Abstract
DNA methylation patterns are important for establishing cell, tissue, and organism phenotypes, but little is known about their contribution to natural human variation. To determine their contribution to variability, we have generated genome-scale DNA methylation profiles of three human populations (Caucasian-American, African-American, and Han Chinese-American) and examined the differentially methylated CpG sites. The distinctly methylated genes identified suggest an influence of DNA methylation on phenotype differences, such as susceptibility to certain diseases and pathogens, and response to drugs and environmental agents. DNA methylation differences can be partially traced back to genetic variation, suggesting that differentially methylated CpG sites serve as evolutionarily established mediators between the genetic code and phenotypic variability. Notably, one-third of the DNA methylation differences were not associated with any genetic variation, suggesting that variation in population-specific sites takes place at the genetic and epigenetic levels, highlighting the contribution of epigenetic modification to natural human variation.
Phenotypic differences between individuals cannot entirely be explained by genetic differences. Considering the transcriptome as a mirror of the sum of regulatory events suggests that nongenetic mechanisms have a profound influence on the phenotype. Epigenetics is responsible for part of this additional layer of control (Feinberg 2007; Portela and Esteller 2010). For example, genetically identical individuals, such as monozygotic twins (Fraga et al. 2005; Kaminsky et al. 2009), cloned animals (Rideout et al. 2001), and Agouti mice (Michaud et al. 1994; Waterland and Jirtle 2003), show DNA methylation and phenotypical differences. Hence, epigenetic variations, and in particular DNA methylation, might participate not only in differences between individuals, but also between human populations, and could contribute to the observed differences in distinct physical appearance, behavior, and response to environmental agents and drugs. In this regard, the presence of DNA methylation differences between an African and an European population using a 27,000-CpG-site microarray platform has been previously reported (Fraser et al. 2012).
DNA methylation at gene promoters is important for transcriptional regulation, with dense promoter hypermethylation around the transcription start site being associated with gene repression (Feinberg 2007; Portela and Esteller 2010). The picture is, in fact, more complex, and, recently, intragenic methylation has been linked to transcriptional and splicing activities (Jones 2012), suggesting a sophisticated regulatory potential for this epigenetic modification. DNA methylation levels are closely related to the genomic context, with CpG-rich regions (CpG islands) located in the 5′ end of genes being predominantly unmethylated. Interindividual variation in DNA methylation at distinct CpG sites has consistently been linked to genetic variation in terms of single nucleotide polymorphisms (SNPs) and defined as methylation quantitative trait loci (meQTL) (Bell et al. 2011, 2012). However, the causal chain of events establishing DNA methylation variability, which is currently under debate, is likely to be mediated in a network of genomic contexts, transcriptional activity, and additional epigenetic layers of regulation, such as histone modifications, DNA binding and modifying factors, nucleosome positioning, and noncoding RNAs.
Many genome-wide association studies (GWAS) have attempted to establish genetic associations with differences between distinct populations (Li et al. 2008a; Lachance et al. 2012), diseases (Kamatani et al. 2009), and the response to external stimuli (Li et al. 2008b; Niu et al. 2010). However, fewer associations were observed than expected, and direct genotype–phenotype relations were not easily explicable since the majority of variant sites are located in noncoding loci (Kilpinen and Dermitzakis 2012). In this context, the epigenetic network is expected to add layers of regulation, suggesting an interplay between the genotype and epitype in gene regulation and phenotypic variation (The ENCODE Project Consortium 2012).
In this study, we performed differential DNA methylation analysis of around 300 individuals from three human populations. In particular, we analyzed B-lymphocytes obtained from Caucasian-American, African-American, and Han Chinese-American individuals at a genome-scale resolution of around 450,000 CpG sites (Dedeurwaerder et al. 2011; Sandoval et al. 2011). DNA methylation levels at distinct loci enabled their separation with respect to geographic origin and could help explain natural human variation. A subsequent integration of genotype data enabled the precise separation into genetically dependent and independent variation.
Results
DNA methylation differences are present in distinct human populations
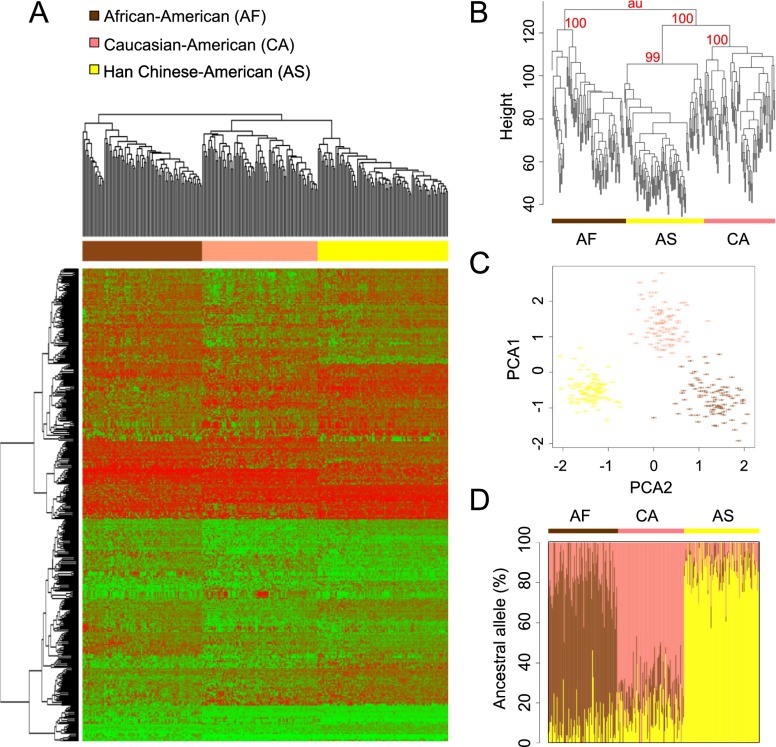
We determined differences in DNA methylation between three populations extensively characterized in the Human Variation Panel in terms of single nucleotide polymorphisms (SNPs) and gene expression (Li et al. 2008b; Niu et al. 2010). The DNA methylation profile was assessed for 288 B-cell lymphoblastoid cell lines (LCL) representing 96 Caucasian-American (CA), 96 African-American (AF), and 96 Han Chinese-American (AS) individuals. After sample randomization, DNA samples were hybridized on the Infinium DNA methylation BeadChip platform (Illumina), which analyzes more than 450,000 CpG sites in the genome (Dedeurwaerder et al. 2011; Sandoval et al. 2011). After normalization, we filtered out poor-quality probes and those containing single nucleotide polymorphisms (SNPs; >1%) (The 1000 Genomes Project Consortium 2010) and copy number variations (CNV; >5%) (Redon et al. 2006) in the detection sequence: Following these filters, from the originally printed 485,577 CpG sites in the microarray, we maintained 406,021 probes for subsequent analysis. Any DNA methylation variation introduced by Epstein-Barr virus (EBV) immortalization of the LCLs was also excluded by positive filtering for variant CpG sites between 10 Caucasian, 10 Asian, and 10 African naive peripheral blood cell samples. Nonetheless, 1373 differentially methylated CpG sites (delta mean β-values ≥0.12; ANOVA, FDR < 0.01) separately generated only from LCL samples were able to cluster naive blood samples perfectly according to their geographical origin, underscoring the capability of LCL samples to determine epigenetic differences between ethnicities (Supplemental Fig. S1). Herein, we determined 439 CpG sites to be differentially methylated between the populations both in LCL and naive blood samples (delta mean β-values ≥ 0.12; ANOVA with Tukey HSD post hoc test, FDR < 0.01) (Fig. 1A; Supplemental Table S1) and were able to cluster them separately (multiscale bootstrap resampling, n = 10,000, approximately unbiased P-value > 0.99) (Fig. 1B). The DNA methylation classification originated a separate branch for AF, and in the other arm of the cluster, two subbranches were obtained corresponding to CA and AS, consistent with the accepted prior genetically defined proximities (Fig. 1A; Li et al. 2008a). These CpG sites with population-specific differential methylation were termed pop-CpGs. Particularly, 172, 129, and 138 CpG sites revealed DNA methylation that differed significantly in AF, CA, and AS samples, respectively. Four hundred thirty-nine randomly selected probes were not able to separate the individuals with respect to their population identity (multiscale bootstrap resampling, n = 10,000).
Figure 1.
DNA methylation separates African-American (AF, brown), Caucasian-American (CA, pink), and Han Chinese-American (AS, yellow) individuals. (A) Hierarchical clustering of 439 pop-CpG sites separating the three populations using absolute DNA methylation levels (low: green; high: red). (B) Multiscale bootstrap resampling (n = 10,000) of the 439 pop-CpG sites significantly differentially methylated between African, Asian, and Caucasian individuals. The three populations cluster separately and consistently with prior genetically defined proximities (approximately unbiased P-value > 0.99). (C) Principal component analysis (PCA) of pop-CpGs displaying the first two principal components. (D) ADMIXTURE analysis of pop-CpGs-defined ancestral DNA methylation status. Each individual is represented by a vertical line, with the lengths corresponding to the ancestry coefficients in up to three inferred ancestral groups.
PCA with the pop-CpGs to apportion the majority of the variation clearly separated the samples with respect to their population identity and identified the ethnic relationship as the strongest component (Fig. 1C). We also performed surrogate variable analysis (SVA) to exclude covariates, other than population association, that drove the separation of the samples. SVA did not detect any surrogate variables, such as batch effects, excluding unknown, unmodeled, or latent source of noise influencing our results. To confirm the common ancestral DNA methylation status within the populations, we performed ADMIXTURE, applying three DNA methylation scenarios (unmethylated: <0.33; hemimethylated: ≥0.33, ≤0.66; methylated >0.66). Here, the analyzed individuals segregated into their assigned populations, displaying common ancestral DNA methylation levels within populations and distinct levels between populations (Fig. 1D).
Out of 439 pop-CpGs, 178 were located in gene promoter regions (including 51 noncoding RNA promoters, GENCODE v13) (Supplemental Table S2), 147 in gene bodies, and 114 in intergenic regions (Supplemental Fig. S2A; Supplemental Table S1). Considering the regional CpG composition and density, 104 pop-CpGs mapped to CpG islands, 124 to CpG island shores, 36 to CpG island shelves, and 175 CpGs outside of the island context (“open sea”) (Supplemental Fig. S2B). Furthermore, we analyzed histone occupancy frequencies extracted from the genome-wide epigenetic mapping determined for an LCL (GM12878) within the ENCODE project at the 439 pop-CpGs (Ernst et al. 2011). Separately analyzing promoter, gene body, and intergenic frequencies revealed an enrichment of pop-CpGs in respect to their representation on the array platform within the enhancer marks H3K27ac and H3K4me1 in intragenic regions and within the heterochromatin mark H3K9me3 at intergenic loci (Fisher's exact test; P < 0.05) (Supplemental Table S3). In addition to intragenic enhancer regions, pop-CpGs revealed enrichment in insulator sites marked by CTCF and underlining the importance of regulatory elements outside the promoter context for natural human variation. Multiple hypotheses testing highlighted the significant enrichment of pop-CpGs for H3K4me1 (FDR = 0.0324) and CTCF (FDR = 0.0378) in intragenic regions (Supplemental Table S3).
To further dissect functional consequences related to differentially methylated pop-CpG sites, we performed enrichment analysis for DNA sequence motifs (Supplemental Fig. S3). Outside the promoter context, we identified an enrichment of DNA motifs related to the transcription factor RREB1 (ras responsive element binding protein 1) in respect to the representation on the DNA methylation array (Hypergeometric test; P < 0.01). Interestingly, intragenic pop-CpGs also revealed an enrichment of binding sites for the enhancer-associated factor TFAP2A (transcription factor activating enhancer binding protein 2α) among others, underscoring the previous identified enrichment for the enhancer-associated histone marks H3K27ac and H3K4me1 in gene bodies. Within gene promoter regions, we identified significant enrichment of the hematopoietic transcription factors IRF1 and SPIB (Hypergeometric test; P < 0.01) among others. Thus, pop-CpGs are associated with histone modifications and transcription factor binding that actively regulate gene expression, suggesting a regulatory network that contributes to the variance observed between populations.
Furthermore, we aimed to determine the relationship between DNA methylation and transcriptional regulation. Therefore, we integrated DNA methylation and gene expression levels and observed significantly decreased gene expression associated to promoter hypermethylation in 12.9% (13 out of 101) of pop-CpG-related genes in the analyzed context (Pearson's correlation test; FDR < 0.01) (Supplemental Fig. S4A). This is in line with previous studies reporting rather low overall associations between promoter DNA methylation and gene expression (Kulis et al. 2012). It is of note that gene body methylation was significantly associated to gene expression in 23.9% of intragenic pop-CpGs (27 out of 113; Pearson's correlation test; FDR < 0.01) (Supplemental Fig. S4B). Here, a gain of DNA methylation was associated to gene repression and activation in 63.0% (17 out of 27) and 37.0% (10 out of 27) of cases, respectively.
Population-specific differential methylation contributes to natural human variation
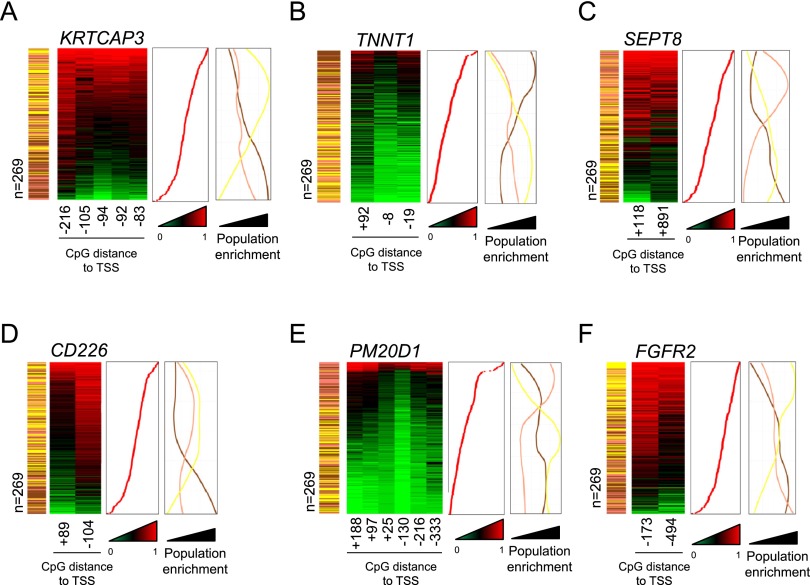
We expected DNA methylation in gene promoters to be directly associated with gene expression and hence phenotype formation, so we extracted promoter-associated pop-CpGs. We wondered whether genes harboring pop-CpGs in their promoter could help explain the well-known phenotypic variation between CA, AS, and AF human populations. In this regard, pop-CpGs were located in genes associated with natural human variation, such as xenobiotic metabolism and transport (GSTT1, GSTM5, ABCB11, SPATC1L), taste transducers (TAS1R3), environmental information processing and adaptation (ARNTL, PRSS3, CNR2), immune response factors (CERK, LCK, CD226, SEPT8), growth factors (FGFR2), keratinocyte-associated genes (KRTCAP3), and melanogenesis (CREB3L3). We also observed the presence of pop-CpGs in genes related to the different penetrance of diseases among distinct human populations, such as diabetes (HLA-B/C, PRKCZ), Parkinson's disease onset (PM20D1), HIV infection (HIVEP3, HTATIP2, CDK11B), enteropathogenic Escherichia coli and measles virus infection (FYN), and hepatitis B virus infection (HLA-DPA1). Illustrative examples of these genes with their DNA methylation differences among human populations are shown in Figure 2.
Figure 2.
Differentially methylated gene promoters of KRTCAP3 (A), TNNT1 (B), SEPT8 (C), CD226 (D), PM20D1 (E), and FGFR2 (F) in Han Chinese-American (yellow), Caucasian-American (pink), and African-American (brown) individuals. Absolute DNA methylation levels at population-specific CpG sites in gene promoters (low: green; high: red) are displayed for single individuals (n = 269). The distance to the gene transcription start site is indicated. The samples are ranked according to their average DNA methylation levels (middle panel) at displayed pop-CpGs. Population enrichment (right panel) is illustrated using absolute sample numbers in a 10-sample window.
To distinguish between random drift at neutral loci and CpG sites that may have experienced accelerated divergence due to local selection, we included data from 14 chimpanzee individuals from three subspecies (Pan troglodytes troglodytes, Pan troglodytes verus, Pan troglodytes schweinfurthii) as the outgroup in the analysis, since this is our closest living relative. Positively selected CpG sites were assessed by determining common ancestral sites between humans and chimpanzees that had a single human outlier population (ANOVA with Tukey HSD post hoc test, P < 0.05) (Supplemental Table S4). We identified 39 CpG sites that could have evolved under local selection pressure. These CpG sites of local adaptation include immune (CERK, CDK11B, HTATIP2) and xenobiotic (GSTT1, SPATC1L) response factors, suggesting that they might be driven by differences in local pathogen and environmental pressure. In particular, SPATC1L (spermatogenesis and centriole associated 1-like) is an interesting case, because it was previously related to the response to alkylating agents, suggesting that epivariation contributes to the variable response to chemotherapeutic treatment (Fry et al. 2008). It is also tempting to speculate that the selective pressure that gives rise to the polymorphisms originates from carcinogens such as nitrosamides, which introduce alkyl groups on guanine bases, the mechanism used by alkylating drugs.
Cross talk between population-specific epigenetic and genetic variants
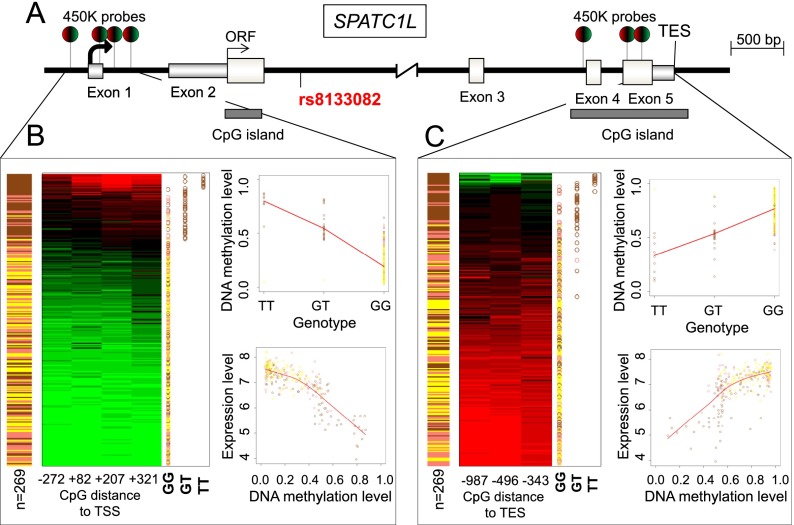
We wondered about the effect of underlying genetic variants on the characterized pop-CpGs. An example of the connection between ethnicity-associated epigenetic and genetic marks is the gene SPATC1L: It was previously identified as a CpG methylation quantitative trait loci (meQTL) and expression quantitative trait loci (eQTL) in a lower-resolution (27,000 CpG sites) screening using Yoruba individuals (Bell et al. 2011). QTLs describe the direct association between single nucleotide polymorphisms (SNPs) and methylation or expression events. Accordingly, the gene expression or CpG site methylation (epitype) are directly correlating to the underlying genetic sequence (genotype). In the present study, we did not only validate the association between methylation and gene expression of SPATC1L, but also established that the entire promoter region is differentially methylated in African-Americans with a high correlation between gene expression and the underlying genotype (Fig. 3). To do this, we identified four pop-CpGs located in the 5′ end that were differentially methylated in African-Americans and directly correlated with gene expression (Fig. 3; Supplemental Fig. S5A). All four promoter-associated pop-CpGs of SPATC1L were assigned as meQTLs associated with a single SNP position (rs8133082) (Fig. 3; Supplemental Fig. S6A). Interestingly, we identified three pop-CpGs in the CpG island that overlapped the transcription end site (TES) of SPATC1L with complete inverse correlation of DNA methylation, gene expression, and genotype association with the promoter region (Fig. 3; Supplemental Figs. S5B, S6B). Individuals with hypomethylated and active promoters revealed hypermethylation at the TES (Pearson's product-moment correlation, rho = 0.89) (Supplemental Fig. S7). Hypermethylation at the TES of actively transcribed SPATC1L might impair the high frequencies of the previously identified antisense transcription that takes place at terminator sites (He et al. 2008).
Figure 3.
Genotype and DNA methylation regulate gene expression of SPATC1L in a conjoined manner and inversely correlate at the transcription start (TSS) and end site (TES). (A) Schematic overview of the gene structure of SPATC1L. (B) AF individuals have high levels of promoter DNA methylation, a TT phenotype enriched in rs8133082, and reduced expression of SPATC1L. The figure displays the absolute DNA methylation levels (low: green; high: red) for four promoter-related pop-CpGs of African-American (brown), Caucasian-American (pink), and Han Chinese-American (yellow) individuals; the genotype of rs8133082 for the individual samples (GG, GT, TT); the correlation between DNA methylation (cg08742575) and the genotype (rs8133082); and the gene expression level (SPATC1L). Samples are ranked by mean CpG methylation values. (C) Unlike the promoter, AF individuals have CpG hypomethylation, which is positively correlated with SPATC1L gene expression (cg11766577).
From the original observation in SPATC1L, we extended the search of meQTLs to the entire set of 439 pop-CpGs. We found 68% (298 of 439) of differentially methylated CpG sites were significantly related to underlying genetic variation (596 SNPs, random forest selection frequencies, FDR < 0.05) (Supplemental Table S5). Because only 13% of the randomly selected 439 CpG sites revealed genetic association, we excluded the common event of epitype–genotype association of CpG sites interrogated in this study (Fisher's exact test, P < 0.01). To exclude the idea that confounding effects based on genetic differences between the populations were driving the association of pop-CpG methylation levels with the underlying genotype, we performed meQTL analysis separately for the three populations. Importantly, we confirmed an association to the genetic background in at least one of the three populations, for 91.6% (273 out of 298) of the meQTLs, suggesting that the detected associations are mainly independent from ethnic variation. Furthermore, we aimed to interrogate the tissue specificity of epitype–genotype associations by analyzing 159 normal primary specimens representing three different solid-tissue types. Taking advantage of genotype and DNA methylation data available from The Cancer Genome Atlas (TCGA), we determined the associations of pop-CpG sites to the underlying genetic background to be maintained across tissue types in 60.7% (181 out of 298) of the meQTLs, suggesting that the interplay between both layers is partially independent of the cell-type context. In particular, we confirmed the presence of blood-related meQTLs in 40.6% of breast, 38.9% of colon, and 32.6% of lung-tissue samples. It is of note that meQTLs are located close to the correlated SNP site, with 69% (412 out of 596) present within a 15-kb window and 38% (227 out of 596) within 5 kb (Supplemental Fig. S8).
However, for the remaining 32% of pop-CpGs, no direct relationship could be detected between the genetic background and variability of CpG sites. We excluded the possibility that the absence of genetic association was a consequence of an uneven number of SNPs in the ±1-Mb region surrounding the pop-CpGs by showing that both groups harbored similar numbers of SNPs in the analyzed regions (Supplemental Fig. S9). The analyzed windows also harbored equal coverage of repetitive elements (SINE, LINE, LTR), and, thus, the same power to identify SNPs (Supplemental Fig. S10). Interestingly, gene promoters associated with potentially epigenetically inherited CpG sites were related to immune response components (FYN, CD226, HIVEP3) (Supplemental Table S1) and enriched for transcription factor binding sites for NFKB (z-test; TRANSFAC: FDR = 0.0195; JASPAR: FDR = 0.0057), a transcription factor involved in the immune defense system, protecting against, for example, viral (i.e., hepatitis B) and bacterial (i.e., Shigellosis) pathogen infection. This raises the possibility that the pathogens in the environment leave stable fingerprints in the epigenomes of the human host population.
Noteworthy, when we interrogated the three aforementioned primary tissue types, we found no further genetic associations of potentially epigenetic inherited CpG sites in 66.0% of cases (93 out of 141), supporting their genetic independence. In this respect, proportional analysis detected no differences in the power to detect genotype–epitype associations between the initial LCL sample set and the primary specimens (Chi-square test; OR: 1.13, P = 0.51), underlining the reliability of the study design.
Population-specific differential methylation could be useful in genome-wide association studies of genetic variants
Only 13% (79 out of 596) of SNPs related to differentially methylated pop-CpGs are located in exonic regions and hence can be directly associated with functional gene alterations (Supplemental Table S6). Therefore, we postulated that DNA methylation presents a little-recognized but highly informative level in the interpretation of genetic variants between human individuals. Comprehensive analysis of genetic variation in genome-wide association studies (GWAS) often reveals genotype–phenotype connections. These include many polymorphisms that are directly connected with phenotypes, but are located within intergenic or intronic regions of unknown function. Studying the close interplay between genetic and epigenetic regulation could help interpret these connections, which are otherwise difficult to explain. In this respect, DNA methylation might display a potent intermediate event that could be useful to interpret the GWAS results. For this reason, we performed model-based gene set analysis (Bauer et al. 2011) to compare SNPs directly associated with pop-CpGs and GWA studies available at GWASdb (Li et al. 2012). We found some interesting associations related to age of menarche, and HIV control; in particular, we determined highly significant enrichment for hepatitis B infection through the detection of six SNPs directly related to CpG sites that were differentially methylated between populations (Enrichment score: 0.99) (Supplemental Table S7; Bauer et al. 2011). These six SNPs were located within the HLA-DPA1 and HLA-DPB1 locus, which is strongly associated with chronic hepatitis B virus (HBV) infection (Kamatani et al. 2009). Concordantly, HBV infection risk alleles are more abundant in populations with a higher incidence of disease, such as those with Asian or African ancestry. However, none of the SNPs is located in a coding context, making it difficult to connect genetic variation and risk phenotype. Functionally, the HLA-DPA1- and HLA-DPB1-related SNPs were defined as eQTLs and thus directly connected to gene expression (O'Brien et al. 2011), although the regulatory mechanism remains elusive.
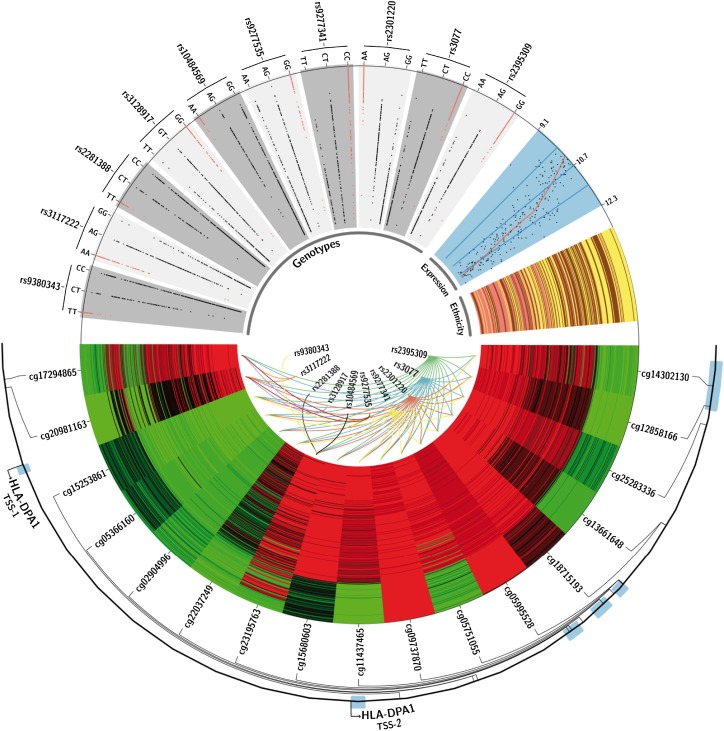
Here, we integrated GWAS results into differential DNA methylation analysis, determining a direct correlation between genetic variation and differences in the methylation of CpGs located in the HLA-DPA1 locus. In particular, the 10 HBV infection–associated SNPs (Kamatani et al. 2009) were significantly correlated with 17 CpG sites in the HLA-DPA1 locus (Pearson's correlation, rho > 0.6) (Fig. 4; Supplemental Table S8). The risk alleles were associated with altered CpG methylation and occurred at high frequencies in Asian and African populations. In detail, the risk alleles were related to DNA hypermethylation in the promoter of HLA-DPA1, which was inversely correlated with gene expression (Fig. 4; Supplemental Fig. S11). Hypermethylation of the promoter of HLA-DPA1 was conjoined with DNA hypomethylation in the gene body, which is also associated with a lower level of gene expression (Jones 2012). Hence, we found DNA methylation associated with HLA-DPA1 gene repression in the Asian and African individuals, identifying the risk alleles for mediating DNA methylation variation and gene repression. These give rise to variation in cell surface receptor presentation and altered HBV binding and infection risk. Consequently, we suggest that DNA methylation could be the unidentified regulatory event connecting genotype (risk alleles), gene expression (eQTLs), and phenotype (infection incidence).
Figure 4.
Genetic polymorphisms related to HBV infection influence DNA methylation and gene expression at the HLA-DPA1 locus. Using Circos (Krzywinski et al. 2009), the figure shows a schematic overview of the HLA-DPA1 locus and DNA methylation, genotype and expression data from African-American (brown), Caucasian-American (pink), and Han Chinese-American (yellow) individuals: DNA methylation levels (low: green; high: red) of CpG sites (n = 17) significantly correlated with the genotype of HBV infection-associated SNPs (n = 10) (Kamatani et al. 2009). Samples are ranked by mean CpG methylation values. SNP-CpG relations are displayed by colored lines. The genotype distribution of SNPs in the HLA-DPA1 locus is shown, which is significantly correlated with the level of CpG methylation (gray boxes; risk alleles are highlighted in red) (Kamatani et al. 2009). The distribution of expression levels of HLA-DPA1 is shown in the blue box.
Discussion
Epigenetic modification, and DNA methylation in particular, provides a layer of gene regulation that has a great influence on phenotypes. It has been widely studied in human cancer. However, outside of the disease context, there are few examples of epigenetic variations that are directly associated with phenotypic differences. Interestingly, a recent genome-wide epigenetic analysis of different casts of honey bees revealed DNA methylation differences between nurse and forager bees at different genomic loci, suggesting that despite being genetically identical, the social behavior of bees is directly associated with their epigenetic profile (Herb et al. 2012). Although done in another species, this study gives an insight into the potential impact of DNA methylation changes on distinct human phenotypes beyond the disease context. Consistently, we aimed to determine DNA methylation differences between human populations and their association with the natural phenotypic variation occurring in the human species. Here, using 439 CpG sites differentially methylated between Caucasian-American, African-American, and Han Chinese-American individuals, we were able to perfectly separate the distinct populations with respect to their geographical origins and to associate them with distinct phenotypic characteristics, such as appearance, drug metabolism, response to external stimuli, sensory perception, and disease susceptibility. Importantly, local selective pressure was shown to induce the manifestation of epigenetic variants, exemplified by immune and xenobiotic response factors and their potential positive selection by differences in local pathogen and environmental pressure.
Taking advantage of the comprehensive characterization of the analyzed samples at the genetic level, we were able to interrogate genotype–epitype associations to distinguish further between the two types of inheritance—genotype dependent and independent. Although, DNA methylation in general reveals low dependence on the genotype (Bell et al. 2011, 2012), two-thirds of population-specific CpG sites were directly associated with the underlying genetic background, suggesting the evolutionarily set genetic context influences DNA methylation, which subsequently functions as an intermediate regulatory event mediating phenotypic differences between populations. Here, the hepatitis B infection risk, which is directly related to underlying genotypes and epitypes, and the consequent enrichment of these in affected populations is an illustrative example of the tight interplay between the two layers of organization. Similar causal connections were concluded by Feinberg and Irizarry from results obtained from high-resolution DNA methylation profiling of normal human and mouse tissues (Feinberg and Irizarry 2010). These investigators suggested that a change in regional CpG density over time was responsible for DNA methylation changes between the species. Genotype–epitype associations in terms of methylation quantitative trait loci were also observed in LCL samples from African and European populations (Bell et al. 2011; Fraser et al. 2012) and human brain samples (Gibbs et al. 2010; Zhang et al. 2010), highlighting their close connection. Accordingly, variation in the DNA methylation of population-specific loci followed similar trends to those seen in genetic variation studies, suggesting that both levels have similar patterns of variation and underlying related selection criteria (Li et al. 2008a).
Interestingly, no direct relation to genetic variation could be detected for one-third of differentially methylated loci, suggesting that the epivariance might be independent of the coding sequence (Rando 2012). In this regard, external stimuli such as toxic xenobiotics (Anway et al. 2005; Zeybel et al. 2012) and differences in dietary or hormone exposure (de Assis et al. 2012) or stress (Seong et al. 2011) are known to induce epigenetic changes with an impact on subsequent generations and may also have been the source of the differences between populations observed here. Detecting immune system–related genes enriched at potentially epigenetically inherited CpG sites suggests that the impact of pathogens on their host's DNA profiles (Paschos and Allday 2010) subsequently leaves footprints in the epigenome of their progeny. However, we cannot entirely exclude that the lack of genetic association for these potentially epigenetically inherited pop-CpG sites is based on unmodeled or unmeasured components in this study. Although, we applied multivariate methodology and interrogated large genomic regions, trans-acting polymorphisms or those not present on the SNP array platform might reveal epitype associations not reported as meQTLs. Herein, using stringent thresholds to identify differentially methylated CpG sites between the populations, the study design favored the selection of high-frequency variables and therefore unmeasured low-frequency polymorphisms were not the scope of this study, and their absence should have influenced the identification of meQTLs only minimally. It is of note that environmental variations between populations (e.g., diet preferences) could have influenced the epitype of germline cells, but might have also directly affected the DNA methylation levels of the somatic cell type analyzed here.
In conclusion, we have identified DNA methylation differences that distinguish three major human ethnic groups. Although many of them are associated with underlying genetic changes, suggesting a direct relationship between the genetic and epigenetic codes, others stand alone as epigenetic markers and CpG methylation quantitative trait loci associated with natural variation in our species. Thus, DNA methylation is a likely contributor to the different phenotypic characteristics, such as differences in drug response and disease frequencies that occur between human populations. From the regulatory point of view, we suggest a scenario wherein the genetic and epigenetic backgrounds set the evolutionarily established blueprint, resulting in phenotypic variation. Detecting distinct pop-CpG sites within populations suggests that divergence and selection pressure not only shape the genetic code, but also the DNA methylation landscape.
Methods
Sample description
DNA samples of 96 unrelated healthy Caucasian-Americans, 96 African-Americans, and 96 Asian-Americans were obtained from lymphoblastoid cell lines included in the Human Variation panel (sample sets HD100AA, HD100CAU, HD100CHI; Coriell Cell Repositories). These samples had been collected and anonymized by the National Institute of General Medical Science (NIGMS), and all subjects had provided written consent for their experimental use. Sample processing was performed in a randomized manner to avoid batch effects. Gender distribution (male frequency) and mean age (±standard deviation) for each ethnicity were 0.5 and 37.3 ± 16.2 yr for Caucasian-American samples, 0.2 and 29.4 ± 9.9 yr for African-Americans, 0.5 and 36.2 ± 15.7 yr for the Han Chinese-Americans, respectively. DNA from naive blood samples was extracted from peripheral blood mononuclear cells of unrelated healthy Caucasian and Asian donors. These samples had been collected and anonymized by the Bellvitge Biomedical Research Institute (IDIBELL) and the National Research Institute for Child Health and Development (NRICHD), and all subjects had provided written consent for their experimental use. DNA methylation data from naive blood samples of African individuals have been previously published (Alisch et al. 2012). Gender distribution (male frequency) and mean age (±standard deviation) for each ethnicity of naive samples were 0.5 and 27.7 ± 2.8 yr for Caucasian samples, 1.0 and 3.8 ± 3.7 yr for Africans, and 0.3 and 33.0 ± 3.4 yr for Asians, respectively.
Genotype and DNA methylation data for primary samples of normal breast (n = 78), colon (n = 38), and lung (n = 32) tissues were obtained from The Cancer Genome Atlas (TCGA) data portal (https://tcga-data.nci.nih.gov/tcga).
Infinium HumanMethylation450 BeadChip
All DNA samples were assessed for integrity, quantity, and purity by electrophoresis in a 1.3% agarose gel, picogreen quantification, and nanodrop measurements. All samples were randomly distributed into 96-well plates. Bisulfite conversion of 500 ng of genomic DNA was performed using the EZ DNA Methylation Kit (Zymo Research) following the manufacturer's instructions. Two hundred nanograms of bisulfite-converted DNA was used for hybridization on the HumanMethylation450 BeadChip (Illumina). Briefly, samples were whole-genome amplified followed by enzymatic end-point fragmentation, precipitation, and resuspension. The resuspended samples were hybridized onto the BeadChip for 16 h at 48°C, then washed. A single nucleotide extension with labeled dideoxy-nucleotides was performed, and repeated rounds of staining were applied with a combination of labeled antibodies differentiating between biotin and DNP.
A three-step normalization procedure was performed using the lumi (Du et al. 2008) package available for Bioconductor (Gentleman et al. 2004), within the R statistical environment (R Development Core Team 2009), consisting of color bias adjustment, background level adjustment, and quantile normalization across arrays (Du et al. 2008). The methylation level (β-value) for each of the 485,577 CpG sites was calculated as the ratio of methylated signal divided to the sum of methylated and unmethylated signals plus 100. After the normalization step, we removed probes related to X and Y chromosomes, and those with a SNP frequency >1% (The 1000 Genomes Project Consortium 2010) in the probe sequence or interrogated CpG site. All analyses were performed in human genome version 18 or 19 (hg18/19). However, to exclude regions of potential copy number variation, we integrated a data set (Redon et al. 2006) that was available only at the version hg17. Therefore, we transformed the loci represented on the array platform into the coordinates of the older genome version ( hg19 to hg17) using the LiftOver tool from UCSC. Subsequently, all probes associated to CNV with a frequency higher than 5% were excluded from the study.
Hierarchical clustering
Samples were organized by hierarchical clustering using the complete agglomeration method for Manhattan distances. The strength of the cluster was assessed from 10,000 bootstrap samples using the pvclust (Suzuki and Shimodaira 2006) package available for the R statistical environment. We also calculated 10,000 clusters using 439 random CpG sites to ensure that the cluster was not generated based on genome-wide differences between the samples and populations.
Surrogate variable identification
To identify possible latent factors that influence the methylation levels, we did a surrogate variable analysis as described in Leek and Storey (2007). We used the SVA11 R package provided by these investigators and ran the two-step protocol to identify the number of latent factors that need to be estimated and to identify the surrogate variables.
Admixture
To describe the ancestral allele methylation status of each individual, we discretized the β-values into three stages, simulating the diploid structure of the human genome. Loci with β-value < 0.33 were annotated as having AA alleles. Those with a β-value > 0.66 were designated BB alleles. All other loci were considered to be AB alleles. We ran the PLINK software (Purcell et al. 2007) with these data to store the pedigree/phenotype information in PLINK format, and then used the ADMIXTURE program to perform maximum likelihood estimation of individual ancestries from multilocus SNPs (with discretized β-values), as described in Alexander et al. (2009).
Identification of population-specific DNA methylation (pop-CpGs sites)
To define CpG sites that are differentially methylated between populations, we excluded outlier samples with an abnormal genome-wide DNA methylation profile to avoid the influence of unmodeled sources on the results. By performing PCA of CpG sites that do not overlap SNPs or CNVs, or that are located in gender-specific chromosomes (with the first two principal components explaining 27% of the variance), we excluded 11 Caucasian and eight African samples that fell outside the 95% confidence interval. Differential methylation analysis of individual CpG sites for the three populations was measured by selecting CpG sites with an absolute difference of the average methylation between two populations above 0.12, in terms of β-values (ranging from 0 to 1). An ANOVA with a Tukey HSD post hoc test was performed, and CpG sites showing an FDR-adjusted P-value < 0.01 were considered to be differentially methylated between populations. Subsequently, a set of naive blood samples from 10 Caucasians, 10 Africans, and 10 Asians was used as a filter to rule out side effects introduced by EBV immortalization of the lymphoblastoid cell lines. Therefore, the arrays were quantile-normalized, and CpG sites with an absolute difference in the average methylation between populations >0.12 were considered to be population-specific CpG sites (pop-CpGs).
Identification of pop-CpGs sites under local selective pressure
To define pop-CpG sites derived by local selective pressure, we integrated DNA methylation data (Infinium HumanMethylation450 BeadChip) from DNA obtained from whole blood of 14 chimpanzee individuals (four P. t. schweinfurthii; four P. t. verus; three P. t. troglodytes; and three of unknown origin), using these as the outgroup.
Because the Infinium HumanMethylation450 BeadChip is designed for use with the human reference genome, we first mapped the probes to the chimpanzee genome (Pan_troglodytes-2.1.4), using BWA (Li and Durbin 2009) and allowing a three edit distance. We retained only the autosomal probes that unambiguously mapped to a single location in the reference chimpanzee genome and probes with either a perfect match, or one mismatch in the 5′ 45 bp and with no mismatches in the 3′ 5 bp (closest to the CpG sites that are being assayed). We also excluded probes with a detection P-value > 0.01 in at least one individual. This filtering step resulted in the retention of 299,924 probes (66.6%). Using this conservative subset of CpG sites, we performed two-color channel signal adjustment and quantile normalization of human and chimpanzee samples on the pooled signals from both channels and recalculated the average β-values (Du et al. 2008). CpG sites with a single human outlier population (ANOVA with Tukey HSD post hoc test, P < 0.05) were considered to have evolved under local selective pressure.
Expression analysis
Expression data for the analyzed samples were obtained using the Human Genome U133 Plus 2.0 expression microarray (Affymetrix), available in the GEO database (GSE24277). A total of 24 Caucasian-American, six African-American, and one Han Chinese-American samples were excluded because expression data were not available. Expression arrays were loaded into the R statistical environment using the affy (Gautier et al. 2004) package, and normalized using the RMA method as described in Irizarry et al. (2003). Associations between DNA methylation and expression were calculated using Pearson's correlation coefficients. P-values below 0.01 after correction for multiple hypothesis testing were considered as significant.
meQTL identification
Genotype information for the analyzed samples was obtained from a set of HumanHap550k and HumanHap650k SNP arrays (Illumina), available in the GEO database (GSE24260, GSE24274). Data sets were combined into a single table containing 660,919 unique SNPs. A total of 24 Caucasians, seven African-American, and two Han Chinese samples were excluded from subsequent analysis because genotyping data were not available.
meQTLs for the 439 differentially methylated sites were identified by interrogating SNPs located in a ±1-Mb window flanking the CpG sites. The window was reduced in 100-kb intervals if it contained more than 1000 SNPs. We used the Random Forest Selection Frequency (RFSF) multivariate method, as described in Michaelson et al. (2010), to identify unique SNPs or additive effects of multiple SNPs on single CpG sites. This method performs well compared with other methods and enables us to identify multiple SNPs acting on a feature (Michaelson et al. 2009, 2010).
The Random Forest algorithm is implemented in R in the randomForest package (Liaw and Wiener 2002). First, we called the Random Forest algorithm to generate 2000 trees for regression and calculated the selection frequency (SF) of the variables (SNPs) used in the construction of the regression model. Bias correction was then applied to the frequencies by subtracting the deviation between the SF of the variable under the null hypothesis (no association between the SNPs and the methylation value) and the average SF of all variables under the null hypothesis; we used 1000 forests of 10 trees to obtain the SF under the null hypothesis, generating a methylation matrix from a random distribution, and applied the correction to the original SF. Eventually, in order to get a metric of how associated with the epitype the SF of a polymorphic site was, we constructed an empirical distribution from the SFs of 10 forests of 2000 trees by permuting the SNP values of our samples, reporting a Q-value for every SNP by comparing its SF with the ones under the null hypothesis.
Enrichment analysis of GWAS-associated polymorphisms
To establish a causal relationship between genetic variability, DNA methylation level, and distinct phenotypes, we determined enrichment of meQTL-related SNPs in the entire set of GWA studies available at GWASdb (Li et al. 2012) using model-based gene set analysis (Bauer et al. 2011). The method analyzes all categories at once by embedding them in a Bayesian network. Probabilistic inference is used to identify the active categories, giving a score that is simply the probability that associates a natural weight to each term, reflecting a measure of certainty of its involvement in the process.
Sequence motif enrichment analysis
De novo motif discovery of promoter-related, intragenic, or intergenic pop-CpGs was performed using GADEM software (Li 2009), using a window of maximal 50 bp flanking the CpG of interest (E-value < 0.05). Subsequently, sequence motifs were assigned to human transcription factor binding sites using JASPAR (Bryne et al. 2008), while removing artifactual and background motifs using MotIV (Motif Identification and Validation) (Mercier et al. 2011). We calculated the enrichment P-values based on the hypergeometric distribution of motif matches at pop-CpG sites, relative to their abundance in the search space (DNA methylation BeadChip). The hypergeometric test reports the probability to obtain the number of motif hits in the pop-CpG set compared with the number present in all CpG sites represented on the BeadChip. The P-values were calculated separately for promoter-, gene body-related, and intergenic CpG sites.
Transcription factor enrichment analysis for pop-CpG sites without genetic association located in promoters was performed using PSCAN (Zambelli et al. 2009). Transcription factor binding annotations defined by TRANSFAC or JASPAR were used independently. The gene promoter region was defined as −950 and +50 bp to the gene transcription start site. Z-test P-values were corrected for multiple hypotheses testing using the Bonferroni method.
Data access
The DNA methylation data generated for this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE36369.
Acknowledgments
The research leading to these results received funding from the European Research Council (ERC) grant EPINORC under agreement number 268626, ERC Starting Grant (260372), NIH grants CA138461 and GM61388 (Pharmacogenomics Research Network), the MICINN Projects SAF2011-22803 and BFU2011-28549, the Cellex Foundation, the European Community's Seventh Framework Programme (FP7/2007-2013) from grant HEALTH-F5-2011-282510 (BLUEPRINT), and the Health and Science Departments of the Generalitat de Catalunya. I.H.H. is a fellow of the Generalitat de Catalunya (FI 2011). T.M.B. and M.E. are ICREA Research Professors.
Author contributions: H.H., S.M., and M.E. conceived and designed the experiments, analyzed the data, and wrote the manuscript. I.H.-H., S.S., A.G., J.S., D.M., K.H., T.M.-B., and L.W. contributed analytical tools and selected the subjects for the DNA methylation analyses. All authors contributed to the final manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.154187.112.
Freely available online through the Genome Research Open Access option.
References
- The 1000 Genomes Project Consortium 2010. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19: 1655–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alisch RS, Barwick BG, Chopra P, Myrick LK, Satten GA, Conneely KN, Warren ST 2012. Age-associated DNA methylation in pediatric populations. Genome Res 22: 623–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anway MD, Cupp AS, Uzumcu M, Skinner MK 2005. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science 308: 1466–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer S, Robinson PN, Gagneur J 2011. Model-based gene set analysis for Bioconductor. Bioinformatics 27: 1882–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JT, Pai AA, Pickrell JK, Gaffney DJ, Pique-Regi R, Degner JF, Gilad Y, Pritchard JK 2011. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol 12: R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JT, Tsai P-C, Yang T-P, Pidsley R, Nisbet J, Glass D, Mangino M, Zhai G, Zhang F, Valdes A, et al. 2012. Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLoS Genet 8: e1002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryne JC, Valen E, Tang M-HE, Marstrand T, Winther O, da Piedade I, Krogh A, Lenhard B, Sandelin A 2008. JASPAR, the open access database of transcription factor-binding profiles: New content and tools in the 2008 update. Nucleic Acids Res 36: D102–D106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Assis S, Warri A, Cruz MI, Laja O, Tian Y, Zhang B, Wang Y, Huang TH-M, Hilakivi-Clarke L 2012. High-fat or ethinyl-oestradiol intake during pregnancy increases mammary cancer risk in several generations of offspring. Nat Commun 3: 1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedeurwaerder S, Defrance M, Calonne E, Denis H, Sotiriou C, Fuks F 2011. Evaluation of the Infinium Methylation 450K technology. Epigenomics 3: 771–784 [DOI] [PubMed] [Google Scholar]
- Du P, Kibbe WA, Lin SM 2008. lumi: A pipeline for processing Illumina microarray. Bioinformatics 24: 1547–1548 [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. 2011. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473: 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP 2007. Phenotypic plasticity and the epigenetics of human disease. Nature 447: 433–440 [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Irizarry RA 2010. Evolution in health and medicine Sackler colloquium: Stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proc Natl Acad Sci (Suppl 1) 107: 1757–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suñer D, Cigudosa JC, Urioste M, Benitez J, et al. 2005. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci 102: 10604–10609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser HB, Lam L, Neumann S, Kobor MS 2012. Population-specificity of human DNA methylation. Genome Biol 13: R8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry RC, Svensson JP, Valiathan C, Wang E, Hogan BJ, Bhattacharya S, Bugni JM, Whittaker CA, Samson LD 2008. Genomic predictors of interindividual differences in response to DNA damaging agents. Genes Dev 22: 2621–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA 2004. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20: 307–315 [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. 2004. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs JR, van der Brug MP, Hernandez DG, Traynor BJ, Nalls MA, Lai S-L, Arepalli S, Dillman A, Rafferty IP, Troncoso J, et al. 2010. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet 6: e1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW 2008. The antisense transcriptomes of human cells. Science 322: 1855–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herb BR, Wolschin F, Hansen KD, Aryee MJ, Langmead B, Irizarry R, Amdam GV, Feinberg AP 2012. Reversible switching between epigenetic states in honeybee behavioral subcastes. Nat Neurosci 15: 1371–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Jones PA 2012. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat Rev Genet 13: 484–492 [DOI] [PubMed] [Google Scholar]
- Kamatani Y, Wattanapokayakit S, Ochi H, Kawaguchi T, Takahashi A, Hosono N, Kubo M, Tsunoda T, Kamatani N, Kumada H, et al. 2009. A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nat Genet 41: 591–595 [DOI] [PubMed] [Google Scholar]
- Kaminsky ZA, Tang T, Wang S-C, Ptak C, Oh GHT, Wong AHC, Feldcamp LA, Virtanen C, Halfvarson J, Tysk C, et al. 2009. DNA methylation profiles in monozygotic and dizygotic twins. Nat Genet 41: 240–245 [DOI] [PubMed] [Google Scholar]
- Kilpinen H, Dermitzakis ET 2012. Genetic and epigenetic contribution to complex traits. Hum Mol Genet 21: R24–R28 [DOI] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA 2009. Circos: An information aesthetic for comparative genomics. Genome Res 19: 1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulis M, Heath S, Bibikova M, Queirós AC, Navarro A, Clot G, Martínez-Trillos A, Castellano G, Brun-Heath I, Pinyol M, et al. 2012. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet 44: 1236–1242 [DOI] [PubMed] [Google Scholar]
- Lachance J, Vernot B, Elbers CC, Ferwerda B, Froment A, Bodo J-M, Lema G, Fu W, Nyambo TB, Rebbeck TR, et al. 2012. Evolutionary history and adaptation from high-coverage whole-genome sequences of diverse African hunter-gatherers. Cell 150: 457–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, Storey JD 2007. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet 3: e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L 2009. GADEM: A genetic algorithm guided formation of spaced dyads coupled with an EM algorithm for motif discovery. J Comput Biol 16: 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, et al. 2008a. Worldwide human relationships inferred from genome-wide patterns of variation. Science 319: 1100–1104 [DOI] [PubMed] [Google Scholar]
- Li L, Fridley B, Kalari K, Jenkins G, Batzler A, Safgren S, Hildebrandt M, Ames M, Schaid D, Wang L 2008b. Gemcitabine and cytosine arabinoside cytotoxicity: Association with lymphoblastoid cell expression. Cancer Res 68: 7050–7058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MJ, Wang P, Liu X, Lim EL, Wang Z, Yeager M, Wong MP, Sham PC, Chanock SJ, Wang J 2012. GWASdb: A database for human genetic variants identified by genome-wide association studies. Nucleic Acids Res 40: D1047–D1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw A, Wiener M 2002. Classification and regression by randomForest. R News 2: 18–22 [Google Scholar]
- Mercier E, Droit A, Li L, Robertson G, Zhang X, Gottardo R 2011. An integrated pipeline for the genome-wide analysis of transcription factor binding sites from ChIP-Seq. PLoS ONE 6: e16432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson JJ, Loguercio S, Beyer A 2009. Detection and interpretation of expression quantitative trait loci (eQTL). Methods 48: 265–276 [DOI] [PubMed] [Google Scholar]
- Michaelson JJ, Alberts R, Schughart K, Beyer A 2010. Data-driven assessment of eQTL mapping methods. BMC Genomics 11: 502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud EJ, van Vugt MJ, Bultman SJ, Sweet HO, Davisson MT, Woychik RP 1994. Differential expression of a new dominant agouti allele (Aiapy) is correlated with methylation state and is influenced by parental lineage. Genes Dev 8: 1463–1472 [DOI] [PubMed] [Google Scholar]
- Niu N, Qin Y, Fridley BL, Hou J, Kalari KR, Zhu M, Wu T-Y, Jenkins GD, Batzler A, Wang L 2010. Radiation pharmacogenomics: A genome-wide association approach to identify radiation response biomarkers using human lymphoblastoid cell lines. Genome Res 20: 1482–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien TR, Kohaar I, Pfeiffer RM, Maeder D, Yeager M, Schadt EE, Prokunina-Olsson L 2011. Risk alleles for chronic hepatitis B are associated with decreased mRNA expression of HLA-DPA1 and HLA-DPB1 in normal human liver. Genes Immun 12: 428–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschos K, Allday MJ 2010. Epigenetic reprogramming of host genes in viral and microbial pathogenesis. Trends Microbiol 18: 439–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portela A, Esteller M 2010. Epigenetic modifications and human disease. Nat Biotechnol 28: 1057–1068 [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. 2007. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. 2009. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Rando OJ 2012. Daddy issues: Paternal effects on phenotype. Cell 151: 702–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, et al. 2006. Global variation in copy number in the human genome. Nature 444: 444–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rideout WM III, Eggan K, Jaenisch R 2001. Nuclear cloning and epigenetic reprogramming of the genome. Science 293: 1093–1098 [DOI] [PubMed] [Google Scholar]
- Sandoval J, Heyn H, Moran S, Serra-Musach J, Pujana MA, Bibikova M, Esteller M 2011. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics 6: 692–702 [DOI] [PubMed] [Google Scholar]
- Seong K-H, Li D, Shimizu H, Nakamura R, Ishii S 2011. Inheritance of stress-induced, ATF-2-dependent epigenetic change. Cell 145: 1049–1061 [DOI] [PubMed] [Google Scholar]
- Suzuki R, Shimodaira H 2006. Pvclust: An R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22: 1540–1542 [DOI] [PubMed] [Google Scholar]
- Waterland RA, Jirtle RL 2003. Transposable elements: Targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol 23: 5293–5300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambelli F, Pesole G, Pavesi G 2009. Pscan: Finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic Acids Res 37: W247–W252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeybel M, Hardy T, Wong YK, Mathers JC, Fox CR, Gackowska A, Oakley F, Burt AD, Wilson CL, Anstee QM, et al. 2012. Multigenerational epigenetic adaptation of the hepatic wound-healing response. Nat Med 18: 1369–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Cheng L, Badner JA, Chen C, Chen Q, Luo W, Craig DW, Redman M, Gershon ES, Liu C 2010. Genetic control of individual differences in gene-specific methylation in human brain. Am J Hum Genet 86: 411–419 [DOI] [PMC free article] [PubMed] [Google Scholar]