Senescence is a stable proliferation arrest implicated in tumor suppression and aging. While recent studies have implicated changes in chromatin and lamin B1 regulation in senescence, functional interactions between them are poorly understood. Here, genome-wide mapping of histone modifications in proliferating and senescent primary human lung fibroblasts reveal profound changes in the senescent chromatin landscape. The data suggest that lamin B1 down-regulation in senescence is a key trigger of chromatin changes that impact gene expression, aging, and cancer.
Keywords: senescence, lamin B1, chromatin, gene expression
Abstract
Senescence is a stable proliferation arrest, associated with an altered secretory pathway, thought to promote tumor suppression and tissue aging. While chromatin regulation and lamin B1 down-regulation have been implicated as senescence effectors, functional interactions between them are poorly understood. We compared genome-wide Lys4 trimethylation on histone H3 (H3K4me3) and H3K27me3 distributions between proliferating and senescent human cells and found dramatic differences in senescence, including large-scale domains of H3K4me3- and H3K27me3-enriched “mesas” and H3K27me3-depleted “canyons.” Mesas form at lamin B1-associated domains (LADs) in replicative senescence and oncogene-induced senescence and overlap DNA hypomethylation regions in cancer, suggesting that pre-malignant senescent chromatin changes foreshadow epigenetic cancer changes. Hutchinson-Gilford progeria syndrome fibroblasts (mutant lamin A) also show evidence of H3K4me3 mesas, suggesting a link between premature chromatin changes and accelerated cell senescence. Canyons mostly form between LADs and are enriched in genes and enhancers. H3K27me3 loss is correlated with up-regulation of key senescence genes, indicating a link between global chromatin changes and local gene expression regulation. Lamin B1 reduction in proliferating cells triggers senescence and formation of mesas and canyons. Our data illustrate profound chromatin reorganization during senescence and suggest that lamin B1 down-regulation in senescence is a key trigger of global and local chromatin changes that impact gene expression, aging, and cancer.
Cellular senescence is a stable proliferation arrest implicated in aging and tumor suppression and can be induced by telomere attrition, activation of oncogenes, and other cellular stresses (Campisi 2005; Adams 2009). Senescent cells are further characterized by altered physical morphology and transcription, including up-regulation of genes involved in cellular defense and the inflammatory response, many of which are secreted and are a part of the senescence-associated secretory phenotype (SASP) (Acosta et al. 2008; Coppe et al. 2008; Kuilman et al. 2008). The SASP is thought to promote immune clearance of pre-malignant senescent cells to facilitate tumor suppression (Xue et al. 2007; Krizhanovsky et al. 2008; Kang et al. 2011; Sagiv et al. 2013). However, a chronic SASP is also suggested to promote tissue aging, and, accordingly, clearing of senescent cells in mice delays the onset of age-related pathology (Krtolica et al. 2001; Baker et al. 2011). Recent studies have also described changes in chromatin during cell senescence. Many mammalian cell types develop regions of condensed chromatin, called senescence-associated heterochromatin foci (SAHFs), suggestive of large-scale changes in senescent chromatin (Funayama et al. 2006; Narita et al. 2006; Zhang et al. 2007). Also, a deficiency in the Polycomb-repressive protein EZH2 and subsequent decrease of repression-associated Lys27 trimethylation on histone H3 (H3K27me3) leads to rapid senescence in primary human cells, in part through up-regulation of p16INK4a (hereafter “p16”) (Bracken et al. 2007). In addition, histone loss leads to premature aging of yeast, and overexpression of histones results in life span extension (Dang et al. 2009; Feser et al. 2010), suggesting that the impact of chromatin on cell senescence extends to an impact on cell and organismal aging.
Longevity is also associated with alterations in specific histone post-translational modifications. In yeast, Lys16 acetylation on histone H4 (H4K16ac) increases with age at heterochromatic regions, including telomeres, due to reduction of histone deacetylase Sir2 protein levels (Dang et al. 2009), providing a potential mechanism for the observation that Sir2 overexpression extends yeast life span (Kaeberlein et al. 1999). Likewise, in mice, the histone deacetylase SIRT6 regulates senescence and longevity through Lys9 deacetylation on histone H3 (H3K9ac) (Michishita et al. 2008; Kawahara et al. 2009), and premature aging is linked to altered chromatin at telomeres in both yeast (Dang et al. 2009) and mice (Michishita et al. 2008; Dang et al. 2009). Observations in Caenorhabditis elegans link the loss of the Trithorax-mediated active transcription histone modification H3K4me3 and gain of repressed transcription modification H3K27me3 to extended longevity through an effect that may be inherited transgenerationally (Greer et al. 2010, 2011). Alterations in heterochromatin factors have also been described in prematurely aging cells from Hutchinson-Gilford progeria syndrome (HGPS) patients; namely, decreased levels of heterochromatin protein 1 (HP1), H3K9me3, and H3K27me3 and increased levels of H4K20me3 (Scaffidi and Misteli 2005; Shumaker et al. 2006; Taimen et al. 2009; McCord et al. 2013).
Results
These studies highlight a relationship between chromatin regulation in cell senescence, cancer, and aging; however, there is limited understanding of specific chromatin changes that occur on a genome-wide scale. Here we report genome-wide chromatin changes during senescence in IMR90 primary human lung fibroblasts. The cells were serially passaged in culture at physiological oxygen (3%) until replicative senescence and maintained in culture in a senescent state for 2 wk prior to analysis (Supplemental Fig. 1A). As expected, the early passage cells (population doubling [PD] 24; hereafter “proliferating cells”) exhibit hallmarks of proliferation, including few senescence-associated β-galactosidase (SA-β-gal)-positive cells and low levels of p16 (Supplemental Fig. 1B–D); comparatively, late passage senescent cells (PD87; hereafter “senescent cells”) show nearly 100% SA-β-gal-positive cells, up-regulated p16 levels (Supplemental Fig. 1B–D), and shortened telomeres (data not shown).
To survey chromatin changes that occur during senescence, we performed chromatin immunoprecipitation (ChIP) followed by genome-wide parallel sequencing (ChIP-seq) for total histone H3 and two H3 modifications—H3K4me3 and H3K27me3—in proliferating cells and senescent cells. Trithorax-mediated H3K4me3 is canonically associated with promoters of transcriptionally active genes (Barski et al. 2007; Guenther et al. 2007; Shilatifard 2012), whereas Polycomb-mediated H3K27me3 is associated with facultative heterochromatin (Lee et al. 2006a; Schwartz et al. 2006; Barski et al. 2007; Schuettengruber et al. 2009). We also performed a transcriptome analysis using microarrays, assessing RNA levels at 33,288 RefSeq transcripts from the same cell samples used for ChIP (Supplemental Text 1; Supplemental Fig. 2; Supplemental Tables 1, 2). Our microarray data largely agree with other previously published data sets (Shelton et al. 1999; Zhang et al. 2003) and were further validated by quantitative RT–PCR (qRT–PCR) of >50 randomly selected genes that show altered expression, including known down-regulated cell cycle genes and up-regulated SASP genes (e.g., Supplemental Fig. 2B,C). Hence, by several independent assays, the proliferating and senescent cells show expected patterns of physiology and gene expression.
We mapped ChIP-seq data for the histone modifications to the human genome, quantified binding enrichment by normalization to total histone H3, and subsequently assessed each resulting enrichment map for regions of significant binding. We validated these maps by performing qPCR across >100 genomic loci; indeed, qPCR strongly correlated with ChIP-seq results (R = 0.83) (e.g., Supplemental Fig. 3). It is important to note that while total histone H3 decreases significantly during senescence as measured by Western blot (Supplemental Fig. 4A, lysates normalized by cell number; O'Sullivan et al. 2010), the relative levels of H3K4me3 and H3K27me3 (normalized to histone H3) do not significantly change between proliferating and senescent cells (senescent sample concentrated ∼13-fold for equivalent loading of H3 level) (Supplemental Fig. 4B). Furthermore, ChIP-seq and ChIP-qPCR data were normalized to total histone H3 ChIP, which accounted for any regional differences in histone occupancy that could affect modification levels, thereby providing a platform to specifically identify regions of differential histone modifications (see Supplemental Fig. 4C for track views of total H3 and the modifications).
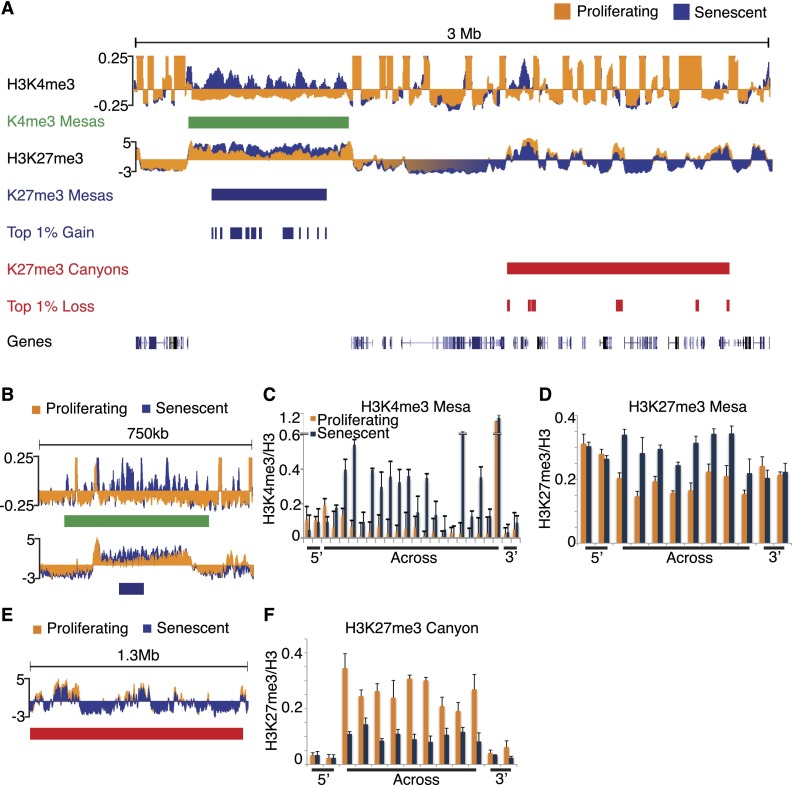
Both modifications (normalized to total H3) show altered patterns genome-wide in senescence (Supplemental Fig. 5). By visual inspection, both histone modifications appear to be changed in large domains in senescence. Notably, H3K4me3 is surprisingly enriched across the genome in extremely large domains, often hundreds of kilobases. We developed a new algorithm to identify large, differentially enriched H3K4me3 regions in senescent cells (H3K4me3-enriched mesas; hereafter “K4me3 mesas”) (Fig. 1A, H3K4me3 shown in the top track, proliferating tracks in orange and senescent tracks in blue; see the Supplemental Material for detailed analysis description). We identified 648 mesas spanning 50 kb (minimal size) to several hundred kilobases in length, occupying ∼17% of the senescent genome (Fig. 1A, green bars representing computationally defined H3K4me3 mesas). Unlike the canonical, sharp, highly enriched peaks of H3K4me3 observed over gene promoters (Roh et al. 2006), H3K4me3 enrichment in mesas is broad, covering the entire region with few, if any, peaks of enrichment that stand out above the general elevated signal.
Figure 1.
Large-scale chromatin changes occur in the senescence genome for both histone modifications. (A) Sample tracks of H3K4me3 and H3K27me3 ChIP-seq data over a 3-Mb region of chromosome 1 (Chr1: 38,016,703–41,116,920) show major regions of gains and losses for both histone modifications. Proliferating tracks are shown in orange, and senescence tracks are shown in blue. K4me3 mesas are shown in green blocks, the top 1% gain regions and K27me3 mesas are shown in blue blocks, and the top 1% loss regions and K27me3 canyons are shown in red blocks. (B) Example of overlapped K4me3 and K27me3 mesas over a 750-kb region of chromosome 7 (Chr7: 35,060,526–35,814,473). Proliferating tracks are shown in orange, and senescence tracks are shown in blue. (C,D) ChIP-qPCR validation of the K4me3 (C) and K27me3 (D) mesas shown in B. Proliferating data are shown in orange, and senescence data are shown in blue. ChIP-qPCR primers are tiled across the entire region of the mesa and include 3′ and 5′ flanking primers. ChIP-qPCR data are shown as ratios of modification to total histone H3. ChIP-qPCR data are the average of three biological replicates, and error bars represent standard deviation from the mean. (E) Example of K27me3 canyon over a 1.3-Mb region of chromosome 6 (Chr6: 36,239,850–37,570,928). Proliferating tracks are shown in orange, and senescence tracks are shown in blue. (F) ChIP-qPCR validation of the K27me3 canyon shown in E. Proliferating data are shown in orange, and senescence data are shown in blue. ChIP-qPCR primers are tiled across the entire region of the canyon and include 3′ and 5′ flanking primers. ChIP-qPCR data are shown as ratios of modification to total histone H3. ChIP-qPCR data are the average of three biological replicates, and error bars represent the standard deviation from the mean.
The ChIP-seq analysis also showed large-scale changes in the senescence-associated H3K27me3 profile, notably both regions of gain and loss, with varying sizes. H3-normalized H3K27me3 sequence reads were first analyzed in a sliding window analysis to define regions of significant gain or loss. The top 1% gain and loss regions were then “stitched” together to define contiguous regions of gain or loss across the genome for H3K27me3 (see the Supplemental Material for detailed analysis description). Using this method, we computationally defined 1440 H3K27me3-enriched mesas (hereafter “K27me3 mesas”) across the senescence genome spanning 4.3 kb to 27.5 Mb (average size of 394 kb), which showed striking overlap with the previously defined K4me3 mesas (Fig. 1A, blue bars representing computationally defined K27me3 mesas).
Analysis also revealed large domain losses of H3K27me3 (H3K27me3-depleted canyons; hereafter “K27me3 canyons”) (Fig. 1A, red bars representing computationally defined canyons). We defined 1374 K27me3 canyons across the senescent genome spanning 7 kb to 19 Mb (average size of 730.9 kb). Interestingly, the canyon phenomenon was limited to H3K27me3, as H3K4me3 did not show any significant loss across broad domains.
To validate the discovery methods, we performed ChIP-qPCR in triplicate (using three independently senesced sets of IMR90 cells) at varying intervals across and outside (upstream and downstream) multiple mesas and canyons from different chromosomes (Fig. 1B [example of overlapped K4me3 mesa and K27me3 mesa is validated by ChIP-qPCR in C,D], E [example K27me3 canyon is validated by ChIP-qPCR in F]). We also discovered mesa and canyon trends by ChIP-qPCR analysis in a different fibroblast strain (senescent MRC5), suggesting that these large-scale chromatin changes occur in other senescent cells (Supplemental Fig. 6A–C). We further confirmed K27me3 canyons by using a second, distinct antibody against H3K27me3 (Supplemental Fig. 6D). Moreover, we validated the K27me3 mesas and canyons by generating additional H3 and H3K27me3 ChIP-seq maps using three biological replicates of proliferating and senescent IMR90s (Supplemental Fig. 7). These repeated ChIP-seq data confirmed our observation that K27me3 mesas and canyons form across the senescent genome (Supplemental Fig. 7). Finally, we also observed K27me3 mesa and canyon trends in another published data set (Chandra et al. 2012), analyzing H3K27me3 enrichment or loss in that data set using our defined mesa and canyon regions, albeit to a lesser degree than in our study (Supplemental Fig. 7, far right boxes).
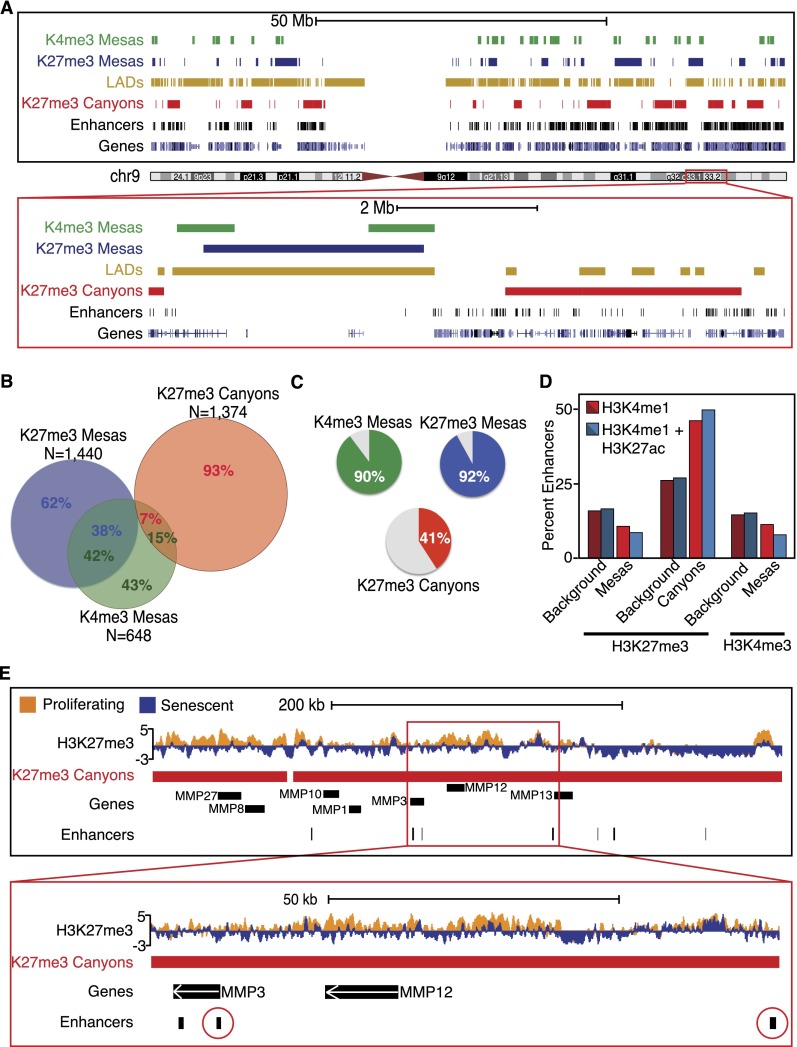
Thus, we defined three major domains of chromatin change in senescence: K4me3 mesas, K27me3 mesas, and K27me3 canyons. Interestingly, both types of mesas often overlap, whereas K27me3 canyons typically occupy different regions of the genome (Fig. 2A, K4me3 mesas in green, K27me3 mesas in blue, and K27me3 canyons in red). By nucleotide measure, ∼38% of K27me3 mesas overlap with K4me3 mesas, and 42% of K4me3 mesas overlap with K27me3 mesas, showing a high degree of overlap (Fig. 2B, blue and green circles). In contrast, by nucleotide measure, only 15% of K4me3 mesas overlap with K27me3 canyons, and only 7% of K27me3 canyons overlap with K4me3 mesas, highlighting the separation of canyons from the mesas (Fig. 2B, green and red circles). The observation of overlap between mesas, but not with canyons, is also reflected when measuring the overlap between features at an 80% cutoff parameter (Supplemental Fig. 8). Thus, the computational measure underscores the visual overlaps and differences observed in the track views (Fig. 2A), highlighting the greater concordance of the two types of mesas compared with mesas and canyons, which appear to occupy different regions of the genome.
Figure 2.
Mesas are overlapped features that are enriched for LADs; canyons are outside of LADs and enriched for enhancers. (A, top) Representation of all K4me3 (green blocks) and K27me3 (blue blocks) mesas and K27me3 canyons (red blocks) across the whole of chromosome 9. LADs are shown in gold blocks, enhancers are shown in black blocks, and genes are shown in blue blocks at the bottom of the image. Whole-chromosome views show the enrichment for mesas within LADs and canyon formation adjacent to LADs or in general LAD-poor regions. Moreover, canyons appear to be enriched for enhancers, which are also located outside of LADs and in gene-rich regions. (Bottom) Closer view of a section of a 9.2-Mb region of chromosome 9 (Chr9: 118,000,000–127,200,000) highlights the relationship of mesas with LADs and canyons with enhancer regions. (B) Venn diagram representation of the nucleotide overlap between K4me3 mesas, K27me3 mesas, and K27me3 canyons shows a high degree of overlap between the two mesas but little overlap between canyons and mesas, highlighting the difference between the large-scale changes that are visually depicted in A. (C) Pie chart representations of the percent of nucleotides in K4me3 mesas (top left panel in green), K27me3 mesas (top right panel in blue), and K27me3 canyons (bottom panel in red) that overlap with LADs. Most nucleotides in mesas overlap with LADs (90% for K4me3 mesas and 92% for K27me3 mesas), but only 41% of canyon nucleotides overlap with LADs, highlighting the difference between the large-scale changes that are visually depicted in A. (D) Column plot representation of the overlap between K4me3 mesas, K27me3 mesas, and K27me3 canyons with two categories of enhancers (H3K4me1 shown in red and H3K4me1+H3K27ac shown in blue) highlights the enrichment of enhancers in K27me3 canyons over background measure. Mesas, as expected, are not enriched for enhancers. (E) Example track view of the MMP cluster on chromosome 11 that shows H3K27me3 loss over genes and enhancers. The bottom view is a closeup of the MMP3 and MMP12 genes (up-regulated in senescence; Chr11: 102,017,147–102,489,087) with a cluster of enhancers (circled in red) nearby that are also contained within the K27me3 canyon.
The large-scale and distinct nature of mesas and canyons strongly indicates a profound change in global chromatin organization during senescence. DNA–lamin interactions at the nuclear membrane are a foundation for chromatin organization, and DNA at the nuclear periphery is strongly associated with heterochromatin and transcriptional silencing (Kourmouli et al. 2000; Mattout-Drubezki and Gruenbaum 2003; Shaklai et al. 2007; Finlan et al. 2008; Reddy et al. 2008; Towbin et al. 2010). Given the key role of lamins in controlling chromatin organization (Peric-Hupkes et al. 2010) and down-regulation of lamins in senescent cells (Shimi et al. 2011; Freund et al. 2012), we hypothesized that disruption of nuclear lamin interaction in senescent cells may contribute to the large-scale chromatin changes.
To investigate this possibility, we first investigated the proximity of canyons and mesas to lamin-associated domains (LADs). We performed ChIP-seq of lamin B1 in proliferating cells and defined 2054 LADs across the genome (see the Supplemental Material), which largely confirms a previous LAD DamID ChIP–chip data set in proliferating IMR90 cells (Supplemental Fig. 9; Guelen et al. 2008). The median size of the LADs is 450 kb, and by nucleotide measurement, LADs cover ∼61% of the genome in proliferating cells. Interestingly, by visual inspection, K4me3 and K27me3 mesas appear to be strongly associated with LADs, whereas K27me3 canyons appear to form in between LADs or over “gaps” in the LAD profile (Fig. 2A, LADs shown in gold).
We computationally defined the mesa–LAD and canyon–LAD relationship by assessing overlapping nucleotides (Fig. 2C). Approximately 60% of LAD nucleotides have overlap with mesas or canyons (Supplemental Fig. 10A). Indeed, 90% of K4me3 mesa nucleotides and 92% of K27me3 nucleotides overlap with LADs (Fig. 2C, green and blue pie charts at the top), underscoring the occurrence of mesas within LADs. In contrast, only 41% of K27me3 canyon nucleotides overlap with LADs (Fig. 2C, red pie chart at the bottom), highlighting the visual observation of canyon formation adjacent to and outside of LADs. The high overlap of mesas with LADs, but not with canyons, is also reflected when measuring the overlap between features at an 80% cutoff parameter (Supplemental Fig. 10B). We further analyzed the mesa–LAD and canyon–LAD relationship by assessing the size of the LADs that overlap these features (Supplemental Fig. 10C); LADs that overlap K27me3 canyons are smaller than the LADs overlapping K27me3 mesas (Supplemental Fig. 10C, cf. red vs. gray LAD size distribution), again emphasizing the incidence of canyons outside of the large LADs that overlap mesas. Together, this analysis highlights a nonrandom nature of mesa and canyon formation in the genome.
LADs are generally gene-poor relative to their genomic occupancy (Pickersgill et al. 2006; Guelen et al. 2008; Peric-Hupkes et al. 2010; van Bemmel et al. 2010), suggesting that K27me3 canyons, often outside of LADs, may be enriched for genes and their regulatory regions. Strikingly, we found both inactive enhancers (as defined by regions enriched for H3K4me1) and active enhancers (as defined by regions enriched for both H3K4me1 and H3K27ac) strongly overlapping with K27me3 canyons (Fig. 2D). Specifically, 51% of inactive enhancers and 49% of active enhancers are within canyons, and this association is nonrandom and significant (P = 0.001 for enrichment over background), as shown by permutation analysis (Fig. 2D). Thus, K27me3 canyons, mostly outside of LADs, are strongly enriched for enhancers. Furthermore, enhancers near key senescence genes are often contained within K27me3 canyons, where the H3K27me3 loss is also associated with up-regulated gene expression (65.1% of SASP genes are in canyons), as seen in the MMP cluster on chromosome 11 and specifically for the MMP3 gene (Fig. 2E). Hence, H3K27me3 loss at enhancer regions may play an important role in regulating transcriptional changes in senescence.
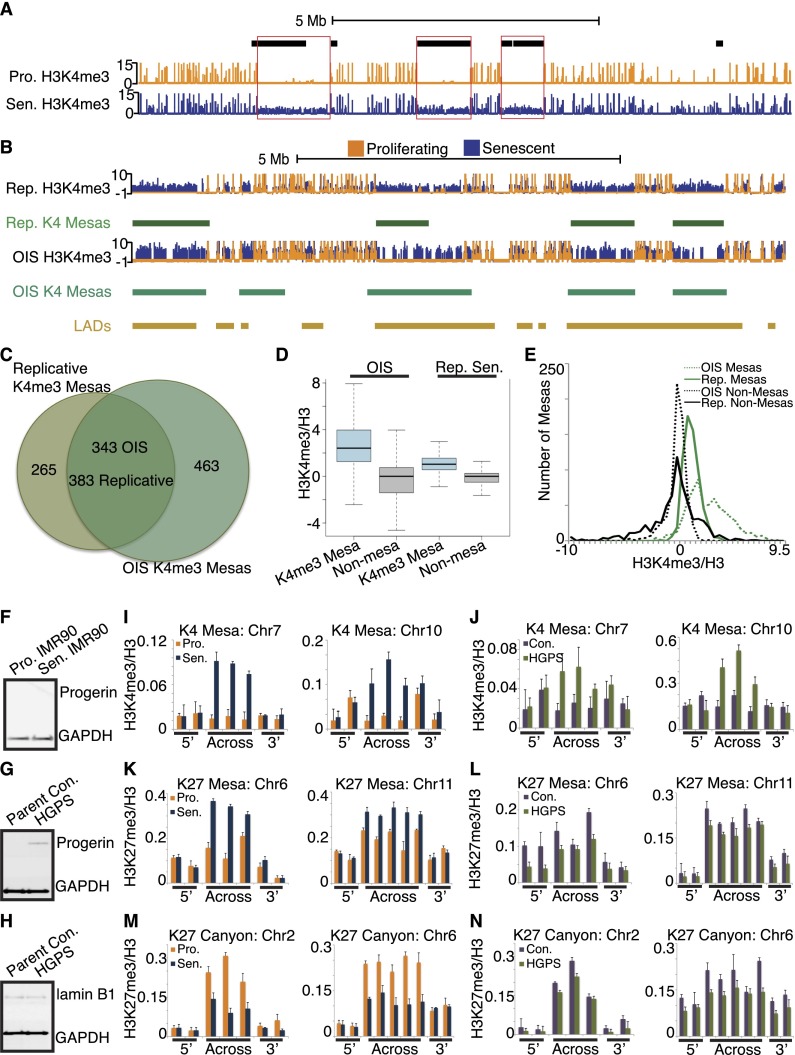
Similar profound alterations of chromatin have also been reported in genome-wide analyses of DNA methylation in cancer cells (Schuster-Bockler and Lehner 2012). Since senescence is a tumor suppression mechanism, we examined the intersection of senescent canyons and mesas with DNA differentially methylated regions (DMRs) in cancer. We found that 60% of K4me3 mesas overlap at least 50% with cancer-specific DNA hypomethylated DMRs (P < 2.2 × 10−16) in human primary colon adenocarcinoma (Berman et al. 2012; data not shown). Comparison with colorectal cancer reveals that 76% of K4me3 mesas overlap at least 50% with large (≥100 kb) DNA hypomethylated DMRs (example in Fig. 3A; Hansen et al. 2011). The data indicate similarities between chromatin regulation in senescent cells and cancer cells.
Figure 3.
H3K4me3 mesas overlap hypomethylated regions in cancer; mesas form in an OIS model and HGPS. (A) Example track view of a cluster of H3K4me3 mesas on chromosome 1 (Chr1: 30,500,000–40,000,000) shows a strong degree of overlap with cancer hypomethylated regions (black blocks). Proliferating tracks are shown in orange, and senescence tracks are shown in blue. (B) Example track view of the cluster of H3K4me3 mesas between replicative senescence and OIS shows a strong degree of overlap between the mesas of the two different senescence models. Proliferating tracks are shown in orange, and senescence tracks are shown in blue. Replicative senescence mesas are shown in green blocks, OIS mesas are shown in teal blocks, and LADs are shown in gold blocks. (C) Venn diagram representing the degree of overlap between the OIS and replicative senescence mesas shows a high degree of overlap between the two sets. Note that the OIS model has almost 200 more mesas than the replicative senescence. (D) Box plot analysis of the K4me3 mesas in replicative senescence and OIS (blue boxes) compared with nonmesa control regions (gray) shows an even higher enrichment for H3K4me3 gain in OIS mesas than replicative senescence mesas. (E) Histogram analysis of the OIS and replicative senescence mesas shows a bimodal pattern for the OIS mesas (green dotted line) compared with the single mode of replicative senescence mesas (green solid line). This suggests that a subset of OIS mesas is particularly enriched for H3K4me3, even more than the replicative senescence mesas. Control nonmesa regions are shown in black solid (replicative senescence) and dotted (OIS) lines. (F,G) Western blot analysis of progerin expression in proliferating and senescent IMR90s (F) and parental control and HGPS cell strains (G) shows progerin expression only in HGPS cells, not in IMR90 or parental control cells. (H) Western blot analysis of lamin B1 expression in parental control and HGPS cell strains shows similar levels of lamin B1 in both cell populations at the time of experimentation, underscoring the proliferating state of the HGPS cells at this point. (I,J) ChIP-qPCR evidence for K4me3 mesa formation in HGPS. Replicative senescence qPCR mesa validation shown in I (proliferating shown in orange and senescence shown in blue). The same K4me3 mesa analysis shown in J (parent control shown in purple and HGPS shown in green) indicates that K4me3 mesas may be a shared feature of HGPS. ChIP-qPCR primers are tiled across the region of the mesas and include 3′ and 5′ flanking primers. ChIP-qPCR data are shown as ratios of modification to total histone H3. ChIP-qPCR data are the average of three biological replicates, and error bars represent the standard deviation from the mean. (K,L) ChIP-qPCR test for K27me3 mesa formation in HGPS suggests that these features may not be forming in proliferating HGPS. Replicative senescence qPCR mesa validation shown in K (proliferating shown in orange and senescence shown in blue). The same K4me3 mesa analysis shown in L (parent control shown in purple and HGPS shown in green) indicates that K27me3 mesas may not be a shared feature of HGPS. Note that the K27me3 signal in HGPS is generally lower than the parental control, even at flanking regions. ChIP-qPCR primers are tiled across the region of the mesas and include 3′ and 5′ flanking primers. ChIP-qPCR data are shown as ratios of modification to total histone H3. ChIP-qPCR data are the average of three biological replicates, and error bars represent the standard deviation from the mean. (M,N) ChIP-qPCR test for K27me3 canyon formation in HGPS suggests that these features may not be forming in proliferating HGPS. Replicative senescence qPCR canyon validation shown in M (proliferating shown in orange and senescence shown in blue). The same K4me3 canyon analysis shown in N (parent control shown in purple and HGPS shown in green) indicates that K27me3 canyons are maybe not forming in still-proliferating HGPS cells. Note that the K27me3 signal in HGPS is generally lower than the parental control, even at flanking regions. ChIP-qPCR primers are tiled across the region of the canyons and include 3′ and 5′ flanking primers. ChIP-qPCR data are shown as ratios of modification to total histone H3. ChIP-qPCR data are the average of three biological replicates, and error bars represent the standard deviation from the mean.
To further investigate this connection, we examined K4me3 mesa formation in the cancer-related senescence model of oncogene-induced senescence (OIS) (Cosme-Blanco et al. 2007; Feldser and Greider 2007). IMR90 cells harboring a tamoxifen-inducible RAS oncogene undergo rapid senescence (Young et al. 2009). H3 and H3K4me3 ChIP-seq was performed for control and OIS cells, leading to identification of 806 K4me3 mesas, more than observed in replicative senescence (648 mesas). There is a striking overlap of OIS mesas with the original K4me3 mesas in replicative senescence (Fig. 3B, replicative senescence mesas shown in green and OIS mesas shown in teal): Sixty percent of replicative senescence K4me3 mesas overlap with OIS mesas (Fig. 3C), highlighting the obvious visual overlap in the tracks. Interestingly, the H3K4me3 gains in mesas in OIS seem more pronounced than in replicative senescence (Fig. 3D), and further analysis revealed a subset that is particularly enriched for H3K4me3 (Fig. 3E, cf. green dotted line for OIS mesas and the solid green line for replicative senescence mesas).
We further examined mesa and canyon formation in primary skin fibroblasts from HGPS patients, a segmental premature aging syndrome associated with accelerated cell senescence. HGPS is most commonly associated with a lamin A splicing mutation (to generate progerin), and we reasoned that lamin A mutation might phenocopy the major chromatin changes associated with lamin B1 down-regulation in senescence. We performed ChIP-qPCR on patient-derived proliferating HGPS cells across regions corresponding to the K4me3 mesas, K27me3 mesas, and K27me3 canyons of senescent IMR90 cells. Importantly, the patient-derived HGPS cell strain was progerin-positive, compared with progerin-negative IMR90 and parental control cells (Fig. 3F,G). Moreover, the HGPS cells were proliferating and not senescent at the time of the experimentation (at passage doubling 14; both cyclin A-positive and p16-negative measured by qRT–PCR) (data not shown), and the parental and HGPS lamin B1 levels were equivalent at the time cells were harvested for experiments (Fig. 3H).
We performed ChIP-qPCR across regions corresponding to K4me3 and K27me3 mesas and to K27me3 canyons of senescent IMR90 cells. As a control, we used passage-matched cells from an unaffected progerin-negative parent. We did not detect either K27me3 mesas or K27me3 canyons in HGPS across the regions tested (Fig. 3K–N); we note that, in general, our results show that H3K27me3 is broadly reduced in HGPS compared with the parental control, as previously observed (Shumaker et al. 2006; McCord et al. 2013). Hence, it appears that lowered H3K27me3 in HGPS may extend across the genome prior to senescence. Interestingly, in contrast, K4me3 mesas arise in the HGPS cell line compared with parent control across two different K4me3 mesa regions (Fig. 3I,J [IMR90 proliferating and senescent control shown in I, and HGPS and parent control shown in J]). Taken together, the data indicate that large-scale chromatin changes, particularly K4me3 mesas, may be a shared feature in prematurely aging HGPS cells and senescent cells. In particular, premature formation of H3K4me3 mesas might contribute to premature senescence of the HGPS cells.
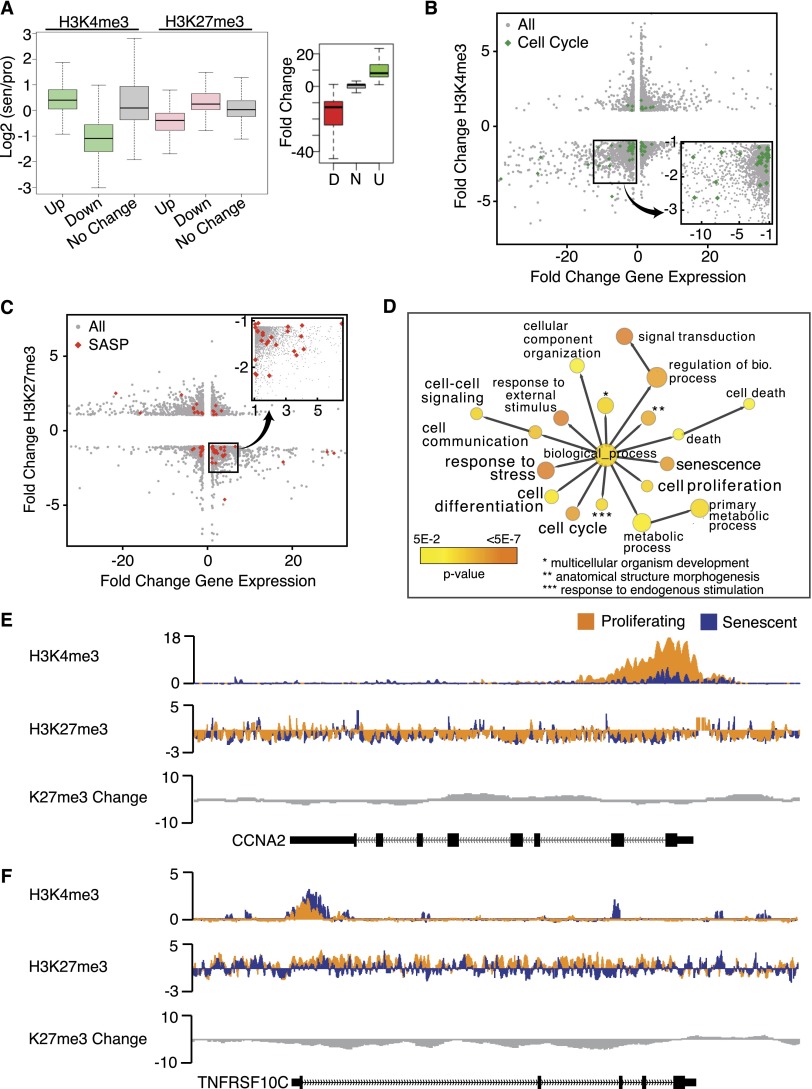
The analysis above focused on large-scale chromatin changes that occur in senescence. Given the overlap of canyons with enhancer and gene regions, we next compared chromatin alteration with gene expression. First, we quantified H3K4me3 and H3K27me3 enrichment at the top 500 most up-regulated and down-regulated genes in senescence (Fig. 4A, gene expression fold changes between −40-fold and 20-fold as shown in the right panel). H3K4me3 is enriched at up-regulated genes and is notably depleted at down-regulated genes compared with randomly selected genes with no transcriptional change (Fig. 4A, green boxes). H3K27me3 shows the opposite pattern, where the modification is depleted at up-regulated genes and enriched at down-regulated genes. To assess specific genes, we performed a scatter plot analysis, comparing gene expression with changes in H3K4me3 (Fig. 4B) and H3K27me3 (Fig. 4C). We found that H3K4me3 depletion is correlated to cell cycle and other proliferation genes that are transcriptionally down-regulated in senescence (Fig. 4B, cell cycle genes marked in green diamonds, expanded in the insert; Supplemental Fig. 11A shows the gene ontology [GO] analysis for down-regulated genes that lose H3K4me3; Chicas et al. 2012). Consistently, the OIS cells also display H3K4me3 loss at down-regulated cell cycle genes (Supplemental Fig. 11B).
Figure 4.
Changes in H3K4me3 and H3K27me3 are correlated to gene expression changes in senescence. (A) Box plot representation of H3K4me3 (green) and H3K27me3 (pink) at the top 500 up-regulated and down-regulated genes in senescence shows loss of H3K4me3 at down-regulated genes and loss of H3K27me3 at up-regulated genes. Control genes (no change) are shown in gray boxes. Box plot on the right depicts the range of expression changes at the top 500 up-regulated and down-regulated genes. Enrichment is reported as a ratio of senescence/proliferating, and all ChIP-seq signal is normalized to total histone H3. (B) Scatter plot analysis of H3K4me3-mediated gene regulation in senescence; fold change H3K4me3 is shown on the Y-axis, and fold change gene expression is shown on the X-axis. Cell cycle genes are marked in green. The bottom left quadrant contains down-regulated genes that also lose H3K4me3. The boxed region in the bottom left quadrant is shown in the closeup on the right to highlight the enrichment of cell cycle genes for H3K4me3 loss. (C) Scatter plot analysis of H3K27me3-mediated gene regulation in senescence; the fold change H3K27me3 is shown on the Y-axis, and the fold change gene expression is shown on the X-axis. SASP genes are marked in red. The bottom right quadrant contains up-regulated genes that also lose H3K27me3. The boxed region in the bottom right quadrant is shown in the closeup on top to highlight the enrichment of SASP genes for H3K27me3 loss. (D) GO analysis of up-regulated genes that lose H3K27me3 shows a strong enrichment for genes that are involved in the senescence pathway, including senescence genes, anti-proliferation genes, and cell death/stress genes. Each GO category color indicates the level of significance (yellow to orange, as shown on the key in the bottom left). (E) Sample tracks of H3K4me3 and H3K27me3 ChIP-seq data over a down-regulated cell cycle gene, CCNA2 (Chr4: 122,955,000–122,966,500), show pronounced loss of H3K4me3 at the promoter and TSS of the gene. Proliferating tracks are shown in orange, senescence tracks are shown in blue, and H3K27me3 change is shown in gray. The gene location is shown on the bottom track. (F) Sample tracks of H3K4me3 and H3K27me3 ChIP-seq data over the up-regulated SASP gene TNFRSF10c (Chr8: 23,012,616–23,034,551) show pronounced loss of H3K27me3 across the gene. Proliferating tracks are shown in orange, senescence tracks are shown in blue, and H3K27me3 change is shown in gray. The gene location is shown on the bottom track.
Strikingly, scatter plot analysis revealed that H3K27me3 loss is strongly correlated to a number of genes involved in the senescence response, including key SASP genes up-regulated in senescence (Fig. 4C, SASP genes marked in red diamonds, expanded in the insert; SASP and other senescence-related genes listed in Supplemental Table 3). GO analysis of the lower right quadrant of the H3K27me3 scatter plot (H3K27me3 loss and increased gene expression) shows significant enrichment for senescence-related categories, including senescence, anti-proliferation, and stress response (Fig. 4D; Supplemental Table 4 for full gene list of all GO terms and genes and statistics for H3K4me3 and H3K27me3; Supplemental Text 2 for GO and scatter plot analysis). It is important to note that, while some of the genes enriched in this quadrant also gain H3K4me3 in the senescent state, the key defining chromatin change correlated to this category of genes is the loss of H3K27me3.
As predicted by the scatter plot analysis, overlaid proliferating and senescence ChIP-seq tracks clearly show H3K4me3 loss at the promoter and transcriptional start site (TSS) of the senescence-down-regulated CCNA (cyclin A) gene and little change of H3K27me3 (Fig. 4E, proliferating tracks in orange, senescent tracks in blue, and H3K27me3 difference track in gray). Tumor necrosis factor receptor 10c (TNFRSF10c) is an up-regulated SASP gene (18-fold up-regulated) and is enriched in the scatter plot analysis for H3K27me3 loss. ChIP-seq tracks show H3K27me3 loss across the entire TNFRSF10c locus as well as H3K4me3 increase at the promoter and TSS (Fig. 4F). Together, these analyses highlight a second type of chromatin change in senescent cells, where localized chromatin changes are correlated to the transcriptional change in senescence. Remarkably, these local changes are typically encompassed within large-scale features; namely, the K27me3 canyons.
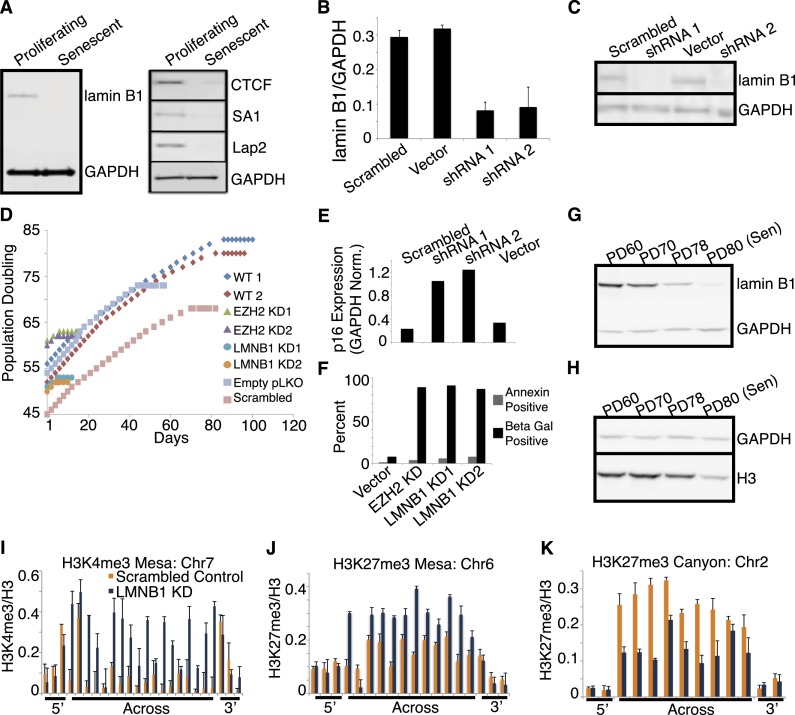
Our observations above show large-scale chromatin changes, including mesas that overlap almost entirely with LADs as well as canyons, typically located in between LADs and associated with localized chromatin changes correlated to gene expression. In light of this and knowing that lamin B1 is down-regulated in senescent cells (Shimi et al. 2011; Freund et al. 2012), we investigated whether down-regulation of lamin B1 is a trigger of chromatin changes in senescent cells. As previously shown, we detected a significant reduction in lamin B1 transcript and protein levels to nearly undetectable levels in senescent cells (Fig. 5A), whereas lamins A and C do not change (data not shown). Interestingly, we also detected reduced protein levels of other lamin interaction and nuclear organizational components, including the cohesin component SA1, the boundary element CTCF, and the lamin-interacting deacetylase Lap2 (lamin-associated deacetylase) (Fig. 5B). Importantly, the loss of lamin B1 was not due to the nonproliferative state in senescent cells, as quiescent cells achieved by serum starvation did not exhibit lamin B1 loss (Supplemental Fig. 12A; Shimi et al. 2011), and quiescent cells caused by contact inhibition did not exhibit canyon and mesa formation (Supplemental Fig. 12B,C).
Figure 5.
Lamin B1 is significantly decreased in senescence, and LMNB1 knockdown in proliferating cells causes formation of canyons and mesas. (A, left panel) Western blot analysis of total lamin B1 in proliferating and senescent cells shows lamin B1 loss in senescent cells. Lysates were normalized by total cell count; GAPDH was used as a loading control. (Right panel) CTCF (boundary element), SA1 (cohesin component), and Lap2 are decreased in senescent lysates compared with proliferating cells. Lysates were normalized by total cell count with GAPDH as a loading control. (B) qRT–PCR measure of lamin B1 mRNA expression in knockdown compared with control. LMNB1 expression is reduced by >60% by two different shRNA constructs (shRNA 1 and shRNA 2) compared with scrambled hairpin and vector-only control infected cells. (C) Western blot analysis of total protein in LMNB1 knockdown compared with control. Total lamin B1 is significantly reduced at the protein level in two knockdowns (shRNA1 and shRNA 2) compared with scrambled hairpin and vector-only control infected cells. Lysates were normalized by total cell count with GAPDH used as a loading control. (D) Life span analysis of LMNB1 knockdown shows rapid senescence within two PDs following cell infection compared with controls. Life span is visualized by plotting PDs (Y-axis) to days of growth in culture (X-axis). Two life spans of wild-type (WT), uninfected IMR90 show senescence after 78 (red diamonds) and 81 (blue diamonds) PDs. EZH2 knockdown causes senescence after only two cell passages following infection (green and purple triangles). LMNB1 knockdown by two different shRNA constructs shows the same kinetics of rapid senescence as EZH2 (turquoise and orange circles). The rapid senescence kinetics are specific to EZH2 and LMNB1 knockdown, as empty vector-only and scrambled hairpin infections (blue and pink squares) have a nearly normal life span, although not as long as wild-type controls (red and blue diamonds). The flattening of the growth curves indicates the length of time that cells were kept in a senescent state prior to harvesting for further experimentation. (E) LMNB1 knockdown results in up-regulation of p16. qRT–PCR measure of p16 expression in LMNB1 knockdown cells compared with control shows expected senescence up-regulation of p16 following LMNB1 knockdown. Relative p16 expression is up-regulated in two knockdowns (shRNA1 and shRNA 2) compared with scrambled hairpin and vector-only control infected cells; GAPDH was used as a control. (F) LMNB1 and EZH2 knockdown causes cellular senescence, not cell death. EZH2 and LMNB1 knockdown does not cause significant cell death, as measured by annexin staining compared with empty vector control. EZH2 and LMNB1 knockdown cells undergo rapid senescence, within two PDs, as measured by SA-β-gal staining, compared with empty vector control. For both types of knockdown, stably infected cells were maintained in culture for 2–3 d to ensure growth cessation prior to experimentation. (G,H) Western blot analysis of lamin B1 expression (G) and histone H3 expression (H) in a time course of IMR90 cells approaching senescence. The PD60, PD70, and PD78 time points are all proliferating states compared with senescence at PD80. Lamin B1 levels appear to be decreasing in proliferating cells prior to achievement of senescence compared with GAPDH control (G), whereas histone H3 decrease does not appear to occur until cells are in a senescent state compared with GAPDH control (H). (I–K) ChIP-qPCR evidence for K4me3 mesa (Chr7: 61,558,937–64,197,991), K27me3 mesa (Chr6: 167,743,380–169,126,883), and K27me3 canyon (Chr2: 85,956,542–86,782,134) formation in LMNB1 knockdown (KD; regions defined in replicative senescence). Control knockdown data are shown in orange, and LMNB1 knockdown is shown in blue. ChIP-qPCR primers are tiled across the entire region of the mesas and canyon and include 3′ and 5′ flanking primers. ChIP-qPCR data are shown as ratios of modification to total histone H3. ChIP-qPCR data are the average of three biological replicates, and error bars represent standard deviation from the mean.
To assess the functional link between lamin B1 loss and senescence, expression of the lamin B1 gene LMNB1 was reduced using lentiviral expression of targeted shRNA in proliferating cells. Two different shRNAs lowered lamin B1 RNA levels >50% (Fig. 5B), and protein levels significantly declined (Fig. 5C). Compared with scrambled shRNA or vector controls, the LMNB1 knockdown cells exhibited rapid cessation of cell division within two passages following infection (Fig. 5D, blue and orange circles) and became senescent, exhibiting up-regulated p16 expression (Fig. 5E) and high SA-β-gal-positive cells (senescence marker) compared with low annexin-positive cells (apoptosis marker) (Fig. 5F). The premature senescence phenotype was specific to the LMNB1 knockdown cells, as vector control cells showed almost normal life span (Fig. 5D, blue and pink squares) and little cell death or senescence following infection (Fig. 5E,F). We further note that lamin B1 loss appears to occur in nearly senescent cells across a time course of cells approaching senescence (Fig. 5G). Compared with midlife PD60 cells, lamin B1 appears to be slightly reduced at PD70 and largely reduced at PD78, which is two PDs before senescence at PD80. In contrast, the major loss of total histone associated with senescence does not appear until after lamin B1 is reduced, with slight reduction of histone H3 at PD78 and significant reduction at PD80, when the cells reach senescence (Fig. 5H).
The rapidity of premature senescence after LMNB1 knockdown was similar to the senescence following knockdown of the H3K27me3 methylase EZH2 (Fig. 5D, green and purple triangles) as previously described (Bracken et al. 2007). This and the observation that lamin B1 loss appears to be occurring in nearly senescent cells prior to senescence suggested that a functional link may exist between lamin disruption and the large-scale chromatin changes that we detected in senescent cells. To address this possibility, we performed ChIP-qPCR for H3K4me3 and H3K27me3 in the proliferating cells following lamin B1 knockdown (Fig. 5I–K). Strikingly, LMNB1 knockdown led to development of specific K4me3 mesas, K27me3 mesas, and K27me3 canyons across the genome (canyon and mesa regions were selected at random from the originally defined set and were remarkably similar to these features in senescent cells) (cf. Figs. 5I–K and 1C,D,F). The observation of mesa and canyon formation in LMNB1 knockdown strongly suggests that disruption of nuclear lamina-mediated organization may facilitate global chromatin changes during senescence.
Discussion
Here we show profound reorganization in the human epigenome upon senescence, with acquisition of large, contiguous stretches of altered chromatin—K4me3 mesas, K27me3 mesas, and K27me3 canyons—focused in and around the location of LADs in proliferating cells (Figs. 1A, 2A). We note that correlated K4me3 and K27me3 mesas over LADs may represent unusually large regions of “bivalent” chromatin (that is, marked by the presence of both active and repressive chromatin modifications) (Bernstein et al. 2006); whether the bivalency represents chromatin instability remains to be determined. Importantly, several lines of evidence indicate that mesa and canyon chromatin changes in senescence are closely linked to altered structure of LADs in senescence: First, lamin B1 declines to undetectable levels during senescence (Fig. 5A; Shimi et al. 2011; Freund et al. 2012), and the initial lamin B1 decline is prior to reduction of histone levels (Fig. 5G,H). Second, forced reduction of lamin B1, which leads to premature senescence, concomitantly results in mesas and canyons (Fig. 5I–K). Third, K4me3 mesas are detected in patient-derived HGPS cells (Fig. 3I,J), which harbor a lamin A/progerin mutation and undergo premature senescence.
HGPS is a segmental progeroid syndrome; thus, our results indirectly link formation of H3K4me3 mesas to accelerated tissue and organismal aging, perhaps due to acquisition of premature senescence chromatin changes. We also connect the senescence-associated chromatin features to alterations in cancer genomes. Specifically, mesas strongly correlate to the broad-scale, differentially DNA hypomethylated regions in colorectal cancer (Fig. 3A). Additionally, we detected H3K4me3 mesas in OIS (Fig. 3B,C). Together, these findings implicate the large-scale changes in senescent chromatin to both cancer and aging.
The functional consequence of the large-scale alterations in the senescence epigenome is not yet known; however, several lines of evidence are consistent with the notion that these large-scale chromatin changes are effectors of senescence. First, knockdown of lamin B1 triggers formation of mesas, overlapping with LADs, and accelerated senescence (Fig. 5D,I–K). Second, mesas form prematurely in proliferating HGPS cells (Fig. 3H–J) and thus are not simply a consequence of premature senescence in these cells. Third, localized H3K27me3 loss in canyons is strongly connected to up-regulation of key senescence and anti-proliferation genes, including the canonical SASP genes (Fig. 4C). Many of these genes are located within and near K27me3 canyons. Moreover, K27me3 canyons are enriched for inactive and active enhancers (Fig. 2D), strongly suggesting that canyons may contribute to the senescent state by altering gene expression via regulation of enhancer activity.
Interestingly, the K4me3 mesa and K27me3 canyon alterations that we observed are consistent with acquisition of a more open chromatin state, which supports previous findings that, in general terms, histone modifications characteristic of open chromatin tend to oppose longevity (Michishita et al. 2008; Dang et al. 2009; Kawahara et al. 2009; Greer et al. 2010, 2011). These results are in apparent contrast to the description of SAHFs in some senescent cell types (Funayama et al. 2006; Narita et al. 2006; Zhang et al. 2007). In a recent study, the higher-order heterochromatin organization reflected in SAHFs in oncogene-induced senescent cells was reported to be independent of regional changes in chromatin modifications (Chandra et al. 2012); however, since the genomic locations of SAHFs are not yet defined, the relationship between mesas, canyons, and SAHFs remains to be determined.
Our analysis provides a framework for understanding how down-regulation of lamin B1 in senescent cells initiates global chromatin changes, including reduction of histone levels, K4me3 and K27me3 mesas, and K27me3 canyons. Importantly, some of these global chromatin changes (K27me3 canyons) encompass local changes at gene enhancers that are likely to drive expression of SASP genes, key effectors of the senescence program. Indeed, lamin B1 is reduced in many normal cell types undergoing senescence, and overexpression delays senescence (Shimi et al. 2011; Freund et al. 2012). We note that certain cell types and disease states exhibit opposite effects of lamin B1 alteration (Barascu et al. 2012; Dreesen et al. 2013). In these diseased cells types, there may be other chromatin changes that alter the nuclear lamin organization and/or affect the cell cycle, perhaps leading to the opposite response to lamin B1 fluctuations compared with the nondiseased IMR90 model system used in our study. This is particularly interesting given our observation that proliferating but prematurely aging HGPS cells exhibit a subset of the global chromatin changes that we observed in senescent normal fibroblast cells. Moreover, we also link global chromatin changes in normal senescent cells to epigenetic alterations in cancer. In sum, we identified a lamin B1-regulated program of global chromatin reconfiguration in senescence that extends to local gene regulatory changes and to both aging and cancer.
Materials and methods
Antibodies
All antibodies used for this work are listed in Supplemental Table 5.
Cell culture
IMR90 cells (obtained from Coriell Institute for Medical Research, Camden, NJ) were grown in standard tissue culture medium (DMEM with 10% FBS and 1% penicillin/streptomycin) at 3% oxygen. For senescence studies, cells were replicatively passaged until growth ceased. Senescent cells were maintained in dishes for 2 wk to ensure growth termination. For knockdown experiments, midlife IMR90 cells were infected with lentiviral hairpin constructs (TRC collection) designed against lamin B1 in medium containing 8 μg/mL polybrene and 5% FBS for 24 h. Cells were then maintained in standard tissue culture medium with addition of 0.5 μg/mL puromycin to select a stably infected population. For lamin B1 knockdown studies, stably infected cells were established within two PDs, soon after which replication ceased. Senescence cells were maintained in dishes for 2–3 d to ensure growth termination prior to downstream experiments. For all cells, senescence was determined by monitoring p16 up-regulation and down-regulation of cyclin genes and by SA-β-gal staining. HGPS cells (AGO3199 and AG11498) and the parent control (AGO3257) (obtained from Coriell Institute for Medical Research, Camden, NJ) were grown in Eagle's MEM with Earle's salts, nonessential amino acids, 15% FBS, and 1% penicillin/streptomycin at 3% oxygen. For the experiment with HGPS and control, both cell lines were harvested for experiments at passage 14.
Whole-cell extract preparation and Western blot analysis
Medium from cells growing in 10-cm2 dishes was removed, and cells were washed in 1× PBS. Cells were scraped into 5 mL of 1× PBS, collected by centrifugation at 1200 rpm for 3 min at 4°C, and resuspended in 500 μL to 1 mL of standard SDS-PAGE loading buffer. Volumes were adjusted based on cell count, resulting in cell number-normalized lysate. Samples were boiled for 10 min, vortexed, and spun down to remove debris. Extracts were resolved on standard SDS-PAGE gels, transferred to nitrocellulose, and blotted for proteins of interest. All blots were blocked in 5% milk in TBST (1× TBS + 0.1% Tween). Antibodies were diluted in the same blocking solution. The antibody dilutions are listed in Supplemental Table 4. Washes were done in TBST. Visualization of HRP-conjugated secondary antibodies was achieved by standard chemiluminescence. Images of Western blots were taken with a Fujifilm LAS-4000 imager.
RNA preparation and qRT–PCR
RNA was purified using Trizol extraction. RT was performed using the Applied Biosystems High-Capacity RNA-to-cDNA kit (4387406). cDNA was quantified by qRT–PCR using standard procedures on a 7900HT Fast-Real-Time PCR (ABI). Primer sequences are available on request.
ChIP-qPCR or ChIP-seq
Cells in 10-cm2 dishes were fixed in 1% formaldehyde for 10 min, and fixation was quenched with addition of glycine to 125 mM for an additional 5 min. Cells were harvested by scraping from plates and washed twice in 1× PBS before storage at −80°C. ChIP was performed as described in the Young laboratory protocol (Lee et al. 2006b), except that extracts were sonicated twice for 9 min each round (30 sec of sonication with intermediate incubation of 30 sec per round) using a Bioruptor (Diagenode). All ChIPs were performed using 500 μg of extract and 2 μg of antibody per sample. Thirty microliters of Protein G Dynabeads (Invitrogen, 100.02D) was used per ChIP. Controls with IgG and no antibody controls were routinely performed, and all antibodies were tested by titration to be functioning within the linear range of the protocol. Following elution, ChIP DNA was analyzed by standard qPCR methods on a 7900HT Fast-Real-Time PCR (ABI). Primer sequences are available on request. For sequencing, 10 ng of ChIP DNA was used to make sequencing libraries using standard Illumina library single-end construction procedures. Sequencing was performed on either Illumina GAIIx (36 base pairs [bp], single-end reads) or Hi-Seq (100 bp, paired-end or single-end reads) platforms.
Computational methods
ChIP-seq analysis and computational methods used to characterize canyons and mesas are in the Supplemental Material.
Acknowledgments
We thank the NIH Roadmap Epigenomics Initiative (http://nihroadmap.nih.gov/epigenomics) and ENCODE for the use of their H3K4me1 and H3K27ac ChIP-seq data sets in proliferating IMR90 cells, the IDOM Functional Genomics Core Sequencing Facility and the Penn Genome Frontiers Institute for ChIP-seq, the Beatson Institute microarray core facility for microarray hybridization, and the Protein Expression Core (Epigenetics of Aging P01) for various support. This work was supported by an NIA P01 grant (P01AG031862) awarded to P.D.A. and S.L.B., and a CRUK program grant (A10250) awarded to P.D.A.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.223834.113.
References
- Acosta JC, O'Loghlen A, Banito A, Guijarro MV, Augert A, Raguz S, Fumagalli M, Da Costa M, Brown C, Popov N, et al. 2008. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell 133: 1006–1018 [DOI] [PubMed] [Google Scholar]
- Adams PD 2009. Healing and hurting: Molecular mechanisms, functions, and pathologies of cellular senescence. Mol Cell 36: 2–14 [DOI] [PubMed] [Google Scholar]
- Baker DJ, Wijshake T, Tchkonia T, LeBrasseur NK, Childs BG, van de Sluis B, Kirkland JL, van Deursen JM 2011. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature 479: 232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barascu A, Le Chalony C, Pennarun G, Genet D, Imam N, Lopez B, Bertrand P 2012. Oxidative stress induces an ATM-independent senescence pathway through p38 MAPK-mediated lamin B1 accumulation. EMBO J 31: 1080–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Berman BP, Weisenberger DJ, Aman JF, Hinoue T, Ramjan Z, Liu Y, Noushmehr H, Lange CP, van Dijk CM, Tollenaar RA, et al. 2012. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet 44: 40–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, Theilgaard-Monch K, Minucci S, Porse BT, Marine JC, et al. 2007. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev 21: 525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J 2005. Senescent cells, tumor suppression, and organismal aging: Good citizens, bad neighbors. Cell 120: 513–522 [DOI] [PubMed] [Google Scholar]
- Chandra T, Kirschner K, Thuret JY, Pope BD, Ryba T, Newman S, Ahmed K, Samarajiwa SA, Salama R, Carroll T, et al. 2012. Independence of repressive histone marks and chromatin compaction during senescent heterochromatic layer formation. Mol Cell 47: 203–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicas A, Kapoor A, Wang X, Aksoy O, Evertts AG, Zhang MQ, Garcia BA, Bernstein E, Lowe SW 2012. H3K4 demethylation by Jarid1a and Jarid1b contributes to retinoblastoma-mediated gene silencing during cellular senescence. Proc Natl Acad Sci 109: 8971–8976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppe JP, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J, Nelson PS, Desprez PY, Campisi J 2008. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol 6: 2853–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosme-Blanco W, Shen MF, Lazar AJ, Pathak S, Lozano G, Multani AS, Chang S 2007. Telomere dysfunction suppresses spontaneous tumorigenesis in vivo by initiating p53-dependent cellular senescence. EMBO Rep 8: 497–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, Kaeberlein M, Kennedy BK, Berger SL 2009. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature 459: 802–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreesen O, Chojnowski A, Ong PF, Zhao TY, Common JE, Lunny D, Lane EB, Lee SJ, Vardy LA, Stewart CL, et al. 2013. Lamin B1 fluctuations have differential effects on cellular proliferation and senescence. J Cell Biol 200: 605–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldser DM, Greider CW 2007. Short telomeres limit tumor progression in vivo by inducing senescence. Cancer Cell 11: 461–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feser J, Truong D, Das C, Carson JJ, Kieft J, Harkness T, Tyler JK 2010. Elevated histone expression promotes life span extension. Mol Cell 39: 724–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA 2008. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet 4: e1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund A, Laberge RM, Demaria M, Campisi J 2012. Lamin B1 loss is a senescence-associated biomarker. Mol Biol Cell 23: 2066–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funayama R, Saito M, Tanobe H, Ishikawa F 2006. Loss of linker histone H1 in cellular senescence. J Cell Biol 175: 869–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Maures TJ, Hauswirth AG, Green EM, Leeman DS, Maro GS, Han S, Banko MR, Gozani O, Brunet A 2010. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature 466: 383–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Maures TJ, Ucar D, Hauswirth AG, Mancini E, Lim JP, Benayoun BA, Shi Y, Brunet A 2011. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature 479: 365–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. 2008. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453: 948–951 [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA 2007. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130: 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KD, Timp W, Bravo HC, Sabunciyan S, Langmead B, McDonald OG, Wen B, Wu H, Liu Y, Diep D, et al. 2011. Increased methylation variation in epigenetic domains across cancer types. Nat Genet 43: 768–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L 1999. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev 13: 2570–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang TW, Yevsa T, Woller N, Hoenicke L, Wuestefeld T, Dauch D, Hohmeyer A, Gereke M, Rudalska R, Potapova A, et al. 2011. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature 479: 547–551 [DOI] [PubMed] [Google Scholar]
- Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, Lin M, McCord RA, Ongaigui KC, Boxer LD, Chang HY, et al. 2009. SIRT6 links histone H3 lysine 9 deacetylation to NF-κB-dependent gene expression and organismal life span. Cell 136: 62–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourmouli N, Theodoropoulos PA, Dialynas G, Bakou A, Politou AS, Cowell IG, Singh PB, Georgatos SD 2000. Dynamic associations of heterochromatin protein 1 with the nuclear envelope. EMBO J 19: 6558–6568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizhanovsky V, Yon M, Dickins RA, Hearn S, Simon J, Miething C, Yee H, Zender L, Lowe SW 2008. Senescence of activated stellate cells limits liver fibrosis. Cell 134: 657–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J 2001. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: A link between cancer and aging. Proc Natl Acad Sci 98: 12072–12077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuilman T, Michaloglou C, Vredeveld LC, Douma S, van Doorn R, Desmet CJ, Aarden LA, Mooi WJ, Peeper DS 2008. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell 133: 1019–1031 [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. 2006a. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125: 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Johnstone SE, Young RA 2006b. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc 1: 729–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattout-Drubezki A, Gruenbaum Y 2003. Dynamic interactions of nuclear lamina proteins with chromatin and transcriptional machinery. Cell Mol Life Sci 60: 2053–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCord RP, Nazario-Toole A, Zhang H, Chines PS, Zhan Y, Erdos MR, Collins FS, Dekker J, Cao K 2013. Correlated alterations in genome organization, histone methylation, and DNA-lamin A/C interactions in Hutchinson-Gilford progeria syndrome. Genome Res 23: 260–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, et al. 2008. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 452: 492–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M, Krizhanovsky V, Nunez S, Chicas A, Hearn SA, Myers MP, Lowe SW 2006. A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell 126: 503–514 [DOI] [PubMed] [Google Scholar]
- O'Sullivan RJ, Kubicek S, Schreiber SL, Karlseder J 2010. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat Struct Mol Biol 17: 1218–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Graf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. 2010. Molecular maps of the reorganization of genome–nuclear lamina interactions during differentiation. Mol Cell 38: 603–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B 2006. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet 38: 1005–1014 [DOI] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H 2008. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 452: 243–247 [DOI] [PubMed] [Google Scholar]
- Roh TY, Cuddapah S, Cui K, Zhao K 2006. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci 103: 15782–15787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagiv A, Biran A, Yon M, Simon J, Lowe SW, Krizhanovsky V 2013. Granule exocytosis mediates immune surveillance of senescent cells. Oncogene 32: 1971–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T 2005. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nat Med 11: 440–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Ganapathi M, Leblanc B, Portoso M, Jaschek R, Tolhuis B, van Lohuizen M, Tanay A, Cavalli G 2009. Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol 7: e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster-Bockler B, Lehner B 2012. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature 488: 504–507 [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V 2006. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet 38: 700–705 [DOI] [PubMed] [Google Scholar]
- Shaklai S, Amariglio N, Rechavi G, Simon AJ 2007. Gene silencing at the nuclear periphery. FEBS J 274: 1383–1392 [DOI] [PubMed] [Google Scholar]
- Shelton DN, Chang E, Whittier PS, Choi D, Funk WD 1999. Microarray analysis of replicative senescence. Curr Biol 9: 939–945 [DOI] [PubMed] [Google Scholar]
- Shilatifard A 2012. The COMPASS family of histone H3K4 methylases: Mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem 81: 65–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T, Butin-Israeli V, Adam SA, Hamanaka RB, Goldman AE, Lucas CA, Shumaker DK, Kosak ST, Chandel NS, Goldman RD 2011. The role of nuclear lamin B1 in cell proliferation and senescence. Genes Dev 25: 2579–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumaker DK, Dechat T, Kohlmaier A, Adam SA, Bozovsky MR, Erdos MR, Eriksson M, Goldman AE, Khuon S, Collins FS, et al. 2006. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc Natl Acad Sci 103: 8703–8708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taimen P, Pfleghaar K, Shimi T, Moller D, Ben-Harush K, Erdos MR, Adam SA, Herrmann H, Medalia O, Collins FS, et al. 2009. A progeria mutation reveals functions for lamin A in nuclear assembly, architecture, and chromosome organization. Proc Natl Acad Sci 106: 20788–20793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin BD, Meister P, Pike BL, Gasser SM 2010. Repetitive transgenes in C. elegans accumulate heterochromatic marks and are sequestered at the nuclear envelope in a copy-number- and lamin-dependent manner. Cold Spring Harb Symp Quant Biol 75: 555–565 [DOI] [PubMed] [Google Scholar]
- van Bemmel JG, Pagie L, Braunschweig U, Brugman W, Meuleman W, Kerkhoven RM, van Steensel B 2010. The insulator protein SU(HW) fine-tunes nuclear lamina interactions of the Drosophila genome. PLoS ONE 5: e15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW 2007. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445: 656–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AR, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JF, Tavare S, Arakawa S, Shimizu S, Watt FM 2009. Autophagy mediates the mitotic senescence transition. Genes Dev 23: 798–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Pan KH, Cohen SN 2003. Senescence-specific gene expression fingerprints reveal cell-type-dependent physical clustering of up-regulated chromosomal loci. Proc Natl Acad Sci 100: 3251–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Chen W, Adams PD 2007. Molecular dissection of formation of senescence-associated heterochromatin foci. Mol Cell Biol 27: 2343–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]