease progression in Alzheimer disease. Neurology. 2009;73:768–774. doi: 10.1212/WNL.0b013e3181b6bb95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong MG, Myers AJ, Magnusson PK, Prince JA. Transcriptome-wide assessment of human brain and lymphocyte senescence. PLoS One. 2008;3:e3024. doi: 10.1371/journal.pone.0003024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutcheson HB, Olson LM, Bradford Y, Folstein SE, Santangelo SL, Sutcliffe JS, Haines JL. Examination of NRCAM, LRRN3, KIAA0716, and LAMB1 as autism candidate genes. BMC Med Genet. 2004;5:12. doi: 10.1186/1471-2350-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii N, Wanaka A, Tohyama M. Increased expression of NLRR-3 mRNA after cortical brain injury in mouse. Brain Res Mol Brain Res. 1996;40:148–152. doi: 10.1016/0169-328x(96)00090-3. [DOI] [PubMed] [Google Scholar]
- Jahanshad NPK, Sprooten E, Mandl R, Nichols T, Almasy L, Blangero J, Brouwer RM, Curran J, Zubicaray C, Duggirala R, Fox P, Hong E, Landman BA, Nicholas M, McMahon FJ, Medland SE, Mitchell B, Olvera R, Peterson C, Starr JM, Sussmann J, AW T, Wardlaw JM, Wright AC, Pol H, Bastin ME, McIntosh AM, Deary IJDCG. Multi-Site Genetic Analysis of Diffusion Images and Voxelwise Heritability Analysis: A Pilot Project of the ENIGMA-DTI Working Group. Neuroimage. 2013 doi: 10.1016/j.neuroimage.2013.04.061. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelsing J, Rostrup E, Markenroth K, Paulson OB, Gundersen HJ, Hemmingsen R, Pakkenberg B. Assessment of in vivo MR imaging compared to physical sections in vitro--a quantitative study of brain volumes using stereology. Neuroimage. 2005;26:57–65. doi: 10.1016/j.neuroimage.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Kalluri HS, Dempsey RJ. IGFBP-3 inhibits the proliferation of neural progenitor cells. Neurochem Res. 2011;36:406–411. doi: 10.1007/s11064-010-0349-2. [DOI] [PubMed] [Google Scholar]
- Kamboh MI, Demirci FY, Wang X, Minster RL, Carrasquillo MM, Pankratz VS, Younkin SG, Saykin AJ, Jun G, Baldwin C, Logue MW, Buros J, Farrer L, Pericak-Vance MA, Haines JL, Sweet RA, Ganguli M, Feingold E, Dekosky ST, Lopez OL, Barmada MM. Genome-wide association study of Alzheimer's disease. Transl Psychiatry. 2011;2:e117. doi: 10.1038/tp.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamboh MI, Demirci FY, Wang X, Minster RL, Carrasquillo MM, Pankratz VS, Younkin SG, Saykin AJ, Jun G, Baldwin C, Logue MW, Buros J, Farrer L, Pericak-Vance MA, Haines JL, Sweet RA, Ganguli M, Feingold E, Dekosky ST, Lopez OL, Barmada MM. Genome-wide association study of Alzheimer’s disease. Transl Psychiatry. 2012;2:e117. doi: 10.1038/tp.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent JW, Jr., Goring HH, Charlesworth JC, Drigalenko E, Diego VP, Curran JE, Johnson MP, Dyer TD, Cole SA, Jowett JB, Mahaney MC, Comuzzie AG, Almasy L, Moses EK, Blangero J, Williams-Blangero S. Genotypexage interaction in human transcriptional ageing. Mech Ageing Dev. 2012;133:581–590. doi: 10.1016/j.mad.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara T, Shinohara S, Fujikawa R, Sugimoto Y, Murata M, Miyake J. Regulation of cysteine-rich protein 2 localization by the development of actin fibers during smooth muscle cell differentiation. Biochem Biophys Res Commun. 2011;411:96–101. doi: 10.1016/j.bbrc.2011.06.100. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick B, Messias E, Harvey PD, Fernandez-Egea E, Bowie CR. Is schizophrenia a syndrome of accelerated aging? Schizophr Bull. 2008;34:1024–1032. doi: 10.1093/schbul/sbm140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizil C, Kyritsis N, Dudczig S, Kroehne V, Freudenreich D, Kaslin J, Brand M. Regenerative Neurogenesis from Neural Progenitor Cells Requires Injury-Induced Expression of Gata3. Dev Cell. 2011 doi: 10.1016/j.devcel.2012.10.014. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Takahashi H, Inoue A, Harada H, Toshimori S, Kobayashi Y, Goto K, Sugimoto K, Yano H, Ohnishi T, Tanaka J. Oct-3/4 promotes migration and invasion of glioblastoma cells. Journal of cellular biochemistry. 2012;113:508–517. doi: 10.1002/jcb.23374. [DOI] [PubMed] [Google Scholar]
- Kochunov P, Glahn D, Lancaster J, Wincker P, Smith S, Thompson P, Almasy L, Duggirala R, Fox P, Blangero J. Genetics of microstructure of cerebral white matter using diffusion tensor imaging. Neuroimage. 2010a;15:1109–1116. doi: 10.1016/j.neuroimage.2010.01.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Glahn D, Lancaster J, Winkler A, Kent J, Olvera R, Cole S, Dyer T, Almasy L, Duggirala R, Fox P, Blangero J. Whole Brain and Regional Hyperintense White Matter Volume and Blood Pressure: Overlap of Genetic Loci produced by Bivariate, Whole-Genome Linkage Analyses. Stroke. 2010b;41:2137–2142. doi: 10.1161/STROKEAHA.110.590943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Glahn D, Nichols T, Winkler A, Hong E, Holcomb H, Stein J, Thompson P, Curran J, Carless M, Olvera R, Johnson M, Cole S, Kochunov V, Kent J, Blangero J. Genetic Analysis of Cortical Thickness and Fractional Anisotropy of Water Diffusion in the Brain. Frontiers in Neuroscience. 2011a;5:1–15. doi: 10.3389/fnins.2011.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Glahn D, Winkler A, Duggirala R, Olvera RL, Cole S, Dyer TD, Almasy L, Fox PT, Blangero J. Analysis of genetic variability and whole genome linkage of whole-brain, subcortical, and ependymal hyperintense white matter volume. Stroke. 2009a;40:3685–3690. doi: 10.1161/STROKEAHA.109.565390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Glahn DC, Hong LE, Lancaster J, Curran JE, Johnson MP, Winkler AM, Holcomb HH, Kent JW, Jr., Mitchell B, Kochunov V, Olvera RL, Cole SA, Dyer TD, Moses EK, Goring H, Almasy L, Duggirala R, Blangero J. P-selectin Expression Tracks Cerebral Atrophy in Mexican-Americans. Front Genet. 2012;3:65. doi: 10.3389/fgene.2012.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Glahn DC, Lancaster J, Thompson PM, Kochunov V, Rogers B, Fox P, Blangero J, Williamson DE. Fractional anisotropy of cerebral white matter and thickness of cortical gray matter across the lifespan. Neuroimage. 2011b;58:41–49. doi: 10.1016/j.neuroimage.2011.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Glahn DC, Lancaster J, Winkler A, Karlsgodt K, Olvera RL, Curran JE, Carless MA, Dyer TD, Almasy L, Duggirala R, Fox PT, Blangero J. Blood pressure and cerebral white matter share common genetic factors in Mexican Americans. Hypertension. 2011c;57:330–335. doi: 10.1161/HYPERTENSIONAHA.110.162206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Lancaster JL, Glahn DC, Purdy D, Laird AR, Gao F, Fox P. Retrospective motion correction protocol for high-resolution anatomical MRI. Hum Brain Mapp. 2006;27:957–962. doi: 10.1002/hbm.20235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Robin D, Royall D, Lancaster J, Kochunov V, Coyle T, Schlosser A, Fox P. Can structural MRI cerebral health markers track cognitive trends in executive control function during normal maturation and adulthood? Hum Brain Mapp. 2009b;30:2581–2594. doi: 10.1002/hbm.20689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Rogers W, Mangin JF, Lancaster J. A Library of Cortical Morphology Analysis Tools to Study Development, Aging and Genetics of Cerebral Cortex. Neuroinformatics. 2011 Jun 23; doi: 10.1007/s12021-011-9127-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Thompson PM, Coyle TR, Lancaster JL, Kochunov V, Royall D, Mangin JF, Riviere D, Fox PT. Relationship among neuroimaging indices of cerebral health during normal aging. Hum Brain Mapp. 2008;29:36–45. doi: 10.1002/hbm.20369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Thompson PM, Lancaster JL, Bartzokis G, Smith S, Coyle T, Royall DR, Laird A, Fox PT. Relationship between white matter fractional anisotropy and other indices of cerebral health in normal aging: tract-based spatial statistics study of aging. Neuroimage. 2007;35:478–487. doi: 10.1016/j.neuroimage.2006.12.021. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fievet N, Barberger-Gateau P, Engelborghs S, De Deyn P, Mateo I, Franck A, Helisalmi S, Porcellini E, Hanon O, de Pancorbo MM, Lendon C, Dufouil C, Jaillard C, Leveillard T, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossu P, Piccardi P, Annoni G, Seripa D, Galimberti D, Hannequin D, Licastro F, Soininen H, Ritchie K, Blanche H, Dartigues JF, Tzourio C, Gut I, Van Broeckhoven C, Alperovitch A, Lathrop M, Amouyel P. Genome-wideassociation study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- Lazarov O, Mattson MP, Peterson DA, Pimplikar SW, van Praag H. When neurogenesis encounters aging and disease. Trends Neurosci. 2011;33:569–579. doi: 10.1016/j.tins.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet. 2011;88:294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengi AJ, Corl BA. Identification and characterization of a novel bovine stearoyl-CoA desaturase isoform with homology to human SCD5. Lipids. 2007;42:499–508. doi: 10.1007/s11745-007-3056-2. [DOI] [PubMed] [Google Scholar]
- Lerch JP, Evans AC. Cortical thickness analysis examined through power analysis and a population simulation. Neuroimage. 2005;24:163–173. doi: 10.1016/j.neuroimage.2004.07.045. [DOI] [PubMed] [Google Scholar]
- Leritz EC, Salat DH, Williams VJ, Schnyer DM, Rudolph JL, Lipsitz L, Fischl B, McGlinchey RE, Milberg WP. Thickness of the human cerebral cortex is associated with metrics of cerebrovascular health in a normative sample of community dwelling older adults. Neuroimage. 2011;54:2659–2671. doi: 10.1016/j.neuroimage.2010.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang Y, Wu G, Shi F, Zhou L, Lin W, Shen D. Discriminant analysis of longitudinal cortical thickness changes in Alzheimer's disease using dynamic and network features. Neurobiol Aging. 2012;33:427. doi: 10.1016/j.neurobiolaging.2010.11.008. e415–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunnon K, Sattlecker M, Furney SJ, Coppola G, Simmons A, Proitsi P, Lupton MK, Lourdusamy A, Johnston C, Soininen H, Kloszewska I, Mecocci P, Tsolaki M, Vellas B, Geschwind D, Lovestone S, Dobson R, Hodges A. A Blood Gene Expression Marker of Early Alzheimer's Disease. J Alzheimers Dis. 2011 doi: 10.3233/JAD-2012-121363. [DOI] [PubMed] [Google Scholar]
- Magnotta VA, Andreasen NC, Schultz SK, Harris G, Cizadlo T, Heckel D, Nopoulos P, Flaum M. Quantitative in vivo measurement of gyrification in the human brain: changes associated with aging. Cereb Cortex. 1999;9:151–160. doi: 10.1093/cercor/9.2.151. [DOI] [PubMed] [Google Scholar]
- Maloney B, Sambamurti K, Zawia N, Lahiri DK. Applying epigenetics to Alzheimer’s disease via the latent early-life associated regulation (LEARn) model. Curr Alzheimer Res. 2011;9:589–599. doi: 10.2174/156720512800617955. [DOI] [PubMed] [Google Scholar]
- McGeer PL, Rogers J, McGeer EG. Inflammation, anti-inflammatory agents and Alzheimer disease: the last 12 years. J Alzheimers Dis. 2006;9:271–276. doi: 10.3233/jad-2006-9s330. [DOI] [PubMed] [Google Scholar]
- Melville SA, Buros J, Parrado AR, Vardarajan B, Logue MW, Shen L, Risacher SL, Kim S, Jun G, DeCarli C, Lunetta KL, Baldwin CT, Saykin AJ, Farrer LA. Multiple loci influencing hippocampal degeneration identified by genome scan. Ann Neurol. 2012;72:65–75. doi: 10.1002/ana.23644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell BD, Kammerer CM, Blangero J, Mahaney MC, Rainwater DL, Dyke B, Hixson JE, Henkel RD, Sharp RM, Comuzzie AG, VandeBerg JL, Stern MP, MacCluer JW. Genetic and environmental contributions to cardiovascular risk factors in Mexican Americans. The San Antonio Family Heart Study. Circulation. 1996;94:2159–2170. doi: 10.1161/01.cir.94.9.2159. [DOI] [PubMed] [Google Scholar]
- Moses EK, Curran JE, Johnson MP, Goring HH, Dyer TD, Charlesworth J, Cole SA, Jowett JB, Abraham LA, Rainwater DL, Mahaney MC, Almasy L, MacCluer JW, Kissebah AH, Collier GR, Blangero J. Identification of the VNN1 gene as a novel CVD risk factor. Circulation. 2007;116:57–57. [Google Scholar]
- Muenzer J, Bodamer O, Burton B, Clarke L, Frenking GS, Giugliani R, Jones S, Rojas MV, Scarpa M, Beck M, Harmatz P. The role of enzyme replacement therapy in severe Hunter syndrome-an expert panel consensus. Eur J Pediatr. 2012;171:181–188. doi: 10.1007/s00431-011-1606-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers JP, Santiago-Medina M, Gomez TM. Regulation of axonal outgrowth and pathfinding by integrin-ECM interactions. Dev Neurobiol. 2011;71:901–923. doi: 10.1002/dneu.20931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narr KL, Woods RP, Thompson PM, Szeszko P, Robinson D, Dimtcheva T, Gurbani M, Toga AW, Bilder RM. Relationships between IQ and regional cortical gray matter thickness in healthy adults. Cerebral cortex. 2007;17:2163–2171. doi: 10.1093/cercor/bhl125. [DOI] [PubMed] [Google Scholar]
- Nuutinen T, Suuronen T, Kauppinen A, Salminen A. Clusterin: a forgotten player in Alzheimer's disease. Brain Res Rev. 2009;61:89–104. doi: 10.1016/j.brainresrev.2009.05.007. [DOI] [PubMed] [Google Scholar]
- Panizzon MS, Fennema-Notestine C, Kubarych TS, Chen CH, Eyler LT, Fischl B, Franz CE, Grant MD, Hamza S, Jak A, Jernigan TL, Lyons MJ, Neale MC, Prom-Wormley EC, Seidman L, Tsuang MT, Wu H, Xian H, Dale AM, Kremen WS. Genetic and environmental influences of white and gray matter signal contrast: a new phenotype for imaging genetics? Neuroimage. 2011;60:1686–1695. doi: 10.1016/j.neuroimage.2012.01.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry VH, Cunningham C, Holmes C. Systemic infections and inflammation affect chronic neurodegeneration. Nat Rev Immunol. 2007;7:161–167. doi: 10.1038/nri2015. [DOI] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz N, Gunning FM, Head D, Dupuis JH, McQuain J, Briggs SD, Loken WJ, Thornton AE, Acker JD. Selective aging of the human cerebral cortex observed in vivo: differential vulnerability of the prefrontal gray matter. Cerebral cortex. 1997;7:268–282. doi: 10.1093/cercor/7.3.268. [DOI] [PubMed] [Google Scholar]
- Rijsdijsk FV, Viding E, De Brito S, Forgiarini M, Mechelli A, Jones AP, McCrory E. Heritable variations in gray matter concentration as a potential endophenotype for psychopathic traits. Arch Gen Psychiatry. 2011;67:406–413. doi: 10.1001/archgenpsychiatry.2010.20. [DOI] [PubMed] [Google Scholar]
- Risacher SL, Saykin AJ, West JD, Shen L, Firpi HA, McDonald BC. Baseline MRI predictors of conversion from MCI to probable AD in the ADNI cohort. Curr Alzheimer Res. 2009;6:347–361. doi: 10.2174/156720509788929273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivest S. Regulation of innate immune responses in the brain. Nat Rev Immunol. 2009;9:429–439. doi: 10.1038/nri2565. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Rodriguez E, Sanchez-Juan P, Vazquez-Higuera JL, Mateo I, Pozueta A, Berciano J, Cervantes S, Alcolea D, Martinez-Lage P, Clarimon J, Lleo A, Pastor P, Combarros O. Genetic risk score predicting accelerated progression from mild cognitive impairment to Alzheimer's disease. J Neural Transm. 2012 doi: 10.1007/s00702-012-0920-x. [DOI] [PubMed] [Google Scholar]
- Rogers J. The inflammatory response in Alzheimer's disease. J Periodontol. 2008;79:1535–1543. doi: 10.1902/jop.2008.080171. [DOI] [PubMed] [Google Scholar]
- Rogers J, Cooper NR, Webster S, Schultz J, McGeer PL, Styren SD, Civin WH, Brachova L, Bradt B, Ward P, et al. Complement activation by beta-amyloid in Alzheimer disease. Proc Natl Acad Sci U S A. 1992;89:10016–10020. doi: 10.1073/pnas.89.21.10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royall DR, Palmer RF, O’Bryant SE. Validation of a latent variable representing the dementing process. J Alzheimers Dis. 2011;30:639–649. doi: 10.3233/JAD-2012-120055. [DOI] [PubMed] [Google Scholar]
- Salat DH, Lee SY, van der Kouwe AJ, Greve DN, Fischl B, Rosas HD. Age-associated alterations in cortical gray and white matter signal intensity and gray to white matter contrast. Neuroimage. 2009;48:21–28. doi: 10.1016/j.neuroimage.2009.06.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salthouse TA. When does age-related cognitive decline begin? Neurobiol Aging. 2009;30:507–514. doi: 10.1016/j.neurobiolaging.2008.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schillerstrom JE, Royall DR, Palmer RF. Depression, disability and intermediate pathways: a review of longitudinal studies in elders. J Geriatr Psychiatry Neurol. 2008;21:183–197. doi: 10.1177/0891988708320971. [DOI] [PubMed] [Google Scholar]
- Selemon LD, Rajkowska G, Goldman-Rakic PS. Abnormally high neuronal density in the schizophrenic cortex. A morphometric analysis of prefrontal area 9 and occipital area 17. Arch Gen Psychiatry. 1995;52:805–818. doi: 10.1001/archpsyc.1995.03950220015005. discussion 819–820. [DOI] [PubMed] [Google Scholar]
- Sequeira PA, Martin MV, Vawter MP. The first decade and beyond of transcriptional profiling in schizophrenia. Neurobiol Dis. 2012;45:23–36. doi: 10.1016/j.nbd.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpless NE, DePinho RA. How stem cells age and why this makes us grow old. Nat Rev Mol Cell Biol. 2007;8:703–713. doi: 10.1038/nrm2241. [DOI] [PubMed] [Google Scholar]
- Shook BA, Manz DH, Peters JJ, Kang S, Conover JC. Spatiotemporal changes to the subventricular zone stem cell pool through aging. J Neurosci. 2011;32:6947–6956. doi: 10.1523/JNEUROSCI.5987-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AR, Grinberg LT, Farfel JM, Diniz BS, Lima LA, Silva PJ, Ferretti RE, Rocha RM, Filho WJ, Carraro DM, Brentani H. Transcriptional alterations related to neuropathology and clinical manifestation of Alzheimer's disease. PLoS One. 2011;7:e48751. doi: 10.1371/journal.pone.0048751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleegers K, Lambert JC, Bertram L, Cruts M, Amouyel P, Van Broeckhoven C. The pursuit of susceptibility genes for Alzheimer's disease: progress and prospects. Trends Genet. 2010;26:84–93. doi: 10.1016/j.tig.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Sowell ER, Peterson BS, Thompson PM, Welcome SE, Henkenius AL, Toga AW. Mapping cortical change across the human life span. Nat Neurosci. 2003;6:309–315. doi: 10.1038/nn1008. [DOI] [PubMed] [Google Scholar]
- Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM, Toro R, Appel K, Bartecek R, Bergmann O, Bernard M, Brown AA, Cannon DM, Chakravarty MM, Christoforou A, Domin M, Grimm O, Hollinshead M, Holmes AJ, Homuth G, Hottenga JJ, Langan C, Lopez LM, Hansell NK, Hwang KS, Kim S, Laje G, Lee PH, Liu X, Loth E, Lourdusamy A, Mattingsdal M, Mohnke S, Maniega SM, Nho K, Nugent AC, O’Brien C, Papmeyer M, Putz B, Ramasamy A, Rasmussen J, Rijpkema M, Risacher SL, Roddey JC, Rose EJ, Ryten M, Shen L, Sprooten E, Strengman E, Teumer A, Trabzuni D, Turner J, van Eijk K, van Erp TG, van Tol MJ, Wittfeld K, Wolf C, Woudstra S, Aleman A, Alhusaini S, Almasy L, Binder EB, Brohawn DG, Cantor RM, Carless MA, Corvin A, Czisch M, Curran JE, Davies G, de Almeida MA, Delanty N, Depondt C, Duggirala R, Dyer TD, Erk S, Fagerness J, Fox PT, Freimer NB, Gill M, Goring HH, Hagler DJ, Hoehn D, Holsboer F, Hoogman M, Hosten N, Jahanshad N, Johnson MP, Kasperaviciute D, Kent JW, Jr., Kochunov P, Lancaster JL, Lawrie SM, Liewald DC, Mandl R, Matarin M, Mattheisen M, Meisenzahl E, Melle I, Moses EK, Muhleisen TW, Nauck M, Nothen MM, Olvera RL, Pandolfo M, Pike GB, Puls R, Reinvang I, Renteria ME, Rietschel M, Roffman JL, Royle NA, Rujescu D, Savitz J, Schnack HG, Schnell K, Seiferth N, Smith C, Steen VM, Valdes Hernandez MC, Van den Heuvel M, van der Wee NJ, Van Haren NE, Veltman JA, Volzke H, Walker R, Westlye LT, Whelan CD, Agartz I, Boomsma DI, Cavalleri GL, Dale AM, Djurovic S, Drevets WC, Hagoort P, Hall J, Heinz A, Jack CR, Jr., Foroud TM, Le Hellard S, Macciardi F, Montgomery GW, Poline JB, Porteous DJ, Sisodiya SM, Starr JM, Sussmann J, Toga AW, Veltman DJ, Walter H, Weiner MW, Bis JC, Ikram MA, Smith AV, Gudnason V, Tzourio C, Vernooij MW, Launer LJ, DeCarli C, Seshadri S, Andreassen OA, Apostolova LG, Bastin ME, Blangero J, Brunner HG, Buckner RL, Cichon S, Coppola G, de Zubicaray GI, Deary IJ, Donohoe G, de Geus EJ, Espeseth T, Fernandez G, Glahn DC, Grabe HJ, Hardy J, Hulshoff Pol HE, Jenkinson M, Kahn RS, McDonald C, McIntosh AM, McMahon FJ, McMahon KL, Meyer-Lindenberg A, Morris DW, Muller-Myhsok B, Nichols TE, Ophoff RA, Paus T, Pausova Z, Penninx BW, Potkin SG, Samann PG, Saykin AJ, Schumann G, Smoller JW, Wardlaw JM, Weale ME, Martin NG, Franke B, Wright MJ, Thompson PM. Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet. 2011;44:552–561. doi: 10.1038/ng.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Lai YH, Han XD, Wong PM, Peters LL, Chui DH. Murine Hn1 on chromosome 11 is expressed in hemopoietic and brain tissues. Mamm Genome. 1997;8:695–696. doi: 10.1007/s003359900540. [DOI] [PubMed] [Google Scholar]
- Tanzi RE. A Brief History of Alzheimer's Disease Gene Discovery. J Alzheimers Dis. 2012 doi: 10.3233/JAD-2012-129044. 2012. [DOI] [PubMed] [Google Scholar]
- Tchaicha JH, Reyes SB, Shin J, Hossain MG, Lang FF, McCarty JH. Glioblastoma angiogenesis and tumor cell invasiveness are differentially regulated by beta8 integrin. Cancer research. 2011;71:6371–6381. doi: 10.1158/0008-5472.CAN-11-0991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thambisetty M, An Y, Kinsey A, Koka D, Saleem M, Guntert A, Kraut M, Ferrucci L, Davatzikos C, Lovestone S, Resnick SM. Plasma clusterin concentration is associated with longitudinal brain atrophy in mild cognitive impairment. Neuroimage. 2012a;59:212–217. doi: 10.1016/j.neuroimage.2011.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thambisetty M, Beason-Held LL, An Y, Kraut M, Nalls M, Hernandez DG, Singleton AB, Zonderman AB, Ferrucci L, Lovestone S, Resnick SM. Alzheimer Risk Variant CLU and Brain Function During Aging. Biol Psychiatry. 2012b doi: 10.1016/j.biopsych.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thambisetty M, Simmons A, Velayudhan L, Hye A, Campbell J, Zhang Y, Wahlund LO, Westman E, Kinsey A, Guntert A, Proitsi P, Powell J, Causevic M, Killick R, Lunnon K, Lynham S, Broadstock M, Choudhry F, Howlett DR, Williams RJ, Sharp SI, Mitchelmore C, Tunnard C, Leung R, Foy C, O’Brien D, Breen G, Furney SJ, Ward M, Kloszewska I, Mecocci P, Soininen H, Tsolaki M, Vellas B, Hodges A, Murphy DG, Parkins S, Richardson JC, Resnick SM, Ferrucci L, Wong DF, Zhou Y, Muehlboeck S, Evans A, Francis PT, Spenger C, Lovestone S. Association of plasma clusterin concentration with severity, pathology, and progression in Alzheimer disease. Arch Gen Psychiatry. 2010;67:739–748. doi: 10.1001/archgenpsychiatry.2010.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Hayashi KM, de Zubicaray G, Janke AL, Rose SE, Semple J, Herman D, Hong MS, Dittmer SS, Doddrell DM, Toga AW. Dynamics of gray matter loss in Alzheimer's disease. J Neurosci. 2003;23:994–1005. doi: 10.1523/JNEUROSCI.23-03-00994.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Hayashi KM, Sowell ER, Gogtay N, Giedd JN, Rapoport JL, de Zubicaray GI, Janke AL, Rose SE, Semple J, Doddrell DM, Wang Y, van Erp TG, Cannon TD, Toga AW. Mapping cortical change in Alzheimer’s disease, brain development, and schizophrenia. Neuroimage. 2004;23(Suppl 1):S2–S18. doi: 10.1016/j.neuroimage.2004.07.071. [DOI] [PubMed] [Google Scholar]
- Turner JA, Calhoun VD, Michael A, van Erp TG, Ehrlich S, Segall JM, Gollub RL, Csernansky J, Potkin SG, Ho BC, Bustillo J, Schulz SC, Fbirn Wang L. Heritability of multivariate gray matter measures in schizophrenia. Twin Res Hum Genet. 2012;15:324–335. doi: 10.1017/thg.2012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner ST, Fornage M, Jack CR, Jr., Mosley TH, Kardia SL, Boerwinkle E, de Andrade M. Genomic susceptibility loci for brain atrophy in hypertensive sibships from the GENOA study. Hypertension. 2005;45:793–798. doi: 10.1161/01.HYP.0000154685.54766.2d. [DOI] [PubMed] [Google Scholar]
- Van Balkom ID, Vuijk PJ, Franssens M, Hoek HW, Hennekam RC. Development, cognition, and behaviour in Pitt-Hopkins syndrome. Dev Med Child Neurol. 2011;54:925–931. doi: 10.1111/j.1469-8749.2012.04339.x. [DOI] [PubMed] [Google Scholar]
- van Praag H, Shubert T, Zhao C, Gage FH. Exercise enhances learning and hippocampal neurogenesis in aged mice. J Neurosci. 2005;25:8680–8685. doi: 10.1523/JNEUROSCI.1731-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Veen T, Nielsen J, Berkhof J, Barkhof F, Kamphorst W, Bo L, Ravid R, Verweij CL, Huitinga I, Polman CH, Uitdehaag BM. CCL5 and CCR5 genotypes modify clinical, radiological and pathological features of multiple sclerosis. J Neuroimmunol. 2007;190:157–164. doi: 10.1016/j.jneuroim.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Villeda SA, Luo J, Mosher KI, Zou B, Britschgi M, Bieri G, Stan TM, Fainberg N, Ding Z, Eggel A, Lucin KM, Czirr E, Park JS, Couillard-Despres S, Aigner L, Li G, Peskind ER, Kaye JA, Quinn JF, Galasko DR, Xie XS, Rando TA, Wyss-Coray T. The ageing systemic milieu negatively regulates neurogenesis and cognitive function. Nature. 2011;477:90–94. doi: 10.1038/nature10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitner EB, Farfel-Becker T, Eilam R, Biton I, Futerman AH. Contribution of brain inflammation to neuronal cell death in neuronopathic forms of Gaucher's disease. Brain. 2012;135:1724–1735. doi: 10.1093/brain/aws095. [DOI] [PubMed] [Google Scholar]
- Walhovd KB, Fjell AM, Brewer J, McEvoy LK, Fennema-Notestine C, Hagler DJ, Jr., Jennings RG, Karow D, Dale AM. Combining MR imaging, positron-emission tomography, and CSF biomarkers in the diagnosis and prognosis of Alzheimer disease. AJNR Am J Neuroradiol. 2010;31:347–354. doi: 10.3174/ajnr.A1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei TC, Lin HY, Lu CC, Chen CM, You LR. Expression of Crip2, a LIM-domain-only protein, in the mouse cardiovascular system under physiological and pathological conditions. Gene Expr Patterns. 2011;11:384–394. doi: 10.1016/j.gep.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Weinstein G, Wolf PA, Beiser AS, Au R, Seshadri S. Risk Estimations, Risk Factors, and Genetic Variants Associated with Alzheimer's Disease in Selected Publications from the Framingham Heart Study. J Alzheimers Dis. 2011 doi: 10.3233/JAD-2012-129040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler AM, Kochunov P, Blangero J, Almasy L, Zilles K, Fox PT, Duggirala R, Glahn DC. Cortical thickness or grey matter volume? The importance of selecting the phenotype for imaging genetics studies. Neuroimage. 2010;53:1135–1146. doi: 10.1016/j.neuroimage.2009.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolkowitz OM, Reus VI, Mellon SH. Of sound mind and body: depression, disease, and accelerated aging. Dialogues Clin Neurosci. 2011;13:25–39. doi: 10.31887/DCNS.2011.13.1/owolkowitz. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood SH, Craig T, Li Y, Merry B, de Magalhaes JP. Whole transcriptome sequencing of the aging rat brain reveals dynamic RNA changes in the dark matter of the genome. Age (Dordr) 2012 doi: 10.1007/s11357-012-9410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss-Coray T. Inflammation in Alzheimer disease: driving force, bystander or beneficial response? Nat Med. 2006;12:1005–1015. doi: 10.1038/nm1484. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.