Poly-ADP-ribosylation post-translational modifications play important roles in many biological processes, including the DNA damage response. Yu and colleagues now identify a set of Forkhead-associated (FHA) and BRCA1 C-terminal (BRCT) domains that recognize poly(ADP-ribose) (PAR). They show that the interaction between PAR and the BRCT domain of the Nijmegan breakage syndrome protein NBS1 is important for the activation of ATM in response to DNA damage. This study thus demonstrates how two novel PAR-binding modules mediate the DNA damage response.
Keywords: poly(ADP-ribose), PAR binding domain, DNA damage
Abstract
Poly-ADP-ribosylation is a unique post-translational modification participating in many biological processes, such as DNA damage response. Here, we demonstrate that a set of Forkhead-associated (FHA) and BRCA1 C-terminal (BRCT) domains recognizes poly(ADP-ribose) (PAR) both in vitro and in vivo. Among these FHA and BRCT domains, the FHA domains of APTX and PNKP interact with iso-ADP-ribose, the linkage of PAR, whereas the BRCT domains of Ligase4, XRCC1, and NBS1 recognize ADP-ribose, the basic unit of PAR. The interactions between PAR and the FHA or BRCT domains mediate the relocation of these domain-containing proteins to DNA damage sites and facilitate the DNA damage response. Moreover, the interaction between PAR and the NBS1 BRCT domain is important for the early activation of ATM during DNA damage response and ATM-dependent cell cycle checkpoint activation. Taken together, our results demonstrate two novel PAR-binding modules that play important roles in DNA damage response.
Post-translational modifications, such as protein phosphorylation, ubiquitination, acetylation, and methylation, are important for numerous biological processes. Protein ADP-ribosylation is a unique post-translational modification that has been shown to play critical roles in many cellular events, especially DNA damage response (Schreiber et al. 2006; Gibson and Kraus 2012).
Poly-ADP-ribosylation is catalyzed by poly-ADP-ribosylation polymerases (PARPs) (Ame et al. 2004; Hottiger et al. 2010). Using NAD+ as the donor, ADP-ribose is covalently linked to the side chains of arginine, aspartic acid, and glutamic acid residues in target proteins. After catalyzing the first ADP-ribose on the target, other ADP-riboses can be covalently linked onto the first ADP-ribose to form both liner and branched polymers, known as poly(ADP-ribose) (PAR) (Schreiber et al. 2006; Luo and Kraus 2012). Following DNA damage, massive poly-ADP-ribosylation is synthesized at DNA lesions within a few seconds (D'Amours et al. 1999; Kim et al. 2005). To date, two major nuclear substrates of poly-ADP-ribosylation are known: the PARP1 enzyme itself and histones (Schreiber et al. 2006; Messner and Hottiger 2011). Once PAR is synthesized at the DNA damage site, it is also quickly recognized and hydrolyzed by PAR glycosylase (PARG) (D'Amours et al. 1999; Gagne et al. 2006; Kim et al. 2007a). Thus, the half-life of PAR at DNA damage sites is very short, and the biological function of this dynamic post-translation modification at DNA damage sites remains elusive.
Recently, several PAR-binding proteins have been identified as the “readers” to recognize PAR signals (Karras et al. 2005; Ahel et al. 2008; Wang et al. 2012), suggesting that PAR is likely to function as recruiting signals to induce DNA damage response factors to DNA damage sites. To identify other PAR-binding modules, we examined both the Forkhead-associated (FHA) domain and the BRCA1 C-terminal (BRCT) domain. The FHA and BRCT domains are known as phospho-protein-binding domains. Many FHA domain- or BRCT domain-containing proteins are involved in DNA damage response (Li et al. 2002; Glover et al. 2004; Mahajan et al. 2008; Mohammad and Yaffe 2009). It has been shown that the FHA domains recognize phospho-Thr (pThr) motifs (Sun et al. 1998; Durocher et al. 1999, 2000; Li et al. 2002; Mahajan et al. 2008). For example, the FHA domain of Rad53 recognizes the pThr motif of Rad9 in budding yeast (Sun et al. 1998), the FHA domain of fission yeast NBS1 recognizes the pThr of Ctp1(Williams et al. 2009), and the FHA domain of human NBS1 and RNF8 recognizes the pThr motifs of MDC1 (Chapman and Jackson 2008; Spycher et al. 2008; Wu et al. 2008; Lloyd et al. 2009). While the BRCT domains have been shown to recognize phospho-Ser (pSer) motifs (Manke et al. 2003; Yu et al. 2003), the BRCA1 BRCT domain binds pSer motifs in several downstream partners (Yu et al. 2003; Yu and Chen 2004; Kim et al. 2007b; Liu et al. 2007; Wang et al. 2007). The MDC1 BRCT domain recognizes the pS139 site of H2AX (Stucki et al. 2005). These phospho-protein-dependent interactions are important for DNA damage checkpoint activation and DNA damage repair. However, based on the peptide screening, not all of the FHA and BRCT domains have high affinity to phospho-proteins (Durocher et al. 2000; Rodriguez et al. 2003). In particularly, our recent study suggests that the BARD1 BRCT domain recognizes ADP-ribose (Baer 2013; Li and Yu 2013). Thus, we asked whether these domains have other binding partners besides phospho-proteins.
Following the DNA damage, PAR is quickly synthesized at the DNA damage sites. Interestingly, one ADP-ribose residue contains two phosphate groups. Thus, massive PAR synthesized upon DNA damage brings huge amounts of phosphate moieties at DNA damage sites in a very short period. We therefore wondered whether the FHA and BRCT domains could recognize PAR. In this study, we screened 19 FHA and BRCT domains and found that five of them bind PAR both in vitro and in vivo. Moreover, the interaction with PAR facilitates the fast recruitment of these FHA or BRCT domain-containing proteins to DNA lesions and the relevant DNA damage repair process.
Results
A set of BRCT and FHA domains binds PAR
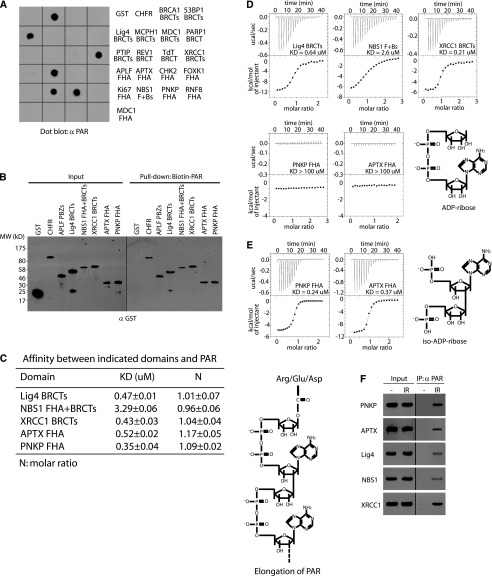
Since CHFR and APLF are known PAR-binding proteins, which bind PAR via their PBZ motifs (Ahel et al. 2008; Li et al. 2010; Oberoi et al. 2010), we used GST, recombinant CHFR, and APLF PBZ motifs as negative and positive controls, respectively, to screen PAR-binding domains. We examined 19 FHA or BRCT domains and found that two FHA domains (from PNKP and APTX), two BRCT domains (from Ligase4 and XRCC1), and an FHA–BRCT fusion domain (from NBS1) interacted with PAR (Fig. 1A). Reverse pull-down assays confirmed the direct interaction between PAR and these FHA/BRCT domains (Fig. 1B). Using isothermal titration calorimetry (ITC) assays, we measured the affinity between PAR and these FHA/BRCT domains (Fig. 1C), which is in the physiologically relevant range and is similar to that between PAR and other PAR-binding domains (Karras et al. 2005; Wang et al. 2012). Since PAR is the ADP-ribose polymer with mixed length and contains both linear and branched forms, the affinity between the PAR-binding domain and PAR could not be accurately measured. We then measured the affinity between these FHA/BRCT domains and ADP-ribose, the basic unit of PAR. The ITC results show that the BRCT domains of Ligase4 and XRCC1 and the FHA–BRCT fusion domain of NBS1 recognize ADP-ribose. However, the FHA domains of PNKP and APTX do not interact with ADP-ribose (Fig. 1D). Since the FHA domains of PNKP and APTX recognize PAR, we wonder whether these FHA domains interact with the linkage between each ADP-ribose in PAR. We used phosphodiesterase to digest PAR into iso-ADP-ribose, the linkage between two individual ADP-riboses, and found that the FHA domains of PNKP and APTX have high affinity with iso-ADP-ribose (Fig. 1E). Following DNA damage, PAR is heavily synthesized at DNA lesions (D'Amours et al. 1999; Kim et al. 2005). It has been reported that PNKP, APTX, Ligase4, XRCC1, and NBS1 all participate in DNA damage response (Su 2006; Polo and Jackson 2011). We examined the in vivo interactions between PAR and PNKP, APTX, Ligase4, XRCC1, or NBS1. With ionizing radiation (IR) treatment, PAR was significantly synthesized in the cells and interacted with PNKP, APTX, Ligase4, XRCC1, or NBS1 (Fig. 1F). Taken together, these results demonstrate that a set of FHA/BRCT domains interacts with PAR.
Figure 1.
The BRCT and FHA domains bind PAR. (A) PAR-binding screening among the BRCT and FHA domains. The interactions between PAR and GST (negative control), GST-CHFR (positive control), or the indicated domains was examined by dot blot using anti-PAR antibody. (B) The interactions between biotin-PAR and recombinant proteins were examined by pull-down with Streptavidin beads and blotted with anti-GST antibody. (C) The affinity between PAR and the recombinant proteins was measured by ITC. (D) The affinity between ADP-ribose and the recombinant proteins in B was measured by ITC. (E) The affinity between iso-ADP-ribose and the recombinant PNKP FHA or APTX FHA was measured by ITC. Titration of ligand (PAR, ADP-ribose, or iso-ADP-ribose) into a solution containing the purified protein was performed. The fit of the data to an equilibrium-binding isotherm is shown. (C–E) The fit provides an equilibrium dissociation constant (KD) for the binding of the ligand to the protein. (F) The in vivo interaction between PAR and PNKP, APTX, Ligase4, NBS1, or XRCC1 was measured by coimmunoprecipitation (co-IP) with the indicated antibodies. Whole-cell lysates were blotted and are shown as the input.
PAR-binding pockets are conserved in the BRCT and FHA domains
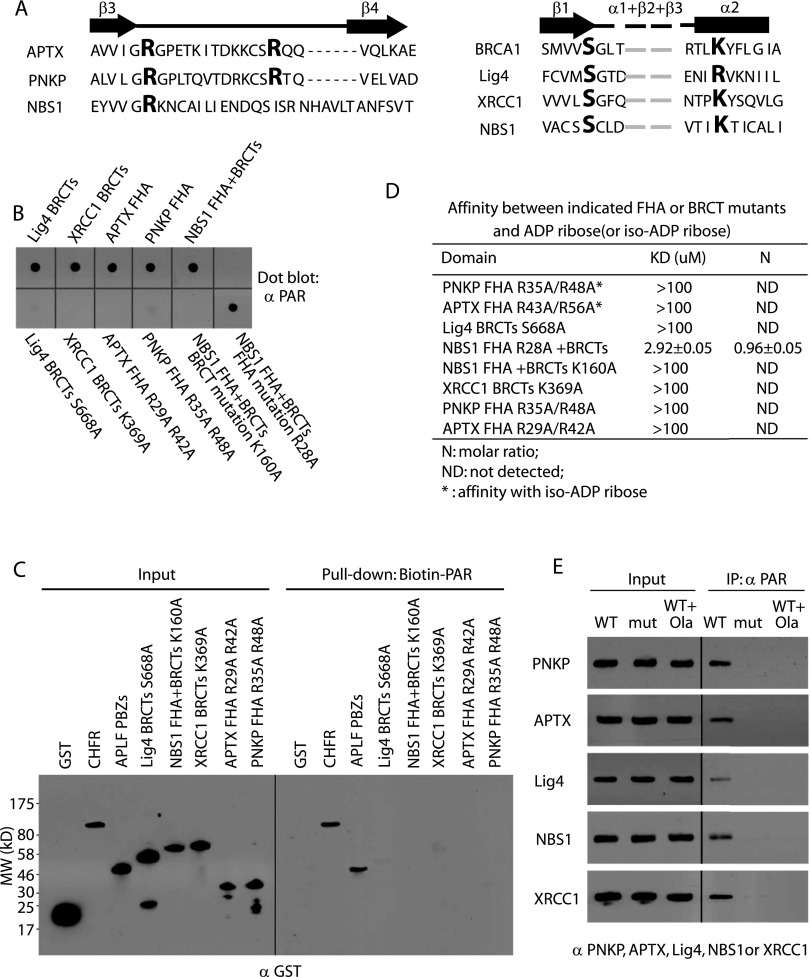
Next, we examined the PAR-binding sites in these FHA/BRCT domains. Different FHA or BRCT domains are predicted to fold into similar secondary structures, respectively, with binding pockets that can recognize pThr or pSer motifs (Fig. 2A; Glover et al. 2004; Mahajan et al. 2008; Mohammad and Yaffe 2009). We asked whether the similar binding pockets recognize iso-ADP-ribose and ADP-ribose, respectively. For the FHA domains of PNKP and APTX, two conserved arginine residues in each binding pocket were mutated into alanines. Both mutants abolished the interaction with PAR in the pull-down and reciprocal pull-down assays (Fig. 2B,C). Moreover, we did not detect any affinity between the mutant FHA domains and iso-ADP-ribose using ITC assays (Fig. 2D). Following IR treatment, the FHA domain mutations abolished the interaction between PAR and PNKP or APTX in vivo (Fig. 2E). Single mutation in the binding pocket of the BRCA1 BRCT domain abolished the interaction between the pSer motif and the BRCA1 BRCT domain (Shiozaki et al. 2004). Based on the similarity of the secondary structure in the BRCT domain, we mutated the conserved Ser or Lys residues to disrupt the putative binding pocket in the BRCT domain of Ligase4 and XRCC1 (Fig. 2A). Like those FHA domain mutants, these BRCT domain mutants abolished the interaction with PAR in vitro (Fig. 2B and C). The full-length proteins bearing these BRCT domain mutations failed to interact with PAR in vivo (Fig. 2E). In addition to these FHA domains and BRCT domains, NBS1 has a FHA domain and a BRCT domain that are fused together. This FHA–BRCT domain also recognizes PAR. To study which subdomain of this FHA–BRCT fusion domain recognizes ADP-ribose, we mutated the conserved residues in each binding pocket. As shown in Figure 2, B and D, mutation of conserved Lys residue in the FHA fold did not significantly affect the interaction between the FHA–BRCT fusion domain and PAR or ADP-ribose, whereas mutation in the BRCT fold abolished the interaction (Fig. 2B,D), suggesting that the BRCT fold in NBS1 mediates the interaction with PAR. This result is further confirmed by the in vivo coimmunoprecipitation (co-IP) assay (Fig. 2E). Moreover, when cells were treated with olaparib to suppress the DNA damage-induced PAR synthesis, these FHA/BRCT domain-containing proteins no longer interacted with PAR in vivo (Fig. 2E).
Figure 2.
PAR-binding pockets in the FHA and BRCT domains. (A) Based on the secondary structure, amino acid sequences of FHA (left) and BRCT (right) are compared to show the conserved residues (similarity to the R28 of the NBS1 FHA domain and the S1655/K1702 of the BRCA1 BRCT domain) that form the potential PAR-binding pocket. The β strand is shown as arrow, and the α helix is shown as box. Conserved residues are marked in red. (B) Mutations of conserved residues (Lig4 BRCT domain S668A, XRCC1 BRCT domain K369A, APTX FHA R29A/R42A, PNKP FHA R35A/R48A, and NBS1 FHA+BRCT domain K160A) in the potential binding pockets of the FHA and BRCT domains abolish the PAR binding. Dot blot was used to examine the interaction with anti-PAR antibody. The intact domains were used as the positive control. (C) The interactions between biotin-PAR and the indicated GST tagged FHA and BRCT domains were examined by pull-down with Streptavidin beads and blotting with anti-GST antibody. (D) The affinity between iso-ADP-ribose or ADP-ribose and the indicated mutant domains was measured by ITC. (E) The in vivo interaction between PAR and wild-type or mutants (Lig4 S668A, XRCC1 K369A, APTX R29A/R42A, PNKP R35A/R48A, and NBS1 K160A) was examined by co-IP with the indicated antibodies in the presence or absence of olaparib pretreatment. Whole-cell lysates were blotted and are shown as the input.
Computational analysis of the PAR-binding pockets in the FHA and BRCT domains
The binding between these FHA domains and iso-ADP-ribose was further analyzed by computational modeling. The structure of several FHA domains, including the FHA domains of PNKP, APTX, CHK2, and RNF8, has been solved (Li et al. 2002; Huen et al. 2007; Ali et al. 2009; Becherel et al. 2010). We examined the binding pockets of these FHA domains. The electrostatic potential of these four proteins, calculated by the MM-PBSA program, indicated that the peptide-binding sites are primarily positively charged that are complementary and primed for the recognizing negatively charged molecules such as phosphate moieties. The FHA domain of PNKP has been shown to recognize an adjacent pS–pT motif derived from XRCC1 (Whitehouse et al. 2001; Loizou et al. 2004; Ali et al. 2009). However, the affinity between the PNKP FHA domain and iso-ADP-ribose is even slightly higher than that between the PNKP FHA domain and the pS–pT peptide (Supplemental Fig. 1). Consistently, the binding pocket formed by the R35 and R48 of the PNKP FHA domain well accommodates iso-ADP-ribose, in which two phosphate groups mimic the two phosphate groups in the pS–pT peptide and form salt bridges with Arg residues (Supplemental Fig. 1A). The binding model was further validated by a rigorous binding free energy calculation; namely, the MM-PBSA method (Rastelli et al. 2010), which has been employed for estimating the relative binding free energy of protein with peptide and protein with small molecules (Kollman et al. 2000), including the BRCA1 with phospho-peptides (Anisimov et al. 2011) and viruses with ADP-ribose (Rungrotmongkol et al. 2010) reported recently. Of note, such calculations provide estimates of the relative binding free energy but not the absolute binding free energy of the protein with a ligand. Thus, the calculated binding free energies are ideal to be compared with experimental binding affinities of different ligands with the same protein. The binding free energy between the PNKP FHA domain and iso-ADP-ribose, calculated by the MM-PBSM method, is lower than that between the PNKP FHA domain and pS–pT peptide (Supplemental Fig. 1B), indicating a stronger binding affinity between the PNKP FHA domain and iso-ADP-ribose. These results are consistent with the binding affinities measured by ITC assays (Supplemental Fig. 1D). The binding pocket formed by R29 and R42 in the FHA domain of APTX is similar to that of PNKP (Supplemental Fig. 2A). It has been reported that the FHA domain of APTX recognizes the pS-D–pT-D motif of MDC1 (Becherel et al. 2010). The affinity between the FHA domain of ATPX and the pS-D–pT-D peptide is consistent with previous published results. However, the binding between the FHA domain of ATPX and iso-ADP-ribose is much stronger. Again, based on the structure of the FHA domain of APTX (Becherel et al. 2010), the analyses by the MM-PBSM method suggest that the binding free energy between the APTX FHA and iso-ADP-ribose is much lower than that between the APTX FHA domain and the pS-D–pT-D peptide, which is consistent with the binding affinity measured by ITC assays (Supplemental Fig. 2B–D). Moreover, the R44 in PNKP is replaced by the more flexible K38 in APTX. The mobility of the K38 in APTX may contribute to a less favorable binding to the phospho-peptide than iso-ADP-ribose. As indicated by the models in Supplemental Figure 2A, K38 in APTX moves away from the peptide, whereas it cooperates with R29 and R42 to interact with the phosphate group in iso-ADP-ribose. Moreover, the two phosphate groups in iso-ADP-ribose are on the surface of FHA domains in PNKP and APTX and can connect with additional units of poly-(ADP-ribose) in PAR (Supplemental Figs. 1A, 2A).
The structure of the FHA domain of RNF8 and CHK2 is quite different from that of PNKP and APTX (Li et al. 2002; Huen et al. 2007). Both FHA domains failed to interact with iso-ADP-ribose or PAR (Supplemental Fig. 3). The FHA domain of RNF8 recognizes the pT-Q motifs in MDC1 (Huen et al. 2007; Kolas et al. 2007; Mailand et al. 2007). The binding pocket in the FHA domain of RNF8 includes two hydrophobic residues (L57 and L82), creating a narrower and electro-neutral binding site that favors binding Glu residue in the peptide. Although the first phosphate group in iso-ADP-ribose could form salt bridge with R61 of RNF8, the second phosphate group does not mimic the Glu of the binding peptide, which can be attributed to the steric hindrance caused by L57 in RNF8 (Supplemental Fig. 3A). Similarly, only one phosphate group could be fitted into the binding pocket in the FHA domain of CHK2 (Supplemental Fig. 3A). The calculated binding free energy suggests that the FHA domains of RNF8 and CHK2 favor interacting with pT motifs over iso-ADP-ribose, which is also consistent with our ITC analyses (Supplemental Fig. 3B,C).
We also examined the binding pocket in the BRCT domains of Ligase4, XRCC1, BRCA1, and MDC1 because the structure of these BRCT domains has been solved (Zhang et al. 1998; Sibanda et al. 2001; Shiozaki et al. 2004; Stucki et al. 2005; Wu et al. 2009; Campbell et al. 2010; Cuneo et al. 2011). However, the binding pockets in the BRCT domains of BRCA1 and MDC1 are much larger compared with those in the FHA domain. Although one phosphate group in ADP-ribose can form salt bridges with K1936 and T1898 in MDC1 or with K1702 and S1655 in BRCA1, the pockets form few contacts with ribose surges and the other phosphate in ADP-ribose. Once the adenine of ADP-ribose is docked into the binding pockets, one ribose sugar could not be linked with other unit of PAR (Supplemental Fig. 4A). Moreover, the binding free energy suggests that both BRCT domains favor binding to their phospho-protein partners over ADP-ribose, which once again is consistent with our results from ITC assays (Supplemental Fig. 4B,C). The phospho-peptide-binding partners of the BRCT domains of Ligase4 and XRCC1 have not been identified. However, both BRCT domains recognize ADP-ribose. Thus, we could not compare their binding affinities with phospho-proteins as well as the binding models. Based on the structure of the unbound BRCT domains and mutation analyses in Supplemental Figure 5, we showed that the phosphate groups in ADP-ribose interact with K675 in Ligase4 and K369 in XRCC1. Based on the binding models, the ADP-ribose adopts an extended conformation to bind to the binding site, allowing two sugar groups to connect with the remaining units of PAR. Due to lacking the structure of the human NBS1 FHA–BRCT domain, we are currently unable to examine the details of the interaction between the NBS1 BRCT domain and ADP-ribose.
Collectively, the model analyses allow us to examine the details of the interaction between the FHA/BRCT domains and PAR, which might provide the molecular basis of the interactions.
PAR mediates the early recruitment of NBS1 during DNA damage response
Massive poly-ADP-ribosylation occurs at DNA lesions, suggesting that PAR could provide critical signals to recruit PAR-binding proteins to DNA lesions and allow these PAR-binding proteins to fulfill their DNA damage repair missions. To study the biological function of these novel PAR-binding domains, we used laser microirradiation to examine whether PAR recruits the FHA and BRCT domains to DNA damage sites. The advantage of the laser microirradiation approach is that we can monitor the early DNA damage response in live cells, since the half-life of PAR is very short at DNA lesions. The times of cell exposure to the laser beam and the pulse energy were strictly equal in every operation (see the Materials and Methods for details), which ensured the same laser microirradiation treatment to all the cells. We first examined NBS1, since NBS1 is an important subunit in the MRN complex that activates ATM in response to DNA damage (Horejsi et al. 2004; Lee and Paull 2004; Stracker and Petrini 2011). As shown in Figure 3A, NBS1 was quickly recruited to DNA damage sites within 20 sec. The quick loading of NBS1 at DNA damage sites is consistent with the quick PAR synthesis at DNA damage sites (Fig. 3B), suggesting that PAR at DNA damage sites mediates the fast recruitment of NBS. Next, we treated cells with olaparib, the PARP inhibitor, to suppress PAR synthesis. Olaparib treatment abolished the early recruitment of NBS1 to DNA damage, although NBS1 still slowly accumulated at DNA damage sites (Fig. 3A), confirming that PAR is required for the fast relocation of NBS to DNA damage sites. The half-life of PAR is relatively short at DNA damage sites because PAR is quickly hydrolyzed by PARG at DNA damage sites. However, NBS1 was still retained at DNA damage sites even after PAR was digested (Fig. 3B). These results suggest that PAR mediates the fast recruitment of NBS1 to DNA damage sites, while other signals at DNA damage sites are important for the prolonged retention of NBS1 at DNA damage sites. Following DNA damage, γH2AX plays an important role in retaining DNA damage response factors at DNA damage sites (Celeste et al. 2003; Bonner et al. 2008; Lukas et al. 2011; Polo and Jackoson 2011). Thus, we examined whether γH2AX was associated with the prolonged retention of NBS1. Interestingly, in H2AX−/− cells, the stable retention of NBS1 at DNA damage sites was impaired. Following the hydrolysis of PAR, NBS1 was dropped from DNA damage sites (Fig. 3A). Moreover, additional olaparib treatment in H2AX−/− cells abolished both the fast recruitment and slow accumulation of NBS1 to DNA damage sites. These results suggest that there are two stages of the recruitment of NBS1 to DNA damage sites. The quick relocation of NBS1 to DNA damage sites is mediated by PAR at DNA damage sites, whereas the slow accumulation or stable retaining of NBS1 at DNA damage sites is regulated by H2AX. The FHA–BRCT fusion domain is essential for the relocation of NBS1 to DNA damage sites (Kobayashi et al. 2004; Stracker and Petrini 2011). The BRCT fold of NBS1 recognizes PAR, whereas the FHA fold of NBS1 could recognize the pThr motifs in MDC1 (Chapman and Jackson 2008; Spycher et al. 2008; Wu et al. 2008; Lloyd et al. 2009), a functional partner of γH2AX (Stucki et al. 2005; Lou et al. 2006). Again, in MDC1−/− cells, the stable retention of NBS1 to DNA damage sites is impaired, and olaparib treatment abolished the relocation of NBS1 to DNA damage sites in MDC1−/− cells (Fig. 3A). Similar results were observed when we examined the relocation of the FHA+BRCT domain of NBS1 to DNA damage sites (Supplemental Fig. 6), and olaparib treatment did not affect the relocation kinetics of γH2AX or MDC1 at DNA damage sites in wild-type cells (Supplemental Fig. 7A). Thus, these results suggest that PAR is the initial signal that induces the recruitment of NBS1 to DNA damage sites, while MDC1 retains NBS1 at DNA damage sites for the prolonged period. To validate this model, cells were treated with wortmannin, a PI3 kinase inhibitor, to suppress γH2AX and the recruitment of MDC1 to DNA lesions (Paull et al. 2000; Goldberg et al. 2003) but not affect the DNA damage-induced PAR synthesis at DNA damage sites (Supplemental Fig. 7B). With wortmannin treatment, NBS1 could still be recruited to DNA damage sites. However, it was dropped off from DNA damage sites when PAR was hydrolyzed (Fig. 3C). When cells were treated with both wortmannin and olaparib to suppress both the γH2AX-dependent pathway and PAR synthesis, NBS1 failed to be recruited to DNA damage sites (Fig. 3C). Moreover, with the treatment of gallotannin (GLTN), a cell-permeable PARG inhibitor (Ying et al. 2001; Fathers et al. 2012), to suppress PARG-dependent PAR hydrolysis, the half-life of PAR at DNA damage sites was significantly prolonged (Supplemental Fig. 7C). With both wortmannin and GLTN treatment to suppress γH2AX and prolong the half-life of PAR, NBS1 was still stably retained by PAR at DNA damage sites for the prolonged time (Fig. 3C). Collectively, these results demonstrate that PAR synthesis at DNA damage sites is critical for the recruitment of NBS1.
Figure 3.
PAR mediates the function of NBS1 during early DNA damage response. (A) PAR is required for the fast recruitment of NBS1 to DNA damage sites. The NBS1-GFP was expressed in wild-type, H2AX−/−, or MDC1−/− MEFs treated with or without olaparib. The relocation of NBS1-GFP to DNA damage sites was monitored in a time course following laser microirradiation. (B) The kinetics of the accumulation of PAR and endogenous NBS1 at DNA damage sites was examined. (C) The effect of wortmannin, olaparib, or GLTN on the recruitment of NBS1 to DNA damage sites. NBS1-GFP was expressed in U2OS cells. The relocation of NBS1-GFP to DNA damage sites was monitored in a time course following laser microirradiation. GFP/RFP fluorescence at the laser line was converted into a numerical value using Axiovision software (version 4.5). Normalized fluorescent curves from 50 cells from three independent experiments were averaged. (A–C) The error bars represent the standard deviation.
The interaction between PAR and the BRCT domain of NBS1 is important for the recruitment of NBS1 to DNA damage sites
Since the BRCT fold of NBS1 is a PAR-binding motif and the FHA fold of NBS1 is a phospho-protein-binding motif, we further dissected the functions of these two motifs in vivo. We mutated Arg28 into alanine (R28A) in the FHA fold of NBS1, which does not affect the interaction between NBS1 and PAR. As shown in Figure 4A, in the wild-type mouse embryonic fibroblasts (MEFs), the R28A mutant could still be recruited to DNA damage sites but were not able to stay at DNA damage sites for the prolonged time, since this mutant abolished the interaction with MDC1. Olaparib treatment abolished the recruitment of NBS1 to DNA damage, since there is no PAR signal at DNA damage sites to recruit the R28A mutant. GLTN treatment that kept PAR at DNA damage sites for the prolonged time stabilized NBS1 at DNA damage sites. Moreover, the status of MDC1 did not affect the fast recruitment of the R28A mutant of NBS1 (Fig. 4A). In contrast, the K160A mutant of NBS1 that disrupts the interaction with PAR abrogated the early recruitment of NBS1 to DNA damage sites in wild-type cells. Loss of MDC1 further abolished the slow accumulation of NBS1 to DNA damage sites (Fig. 4B). Taken together, the FHA–BRCT fusion domain of NBS1 recognizes both PAR and MDC1. PAR is important for the initial recruitment of NBS1 to DNA damage sites, whereas MDC1 is required for the retention of NBS1 at DNA damage sites for the prolonged time.
Figure 4.
The relocation kinetics of NBS1 mutants during DNA damage response. (A) The R28A mutation in the FHA domain of NBS1 abolishes the stable retention but not the early recruitment of NBS1 to DNA damage sites. The NBS1 R28A-GFP was expressed in wild-type or MDC1−/− MEFs treated with or without olaparib or GLTN. The relocation of the R28A-GFP to DNA damage sites was monitored in a time course following laser microirradiation. (B) K160A mutation in the BRCT domain of NBS1 abolishes the early recruitment but not the stable retention of NBS1 at DNA damage sites. The NBS1 K160A-GFP was expressed in wild-type or MDC1−/− MEFs treated with or without olaparib. The relocation of NBS1 K160A-GFP to DNA damage sites was monitored in a time course following laser microirradiation. GFP fluorescence at the laser line was converted into a numerical value using Axiovision software (version 4.5). Normalized fluorescent curves from 50 cells from three independent experiments were averaged. (A,B) The error bars represent the standard deviation.
PAR is important for the NBS1-mediated early ATM activation during DNA damage response
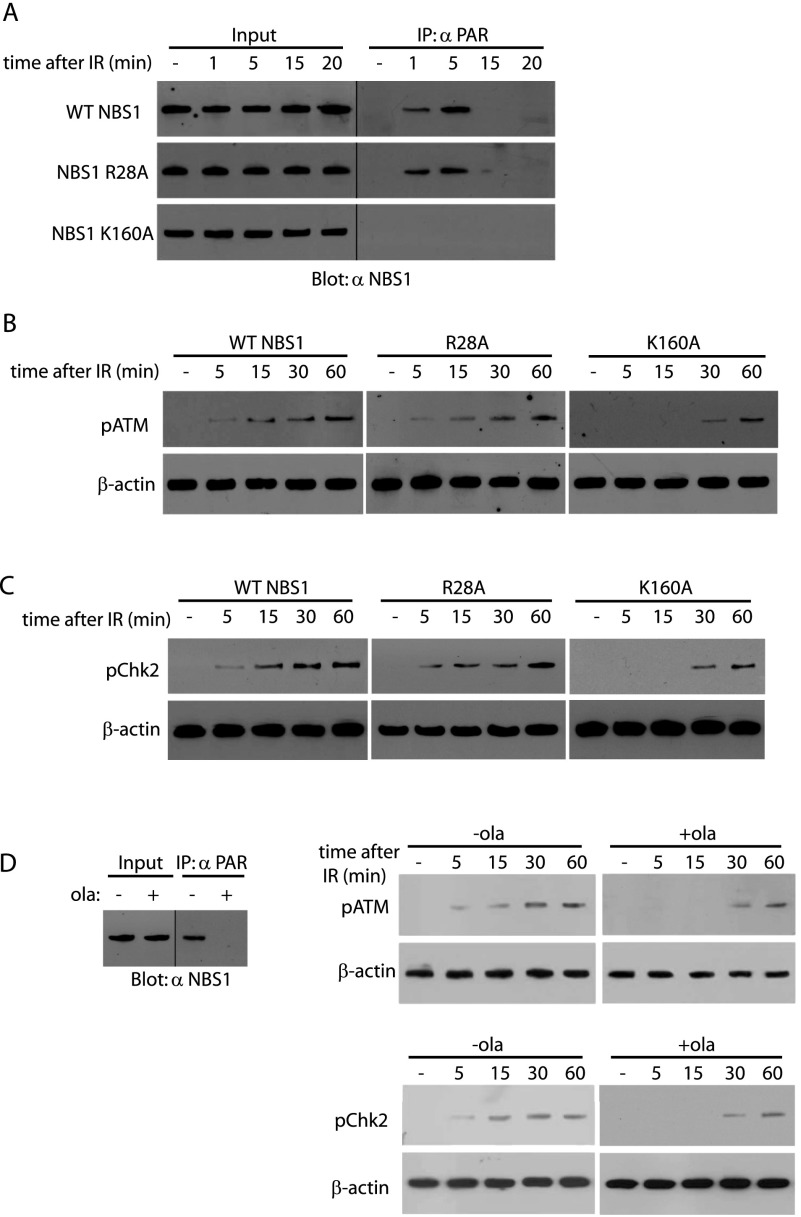
Since the MRN complex is important for the ATM activation in response to DNA damage (Horejsi et al. 2004; Lee and Paull 2004), we asked whether PAR-mediated relocation of NBS1 to DNA damage sites is important for the early activation of ATM. First, we examined the in vivo binding between PAR and wild-type NBS1 or the mutants following IR treatment. Cells with endogenous NBS1 depletion were reconstituted by siRNA-resistant wild-type NBS1, the K160A mutant, or the R28A mutant (Supplemental Fig. 8). As shown in Figure 5A, both wild-type NBS1 and the R28A mutant interacted with PAR within 5 min following IR. Since PAR is quickly digested by PARG in vivo, the interaction was disrupted at ∼15 min after DNA damage. As the K160A mutation abolishes the binding with PAR, we could not detect any interaction between PAR and the K160A mutant in vivo. With this interaction kinetics, we examined the activation of ATM. In the presence of wild-type NBS1 or the R28A mutant, we found that ATM was activated within 5 min following DNA damage response by examining the ATM Ser1981 phosphorylation (Fig. 5B), a surrogate maker of ATM activation (Bakkenist and Kastan 2003; Pellegrini et al. 2006). Moreover, we examined the phosphorylation of Chk2, the downstream functional partner of ATM (Falck et al. 2001; Smith et al. 2010), for the detection of ATM activation. Although the R28A mutant could not be stably retained at DNA damage sites, the mutant does not affect the late ATM activation. It is possible that activation of other PI3 kinases such as ATR and DNA-PKcs facilitates the late ATM activation (Yang et al. 2003; Stiff et al. 2006). However, in the presence of the K160A mutant, the activation of ATM was significantly delayed (Fig. 5B,C). Moreover, after IR treatment, the interaction between PAR and NBS1 and the early activation of ATM was also impaired when cells were pretreated with olaparib (Fig. 5D). Thus, these results show that PAR is important for the MRN complex-mediated early activation of ATM.
Figure 5.
PAR is important for the NBS1-mediated early activation of ATM in response to DNA damage. (A) NBS1 binds PAR during the early DNA damage response. U2OS cells with endogenous NBS1 knockdown were reconstituted by siRNA-resistant wild-type NBS1, the R28A mutant, or the K160A mutant. Cells were lysed at the indicated time points after IR. The in vivo interaction between PAR and wild-type NBS1 or the mutants was measured by co-IP. Whole-cell lysates were blotted and are shown as the input. (B,C) PAR is important for the NBS1-mediated early activation of ATM and Chk2 during DNA damage response. U2OS cells with endogenous NBS1 knockdown were reconstituted by siRNA-resistant wild-type NBS1, the R28A mutant, or the K160A mutant. Following IR treatment, cells were lysed at the indicated time points and subjected to Western blot detected by anti-pATM (S1981) and anti-pChk2 (T68) antibodies. (D) The NBS1–PAR interaction and the early activation of ATM and Chk2 during DNA damage response were abolished by the PARP inhibitor olaparib. Following IR treatment, cells pretreated by olaparib were lysed at 5 min (left) or the indicated time points (right). (Left) The NBS1–PAR binding was detected by co-IP. (Right) The early activation of ATM and Chk2 was detected by Western blot.
The interaction between PAR and NBS1 mediates the G2/M checkpoint activation and early DNA damage repair
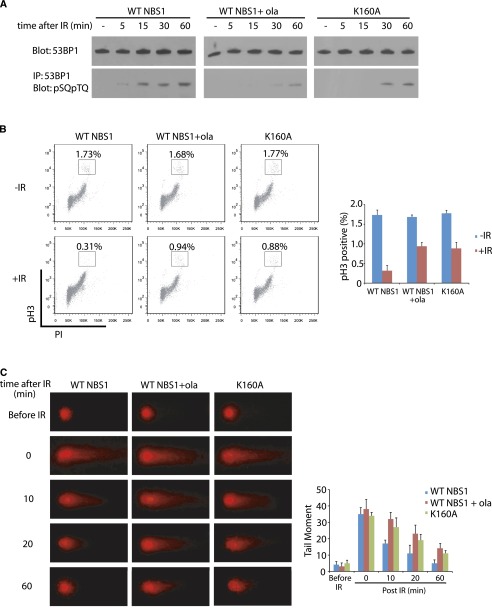
ATM kinase is a master kinase that phosphorylates numerous substrates and governs DNA damage-induced checkpoint activation and DNA damage repair (Kurz and Lees-Miller 2004; Shiloh and Ziv 2013). Thus, we continued to examine the role of PAR in ATM-dependent cell cycle checkpoint activation and DNA damage repair. During DNA damage response, 53BP1 is a key downstream effector of the ATM pathway (Anderson et al. 2001; Rappold et al. 2001; Ward et al. 2003). Accumulated evidence suggests that ATM-dependent 53BP1 phosphorylation activates both the G2/M checkpoint and DNA damage repair (DiTullio et al. 2002; Fernandez-Capetillo et al. 2002; Callen et al. 2013; Chapman et al. 2013). Thus, we asked whether PAR regulates the ATM-dependent 53BP1 phosphorylation. Cells pretreated with olaparib to suppress PAR synthesis were irradiated to induce double-strand breaks (DSBs). Olaparib treatment clearly suppressed early phosphorylation of 53BP1 during DNA damage response. To examine whether early phosphorylation of 53BP1 is also mediated by the interaction between PAR and the BRCT domain of NBS1, we examined the cells only expressing the K160A mutant, in which the early activation of ATM was suppressed. Consistently, the early phosphorylation of 53BP1 was suppressed in the presence of the K160A mutant of NBS1 compared with that in the presence of wild-type NBS1 (Fig. 6A). 53BP1 has been shown to mediate the G2/M checkpoint activation and DNA damage repair. (DiTullio et al. 2002; Fernandez-Capetillo et al. 2002; Callen et al. 2013; Chapman et al. 2013) Here, we examined both the G2/M checkpoint activation and DNA damage repair. Following DSBs, cells are transiently arrested before entering mitosis to provide enough time for DNA damage repair. This short and transient cell cycle arrestment at the G2/M boundary is named as the G2/M checkpoint (Lukas et al. 2004). To examine the G2/M checkpoint, we monitored mitotic population by examining the phospho-histone H3 population following DSBs, which is a standard assay for studying the transient G2/M checkpoint (Wang et al. 2002; Wu et al. 2011). As shown in Figure 6B, with IR treatment, normal cells were arrested before mitosis, as phospho-histone H3 positively stained cells were significantly reduced. However, with olaparib treatment, cells could not be fully arrested at the G2/M boundary, suggesting the loss of the transient G2/M checkpoint. Moreover, compared with wild-type NBS1, the K160A mutant also abrogated the G2/M checkpoint following IR-induced DSBs. Next, we examined the early DNA damage repair using comet assays. Again, DNA damage repair was significantly impaired when cells were treated with olaparib or only expressed the K160A mutant of NBS1 (Fig. 6C). Taken together, these results suggest that the PAR-mediated ATM activation is likely to be critical for the early checkpoint activation and DNA damage repair.
Figure 6.
The interaction between PAR and NBS1 is important for the G2/M checkpoint activation and early DNA damage repair. (A) The DNA damage-induced early phosphorylation of 53BP1 is dependent on the interaction between PAR and NBS1. U2OS cells with endogenous NBS1 knockdown were reconstituted by siRNA-resistant wild-type NBS1 or the K160A mutant and were pretreated with or without olaparib. Following IR treatment, cells were lysed at the indicated time points and subjected to immunoprecipitation with anti-53BP1 antibody and Western blot with anti-pSQpTQ antibody. Western blot with anti-53BP1 was used as the input control. (B) Olaparib treatment or the K160A mutant abrogates IR-induced G2/M checkpoint activation. The phospho-histone 3-positive population was examined by flow cytometry, and mean values were calculated from three independent experiments. Error bars represent standard error of the mean. (C) Representative images of comet assay at the indicated time points following 4 Gy of IR treatment. Cells were subjected to neutral comet assays. Tail moments were summarized from three independent experiments with at least 30 cells in single time point per sample. The error bars represent the standard deviation.
PAR mediates the early recruitment of PNKP, APTX, Ligase4, and XRCC1 to DNA damage sites
Next, we examined the role of PAR for the recruitment of other FHA and BRCT domain-containing proteins. For PNKP, it was recruited to DNA damage sites, and olaparib treatment impaired the early recruitment of PNKP (Supplemental Fig. 9A). Like NBS1, PNKP could not be stably retained at DNA damage sites in the H2AX−/− cells. Lacking both PAR and H2AX totally abolished the relocation of PNKP to DNA damage sites. Interestingly, the R35A/R48A mutant PNKP also abolished the relocation of PNKP to DNA damage sites, suggesting that the FHA domain of PNKP is important not only for the early recruitment of PNKP, but also for the PNKP retention at DNA damage sites. A similar phenomenon was observed on the relocation of the PNKP FHA domain to DNA damage sites (Supplemental Fig. 9B). Thus, it is likely that the FHA domain of PNKP recognizes phosphate groups in other molecule besides PAR, which is important for the stability of PNKP at DNA damage sites. It has been reported that PNKP interacted with XRCC1 (Whitehouse et al. 2001; Mani et al. 2007), which also recognizes PAR at DNA damage sites. However, the recruitment of PNKP to DNA damage sites is independent of XRCC1 (Supplemental Fig. 9C). Moreover, when cells were treated with wortmannin to suppress DNA damage–induced phosphorylation signal (Sarkaria et al. 1998), the retention of PNKP at DNA damage sites was significantly impaired. Wortmannin and olaparib treatment together additively abolished the recruitment of PNKP to DNA damage sites (Supplemental Fig. 9D). Thus, it is likely that the recruitment of PNKP to DNA damage sites is via the direct interaction between the FHA domain of PNKP and PAR, and the retention of PNKP at DNA damage sites is mediated by the interaction between the FHA domain and its other phospho-binding partners. Moreover, we observed a very similar phenomenon on APTX (Supplemental Fig. 10A).
For DNA Ligase4, the olaparib treatment impaired the early recruitment of DNA Ligase4 to DNA damage sites. Lacking H2AX significantly affected the stability of Ligase4 at DNA damage sites (Supplemental Fig. 10B). However, the S668A mutant in the BRCT domain did not affect the slow accumulation of Ligase4 at DNA damage sites, suggesting that other motifs of Ligase4 mediate the H2AX-dependent retention of Ligase4 at DNA damage sites. Moreover, olaparib treatment did not show an additional delay for the recruitment of the S668A mutant (Supplemental Fig. 10B). It has been shown that Ligase4 forms a complex with other partners, such as XRCC4 and XLF, via other regions in the BRCT domain (Chen et al. 2000; Sibanda et al. 2001; Riballo et al. 2009). It is possible that other partners of Ligase4 facilitate the prolonged retention of Ligase4 at the sites of DNA damage.
For XRCC1, both the olaparib treatment and the BRCT domain mutation abolished fast recruitment of XRRC1 to DNA damage sites. Although lacking H2X mildly impaired the retention of XRCC1 at DNA damage sites, the olaparib treatment in H2AX−/− cells did not totally abolish the slow accumulation of XRCC1 to DNA damage sites (Supplemental Fig. 10C). These results suggest that PAR and the BRCT domain of XRCC1 are essential for the fast recruitment of XRCC1 to DNA damage sites. However, DNA damage-induced signals other than γH2AX are required for the retention of XRCC1 at DNA damage sites (Supplemental Fig. 10C). Collectively, our results demonstrate that PAR is a bona fide signal for the fast recruitment of various DNA damage response factors to DNA lesions. The retention of these DNA damage response factors at DNA damage sites is mediated by different mechanisms.
To examine the biological significance of PAR in response to DNA damage, we treated wild-type or H2AX−/− MEFs with olaparib followed by a low dose of IR. Lacking either PAR or H2AX, cells could still be resistant to the IR-induced DNA damage. However, after loss of both PAR and H2AX, cells were hypersensitive to a low dose of IR (Supplemental Fig. 10D). These results suggest that PAR synergizes with H2AX to recruit a set of DNA damage factors to DNA lesions for damage repair.
Discussion
Taken together, we identified two novel classes of PAR-binding module that are involved in DNA damage response. Although both the FHA and BRCT domains are known as phospho-protein-binding domains, here we found that a set of FHA and BRCT domains recognize PAR. Interestingly, the PAR-binding pocket coincides with the phospho-amino acid-binding pocket in the FHA or BRCT domain. Since both ADP-ribose and iso-ADP-ribose have two phosphate groups, it is likely that the PAR-binding pockets of the FHA or BRCT domains recognize the phosphate groups in the ADP-ribose or iso-ADP-ribose. In particular, the FHA domain of PNKP is also known to recognize the pS–pT peptide. The two phosphate groups in iso-ARP-ribose might mimic the two phosphate groups on the Ser and Thr residues in the pS–pT peptide and mediate the interaction with the FHA domain of PNKP (Supplemental Fig. 1). Moreover, the FHA domain of APTX can interact with the pS-D–pT-D peptide, in which the Asp residues are also negatively charged and mimic the phosphate group. Thus, the binding mode between the APTX FHA domain and iso-ADP-ribose could be very similar to that between the FHA and phospho-peptide.
Different from the FHA domains of PNKP and APTX, the BRCT domains of Ligase4, XRCC1, and NBS1 recognize ADP-ribose. It is possible that these BRCT domains might also recognize site-specific mono-ADP-ribose. However, in PARP1−/− cells where PAR synthesis is largely suppressed in response to DNA damage, the relocation of these BRCT domain-containing proteins to DNA lesions is also significantly suppressed (Supplemental Fig. 11). It suggests that, at least in the DNA damage context, these BRCT domains recognize PAR at DNA damage sites. Moreover, the binding pockets in these BRCT domains are associated with protein PARylation and PARPs during evolution. For example, PARylation and PARPs only existed in multicellular eukaryotes. Coincidently, XRCC1 does not exist in prokaryotes and yeast. Although both NBS1 and Ligase4 exist in yeast, the key residues in the binding pockets of the BRCT domains are missing in their yeast orthologs (Supplemental Fig. 12), and the function of the BRCT fold of yeast NBS1 is likely to recognize other signals or facilitate the FHA fold recognizing the phospho-amino acid (Lloyd et al. 2009). Thus, PAR synthesis at DNA damage sites could be very important for the recruitment of these BRCT domain-containing proteins to DNA lesions for DNA damage repair in other multicellular eukaryotes.
A previous study indicated that a region within XRCC1 (amino acids 379–400) interacted with PAR (Pleschke et al. 2000). We generated the peptide of this region but did not detect any binding between the peptide with PAR in a dot blot, pull-down, or ITC assay (data not shown). Indeed, this 22-amino-acid region in XRCC1 is too short to be correctly folded based on the structural analysis (Zhang et al. 1998) and does not directly contribute to the PAR-binding pocket of XRCC1. Moreover, not all of the FHA and BRCT domains could interact with PAR. In particular, the FHA domain of RNF8 and CHK2 and the BRCT domain of MDC1 and BRCA1 do not have the affinity with PAR. Computational model analyses also do not support the binding between the phospho-peptide-binding pockets in these domains and ADP-ribose/iso-ADP-ribose. Additional structural analysis will reveal the details of these PAR-mediated interactions.
Following DNA damage, PAR is massively synthesized at the DNA damage sites, which provides the platform to recruit DNA damage response proteins to lesions (D'Amours et al. 1999; Kim et al. 2005; Gibson and Kraus 2012). Here, we analyzed the biological function of the interaction between PAR and NBS1 because it may partially explain the molecular mechanism by which the MRN complex recognizes the DSBs and induces the early ATM activation. Our study demonstrates that both DNA damage-induced PAR synthesis and the ADP-ribose-binding pocket in NBS1 are important for early ATM activation, which controls the transient G2/M checkpoint and early DNA damage repair via the ATM-dependent signal transduction pathway. Accumulated evidence suggests that 53BP1 is a key downstream effector in the ATM-dependent pathway and could be phosphorylated by ATM and CHK2 (Anderson et al. 2001; Rappold et al. 2001; Ward et al. 2003). Phosphorylated 53BP1 is known to regulate its functional partners to govern the transient G2/M checkpoint and DNA damage repair (DiTullio et al. 2002; Fernandez-Capetillo et al. 2002; Callen et al. 2013; Chapman et al. 2013), which is consistent with our observations. Moreover, the PAR-dependent early ATM activation controls the transient G2/M checkpoint, which plays an important role in maintaining genomic stability. The transient G2/M checkpoint allows the completion of quick DNA damage repair before entering into mitosis so that the DNA lesions would not be transmitted from mother cells to daughter cells (Chen et al. 2000; Abraham 2001). However, the G2/M checkpoint only transiently exists. Prolonged arresting at the G2/M boundary will cause the mitotic exit and genomic instability (Hirose et al. 2001; Chiu et al. 2005; Yang et al. 2010). Meanwhile, DNA damage repair should also be completed quickly if cells are at the G2/M transition, since the G2/M checkpoint only transiently exists. Thus, the early activation of ATM and the ATM-dependent pathway at least plays an important role in maintaining the genomic stability of cells during the G2/M transition period. Moreover, DNA damage-induced PAR synthesis may regulate multiple layers of DNA damage repair, since PAR recruits many other repair machineries—including these FHA and BRCT domain-containing proteins—to DNA damage sites. These repair machineries may function together with ATM in a complicated network for early DNA damage repair.
Interestingly, PAR is also quickly degraded within a few minutes following DNA damage (D'Amours et al. 1999; Gagne et al. 2006; Kim et al. 2007a). Without other DNA damage response signals or other functional partners such as γH2AX, these DNA damage response factors could not stay at DNA lesions for the prolonged time (Su 2006; Polo and Jackoson 2011). Thus, DNA damage response signals such as γH2AX provide the selection for retaining DNA damage response factors at the DNA lesions for different repair mechanisms. Loss of both PAR and γH2AX, a portion of DNA damage repair proteins would not be able to reach DNA lesions, which causes cell lethality. Similar mechanism has been implicated in the cancer clinical trials, which combined PARP inhibitors and PI3 kinase inhibitors to boost the efficacy of cancer chemotherapy (Ibrahim et al. 2012; Juvekar et al. 2012).
Materials and methods
Plasmids and antibodies
For GST fusion proteins, the BRCA1 BRCT domain, 53BP1 BRCT domain, Ligase4 BRCT domain, MCPH1 BRCT2+3, MDC1 BRCT domain, PARP1 BRCT, PTIP BRCT3+4, REV1 BRCT, TdT BRCT, XRCC1 BRCT domain, APLF FHA, APTX FHA, CHK2 FHA, FOXK1 FHA, Ki67 FHA, NBS1 FHA+BRCT domain, PNKP FHA, RNF8 FHA, and MDC1 FHA were cloned into the pGEX-4T1 vector, respectively. CHFR with an N-terminal GST tag and the NBS1 FHA+BRCT domain with a C-terminal GST tag were cloned into pFastBac1 vector. For the constructs used in microirradiation experiments, PNKP, APTX, Ligase4, NBS1, XRCC1, and their indicated domains were cloned into pEGFP vector to generate plasmids encoding GFP fusion proteins (or domains). The mutations in the proteins or domains above were generated using the QuikChange site-directed mutagenesis kit (Stratagene). For the constructs used to establish stable cell lines, the siRNA-resistant full-length cDNA of NBS1 and NBS1 K160A were cloned into pCMV-Tag 4A vector. The siRNA duplexes were purchased from Dharmacon Research. The sequences of NBS1 and XRCC1 siRNA used were 5′-GTACGTTGTTGGAAGGAAAdTdT-3′ and 5′-GGGAAGAGGAAGTTGGATTdTdT-3′, respectively. siRNAs were transfected into cells using Oligofectamine (Invitrogen) according to the manufacturer's instructions. Anti-pATM, anti-pCHK2, anti-pSQpTQ, anti-XRCC1, and anti-NBS1 antibodies were purchased from Cell Signaling; anti-phosphorylated histone H3 antibody was purchased from Upstate Biotechnology; anti-Flag and anti-β-actin antibodies were purchased from Sigma; and anti-PAR antibody was purchased from Trevigen.
Immunoprecipitation and Western blot
U2OS cells were lysed with NETN-100 buffer (20 mM Tris-HCl at pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5% nonidet P-40) on ice. Soluble fractions were subjected to immunoprecipitation and Western blot and probed with antibodies as indicated.
Immunofluorescence
Cells were fixed in 3% paraformaldehyde for 25 min and permeabilized in 0.5% Triton X-100 for 20 min at room temperature. Samples were blocked with 5% goat serum and then incubated in primary antibody for 60 min. Samples were then washed with PBS three times and incubated with secondary antibody for 30 min. After PBS wash, the nuclei were stained by DAPI. The signals were visualized by fluorescence microscope.
Generation and purification of PAR and iso-ADP-ribose
PAR (or biotin-PAR) was synthesized and purified in vitro according to the previous work as described (Fahrer et al. 2007) with some modifications. Briefly, PAR was synthesized in a 15-mL incubation mixture comprising 100 mM Tris-HCl (pH 7.8), 10 mM MgCl2, 1 mM NAD+, 10 mM DTT, 60 mg/mL histone H1, 60 mg/mL histone type IIa, 50 mg/mL octameric oligonucleotide GGAATTCC, and 150 nM human PARP-1. The reaction was stopped after 60 min by addition of 20 mL of ice-cold 20% TCA. Following precipitation, the pellet was washed with ice-cold 99.8% ethanol. Polymer was detached using 0.5 M KOH/50 mM EDTA and was purified by phenol-chloroform extraction and isopropanol precipitation. Purified PAR was fractionated according to chain length by anion exchange high-pressure liquid chromatography (HPLC) protocol. Iso-ADP-ribose was generated and purified in vitro according to the procedure by Wang et al. (2012). Briefly, the purified PAR was digested by 50 U of snake venom phosphodiesterase (Worthington) with 15 mM MgCl2 overnight at room temperature. The product of the phosphodiesterase digestion, iso-ADPR, was further purified by ion exchange chromatography and Superdex 75 on fast protein liquid chromatography (FPLC). Purified iso-ADP-ribose was dried in air, dissolved by ddH2O to 50 mM, stored at −20°C.
Dot blot
Recombinant proteins (10 pmol) were conjugated to the glutathione beads and incubated with PAR (100 pmol, calculated as the ADP-ribose unit) for 2 h at 4°C. The beads were washed four times with NETN-100 buffer. GST fusion proteins were eluted from beads by glutathione and spotted onto a nitrocellulose membrane. The membrane was blocked with TBST buffer (0.15 M NaCl, 0.01 M Tris-HCl at pH 7.4, 0.1% Tween 20) supplemented with 5% milk and extensively washed with TBST. After drying in the air, the membrane was examined by anti-PAR antibody.
GST fusion protein expression and pull-down assay
GST fusion proteins were expressed in Escherichia coli or using the Bac-to-Bac baculovirus expression system (for recombinant CHFR and NBS1) (Invitrogen) and purified under standard procedures. Purified GST fusion proteins (1 pmol) were incubated with biotin-labeled PAR (5 pmol) and streptavidin beads for 2 h at 4°C. After washing with NETN-100 buffer four times, the samples were boiled in the SDS sample buffer. The elutes were analyzed by Western blot with anti-GST antibody.
ITC
ITC was carried out at 16°C with an ITC 200 Microcalorimeter (GE Healthcare). Proteins were dialyzed extensively into the buffer containing 10 mM Na2HPO4 (pH 7.5), and 100 mM NaCl at the final concentration of 20∼60 μM. Ligands (PAR, ADP-ribose, or iso-ADP-ribose) in the injection syringe were also diluted by the same buffer at the final concentration of 150∼750 μM (the concentration of PAR was calculated as the ADP-ribose unit). A typical titration consisted of 19 consecutive 2 μL injections of ligands following a preinjection of 0.4 μL of ligands into the protein solution at time intervals of 120 sec while stirring at 1000 rpm. Binding isotherms were integrated and analyzed using the software Origin 7.0 (OriginLab) provided by the manufacturer.
Molecular modeling
For the FHA domain proteins, crystal structures of CHK2/peptide (HFD-pT-YLIR; Protein Data Bank [PDB] ID: 1GXC) (Li et al. 2002), RNF8/peptide (ELK-pT-ERY; PDB ID: 2PIE) (Huen et al. 2007), PNKP/peptide (YAG-pS–pT-DEN; PDB ID:2W3O) (Ali et al. 2009), and APTX (PDB ID:3KT9) (Becherel et al. 2010) were used. To construct the APTX peptide (D-pS–D-pT-DA), APTX was aligned with the PNKP/XRCC1 peptide, and the XRCC1 peptide was mutated into the D-pS–D-pT-DA peptide followed by a structural minimization using the MOE program (Chemical Computing Group). For the comparison of the binding affinity between iso-ADP-ribose, peptides, and the FHA domain, five amino acids neighboring the pSer or pThr (capped with ACE and NME at their N and C termini) from the peptides comparable with the size of the iso-ADP-ribose were used in the binding free energy calculations. For the BRCT domain proteins, crystal structures of MDC1/peptide (pS-QEY; PDBID: 3K05) (Campbell et al. 2010), BRCA1/peptide (ISRST-pS-PTFNK; PDB ID: 1T29) (Shiozaki et al. 2004), NMR (nuclear magnetic resonance) structure of Ligase4 (PDB ID: 2E2W), and XRCC1 (PDB ID: 2D8M) were used.
For docking simulations, all of the protein structures were processed, and the protons were added according to the pH 7.0 using the MOE program (Chemical Computing Group) before they were used in the docking simulations. Two docking programs were used to evaluate and select the best binding poses. They were the GOLD program (version 4.0.1) (Jones et al. 1997) and the Glide module from Schrodinger program suite (Friesner et al. 2006). For the FHA domains of CHK2, RNF8, PNKP, and APTX, iso-ADP-ribose was docked into the binding site, whereas ADP-ribose was docked into the binding sites of MDC1, BRCA1, XRCC1, and the Ligase4 BRCT domain. In the docking simulation using the GOLD program, the centers of the binding sites for the proteins were selected at the residues mutated in the experiments and showed to be important for binding to peptides, iso-ADP-ribose, and ADP-ribose experimentally. The radius of the binding site was defined as 13 Å, large enough to cover the binding pockets. For each genetic algorithm (GA) run, a maximum of 200,000 operations were performed on a population of five islands of 100 individuals. Operator weights for crossover, mutation, and migration were set to 95, 95, and 10, respectively. The docking simulations were terminated after 20 runs for each ligand. GoldScore implemented in Gold 4.0.1 was used as the fitness function to evaluate the docked conformations. In the docking simulation using Glide, the center of the box was selected at the amino acids mutated in each protein, which were found important for binding, and the XP mode was used in docking. All of the top-ranked binding poses of iso-ADP-ribose and ADP-ribose with the proteins from two docking programs were inspected, and the poses with the phosphate groups similar to the phospho-peptides in the crystal structures and compatible with PAR were selected and are shown. The electrostatic potential surfaces of the proteins were calculated using the APBS (Baker et al. 2001) module in the PyMOL program (http://www.pymol.org) based on parameters generated from the PDB2PQR server (Dolinsky et al. 2004).
The selected binding models were then subjected to the MD simulation using the Amberprogram suite (version 12) , and the binding free energy was calculated using the Amber program suite (version 10). The force field parameters for ADP-ribose and iso-ADP-ribose were derived by using the Antechamber module in Amber. The point charge parameters of both ligands were derived from the minimized geometry at the RHF level using a 6-31G* basis set with Gaussian09 and followed by the RESP fitting of the electrostatic field potential generated from the point charges at each atom site to those calculated from Gaussian09.
The topology and coordinate files for each protein–ligand complex were prepared by first adding counter-ions to neutralize the charges of the system before it was solvated in a 12 Å cubic box of the TIP3P (Jorgensen et al. 1983) water. The system was initially minimized by a 1000-step steepest decent and a 2000-step conjugate gradient minimization procedures to the solvents. Then, a 2-psec simulation was performed to raise the temperature of the system to 150K, followed by another 18 psec of simulation to increase the temperature further to 298K, where the protein ligands were fixed using 10 kcal/mol force constants in reference to the initial structure. A second 60-nsec equilibration of the system at 298K was performed by constraining the backbone atoms of the system with a 2 kcal/mol force constant. The production run was 2 nsec. Conformations were saved from the trajectory at intervals of 1 psec. Conformations collected from 0.5–2 nsec were used for the binding affinity prediction calculations. The MD simulations were performed using the GPU accelerated version of the PMEMD program (Götz et al. 2012) in the isothermal isobaric (NTP, T = 298K and P = 1 atm) ensemble. The SHAKE (Ryckaert et al. 1977) algorithm was used to fix bonds involving hydrogen. The PME method (Darden et al. 1993) was used, and the nonbonded cutoff distance was set at 10 Å. The time step was 2 fsec, and the neighboring pairs list was updated every 20 steps.
The MM-PBSA method was used for binding free energy calculations. In the MM-PBSA calculation, the 31 conformations corresponding to 50-psec intervals in the trajectory were used for the molecular mechanics calculations. Eight conformations (taken at intervals of 200 psec) from the 1.5-nsec trajectory were chosen for the normal mode calculations for entropic contribution to the binding free energy. In the normal mode calculations, a distance-dependent dielectric constant of −4r was used, the maximum cycle was set to 60,000, and the convergence tolerance was 0.0002 kcal mol−1Å−. For the solvent-accessible surface area calculation, the default value of 0.0072 kcal/mol × Å2 for the surface tension coefficient was used.
For Ligase4 and XRCC1, we analyzed all 20 conformations (or models) of the NMR structures and performed docking simulations followed by MD simulations to evaluate the stability of binding models. For Ligase4, we found that the first and 18th conformations of the NMR structures are suitable for binding model determination. The binding model of ADP-ribose with the 18th conformation from the NMR structure yielded stable structures in the 4-nsec MD simulations. For XRCC1, we performed molecular dynamics simulations of the protein ligand-binding model based on four different XRCC1 conformations. We found that the second, third, sixth, and 17th conformations of the NMR structures gave well-defined and open binding sites and are suitable for generating the binding models with ADP-ribose for further MD simulations. Only the binding model between ADP-ribose and the 17th conformation of XRCC1 gave stable structures in a 4-nsec MD simulation.
Laser microirradiation and live-cell imaging
U2OS cells and MEFs were plated on glass-bottomed culture dishes (Mat Tek Corporation). Laser microirradiation was performed using an IX 71 microscope (Olympus) coupled with the MicoPoint laser illumination and ablation system (Photonic Instruments, Inc.). A 337.1-nm laser diode (3.4 mW) transmitted through a specific dye cell and then yielded a 365-nm wavelength laser beam that was focused through 60× UPlanSApo/1.35 oil objective to yield a spot size of 0.5–1 μm. The time of cell exposure to the laser beam was ∼3.5 nsec. The pulse energy was 170 μJ at 10 Hz. Images were taken by the same microscope with the CellSens software (Olympus). GFP fluorescence at the laser line was converted into a numerical value using Axiovision software (version 4.5). Normalized fluorescent curves from 50 cells from three independent experiments were averaged. The error bars represent the standard deviation.
Comet assays
Single-cell gel electrophoretic comet assays were performed under neutral conditions according to a previous study (Olive and Banath 2006). Briefly, U2OS cells were treated with or without 4 Gy of IR and recovered in normal culture medium for the indicated time at 37°C. Cells were collected and rinsed twice with ice-cold PBS; 2 × 104 cells per milliliter were combined with 1% LMAgarose at 40°C at the ratio of 1:3 (v/v) and immediately pipetted onto slides. For cellular lysis, the slides were immersed in the neutral lysis solution (2% sarkosyl, 0.5 M Na2EDTA, 0.5 mg/mL proteinase K at pH 8.0) overnight at 37°C in the dark followed by washing in the rinse buffer (90 mM Tris buffer, 90 mM boric acid, 2 mM Na2EDTA at pH 8.5) for 30 min with two repeats. Next, the slides were subjected to electrophoresis at 20 V (0.6 V/cm) for 25 min and stained in 2.5 μg/mL propidium iodide for 20 min. All images were taken with a fluorescence microscope and analyzed by Comet Assay IV software.
IR treatment and colony formation assay
Cells were irradiated with a 137Cs source at a dose of 10 Gy (or at the indicated doses). After irradiation, cells were lysed at the indicated time points for immunoprecipitation or Western blot. For colony formation assay, 500 wild-type or H2AX−/− MEFs were seeded into six-well plates and then treated by various doses of IR with or without olaparib. After a 7-d culture, the viable cells were fixed and stained with crystal violet. The number of colonies (>50 cells for each colony) was calculated.
G2/M checkpoints assay
Cells expressing the wild-type NBS1 or NBS1 K160A pretreated with or without olaparib were treated with or without 2 Gy of IR. After 1 h of recovery, cells were fixed with 70% (v/v) ethanol, stained with rabbit antibody to phospho-histone H3 (pSer10), and then incubated with FITC-conjugated goat secondary antibody to rabbit. The stained cells were treated with RNase A and then incubated with propidium iodide. Samples were analyzed by flow cytometry.
Drug treatment
For live-cell imaging, immunoprecipitation, or Western blot, 100 nM olaparib, 10 μM GLTN, or 10 μM wortmannin was added into the cell culture medium 1 h before laser microirradiation or cell lysis. For colony formation assay, 100 nM olaparib was added into the medium during the culture.
Statistical analyses
All experiments were performed in triplicates unless indicated otherwise. Means and standard deviations were plotted. Student's t-test was used for statistical analyses.
Acknowledgments
We thank Drs. Ming Lei and Feng Zhang for technical support. This work was supported by the National Institute of Health (CA132755 and CA130899 to X.Y.). X.Y. is a recipient of the Era of Hope Scholar Award from the Department of Defense.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.226357.113.
References
- Abraham RT 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev 15: 2177–2196 [DOI] [PubMed] [Google Scholar]
- Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, West SC 2008. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature 451: 81–85 [DOI] [PubMed] [Google Scholar]
- Ali AA, Jukes RM, Pearl LH, Oliver AW 2009. Specific recognition of a multiply phosphorylated motif in the DNA repair scaffold XRCC1 by the FHA domain of human PNK. Nucleic Acids Res 37: 1701–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ame JC, Spenlehauer C, de Murcia G 2004. The PARP superfamily. Bioessays 26: 882–893 [DOI] [PubMed] [Google Scholar]
- Anderson L, Henderson C, Adachi Y 2001. Phosphorylation and rapid relocalization of 53BP1 to nuclear foci upon DNA damage. Mol Cell Biol 21: 1719–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimov VM, Ziemys A, Kizhake S, Yuan Z, Natarajan A, Cavasotto CN 2011. Computational and experimental studies of the interaction between phospho-peptides and the C-terminal domain of BRCA1. J Comput Aided Mol Des 25: 1071–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer R 2013. Luring BRCA1 to the scene of the crime. Cancer Cell 23: 565–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA 2001. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci 98: 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 421: 499–506 [DOI] [PubMed] [Google Scholar]
- Becherel OJ, Jakob B, Cherry AL, Gueven N, Fusser M, Kijas AW, Peng C, Katyal S, McKinnon PJ, Chen J, et al. 2010. CK2 phosphorylation-dependent interaction between aprataxin and MDC1 in the DNA damage response. Nucleic Acids Res 38: 1489–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y 2008. γH2AX and cancer. Nat Rev Cancer 8: 957–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen E, Di Virgilio M, Kruhlak MJ, Nieto-Soler M, Wong N, Chen HT, Faryabi RB, Polato F, Santos M, Starnes LM, et al. 2013. 53BP1 Mediates Productive and Mutagenic DNA Repair through Distinct Phosphoprotein Interactions. Cell 153: 1266–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell SJ, Edwards RA, Glover JN 2010. Comparison of the structures and peptide binding specificities of the BRCT domains of MDC1 and BRCA1. Structure 18: 167–176 [DOI] [PubMed] [Google Scholar]
- Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A 2003. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol 5: 675–679 [DOI] [PubMed] [Google Scholar]
- Chapman JR, Jackson SP 2008. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep 9: 795–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Barral P, Vannier JB, Borel V, Steger M, Tomas-Loba A, Sartori AA, Adams IR, Batista FD, Boulton SJ 2013. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol Cell 49: 858–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Trujillo K, Sung P, Tomkinson AE 2000. Interactions of the DNA ligase IV-XRCC4 complex with DNA ends and the DNA-dependent protein kinase. J Biol Chem 275: 26196–26205 [DOI] [PubMed] [Google Scholar]
- Chiu CC, Li CH, Ung MW, Fuh TS, Chen WL, Fang K 2005. Etoposide (VP-16) elicits apoptosis following prolonged G2-M cell arrest in p53-mutated human non-small cell lung cancer cells. Cancer Lett 223: 249–258 [DOI] [PubMed] [Google Scholar]
- Cuneo MJ, Gabel SA, Krahn JM, Ricker MA, London RE 2011. The structural basis for partitioning of the XRCC1/DNA ligase III-α BRCT-mediated dimer complexes. Nucleic Acids Res 39: 7816–7827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amours D, Desnoyers S, D'Silva I, Poirier GG 1999. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J 342: 249–268 [PMC free article] [PubMed] [Google Scholar]
- Darden TA, York DM, Pedersen L 1993. Particle mesh Ewald: An N-log(N) method for Ewald sums in large systems. J Chem Phys 98: 10089–10092 [Google Scholar]
- DiTullio RA Jr, Mochan TA, Venere M, Bartkova J, Sehested M, Bartek J, Halazonetis TD 2002. 53BP1 functions in an ATM-dependent checkpoint pathway that is constitutively activated in human cancer. Nat Cell Biol 4: 998–1002 [DOI] [PubMed] [Google Scholar]
- Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA 2004. PDB2PQR: An automated pipeline for the setup of Poisson–Boltzmann electrostatics calculations. Nucleic Acids Res 32: W665–W667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durocher D, Henckel J, Fersht AR, Jackson SP 1999. The FHA domain is a modular phosphopeptide recognition motif. Mol Cell 4: 387–394 [DOI] [PubMed] [Google Scholar]
- Durocher D, Taylor IA, Sarbassova D, Haire LF, Westcott SL, Jackson SP, Smerdon SJ, Yaffe MB 2000. The molecular basis of FHA domain:phosphopeptide binding specificity and implications for phospho-dependent signaling mechanisms. Mol Cell 6: 1169–1182 [DOI] [PubMed] [Google Scholar]
- Fahrer J, Kranaster R, Altmeyer M, Marx A, Burkle A 2007. Quantitative analysis of the binding affinity of poly(ADP-ribose) to specific binding proteins as a function of chain length. Nucleic Acids Res 35: e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falck J, Mailand N, Syljuasen RG, Bartek J, Lukas J 2001. The ATM–Chk2–Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature 410: 842–847 [DOI] [PubMed] [Google Scholar]
- Fathers C, Drayton RM, Solovieva S, Bryant HE 2012. Inhibition of poly(ADP-ribose) glycohydrolase (PARG) specifically kills BRCA2-deficient tumor cells. Cell Cycle 11: 990–997 [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Chen HT, Celeste A, Ward I, Romanienko PJ, Morales JC, Naka K, Xia Z, Camerini-Otero RD, Motoyama N, et al. 2002. DNA damage-induced G2–M checkpoint activation by histone H2AX and 53BP1. Nat Cell Biol 4: 993–997 [DOI] [PubMed] [Google Scholar]
- Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT 2006. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein−ligand complexes. J Med Chem 49: 6177–6196 [DOI] [PubMed] [Google Scholar]
- Gagne JP, Hendzel MJ, Droit A, Poirier GG 2006. The expanding role of poly(ADP-ribose) metabolism: Current challenges and new perspectives. Curr Opin Cell Biol 18: 145–151 [DOI] [PubMed] [Google Scholar]
- Gibson BA, Kraus WL 2012. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol 13: 411–424 [DOI] [PubMed] [Google Scholar]
- Glover JN, Williams RS, Lee MS 2004. Interactions between BRCT repeats and phosphoproteins: Tangled up in two. Trends Biochem Sci 29: 579–585 [DOI] [PubMed] [Google Scholar]
- Goldberg M, Stucki M, Falck J, D'Amours D, Rahman D, Pappin D, Bartek J, Jackson SP 2003. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature 421: 952–956 [DOI] [PubMed] [Google Scholar]
- Götz AW, Williamson MJ, Xu D, Poole D, Le Grand S, Walker RC 2012. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized born. J Chem Theory Comput 8: 1542–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y, Berger MS, Pieper RO 2001. p53 effects both the duration of G2/M arrest and the fate of temozolomide-treated human glioblastoma cells. Cancer Res 61: 1957–1963 [PubMed] [Google Scholar]
- Horejsi Z, Falck J, Bakkenist CJ, Kastan MB, Lukas J, Bartek J 2004. Distinct functional domains of Nbs1 modulate the timing and magnitude of ATM activation after low doses of ionizing radiation. Oncogene 23: 3122–3127 [DOI] [PubMed] [Google Scholar]
- Hottiger MO, Hassa PO, Luscher B, Schuler H, Koch-Nolte F 2010. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci 35: 208–219 [DOI] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J 2007. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131: 901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim YH, Garcia-Garcia C, Serra V, He L, Torres-Lockhart K, Prat A, Anton P, Cozar P, Guzman M, Grueso J, et al. 2012. PI3K inhibition impairs BRCA1/2 expression and sensitizes BRCA proficient triple negative breast cancer to PARP inhibition. Cancer Discov 2: 1036–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G, Willett P, Glen RC, Leach AR, Taylor R 1997. Development and validation of a genetic algorithm for flexible docking. J Mol Biol 267: 727–748 [DOI] [PubMed] [Google Scholar]
- Jorgensen WL, Chandrasekha J, Madura JD, Impey RW, Klein ML 1983. Comparison of simple potential functions for simulating liquid water. J Chem Phys 79: 926–935 [Google Scholar]
- Juvekar A, Burga LN, Hu H, Lunsford EP, Ibrahim YH, Balmana J, Rajendran A, Papa A, Spencer K, Lyssiotis CA, et al. 2012. Combining a PI3K inhibitor with a PARP inhibitor provides an effective therapy for a mouse model of BRCA1-related breast cancer. Cancer Discov 2: 1048–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karras GI, Kustatscher G, Buhecha HR, Allen MD, Pugieux C, Sait F, Bycroft M, Ladurner AG 2005. The macro domain is an ADP-ribose binding module. EMBO J 24: 1911–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MY, Zhang T, Kraus WL 2005. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev 19: 1951–1967 [DOI] [PubMed] [Google Scholar]
- Kim H, Chen J, Yu X 2007a. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science 316: 1202–1205 [DOI] [PubMed] [Google Scholar]
- Kim H, Huang J, Chen J 2007b. CCDC98 is a BRCA1–BRCT domain-binding protein involved in the DNA damage response. Nat Struct Mol Biol 14: 710–715 [DOI] [PubMed] [Google Scholar]
- Kobayashi J, Antoccia A, Tauchi H, Matsuura S, Komatsu K 2004. NBS1 and its functional role in the DNA damage response. DNA Repair 3: 855–861 [DOI] [PubMed] [Google Scholar]
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, et al. 2007. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318: 1637–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W, et al. 2000. Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models. Acc Chem Res 33: 889–897 [DOI] [PubMed] [Google Scholar]
- Kurz EU, Lees-Miller SP 2004. DNA damage-induced activation of ATM and ATM-dependent signaling pathways. DNA Repair) 3: 889–900 [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT 2004. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science 304: 93–96 [DOI] [PubMed] [Google Scholar]
- Li M, Yu X 2013. Function of BRCA1 in the DNA damage response is mediated by ADP-ribosylation. Cancer Cell 23: 693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Williams BL, Haire LF, Goldberg M, Wilker E, Durocher D, Yaffe MB, Jackson SP, Smerdon SJ 2002. Structural and functional versatility of the FHA domain in DNA-damage signaling by the tumor suppressor kinase Chk2. Mol Cell 9: 1045–1054 [DOI] [PubMed] [Google Scholar]
- Li GY, McCulloch RD, Fenton AL, Cheung M, Meng L, Ikura M, Koch CA 2010. Structure and identification of ADP-ribose recognition motifs of APLF and role in the DNA damage response. Proc Natl Acad Sci 107: 9129–9134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Wu J, Yu X 2007. CCDC98 targets BRCA1 to DNA damage sites. Nat Struct Mol Biol 14: 716–720 [DOI] [PubMed] [Google Scholar]
- Lloyd J, Chapman JR, Clapperton JA, Haire LF, Hartsuiker E, Li J, Carr AM, Jackson SP, Smerdon SJ 2009. A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage. Cell 139: 100–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loizou JI, El-Khamisy SF, Zlatanou A, Moore DJ, Chan DW, Qin J, Sarno S, Meggio F, Pinna LA, Caldecott KW 2004. The protein kinase CK2 facilitates repair of chromosomal DNA single-strand breaks. Cell 117: 17–28 [DOI] [PubMed] [Google Scholar]
- Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, Rivera MA, Celeste A, Manis JP, van Deursen J, Nussenzweig A, Paull TT, et al. 2006. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell 21: 187–200 [DOI] [PubMed] [Google Scholar]
- Lukas J, Lukas C, Bartek J 2004. Mammalian cell cycle checkpoints: Signalling pathways and their organization in space and time. DNA Repair 3: 997–1007 [DOI] [PubMed] [Google Scholar]
- Lukas J, Lukas C, Bartek J 2011. More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nat Cell Biol 13: 1161–1169 [DOI] [PubMed] [Google Scholar]
- Luo X, Kraus WL 2012. On PAR with PARP: Cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev 26: 417–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan A, Yuan C, Lee H, Chen ES, Wu PY, Tsai MD 2008. Structure and function of the phosphothreonine-specific FHA domain. Sci Signal 1: re12. [DOI] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J 2007. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131: 887–900 [DOI] [PubMed] [Google Scholar]
- Mani RS, Fanta M, Karimi-Busheri F, Silver E, Virgen CA, Caldecott KW, Cass CE, Weinfeld M 2007. XRCC1 stimulates polynucleotide kinase by enhancing its damage discrimination and displacement from DNA repair intermediates. J Biol Chem 282: 28004–28013 [DOI] [PubMed] [Google Scholar]
- Manke IA, Lowery DM, Nguyen A, Yaffe MB 2003. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science 302: 636–639 [DOI] [PubMed] [Google Scholar]
- Messner S, Hottiger MO 2011. Histone ADP-ribosylation in DNA repair, replication and transcription. Trends Cell Biol 21: 534–542 [DOI] [PubMed] [Google Scholar]
- Mohammad DH, Yaffe MB 2009. 14-3-3 proteins, FHA domains and BRCT domains in the DNA damage response. DNA Repair 8: 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberoi J, Richards MW, Crumpler S, Brown N, Blagg J, Bayliss R 2010. Structural basis of poly(ADP-ribose) recognition by the multizinc binding domain of checkpoint with forkhead-associated and RING domains (CHFR). J Biol Chem 285: 39348–39358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive PL, Banath JP 2006. The comet assay: A method to measure DNA damage in individual cells. Nat Protoc 1: 23–29 [DOI] [PubMed] [Google Scholar]
- Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol 10: 886–895 [DOI] [PubMed] [Google Scholar]
- Pellegrini M, Celeste A, Difilippantonio S, Guo R, Wang W, Feigenbaum L, Nussenzweig A 2006. Autophosphorylation at serine 1987 is dispensable for murine Atm activation in vivo. Nature 443: 222–225 [DOI] [PubMed] [Google Scholar]
- Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR 2000. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J Biol Chem 275: 40974–40980 [DOI] [PubMed] [Google Scholar]
- Polo SE, Jackson SP 2011. Dynamics of DNA damage response proteins at DNA breaks: A focus on protein modifications. Genes Dev 25: 409–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappold I, Iwabuchi K, Date T, Chen J 2001. Tumor suppressor p53 binding protein 1 (53BP1) is involved in DNA damage-signaling pathways. J Cell Biol 153: 613–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastelli G, Del Rio A, Degliesposti G, Sgobba M 2010. Fast and accurate predictions of binding free energies using MM-PBSA and MM-GBSA. J Comput Chem 31: 797–810 [DOI] [PubMed] [Google Scholar]
- Riballo E, Woodbine L, Stiff T, Walker SA, Goodarzi AA, Jeggo PA 2009. XLF-Cernunnos promotes DNA ligase IV-XRCC4 re-adenylation following ligation. Nucleic Acids Res 37: 482–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M, Yu X, Chen J, Songyang Z 2003. Phosphopeptide binding specificities of BRCA1 COOH-terminal (BRCT) domains. J Biol Chem 278: 52914–52918 [DOI] [PubMed] [Google Scholar]
- Rungrotmongkol T, Nunthaboot N, Malaisree M, Kaiyawet N, Yotmanee P, Meeprasert A, Hannongbua S 2010. Molecular insight into the specific binding of ADP-ribose to the nsP3 macro domains of chikungunya and Venezuelan equine encephalitis viruses: Molecular dynamics simulations and free energy calculations. J Mol Graph Model 29: 347–353 [DOI] [PubMed] [Google Scholar]
- Ryckaert J-P, Ciccotti G, Berendsen HJC 1977. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J Comput Phys 23: 327–341 [Google Scholar]
- Sarkaria JN, Tibbetts RS, Busby EC, Kennedy AP, Hill DE, Abraham RT 1998. Inhibition of phosphoinositide 3-kinase related kinases by the radiosensitizing agent wortmannin. Cancer Res 58: 4375–4382 [PubMed] [Google Scholar]
- Schreiber V, Dantzer F, Ame JC, de Murcia G 2006. Poly(ADP-ribose): Novel functions for an old molecule. Nat Rev Mol Cell Biol 7: 517–528 [DOI] [PubMed] [Google Scholar]
- Shiloh Y, Ziv Y 2013. The ATM protein kinase: Regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol 14: 197–210 [PubMed] [Google Scholar]
- Shiozaki EN, Gu L, Yan N, Shi Y 2004. Structure of the BRCT repeats of BRCA1 bound to a BACH1 phosphopeptide: Implications for signaling. Mol Cell 14: 405–412 [DOI] [PubMed] [Google Scholar]
- Sibanda BL, Critchlow SE, Begun J, Pei XY, Jackson SP, Blundell TL, Pellegrini L 2001. Crystal structure of an Xrcc4–DNA ligase IV complex. Nat Struct Biol 8: 1015–1019 [DOI] [PubMed] [Google Scholar]
- Smith J, Tho LM, Xu N, Gillespie DA 2010. The ATM–Chk2 and ATR–Chk1 pathways in DNA damage signaling and cancer. Adv Cancer Res 108: 73–112 [DOI] [PubMed] [Google Scholar]
- Spycher C, Miller ES, Townsend K, Pavic L, Morrice NA, Janscak P, Stewart GS, Stucki M 2008. Constitutive phosphorylation of MDC1 physically links the MRE11-RAD50-NBS1 complex to damaged chromatin. J Cell Biol 181: 227–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiff T, Walker SA, Cerosaletti K, Goodarzi AA, Petermann E, Concannon P, O'Driscoll M, Jeggo PA 2006. ATR-dependent phosphorylation and activation of ATM in response to UV treatment or replication fork stalling. EMBO J 25: 5775–5782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker TH, Petrini JH 2011. The MRE11 complex: Starting from the ends. Nat Rev Mol Cell Biol 12: 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP 2005. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 123: 1213–1226 [DOI] [PubMed] [Google Scholar]
- Su TT 2006. Cellular responses to DNA damage: One signal, multiple choices. Annu Rev Genet 40: 187–208 [DOI] [PubMed] [Google Scholar]
- Sun Z, Hsiao J, Fay DS, Stern DF 1998. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science 281: 272–274 [DOI] [PubMed] [Google Scholar]
- Wang B, Matsuoka S, Carpenter PB, Elledge SJ 2002. 53BP1, a mediator of the DNA damage checkpoint. Science 298: 1435–1438 [DOI] [PubMed] [Google Scholar]
- Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ 2007. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science 316: 1194–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Michaud GA, Cheng Z, Zhang Y, Hinds TR, Fan E, Cong F, Xu W 2012. Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination. Genes Dev 26: 235–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward IM, Minn K, van Deursen J, Chen J 2003. p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol Cell Biol 23: 2556–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse CJ, Taylor RM, Thistlethwaite A, Zhang H, Karimi-Busheri F, Lasko DD, Weinfeld M, Caldecott KW 2001. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell 104: 107–117 [DOI] [PubMed] [Google Scholar]
- Williams RS, Dodson GE, Limbo O, Yamada Y, Williams JS, Guenther G, Classen S, Glover JN, Iwasaki H, Russell P, et al. 2009. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell 139: 87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Luo K, Lou Z, Chen J 2008. MDC1 regulates intra-S-phase checkpoint by targeting NBS1 to DNA double-strand breaks. Proc Natl Acad Sci 105: 11200–11205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu PY, Frit P, Meesala S, Dauvillier S, Modesti M, Andres SN, Huang Y, Sekiguchi J, Calsou P, Salles B, et al. 2009. Structural and functional interaction between the human DNA repair proteins DNA ligase IV and XRCC4. Mol Cell Biol 29: 3163–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Chen Y, Lu LY, Wu Y, Paulsen MT, Ljungman M, Ferguson DO, Yu X 2011. Chfr and RNF8 synergistically regulate ATM activation. Nat Struct Mol Biol 18: 761–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Yu Y, Hamrick HE, Duerksen-Hughes PJ 2003. ATM, ATR and DNA-PK: Initiators of the cellular genotoxic stress responses. Carcinogenesis 24: 1571–1580 [DOI] [PubMed] [Google Scholar]
- Yang L, Besschetnova TY, Brooks CR, Shah JV, Bonventre JV 2010. Epithelial cell cycle arrest in G2/M mediates kidney fibrosis after injury. Nat Med 16: 535–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying W, Sevigny MB, Chen Y, Swanson RA 2001. Poly(ADP-ribose) glycohydrolase mediates oxidative and excitotoxic neuronal death. Proc Natl Acad Sci 98: 12227–12232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Chen J 2004. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol Cell Biol 24: 9478–9486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Chini CC, He M, Mer G, Chen J 2003. The BRCT domain is a phospho-protein binding domain. Science 302: 639–642 [DOI] [PubMed] [Google Scholar]
- Zhang X, Morera S, Bates PA, Whitehead PC, Coffer AI, Hainbucher K, Nash RA, Sternberg MJ, Lindahl T, Freemont PS 1998. Structure of an XRCC1 BRCT domain: A new protein-protein interaction module. EMBO J 17: 6404–6411 [DOI] [PMC free article] [PubMed] [Google Scholar]