Abstract
Somatic mutations in kinase genes are associated with sensitivity of solid tumors to kinase inhibitors, but patients with metastatic cancer eventually develop disease progression. In EGFR mutant lung cancer, modeling of acquired resistance (AR) with drug-sensitive cell lines has identified clinically relevant EGFR tyrosine kinase inhibitor (TKI) resistance mechanisms such as the second-site mutation, EGFR T790M, amplification of the gene encoding an alternative kinase, MET, and epithelial–mesenchymal transition (EMT). The full spectrum of DNA changes associated with AR remains unknown. We used next-generation sequencing to characterize mutational changes associated with four populations of EGFR mutant drug-sensitive and five matched drug-resistant cell lines. Comparing resistant cells with parental counterparts, 18–91 coding SNVs/indels were predicted to be acquired and 1–27 were lost; few SNVs/indels were shared across resistant lines. Comparison of two related parental lines revealed no unique coding SNVs/indels, suggesting that changes in the resistant lines were due to drug selection. Surprisingly, we observed more CNV changes across all resistant lines, and the line with EMT displayed significantly higher levels of CNV changes than the other lines with AR. These results demonstrate a framework for studying the evolution of AR and provide the first genome-wide spectrum of mutations associated with the development of cellular drug resistance in an oncogene-addicted cancer. Collectively, the data suggest that CNV changes may play a larger role than previously appreciated in the acquisition of drug resistance and highlight that resistance may be heterogeneous in the context of different tumor cell backgrounds.
Over the past several decades, somatic mutations in genes encoding kinases have become associated with increased sensitivity of different solid tumors to kinase inhibitors. Examples include the gene products of specific “driver oncogenes” including EGFR, ALK, BRAF, and KIT, which are effectively targeted with gefitinib/erlotinib (Maemondo et al. 2010; Mitsudomi et al. 2010), crizotinib (Kwak et al. 2010), vemurafenib (Sosman et al. 2012), and imatinib (Demetri et al. 2002), in lung cancer (EGFR, ALK), melanoma (BRAF), and gastrointestinal stromal tumors (KIT), respectively. Unfortunately, virtually all patients with metastatic cancer eventually develop disease progression, limiting the effectiveness of these agents. Common mechanisms of acquired resistance include the development of second-site gene mutations (e.g., “gatekeeper mutations”) that alter binding of drug to target and re-activation of the original oncogene-driven kinase signaling pathway through the up-regulation of alternative kinases. For example, in patients with EGFR mutant lung adenocarcinomas harboring drug-sensitive mutations (deletions in exon 19 or the L858R point mutation in exon 21), tumor cells in more than half develop a second-site EGFR T790M mutation (Kobayashi et al. 2005; Pao et al. 2005), while 5%–10% acquire MET amplification (Bean et al. 2007; Engelman et al. 2007). Occasionally, changes in tumor histology have also been observed, with tumor cells displaying features of small-cell lung cancer or epithelial–mesenchymal transition (EMT) (Sequist et al. 2011).
A common laboratory method used to model acquired resistance involves the development of isogenic pairs of drug-sensitive and drug-resistant human tumor cell lines. Parental drug-sensitive cells are cultured in stepwise fashion with increasing concentrations of drug until cells emerge that are 50-fold to 100-fold less sensitive to growth inhibition. Cells are initially treated with a drug concentration at which 30% of the cells are growth inhibited or killed (GI30), and when cells resume normal growth patterns, the drug concentration is increased (Chmielecki et al. 2011; Ohashi et al. 2012). In EGFR mutant lung cancer, this type of modeling using EGFR tyrosine kinase inhibitors (TKIs) has reliably identified clinically relevant resistance mechanisms such as EGFR T790M, MET amplification, and EMT (Chmielecki et al. 2011; Ohashi et al. 2012). To date, the full spectrum of DNA mutations and copy number changes associated with such resistance mechanisms remains to be determined.
Next-generation sequencing (NGS) technologies augmented with bioinformatics analyses provide powerful approaches to screen for genome-wide genetic alterations in matched samples to identify various types of mutations associated with drug resistance. In a recent study, RNA sequencing (RNA-seq) was applied to detect mutations in drug-resistant clones developed from parental cell lines (Wacker et al. 2012). To our knowledge, the use of genome-wide DNA sequencing to compare drug-sensitive and drug-resistant cell lines has not yet been reported. Here, we used whole-genome sequencing (WGS) or whole-exome sequencing (WES) and bioinformatics analysis to characterize mutational changes associated with four populations of parental EGFR mutant drug-sensitive lines and five corresponding drug-resistant lines that were already known to harbor EGFR T790M mutations, MET amplification, or EMT, respectively (Fig. 1). These studies illustrate the power of NGS technologies to uncover genome-wide changes associated with drug resistance.
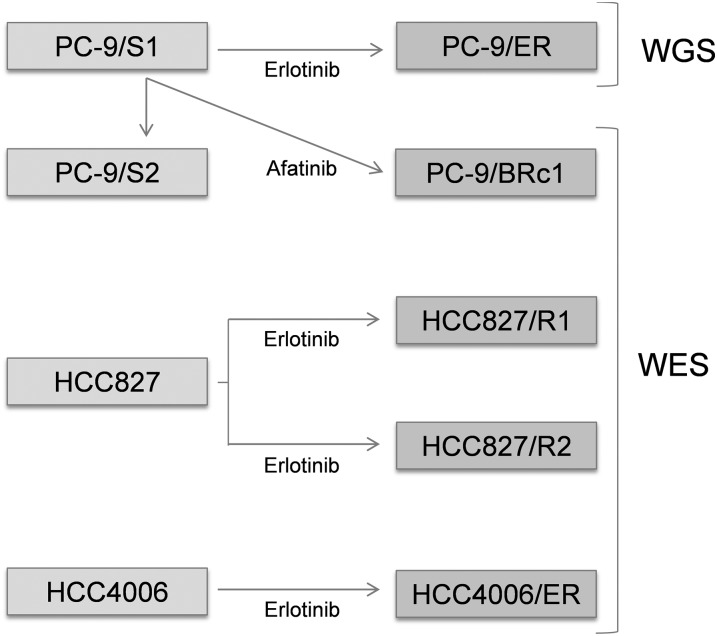
Figure 1.
Description of cell lines examined. (TKI) Tyrosine kinase inhibitor; (WGS) whole-genome sequencing; (WES) whole-exome sequencing. Cell lines in the left boxes are drug-sensitive, while those in the right boxes are drug-resistant. PC-9/S2, PC-9/ER, and PC-9/BRc1 were derived from PC-9/S1 cells; HCC827/R1 and HCC827/R2 were derived from HCC827 cells; HCC4006/ER were derived from HCC4006 cells. Comparisons between PC-9/S1 and PC-9/S2, PC-9/S1 and PC-9/ER, PC-9/S1 and PC-9/BRc1, HCC827 and HCC827/R1, HCC827 and HCC827/R2, and HCC4006 and HCC4006/ER were performed as detailed in the text.
Results
Spectrum of genetic alterations associated with an isogenic pair of drug-sensitive and drug-resistant cells: PC-9/S1 versus PC-9/ER (T790M)
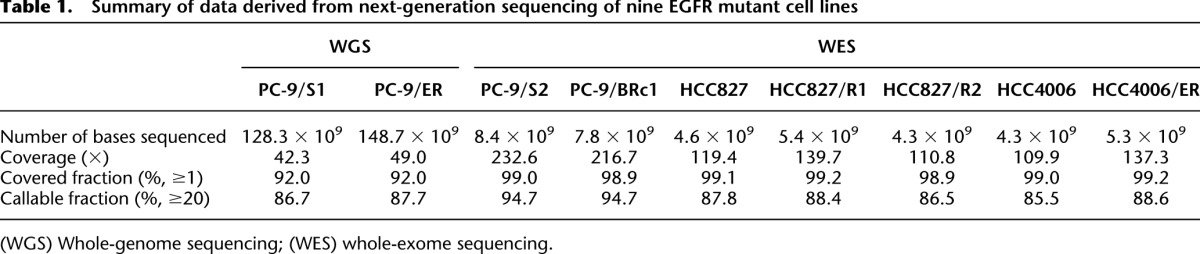
PC-9/S1 parental cells are known to harbor a drug-sensitive EGFR exon 19 deletion, while polyclonal PC-9/ER cells, developed after long-term culture in the EGFR TKI erlotinib, contain a second-site EGFR T790M mutation (Fig. 1; Chmielecki et al. 2011). PC-9/ER cells display sensitivity to the T790M-specific inhibitor WZ4002 (Zhou et al. 2009), suggesting that they remain dependent on EGFR signaling for survival (Supplemental Fig. S1). To determine the full spectrum of mutations associated with erlotinib sensitivity and resistance, we performed WGS on genomic DNA from both lines. We denote this parental cell line sequenced by WGS as PC-9/S1, to distinguish it from a second set of parental cells (PC-9/S2) sequenced by WES after multiple passages in culture in the absence of TKI selection (see next section and Fig. 1). For PC-9/S1 cells, a total of 128.3 × 109 bases were covered by short reads (100 bp, paired-end), with an average of 42.3× coverage of the human genome, and for PC-9/ER cells, 148.7 × 109 bases of short reads were obtained, with an average of 49.0× coverage (Table 1). These sequence reads covered ∼92.0% bases of the human reference genome (hg19) by at least one read and ∼87.2% bases by a depth of at least 20×. We then compared data from the resistant and parental lines to identify mutations that occurred at >20% allele frequency that were unique to each cell population. As expected, both parental and resistant cells were found to harbor the same EGFR exon 19 deletion (c.2235_2249del, p.E746_A750del, at chr7: 55242465–55242479), while only the resistant cells harbored EGFR T790M (c.C2369T, p.T790M, at chr7: 55249071).
Table 1.
Summary of data derived from next-generation sequencing of nine EGFR mutant cell lines
Using a set of optimized filtering criteria for high prediction accuracy (see Methods and Supplemental Fig. S2), we identified a total of 7060 novel single nucleotide variants (SNVs) and 7442 small insertions/deletions (indels) that were unique to PC-9/ER versus parental PC-9/S1 cells. Thirty-three SNVs (including 19 missense, three stop-gain, and 11 synonymous SNVs) and 11 indels were predicted to occur in exonic regions (Table 2). We chose for validation by direct sequencing the predicted exonic SNVs/indels that did not fail our manual review (see Methods) and were amenable to primer design. All selected SNVs (n = 15, 100%) and 86% of selected indels (n = 7) for validation were verified to be present only in PC-9/ER cell DNA by direct sequencing (Table 3; Supplemental Table S1). In the reverse comparison, nine and four predicted exonic SNVs and deletions, respectively, were unique to PC-9/S1 parental cells (Table 2); all selected SNVs (n = 4) and 50% of indels (n = 2) were validated (Fig. 2; Tables 3, 4). These data indicate that in this isogenic pair of cells, exonic mutations are both acquired and lost during the selection process for resistance, with more mutations being acquired than lost.
Table 2.
Summary of single nucleotide variants (SNVs) and small insertions/deletions (indels) unique to each cell line
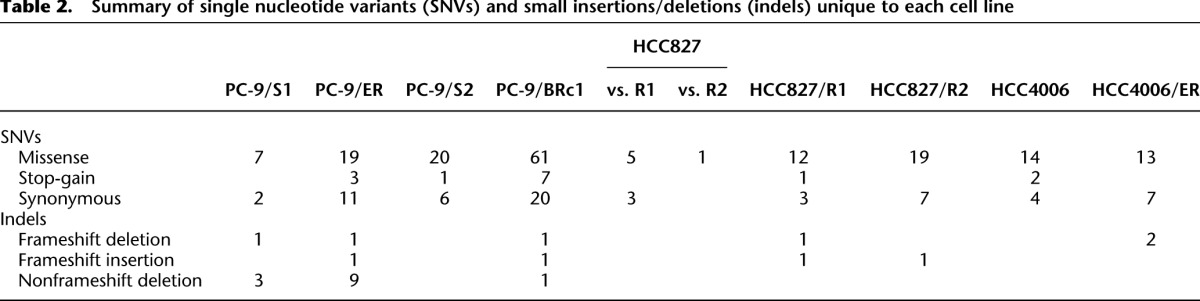
Table 3.
Summary of validation studies on putative SNVs and indels
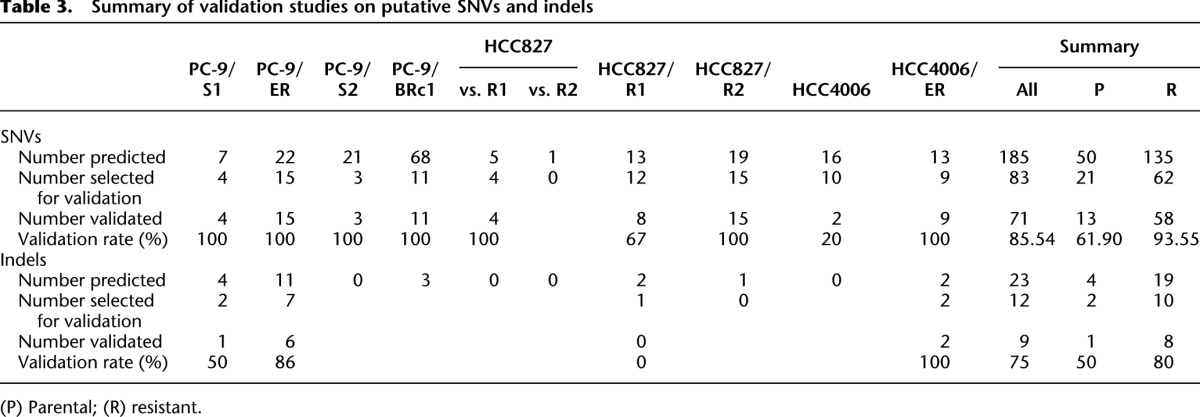
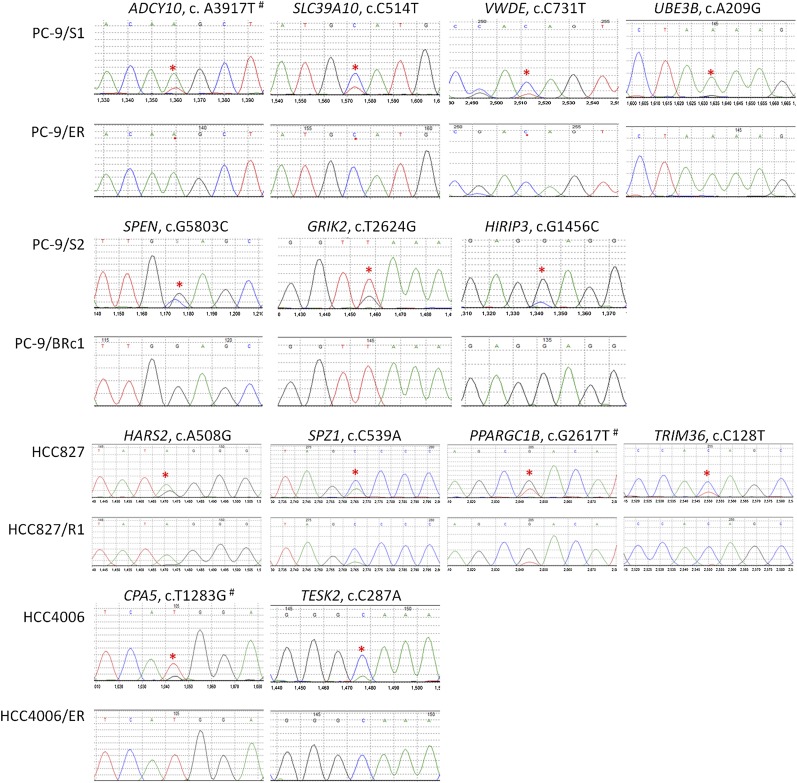
Figure 2.
Sanger sequencing chromatograms of mutations “lost” in drug-resistant cell lines compared with matched drug-sensitive cell lines. For each panel, the mutation marked by a red asterisk is shown in the sensitive line (top) and resistant line (bottom). (#) The mutation occurs in multiple transcripts with different nucleotide positions and/or amino acid positions. Detailed information is available in Table 4.
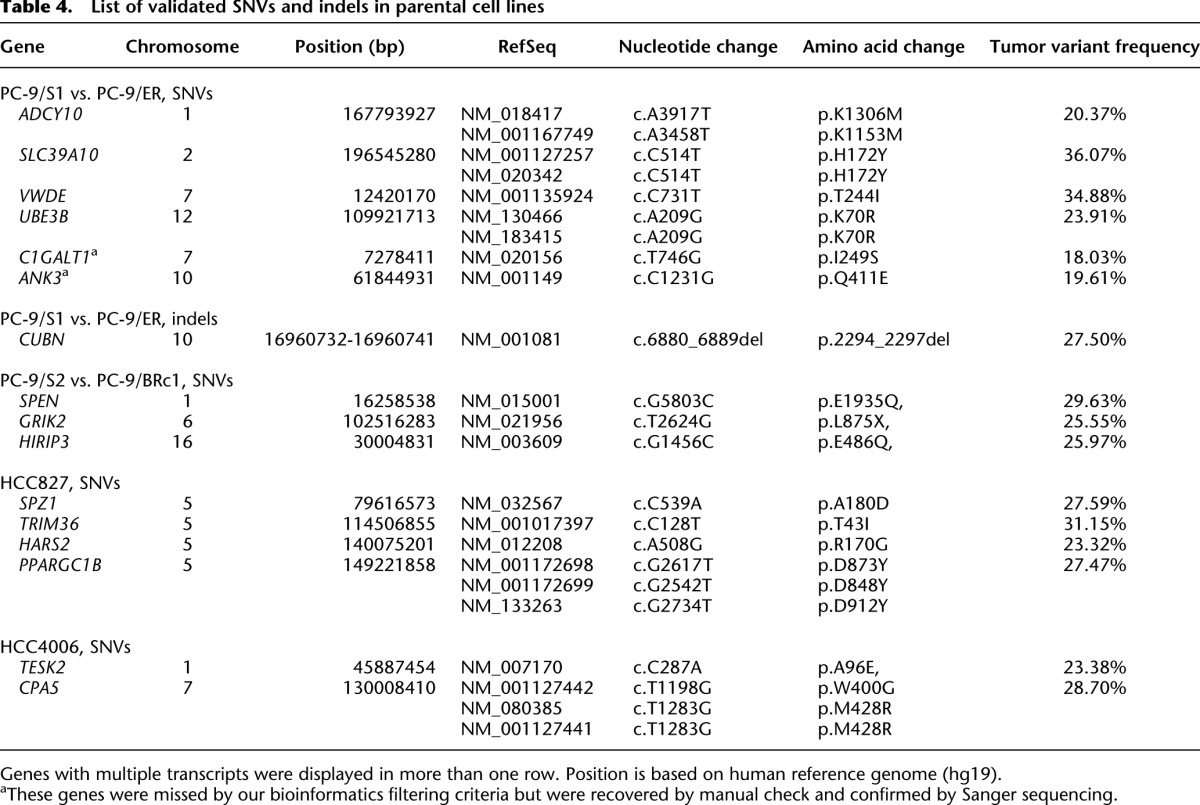
Table 4.
List of validated SNVs and indels in parental cell lines
We next applied the software tool Control-FREEC (Boeva et al. 2011, 2012) to detect CNVs uniquely aberrant in PC-9/ER cells compared with PC-9/S1 parental cells. While many small amplified/deleted regions were detected across the genome, there were three large blocks of amplifications (spanning >1 Mb) involving chromosomes 5, 7, and 22 (Supplemental Fig. S3; Supplemental Table S2). The 5p15.1–5p15.2 locus overlapped with a region we previously reported in tumor samples from patients with EGFR mutant lung cancer and acquired resistance to EGFR TKIs (Bean et al. 2007); the region spans ∼3.7 Mb and encompasses cancer genes collected from the Cancer Gene Census (CGC) database (Futreal et al. 2004) such as ANKH, CTNND2, DNAH5, FAM105A, FAM105B, and TRIO. The locus 7p11.2–7p13 involves EGFR, the amplification of which has been frequently reported in patients with acquired resistance (Balak et al. 2006). The third locus, which is at 22q12.3–22q13.1, spans ∼3.2 Mb and involves many genes including the CGC cancer gene MYH9. Large blocks of deletions were detected in 2q32–2q34, 7q31.1–7q35, 10p11.21–10p15.3, 22q11.21, and Xp21.1 (Supplemental Table S2). Loss of copy number in these loci involved multiple CGC genes such as IDH1, MET, SMO, and BRAF. Taken together with the SNV/indel data, these analyses show that more genes were affected by copy number changes than exonic SNVs/indels during the development of drug resistance.
Spectrum of genetic alterations associated with an isogenic pair of drug-sensitive and drug-resistant cells: PC-9/S2 versus PC-9/BRc1 (T790M)
Polyclonal PC-9 parental cells were treated with a different EGFR TKI, afatinib, and used to select for a T790M-harboring resistant line, PC-9/BRc1, which was derived from a single-cell clone (Chmielecki et al. 2011). Through WES, we compared the exomes in PC-9/BRc1 and PC-9/S2 cells (Fig. 1; see below). We obtained 8.4 × 109 bases of short reads (74-bp paired-end) for PC-9/S2 cells with an average of 232.6× coverage, and 7.8 × 109 bases of short reads for PC-9/BRc1 (216.7× coverage). These sequence reads covered ∼99.0% of bases of the targeted regions (NimbleGen SeqCap EZ Exome Library kit v2) by at least one read and ∼94.7% of bases by a depth of at least 20× (Table 1). A total of 88 SNVs and three indels were detected in exonic regions that were unique to PC-9/BRc1 cells, while 27 SNVs were unique to PC-9/S2 (Table 2). For validation, we selected mutations likely to have high functional impact (e.g., those indicated as “probably damaging” by PolyPhen-2 software) (Adzhubei et al. 2010). All of the selected SNVs (11 for PC-9/BRc1 and three for PC-9/S2) were validated (Tables 3, 4; Supplemental Table S3). Thus, again, we predicted more coding SNVs/indels in the resistant cell population compared with the parental cells. A greater number of changes may have been observed in PC-9/BRc1 cells than PC-9/ER cells, since the former were derived from a single cell clone, while the latter were polyclonal.
CNV detection using WES data can be variable, since interpretations can be affected by the non-uniform nature of exome capture reactions (Krumm et al. 2012). We therefore applied two software tools, VarScan 2 (Koboldt et al. 2012) and ExomeCNV (Sathirapongsasuti et al. 2011), to determine the CNVs in PC-9/BRc1 cells. To get more reliable results, we focused only on the regions detected by both tools. When using VarScan 2, we selected regions with >1000 bp and log ratios >0.25 or <−0.25. For ExomeCNV, we selected regions with >1000 bp with abnormal copy numbers (e.g., ≠2) (Supplemental Fig. S4; Supplemental Table S4). Large amplified regions encompassing CGC genes included 1p36.21–1p36.33 (CAMTA1, PRDM16, RPL22, TNFRSF14), 3q13.13–3q27.3 (BCL6, EIF4A2, ETV5, FOXL2, GATA2, GMPS, MECOM, MLF1, PIK3CA, RPN1, SOX2, WWTR1), and 21p11.1–21q22.3 (ERG, OLIG2, RUNX1, TMPRSS2, U2AF1) among others. PC-9/BRc1 cells also had regions of copy number loss, affecting cancer-related genes such as ABI1, GATA3, KIF5B, KLF6, and MLLT10 on 10p11.1–10p15.3, AKT1 on 14q32.33, and MYH9 and PDGFB on 22q12.3–22q13.1. More details are provided in Supplemental Table S4. Collectively, similar to PC-9/ER cells, we predicted that PC-9/BRc1 cells harbored more copy number changes than SNVs and indels when compared with their parental cell counterparts.
Spectrum of genetic alterations associated with the two PC-9 parental cell populations: PC-9/S1 versus PC-9/S2
To determine whether the DNA changes associated with acquired resistance in PC-9/ER and PC-9/BRc1 cells were random or due to drug selection, we compared the profiles of the two parental cell populations, PC-9/S1 and PC-9/S2. These cells were passaged about six to eight times (1.5 mo) in media without drug selection (Fig. 1). Using the same pipeline for paired samples and our standard cutoff of >20% mutation allele frequency for a called mutation, we did not find any coding SNVs/indels that uniquely occurred in either cell line, even allowing for differences in sequencing coverage (42.3× for PC-9/S1 and 232.6× for PC-9/S2 cells). Thus, the SNVs detected in the resistant cell lines were likely due to drug treatment and did not arise from the normal culturing process. We did not compare CNV differences because the significantly different depth of coverage provided by WGS and WES would strongly affect the CNV calling.
Spectrum of genetic alterations associated with an isogenic pair of drug-sensitive and drug-resistant cells: HCC827 versus HCC827/R1 (EGFR T790M) and HCC827/R2 (MET amplification)
We used WES to characterize the spectrum of mutations associated with a different set of isogenic pairs of cell lines. HCC827 cells, harboring an exon 19 deletion, are sensitive to erlotinib; drug selection in vitro led to two polyclonal resistant lines: HCC827/R1, which harbor the T790M mutation and lack MET amplification, and HCC827/R2, which lack T790M and display MET amplification (Ohashi et al. 2012). HCC827/R1 but not HCC827/R2 cells further display sensitivity to the T790M-specific TKI, WZ4002 (Supplemental Fig. S5); conversely, HCC827/R2 but not HCC827/R1 cells display sensitivity to a MET TKI, SGX-532 (Ohashi et al. 2012; data not shown). Details of the sequencing data are listed in Tables 1–3. As expected, all three lines harbored the same EGFR exon 19 deletion (c.2235_2249del, p.E746_A750del, at chr7: 55242465–55242479). In HCC827/R1 cells, the EGFR T790M point mutation (c.C2369T) was found manually at low allele frequency (7%), while HCC827/R2 cells did not contain any alleles with T790M.
We detected 16 exonic SNVs (12 missense, one stop-gain, and three synonymous) and two indels (Table 2) that were unique to HCC827/R1 cells compared with parental cells. Conversely, eight SNVs were found only in parental cells. In HCC827/R2 cells, 26 SNVs (19 missense and seven synonymous), and one indel (Table 2) were predicted to be unique, while only one SNV was detected as significant in the parental line (Table 2). Validation rates are shown in Table 3, and the validated SNVs/indels are shown in Supplemental Tables S5 and S7 for HCC827/R1 and HCC827/R2, respectively. Thus, as in PC-9 cells, HCC827 resistant cells harbored more genetic changes than parental cells.
We applied the same pipeline as in PC-9/BRc1 to detect CNVs for HCC827/R1 and for HCC827/R2, both of which were compared with HCC827 parental cells. In HCC827/R1, large amplifications were found in chromosomes 7 and 18, where the cancer genes BRAF (7q34) and BCL2 (18q21.33–18q22.1) are located, respectively (Supplemental Fig. S6). We also found an amplified region on 21p11.1–21q22.3 of unknown significance (Supplemental Table S6). Regions with fewer copies compared with parental cells were found in 5p11–5q35.3 (involving the CGC gene PDGFRB), 7p11.2–7p12.1 (involving the gene EGFR), and 12p12.2–12p13.33.
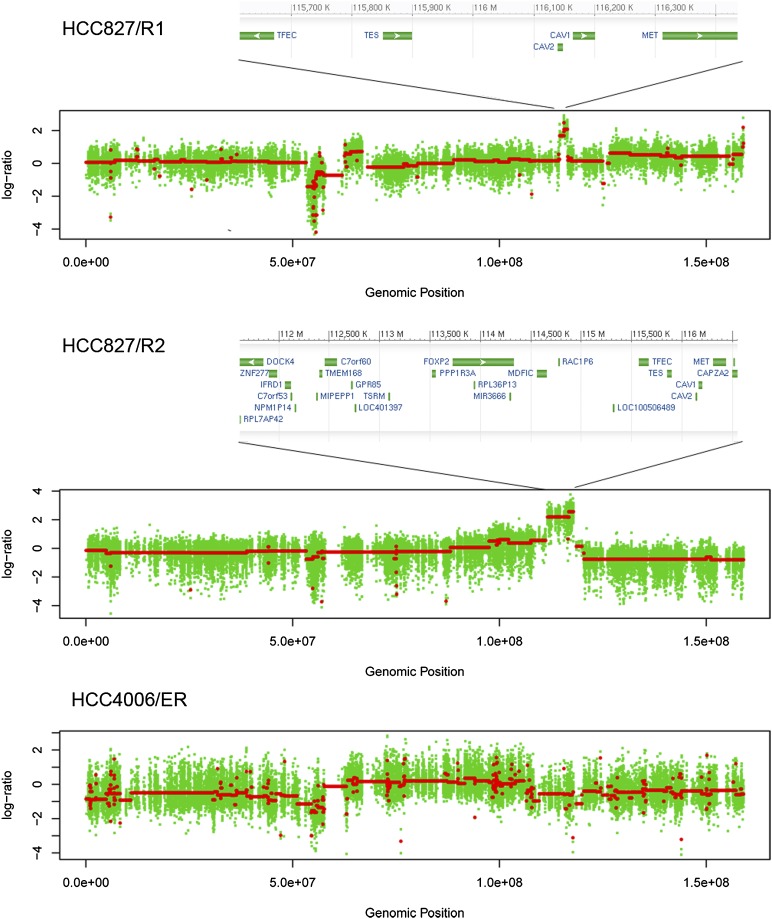
In HCC827/R2, amplifications were mainly detected in 5p15.2–5p15.33, 7q21.3–7q31.1, and 18q11.2 (Supplemental Fig. S7; Supplemental Table S8). On chromosome 7, there was a 6-Mb block encompassing MET (Fig. 3; Supplemental Fig. S7); this gene was known to be amplified by fluorescent in situ hybridization (FISH) (Ohashi et al. 2012). Interestingly, in the HCC827/R1 cell line, the same region displayed a different pattern: We found a sharp low-level peak spanning only ∼1.9 Mb (Fig. 3; Supplemental Fig. S6). Consistent with these data, HCC827/R1 cells did not display MET amplification by FISH (Ohashi et al. 2012). We specifically examined the amplified regions in both HCC827/R1 and R2 cells and found that the amplicons in both covered all MET coding regions. In HCC827/R1, the amplified region was predicted to have a low copy number, while in HCC827/R2, the amplified region was large and with a high copy number (Fig. 3; Supplemental Fig. S6).
Figure 3.
Copy number variation (CNV) regions on chromosome 7 for HCC827/R1, HCC827/R2, and HCC4006/ER cells. (X-axis) Genomic position; (y-axis) log2 ratio of CNVs in resistant versus sensitive cells. Red lines indicate the segments. The size of the MET amplicon is different in HCC827/R1 and HCC827/R2 cells. See text for details.
To validate these CNV changes further and to examine whether there were other structural variants that may affect MET, we conducted RNA-seq of these three cell lines and systematically searched for gene fusion events involving MET using FusionMap (Ge et al. 2011). We did not find any evidence for structural variations involving MET. We also examined exon-level and transcript-level expression intensities as measured by the fragments per kilobase of transcript per million fragments mapped (FPKM) algorithm. We found that all exons of MET were expressed in all three cell lines, with the highest expression in HCC827/R2 cells. Previous studies have shown that some lung adenocarcinomas harbor mutations in MET that result in skipping of exon 14 (Onozato et al. 2009). Analysis of our RNA-seq data indicated no evidence for exon skipping. Collectively, these data show that MET is amplified and expressed at different levels in HCC827/R1 and HCC827/R2 cells, with the highest amplification/expression in HCC827/R2 cells.
Spectrum of genetic alterations associated with an isogenic pair of drug-sensitive and drug-resistant cells: HCC4006 versus HCC4006/ER cells (EMT)
We next used WES to identify mutations associated with HCC4006 parental and polyclonal resistant cells, the latter of which developed features consistent with EMT (i.e., loss of E-cadherin, increased expression of vimentin, and spindle-like morphology) (Ohashi et al. 2012). Both cell lines harbored the same known EGFR mutation (i.e., 9-bp nonframeshift deletion [c.2239_2247del, p.747_749del, at chr7: 55242469–55242477] coupled with c.G2248C, p.A750P, chr7: 55242478). Neither harbored the T790M mutation as expected, and HCC4006/ER cells are resistant to the T790M-specific TKI, WZ4002 (Supplemental Fig. S8). We found 20 exonic SNVs (13 missense and seven synonymous SNVs) and two frameshift deletions unique to HCC4006/ER cells (Table 2), most of which were validated (Table 3; Supplemental Table S9). In contrast, 20 SNVs (14 missense, two stop-gain, and four synonymous SNVs) were predicted to be unique to parental cells; two of 10 coding SNVs were confirmed by direct sequencing (Table 4). Similar to our observations in the other resistant cell lines, more mutations were “selected for” during drug treatment, while fewer mutations were “selected against.”
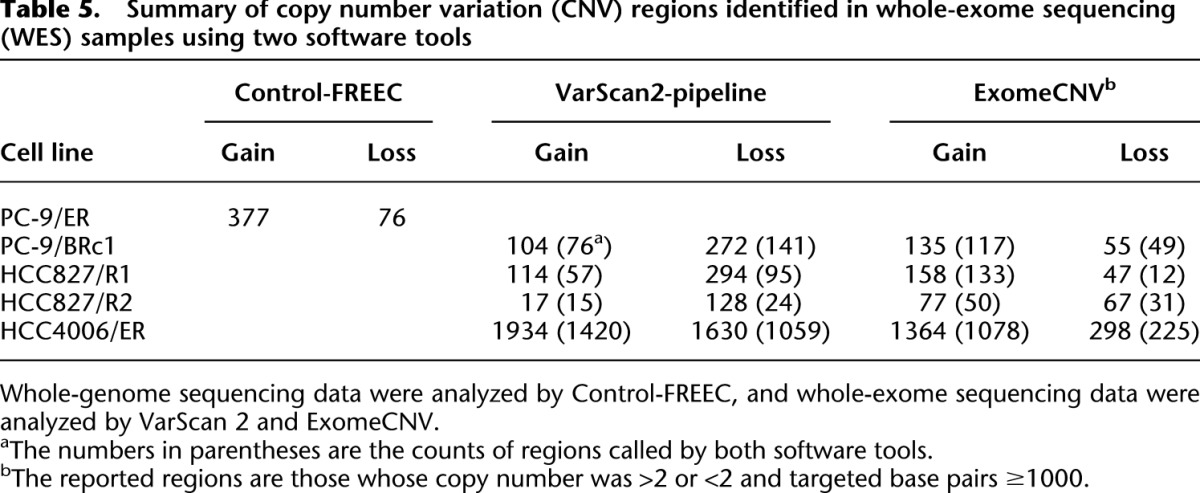
Our CNV analysis revealed that compared with parental cells, HCC4006/ER cells displayed a large number of duplications/deletions across the whole genome (Supplemental Fig. S9). Surprisingly, the number of CNV gains and losses were at least 10-fold greater than that seen in the other cell line comparisons (Table 5). Although the numbers of regional gains/losses might be significantly affected by the size of CNVs and the segmentation methods adopted by different software tools, the observed trend of many more aberrant CNVs in HCC4006/ER was clearly supported by the actual depth of coverage at CNV regions, regardless of software tools (data not shown). The most significantly altered region involved a deletion on chromosome 11, spanning 7.7 Mb in 11p13–11p12 and encompassing the cancer genes WT1 and LMO2.
Table 5.
Summary of copy number variation (CNV) regions identified in whole-exome sequencing (WES) samples using two software tools
Sample relatedness
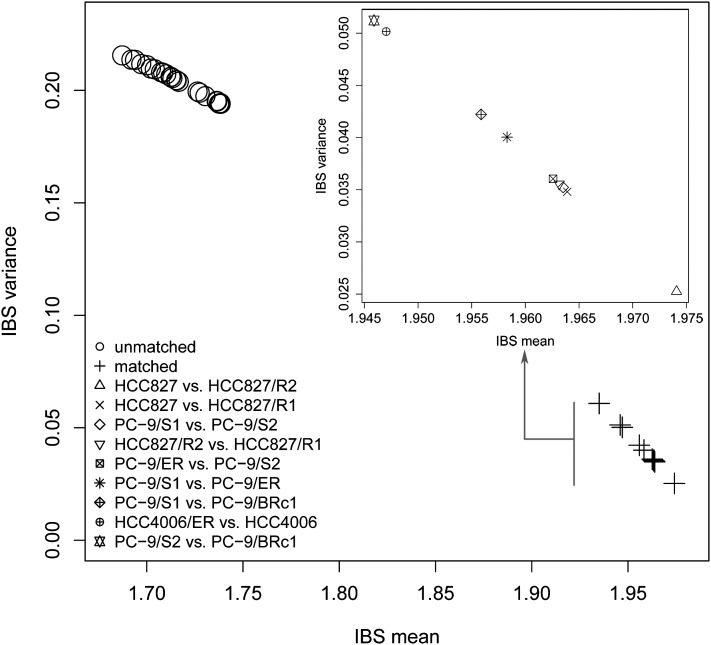
To quantitatively assess genetic relationships between parental/resistant cell line pairs, we adapted the genetic concept of measuring relatedness between individuals based on their shared genetic information. We hypothesized that even though each cell line displayed unique mutations, truly matched parental and resistant cell line samples should share more common SNVs than with unmatched lines. To test this, we computed pairwise identity-by-state (IBS) (Goh et al. 2011) based on the called SNVs for all pairs of cell lines formed by any two of the nine cell lines, regardless of whether they were matched or unmatched. As expected, the four PC-9-related cell lines, the three HCC827-related cell lines, and the two HCC4006-related cell lines each grouped together (Fig. 4), while all the other pairs did not. These data support the notion that even though each line acquired mutations during drug selection, cell lines generated from the same parental cell line remained more closely related to each other than to the other lines. Moreover, the data internally confirm that the samples were not contaminated with each other throughout the process of drug selection and sequencing.
Figure 4.
Pairwise comparison of samples. Identity-by-state (IBS) analysis was applied to compute the shared alleles for each pair of cell lines, with the mean on the x-axis and the variance on the y-axis. On the main panel, each point represents a pair of cell lines, regardless of whether they were matched (denoted by +) or not (denoted by a circle dot). In the internal panel, the truly matched sensitive-resistant pairs were enlarged to show the details.
Mutation patterns
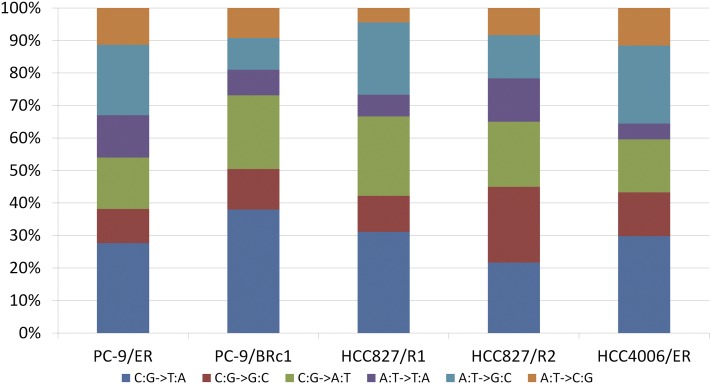
We compared mutation patterns in each of the cell lines (Fig. 5). In both PC-9/ER and PC-9/BRc1 resistant cells, the most prevalent mutations were C:G → T:A transitions, followed by C:G → A:T transversions. C:G → T:A transitions are predominantly seen in lung cancers from never/light smokers, while C:G → A:T transversions are more predominant in smokers (Govindan et al. 2012). Similar data were obtained for HCC827/R1, HCC827/R2, and HCC4006/ER cells, although the numbers of mutations were low. The transition over transversion ratio (Ti/Tv) was 2.01 for both PC-9/S1 and PC-9/ER cells (WGS) and varied slightly for each pair of WES samples (HCC827: 2.42, HCC827/R1: 2.37, HCC827/R2: 2.46, HCC4006: 2.51, and HCC4006/ER: 2.40). Since there was not much difference in the Ti/Tv ratios among the drug-sensitive and drug-resistant lines, we could not discern whether TKI treatment selected for certain types of mutations over others.
Figure 5.
Patterns of mutations that uniquely occurred in each resistant cell line.
We then investigated these SNVs in greater detail to identify any genome-wide patterns of their location. For the exome mutations, we surveyed the following genomic features: GC content (Karolchik et al. 2003), DNA replication timing (Hansen et al. 2010), presence of lamina-associated domains (Guelen et al. 2008), chromosome banding (Furey and Haussler 2003), and recombination rate (Kong et al. 2002). We found that there were no mutations in repeat elements (Jurka 2000) and CpG islands (Irizarry et al. 2009). The distribution of GC content at a resolution of 200 bp around each SNV did not display a different pattern compared with the genome-wide background, with mean GC content of 0.4. For chromosome banding, recombination rate, DNA replication timing, and lamina-associated domains, we did not detect any significant enrichment. For instance, almost half of the SNVs were located in “gneg” regions, while the other half resided in “gpos” regions (i.e., recognized stain values from Giemsa stains).
We next surveyed whether the SNVs were preferentially located in specific regions within genes, for instance, the C or N terminus. We found an excess of these mutations located in the 5′-UTR regions of genes (Supplemental Table S10). This observation might be due to the limited number of SNVs available and will be confirmed in future studies.
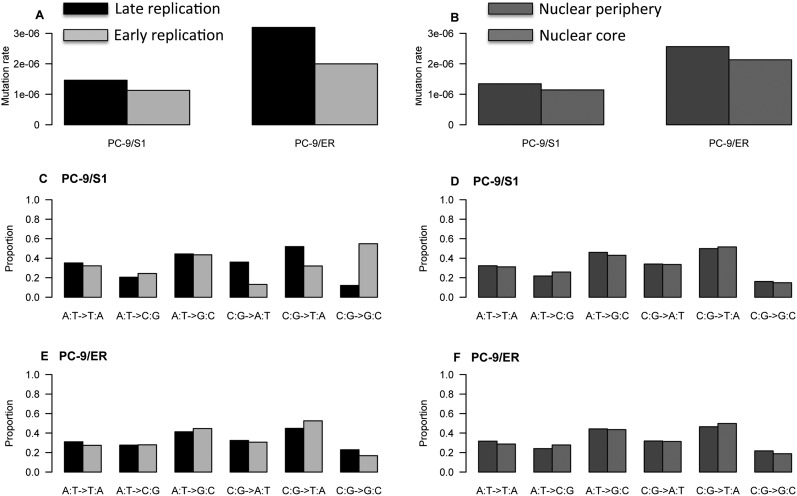
We then performed similar analyses for the WGS data of PC-9/ER and PC-9/S1 cells. We overlaid the SNVs with information on DNA replication timing and lamina-associated domains (Fig. 6). We found that there was a higher frequency of SNVs in “constant late” replication timing zones as compared with “constant early” replication timing zones (χ2 P-value < 10−5). These replication timing zones were identified based on consistency in the patterns across eight different cell types (Hansen et al. 2010). These findings are consistent with previous data showing an enrichment of mutation frequencies in late replication domains across multiple different cell types (Liu et al. 2013). We further identified an enrichment of SNV frequencies in genomic regions harboring lamina-associated domains compared with the remainder of the nucleus (χ2 P-value < 10−5).
Figure 6.
Patterns of SNV frequencies (A,B) and signatures (C–F) across different stratifications of genomic material.
Finally, in each case, we examined the mutation signatures, i.e., the six different types of nucleotide substitutions that might arise (AT|TA, AT|CG, AT|GC, CG|AT, CG|TA, and CG|GC) (Fig. 6). The mutation signatures stratified by lamina-associated domains were quite similar in the two samples, with a correlation of 0.98, whereas the correlation of mutation transversion patterns stratified by DNA replication timing was less similar, with a correlation of only 0.27.
Mutations shared across resistant cells
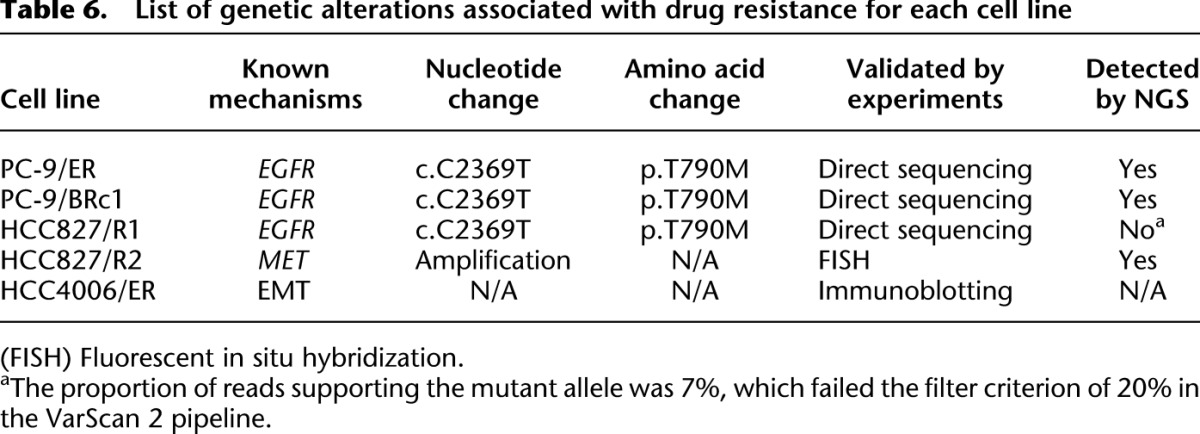
We compared data from all the resistant lines to determine if there were mutations shared by cells regardless of the known mechanisms of acquired resistance (i.e., EGFR T790M, MET, or EMT). While EGFR T790M mutations were found as expected in three of five resistant lines (PC-9/ER, PC-9/BRc1, HCC827/R1) (Table 6), surprisingly, only one gene was observed to be mutated in more than one line. LRP1B mutations were found in two cell lines, with three mutations in PC-9/BRc1 and one in HCC827/R1 cells.
Table 6.
List of genetic alterations associated with drug resistance for each cell line
Among CNV changes across drug-resistant cell lines, chromosome 7 harbored the most frequent co-occurring regions. Amplified regions on 7q around MET in HCC827/R1 and HCC827/R2 overlapped with each other as discussed above (Fig. 3). Changes in regions on 7p, especially involving EGFR, were also observed; e.g., amplification in PC-9/ER (Supplemental Table S2) and amplification/deletion in HCC827/R1 (Supplemental Table S6). Adjacent regions on 5p, which have been frequently reported in lung cancer samples (Bean et al. 2007; Weir et al. 2007), were detected both in PC-9/ER (Supplemental Table S2) and HCC827/R2 cells (Supplemental Table S8).
Driver gene specification
Finally, to determine whether there were other potential driver genes, we systematically searched for non-silent SNVs/indels located in kinase genes, especially those that might impact the three key phosphorylation residues; i.e., serine, threonine, and tyrosine. We assessed the functional impact of SNVs using PolyPhen-2 (Adzhubei et al. 2010) and SIFT (Kumar et al. 2009) algorithms, which predict damage to protein function or structure based on amino acid conservation and structural features. A total of six kinase genes were found to harbor non-silent SNVs/indels occurring in six cell lines (Supplemental Table S11). Among them, only three variants were located within kinase domains; C2369T (T790M) in EGFR in PC-9/ER, PC-9/BRc1, and HCC827/R1 cells, A1491T (E497D) in HIPK3 in HCC827/R2 cells, and C287A (A96E) in TESK2 in HCC4006 cells (Supplemental Fig. S10). Both the C2369T (T790M) mutation in EGFR and the A1491T (E497D) mutation in HIPK3 are predicted to be “deleterious” (SIFT score < 0.05 or PolyPhen-2 ≥ 0.5), while C287A (A96E) in TESK2 occurred in parental cells and is predicted to be “benign.” However, only the C2369T (T790M) mutation in EGFR impacts phosphorylation sites. Put together, these results suggest that the T790M in EGFR is the most likely mutation affecting drug resistance in the cells in which it was detected.
Discussion
In the past decade, multiple new targeted therapies have shown remarkable anti-tumor activity in genetically defined “oncogene-addicted” cancers (Demetri et al. 2002; Kwak et al. 2010; Maemondo et al. 2010; Mitsudomi et al. 2010; Sosman et al. 2012). However, acquired resistance remains a significant obstacle limiting the survival of patients with metastatic disease. Many mechanisms have been identified, but comprehensive genomic profiles of resistant tumor cells have not yet been established. Here, we used a model system of “oncogene addiction”—isogenic pairs of drug-sensitive and drug-resistant EGFR mutant lung cancer cells—and next-generation sequencing to characterize genome-wide changes associated with the acquisition of drug resistance in vitro. Importantly, the study of these EGFR mutant cells has already identified mechanisms of resistance found in human patient samples (i.e., secondary EGFR mutations, MET amplification, and EMT) (Chmielecki et al. 2011; Ohashi et al. 2012), suggesting that additional genetic changes identified are likely to have clinical relevance as well. To our knowledge, this is the first comprehensive analysis using WGS or WES of isogenic pairs of drug-sensitive and drug-resistant cell lines.
Comparing resistant cells with their matched parental counterparts, we identified 18–91 coding SNVs/indels that were acquired and 1–27 that were lost during drug treatment. While the secondary EGFR T790M mutation was found appropriately in the two resistant lines known to harbor this mutation, very few exonic SNVs/indels were shared across resistant lines, and many of the additional mutations identified did not have obvious biological significance. Analysis of mutation spectra across parental cells sequenced at different times and the resistant cells treated with either erlotinib or afatinib suggest that the SNVs/indels that were acquired or lost were due to drug selection, not just random mutation during in vitro culturing. These data illustrate five important principles. First, WGS/WES can be used to detect resistance mechanisms in isogenic pairs of lines. Second, the number of exonic SNVs/indels that differ among isogenic pairs of lines is relatively low (magnitude of only 102). Third, additional biological studies are needed to determine if many mutations are just “passengers” or, indeed, contribute to gain of fitness in the acquisition of acquired resistance. Fourth, analysis of the Ti/Tv ratios in treated cells suggested that TKI treatment does not significantly alter the ratio of transition/transversion mutations induced in cells. Finally, by extrapolating the findings from cell lines to human tumors, the acquisition of resistance may be unique in each individual patient and even within individual tumors within patients.
When investigating genome-wide patterns of these SNVs, we identified some interesting trends. For instance, we found that the SNVs were enriched in 5′ UTRs within protein-coding genes as compared with 3′ UTRs based on exome sequencing data. When investigating WGS data, we found that there was a significant difference in SNV frequency as well as the mutation signatures in genomic material with late replication timing as well as those containing nuclear lamina-associated domains. These data suggest that certain areas of the genome might be more prone to accumulation of SNVs.
Surprisingly, we observed more CNV changes than SNV/indel changes across all resistant lines, and the one line that had an EMT phenotype displayed significantly higher levels of CNV changes than the other lines with acquired resistance. These observations suggest that CNV changes may play a larger role than previously appreciated in the acquisition of drug resistance and again highlight that resistance may be heterogeneous in the context of different tumor cell backgrounds.
This study has some limitations. For example, WGS was performed on one isogenic pair of lines, while WES was used for the remaining pairs. WGS enables detection of all types of possible mutations, including SNVs, indels, CNVs, and structural variants (SVs), while WES has limited ability to identify SVs. However, WES generally delivers higher coverage than WGS (>100× vs. ∼40×; Illumina HiSeq 2000 platform), which allows for greater power in discovering SNVs/indels that have low allele frequency in a cell population. Here, to enable comparison of WGS and WES data, we focused on detecting SNVs/indels with >20% allele frequency. Furthermore, for most of the cell lines, we did not perform whole transcriptome sequencing, which could enable the detection of changes at the RNA level, such as alternative splicing, gene-fusion events, etc. (Liu et al. 2012). In future studies, we plan to explore the significance of mutations that occur at both DNA (e.g., lower allele frequency) and RNA levels (e.g., transcriptional level).
A second limitation involves the use of WES data to call CNVs. While CNV detection using WGS data has been successfully applied in cancer (Campbell et al. 2008; Chiang et al. 2009; Dahlman et al. 2012), WES data have only recently been proven to be practically workable. Since WES data are vulnerable to various biases such as GC content, target capture reactions, and non-uniform data distribution, caution should still be taken when detecting CNV changes from WES data. Because the false discovery rate in CNV calls can be high, especially in whole-exome sequencing data, we applied two computational tools for CNV detection and focused on the consistent regions called by both tools to improve data quality. Note that amplification of the entire MET gene in HCC827/R2 and in HCC827/R1 was detected by both tools, providing evidence of the quality of the CNV changes we detected.
A third limitation involves the various cell lines examined. All of the parental lines were derived from polyclonal populations, and only the PC-9/BRc1 resistant line was derived from a single-cell clone. To determine if the identified SNVs/indels coexist in all or only some of the resistant cells, we would need to perform single-cell sequencing from multiple clonally derived cell populations. In addition, the PC-9/S1 and PC-9/S2 control cells were just two splits from starting polyclonal population of cells grown separately for ∼1.5 mo, making them a less compelling control than if we had examined cells cultured for longer periods of time in the absence of drug selection. These issues can be addressed in future studies.
In summary, these results demonstrate a framework for studying the evolution of drug-related genetic variants over time and provide the first genome-wide spectrum of mutations associated with the development of cellular drug resistance in an oncogene-addicted cancer. In future studies, we plan to use this framework to examine the effect of different types and doses of targeted therapies on the evolution of drug resistance and to extend these analyses to mechanisms of acquired resistance to cytotoxic chemotherapies and radiation.
Methods
Cell culture
EGFR-mutant TKI-sensitive parental cell lines PC-9, HCC827, and HCC4006 were cultured in erlotinib or afatinib following well-established TKI dose-escalation protocols to develop PC-9/ER, PC-9/BRc1, HCC827/R1, HCC827/R2, and HCC4006/ER cells (Chmielecki et al. 2011; Ohashi et al. 2012). Details of cell culture conditions and treatments were described in Ohashi et al. (2012).
Next-generation sequencing
DNAs were extracted from each cell line using a DNeasy kit (Qiagen). PC-9/S1 and PC-9/ER DNA samples were submitted for whole-genome sequencing on an Illumina Genome Analyzer IIx platform. Whole-exome sequencing of PC-9/S2 and PC-9/BRc1 samples was conducted on an Illumina HiSeq 2000 platform using the NimbleGen SeqCap EZ Exome Library kit v2. HCC827, HCC827/R1, HCC827/R2, HCC4006, and HCC4006/ER DNA samples were submitted for whole-exome sequencing on an Illumina HiSeq 2000 platform using the Agilent SureSelect 38-Mb Kit.
Read mapping and alignment
Quality-control analysis of sequence reads was performed using FastQC software (FastQC; http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads that failed to pass quality control were removed from further analysis. We mapped the reads of each sample to the human reference genome (hg19) using BWA (version 0.5.9-r16) (Li and Durbin 2009). Local realignment was performed around small indels using the Genome Analysis Toolkit (GATK) (DePristo et al. 2011). The base quality scores initially reported by Illumina platform were recalibrated based on covariates of the read group, the reported base quality score, the machine cycle, and the combination of the base and its ahead base. After post-alignment refinement and removal of duplicate reads, we called somatic variants using VarScan 2 (Koboldt et al. 2012). The pipeline is shown in Supplemental Figure S2.
Detection of unique variants
To search for cell-line-specific variants, we performed the following comparisons: (1) PC-9/S1 unique variants compared with PC-9/ER, (2) PC-9/ER unique variants compared with PC-9/S1, (3) PC-9/S2 unique variants compared with PC-9/BRc1, (4) PC-9/BRc1 unique variants compared with PC-9/S2, (5) PC-9/S1 unique variants compared with PC-9/S2, (6) PC-9/S2 unique variants compared with PC-9/S1, (7) HCC827 unique variants compared with HCC827/R1, (8) HCC827 unique variants compared with HCC827/R2, (9) HCC827/R1 unique variants compared with HCC827, (10) HCC827/R2 unique variants compared with HCC827, (11) HCC4006 unique variants compared with HCC4006/ER, and (12) HCC4006/ER unique variants compared with HCC4006. In each case, the VarScan 2 “somatic” model was executed designating the targeted cell line as “tumor” and the cell line to be compared as “normal.”
To select high-confidence SNVs, we started with the somatic SNVs classified as “high confidence” by VarScan 2 and performed the following filtering: (1) at least 15 supporting reads in the tumor sample at the position; (2) at least five reads supporting the mutation allele; (3) supporting reads for the mutation allele in both the forward and reverse strands; (4) somatic P-values < 0.05; (5) the average base quality for variant-supporting reads was >20; and (6) if there were three SNVs within a 10-bp window, all of them were removed. We further removed SNVs that occurred in dbSNP build 131 or the 1000 Genomes Project data set and denoted what remained as novel “somatic” SNVs (The 1000 Genomes Project Consortium 2010). The functional impact of non-silent SNVs was assessed using the PolyPhen-2 (Adzhubei et al. 2010) and SIFT (Kumar et al. 2009) algorithms, which predict the effects on protein functions based on the degree of amino acid conservation and structural information. For high-confidence indels, we implemented similar filtering criteria as for SNVs.
Copy number variations (CNVs)
WGS data and WES data could behave differently in that WES data are more vulnerable to system biases such as the exome capture reaction. We therefore applied different analysis pipelines to detect CNVs in these two data types. For WGS samples, we detected CNVs using the software tool Control-FREEC (Boeva et al. 2011, 2012) with all default parameters. For CNVs in WES data, due to the non-uniform nature of the exome capture reaction, we applied two software tools and focused on the consensus calls by both tools in order to obtain high-confidence results. We first executed the “copynumber” function in VarScan 2 in the four resistant cell lines versus their respective parental cell lines, i.e., (1) PC-9/BRc1 versus PC-9/S2, (2) HCC827/R1 versus HCC827, (3) HCC827/R2 versus HCC827, and (4) HCC4006/ER versus HCC4006. The uniquely mapped reads (e.g., through SAMtools view -q 1) were used for this analysis. To adjust the potential biases introduced by different sample depth, we included a data ratio computed based on the uniquely mapped reads and the read length in the normal and tumor samples following the instruction of VarScan 2. The candidate CNV regions were filtered using the “copyCaller” option of VarScan 2 and then smoothed and segmented by the DNAcopy package (Seshan VE, Olshen A. Cited August 2012. DNAcopy: DNA copy number data analysis. R package version 1.24.20) from the Bioconductor project (Reimers and Carey 2006). Secondly, we applied the R package ExomeCNV (Sathirapongsasuti et al. 2011) to detect CNVs from the WES samples. ExomeCNV takes the targeted intervals as units and determines a log ratio for each interval based on the mapped reads in a pair of matched samples.
Direct dideoxynucleotide-based sequencing
Parental or TKI-resistant specific SNPs and short indels were validated by direct sequencing. Cell line DNAs were used as template for PCR amplification. M13-tagged gene-specific primers were designed using Primer3 software (Rozen and Skaletsky 2000). Sequence chromatograms were analyzed using Mutation Surveyor software (SoftGenetics, LLC) and manual inspection.
Sample relatedness
To assess the correlations among samples, we adopted the calculation of pairwise identity-by-state (IBS) (Goh et al. 2011) based on the called SNVs. For the nine cell lines sequenced in this study, we iteratively compared any two of them, regardless of whether they were matched or unmatched. This resulted in 9 × 8/2 = 36 pairs of cell lines. For each pair, we first obtained the overlapping positions where a SNV is reported in both cell lines and calculated the number of shared alleles at each position. The average value and standard deviation (SD) of the number of shared alleles for all positions were calculated for each pair of cell lines, which were then used to assess the correlations among samples. A higher average number and a lower scale of SD of the shared alleles indicate that the two cell lines share more identical SNVs and, thus, are more likely related to each other than to others.
RNA-seq data analysis
Total RNAs were extracted from HCC827, HCC827/R1, and HCC827/R2 cell lines using a Qiagen RNeasy mini kit. The Illumina Tru-Seq RNA sample prep kit was used for library preparation. Then, RNA sequencing was performed in the Vanderbilt Technologies for Advanced Genomics (VANTAGE) core. Paired-end reads with 50 bp in length were generated by an Illumina HiSeq 2500 and were initially mapped to the human reference genome and human transcriptome using the software TopHat v2.0.8 (Trapnell et al. 2009). We used FusionMap (Ge et al. 2011) to search for potential gene fusion events that might be involved in MET. Gene expression levels were measured by the fragments per kilobase of transcript per million fragments mapped (FPKM) algorithm (Trapnell et al. 2010).
Data access
All predicted variants are available in Supplemental Table S12. The sequencing data from this study can be accessed at the NCBI Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra) under accession numbers SRP022942 and SRP022943.
Competing interest statement
W.P. has received consulting fees from MolecularMD, AstraZeneca, Bristol-Myers Squibb, Symphony Evolution, and Clovis Oncology and research funding for other projects from Enzon, Xcovery, AstraZeneca, and Symphogen. W.P. and K.P. are part of a patent regarding EGFR T790M mutation testing that was licensed by Memorial Sloan-Kettering Cancer Center to MolecularMD. K.B.D. received an honorarium from Illumina, Inc.
Acknowledgments
This work was supported by funds from the National Institutes of Health (NIH) NCI grants R01CA121210, P01CA129243, and U54CA143798 and American Association for Cancer Research, Stand Up to Cancer Innovative Research Grant (SU2C) SU2C-AACR-IRG0109. W.P. received additional support from Vanderbilt’s Specialized Program of Research Excellence in Lung Cancer grant (P50CA90949) and from the VICC Cancer Center Core grant (P30CA68485). P.J. and Z.Z. received support from SU2C-AACR-IRG0109. K.P. received support from NIH/NCI grants R01CA120247 and R00CA131488, the Labrecque Foundation, and Uniting Against Lung Cancer. Z.Z. received additional support from NIH grants R01LM011177 and P30CA68485 and Ingram Professorship Funds. L.L., F.M., and W.P. received support from NIH grant U54CA143798.
Author contributions: The study was designed by W.P. and Z.Z. Analyses were carried out by P.J., J.X., Z.Z., L.L., F.M., K.B.D., and W.P. Experimental work was carried out by H.J., C.B.M., V.P., K.O., and K.P. This manuscript was written by P.J., W.P., F.M., and Z.Z., with contributions from the other authors.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.152322.112.
References
- The 1000 Genomes Project Consortium 2010. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR 2010. A method and server for predicting damaging missense mutations. Nat Methods 7: 248–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balak MN, Gong Y, Riely GJ, Somwar R, Li AR, Zakowski MF, Chiang A, Yang G, Ouerfelli O, Kris MG, et al. 2006. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin Cancer Res 12: 6494–6501 [DOI] [PubMed] [Google Scholar]
- Bean J, Brennan C, Shih JY, Riely G, Viale A, Wang L, Chitale D, Motoi N, Szoke J, Broderick S, et al. 2007. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci 104: 20932–20937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeva V, Zinovyev A, Bleakley K, Vert JP, Janoueix-Lerosey I, Delattre O, Barillot E 2011. Control-free calling of copy number alterations in deep-sequencing data using GC-content normalization. Bioinformatics 27: 268–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeva V, Popova T, Bleakley K, Chiche P, Cappo J, Schleiermacher G, Janoueix-Lerosey I, Delattre O, Barillot E 2012. Control-FREEC: A tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics 28: 423–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PJ, Stephens PJ, Pleasance ED, O’Meara S, Li H, Santarius T, Stebbings LA, Leroy C, Edkins S, Hardy C, et al. 2008. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet 40: 722–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang DY, Getz G, Jaffe DB, O’Kelly MJ, Zhao X, Carter SL, Russ C, Nusbaum C, Meyerson M, Lander ES 2009. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat Methods 6: 99–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmielecki J, Foo J, Oxnard GR, Hutchinson K, Ohashi K, Somwar R, Wang L, Amato KR, Arcila M, Sos ML, et al. 2011. Optimization of dosing for EGFR-mutant non-small cell lung cancer with evolutionary cancer modeling. Sci Transl Med 3: 90ra59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlman KB, Xia J, Hutchinson K, Ng C, Hucks D, Jia P, Atefi M, Su Z, Branch S, Lyle PL, et al. 2012. BRAFL597 mutations in melanoma are associated with sensitivity to MEK inhibitors. Cancer Discov 2: 791–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demetri GD, von Mehren M, Blanke CD, Van den Abbeele AD, Eisenberg B, Roberts PJ, Heinrich MC, Tuveson DA, Singer S, Janicek M, et al. 2002. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med 347: 472–480 [DOI] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43: 491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, et al. 2007. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 316: 1039–1043 [DOI] [PubMed] [Google Scholar]
- Furey TS, Haussler D 2003. Integration of the cytogenetic map with the draft human genome sequence. Hum Mol Genet 12: 1037–1044 [DOI] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR 2004. A census of human cancer genes. Nat Rev Cancer 4: 177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H, Liu K, Juan T, Fang F, Newman M, Hoeck W 2011. FusionMap: Detecting fusion genes from next-generation sequencing data at base-pair resolution. Bioinformatics 27: 1922–1928 [DOI] [PubMed] [Google Scholar]
- Goh L, Chen GB, Cutcutache I, Low B, Teh BT, Rozen S, Tan P 2011. Assessing matched normal and tumor pairs in next-generation sequencing studies. PLoS ONE 6: e17810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindan R, Ding L, Griffith M, Subramanian J, Dees ND, Kanchi KL, Maher CA, Fulton R, Fulton L, Wallis J, et al. 2012. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell 150: 1121–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. 2008. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453: 948–951 [DOI] [PubMed] [Google Scholar]
- Hansen RS, Thomas S, Sandstrom R, Canfield TK, Thurman RE, Weaver M, Dorschner MO, Gartler SM, Stamatoyannopoulos JA 2010. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc Natl Acad Sci 107: 139–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Wu H, Feinberg AP 2009. A species-generalized probabilistic model-based definition of CpG islands. Mamm Genome 20: 674–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J 2000. Repbase update: A database and an electronic journal of repetitive elements. Trends Genet 16: 418–420 [DOI] [PubMed] [Google Scholar]
- Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ, et al. 2003. The UCSC Genome Browser Database. Nucleic Acids Res 31: 51–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, Halmos B 2005. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 352: 786–792 [DOI] [PubMed] [Google Scholar]
- Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK 2012. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22: 568–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, et al. 2002. A high-resolution recombination map of the human genome. Nat Genet 31: 241–247 [DOI] [PubMed] [Google Scholar]
- Krumm N, Sudmant PH, Ko A, O’Roak BJ, Malig M, Coe BP, Quinlan AR, Nickerson DA, Eichler EE 2012. Copy number variation detection and genotyping from exome sequence data. Genome Res 22: 1525–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC 2009. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4: 1073–1081 [DOI] [PubMed] [Google Scholar]
- Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, Ou SH, Dezube BJ, Janne PA, Costa DB, et al. 2010. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 363: 1693–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lee W, Jiang Z, Chen Z, Jhunjhunwala S, Haverty PM, Gnad F, Guan Y, Gilbert H, Stinson J, et al. 2012. Genome and transcriptome sequencing of lung cancers reveal diverse mutational and splicing events. Genome Res 22: 2315–2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, De S, Michor F 2013. DNA replication timing and higher-order nuclear organization determine single-nucleotide substitution patterns in cancer genomes. Nat Commun 4: 1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I, et al. 2010. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 362: 2380–2388 [DOI] [PubMed] [Google Scholar]
- Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J, Seto T, Satouchi M, Tada H, Hirashima T, et al. 2010. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): An open label, randomised phase 3 trial. Lancet Oncol 11: 121–128 [DOI] [PubMed] [Google Scholar]
- Ohashi K, Sequist LV, Arcila ME, Moran T, Chmielecki J, Lin YL, Pan Y, Wang L, de Stanchina E, Shien K, et al. 2012. Lung cancers with acquired resistance to EGFR inhibitors occasionally harbor BRAF gene mutations but lack mutations in KRAS, NRAS, or MEK1. Proc Natl Acad Sci 109: E2127–E2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onozato R, Kosaka T, Kuwano H, Sekido Y, Yatabe Y, Mitsudomi T 2009. Activation of MET by gene amplification or by splice mutations deleting the juxtamembrane domain in primary resected lung cancers. J Thorac Oncol 4: 5–11 [DOI] [PubMed] [Google Scholar]
- Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H 2005. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med 2: e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimers M, Carey VJ 2006. Bioconductor: An open source framework for bioinformatics and computational biology. Methods Enzymol 411: 119–134 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365–386 [DOI] [PubMed] [Google Scholar]
- Sathirapongsasuti JF, Lee H, Horst BA, Brunner G, Cochran AJ, Binder S, Quackenbush J, Nelson SF 2011. Exome sequencing-based copy-number variation and loss of heterozygosity detection: ExomeCNV. Bioinformatics 27: 2648–2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, Bergethon K, Shaw AT, Gettinger S, Cosper AK, et al. 2011. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 3: 75ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, McArthur GA, Hutson TE, Moschos SJ, Flaherty KT, et al. 2012. Survival in BRAF V600–mutant advanced melanoma treated with vemurafenib. N Engl J Med 366: 707–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL 2009. TopHat: Discovering splice junctions with RNA-seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L 2010. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker SA, Houghtaling BR, Elemento O, Kapoor TM 2012. Using transcriptome sequencing to identify mechanisms of drug action and resistance. Nat Chem Biol 8: 235–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, et al. 2007. Characterizing the cancer genome in lung adenocarcinoma. Nature 450: 893–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Ercan D, Chen L, Yun CH, Li D, Capelletti M, Cortot AB, Chirieac L, Iacob RE, Padera R, et al. 2009. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature 462: 1070–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]