The interdependence of p53 and MDM2 is critical for proper cell survival and cell death. Zhu et al. find that TAB1, an activator of TAK1 and p38α, inhibits the E3 ligase activity of MDM2 toward p53 and its homolog, MDMX. Cisplatin-induced cell death is mitigated by TAB1 knockdown. TAB1 stabilizes MDMX and activates p38α to phosphorylate p53, allowing p53 target induction. TAB1 levels are relatively low in cisplatin-resistant clones of ovarian cells and in ovarian tumors, implicating TAB1 as a tumor suppressor.
Keywords: p53, MDM2, MDMX, TAB1, p38α, cisplatin
Abstract
The interdependence of p53 and MDM2 is critical for proper cell survival and cell death and, when altered, can lead to tumorigenesis. Mitogen-activated protein kinase (MAPK) signaling pathways function in a wide variety of cellular processes, including cell growth, migration, differentiation, and death. Here we discovered that transforming growth factor β-activated kinase 1 (TAK1)-binding protein 1 (TAB1), an activator of TAK1 and of p38α, associates with and inhibits the E3 ligase activity of MDM2 toward p53 and its homolog, MDMX. Depletion of TAB1 inhibits MDM2 siRNA-mediated p53 accumulation and p21 induction, partially rescuing cell cycle arrest induced by MDM2 ablation. Interestingly, of several agents commonly used as DNA-damaging therapeutics, only cell death caused by cisplatin is mitigated by knockdown of TAB1. Two mechanisms are required for TAB1 to regulate apoptosis in cisplatin-treated cells. First, p38α is activated by TAB1 to phosphorylate p53 N-terminal sites, leading to selective induction of p53 targets such as NOXA. Second, MDMX is stabilized in a TAB1-dependent manner and is required for cell death after cisplatin treatment. Interestingly TAB1 levels are relatively low in cisplatin-resistant clones of ovarian cells and in ovarian patient's tumors compared with normal ovarian tissue. Together, our results indicate that TAB1 is a potential tumor suppressor that serves as a functional link between p53–MDM2 circuitry and a key MAPK signaling pathway.
The protein product of the p53 gene that is frequently mutated in human tumors plays a central role in coordinating the cellular responses to a wide variety of stress signals (Vousden and Prives 2009). In those tumors that harbor wild-type p53 protein, the functions of p53 are often compromised by overexpression of MDM2 or its homolog, MDMX (Marine et al. 2006). Both MDM2 and MDMX are crucial negative regulators of p53. They bind to the N-terminal transactivation domain of p53 and can thereby inhibit p53 target gene transcription. MDM2 also functions as an E3 ubiquitin ligase promoting ubiquitination and proteasomal degradation of p53, MDMX, and itself (Marine and Lozano 2010; Wade et al. 2010). MDMX is not active as an E3 ligase but can form heteroligomers with MDM2 and regulate the E3 ligase function of MDM2 (Shadfan et al. 2012). Both MDM2 and MDMX are crucial for keeping p53 in check under nonstressed conditions in mice, since deletion of either gene leads to embryonic lethality unless mice also lack functional p53 alleles (Parant et al. 2001; Migliorini et al. 2002; Marine et al. 2006). This suggests that MDM2 and MDMX play nonredundant roles in the regulation of p53. Interplay between p53, MDM2, and MDMX may determine whether a cell responds to p53 activation with growth arrest or apoptosis (Wade et al. 2010). In response to various sources of cellular stress, multiple post-translational modifications, including phosphorylation, regulate the association between MDM2/MDMX and p53, and, in turn, such modifications lead to p53 stabilization and activation (Kruse and Gu 2009; Waning et al. 2010). Most relevant to this study, numerous protein kinases—including members of the mitogen-activated protein kinase (MAPK) family such as p38α, JNK (c-Jun N-terminal kinase), and ERK—have been shown to functionally interact with as well as phosphorylate and activate p53, leading to p53-mediated cellular responses in response to stress stimuli (Wu 2004). Modulation of MDM2 and MDMX phosphorylation also affects their respective functions, especially toward p53 (Chen 2012). Furthermore, several members of the dual specificity phosphatase family—such as Wip1, MKP1, PAC1, and DUSP5—that negatively regulate MAPK signaling have been shown to be p53 targets (Janicke et al. 2008), while MDM2 and MDMX have also been shown to be substrates of MAPKs as well as phosphatases such as Wip1 (Lu et al. 2007b; Malmlof et al. 2007; Gilkes et al. 2008; Zhang et al. 2009). Both p53 and MAPK signaling pathways regulate a wide variety of cellular processes, and alterations in either are often associated with cancer (Dhillon et al. 2007; Vousden and Prives 2009). A better understanding of the interplay between these two critical networks would provide valuable insights for tumorigenesis and therapy resistance.
Transforming growth factor β (TGFβ)-activated kinase 1 (TAK1)-binding protein 1 (TAB1, also known as MAP3K7IP1) is a modular adapter protein that was initially described as an activator of TAK1 (also known as MAP3K7) in response to TGFβ (Shibuya et al. 1996). TAK1 acts as an upstream kinase for NF-κB and MAPK activation in response to multiple stress signals (Ono et al. 2001; Johnson 2002). TAB1 also regulates p38α autoactivation through direct binding (Ge et al. 2002; Johnson 2002). Furthermore, AMP-activated protein kinase (AMPK) binds to TAB1, increasing recruitment and activation of p38α in the ischemic heart (Li et al. 2005). It was also documented that TAB1 and IKKβ (IκB kinase β) form complexes albeit solely in breast cancer cells or normal mammary epithelial cells following the induction of the epithelial–mesenchymal transition by TGF-β (Neil and Schiemann 2008). TAB1 possesses several seemingly distinct protein-binding regions. In addition to the extreme C-terminal 68 amino acids that are sufficient for binding to and activation of TAK1, the C-terminal portion of TAB1 also contains a p38α-binding domain that precedes the TAK1-binding region. Within its N-terminal portion, TAB1 contains a PP2C (protein phosphatase 2C)-like domain, and structural and biochemical analysis suggests that TAB1 may function as a pseudophosphatase (Conner et al. 2006). This PP2C-like region was also found to interact with XIAP, a member of the inhibitor of apoptosis proteins (IAP) family, which has been shown to play roles in signaling to NF-κB and MAPK activation (Yamaguchi et al. 1999; Lu et al. 2007a). Therefore, through its association with several protein complexes, TAB1 is involved in multiple signaling pathways.
Although TAB1 and TAK1 are believed to be functionally entwined, each has distinct roles in the MAPK signaling cascade, and they may not always function together as a complex. TAK1-deficient mouse embryos die at around embryonic day 10 (E10) and exhibit abnormal development of the neural tube, while TAB1-deficient mice die at a later stage of gestation due to abnormal cardiovascular and lung morphogenesis (Komatsu et al. 2002; Shim et al. 2005; Inagaki et al. 2008). Additional observations have disputed an essential role for TAB1 in TAK1 activation (Shim et al. 2005; Bertelsen and Sanfridson 2007; Mendoza et al. 2008).
Here, we identified TAB1 as an MDM2-binding partner that modulates the E3 ligase activity of MDM2, leading to stabilization of MDM2, MDMX, and p53. We discovered that TAB1 regulates p53-mediated outcomes such as cell cycle arrest under some conditions and cell death initiated by cisplatin treatment. Furthermore, our data implicate both TAB1 activation of p38α and stabilization of MDMX in facilitating apoptosis in response to cisplatin. We hypothesize that TAB1 serves as a functional link to regulate cross-talk between p53 and MAPK signaling pathways.
Results
TAB1 interacts with MDM2
A high-throughput yeast two-hybrid screen was carried out to identify MDM2-interacting proteins (see Zhu et al. 2009 for more information), and multiple interacting clones encoding TAB1 were identified from three different cDNA libraries. The association of endogenous TAB1 and MDM2 in human cells was then demonstrated using cell lysates from U2OS cells (Fig. 1A) and HCT116 cells (Supplemental Fig. S1A); the specificity of the interaction was confirmed by the diminished signal seen when the two proteins were coimmunoprecipitated from extracts of U2OS cells treated with either TAB1 or MDM2 siRNA (Fig. 1A).
Figure 1.
TAB1 stabilizes p53 by inhibiting MDM2. (A) Specificity of binding between endogenous MDM2 and TAB1. (Left panel) Whole-cell lysates (500 μg) from U2OS cells were immunoprecipitated with a rabbit polyclonal anti-TAB1 antibody (α-T) or control rabbit IgG and then subjected to immunoblotting with anti-MDM2 (3G5+4B11+5B10) and anti-TAB1 antibodies. (Right panel) U2OS cells were transfected with siRNAs targeting luciferase (C), TAB1 (T1), or MDM2 (M). After 48 h, whole-cell lysates (500 μg) were prepared and subjected to immunoprecipitation with rabbit polyclonal anti-TAB1 (α-T) followed by immunoblotting with anti-MDM2 (3G5+4B11+5B10), anti-TAK1, and anti-TAB1 antibodies. Short (SE) and longer (LE) exposures of the TAB1 immunoblot are shown. (B) TAB1 blocks degradation of p53 by coexpressed MDM2. U2OS cells were transfected with Myc-TAB1 (1.5 μg), Flag-MDM2 (1.5 μg), and HA-p53 (0.35 μg) constructs as indicated. Cell lysates were used for immunoblotting with anti-HA, and anti-Flag antibodies. A coexpressed GFP construct was added in each case to control for transfection efficiency and loading. (C) Ectopic expression of TAB1 stabilizes endogenous MDM2 and p53. U2OS cells were transfected with increasing amounts of Myc-TAB1 (0.5, 1, 2, and 3 μg) as indicated. Cell lysates were used for immunoblotting with anti-MDM2 (3G5+4B11+5B10), anti-p53 (DO-1), anti-p21, anti-Myc, and anti-Actin antibodies. (D) TAK1 cooperates with TAB1 to inhibit MDM2-mediated p53 degradation. U2OS cells were transfected with HA-p53 (0.3 μg) and combinations of Flag-MDM2 (1.5 μg), Myc-TAB1 (0.5 or 1 μg), pc-TAK1 (0.5 or 1 μg), or kinase-defective pc-TAK1 K63W (TAK1KW; 0.5 or 1 μg) as indicated. Cell lysates were used for immunoblotting with anti-Flag, anti-HA, anti-Myc (9E10), and anti-TAK1 antibodies. A GFP construct was cotransfected in each case to control for transfection efficiency and loading.
To map the region of MDM2 that is required for TAB1 binding, a series of Flag-tagged MDM2 deletion and truncation mutants was constructed and transiently overexpressed in H1299 cells together with Myc-tagged TAB1. Cell extracts were subjected to immunoprecipitation with an anti-Myc antibody followed by immunoblotting with an anti-Flag antibody (Supplemental Fig. S1). Our results indicate that amino acids 223–339 span the primary binding sites for TAB1. However, the MDM2 N terminus (1–222) and MDM2 C-terminal RING-containing regions also interacted with TAB1 albeit weakly, indicating that either TAB1 interacts with multiple surfaces on MDM2 or its interactions are dependent on the tertiary structure of MDM2.
TAB1 inhibits MDM2-mediated p53 degradation and ubiquitination
To determine the functional consequences of the TAB1–MDM2 association, we coexpressed TAB1 with MDM2 and p53 in U2OS cells. MDM2-mediated degradation of p53 was markedly inhibited by TAB1 (Fig. 1B). More significantly, the levels of endogenously expressed p53 protein and two p53 targets (MDM2 and p21) in U2OS cells were elevated following ectopic TAB1 expression (Fig. 1C). Ectopic TAK1 and TAB1 cooperated to inhibit MDM2-mediated p53 degradation (Fig. 1D), and TAB1 and TAK1 mutually stabilized each other (Fig. 1D, cf. lanes 3 and 5 for TAK1 stabilization of TAB1 and lanes 5 and 6 for TAB1 stabilization of TAK1). It is therefore possible that the inhibitory effect of TAK1 on MDM2 was mediated by TAB1. A kinase-dead mutant of TAK1 (K63W) (Yamaguchi et al. 1995) that stabilized TAB1 to a lesser extent (Fig. 1D, cf. lanes 5 and 8) also inhibited p53 degradation to a correspondingly reduced extent. Note that both wild-type TAK1 and its kinase-dead derivative by themselves had no inhibitory effect on MDM2 degradation of p53.
In keeping with its ability to stabilize p53, TAB1 expression inhibited the ability of MDM2 to ubiquitinate p53 (Fig. 2A). Ectopic TAB1 also modestly repressed MDM2 autoubiquitination (Fig. 2B) and, more significantly, inhibited MDM2 ubiquitination of MDMX, one of its well-known E3 ligase substrates (Fig. 2C). These results suggest that TAB1 functions as a general inhibitor of the E3 ligase activity of MDM2 albeit to varying extents. Note that TAB1 is not a universal inhibitor of E3 ligase/proteasome machinery, as it had no inhibitory effect on E2F1 ubiquitination, which has been shown to be mediated by the E3 ligase Skp2 (Supplemental Fig. S2; Marti et al. 1999).
Figure 2.
Ectopic expression of TAB1 inhibits E3 ligase activity of MDM2. (A) TAB1 inhibits MDM2-mediated p53 polyubiquitination. H1299 cells were transfected with the indicated combinations of Myc-TAB1 (0.6 or 1.2 μg), Flag-MDM2 (1.2 μg), and p53 (0.3 μg) plasmids along with an HA-ubiquitin (HA-Ub; 1.2 μg) plasmid as indicated. The cells were treated with MG132 (20 μM) for 4 h before harvesting. Whole-cell lysates were subjected to immunoprecipitation with anti-p53 (FL-393-G) antibody followed by immunoblotting with anti-HA antibody to detect ubiquitinated p53. (B) TAB1 inhibits MDM2 autoubiquitination. H1299 cells were transfected with combinations of Myc-TAB1 (1.2 μg) and Flag-MDM2 (1.2 μg) plasmids in the presence of an HA-ubiquitin (HA-Ub; 1.2 μg) plasmid as indicated. The cells were treated with MG132 as in A before harvesting. Whole-cell lysates were subjected to immunoprecipitation with anti-Flag antibody followed by immunoblotting with anti-HA antibody to detect ubiquitinated MDM2. (C) TAB1 inhibits MDM2-mediated MDMX ubiquitination. H1299 cells were transfected with combinations of HA-MDMX (0.6 μg), Flag-MDM2 (0.8 or 1.6 μg), and Myc-TAB1 (0.5 or 1.0 μg) plasmids in the presence of a His-ubiquitin (His-Ub; 1.4 μg) plasmid as indicated. The cells were treated with MG132 as in A before harvesting. Cells were lysed in denaturing buffer and subjected to Ni-NTA bead binding as described in the Materials and Methods. Ubiquitinated MDMX was detected by an anti-MDMX antibody.
TAB1 ablation attenuates p53 activation that results from knockdown of MDM2
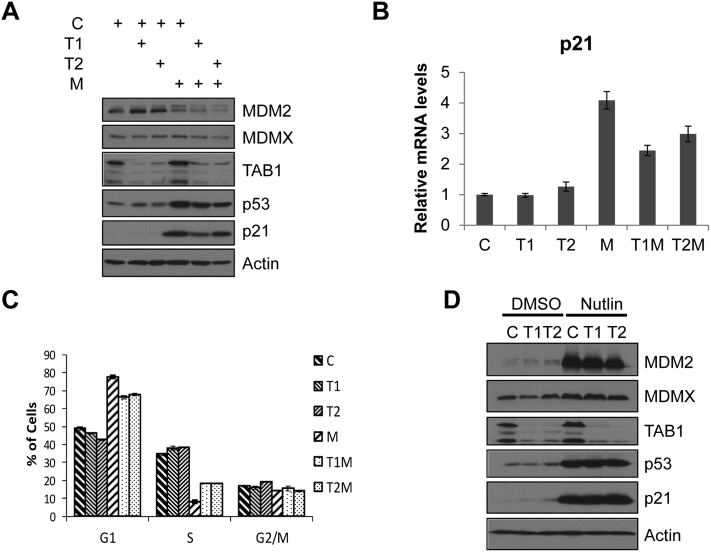
To evaluate how TAB1 regulates MDM2 when expressed at normal endogenous levels, we used two different siRNAs to examine the effect of TAB1 down-regulation on the functions of MDM2 and p53. Depletion of TAB1 in U2OS cells did not affect the cellular levels of p53 and MDM2 (Fig. 3A). When we introduced MDM2 siRNA into U2OS cells, as expected, p53 was stabilized, and p21 expression was increased. Interestingly, upon down-regulation of TAB1, MDM2 knockdown-mediated p53 stabilization was attenuated, and the levels of p21 protein (Fig. 3A) and RNA (Fig. 3B) were also markedly decreased. In line with this, ablation of TAB1 partially rescued cell cycle arrest resulting from MDM2 knockdown in U2OS cells (Fig. 3C). Similar restoration of the cell cycle was observed when MDM2 and/or TAB1 were ablated in HCT116 or RKO cells (Supplemental Fig. S3). Since p53 stabilization and p21 expression induced by Nutlin (a small molecule that disrupts the p53–MDM2 interaction) were not attenuated upon TAB1 ablation, we surmise that TAB1 modulates p53 function through MDM2 (Fig. 3D).
Figure 3.
TAB1 is required for full p53 activation upon ablation of MDM2. (A) Down-regulation of TAB1 attenuates p53 activation in cells depleted of MDM2. U2OS cells were transfected with 25 nM each control luciferase siRNA (C), two different TAB1 siRNAs (T1 and T2), or MDM2 siRNA (M) as indicated to a total siRNA concentration of 50 nM in all samples (balanced with control luciferase siRNA). Forty-eight hours after transfection, cells were harvested, and cell lysates were prepared and then subjected to immunoblotting with anti-MDM2 (3G5+4B11+5B10), anti-MDMX, anti-TAB1, anti-p53 (DO-1), anti-p21, or anti-Actin antibodies. (B) Down-regulation of TAB1 attenuates p53-induced p21 transcription in response to MDM2 ablation. U2OS cells were transfected with 25 nM each control luciferase siRNA (C), MDM2 siRNA (M), TAB1 siRNA (T1 or T2), or both MDM2 and TAB1 siRNAs (T1M or T2M) as indicated to a total siRNA concentration of 50 nM as in A. Forty-eight hours after transfection, cells were harvested, and total mRNAs were extracted. cDNAs were then reverse-transcribed, and quantitative real-time PCR was performed using p21 gene-specific primers. (C) Reducing TAB1 levels attenuates cell cycle arrest induced by MDM2 ablation. U2OS cells were transfected with siRNAs as in A. Cells were trypsinized and fixed followed by cell cycle analysis. The plot was obtained from three separate experiments. (D) Attenuation of p53-induced response by ablation of TAB1 requires p53–MDM2 interaction. U2OS cells were transfected with 25 nM each control luciferase siRNA (C) or two different TAB1 siRNAs (T1 and T2). Forty-two hours after transfection, cells were treated with DMSO or Nutlin (5 μM). Six hours later, cells were harvested, and cell lysates were prepared and then immunoblotted with anti-MDM2 (3G5+4B11+5B10), anti-p53 (DO-1), anti-MDMX, anti-TAB1, anti-p21, or anti-Actin antibodies.
TAB1 mediates the apoptotic response of p53 to cisplatin by selective induction of NOXA expression
Based on its ability to prevent MDM2 from degrading p53, we anticipated that TAB1 would be required to regulate the MDM2–p53 circuit under a variety of stress conditions. To test this, we examined the impact of TAB1 knockdown on a variety of different genotoxic assaults—including 5-fluorouracil, actinomycin D, cisplatin, daunorubicin (Dauno), deferoxamine mesylate, doxorubicin, etoposide (ETP), hydoxyurea, neocarzinostatin (NCS), and taxol—in U2OS cells. Unexpectedly, reduction of TAB1 levels had little or no effect on cell cycle arrest or cell death caused by all but one of these various agents (Supplemental Table S1). The one striking exception was cisplatin, where ablation of TAB1 markedly attenuated cell death induced by this agent as measured by analysis of cells with sub-G1 DNA content (Fig. 4A) or PARP cleavage (Fig. 4B). In both apoptosis assays, siRNA-mediated knockdown of TAK1 also impacted cisplatin-mediated cell death (Fig. 4A,B). The effects of cisplatin were completely abolished by the caspase inhibitor z-VAD-fmk, indicating that in this setting, cell death occurred by the process of apoptosis (Supplemental Fig. S4). As a control, ETP-induced cell death, which, as mentioned above, was not significantly attenuated by ablation of TAB1, was also abolished by z-VAD-fmk treatment (Supplemental Fig. S4). As previously reported (Bragado et al. 2007), cisplatin treatment resulted in substantially more cell death in HCT116 (p53+/+) than in HCT116 (p53−/−) cells, indicating that the drug requires p53 to produce apoptosis (Fig. 4C). Correspondingly, ablation of TAB1 significantly reduced cisplatin-induced cell death in HCT116 (p53+/+) cells but not in HCT116 (p53−/−) cells, indicating that TAB1 is involved in the p53-mediated apoptotic response (Fig. 4C). TAB1 knockdown also modulated cisplatin-induced cell death in SkHep1 liver cancer cells containing wild-type p53 but not in the p53-null Saos2 osteosarcoma cell line (Supplemental Fig. S5). Consistent with these findings, a colony formation assay confirmed that TAB1-ablated cells have a higher survival rate after cisplatin treatment compared with control cells (Fig. 4D).
Figure 4.
TAB1 ablation attenuates the cellular p53 response to cisplatin treatment. (A) TAB1 knockdown reduces cisplatin-mediated cell death. U2OS cells were transfected with 25 nM each control luciferase siRNA (C), TAB1 siRNAs (T1 and T2), or TAK1 siRNA (TK). Forty-eight hours after transfection, cells were treated with vehicle (DMSO) or cisplatin (CDDP; 5 μg/mL) for 24 h and then trypsinized and fixed followed by cell cycle analysis. The plot was obtained from three separate experiments. (B) Cisplatin-induced Parp cleavage is inhibited by TAB1 ablation. U2OS cells were transfected with siRNAs and treated with either vehicle (DMSO) or cisplatin (CDDP) as in A. Total cell lysates were prepared and then immunoblotted with anti-Parp antibody and anti-Actin antibodies. (C) Cisplatin-induced cell death is a p53-mediated response. HCT116 (p53+/+) and HCT116 (p53−/−) cells were transfected with 25 nM each control luciferase siRNA (C) or TAB1 siRNA (T1). Forty-eight hours after transfection, cells were treated with vehicle (DMSO) or cisplatin (CDDP; 10 μg/mL) for 24 h. Cells were then trypsinized and fixed, followed by FACS analysis. The plot was derived from three separate experiments. (D) TAB1 depletion enhances cell survival in response to cisplatin treatment. U2OS cells were transfected with 25 nM each control luciferase (C) or TAB1 siRNA (T1). Eighteen hours after transfection, cells were trypsinized, and 8 × 104 cells were seeded in a 35-mm dish. Twenty-four hours later, cells were treated with cisplatin for 24 h. Cells were then washed with PBS three times and incubated in fresh Dulbecco modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS). Six days later, viable cells were assayed for colony formation by cystal violet staining. Stained plates were photographed, and the number of colonies formed was manually scored using a 1-cm × 1-cm grid system and graphed as described in the Materials and Methods. (E) TAB1 ablation inhibits NOXA expression. U2OS cells were transfected with 25 nM each luciferase control siRNA (C), TAB1 siRNAs (T1 and T2), or TAK1 siRNA (TK) as indicated to a total siRNA concentration of 50 nM and then treated with either vehicle (DMSO) or cisplatin (CDDP) as in A. Total cell lysates were prepared and immunoblotted with anti-MDM2 (3G5+4B11+5B10), anti-p53 (DO-1), anti-TAK1, anti-TAB1, anti-p21, anti-NOXA, or anti-Actin antibodies. (F) TAB1 ablation has a selective effect on expression of p53 target genes. U2OS cells were transfected with 25 nM each luciferase control siRNA (C), TAB1 siRNAs (T1 and T2), or TAK1 siRNA (TK) and treated with vehicle (DMSO) or cisplatin (CDDP) as in E. Total mRNAs were extracted and reverse-transcribed, and then cDNAs were analyzed by quantitative real-time PCR using primers targeting NOXA, PUMA, and p21 genes. (G) NOXA ablation reduces cisplatin-induced apoptosis. U2OS cells were transfected with 25 nM each luciferase control siRNA (C), NOXA siRNAs (N1 and N2), or TAB1 siRNA (T1). Forty-eight hours after transfection, cells were treated with vehicle (DMSO) or cisplatin (CDDP; 5 μg/mL) for 24 h and then trypsinized and fixed, followed by cell cycle analysis.
Not only was TAB1 regulation of the p53 response stimulus-specific, but we also found that p53 targets varied in their response to cisplatin. In particular, NOXA protein levels were increased after cisplatin treatment in a manner that required full expression of TAB1, while protein levels of p21 protein remained unchanged in the absence or presence of the drug with or without TAB1 knockdown (Fig. 4E). The inability of cisplatin treatment to increase levels of p21 protein has been previously reported (Cuadrado et al. 2007). The inhibitory effect of TAB1 ablation also selectively affected NOXA mRNA levels, since it had no impact on either p21 or Puma (another p53 proapoptotic target) mRNA induction by p53 (Fig. 4F). Similar inhibition of NOXA induction in response to cisplatin treatment upon TAB1 ablation was observed in SkHep1 cells (Supplemental Fig. S6). Importantly, NOXA ablation by siRNA reduced cisplatin-induced U2OS cell death to an extent similar to TAB1 knockdown (Fig. 4G). p21 induction has been correlated with protection from cell death (Gartel and Tyner 2002), and we cannot rule out that unchanged p21 protein levels upon cisplatin treatment may be an additional contributing factor to the apoptotic response. Taken together, we delineated a pathway whereby TAB1 leads to apoptosis in cells treated with cisplatin that requires selective induction of the p53 target NOXA.
TAB1 is required for p38α-mediated p53 phosphorylation
Intriguingly, despite the impact on p53-mediated cell death in cisplatin-treated cells, ablating TAB1 (or TAK1) had little effect on p53 protein levels under the conditions used; i.e., after 24 h of treatment with this drug (Fig. 4E). Since our data in Figures 1 and 2 implicated TAB1 in regulating p53 stability and ubiquitination, we examined the kinetics of p53 induction after cisplatin treatment and found that at an earlier time point (4 h after cisplatin treatment), TAB1 or TAK1 ablation led to a modest but reproducible attenuation of steady-state p53 levels (Supplemental Fig. S7). In line with this, p53 in cells with TAB1 depletion had a shorter half-life in the presence of cisplatin compared with cells transfected with control siRNA (Fig. 5A). Moreover, more ubiquitinated p53 species were identified in TAB1-ablated cells in both in vivo ubiquitination (Fig. 5B) and in vitro degradation (Fig. 5C) assays. At later time points after drug treatment when p53 levels are not affected, there may be other processes affected by cisplatin, such as levels or translation of p53 mRNA. Importantly, despite its only subtle effect on p53 protein levels, there was a significant impact of TAB1 knockdown on p53 post-translational modifications, specifically on phosphorylation of S15 and S46 (Fig. 5D).
Figure 5.
TAB1 regulates p53 phosphorylation via p38α signaling in cisplatin-treated cells. (A) TAB1 ablation accelerates p53 turnover. U2OS cells were transfected with 25 nM each luciferase control siRNA (C) or TAB1 siRNA (T1). Forty-eight hours after transfection, cells were treated with vehicle (DMSO) or cisplatin (CDDP; 5 μg/mL) for 5 h. Next, cyclohexamide (CHX; 100 μg/mL) was added, and cells were harvested at the indicated times. (Top panel) Cell lysates were subjected to immunoblotting with the indicated antibodies. (Bottom panel) Quantification of the immunoblot data was carried out using Image J software. (B) Endogenous p53 ubiquitination is increased upon TAB1 depletion in response to cisplatin treatment. U2OS cells (∼8 × 105 cells in 10-cm dishes) were transfected with 25 nM each luciferase control siRNA (C), TAB1 siRNA (T), or MDM2 siRNA (M). Six hours later, each plate of cells was trypsinized and replated into three 60-mm dishes. Twenty-four hours after the initial siRNA transfection, cells were transfected with His-tagged ubiquitin (His-Ub; 4 μg) for 24 h and treated with vehicle (DMSO), cisplatin (CDDP; 5 μg/mL), or ETP (15 μM) for 5 h before harvesting. Cells were lysed in denaturing buffer and bound to Ni-NTA beads as described in the Materials and Methods. Ubiquitinated endogenous p53 was detected by an anti-p53 antibody (DO-1). (C) TAB1 plays a role in ubiquitination of endogenously expressed p53 in response to cisplatin treatment. U2OS cells were transfected with 25 nM each luciferase control siRNA (C), TAB1 siRNA (T1), or MDM2 siRNA (M). Forty-eight hours after transfection, cells were treated with cisplatin (CDDP; 5 μg/mL) for 5 h before harvesting. In vitro degradation assays were performed as described in the Materials and Methods, and aliquots were removed from reaction mixtures at the indicated time points and subjected to immunoblotting with anti-p53 (DO-1) antibody. (D) A p38α inhibitor attenuates cisplatin-mediated p53 phosphorylation in U2OS cells. U2OS cells were transfected with 25 nM each luciferase control siRNA (C) or TAB1 siRNAs (T1 and T2). Forty-eight hours after transfection, cells were pretreated with the p38α inhibitor SB203580 (SB; 5 μM) or DMSO for 1 h and then treated with vehicle (DMSO), cisplatin (CDDP; 5 μg/mL), or both cisplatin and SB203580 (CDDP+SB) for 24 h. Total cell lysates were prepared and immunoblotted with anti-p53 (S15-p), anti-p53 (S46-p), anti-p53 (DO-1), anti-p38α (T180/Y182-p), and anti-p38α antibodies. (E) A p38α inhibitor attenuates cisplatin-mediated cell death in U2OS cells. U2OS cells were transfected with 25 nM each luciferase control siRNA (C) or TAB1 siRNAs (T1 and T2) and treated with p38α inhibitor SB203580 and cisplatin as in D. Cells were then trypsinized and fixed, followed by cell cycle analysis.
Phosphorylation of p53 at S46 has been well documented to facilitate the apoptotic response (Oda et al. 2000; D'Orazi et al. 2002; Mayo et al. 2005), and activation of p38α has been shown to mediate p53 phosphorylation at N-terminal sites, including S15 and S46 (Bulavin et al. 1999; Kim et al. 2002). Consistent with previous reports that TAB1 activates p38α through either activating TAK1 or direct binding to p38α (Ge et al. 2002; Johnson 2002), TAB1 knockdown prevented full phosphorylation of p38α at T180 and Y182. Since treatment with a p38α inhibitor led to reduced p53 phosphorylation at S15 and S46 (Fig. 5D), we believe that the activity of p53 is altered due to its modification by phosphorylation by p38α in a TAB1-dependent manner.
TAB1 has been shown to activate p38α through either direct binding or stimulation of TAK1 (Ge et al. 2002; Johnson 2002). To further investigate which kinase signaling pathway involves TAB1 to affect p53 phosphorylation status and mediate a p53 response, we treated cells with inhibitors of TAK1 or p38α together with cisplatin treatment. A TAK1 kinase inhibitor (5Z-7-oxozeaenol) (Ninomiya-Tsuji et al. 2003) had little effect on cisplatin-induced cell death in U2OS cells treated with control or TAB1 siRNAs (Supplemental Fig. S8), suggesting that the kinase activity of TAK1 is not essential for cisplatin-activated signaling pathways. Relevantly, inhibition of TAK1 downstream pathways (CAY10512 for NF-κB and SP100625 for JNK) in U2OS cells also had little effect on or even enhanced cisplatin-induced cell death (Supplemental Fig. S9). Furthermore, TAB1 knockdown was still able to attenuate cisplatin-induced cell death in the presence of those inhibitors (Supplemental Fig. S9). Therefore, it is not likely that TAB1 works through TAK1 to regulate p53 phosphorylation and activation after cisplatin treatment. On the other hand, when we preincubated U2OS cells with a p38α inhibitor (SB203580), this compound counteracted cisplatin-induced cell death (Fig. 5E). However, since TAB1 knockdown could further reduce the percentage of sub-G1 cells treated with the p38α inhibitor (Fig. 5E), this suggests that in addition to its functional association with p38α, TAB1 employs one or more additional mechanisms to contribute to cisplatin-induced cell death.
TAB1 contributes to p53-mediated intrinsic apoptosis through modulating cellular levels and localization of MDMX in response to cisplatin treatment
Previous studies reported that in cisplatin-treated cells, MDMX has an unexpected function as a mitochondrially associated proapoptotic factor in the p53-intrinsic cell death pathway (Mancini et al. 2004, 2009; Mancini and Moretti 2009). Since these investigators provided evidence that MDMX plays a role in recruiting p53 phosphorylated at S46 to mitochondria in cisplatin-treated cells, we tested whether TAB1 might be involved in this process. Indeed, our results implicate MDMX in the regulation of p53 by TAB1. First, we observed reduced cellular levels of MDMX in cells treated with cisplatin when TAB1 (or TAK1) was ablated by siRNA (Fig. 6A,E). This is consistent with our data (shown in Fig. 2C) that TAB1 inhibited MDM2-mediated ubiquitination of MDMX. Second, it is well established that some DNA damage-inducing agents (via ATM activation) elicit MDM2 degradation of MDMX (Kawai et al. 2003; Chen et al. 2005; Okamoto et al. 2005; Pereg et al. 2005; Jin et al. 2006). Strikingly, we found that MDMX was significantly more resistant to degradation upon cisplatin treatment when compared with other stress stimuli such as Dauno, ETP, and NCS (Fig. 6B). Note that we found that Dauno and NCS induced cell cycle arrest while ETP induced apoptosis in U2OS cells (data not shown), suggesting that the changes in cellular levels of MDMX do not correlate with any specific cell cycle profile. Third, consistent with the above-mentioned reports as well as our data that TAB1 is required for maximal cell death upon cisplatin treatment, ablation of MDMX by different siRNAs attenuated cisplatin-induced cell death similarly to ablation of TAB1, suggesting the possibility that TAB1 may participate in the cisplatin-mediated cellular response through modulating the cellular levels of MDMX (Fig. 6C). Finally, less mitochondrially localized MDMX was detected in U2OS cells treated with TAB1 siRNA compared with control siRNA (Supplemental Fig. S10). Thus, our results suggest that TAB1 contributes to p53-mediated intrinsic apoptosis through maintaining cellular levels of MDMX and modulating its localization in response to cisplatin treatment.
Figure 6.
TAB1 contributes to p53-mediated intrinsic apotptosis through modulating cellular levels of MDMX in response to cisplatin treatment. (A) TAB1 ablation reduces cellular levels of MDMX in response to cisplatin treatment. U2OS cells were transfected with 25 nM each luciferase control siRNA (C), TAB1 siRNAs (T1 and T2), or TAK1 siRNA (TK). Forty-eight hours after transfection, cells were treated with vehicle (DMSO) or cisplatin (CDDP; 5 μg/mL) for 4 h. Total cell lysates were prepared, followed by immunoblotting with anti-MDMX and anti-Actin antibodies. (B) MDMX is resistant to degradation in cisplatin-treated cells. U2OS cells were treated with DMSO (−), cisplatin (CDDP; 5 μg/mL), ETP (30 μM), Dauno (0.22 μM), NCS (320 ng/mL), or actinomycin D (ActD; 5 nM) for 5 h. Cell lysates were prepared for immunoblotting with anti-MDMX and anti-Actin antibodies. (C) MDMX ablation attenuates cisplatin-mediated cell death. U2OS cells were transfected with 25 nM each luciferase control siRNA (C), TAB1 siRNA (T1), or MDMX siRNAs (X4 and X11). Forty-eight hours after transfection, cells were treated with vehicle (DMSO) or cisplatin (CDDP; 5 μg/mL) for an additional 24 h and then trypsinized and fixed for cell cycle analysis. The plot was obtained from three separate experiments.
TAB1 is a potential tumor suppressor, and its ablation contributes to cisplatin resistance
Cisplatin is widely used in treating solid tumors, including testicular, ovarian, cervical, head and neck, and small-cell lung cancers (Basu and Krishnamurthy 2010). However, patients who initially respond to cisplatin therapy often develop resistance to the drug during the course of the treatment (Kelland 2007). To examine the relevance of the pathway that we delineated, we analyzed publicly available data sets. As shown in Figure 7A, levels of TAB1 were markedly lower in clonally derived cisplatin-resistant A2780 ovarian cancer cell lines than in their cisplatin-sensitive counterparts (Gene Expression Omnibus [GEO] data set no. GSE33482). A similar trend was observed in another data set using both cisplatin-sensitive and cisplatin-resistant HeLa cells (data not shown). Furthermore, TAB1 levels in human ovarian serous cystadenocarcinoma tumor samples were relatively low compared with normal ovarian tissue samples (Fig. 7B, The Cancer Genome Atlas [TCGA] data matrix). Interestingly, when we stratified the p53 status in samples, TAB1 levels also tended to be even lower in samples with wild-type p53 compared with samples with p53 having missense mutation (Fig. 7C). Therefore, our results both suggest that TAB1 ablation contributes to cisplatin resistance and implicate TAB1 as a potential tumor suppressor.
Figure 7.
TAB1 levels are relatively low in cisplatin-resistant clones of ovarian cancer cells and in ovarian tumors. (A) Cisplatin-resistant ovarian cancer cell line clones have lower cellular levels of TAB1 compared with their cisplatin-sensitive counterparts. Cellular levels of TAB1 mRNA in clonally derived cisplatin-resistant A2780 ovarian cancer cell lines (A2780-cis) and their cisplatin-sensitive counterparts (A2780) were analyzed based on a GEO publicly available data set (GSE33482). (B) TAB1 levels in human ovarian serous cystadenocarcinoma tumor samples compared with normal ovarian tissue samples. (Top panel) TAB1 levels (TCGA Agilent G4502A; N = 589) were analyzed comparing tumor samples with normal ovarian tissue samples. (Bottom panel) The Student's t-test (two-tailed, unpaired heteroscedastic t-test) was performed to verify statistically significant difference between the tumor (N = 589) and normal (N = 8) samples (P-value < 0.0005). The bars within the box represent the median gene expression. Round circles indicate outliers. The asterisk indicates statistical significance. (C) Comparison of TAB1 levels in ovarian serous cystadenocarcinoma tumor samples harboring wild-type p53 or p53 with missense mutations. Relative expression levels of TAB1 in tumor samples were analyzed based on their p53 status (wild-type, N = 77; mutant, N = 253). (D) Regulation of the p53/MDM2/MDMX circuit by TAB1. TAB1 inhibits MDM2 E3 ligase activity toward p53. (Left panel) Stabilization of p53 leads to cell cycle arrest. In response to cisplatin treatment, TAB1 also activates p38α, which in turn phosphorylates p53 to mediate an apoptotic response. (Right panel) At the same time, TAB1 modulates the cellular levels of MDMX and facilitates MDMX mitochondrial localization, which contributes to the p53-mediated intrinsic apoptotic response.
Discussion
We report here a functional link between p53/MDM2/MDMX circuitry and MAPK signaling through a newfound interaction between TAB1 and MDM2. Our results demonstrate that TAB1, a scaffold protein with multiple binding partners that are involved in different signaling pathways, is critical for p53 activation under specific conditions. TAB1, an inhibitor of MDM2 E3 ligase activity, is required for p53 up-regulation and cell cycle arrest when MDM2 is ablated. TAB1 is also a key mediator of p53-dependent cell death albeit uniquely in cisplatin-treated cells. In response to cisplatin, TAB1 both modulates p53 phosphorylation and activation through its functional interaction with p38α and regulates the cellular level of MDMX to facilitate p53-intrinsic apoptosis (modeled in Fig. 7D). Several aspects of this pathway merit further discussion.
TAB1 regulates p53 levels when MDM2 is ablated
In unstressed cells, p53 is rapidly turned over due in large part to the E3 ligase activity of MDM2. We discovered that reducing TAB1 levels via siRNA affects p53 levels and activity only in the context of codepleted MDM2. One speculation is that the reduced cellular pool of MDM2 upon siRNA knockdown may be a better inhibitory target for TAB1. Alternatively, p53 stabilization upon MDM2 depletion may set the cells into a “stressed” mode, which may trigger downstream events that somehow facilitate the interaction between TAB1 and MDM2. If the association between p53 and MDM2 were not disrupted under this kind of “stressed” condition, ablation of TAB1 would enhance MDM2 E3 ligase function.
TAB1 works through p38α to activate p53 in cisplatin-treated cells
Since TAB1 interacts with two major cellular signaling proteins (TAK1 and p38α) and plays important roles in their respective activations, we tested which of these two proteins is required for TAB1 regulation of p53 after cisplatin treatment. TAK1 is a member of the MAPKKK family that, through activation of downstream kinases, including IKK, JNK and p38α, activates key transcription factors such as AP-1 and NF-κB (Landstrom 2010). TAK1 was also reported to play a role in activation of the Snf1/AMPK family, which is important for metabolic regulation in cells (Momcilovic et al. 2006; Herrero-Martin et al. 2009; Chang et al. 2010). The outcome of the TAK1 signaling cascade is most likely tissue-specific and context-dependent, as TAK1 has been implicated in both tumor suppression (Konishi et al. 2003; Thakur et al. 2009; Bettermann et al. 2010; Inokuchi et al. 2010) and tumorigenesis (Melisi et al. 2011). Unlike TAB1, both wild-type TAK1 and its kinase-dead derivative do not inhibit MDM2-mediated p53 degradation by themselves. However, ectopic TAK1 enhances the inhibitory effect of coexpressed TAB1, most likely due to its ability to stabilize TAB1 (see Fig. 1). TAB1 is itself a substrate of TAK1 kinase (Prickett et al. 2008), and our results also suggest that TAK1 kinase activity is required for its ability to stabilize TAB1 (Figs. 1, 4). Therefore, we surmise that TAK1 modulates the p53–MDM2 circuit through its regulatory effect on TAB1. However, the fact that a TAK1-specific inhibitor (5Z-7-oxozeaenol) has no apparent effect on cisplatin-induced cell death in U2OS cells implies that signaling via this drug does not involve TAK1 activation (Supplemental Fig. S8).
In contrast to TAK1, our data strongly implicate p38α as a TAB1 target in regulating p53 after cisplatin treatment. p38 MAPKs, also known as stress-activated protein kinases, comprise four family members—p38α, p38β, p38γ, and p38δ—that possess both overlapping and specific functions (Cuenda and Rousseau 2007). TAB1 has been shown to specifically interact with p38α but not other p38 family members (Ge et al. 2002). The roles of p38α are complex (Wagner and Nebreda 2009). It can serve as a negative regulator of cell cycle progression and can facilitate induction of apoptosis, indicating that it may function as a tumor suppressor. However, its involvement in invasion, inflammation, and angiogenesis suggests that it may also have oncogenic functions. In response to DNA damage, activation of p38α can mediate apoptosis or induce a G2/M cell cycle checkpoint through p53-dependent and p53-independent mechanisms (Han and Sun 2007). Selective nuclear accumulation of p38α in response to DNA damage has been suggested to increase the phosphorylation of p38α nuclear targets (Wood et al. 2009). Additionally, it has been reported that TAB1 modulates intracellular localization of p38α and its downstream signaling (Lu et al. 2006). Our observations further support the notion that p38α actively participates in the DNA damage-induced cellular response. However, inhibiting p38α catalytic activity by SB203580 reduces but does not completely abolish the inhibitory effect of TAB1 on p53 activation, implying that TAB1 works through additional mechanisms to regulate p53 function.
Cisplatin is distinct among genotoxic agents in requiring TAB1 for activation of p53-mediated apoptosis
It is remarkable that cisplatin treatment-associated cell death appears to be unique among the several stress-inducing agents that we tested in being mitigated by knockdown of TAB1. At this stage, we can only speculate on scenarios that might explain this finding. First, two distinct TAB1-regulated mechanisms (p38α activation and stabilized MDMX levels) are required for p53 to produce apoptosis in cisplatin-treated cells. Genotoxic agents such as ETP that activate p38α (Kurosu et al. 2005) may not be able to mediate MDMX-facilitated p53-intrinsic apoptotic events through TAB1 because they induce MDMX degradation. On the other hand, while MDMX is more resistant to degradation by some agents, such as actinomycin D (Biderman et al. 2012), those agents may not be able to activate p38α. Second, cisplatin but not other treatments may lead to phosphorylation and activation of TAB1 by either p38α or an unidentified protein kinase such that it can prevent MDM2-mediated MDMX degradation. Third, cisplatin may be unique in inhibiting the abililty of the Wip1 phosphatase (a p53 target that negatively regulates p53, ATM, and p38α) to dephosphorylate and thereby inactivate p38α or TAB1. Fourth, cisplatin may be unique in promoting specific modifications on MDMX or MDM2, which affect their interactions with TAB1 and prevent degradation of MDMX, and/or facilitating its mitochondrial function. Finally, TAB1/TAK1 involvement in HIPK2 activation in response to Wnt-1 signaling (Kanei-Ishii et al. 2004) may facilitate the p53 response to cisplatin. HIPK2 has been shown to phosphorylate p53 at S46, and its function and stability are regulated by MDM2 depending on the dosage of the DNA damage agents (Rinaldo et al. 2007). It would be interesting to investigate the interplay between TAB1, MDM2, and HIPK2 in the cellular apoptotic response upon cisplatin treatment as well as determine whether cisplatin regulates relevant modifications in TAB1, MDM2, or MDMX.
NOXA is selectively impacted by TAB1 knockdown in cisplatin-treated cells
Our results suggest that in response to cisplatin, specific genes (such as NOXA) are activated to promote apoptosis. Furthermore, our results suggest that NOXA plays a role in apoptosis in cells treated with this drug. It was previously shown that ERK-dependent but p53-independent NOXA induction is critical to cisplatin-induced cell death in some cell types and that a MEK inhibitor (U0126) attenuates cisplatin-mediated apoptosis (Sheridan et al. 2010). When we tested this inhibitor, we found that it has little effect on cisplatin-induced cell death in U2OS cells, although it greatly attenuates cell death in SkHep1 cells (Supplemental Fig. S11). Thus, the involvement of the ERK signaling cascade in cisplatin-associated cytotoxicity is cell type-specific, and this pathway is not the major player in cisplatin-mediated cell death in U2OS cells. Further studies are needed to investigate how TAB1 selectively regulates p53 transactivation of NOXA. It is noteworthy that a p38α-regulated transcription coactivator, Hamlet (p18), stimulates p53-dependent apoptosis in response to certain types of stresses such as UV and cisplatin but not in response to γ-irradiation (Cuadrado et al. 2007). Also in that study, it was shown that down-regulation of Hamlet specifically prevents NOXA induction. It would be interesting to test whether TAB1 affects the levels or function of Hamlet in response to cisplatin treatment, which in turn would affect p53 transcription.
Cisplatin is a common and effective chemotherapeutic agent for some forms of cancer (Basu and Krishnamurthy 2010). That TAB1 levels are inversely correlated with cisplatin sensitivity (at least in A2780 ovarian cancer cell line-derived clones) suggests that loss of TAB1 may contribute to cisplatin resistance in some cases. The interplay between p53/MDM2/MDMX and TAB1 that we uncovered here may provide new insights into resistance to this drug that occurs in a significant number of cisplatin-treated patients, in particular those whose tumors harbor wild-type p53. It should be noted, however, that some tumors commonly treated with cisplatin, such as ovarian cancers, sustain a very high frequency of p53 mutation. While our analysis revealed significantly lower levels of TAB1 in ovarian tumors with wild-type p53 when compared with those with mutant p53, there was still reduced TAB1 in a subset of the latter tumors as well. This implies that either mutant p53 can be activated by TAB1 to produce cell death, which is unlikely, or TAB1 is playing additional p53-independent roles in cisplatin-treated cells. Further studies will hopefully clarify these points and provide more insight into the many roles of TAB1 in cells.
Materials and methods
Plasmids and cell culture
Flag-MDM2 (wild-type), HA-p53, Myc-MDMX, and HA-ubiquitin were described previously (Poyurovsky et al. 2003; Zhu et al. 2009). HA-MDMX was a gift from Dr. A. Jochemsen (Leiden University Medical Center, Netherlands). His-ubiquitin (pcBH2Ub) was kindly provided by Dr. R. Baer (Columbia University). Construction of a Myc-tagged TAB1 plasmid (Myc-TAB1) and an untagged TAK1 plasmid (pcDNA-TAK1) is described in the Supplemental Material. U2OS cells (osteosarcoma cells expressing wild-type p53), H1299 cells (p53-null lung epithelial carcinoma cells), HCT116 (p53+/+), and derivative HCT116 (p53−/−) cells (colon carcinoma cells; a kind gift from Dr. B. Vogelstein, Johns Hopkins University) were grown in Dulbecco modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS) at 37°C.
Antibodies and drugs
A complete list of the antibodies and drugs used in this study is provided in the Supplemental Material.
Transfection, Western blot, and cell cycle analysis
Transfection, Western blot, and cell cycle analysis were performed as previously described (Zhu et al. 2009). Detailed procedures are provided in the Supplemental Material.
Ubiquitination and degradation assays
In vivo ubiquitination assays and in vitro degradation assays were carried out as previously described (Zhu et al. 2009). Detailed protocols are provided in Supplemental Material.
siRNA interference
siRNA oligonucleotides (T1, J-004770-07 and T2, J-004770-08) targeting human TAB1 were purchased from Dharmacon. Reported siRNA oligonucleotides targeting luciferase (Urist et al. 2004), human TAK1 (Bertelsen and Sanfridson 2007), MDM2 (Jin et al. 2003), and NOXA (N1 [Sheridan et al. 2010]) were obtained from Invitrogen. Second siRNA oligonucleotides targeting NOXA (N2, D-005275-07) were purchased from Dharmacon. siRNA oligonucleotides targeting MDMX (X11 [Chen et al. 2005] and X4 [Hs_MDM4_2 FlexiTube siRNA]) were obtained from Qiagen. Transfection was performed using Dharmafect 1 (Dharmacon) according to the manufacturer's instructions.
RNA extraction and quantitative RT–PCR analysis
RNA was extracted using a Qiagen RNeasy minkit, and cDNA was synthesized with the QuantiTect reverse transcription kit (Qiagen). Samples were analyzed by quantitative real-time PCR on an Applied Biosystems Step One Plus instrument using the SYBR Green dye (Applied Biosystems). RNA expression was normalized to RPL32 mRNA expression. Relative levels were calculated by the comparative Ct method (ΔΔCT method). Graphs are representative of multiple independent experiments, with error bars representing technical PCR replicates. Primer sequences are available on request.
Colony formation assay
U2OS cells (8 × 104) transfected with either control or TAB1 siRNAs were seeded in a 35-mm dish. Twenty-four hours later, cells were treated with cisplatin for 24 h. Cells were then washed with PBS three times and incubated in fresh DMEM medium with 10% FBS. Six days later, cells were fixed and subjected to cystal violet staining. Stained dishes were photographed, and the number of colonies formed in each dish was manually scored using a 1-cm × 1-cm grid system and graphed. A more detailed protocol is provided in the Supplemental Material.
Gene expression analysis
Microarray gene expression data from a GEO publicly available data set (GSE33482) and TCGA human ovarian serous cystadenocarcinoma data matrix (Agilent G4502A, N = 589) were obtained for determining cellular levels of TAB1 in clonally derived cisplatin-resistant A2780 ovarian cancer cell lines (A2780-cis) and their cisplatin-sensitive counterparts (A2780) and comparing TAB1 expression between tumor samples with normal ovarian tissue samples, respectively. The somatic mutation data sets for gene expression of the ovarian serous cystadenocarcinoma tumor samples in the TCGA data matrix were downloaded from the Broad Genome Data Analysis Center (GDAC) Firehose Analytics Platform (https://confluence.broadinstitute.org/display/GDAC/Home) and analyzed to stratify tumor samples on the basis of p53 mutation status (wild-type or missense mutations), and relative TAB1 expression was analyzed. A detailed description of the statistical analysis of these data sets is provided in the Supplemental Material.
Acknowledgments
We thank Ella Freulich for expert technical assistance, and members of the Prives' laboratory for helpful suggestions. This work was supported by grant CA58316 from the NIH.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.212258.112.
References
- Basu A, Krishnamurthy S 2010. Cellular responses to cisplatin-induced DNA damage. J Nucleic Acids doi: 10.4061/2010/201367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertelsen M, Sanfridson A 2007. TAB1 modulates IL-1α mediated cytokine secretion but is dispensable for TAK1 activation. Cell Signal 19: 646–657 [DOI] [PubMed] [Google Scholar]
- Bettermann K, Vucur M, Haybaeck J, Koppe C, Janssen J, Heymann F, Weber A, Weiskirchen R, Liedtke C, Gassler N, et al. 2010. TAK1 suppresses a NEMO-dependent but NF-κB-independent pathway to liver cancer. Cancer Cell 17: 481–496 [DOI] [PubMed] [Google Scholar]
- Biderman L, Poyurovsky MV, Assia Y, Manley JL, Prives C 2012. MdmX is required for p53 interaction with and full induction of the Mdm2 promoter after cellular stress. Mol Cell Biol 32: 1214–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragado P, Armesilla A, Silva A, Porras A 2007. Apoptosis by cisplatin requires p53 mediated p38α MAPK activation through ROS generation. Apoptosis 12: 1733–1742 [DOI] [PubMed] [Google Scholar]
- Bulavin DV, Saito S, Hollander MC, Sakaguchi K, Anderson CW, Appella E, Fornace AJ Jr 1999. Phosphorylation of human p53 by p38 kinase coordinates N-terminal phosphorylation and apoptosis in response to UV radiation. EMBO J 18: 6845–6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang MY, Ho FM, Wang JS, Kang HC, Chang Y, Ye ZX, Lin WW 2010. AICAR induces cyclooxygenase-2 expression through AMP-activated protein kinase-transforming growth factor-β-activated kinase 1–p38 mitogen-activated protein kinase signaling pathway. Biochem Pharmacol 80: 1210–1220 [DOI] [PubMed] [Google Scholar]
- Chen J 2012. The roles of MDM2 and MDMX phosphorylation in stress signaling to p53. Genes Cancer 3: 274–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Gilkes DM, Pan Y, Lane WS, Chen J 2005. ATM and Chk2-dependent phosphorylation of MDMX contribute to p53 activation after DNA damage. EMBO J 24: 3411–3422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner SH, Kular G, Peggie M, Shepherd S, Schuttelkopf AW, Cohen P, Van Aalten DM 2006. TAK1-binding protein 1 is a pseudophosphatase. Biochem J 399: 427–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadrado A, Lafarga V, Cheung PC, Dolado I, Llanos S, Cohen P, Nebreda AR 2007. A new p38 MAP kinase-regulated transcriptional coactivator that stimulates p53-dependent apoptosis. EMBO J 26: 2115–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuenda A, Rousseau S 2007. p38 MAP-kinases pathway regulation, function and role in human diseases. Biochim Biophys Acta 1773: 1358–1375 [DOI] [PubMed] [Google Scholar]
- Dhillon AS, Hagan S, Rath O, Kolch W 2007. MAP kinase signalling pathways in cancer. Oncogene 26: 3279–3290 [DOI] [PubMed] [Google Scholar]
- D'Orazi G, Cecchinelli B, Bruno T, Manni I, Higashimoto Y, Saito S, Gostissa M, Coen S, Marchetti A, Del Sal G, et al. 2002. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat Cell Biol 4: 11–19 [DOI] [PubMed] [Google Scholar]
- Gartel AL, Tyner AL 2002. The role of the cyclin-dependent kinase inhibitor p21 in apoptosis. Mol Cancer Ther 1: 639–649 [PubMed] [Google Scholar]
- Ge B, Gram H, Di Padova F, Huang B, New L, Ulevitch RJ, Luo Y, Han J 2002. MAPKK-independent activation of p38α mediated by TAB1-dependent autophosphorylation of p38α. Science 295: 1291–1294 [DOI] [PubMed] [Google Scholar]
- Gilkes DM, Pan Y, Coppola D, Yeatman T, Reuther GW, Chen J 2008. Regulation of MDMX expression by mitogenic signaling. Mol Cell Biol 28: 1999–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Sun P 2007. The pathways to tumor suppression via route p38. Trends Biochem Sci 32: 364–371 [DOI] [PubMed] [Google Scholar]
- Herrero-Martin G, Hoyer-Hansen M, Garcia-Garcia C, Fumarola C, Farkas T, Lopez-Rivas A, Jaattela M 2009. TAK1 activates AMPK-dependent cytoprotective autophagy in TRAIL-treated epithelial cells. EMBO J 28: 677–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki M, Komatsu Y, Scott G, Yamada G, Ray M, Ninomiya-Tsuji J, Mishina Y 2008. Generation of a conditional mutant allele for Tab1 in mouse. Genesis 46: 431–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inokuchi S, Aoyama T, Miura K, Osterreicher CH, Kodama Y, Miyai K, Akira S, Brenner DA, Seki E 2010. Disruption of TAK1 in hepatocytes causes hepatic injury, inflammation, fibrosis, and carcinogenesis. Proc Natl Acad Sci 107: 844–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicke RU, Sohn D, Schulze-Osthoff K 2008. The dark side of a tumor suppressor: Anti-apoptotic p53. Cell Death Differ 15: 959–976 [DOI] [PubMed] [Google Scholar]
- Jin Y, Lee H, Zeng SX, Dai MS, Lu H 2003. MDM2 promotes p21waf1/cip1 proteasomal turnover independently of ubiquitylation. EMBO J 22: 6365–6377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Dai MS, Lu SZ, Xu Y, Luo Z, Zhao Y, Lu H 2006. 14-3-3γ binds to MDMX that is phosphorylated by UV-activated Chk1, resulting in p53 activation. EMBO J 25: 1207–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson G 2002. Signal transduction. Scaffolding proteins—more than meets the eye. Science 295: 1249–1250 [DOI] [PubMed] [Google Scholar]
- Kanei-Ishii C, Ninomiya-Tsuji J, Tanikawa J, Nomura T, Ishitani T, Kishida S, Kokura K, Kurahashi T, Ichikawa-Iwata E, Kim Y, et al. 2004. Wnt-1 signal induces phosphorylation and degradation of c-Myb protein via TAK1, HIPK2, and NLK. Genes Dev 18: 816–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai H, Wiederschain D, Kitao H, Stuart J, Tsai KK, Yuan ZM 2003. DNA damage-induced MDMX degradation is mediated by MDM2. J Biol Chem 278: 45946–45953 [DOI] [PubMed] [Google Scholar]
- Kelland L 2007. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer 7: 573–584 [DOI] [PubMed] [Google Scholar]
- Kim SJ, Hwang SG, Shin DY, Kang SS, Chun JS 2002. p38 kinase regulates nitric oxide-induced apoptosis of articular chondrocytes by accumulating p53 via NFκB-dependent transcription and stabilization by serine 15 phosphorylation. J Biol Chem 277: 33501–33508 [DOI] [PubMed] [Google Scholar]
- Komatsu Y, Shibuya H, Takeda N, Ninomiya-Tsuji J, Yasui T, Miyado K, Sekimoto T, Ueno N, Matsumoto K, Yamada G 2002. Targeted disruption of the Tab1 gene causes embryonic lethality and defects in cardiovascular and lung morphogenesis. Mech Dev 119: 239–249 [DOI] [PubMed] [Google Scholar]
- Konishi N, Nakamura M, Kishi M, Ishida E, Shimada K, Matsuyoshi S, Nagai H, Emi M 2003. Genetic mapping of allelic loss on chromosome 6q within heterogeneous prostate carcinoma. Cancer Sci 94: 764–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse JP, Gu W 2009. Modes of p53 regulation. Cell 137: 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosu T, Takahashi Y, Fukuda T, Koyama T, Miki T, Miura O 2005. p38 MAP kinase plays a role in G2 checkpoint activation and inhibits apoptosis of human B cell lymphoma cells treated with etoposide. Apoptosis 10: 1111–1120 [DOI] [PubMed] [Google Scholar]
- Landstrom M 2010. The TAK1–TRAF6 signalling pathway. Int J Biochem Cell Biol 42: 585–589 [DOI] [PubMed] [Google Scholar]
- Li J, Miller EJ, Ninomiya-Tsuji J, Russell RR 3rd, Young LH 2005. AMP-activated protein kinase activates p38 mitogen-activated protein kinase by increasing recruitment of p38 MAPK to TAB1 in the ischemic heart. Circ Res 97: 872–879 [DOI] [PubMed] [Google Scholar]
- Lu G, Kang YJ, Han J, Herschman HR, Stefani E, Wang Y 2006. TAB-1 modulates intracellular localization of p38 MAP kinase and downstream signaling. J Biol Chem 281: 6087–6095 [DOI] [PubMed] [Google Scholar]
- Lu M, Lin SC, Huang Y, Kang YJ, Rich R, Lo YC, Myszka D, Han J, Wu H 2007a. XIAP induces NF-κB activation via the BIR1/TAB1 interaction and BIR1 dimerization. Mol Cell 26: 689–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Ma O, Nguyen TA, Jones SN, Oren M, Donehower LA 2007b. The Wip1 Phosphatase acts as a gatekeeper in the p53-Mdm2 autoregulatory loop. Cancer Cell 12: 342–354 [DOI] [PubMed] [Google Scholar]
- Malmlof M, Roudier E, Hogberg J, Stenius U 2007. MEK–ERK-mediated phosphorylation of Mdm2 at Ser-166 in hepatocytes. Mdm2 is activated in response to inhibited Akt signaling. J Biol Chem 282: 2288–2296 [DOI] [PubMed] [Google Scholar]
- Mancini F, Moretti F 2009. Mitochondrial MDM4 (MDMX): An unpredicted role in the p53-mediated intrinsic apoptotic pathway. Cell Cycle 8: 3854–3859 [DOI] [PubMed] [Google Scholar]
- Mancini F, Gentiletti F, D'Angelo M, Giglio S, Nanni S, D'Angelo C, Farsetti A, Citro G, Sacchi A, Pontecorvi A, et al. 2004. MDM4 (MDMX) overexpression enhances stabilization of stress-induced p53 and promotes apoptosis. J Biol Chem 279: 8169–8180 [DOI] [PubMed] [Google Scholar]
- Mancini F, Di Conza G, Pellegrino M, Rinaldo C, Prodosmo A, Giglio S, D'Agnano I, Florenzano F, Felicioni L, Buttitta F, et al. 2009. MDM4 (MDMX) localizes at the mitochondria and facilitates the p53-mediated intrinsic-apoptotic pathway. EMBO J 28: 1926–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marine JC, Lozano G 2010. Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Differ 17: 93–102 [DOI] [PubMed] [Google Scholar]
- Marine JC, Francoz S, Maetens M, Wahl G, Toledo F, Lozano G 2006. Keeping p53 in check: Essential and synergistic functions of Mdm2 and Mdm4. Cell Death Differ 13: 927–934 [DOI] [PubMed] [Google Scholar]
- Marti A, Wirbelauer C, Scheffner M, Krek W 1999. Interaction between ubiquitin–protein ligase SCFSKP2 and E2F-1 underlies the regulation of E2F-1 degradation. Nat Cell Biol 1: 14–19 [DOI] [PubMed] [Google Scholar]
- Mayo LD, Seo YR, Jackson MW, Smith ML, Rivera Guzman J, Korgaonkar CK, Donner DB 2005. Phosphorylation of human p53 at serine 46 determines promoter selection and whether apoptosis is attenuated or amplified. J Biol Chem 280: 25953–25959 [DOI] [PubMed] [Google Scholar]
- Melisi D, Xia Q, Paradiso G, Ling J, Moccia T, Carbone C, Budillon A, Abbruzzese JL, Chiao PJ 2011. Modulation of pancreatic cancer chemoresistance by inhibition of TAK1. J Natl Cancer Inst 103: 1190–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza H, Campbell DG, Burness K, Hastie J, Ronkina N, Shim JH, Arthur JS, Davis RJ, Gaestel M, Johnson GL, et al. 2008. Roles for TAB1 in regulating the IL-1-dependent phosphorylation of the TAB3 regulatory subunit and activity of the TAK1 complex. Biochem J 409: 711–722 [DOI] [PubMed] [Google Scholar]
- Migliorini D, Lazzerini Denchi E, Danovi D, Jochemsen A, Capillo M, Gobbi A, Helin K, Pelicci PG, Marine JC 2002. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol Cell Biol 22: 5527–5538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momcilovic M, Hong SP, Carlson M 2006. Mammalian TAK1 activates Snf1 protein kinase in yeast and phosphorylates AMP-activated protein kinase in vitro. J Biol Chem 281: 25336–25343 [DOI] [PubMed] [Google Scholar]
- Neil JR, Schiemann WP 2008. Altered TAB1:IκB kinase interaction promotes transforming growth factor β-mediated nuclear factor-κB activation during breast cancer progression. Cancer Res 68: 1462–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninomiya-Tsuji J, Kajino T, Ono K, Ohtomo T, Matsumoto M, Shiina M, Mihara M, Tsuchiya M, Matsumoto K 2003. A resorcylic acid lactone, 5Z-7-oxozeaenol, prevents inflammation by inhibiting the catalytic activity of TAK1 MAPK kinase kinase. J Biol Chem 278: 18485–18490 [DOI] [PubMed] [Google Scholar]
- Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T, Nishimori H, Tamai K, Tokino T, Nakamura Y, et al. 2000. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 102: 849–862 [DOI] [PubMed] [Google Scholar]
- Okamoto K, Kashima K, Pereg Y, Ishida M, Yamazaki S, Nota A, Teunisse A, Migliorini D, Kitabayashi I, Marine JC, et al. 2005. DNA damage-induced phosphorylation of MdmX at serine 367 activates p53 by targeting MdmX for Mdm2-dependent degradation. Mol Cell Biol 25: 9608–9620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Ohtomo T, Sato S, Sugamata Y, Suzuki M, Hisamoto N, Ninomiya-Tsuji J, Tsuchiya M, Matsumoto K 2001. An evolutionarily conserved motif in the TAB1 C-terminal region is necessary for interaction with and activation of TAK1 MAPKKK. J Biol Chem 276: 24396–24400 [DOI] [PubMed] [Google Scholar]
- Parant J, Chavez-Reyes A, Little NA, Yan W, Reinke V, Jochemsen AG, Lozano G 2001. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet 29: 92–95 [DOI] [PubMed] [Google Scholar]
- Pereg Y, Shkedy D, de Graaf P, Meulmeester E, Edelson-Averbukh M, Salek M, Biton S, Teunisse AF, Lehmann WD, Jochemsen AG, et al. 2005. Phosphorylation of Hdmx mediates its Hdm2- and ATM-dependent degradation in response to DNA damage. Proc Natl Acad Sci 102: 5056–5061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyurovsky MV, Jacq X, Ma C, Karni-Schmidt O, Parker PJ, Chalfie M, Manley JL, Prives C 2003. Nucleotide binding by the Mdm2 RING domain facilitates Arf-independent Mdm2 nucleolar localization. Mol Cell 12: 875–887 [DOI] [PubMed] [Google Scholar]
- Prickett TD, Ninomiya-Tsuji J, Broglie P, Muratore-Schroeder TL, Shabanowitz J, Hunt DF, Brautigan DL 2008. TAB4 stimulates TAK1–TAB1 phosphorylation and binds polyubiquitin to direct signaling to NF-κB. J Biol Chem 283: 19245–19254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldo C, Prodosmo A, Mancini F, Iacovelli S, Sacchi A, Moretti F, Soddu S 2007. MDM2-regulated degradation of HIPK2 prevents p53Ser46 phosphorylation and DNA damage-induced apoptosis. Mol Cell 25: 739–750 [DOI] [PubMed] [Google Scholar]
- Shadfan M, Lopez-Pajares V, Yuan ZM 2012. MDM2 and MDMX: Alone and together in regulation of p53. Transl Cancer Res 1: 88–89 [PMC free article] [PubMed] [Google Scholar]
- Sheridan C, Brumatti G, Elgendy M, Brunet M, Martin SJ 2010. An ERK-dependent pathway to Noxa expression regulates apoptosis by platinum-based chemotherapeutic drugs. Oncogene 29: 6428–6441 [DOI] [PubMed] [Google Scholar]
- Shibuya H, Yamaguchi K, Shirakabe K, Tonegawa A, Gotoh Y, Ueno N, Irie K, Nishida E, Matsumoto K 1996. TAB1: An activator of the TAK1 MAPKKK in TGF-β signal transduction. Science 272: 1179–1182 [DOI] [PubMed] [Google Scholar]
- Shim JH, Xiao C, Paschal AE, Bailey ST, Rao P, Hayden MS, Lee KY, Bussey C, Steckel M, Tanaka N, et al. 2005. TAK1, but not TAB1 or TAB2, plays an essential role in multiple signaling pathways in vivo. Genes Dev 19: 2668–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur N, Sorrentino A, Heldin CH, Landstrom M 2009. TGF-β uses the E3-ligase TRAF6 to turn on the kinase TAK1 to kill prostate cancer cells. Future Oncol 5: 1–3 [DOI] [PubMed] [Google Scholar]
- Urist M, Tanaka T, Poyurovsky MV, Prives C 2004. p73 induction after DNA damage is regulated by checkpoint kinases Chk1 and Chk2. Genes Dev 18: 3041–3054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Prives C 2009. Blinded by the light: The growing complexity of p53. Cell 137: 413–431 [DOI] [PubMed] [Google Scholar]
- Wade M, Wang YV, Wahl GM 2010. The p53 orchestra: Mdm2 and Mdmx set the tone. Trends Cell Biol 20: 299–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EF, Nebreda AR 2009. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer 9: 537–549 [DOI] [PubMed] [Google Scholar]
- Waning DL, Lehman JA, Batuello CN, Mayo LD 2010. Controlling the Mdm2–Mdmx–p53 circuit. Pharmaceuticals (Basel) 3: 1576–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood CD, Thornton TM, Sabio G, Davis RA, Rincon M 2009. Nuclear localization of p38 MAPK in response to DNA damage. Int J Biol Sci 5: 428–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GS 2004. The functional interactions between the p53 and MAPK signaling pathways. Cancer Biol Ther 3: 156–161 [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Shirakabe K, Shibuya H, Irie K, Oishi I, Ueno N, Taniguchi T, Nishida E, Matsumoto K 1995. Identification of a member of the MAPKKK family as a potential mediator of TGF-β signal transduction. Science 270: 2008–2011 [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Nagai S, Ninomiya-Tsuji J, Nishita M, Tamai K, Irie K, Ueno N, Nishida E, Shibuya H, Matsumoto K 1999. XIAP, a cellular member of the inhibitor of apoptosis protein family, links the receptors to TAB1–TAK1 in the BMP signaling pathway. EMBO J 18: 179–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Lin L, Guo H, Yang J, Jones SN, Jochemsen A, Lu X 2009. Phosphorylation and degradation of MdmX is inhibited by Wip1 phosphatase in the DNA damage response. Cancer Res 69: 7960–7968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Poyurovsky MV, Li Y, Biderman L, Stahl J, Jacq X, Prives C 2009. Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol Cell 35: 316–326 [DOI] [PMC free article] [PubMed] [Google Scholar]