Here, Mohr and colleagues establish a role for the poly(A)-binding protein (PABP) repressor Paip2 in viral infection. The investigators find that human cytomegalovirus (HCMV) infection causes the up-regulation of Paip2 as well as PABP. The data indicate that Paip2 accumulation represents an innate host response to counteract the virus-induced increase in PABP abundance, limit the assembly of translation initiation factor complexes, and restrict viral growth. Paip2 thus plays a significant role in an innate defense mechanism to restrict viral protein synthesis and replication.
Keywords: Paip2, innate defense, HCMV replication, translation control, PABP, mTORC1
Abstract
The capacity of polyadenylate-binding protein PABPC1 (PABP1) to stimulate translation is regulated by its repressor, Paip2. Paradoxically, while PABP accumulation promotes human cytomegalovirus (HCMV) protein synthesis, we show that this is accompanied by an analogous increase in the abundance of Paip2 and EDD1, an E3 ubiquitin ligase that destabilizes Paip2. Coordinate control of PABP1, Paip2, and EDD1 required the virus-encoded UL38 mTORC1 activator and resulted in augmented Paip2 synthesis, stability, and association with PABP1. Paip2 synthesis also increased following serum stimulation of uninfected normal fibroblasts, suggesting that this coregulation may play a role in how uninfected cells respond to stress. Significantly, Paip2 accumulation was dependent on PABP accrual, as preventing PABP1 accumulation suppressed viral replication and inhibited the corresponding Paip2 increase. Furthermore, depleting Paip2 restored the ability of infected cells to assemble the translation initiation factor eIF4F, promoting viral protein synthesis and replication without increasing PABP1. This establishes a new role for the cellular PABP1 inhibitor Paip2 as an innate defense that restricts viral protein synthesis and replication. Moreover, it illustrates how a stress-induced rise in PABP1 triggered by virus infection can counter and surpass a corresponding increase in Paip2 abundance and stability.
The control of gene expression at the level of mRNA translation facilitates swift responses to an ever-changing environment and plays a fundamental role in the response to stress and viral infection (Mohr and Sonenberg 2012). To replicate productively, viruses must seize control of the cellular protein synthesis machinery and the complex networks that regulate its activity. At the same time, host defenses use translational control mechanisms to respond to microbial infection and mobilize innate host defenses (Walsh and Mohr 2011). While some of these responses can affect specific mRNAs, others can globally impact the translational capacity of the cell (Mohr and Sonenberg 2012; Walsh et al. 2013). As such, the steady-state level of translation initiation factors and repressors are poised to play a powerful role in controlling translation in infected and uninfected cells.
Unlike many viruses that impair host mRNA translation or inactivate cellular translation factors, host mRNA translation proceeds in human cytomegalovirus (HCMV)-infected cells (Stinski 1977). To ensure that viral mRNAs (which contain 7-methyl guanosine [m7G]-capped 5′ and 3′ polyadenylated termini) effectively compete for limiting translation factors, HCMV infection triggers an increase in the intracellular concentration of key initiation factors, including the cellular polyadenylate-binding protein PABPC1 (PABP1) and the multisubunit cap-binding complex eIF4F (Kudchodkar et al. 2004; Isler et al. 2005; Walsh et al. 2005; Perez et al. 2011). Comprised of the cap-binding protein eIF4E and the RNA helicase eIF4A bound to the large molecular scaffold eIF4G, eIF4F complex assembly is regulated on the m7GTP cap structure, facilitating 40S ribosome subunit loading onto the mRNA 5′ end (Jackson et al. 2010). PABP1 bound to the mRNA 3′ poly(A) tail stimulates translation by binding to eIF4G and in effect juxtaposes the mRNA 3′ end with the eIF4F complex assembled on the 5′ end, forming a closed loop or circular topology (Kahvejian et al. 2001). While the level of mRNAs encoding the individual eIF4F subunits increases upon infection, PABP1 abundance is controlled at the level of translation and is dependent on the virus-encoded mTORC1 activator encoded by the UL38 gene. Importantly, preventing the HCMV-induced rise in PABP1 levels interferes with eIF4F assembly, viral protein production, and viral replication (McKinney et al. 2012). Thus, successfully increasing the concentration of the host PABP1 represents an important aspect of the productive viral life cycle.
Multiple distinct regulatory pathways maintain PABP1 homeostasis in cells. An adenine-rich autoregulatory region in the PABP1 mRNA 5′ untranslated region (UTR) reportedly represses its synthesis (Wu and Bag 1998; Patel et al. 2005). As a member of an mRNA family that contains a 5′-terminal oligopyrimidine (TOP) region in the 5′ UTR, PABP1 synthesis is coordinately regulated at the level of translation, together with other host ribosomal protein genes and translation factors, and is stimulated in response to mitogenic, growth, and nutritional stimuli (Hornstein et al. 1999a). TOP mRNA translation is also stimulated by HCMV infection (McKinney et al. 2012). PABP1 activity, however, is controlled through its association with interacting protein partners (Derry et al. 2006). Besides associating with eIF4G, PABP1 interacts with Paip1 (an eIF4G-like molecule that stimulates translation) and Paip2 (a potent translation repressor) (Craig et al. 1998; Khaleghpour et al. 2001a; Roy et al. 2002; Martineau et al. 2008).
Paip2 inhibits translation by displacing PABP1 from the poly(A) tail at the mRNA 3′ end and preventing PABP1 from binding eIF4G (Khaleghpour et al. 2001b; Karim et al. 2006). To ensure proper control of gene expression, PABP1 and its repressor, Paip2, are tightly coregulated, as PABP1 depletion by RNAi triggers a corresponding reduction in Paip2. This requires binding of Paip2 to the E3 ubiquitin ligase EDD1, as opposed to PABP1, and illustrates how differential binding to EDD1 and PABP1 control Paip2 levels (Yoshida et al. 2006). A role for the converse scenario—namely, how Paip2 might respond to elevated PABP1 levels—has not been investigated and remains unknown, in part due to the difficulty in overexpressing PABP (Wormington et al. 1996; Wu and Bag 1998; Hornstein et al. 1999b; Ma et al. 2006). Here we show that EDD1 and newly synthesized Paip2 levels surprisingly increased in response to PABP1 accumulation induced upon infection with HCMV. This required expression of the virus-encoded UL38 mTORC1 activator. Paip2 stability also increased in HCMV-infected cells, and Paip2 was detected bound to PABP1 even though EDD1 levels were elevated. Significantly, besides suppressing eIF4F assembly, viral protein production, and replication, preventing the HCMV-induced increase in PABP1 abundance inhibited Paip2 accumulation. Moreover, depleting Paip2 restored virus protein production and replication in cells where the PABP increase was blocked by RNAi. Thus, Paip2 depletion obviates the need to increase PABP1 abundance in HCMV-infected cells. Taken together, this demonstrates that while increasing PABP and Paip2 levels are both dependent on mTORC1 signaling, Paip2 abundance specifically increases in response to PABP accumulation. This not only suggests that the coordinate increase in Paip2 synthesis and stability naturally limits PABP1 activity, but establishes that it can function as a previously unrecognized innate host defense with the capacity to restrict viral protein synthesis and replication. Sufficiently increasing cellular PABP1 levels allows HCMV to overcome this host restriction and effectively counter the potential negative impact of the Paip2 repressor. This is the first example illustrating how a virus can manipulate the PABP1/Paip2 homeostatic switch by increasing PABP1 levels to facilitate productive virus growth.
Results
PABP1 accumulation induced by HCMV infection is accompanied by a corresponding increase in Paip2 and EDD1 abundance
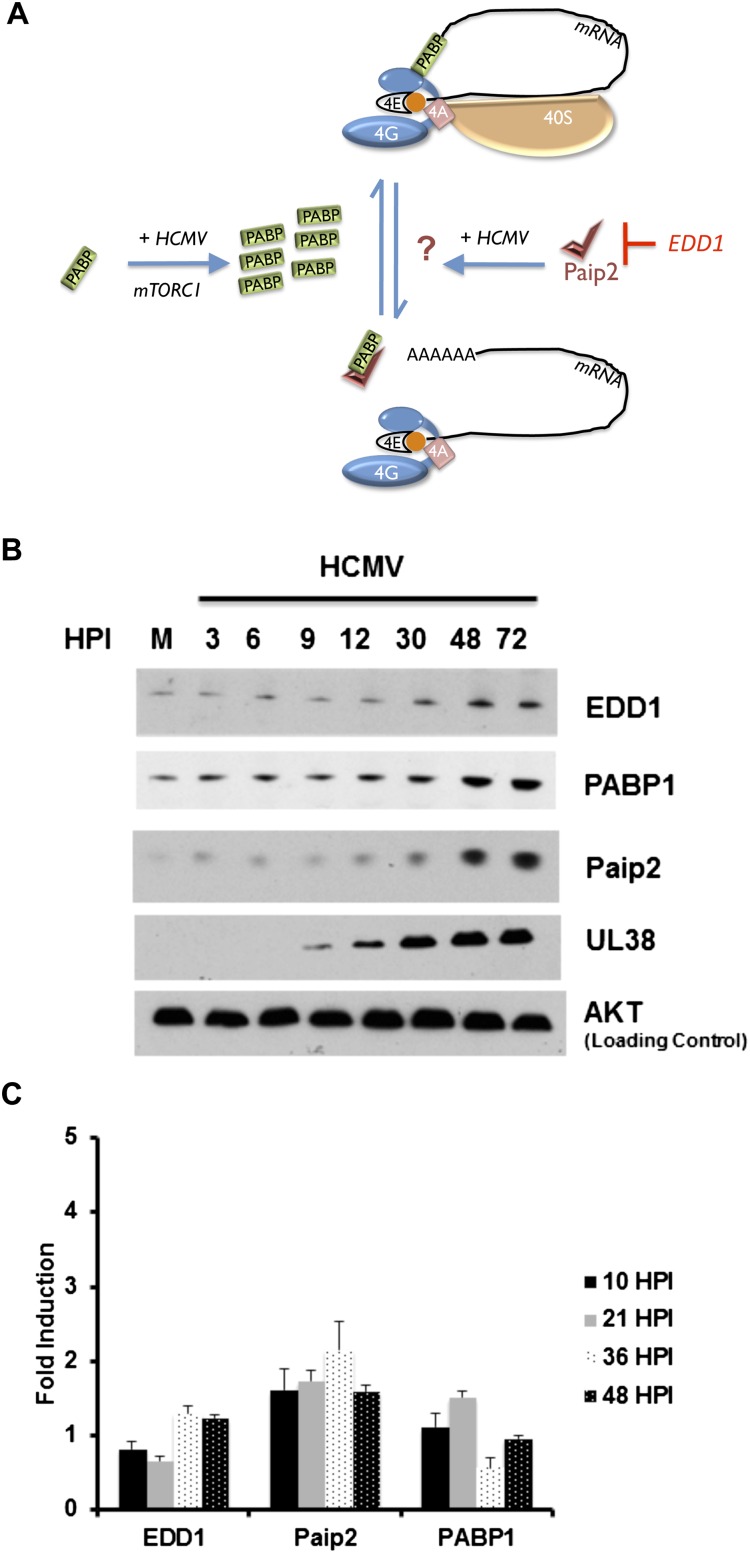
To investigate how PABP1 function is influenced by its endogenous cellular inhibitor, Paip2, in HCMV-infected cells, we first compared the overall abundance of Paip2 mRNA and protein in mock-infected versus HCMV-infected normal human diploid fibroblasts (NHDFs). Given that Paip2 inhibits PABP1 (Fig. 1A) and that the virus-induced increase in PABP1 levels stimulates HCMV replication, it was surprising that Paip2 and EDD1 both accumulated along with PABP1 between 30 and 48 h post-infection (hpi) (Fig. 1B). By 72 hpi, overall levels of Paip2 and EDD1 rose between fourfold and fivefold, and PABP abundance increased between sevenfold and eightfold (Supplemental Fig. S1). The HCMV-induced increase in EDD1 protein abundance was not accompanied by a detectable corresponding increase in steady-state mRNA levels, whereas the increase in Paip2 protein was only accompanied by a modest increase in mRNA (Fig. 1C), suggesting an underlying post-transcriptional control strategy similar to what we previously defined for PABP1 (Perez et al. 2011). Importantly, while reducing PABP1 abundance has been shown to trigger a corresponding decrease in Paip2 and EDD1 levels (Yoshida et al. 2006), this is the first demonstration that these factors are coordinately up-regulated. Since PABP1 function is controlled by the relative abundance of Paip2 and PABP1, the mechanisms underlying the coordinate regulation of PABP1, Paip2, and EDD1 in HCMV-infected cells were investigated.
Figure 1.
Accumulation of the cellular PABP repressor Paip2; its negative regulator, EDD1; and PABP1 in HCMV-infected cells. (A) Cartoon illustrating control of PABP homeostasis in response to serum stimulation or HCMV infection. PABP is encoded by an mRNA containing a TOP element and is repressed in growth-arrested cells (Hornstein et al. 1999a). Serum stimulation or HCMV infection activates mTORC1 and stimulates new PABP synthesis, which in turn promotes eIF4F assembly and 40S ribosome recruitment to the mRNA 5′ end (Perez et al. 2011; McKinney et al. 2012). The PABP repressor Paip2 regulates PABP1 function by (1) preventing its association with the mRNA poly(A) tail and (2) binding to PABP1 and preventing its association with eIF4G, both of which repress translation (Derry et al. 2006). Excess Paip2 that is not bound to PABP interacts with the E3 ubiquitin ligase EDD1, which targets Paip2 for proteolysis (Yoshida et al. 2006). While PABP depletion stimulates Paip2 degradation, how Paip2 responds to PABP accumulation is unknown and is depicted as a question mark. (B) Asynchronous NHDFs were mock-infected (0 hpi) or infected with HCMV at a multiplicity of 3. At the indicated times post-infection, total protein was isolated, fractionated by SDS-PAGE, and analyzed by immunoblotting with the indicated antisera. (C) As in B except at the indicated times post-infection, total RNA was isolated through Trizol extraction and subjected to RT-qPCR using the indicated primer sets. Each reaction product was normalized to the signal obtained using 18S rRNA, and the fold induction upon infection was calculated as described (Perez et al. 2011).
Paip2 is regulated by the HCMV-encoded UL38 mTORC1 activator
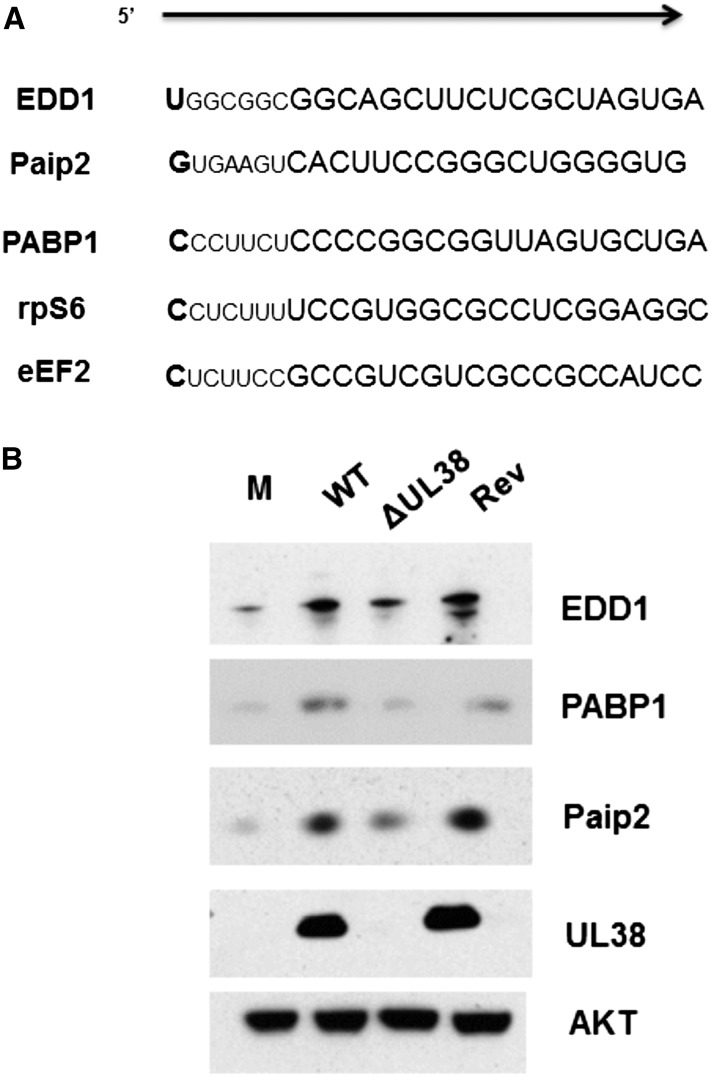
UL38 is a multifunctional HCMV-encoded early gene product that may serve different roles in discrete compartments of the cell (Tenney and Colberg-Poley 1991; Varnum et al. 2004; Qian et al. 2011). One characterized role for UL38 in positively regulating translation is binding to TSC2, inhibiting the tuberous sclerosis complex, constitutively activating mTORC1, and subsequently disabling the 4E-BP1 translation initiation inhibitor (Moorman et al. 2008). We showed that UL38 not only regulates PABP1 accumulation in an mTORC1-dependent manner that requires 4E-BP1 inactivation, but also translationally activates the TOP motif in the 5′ UTR of the mRNA (McKinney et al. 2012). While the TOP motif confers translational repression during nutrient and growth factor starvation and consists of a 5′ terminal cytosine residue followed by a 4- to 13-nucleotide (nt) sequence of pyrimidine residues (Meyuhas 2000), the Paip2 mRNA does not contain a canonical TOP motif in its 5′ UTR (Fig. 2A). To determine whether Paip2 accumulation in HCMV-infected cells was, like PABP1, dependent on UL38, NHDFs were mock-infected or infected with wild-type HCMV, a UL38-deficient HCMV, or a virus in which the UL38 mutation was repaired to wild type, and Paip2, PABP1, and EDD1 abundance was evaluated by immunoblotting. Although Paip2, PABP1, and EDD1 all accumulated in cells infected with wild-type HCMV, their accumulation was impaired in cells infected with a UL38-deficient virus (Fig. 2B). Infection with a revertant virus where the UL38 gene was reintroduced into the UL38-deficient (ΔUL38) HCMV genome restored robust accumulation of PABP1, Paip2, and EDD1 to near wild-type levels, proving that the coordinate accumulation of PABP, Paip2, and EDD1 was indeed UL38-dependent (Fig. 2B).
Figure 2.
Paip2 and EDD1 mRNAs do not contain a canonical TOP element but encode proteins that accumulate in a UL38-dependent manner together with PABP1. (A) Comparison of the Paip2 mRNA 5′ terminus with those of canonical TOP-bearing mRNAs, which contain a 5′-terminal cytosine residue followed by a 4- to 14-nt pyrimidine tract and a GC-rich region. (B) Asynchronous NHDFs were either mock-infected or infected (MOI = 3) with wild-type (WT) HCMV, a UL38-deficient mutant virus (ΔUL38), or a revertant virus where the UL38 deficiency was repaired by reintroducing a wild-type UL38 gene (Rev). At 48 hpi, total protein was isolated, fractionated by SDS-PAGE, and analyzed by immunoblotting with the indicated antisera.
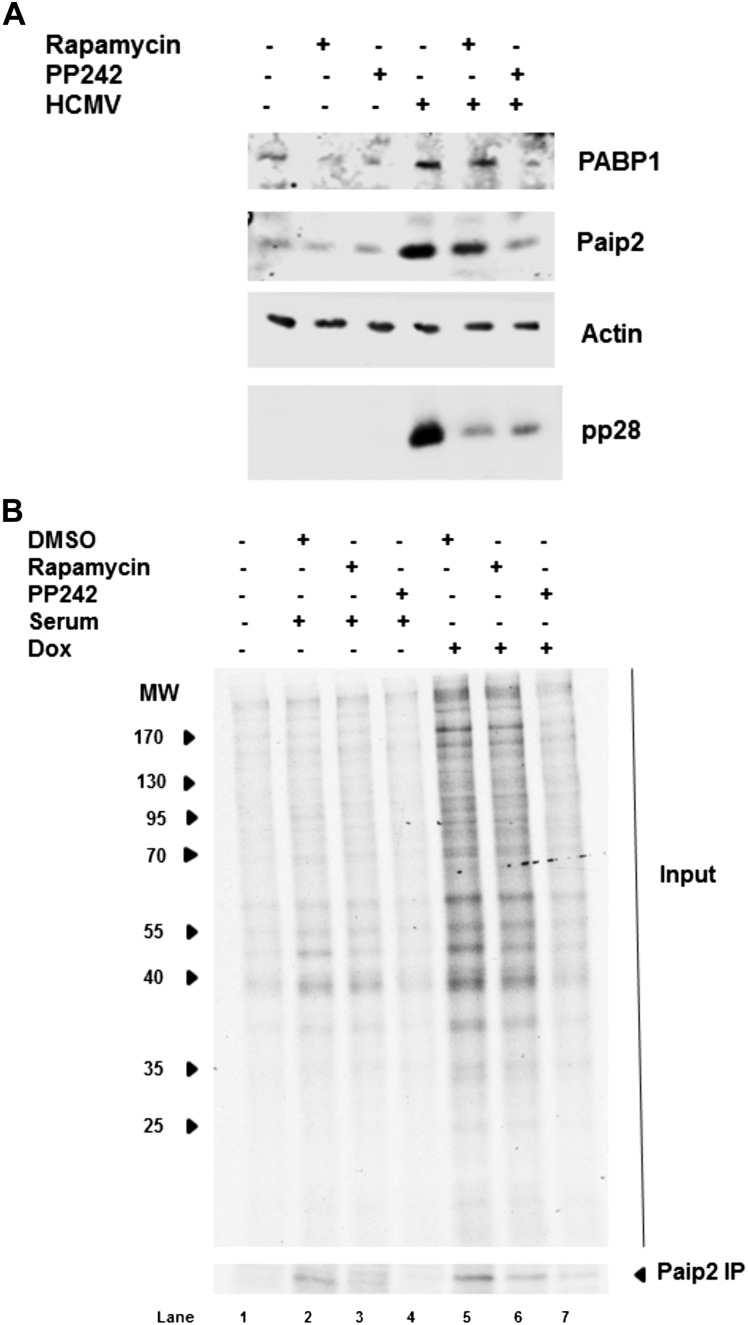
UL38 is a multifunctional protein that not only activates mTORC1, but also regulates apoptosis and the unfolded protein response (Terhune et al. 2007; Qian et al. 2011). In addition, the control of Paip2 abundance is incompletely understood. To determine whether mTOR activity in HCMV-infected cells regulates Paip2 steady-state levels, mock-infected and HCMV-infected NHDFs were treated with the mTORC1-selective inhibitor rapamycin or the mTOR active site inhibitor PP242 (Feldman et al. 2009). The virus-induced increase in PABP1 and Paip2 was subsequently evaluated by immunoblotting. Figure 3A demonstrates that Paip2 accumulation in HCMV-infected cells was reduced by rapamycin treatment and nearly abrogated by PP242. Thus, Paip2 accumulation in response to HCMV infection, similar to PABP accumulation, requires mTOR signaling. The greater reduction in PABP1 and Paip2 achieved with PP242 versus rapamycin could indicate that the levels of both proteins are controlled by 4E-BP1 phosphorylation, which is differentially sensitive to PP242 compared with rapamycin (Feldman et al. 2009). Indeed, 4E-BP1 phosphorylation controls the virus-induced PABP1 increase (Perez et al. 2011). Alternatively, as HCMV infection becomes resistant to rapamycin after 36 hpi, it is possible that mTORC2 could potentially be involved (Kudchodkar et al. 2004, 2006).
Figure 3.
Paip2 abundance in response to HCMV infection, serum stimulation, or UL38 induction is regulated by mTORC1. (A) Asynchronous NHDFs were either mock-infected or HCMV-infected in the presence of DMSO, the mTORC1-selective inhibitor rapamycin (100 nM), or the active site mTOR inhibitor PP242 (2.5 μM). At 48 hpi, total protein was collected, fractionated by SDS-PAGE, and analyzed by immunoblotting with the indicated antisera. (B, lanes 1–4) NHDFs that express UL38 upon induction with dox cells were growth-arrested by serum deprivation; treated for 1 h with DMSO, rapamycin (100 nM), or PP242 (2.5 μM); and subsequently untreated or serum-stimulated with 20% FBS for 20 min in the presence or absence of drug. (Lanes 5–7) Alternatively, cells were dox-treated for 72 h. Subsequently, cultures were metabolically pulse-labeled for 1 h with [35S]Met-Cys, and total protein was collected. A sample of the lysate was immunoprecipitated using anti-Paip2 antisera. Lysates (input panel) and isolated immune complexes (Paip2 panel) were fractionated by SDS-PAGE, and radiolabeled polypeptides were directly visualized by exposing the fixed, dried gel to X-ray film. The migration of molecular weight standards (in kilodaltons) is shown to the left of the top panel.
To determine whether ectopic expression of UL38 is sufficient to induce Paip2 accumulation in uninfected cells, primary NHDFs were transduced with a lentivirus expressing UL38 under the control of a doxycycline (dox)-inducible promoter. Following growth arrest by serum deprivation, the cells were either exposed to serum in the absence of dox or treated with dox in the absence of serum. After metabolic labeling of newly synthesized proteins with 35S amino acids for 1 h, total protein was harvested, fractionated by SDS-PAGE, and visualized by autoradiography (Fig. 3B). In addition, Paip2 synthesis was evaluated by immunoprecipitation. Both serum exposure in the absence of dox and dox treatment in the absence of serum were sufficient to stimulate total cellular protein synthesis and new Paip2 synthesis (Fig. 3B, cf. lanes 2–5). Importantly, UL38 induction was more effective than serum stimulation at stimulating 35S amino acid incorporation into total protein and newly synthesized Paip2. Thus, ectopic UL38 expression in uninfected cells in the absence of any other HCMV-encoded proteins is necessary and sufficient to increase Paip2 abundance.
To determine whether Paip2 synthesis in response to UL38 expression in uninfected NHDFs requires mTOR activity, serum-deprived NHDFs that express inducible UL38 were treated with serum or dox in the presence of rapamyin or PP242. Figure 3B shows that either serum or UL38 induced Paip2 accumulation in a rapamycin or PP242-sensitive manner. Thus, new Paip2 synthesis and accumulation in uninfected NHDFs is induced in response to UL38 expression in a manner dependent on mTORC1 activity. Notably, unlike PABP mRNA, whose translation is also stimulated by mTORC1, Paip2 mRNA does not contain a canonical TOP element, raising the possibility that Paip2 synthesis and accumulation are intrinsically responsive to PABP abundance.
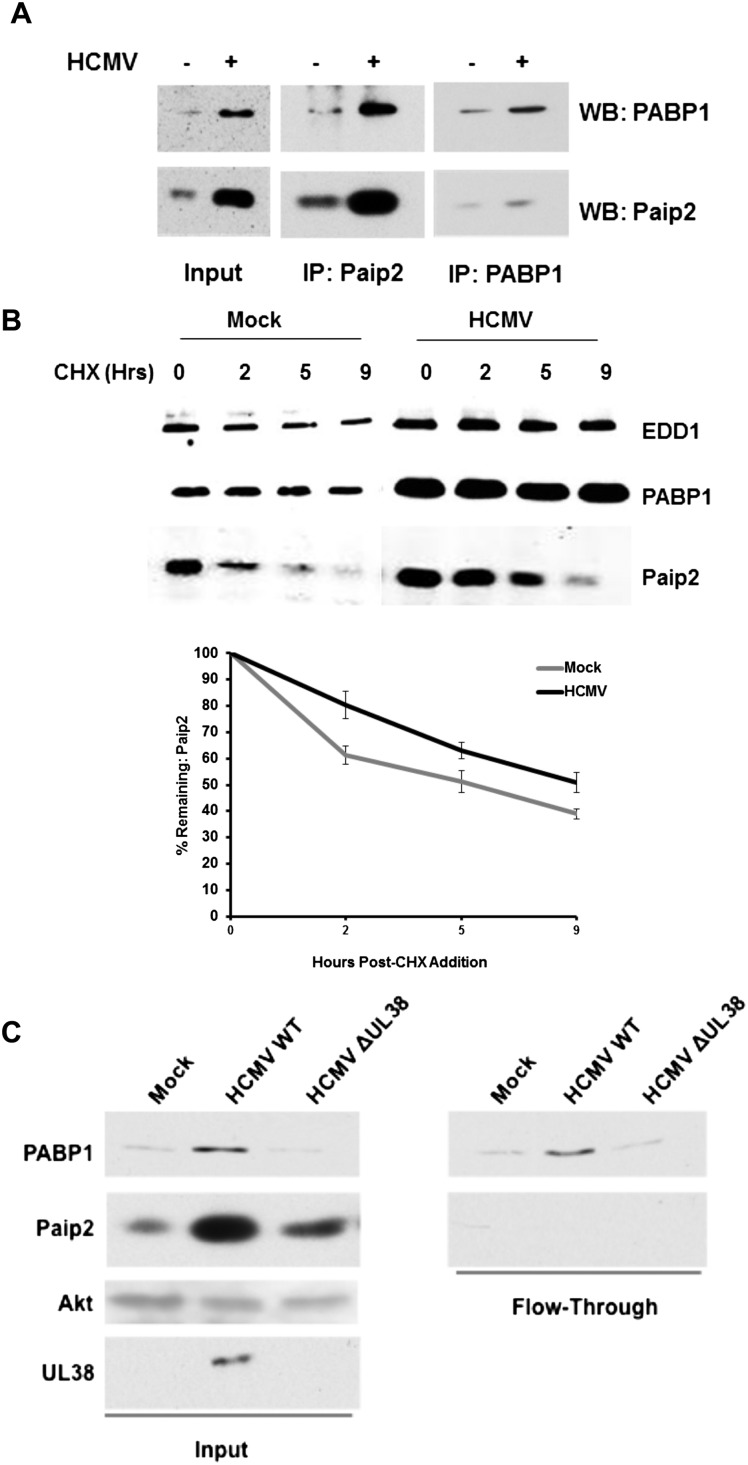
Paip2 stability increases during infection through increased binding to PABP1
By associating with PABP1 in a 2:1 stoichiometry via two conserved PABP-interacting motifs (PAM1 and PAM2), Paip2 inhibits PABP1 (Karim et al. 2006). When PABP1 is limiting, PAM2 binds to the PABC domain of PABP1 or EDD1 (Yoshida et al. 2006). To understand how the virus might benefit from increasing PABP1 while simultaneously stimulating accumulation of Paip2 and EDD1, the possibility that the association of PABP1 with Paip2 during infection might be impaired was investigated. Nonionic detergent lysates prepared from mock-infected or HCMV-infected cultures were immunoprecipitated using anti-Paip2 or anti-PABP1 sera, and the amount of Paip2 and PABP1 present in immune complexes was evaluated by immunoblotting. While the total levels of both proteins increased in HCMV-infected cells (see Input in Fig. 4A), it was surprising that (1) PABP1 and Paip2 remained associated, and (2) the overall amount of associated Paip2 and PABP increased (Fig. 4A). However, it remained possible that the ratio of free PABP1 to free Paip2 could be altered during infection in a manner dependent on their rates of synthesis and turnover. To determine the impact of HCMV infection on the stability of PABP and Paip2, primary NHDFs either mock-infected or infected with HCMV were treated with cycloheximide (CHX) to attenuate new protein synthesis, and total EDD1, Paip2, and PABP1 levels were measured over a 9-h time period by immunoblotting. The efficacy of CHX treatment was verified by monitoring the inhibition of 35S amino acid incorporation into acid-insoluble material (data not shown). While PABP1 and EDD1 levels remained fairly consistent throughout the CHX treatment in mock-infected cells, decreased Paip2 levels were readily detected shortly after treatment (Fig. 4B). In contrast, Paip2 abundance decreased more slowly in HCMV-infected cells than in mock-infected cells. Quantification of this decay revealed that Paip2 half-life nearly doubled in HCMV-infected cells, as Paip2 levels decreased by 50% after 5 h of CHX treatment in uninfected cells versus 9 h in infected cells (Fig. 4B). Since Paip2 homeostasis is dependent on its ability to bind PABP1 (Yoshida et al. 2006), variation in Paip2 stability likely reflects changes in the balance of free PABP1 to free Paip2. In this case, increasing PABP1 concentration in HCMV-infected cells would serve as a sink to stabilize the pool of newly synthesized Paip2.
Figure 4.
Paip2 associates with PABP and is stabilized in HCMV-infected cells. (A) Soluble cell-free extracts were prepared from asynchronous NHDFs that were either mock-infected (−) or HCMV-infected (+) at 48 hpi (MOI = 3). Samples were immunoprecipitated (IP) using anti-Paip2 or anti-PABP1 antisera. Isolated immune complexes and a sample of the input extract were fractionated by SDS-PAGE and analyzed by Western blotting (WB) using antisera specific for Paip2 or PABP1. (B) NHDFs that were mock-infected or infected with HCMV (MOI = 3) were treated with 100 μg/mL CHX at 48 hpi. At the indicated times (hours) following CHX addition, total protein was isolated, fractionated by SDS-PAGE, and analyzed by immunoblotting using the indicated primary antibodies and a secondary antibody covalently linked to an infrared fluorophore. The membrane was scanned, and the fraction of Paip2 remaining at each time point was quantified using an Odyssey infrared imager. Each band was measured for raw intensity value, and each time point was normalized to the amount of Paip2 present at time 0. (C) Paip2 was immunodepleted from soluble cell-free extracts prepared from asynchronous NHDFs that were either mock-infected (Mock) or HCMV-infected (wild type vs. ΔUL38) at 48 hpi (MOI = 3). Samples of the input extracts and the fractions not bound to anti-Paip2 antibody (flow-through) were fractionated by SDS-PAGE and analyzed by immunoblotting using the indicated antisera.
A key prediction of this model is that the virus-induced increase in PABP1 concentration is sufficient to maintain a pool of free PABP not bound to Paip2. To test this hypothesis, the amounts of free PABP not bound to Paip2 in cell-free lysates prepared from NHDFs infected with wild-type HCMV or a UL38-deficient virus (ΔUL38) that is unable to increase PABP abundance were compared. Initial levels of PABP1, Paip2, and UL38 were first evaluated in input lysates prepared from mock-infected versus HCMV-infected (wild type vs. ΔUL38) by immunoblotting. As expected, PABP1 and Paip2 abundance both increased in a UL38-dependent manner (Fig. 4C). However, following immunodepletion of Paip2 from cell-free lysates, more PABP remained in the unbound fraction isolated from wild-type HCMV-infected cells compared with cells infected with ΔUL38 or uninfected cells (Fig. 4C). Thus, the HCMV-induced PABP1 increase is sufficient to increase the concentration of free PABP1 not bound to its inhibitor, Paip2, in a UL38-dependent manner.
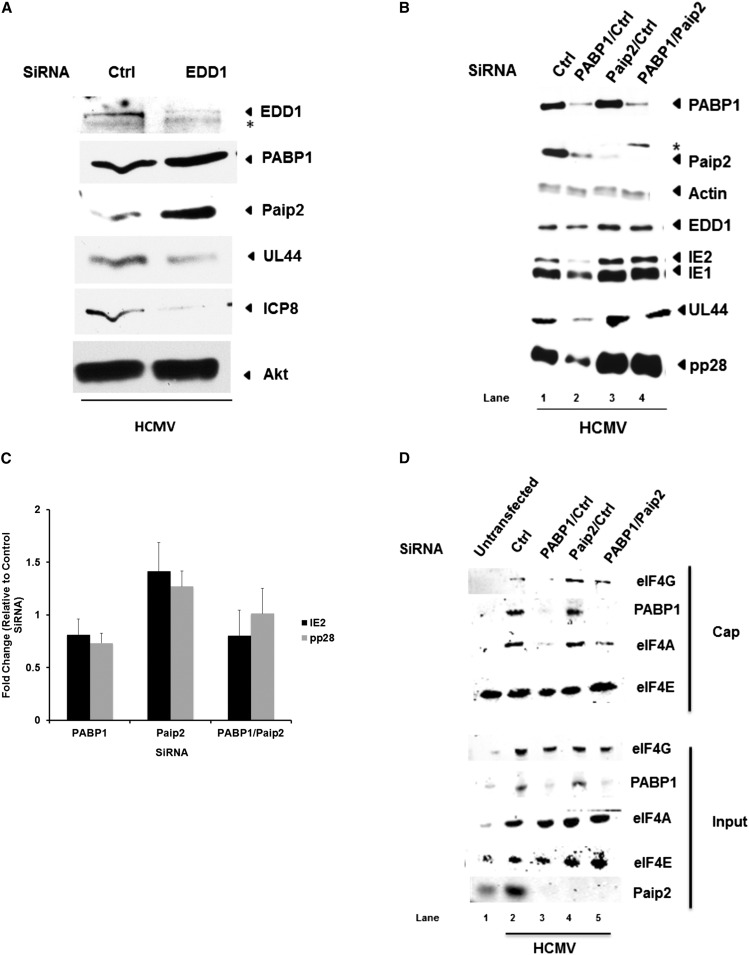
Paip2 accumulation is triggered by the HCMV-induced PABP1 increase and regulates eIF4F assembly in infected cells
Having shown previously that the virus-induced PABP1 increase promoted virus replication, it was puzzling to observe a concomitant rise in the level and stability of its cognate repressor, Paip2, along with its negative regulator, EDD1. Thus, we next considered the hypothesis that HCMV protein accumulation and replication were in fact controlled by the dynamic interplay between PABP1 and Paip2 levels. By reducing the abundance of the E3 ubiquitin ligase EDD1, binding of Paip2 to PABP1 increases, stabilizing Paip2 and inhibiting PABP1 function (Yoshida et al. 2006). Indeed, Paip2 abundance, but not PABP1 abundance, increased upon EDD1 depletion in HCMV-infected cells. (Fig. 5A). Moreover, Paip2 accrual induced by EDD1 depletion coincided with a reduction in overall levels of representative viral proteins (UL44 and ICP8) and suggests that further increasing Paip2 concentration in HCMV-infected cells limits viral protein accumulation (Fig. 5A). Preventing the virus-induced PABP1 increase using RNAi in HCMV-infected cells markedly reduced accumulation of viral gene products representing different temporally expressed classes of viral genes (Fig. 5B, cf. lane 1 vs. 2) in agreement with our earlier work (McKinney et al. 2012). Moreover, we now demonstrate that it also resulted in a corresponding reduction in Paip2 levels. Notably, no differences in PABP1 or viral protein accumulation were detected when Paip2 was depleted with siRNA (Fig. 5B, cf. lane 1 vs. 3). This raised the possibility that PABP1 is produced in vast excess in order to saturate the inhibitory effects of Paip2.
Figure 5.
Increased Paip2 abundance in response to virus-induced PABP accumulation regulates eIF4F assembly and HCMV protein production. (A) NHDFs transfected with the indicated siRNA (control nonsilencing siRNA [ctrl] or EDD1) were infected with HCMV at MOI = 0.1. After 5 d, total protein was collected, fractionated by SDS-PAGE, and analyzed by immunoblotting with the indicated antisera. (*) A nonspecific, cross-reacting band. (B) NHDFs transfected with a control nonsilencing siRNA (ctrl) or the indicated pairs of siRNAs were infected with HCMV at MOI = 0.1. After 5 d, total protein was collected and analyzed as in A. (C) As in B, except that total RNA was isolated using Trizol and subjected to RT-qPCR using primers specific for the HCMV IE2 or pp28 genes. Each reaction product was normalized to the signal obtained using primers specific for 18S rRNA and expressed as the fold change relative to HCMV-infected cells treated with control nonsilencing siRNA. (D) As in B, except that siRNA-treated NHDFs were mock-infected or HCMV-infected (MOI = 3). After 5 d, cell-free extracts prepared using a nonionic detergent were subject to batch chromatography on m7GTP Sepharose. A sample of input extract or m7GTP-bound proteins was fractionated by SDS-PAGE and analyzed by immunoblotting with the indicated antisera.
To test the hypothesis that raising PABP1 abundance in HCMV-infected cells is required to counteract the inhibitory impact of Paip2, the remaining pool of Paip2 was depleted in cells where the HCMV-induced PABP increase was blocked using RNAi. Remarkably, Paip2 depletion in HCMV-infected cells that were unable to increase PABP1 effectively restored viral protein production to levels observed in cultures treated with control, nonsilencing siRNA, or Paip2 siRNA alone (Fig. 5B, cf. lanes 4 and 3 or 1). Thus, similar levels of viral protein accumulation can be achieved by depleting Paip2, effectively obviating the need to increase intracellular PABP concentration. This suggests that rising Paip2 levels function to restrict viral replication, and this is effectively counteracted by raising PABP1 concentration. It further implies that the increase in Paip2 abundance occurs in response to rising PABP levels.
To understand the underlying mechanism whereby Paip2 depletion allows HCMV protein accumulation to proceed without raising intracellular PABP1 levels, the varied ways by which PABP1 post-transcriptionally controls gene expression were examined. PABP1 regulates mRNA stability by protecting mRNAs from deadenylation-mediated decay, and this could account for the observed changes in viral protein accumulation (for review, see Mangus et al. 2003). However, the steady-state abundance of HCMV pp28 and IE2 mRNAs did not detectably change when infected cells were treated with siRNAs targeting PABP, Paip2, or both PABP and Paip2 (Fig. 5C). PABP is also a translation initiation factor (Kahvejian et al. 2005). While HCMV infection, unlike many viruses, does not normally impair ongoing cellular protein synthesis, altering the balance between PABP and Paip2 may impact the efficiency with which viral versus host mRNAs access the translation initiation machinery. Although preventing the HCMV-induced PABP increase modestly reduced global infected cell protein synthesis, the overall profile of protein synthesis in infected cells was not grossly altered by Paip2 depletion in the presence or absence of PABP siRNA treatment (Supplemental Fig. S2). Thus, translation of viral mRNAs was not detectably enriched at the expense of cellular mRNAs.
Besides stimulating protein accumulation, the HCMV-induced PABP increase effectively stimulated assembly of eIF4E, eIF4G, and eIF4A into the heterotrimeric eIF4F translation initiation factor complex (Kudchodkar et al. 2004; Walsh et al. 2005; McKinney et al. 2012). To evaluate the impact of perturbing PABP/Paip2 homeostasis on eIF4F assembly, cell-free lysates prepared from NHDFs treated with siRNAs and subsequently mock-infected or infected with HCMV were subjected to batch chromatography using m7GTP Sepharose. After washing the resin to remove unbound and nonspecifically bound components, bound proteins were fractionated by SDS-PAGE and analyzed by immunoblotting to evaluate binding of eIF4G and eIF4A to the cap-binding protein eIF4E (Fig. 5D). While preventing the HCMV-induced PABP1 increase using RNAi substantially reduced the association of eIF4G and eIF4A with eIF4E bound to the m7GTP cap affinity resin (Fig. 5D, cf. lanes 2 and 3), Paip2 depletion modestly increased eIF4G binding to eIF4E compared with lysates prepared from cultures treated with control, nontargeting siRNA (Fig. 5D, lane 2 vs. 4). Remarkably, Paip2-depletion restored eIF4G and eIF4A binding to eIF4E in cells where the HCMV-induced PABP increase was inhibited by PABP siRNA. This was specific for HCMV-infected cells and not observed in uninfected cells (Supplemental Fig. S3). Thus, the defect in translation initiation factor eIF4F assembly resulting from failure to increase PABP1 concentration in HCMV-infected cells can be effectively corrected by Paip2 depletion. Furthermore, it implies that existing supplies of Paip2 effectively restrict eIF4F assembly, and this can be counteracted by a commensurate virus-induced rise in PABP1 concentration.
Paip2 is a host antiviral restriction factor antagonized by PABP accumulation in HCMV-infected cells
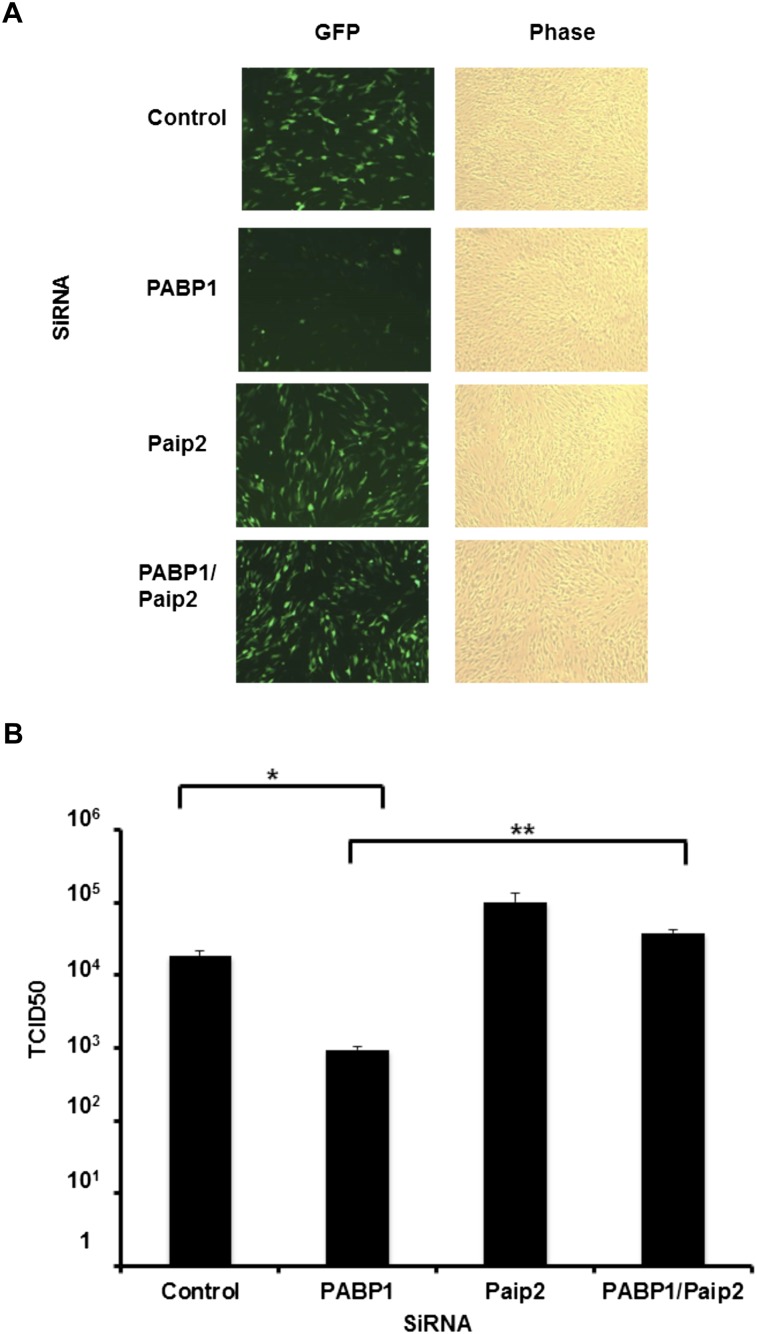
To determine whether Paip2 acts as a restriction factor that limits viral productive growth and spread in cells where the HCMV-induced PABP increase is blocked, NHDFs treated with siRNA (control, PABP1, Paip2, or both PABP and Paip2) were infected with HCMV, and viral replication was evaluated. Under these conditions (multiplicity of infection [MOI] = 0.1), preventing the virus-induced PABP increase using PABP1 siRNA reduced viral replication, whereas Paip2 depletion had a very slight but not significant impact. Significantly, depleting Paip2 in cells where the HCMV-induced PABP increase was blocked resulted in a 40-fold increase of viral replication, restoring viral growth in NHDFs to levels observed in cultures treated with control, nonsilencing siRNA or Paip2 siRNA alone (Fig. 6A,B). Thus, the HCMV-induced increase in PABP1 levels controls viral growth by antagonizing the corresponding increase in the host Paip2 inhibitor. This establishes that the cellular Paip2 can function as a host restriction factor to antagonize viral replication. Furthermore, it provides the first example of a virus that manipulates the PABP/Paip2 axis to specifically counteract Paip2 and foster viral protein accumulation and replication.
Figure 6.
Endogenous Paip2 functions to restrict productive HCMV growth when the virus-induced PABP accumulation is blocked. (A) NHDFs transfected with a control nonsilencing siRNA (ctrl) or the indicated pairs of siRNAs were infected with HCMV (MOI = 0.1) and imaged for EGFP expression 5 d post-infection. (B) Supernatants collected from three independent experiments detailed in A were assayed for viral particle production based on their TCID50 (Heider et al. 2002). Error bars indicate standard error of the mean. (*) P < 0.022 for control versus PABP1 siRNA; (**) P < 0.01 for PABP1 versus PABP1/Paip2 siRNA. Control versus Paip2 was not statistically significant (P > 0.1).
Discussion
To ensure that viral mRNAs effectively compete with cellular mRNAs for limiting translation factors, HCMV uses an unusual strategy and forces its host to increase the overall abundance of several critical initiation factors, one of which is PABP1 (Isler et al. 2005; Walsh et al. 2005). The resulting accumulation of PABP1 in HCMV-infected cells is required to stimulate eIF4F assembly, viral protein production, and replication (McKinney et al. 2012). Here, we show that the PABP1 repressor Paip2 and EDD1, a ubiquitin E3 ligase that regulates Paip2 stability, unexpectedly increase together with PABP upon infection. The coordinate regulation of PABP1, Paip2, and EDD1 is dependent on the HCMV mTORC1 activator encoded by the UL38 gene. Preventing the HCMV-induced increase in PABP1 abundance not only impaired viral protein production and replication, but also inhibited the rise in Paip2 levels. Surprisingly, depleting Paip2 in cells where the HCMV-induced PABP increase was blocked restored their ability to support viral protein production and replication without a commensurate increase in PABP1 levels. This establishes a role for the host PABP inhibitor Paip2 in infection biology, where it can function as an innate defense that restricts viral protein synthesis and replication. Moreover, it provides the first example of how a pathogen can manipulate the cellular PABP1–Paip2 homeostatic axis by raising PABP1 levels to overwhelm the functional capacity of its inhibitor, Paip2.
While previous studies showed that depleting PABP1 coordinately reduced Paip2 stability in a manner requiring the ubiquitin E3 ligase EDD1 (Yoshida et al. 2006), the effect of increasing PABP1 concentration on Paip2 levels was never addressed, in part due to difficulties associated with overexpressing PABP (Wormington et al. 1996; Wu and Bag 1998; Hornstein et al. 1999b; Ma et al. 2006). Indeed, the biological benefit of destabilizing Paip2 when PABP1 levels fall intuitively preserves a pool of functional PABP1, which would otherwise be inhibited by Paip2 under these circumstances (Yoshida et al. 2006). The rise in Paip2 and EDD1 that accompanies the PABP1 increase, however, is more perplexing at first glance, as it has the potential to neutralize the investment involved in making more PABP1. Perhaps the coordinate increase in activator and inhibitor allows for a ready-made supply of inhibitor that can be rapidly enlisted to upset the delicate PABP1–Paip2 balance and thereby antagonize the newly set, elevated PABP1 levels. As PABP1 binds to mRNA in both noncooperative and cooperative modes, the latter involving homophillic protein–protein interactions that promote PABP multimerization on the mRNA poly(A) tail (Kühn and Pieler 1996; Melo et al. 2003; Lin et al. 2012), relatively small changes in the free PABP pool could have a significant impact on binding of the initial subunits to poly(A) mRNA termini. Thus, besides expanding the available pool of free PABP not bound to Paip2, the virus-induced PABP1 increase likely ensures that the intracellular PABP concentration is sufficient for PABP binding to and subsequent multimerization on mRNA. Alternatively, there may also be ways to increase PABP1 without elevating Paip2 or EDD1, and the corresponding rise in Paip2 could be viewed as a host response to infection or stress. In support of this hypothesis, preventing the HCMV-induced PABP increase also suppressed Paip2 accumulation, suggesting that increasing Paip2 abundance is a host response to elevated PABP concentration.
While cellular translation repressors that regulate eIF4F assembly from its component subunits (eIF4E, eIF4G, and eIF4A) are inactivated in herpesvirus-infected cells (Walsh and Mohr 2011), this is the first demonstration of a role for the PABP1–Paip2 axis. Assembly of eIF4F is controlled in part by the availability of its eIF4E subunit, which is regulated by the abundance and phosphorylation status of its repressor protein, 4E-BP1 (Gingras et al. 1999). Remarkably, the levels of the repressors 4E-BP1 and Paip2 are controlled by a homeostatic mechanism that responds to the abundance of their respective cognate initiation factors, eIF4E and PABP1 (Yoshida et al. 2006; Yanagiya et al. 2010). Furthermore, both repressors appear to be regulated by mTORC1 signaling, which is constitutively activated in HCMV-infected cells by the TSC2-binding protein UL38. Notably, antagonizing translation repressors like 4E-BP1 and now Paip2 by virus-encoded factors in infected cells is consistent with the notion that host translation control pathways function as innate defenses with the capacity to restrict productive viral replication. Indeed, the observation that Paip2 depletion restores viral replication in infected cultures where the virus-induced PABP1 increase is blocked implies that Paip2 restricts viral replication by limiting PABP1 availability. Furthermore, restoration of replication to levels that exceed wild type by depleting Paip2 in cells where the HCMV-induced PABP increase was blocked suggests that the scope of PABP1 function can be maintained with far less PABP1 than is induced by HCMV. It also implies that translationally up-regulating PABP1 is meant to overcome the antiviral response of increasing Paip2. Additional virus-encoded functions, including the UL69 RNA-binding protein that associates with PABP and eIF4A (Aoyagi et al. 2010), may also help preserve PABP function by limiting its availability to interact with Paip2.
Although Paip2 clearly is an important inhibitor of PABP1 function, spermatogenesis in mice, growth control in Drosophila, synaptic plasticity, and memory in mice are the only biological process with a documented role for Paip2 (Roy et al. 2002; Yanagiya et al. 2010; Khoutorsky et al. 2013). Our study establishes that Paip2 can play a significant role in infection biology by altering the balance of PABP1 with its cognate repressor. Importantly, infection is a potent stress, and translational control is a major means whereby stress-induced gene expression is controlled. This raises the possibility that Paip2 might play a more global role in controlling PABP1 levels in response to stress in uninfected cells. Future studies in other model systems are required to address this possibility and uncover any parallels with changes to Paip2, EDD1, and PABP1 in HCMV-infected cells.
Even though Paip2 protein synthesis increases in an mTORC1-dependent manner, Paip2 mRNA does not contain a TOP sequence element. While TOP mRNA translation is stimulated in HCMV-infected cells (McKinney et al. 2012), Paip2 represents the first host mRNA whose translation is specifically controlled in infected cells and is not a TOP family member. Thus, while mTORC1 activation controls translation of canonical TOP mRNAs in HCMV-infected cells, non-TOP-containing mRNAs like Paip2 are poised to respond to other translational regulators, like PABP. By not suppressing host mRNA translation, HCMV provides a unique model to evaluate how differential host mRNA translation might impact infection. Translationally regulated host factors might be required, like PABP1, to stimulate viral replication or contribute to host defenses and thereby restrict viral replication like Paip2. Although the extent to which host mRNAs are translationally controlled in HCMV-infected cells and their contribution to virus biology remain to be elucidated, they could reveal a powerful means to control viral replication and allow viruses to access a wealth of functions encoded by host genomes. Likewise, it may also provide infected hosts with a valuable opportunity to attempt to restrict viral replication and identify new innate defense effectors whose synthesis is transitionally regulated. Finally, the HCMV model system may provide valuable insight into how different mRNA populations are either recruited or excluded from polyribosomes.
Materials and methods
Cell culture and viruses
NHDFs (purchased from Clonetics) were routinely subcultured by a 1:3 split and maintained until passage 20 in DMEM supplemented with 5% FBS, 1% L-glutamine, and 1% penicillin/streptomycin (v/v). GFP-expressing HCMV (AD169 strain) wild-type, UL38-null (ΔUL38), or UL38 revertant (REV) viruses were propagated as described (Terhune et al. 2007). Akt inhibitor VIII was purchased from Sigma.
Quantitative PCR (qPCR), antibodies, and Western blotting
RNA was isolated from cell-free lysates and processed into cDNA for qPCR as described (McKinney et al. 2012). The following primers were used for qPCR analysis: PABP1 (fw, CCCAGCTGCTCCTAGACC; and rev, GAGTAGCTGCAGCGGCT), Paip2 (purchased from Origene), EDD1 (UBR5; purchased from Origene), 18s (fw, AGGAATTGACGGAAGGGCACCA; and rev, TTATCGGAATTAACCAGACAAATCG), pp28 (fw, TGCTCTGGGTCGCCAGGTGT; and rev, CAGCCACTACCGCAGAGCC), and IE2 (fw, CGGGTGGATGTGTCACGGGC; and rev, ACGCACCCGCTCTCCCAGA).
Protein lysates were collected and subjected to SDS-PAGE and Western blotting as described (Walsh and Mohr 2004). Anti-PABP1 rabbit polyclonal was a gift from Simon Morley (University Sussex, UK), and anti-UL38 mouse monoclonal was a gift from Tom Shenk (Princeton University). The following primary antibodies were purchased commercially: anti-Paip2 (Sigma, no. P0087), anti-actin (Calbiochem, no. CP01), anti-EDD1 (Bethyl Laboratories, no. A300-573A-2), anti-Akt (Cell Signaling, no. 9272); anti-pp28 (Abcam, no. ab6502), anti-IE1/2 (Millipore, no. MAB810), and anti-UL44 (Virusys, no. CA006). For quantitative immunoblotting, a secondary antibody covalently linked to an infrared fluorophore was used (Li-Cor Biosciences, no. 827-08365) and the membrane was scanned using an Odyssey infrared imager (Li-Cor Biosciences).
Protein half-life analysis, immunoprecipitation, and Paip2 immunodepletion.
Mock-infected or HCMV-infected cells were treated with 100 μg/mL CHX (Research Products International Corp.) for up to 9 h. Western blots were quantified (n = 3) using an Odyssey infrared imager (Li-Cor Biosciences). To determine the efficacy of CHX treatment, cells were pulse-labeled with 35S-labeled amino acids at the indicated times, and the amount of acid-insoluble radioactivity present in cell-free lysates was quantified as described (McKinney et al. 2012).
Paip2 was immunoprecipitated from cell-free lysates prepared from mock-infected or HCMV-infected NHDFs (1.3 × 106 cells per sample) harvested at 48 hpi. After washing with cold PBS, cells were suspended in NP40 lysis buffer (50 mM HEPES at pH 7.4, 100 mM NaCl, 1.5 mM MgCl2, 2 mM EDTA, 0.25% NP40, 1× Roche Phos Stop, protease inhibitor tablets). Lysates were gently rocked for 30 min at 4°C, and, subsequently, lysates were clarified by centrifugation (12,000g) for 10 min at 4°C. CaCl2 (1 mM final concentration) was added to soluble supernatants (0.5 mL), which were subsequently treated with a nuclease cocktail (4 U of micrococcal nuclease [New England Biolabs, no. M0247S], 150 U of RNase T1, 3.75 U of RNase A [Ambion, no. AM2286]) for 20 min at room temperature. Nuclease-treated lysates were precleared by adding 5 μg of purified normal rabbit serum (Invitrogen, no. 17-0780-01) for 1 h at 4°C, and nonspecific binding proteins were subsequently collected by incubation with 10 μL of packed bed volume of protein A Sepharose CL-4B (GE Healthcare, no. 17-780-01) for 1 h at 4°C. An aliquot of soluble, precleared extract (10%) was removed (input fraction), diluted 1:2 in SDS sample buffer, and reserved for later analysis. Anti-Paip2 antibody (5 μg) was added to the remainder of the precleared lysate, and the mixture used to resuspend 10 μL of packed bed volume of protein A Sepharose. After overnight incubation at 4°C, the beads and supernatant were partitioned by brief centrifugation in a microfuge, and the soluble flow-through fraction was removed for analysis by immunoblotting (together with the input fraction). For analysis of Paip2 and Paip-2-associated proteins, the beads were next washed three times (0.5 mL per wash) with NP40 lysis buffer, suspended in SDS-containing sample buffer, and boiled for 3 min. Immune complexes were fractionated by SDS-PAGE and analyzed by immunoblotting. For analysis of PABP and PABP-associated proteins, a similar immunoprecipitation protocol was performed using anti-PABP antibody.
Generating UL38-expressing cells and 35S metabolic labeling
To generate a dox-inducible, UL38-expressing cell line, the 990-base-pair (bp) UL38 gene was PCR-amplified from a pRetro-EBNA-UL38 vector (Moorman et al. 2008) with fw (GACAGGAACTAGTATACCACGCATAGCACT) and rev (ATACGGCCTCGAGCTGACCACCATCTGTAC) primers bearing SpeI and XhoI overhangs, respectively. The product was then digested, purified, and cloned into a pSLIK tet-inducible vector as described (Kobayashi et al. 2012).
Lentiviruses were produced as described (Kobayashi et al. 2012), and a population of transduced cells was selected using 50 μg/mL hygromycin B (Life Technologies).
RNAi, microscopy, and viral replication assay
PABP1, Paip2, and EDD1 siRNA smartpools were purchased from Dharmacon (catalog nos. M019598, M015376, and M007189, respectively). Transfections of siRNAs, low MOI HCMV infections, and viral replication assays were conducted as described (McKinney et al. 2012). For depleting EDD1, 20 nM siRNA was transfected. For depleting two target genes, 20 nM siRNA was transfected for each target (40 nM siRNA total siRNA). EGFP expression was used to measure HCMV spread and was visualized under 5× magnification using a Zeiss Axiovert 200 fluorescent microscope. A paired two-tailed t-test was used to calculate P-values.
Acknowledgments
We thank S. Morley, R. Schneider, and T. Shenk for helpful reagents; D. Walsh and H. Burgess for their critical review of the manuscript; and members of the Mohr laboratory, R. Schneider, and A. Wilson for many helpful discussions. This work was supported by National Institutes of Health (NIH) grants AI073898 and GM056927 to I.M., and CA120768 to D.Y. C.M. was supported by NIH training grant T32 AI007647.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.221341.113.
References
- Aoyagi M, Gaspar M, Shenk T 2010. Human cytomeglovirus UL69 protein facilitates translation by associating with the mRNA cap-binding complex and excluding 4E-BP1. Proc Natl Acad Sci 107: 2640–2645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig AW, Haghighat A, Yu AT, Sonenberg N 1998. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP1 enhances translation. Nature 392: 520–523 [DOI] [PubMed] [Google Scholar]
- Derry MC, Yanagiya A, Martineau Y, Sonenberg N 2006. Regulation of poly(A)-binding protein through PABP-interacting proteins. Cold Spring Harb Symp Quant Biol 71: 537–543 [DOI] [PubMed] [Google Scholar]
- Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D, Shokat KM 2009. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol 7: 371–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N 1999. Regulation of 4E-BP1 phosphorylation: A novel two-step mechanism. Genes Dev 13: 1422–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heider JA, Yu Y, Shenk T, Alwine JC 2002. Characterization of a human cytomegalovirus with phosphorylation site mutations in the immediate-early 2 protein. J Virol 76: 928–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornstein E, Git A, Braunstein I, Avni D, Meyuhas O 1999a. The expression of the poly(A)-binding protein gene is translationally regulated in a growth-dependent fashion through a 5′terminal oligopyrimidine tract motif. J Biol Chem 274: 1708–1714 [DOI] [PubMed] [Google Scholar]
- Hornstein E, Harel H, Levy G, Meyuhas O 1999b. Overexpression of poly(A)-binding protein down-regulates the translation or the abundance of its own mRNA. FEBS Lett 457: 209–213 [DOI] [PubMed] [Google Scholar]
- Isler JA, Skalet AH, Alwine JC 2005. Human cytomegalovirus infection activates and regulates the unfolded protein response. J Virol 79: 6890–6899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CUT, Pestova TV 2010. The mechanisms of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 10: 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahvejian A, Roy G, Sonenberg N 2001. The mRNA closed-loop model: The function of PABP and PABP-interacting proteins in mRNA translation. Cold Spring Harb Symp Quant Biol 66: 293–300 [DOI] [PubMed] [Google Scholar]
- Kahvejian A, Svitkin YV, Sukarieh R, M'Boutchou M, Sonenberg N 2005. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev 19: 104–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim MM, Svitkin YV, Kahvejian A, De Cerscenzo G, Costa-Mattiolo M, Sonenberg N 2006. A mechanism of translational repression by competition of Paip2 with eIF4G for poly(A) binding protein (PABP) binding. Proc Natl Acad Sci 103: 9494–9499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaleghpour K, Svitkin YV, Craig AW, DeMaria CT, Deo RC, Burley SK, Sonenberg N 2001a. Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol Cell 7: 205–216 [DOI] [PubMed] [Google Scholar]
- Khaleghpour K, Kahvejian A, De Crescenzo G, Roy G, Svitkin Y, Imataka H, O'Connor-McCourt M, Sonenberg N 2001b. Dual interactions of the translational repressor Paip2 with poly(A) binding protein. Mol Cell Biol 21: 5200–5213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoutorsky A, Yanagiya A, Gkogkas CG, Fabian MR, Prager-Khoutorsky M, Cao R, Gamache K, Bouthiette F, Parsyan A, Sorge RE, et al. 2013. Control of synaptic plasticity and memory via suppression of poly(A)-binding protein. Neuron 78: 298–311 [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Wilson AC, Chao MV, Mohr I 2012. Control of viral latency in neurons by axonal mTOR signaling and the 4E-BP translation repressor. Genes Dev 14: 1527–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudchodkar SB, Yu Y, Maguire TG, Alwine JC 2004. Human cytomegalovirus infection induces rapamycin-insensitive phosphorylation of downstream effectors of mTOR kinase. J Virol 78: 11030–11039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudchodkar SB, Yu Y, Maguire TG, Alwine JC 2006. Human cytomegalovirus infection alters the substrate specificities and rapamycin sensitivities of raptor- and rictor-containing complexes. Proc Natl Acad Sci 103: 14182–14187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn U, Pieler T 1996. Xenopus poly(A) binding protein: Functional domains in RNA binding and protein–protein interaction. J Mol Biol 256: 20–30 [DOI] [PubMed] [Google Scholar]
- Lin J, Fabian M, Sonenberg N, Meller A 2012. Nanopore detachment kinetics of poly(A) binding proteins from RNA molecules reveals the critical role of C-terminus interactions. Biophys J 102: 1427–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Musa T, Bag J 2006. Reduced stability of mitogen-activated protein kinase-2 mRNA and phosphorylation of poly(A)-binding protein (PABP) in cells overexpressing PABP. J Biol Chem 281: 3145–3156 [DOI] [PubMed] [Google Scholar]
- Mangus DA, Evans MC, Jacobson A 2003. Poly(A)-binding proteins: Multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 4: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martineau Y, Derry MC, Wang X, Yanagiya A, Berlanga JJ, Shyu AB, Imataka H, Gehring K, Sonenberg N 2008. Poly(A)-binding protein-interacting protein 1 binds to eukaryotic translation initiation factor 3 to stimulate translation. Mol Cell Biol 28: 6658–6667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney C, Perez C, Mohr I 2012. Poly(A) binding protein abundance regulates eukaryotic translation initiation factor 4F assembly in human cytomegalovirus-infected cells. Proc Natl Acad Sci 109: 5627–5632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo EO, Dhalia R, Martins de Sa C, Standart N, de Melo Neto OP 2003. Identification of a C-terminal poly(A)-binding protein (PABP)–PABP interaction domain: Role in cooperative binding to poly (A) and efficient cap distal translational repression. J Biol Chem 278: 46357–46368 [DOI] [PubMed] [Google Scholar]
- Meyuhas O 2000. Synthesis of the translational apparatus is regulated at the translational level. Eur J Biochem 267: 6321–6330 [DOI] [PubMed] [Google Scholar]
- Mohr I, Sonenberg N 2012. Host translation at the nexus of infection and immunity. Cell Host Microbe 12: 470–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman NJ, Cristea IM, Terhune S, Rout MP, Chait BT, Shenk T 2008. Human cytomegalovirus protein UL38 inhibits host cell stress responses by antagonizing the tuberous sclerosis protein complex. Cell Host Microbe 3: 253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel GP, Ma S, Bag J 2005. The autoregulatory translational control element of poly(A)-binding protein mRNA forms a heteromeric ribonucleoprotein complex. Nucleic Acids Res 33: 7074–7089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez C, McKinney C, Chuluunbaatar U, Mohr I 2011. Translational control of cytoplasmic poly(A) binding protein in human cytomegalovirus-infected cells. J Virol 85: 156–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Z, Xuan B, Gualberto N, Yu D 2011. The human cytomegalovirus protein pUL38 supresses endoplasmic reticulum stress-mediated cell death independently of its ability to induce mTORC1 activation. J Virol 85: 9103–9113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy G, De Crescenzo G, Khaleghpour K, Kahvejian A, O'Connor-McCourt M, Sonenberg N 2002. Paip1 interacts with poly(A) binding protein through two independent binding motifs. Mol Cell Biol 22: 3769–3782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinski M 1977. Synthesis of proteins and glycoproteins in cells infected with human cytomegalovirus. J Virol 23: 751–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenney DJ, Colberg-Poley AM 1991. Expression of the human cytomegalovirus UL36-38 immediate early region during permissive infection. Virology 182: 199–210 [DOI] [PubMed] [Google Scholar]
- Terhune S, Torigoi E, Moorman N, Silva M, Qian Z, Shenk T, Yu D 2007. Human cytomegalovirus UL38 protein blocks apoptosis. J Virol 81: 3109–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnum SM, Streblow DN, Monroe ME, Smith P, Auberry KJ, Pasa-Tolic L, Wang D, Camp DG 2nd, Rodland K, Wiley S, et al. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: The HCMV proteome. J Virol 78: 10960–10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, Mohr I 2004. Phosphorylation of eIF4E by Mnk-1 enhances HSV-1 translation and replication in quiescent cells. Genes Dev 18: 660–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, Mohr I 2011. Viral subversion of the host protein synthesis machinery. Nat Rev Microbiol 9: 860–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, Perez C, Notary J, Mohr I 2005. Regulation of the translation initiation factor eIF4F by multiple mechanisms in human cytomegalovirus-infected cells. J Virol 79: 8057–8064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, Mathews MB, Mohr I 2013. Tinkering with translation: Protein synthesis in virus-infected cells. Cold Spring Harb Perspect Biol 5: a012351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wormington M, Searfoss AM, Hurney CA 1996. Overexpression of poly(A) binding protein prevents maturation-specific deadenylation and translational inactivation in Xenopus oocytes. EMBO J 15: 900–909 [PMC free article] [PubMed] [Google Scholar]
- Wu J, Bag J 1998. Negative control of the poly(A)-binding protein mRNA translation is mediated by the adenine-rich region of its 5′-untranslated region. J Biol Chem 273: 34535–34542 [DOI] [PubMed] [Google Scholar]
- Yanagiya A, Delbes G, Svitkin YV, Robaire B, Sonenberg N 2010. The poly(A)-binding protein partner Paip2a controls translation during late spermatogenesis in mice. J Clin Invest 120: 3389–3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M, Yoshida K, Kozlov G, Lim NS, Berlanga JJ, Kahvejian A, Gehring K, Wing SS, Sonenberg N 2006. Poly(A) binding protein (PABP) homeostasis is mediated by the stability of its inhibitor, Paip2. EMBO J 25: 1934–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]