Abstract
The functional roles of SNPs within the 8q24 gene desert in the cancer phenotype are not yet well understood. Here, we report that CCAT2, a novel long noncoding RNA transcript (lncRNA) encompassing the rs6983267 SNP, is highly overexpressed in microsatellite-stable colorectal cancer and promotes tumor growth, metastasis, and chromosomal instability. We demonstrate that MYC, miR–17–5p, and miR–20a are up-regulated by CCAT2 through TCF7L2-mediated transcriptional regulation. We further identify the physical interaction between CCAT2 and TCF7L2 resulting in an enhancement of WNT signaling activity. We show that CCAT2 is itself a WNT downstream target, which suggests the existence of a feedback loop. Finally, we demonstrate that the SNP status affects CCAT2 expression and the risk allele G produces more CCAT2 transcript. Our results support a new mechanism of MYC and WNT regulation by the novel lncRNA CCAT2 in colorectal cancer pathogenesis, and provide an alternative explanation of the SNP-conferred cancer risk.
Notwithstanding the considerable advancements in our understanding of the molecular genetic basis of cancer, in the majority of cancer-associated genomic regions, the responsible protein-coding genes have not been identified yet. The discovery of short (19–22 nt), noncoding RNAs (ncRNAs)—called microRNAs (miRNAs) (Ambros 2001)—not only revealed a novel mechanism of gene regulation but also led to the identification of miRNAs directly involved in cancer development (Spizzo et al. 2009). It is therefore plausible that as-yet-unidentified members of the broader category of ncRNA mapping to cancer-associated genomic regions play rate-limiting roles in tumor initiation and/or progression (Rinn and Chang 2012). For instance, we previously reported that highly conserved genomic regions (ultraconserved regions, or UCRs) (Bejerano et al. 2004) are frequently transcribed as long (>200 bp) ncRNAs (lncRNAs) in both normal and tumor tissues (Calin et al. 2007). Furthermore, germline mutations, as well as single nucleotide polymorphisms (SNPs) in ultraconserved ncRNAs, were found to occur more frequently in patients with colon cancer and chronic leukemia than in the general population (Wojcik et al. 2010).
The rs6983267 SNP, mapping to the 8q24.21 chromosomal region, has been consistently associated with an increased risk of colorectal cancer (CRC) (Haiman et al. 2007): The G allele was associated with greater predisposition to CRC than the T allele (odds ratios of 1.27 and 1.47 for heterozygotes and homozygotes, respectively; P = 1.27 × 10−14) (Tomlinson et al. 2007). The increased cancer risk from this SNP variant was also observed in other cancer types, including prostate, ovarian, and inflammatory breast cancer (Ghoussaini et al. 2008; Bertucci et al. 2012). Despite the consistent association between rs6983267 and cancer risk, the underlying molecular and cellular mechanisms remain largely unknown. The genomic region spanning rs6983267 was found to contain DNA enhancer elements (Pomerantz et al. 2009; Tuupanen et al. 2009), and the allelic variants were shown to confer different binding affinity to TCF7L2 (transcription factor 7-like 2 [T-cell specific, HMG-box]), a transcription factor that, together with CTNNB1, plays a central role in the transcriptional activation of WNT target genes. In view of the well-established role of constitutive WNT signaling activation in CRC, these findings suggest that rs6983267 itself resides in a functional element that directly participates in colon cancer pathogenesis.
On the base of the localization of rs6983267 within a highly conserved region of the genome (http://genome.ucsc.edu) (Sotelo et al. 2010) and our previous report of transcription in UCRs (Calin et al. 2007), we hypothesized that an as-yet-unidentified, highly conserved ncRNA could be transcribed from this genomic region. The association between rs6983267 and CRC risk further suggested that this ncRNA is likely to be aberrantly expressed in colon cancer and to play a role in CRC carcinogenesis through WNT signaling.
Results
CCAT2, a novel lncRNA transcript maps to the highly conserved 8q24.21 region encompassing rs6983267 and is overexpressed in microsatellite-stable CRC samples
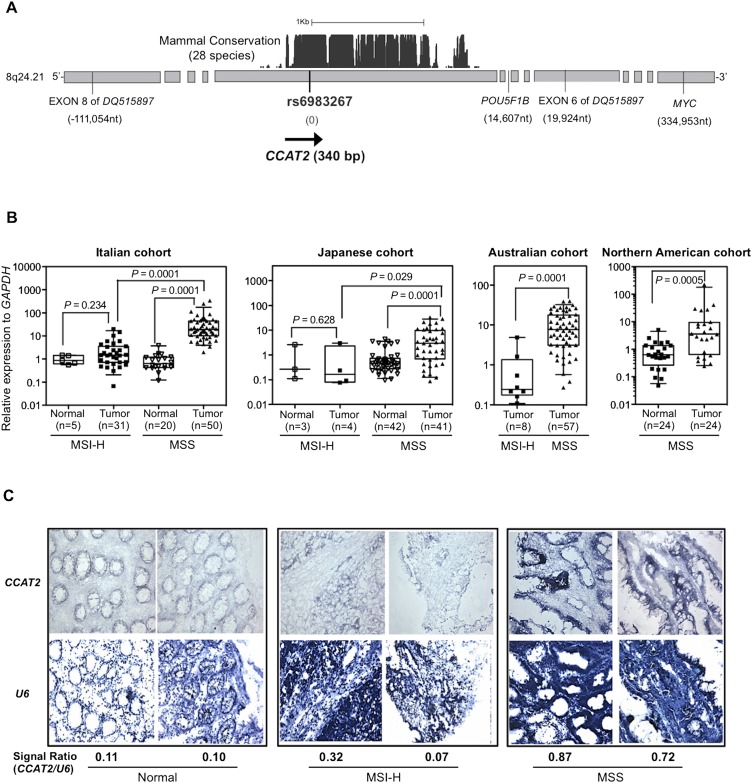
The 8q24.21 genomic region encompasses a highly conserved segment with homologies found in 28 species-conserved tracks (University of California, Santa Cruz Genome Browser; http://genome.ucsc.edu/) (Fig. 1A). We designed primers spanning the rs6983267 SNP and found that this region was transcribed at high levels in human pancreas, lung, prostate, and testis tissues (data not shown). Subsequently, we cloned by RACE (rapid amplification of cDNA ends) a 340-bp nonspliced transcript from bone marrow cDNA (Fig. 1A). To exclude the possibility that this transcript was part of long intergenic noncoding RNAs (lincRNAs) mapped in this region (RP11-382A18.2 and RP11-382A18.1), we performed polymerase chain reaction (PCR) amplification using lincRNA-specific primers combined with primers from within the 340-bp transcript. We did not observe any amplified product in the COLO320 colon cancer cell line that expresses the highest levels of the newly identified transcript, suggesting that it is distinct from the two lincRNAs. We further cloned an intronless 630-bp transcript from mouse testis cDNA. Notably, the sequence overlapping between the transcripts cloned from the human and mouse tissues shared 93% homology and encompassed the rs6983267 SNP.
Figure 1.
CCAT2, a novel lncRNA spanning rs6983267, is overexpressed in colorectal cancer samples. (A) Genomic features of the 8q24.21 region spanning SNP rs6983267 and the genomic location of CCAT2. Numbers in parentheses show the genomic distance relative to rs6983267 (image not to scale). (B) CCAT2 expression in CRC patient samples. CCAT2 expression was quantified using qRT-PCR, and data are presented as box-whisker plots showing the five statistics (lower whisker is 5% minimum, lower box part is the 25th percentile, solid line in box represents the median, upper box part is 75th percentile, and upper whisker is 95% maximum). (C) In situ hybridization of CCAT2 and RNU6-6P (U6) (as normalizer) in CRC patient samples.
Because a previous study using a tiling assay did not find any transcriptional activity in this region (Pomerantz et al. 2009), we sought other independent data to support the existence of this newly identified RNA transcript. By searching the Long RNA-seq data from ENCODE/CSHL (http://genome.ucsc.edu/; Djebali et al. 2012), we found that the rs6983267 region is actively transcribed in the genomic sense orientation in GM12878 lymphoblastoid cells (Supplemental Fig. S1). We next determined whether this transcript has protein-coding potential. The transcript's noncoding nature was suggested by the negative results of the in vitro translation assay, along with the lack of consistent open reading frames (ORFs) by ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html), negative score by PhyloCSF (−271.5366, meaning that CCAT2 is 1027.15366 times more likely to be a noncoding sequence than a coding one), a comparative genomic method differentiating coding and noncoding RNA (Lin et al. 2011), lack of coding potential determined by coding potential assessment tool (CPAT; http://lilab.research.bcm.edu/cpat/) (Wang et al. 2013), and negative data from the small-peptide database (Pepbank; http://pepbank.mgh.harvard.edu/search/blast) (Supplemental Fig. S2A). We further confirmed the existence of this novel RNA by using Northern blot analysis on a panel of colon cancer cell lines probed by sense-specific oligonucleotides spanning rs6983267: A transcript of ∼400 bp in genomic sense orientation (centromere to telomere) was detected with the highest expression in COLO320 cells (Supplemental Fig. S2B). Consistent with this observation, Southern blot analysis revealed that among analyzed samples, COLO320 cells had the highest degree of genomic amplification of the 8q24 region (data not shown). By taking advantage of a cohort of nine paired colon specimens, we determined by quantitative real-time PCR (qRT-PCR) analysis that the newly identified transcript was expressed at much higher levels in tumor tissue than in the adjacent normal mucosa, and confirmed that the transcription occurs in the genomic sense orientation (Supplemental Fig. S2C). We named this novel gene transcript colon cancer–associated transcript 2 (CCAT2).
We further measured CCAT2 expression by qRT-PCR in 215 CRC and 94 paired non-neoplastic mucosal specimens obtained from patients from four different geographical regions (i.e., Italy, Japan, Australia, and North America). We identified significantly higher CCAT2 expression in the CRC tissue as compared with the adjacent mucosae (Fig. 1B). Moreover, microsatellite-stable (MSS) cancers had higher CCAT2 expression (approximately 10 times) than microsatellite-unstable (MSI-H) tumors and adjacent normal colon mucosae (Fig. 1B; Supplemental Fig. S2C). To visualize CCAT2 expression, we performed in situ hybridization by using a locked nucleic acid probe designed against CCAT2 RNA. In agreement with the qRT-PCR findings (Fig. 1B), the epithelial expression pattern observed had stronger staining in MSS than in MSI-H cancers and normal mucosae (Fig. 1C). We did not observe a significant association between CCAT2 expression and genomic DNA amplification in an additional set of CRC samples from Japan (n = 64, P = 0.46) (Supplemental Fig. S2D), suggesting that CCAT2 expression is, at least partially, independent of the genomic DNA copy number alterations in this chromosomal region.
CCAT2 promotes cancer growth and metastasis
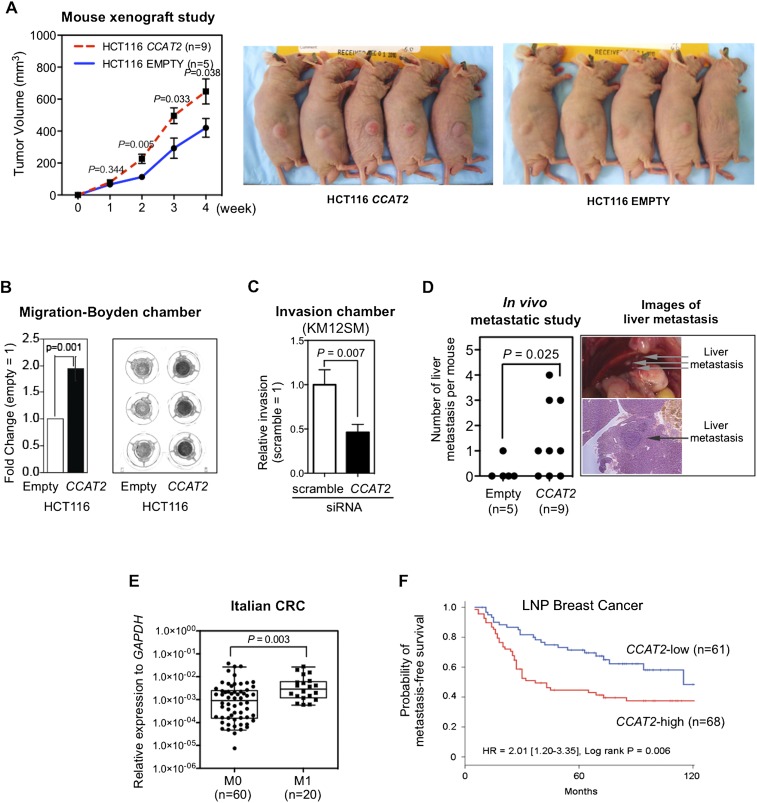
To test the hypothesis that CCAT2 plays an oncogenic role in CRC, we cloned CCAT2 into a retroviral expression vector and transduced it into HCT116 colon cancer cells. This is an MSI-H cell line (MSH6 mutant) with a near-diploid karyotype (Lengauer et al. 1997) that expresses low levels of CCAT2 (when compared with COLO320) (Supplemental Fig. S2B). Retrovirus transfection of the CCAT2 construct caused a 630-times increase in CCAT2 expression compared with the cells transfected with empty vector (Supplemental Fig. S3A). For comparison, the expression difference between high-expresser COLO320 cells and low-expresser HCT116 cells was about 1500 times (Supplemental Fig. S3A). The CCAT2-transduced HCT116 cells did not affect cell proliferation in two-dimensional (2D) culture conditions (Supplemental Fig. S3B), but showed a growth advantage under low-adherence conditions and increased colony-forming potential (Supplemental Fig. S3C,D). Subcutaneous transplantation of CCAT2-overexpressing HCT116 cells resulted in larger xenograft tumors in Swiss nu-nu/Ncr nude mice when compared with empty vector-transduced cells (Fig. 2A). This observed in vivo tumor promotion effect of CCAT2 was independently confirmed by a xenograft study using pcDNA construct–established CCAT2-overexpressing HCT116 cells (data not shown).
Figure 2.
CCAT2 promotes tumor growth and metastasis. (A) CCAT2 increased subcutaneous tumor formation in a mouse xenograft model. Comparison was made between the empty vector and CCAT2 groups at the indicated weekly time points using the t-test. (B) HCT116 cells transduced with CCAT2 showed significantly higher migration ability, as measured by using a migration chamber. Data are presented as mean ± SEM from three independent experiments. (C) Knockdown of CCAT2 reduces invasion ability of KM12SM colon cancer cells. After treatment with CCAT2 siRNA (50 nM) for 24 h, cells were seeded onto an invasion assay chamber for 48 h, and invaded cells were stained and counted. (D) CCAT2 enhances liver metastasis in mice that were incubated by intrasplenic injection with HCT116 cells. A representative image of liver metastasis from the CCAT2 group is shown. (E) CCAT2 expression levels in Italian primary CRC samples from patients with metastasis (M1) were higher than those from without metastasis (M0). Comparison was made using the Mann-Whitney test. Data are presented as box-whisker plots. (F) Breast cancer patients with high CCAT2 had shorter metastasis-free survival. Kaplan-Meier analysis as a function of CCAT2 levels in 129 lymph node–positive breast cancer patients who received adjuvant combination chemotherapy with cyclophosphamide, methotrexate, and 5-fluorouracil. CCAT2 high level to normalizer, >0.0045; CCAT2 low level, ≤0.0045.
We next studied whether CCAT2 is involved in promoting the metastatic phenotype in colon cancer cells. The in vitro migration assay showed a twofold increase in the migration of CCAT2-transduced HCT116 cells (P = 0.001) (Fig. 2B). To validate the role of CCAT2 in the metastatic phenotype, we used KM12SM cells, a CRC cell line established from the spontaneous liver metastasis of KM12C cells (Camps et al. 2004). Knockdown of CCAT2 by siRNA, which reduced the CCAT2 expression by 70%, significantly reduced the invasive ability of KM12SM cells (that express high levels of CCAT2), suggesting the involvement of CCAT2 in metastasis (Fig. 2C; Supplemental Fig. S4). More importantly, spleen injection of CCAT2-overexpressing HCT116 cells into Swiss nu-nu/Ncr nude mice resulted in a higher incidence of liver metastasis (six of nine mice vs. one of five mice) as well as a greater number of metastatic tumors (P = 0.025) than in the control group (Fig. 2D). These studies suggest that modulation of CCAT2 expression levels changes the metastatic capacity of cancer cells. In addition, in the Italian cohort of CRC samples (which was the larger available set), we found significantly higher CCAT2 expression levels in primary CRC tumors from patients with metastasis (TNM category M1) than in those without metastasis (M0; P = 0.003) (Fig. 2E).
To further expand on this issue and in view of the previous report on the correlation between rs6983267 and breast cancer aggressiveness (Bertucci et al. 2012), we measured CCAT2 RNA expression levels in 129 lymph node–positive breast cancer patients who received an adjuvant cyclophosphamide, methotrexate, and 5-fluorouracil combination regimen. High CCAT2 RNA levels were associated with shorter metastasis-free survival when analyzed as both a continuous variable (Supplemental Table S1) and as a dichotomized variable using the median level as a cutoff point (Fig. 2F).
CCAT2 induces chromosomal instability
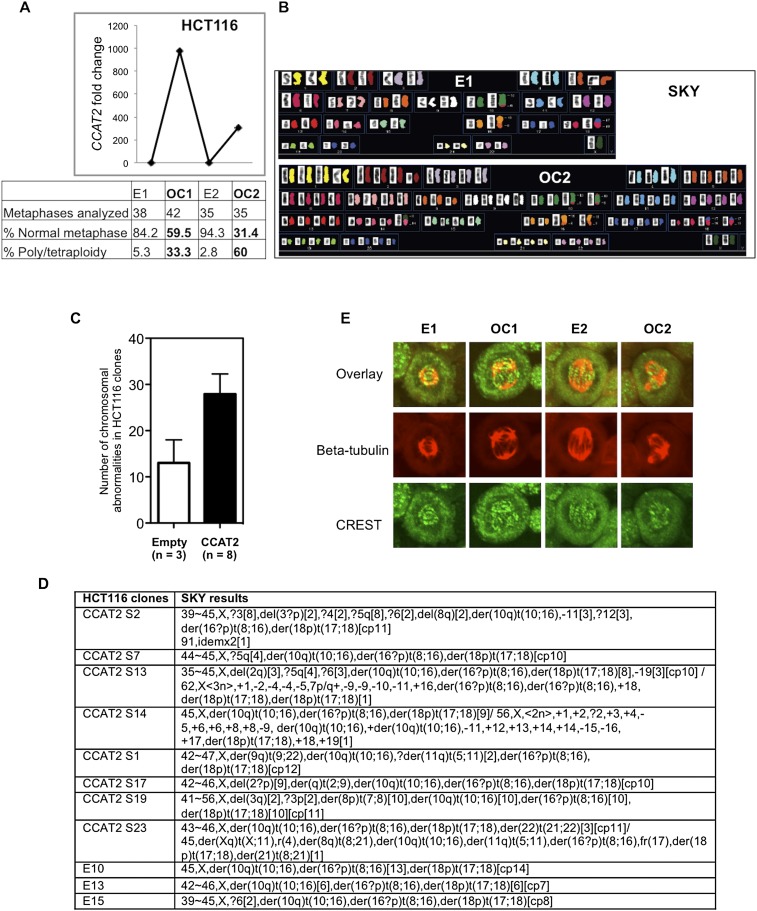
Because chromosomal instability (CIN) represents a main feature of MSS CRCs, and in view of the high CCAT2 expression levels among MSS colon cancers (Fig. 1B,C), we hypothesized that CCAT2 may underlie CIN development. We generated stable HCT116 clones overexpressing CCAT2 (OC1 and OC2) or with basal CCAT2 expression (E1 and E2) by using the pcDNA expression vector (Fig. 3A). Genomic instability analysis showed that the percentage of cells with normal metaphases was markedly lower in OC (59.5% in OC1 and 31.4% in OC2) than in E (84.2% in E1 and 94.3% in E2) clones (Fig. 3A). In the same analysis, CCAT2 dramatically increased the percentage of polyploid cells from 5.3% (E1) and 2.8% (E2) to 33.3% (OC1) and 60.0% (OC2) (Fig. 3A). We detected near-tetraploid status in clone OC2 by spectral karyotype analysis (Fig. 3B) and observed a doubling of nuclear size and DNA content by DAPI staining and flow cytometric analysis (Supplemental Fig. S5A,B). The CIN-inducing ability by CCAT2 was further validated by a second batch of eight overexpressing clones that exhibited numerical and structural chromosomal changes (Fig. 3C,D; Supplemental Fig. S5C). CIN was also observed in HCT116 cells transduced with CCAT2-containing retrovirus (Supplemental Fig. S5D). Stable knockdown of CCAT2 expression in COLO320 cells partially reversed the abnormal metaphase observed in COLO320 cells (Supplemental Fig. S5E). To further identify the mechanism by which CCAT2 overexpression may underlie CIN, we evaluated spindle and CREST/kinetochore structure by immunocytochemistry analysis of the CCAT2 clones (E1, E2, OC1, and OC2) with antibodies directed against beta-tubulin and the centromere. Normal numbers of centrosomes were observed in both E1 and E2 with spindles separating chromosomes from two centromeres in the nucleus (Fig. 3E). However, we detected aberrant numbers of centrosomes pulling chromosomes from three or more different points in the CCAT2-overexpressing clones (Fig. 3E). The observed defects in centrosomes are likely to result in abnormal chromosomal breakage and fusion, and eventually aneuploidy.
Figure 3.
CCAT2 induces chromosomal instability in HCT116 cells. (A) Increased chromosomal abnormalities in CCAT2-overexpressing clones, as identified by genomic instability analysis. (B) Example of spectral karyotypes of HCT116 clones revealed a change from the original diploid (E1) to the tetraploid karyotype in the OC2 clone. (C,D) CCAT2 induces chromosomal instability in an independent set of CCAT2-overexpressing clones. The number of chromosomal abnormalities in each clone was counted; data are expressed as mean ± SD. (E) Abnormal centrosomes in CCAT2-overexpressing clones OC1 and OC2 by immunostaining. Beta-tubulin (red); CREST/kinetochore (green).
Because CIN scores were already available from 218 breast cancer patients with lymph node–negative disease (Smid et al. 2011), we assessed the CCAT2 expression levels in these tumor samples. We identified a significant positive correlation between CCAT2 level and CIN score (Spearman correlation: Rs = 0.17, P = 0.012).
CCAT2 regulates MYC transcription
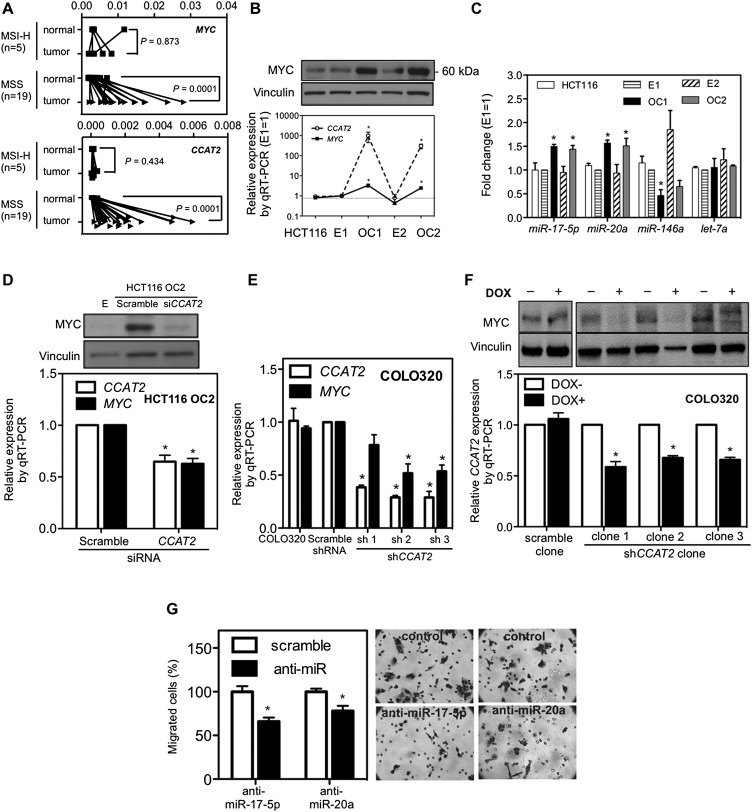
To explore the molecular mechanism underlying CCAT2's oncogenic potential, we characterized the expression of other transcripts located in the same region: MYC and exons 6 and 8 of DQ515897 (but not POU5F1B) showed expression patterns similar to that of CCAT2, although with smaller differences among MSI-H, and MSS cancers and normal colorectal mucosa (Fig. 4A; Supplemental Fig. S6A). In view of these associative data and of previous reports describing a long-range interaction between rs6983267 and MYC (Pomerantz et al. 2009; Sotelo et al. 2010), we asked whether CCAT2 plays a role in regulating MYC expression. Among the HCT116 clones, we found higher MYC expression at both the RNA and protein levels in cells overexpressing CCAT2 (3.4-fold and 2.5-fold increases in OC1 and OC2, respectively; P = 0.030 and P = 0.038, respectively) (Fig. 4B). Higher MYC expression was also found in three additional independent HCT116 clones overexpressing CCAT2 (Supplemental Fig. S6B). Expression of downstream MYC protein-coding gene targets such as BAX (Mitchell et al. 2000), CDC25A (Galaktionov et al. 1996), and CDKN2A (Zindy et al. 1998), and the microRNA targets such as MIR17HG (O'Donnell et al. 2005) and miR-146a (Chang et al. 2008) were also changed accordingly (Fig. 4C; Supplemental Fig. S6C,D). Additionally, the MIR17HG was also up-regulated in retrovirus-transfected cells (Supplemental Fig. S6E) and was significantly reduced by transient MYC down-regulation (Supplemental Fig. S6F), suggesting that these microRNAs are regulated by MYC in our experimental systems. Conversely, transient knockdown of CCAT2 reduced MYC expression (both RNA and protein) in CCAT2-overexpressing clones (Fig. 4D), suggesting that CCAT2 RNA, and not merely its genomic locus, is required for the maintenance of high MYC expression levels. In addition, sustained down-regulation of CCAT2 by shRNA retroviruses decreased MYC expression and MIR17HG expression in COLO320 cells in a CCAT2-dependent fashion (Fig. 4E; Supplemental Fig. S6G). This effect was further validated in Tet-on inducible COLO320 clones, where CCAT2 expression was reduced by specific shRNAs in response to doxycycline (Fig. 4F).
Figure 4.
Regulation of MYC expression by CCAT2. (A) Increased CCAT2 and MYC expression in MSS CRC samples. Expression profile of MYC and CCAT2 in paired CRC/control mucosa samples from Italy using qRT-PCR. (B) Correlation of MYC protein and CCAT2 RNA levels in HCT116 clones with high and basal CCAT2 levels. (C) CCAT2 increased miRNA expression of the MIR17HG, decreased miR-146a expression, and exerted no consistent effect on let-7a (used as negative control). (D) Down-regulation of MYC (upper panel: protein; lower panel: RNA) by CCAT2 siRNA in HCT116 OC2 clone. (E) Reduced MYC expression in COLO320 shCCAT2 stable clones. (F) Reduction of MYC in doxycycline-inducible shCCAT2 clones of COLO320. (G) Reduction of migration in CCAT2-transduced HCT116 cells by anti-miR-17-5p and anti-miR-20a. Twenty-four hours after anti-miR treatment, cells were seeded onto a migration assay chamber for another 24 h, and migrated cells were stained and counted. (*) P < 0.05.
Next, we moved on to determine whether MYC and/or its targets are mediators of the above-observed CCAT2-driven enhanced cell migration. In our experimental system, reducing MYC expression did not significantly decrease the migration of CCAT2-transduced cells (Supplemental Fig. S6H). However, consistent with the consensus that the MIR17HG increases metastasis (Bartley et al. 2011; Zhang et al. 2012), knockdown of miR-17-5p or miR-20a in CCAT2-overexpressing cells led to a significant decrease in cell migration (Fig. 4G; Supplemental Fig. S6I). These findings suggest a not-yet-identified complex network underlying CCAT2-elicited metastatic phenotype.
Mechanisms of CCAT2's regulatory effect
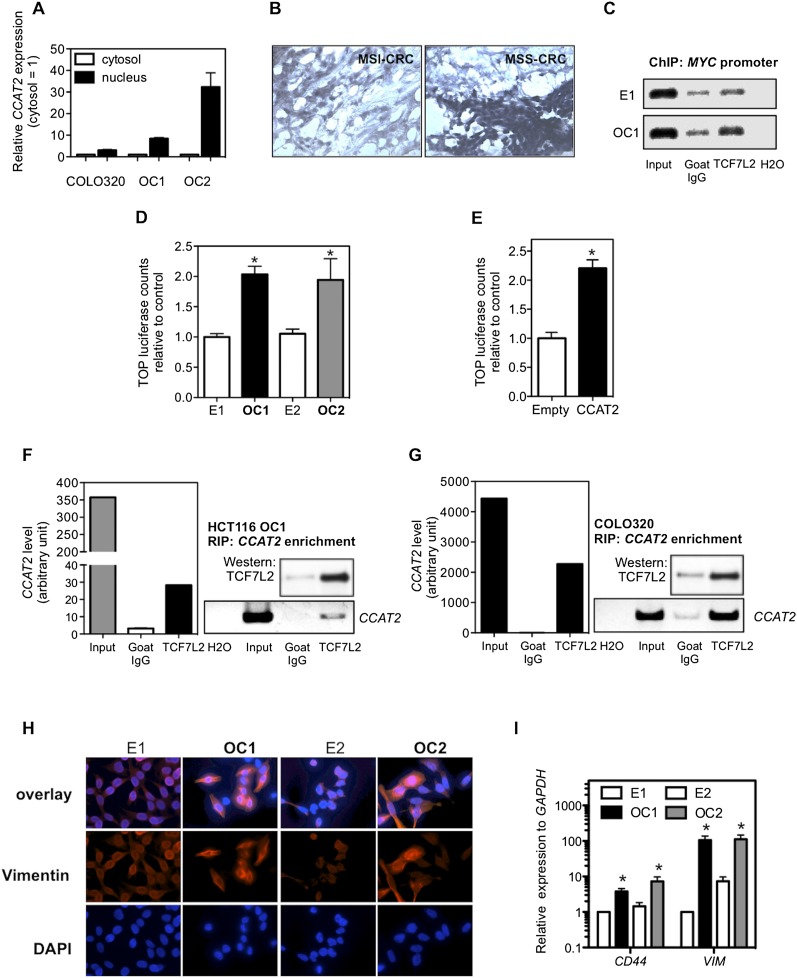
Because CCAT2's subcellular localization may provide additional clues about its functional role, we measured CCAT2 expression in nuclear and cytosolic fractions from CCAT2-expressing HCT116 subclones and the COLO320 cell line by qRT-PCR. The differential enrichments of GAPDH and RNU6-6P RNA were used as fractionation indicators (Supplemental Fig. S7). In these samples, we found a considerable increase in CCAT2 expression in the nucleus versus the cytosol, thus indicating that CCAT2 is mainly localized in the nucleus (Fig. 5A). This was further confirmed by the in situ hybridization findings in CRC samples (Fig. 5B). Consistent with our findings, RNA-seq data showed that the transcript encompassing rs6983267 is exclusively expressed in the nucleus of GM12878 lymphoblastoid cells (Supplemental Fig. S1). We also investigated a potential effect of CCAT2 on MYC mRNA stability (Supplemental Fig. S8), as well as an RNA enhancer effect (Supplemental Fig. S9; Supplemental Results; Orom et al. 2010), but could not find any significant variations related to CCAT2 levels.
Figure 5.
Regulation of TCF7L2 activity by CCAT2. (A) CCAT2 nuclear localization, as identified using qRT-PCR in fractionated COLO320 cells and HCT116 CCAT2 clones. (B) CCAT2 nuclear localization, as identified in CRC samples using in situ hybridization. (C) CCAT2 increased TCF7L2 binding to the MYC promoter. ChIP analysis showed higher binding in CCAT2-overexpressing clone (OC1) than cells with basal CCAT2 expression (E1). Goat IgG was used as a control antibody. (D) Higher WNT activity in CCAT2 stable clones, as identified by TOP-Flash reporter assay. (E) Transient expression of CCAT2 induced a more than twofold increase of TOP-Flash luciferase activity in HEK293 cells. The data in C and D are presented as mean ± SEM (n = 3). (*) P < 0.05. (F) Colocalization of CCAT2 RNA with TCF7L2 protein by RNA immunoprecipitation using anti-TCF7L2 antibodies in OC1 with ectopic CCAT2 overexpression. (G) Colocalization of CCAT2 RNA with TCF7L2 protein by RNA immunoprecipitation using anti-TCF7L2 in COLO320 cells, which have endogenous high CCAT2 expression. A quantitative analysis using qRT-PCR is shown along with the PCR gel images. (H) Immunostaining of HCT116 clones with vimentin antibody or DAPI. Stronger vimentin signal as seen in epithelial–mesenchymal transition was observed in CCAT2-overexpressing clones than in cells with basal CCAT2 expression. (I) CCAT2 increases the mRNA expression levels of CD44 and VIM in HCT116 clones. The data are presented as mean ± SEM from at least three independent experiments. (*) P < 0.05.
Aberrant WNT activation occurs in the majority of CRCs (Clevers 2006), and the risk allele of SNP rs6983267 has been shown to confer enhanced response to WNT signaling through TCF7L2 binding (Tuupanen et al. 2009). Since MYC is an established target of TCF7L2 as confirmed by the presence of TCF7L2-binding sites in its promoter as well as by functional studies (He et al. 1998), it is possible that CCAT2 affects MYC expression through the mediation of TCF7L2. Consistent with previous findings, we identified TCF7L2 binding to the MYC promoter region and observed enhanced binding in the OC1 clone when compared with the E1 clone (Fig. 5C). Next, we sought to determine whether this was due to an increase in nuclear CTNNB1 or TCF7L2 accumulation. Using Western blot analysis, we observed a comparable level of total CTNNB1 expression levels with no increase in nuclear CTNNB1 between CCAT2-overexpressing clones and their control cells (Supplemental Fig. S10). Similarly, TCF7L2 expression levels were comparable in all four clones (Supplemental Fig. S10). Notably, by using a TOP-Flash luciferase assay, we observed a twofold increase in WNT reporter activity in OC1 and OC2 clones with increased CCAT2 expression levels (Fig. 5D). Increased luciferase activity was also observed in an independently generated set of high-CCAT2-expressing HCT116 clones (Supplemental Fig. S11). This result was further confirmed in HEK293 WNT reporter cells transiently transfected with CCAT2 expression vector (Fig. 5E). Consistent with these observations, WNT reporter activity in COLO320 cells was effectively reduced by CCAT2 siRNA and also as expected by TCF7L2 siRNA (Supplemental Fig. S12). These findings suggest that CCAT2 augments TCF7L2 transcriptional activity without increasing the quantity of TCF7L2.
We used RNA immunoprecipitation analysis to examine the physical interaction between CCAT2 and TCF7L2 by pulling down RNAs colocalized with TCF7L2 protein. We found an enrichment of CCAT2 RNA, but not GAPDH and the HOTAIR ncRNA by TCF7L2 antibody compared with the IgG control in the OC1 clone (Fig. 5F; Supplemental Fig. S13). To test whether endogenous CCAT2 binds to TCF7L2 protein, we analyzed the RNA–protein association in COLO320 cells that express high CCAT2 levels. Similar to the findings in the OC1 HCT116 clone, a significant enrichment of CCAT2 was observed in TCF7L2 pull-down RNA samples from COLO320 cells (Fig. 5G; Supplemental Fig. S13).
Overall, these results suggest that the main mechanism through which CCAT2 increases MYC expression relies on activation of WNT signaling by enhancing TCF7L2 transcriptional activity. Accordingly, the expression of specific WNT target genes such as CD44 (Wielenga et al. 1999) and VIM (Gilles et al. 2003) was increased in CCAT2-overexpressing clones (Fig. 5H,I). Conversely, knockdown of CCAT2 expression caused a reduction in the expression level of VIM in both KM12SM (with high endogenous CCAT2 expression) and COLO320 (with very high endogenous CCAT2 expression) cells (Supplemental Fig. S14).
CCAT2 responds to WNT signaling
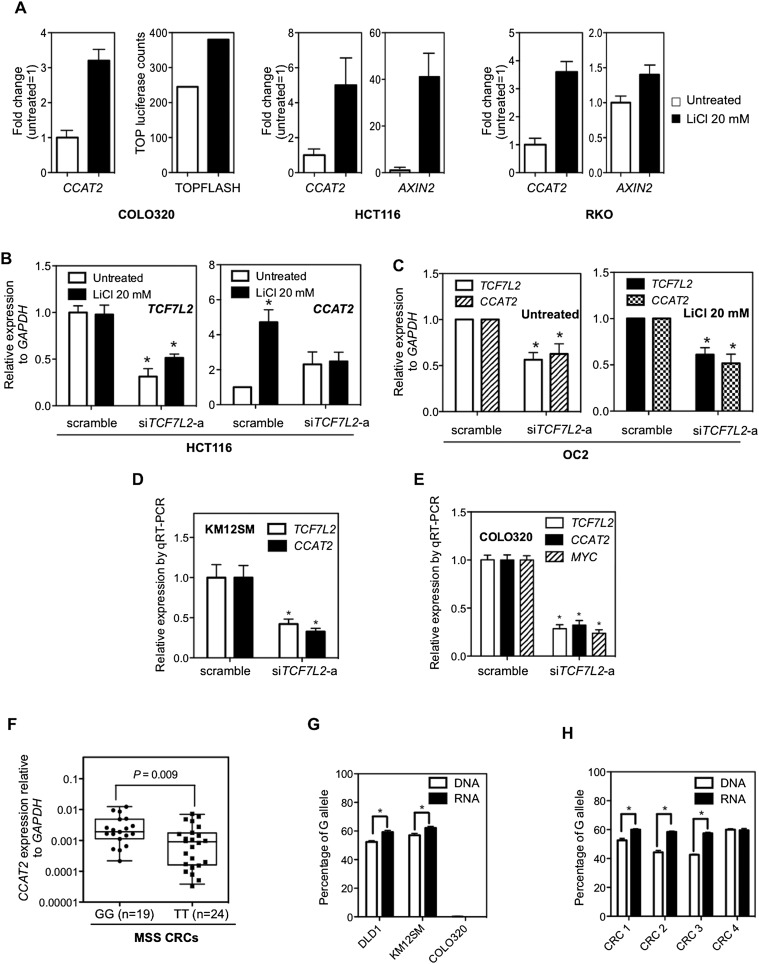
Previously, a TCF7L2-binding element was found to be located within the rs6983267 genomic span (Pomerantz et al. 2009; Tuupanen et al. 2009), which suggested that CCAT2 expression levels might be regulated by this transcription factor. Therefore, we treated three colon cancer cell lines (COLO320, HCT116, and RKO) with lithium chloride (LiCl) in order to further activate canonical WNT signaling through inhibition of glycogen synthesis kinase 3 beta (GSK3B) (Tuupanen et al. 2009). The effect of LiCl was monitored by TOP-Flash luciferase activity and by AXIN2 expression (Jho et al. 2002). In all these cell lines, we observed an increase in CCAT2 expression after LiCl treatment (Fig. 6A). TCF7L2 knockdown using two different siRNAs impaired the LiCl-induced CCAT2 expression increase in HCT116 cells (Fig. 6B; Supplemental Fig. S15), suggesting that the effect on WNT signaling was TCF7L2 dependent. In the CCAT2-overexpressing clones, TCF7L2 siRNA consistently reduced CCAT2 expression, either with or without LiCl stimulation, indicating that TCF7L2 is necessary for the maintenance of high CCAT2 expression levels (Fig. 6C). The endogenous expression of CCAT2 was also significantly down-regulated by TCF7L2 siRNA in KM12SM and COLO320 cells (Fig. 6D,E). These findings suggest the existence of a complex feedback loop of CCAT2 and WNT signaling.
Figure 6.
Regulation of CCAT2 expression by WNT signaling and SNP variance on CCAT2 expression. (A) CCAT2 expression is induced by lithium chloride (LiCl) in three colon cancer cell lines. AXIN2 or TOP-Flash luciferase activity served as a positive control for the activation of WNT signaling. The data are presented as mean ± SD (n = 3). (B) TCF7L2 is indispensable for LiCl-induced CCAT2 expression. HCT116 cells were treated with TCF7L2 siRNA (siGENOME SMARTpool TCF7L2; Dharmacon) for 24 h and then stimulated with LiCl for another 24 h. CCAT2 expression levels were measured by qRT-PCR. (C) TCF7L2 siRNA down-regulates CCAT2 expression in overexpressing OC2 clone, either with or without LiCl stimulation. (D,E) TCF7L2 siRNA (sc43525, Santa Cruz) down-regulates CCAT2 expression in KM12SM cells and COLO320 cells. The data are presented as mean ± SEM (n = 3); (*) P < 0.05 when compared with the respective control. (F) Comparison of CCAT2 expression in cohort of CRC samples from Italy with GG and TT genotype showed higher CCAT2 expression associated with the G allele. The P-value was calculated using the Mann-Whitney test. The data are presented as box-whisker plots. (G,H) The Pyrosequencing data showed a higher percentage of the G allele in the CCAT2 transcripts than its genomic DNA counterpart in heterogeneous rs6983267 cell lines (DLD1 and KM12SM) and CRC patient samples with GT genotype. COLO320, which is TT genotype, served here as a negative control.
rs6983267 alleles affect CCAT2 expression and function
The discovery of CCAT2 and its role in CRC suggests that it may underlie the mechanism of the rs6983267 SNP-conferred cancer risk. We first test if rs6983267 status affects CCAT2 expression in patient samples. We genotyped the rs6983267 SNP in previously profiled MSS samples (Italian, Japan, and Australian cohorts) (Fig. 1B) and separated them into GG, GT, and TT groups. We did not observe a significant difference (P = 0.450) in CCAT2 expression levels between the GG (n = 52) and TT (n = 45) groups (Supplemental Fig. S16). However, further separation of the groups according to the individual cohort revealed that the difference (P = 0.009) is significant in the Italian cohort, which has the largest sample size (Fig. 6F). Inconsistently, there was no significant difference in CCAT2 expression between the GG and TT alleles in 93 CRC patients from the United Kingdom (data not shown). These data suggest that the effect of the rs6983267 allele on CCAT2 expression levels may be very subtle, and for detection better-controlled experimental designs are needed.
To address this question, we took advantage of CRC cell lines (DLD1 and KM12SM cells) with a heterogeneous rs6983267 genotype, to measure the amount of CCAT2 transcript produced from different alleles. We amplified the region spanning the SNP using genomic DNA or RNA as template and sequenced the PCR products by Pyrosequencing, a method that allows for accurate and quantitative analysis of DNA sequence (Fakhrai-Rad et al. 2002). In both DLD1 and KM12SM cells, the percentage of G allele is slightly (∼6%), but significantly, higher in RNA samples than the corresponding genomic DNA samples (Fig. 6G). These data prompted us to determine the allele ratio in genomic DNA and RNA of CRC samples with GT genotype. As shown in Figure 6H, three out of four samples showed consistent findings—that is, significantly higher percentage (7%–15%) of G allele in RNA than in DNA samples. These data suggest that the G allele of rs6983267 produces more CCAT2 transcripts than the T allele.
We next determined whether CCAT2 transcripts containing G or T have different biological functions. We mutated the SNP site of the retrovirus CCAT2 construct from G to T and transfected HCT116 cells with CCAT2-G or CCAT2-T retroviruses. Although we observed higher MYC expression (both RNA and protein) in CCAT2-G- than CCAT2-T-transfected cells (Supplemental Fig. S17A,B), the difference becomes insignificant after normalizing to CCAT2 expression levels (Supplemental Fig. S17C). To test whether the rs6983267 allele affects CCAT2's regulation of MYC in CRC patients, we analyzed CCAT2 and MYC expression correlation in previously genotyped MSS CRC samples. A significant positive correlation between CCAT2 and MYC expression was observed in GG samples (n = 52, coefficient = 0.43; P = 0.001), but not in TT samples (n = 45, coefficient = 0.26; P = 0.078) of CRCs (Supplemental Fig. S18). Furthermore, by using the Vienna RNA Secondary Structure Package (http://www.tbi.univie.ac.at/∼ivo/RNA/), we identified that the CCAT2 G and T alleles have distinct conformations (Supplemental Fig. S19). Although further studies are needed to verify the different biological functions of the rs6983267 G and T alleles, our data indicate that the specific alleles may also differentially affect MYC expression.
Discussion
We report the identification of CCAT2, a novel lncRNA encompassing the cancer-related rs6983267 SNP and located in a highly conserved genomic region enriched for markers of regulatory elements, such as H3K4me1, p300, and H3K27ac (Pomerantz et al. 2009). This lncRNA is expressed at high levels in MSS CRC tumor tissues and influences MYC levels, but has very low-level expression in normal colon tissues. This could explain how this new gene has escaped identification, until now, after genome-wide association studies identified the association between the SNP rs6983267 and cancer risk, as well as why tiling arrays did not identify it. A recent study showed that removal of the region encompassing the rs6983267 SNP confers resistance to intestinal tumors induced by the APCmin mutation in mice (Sur et al. 2012). Because we also cloned a novel transcript encompassing this SNP region from mouse tissue, it can be envisioned that deletion of the region also leads to removal of the CCAT2 transcript. Thus, this study lends further support to the oncogenic function of CCAT2 in CRC pathogenesis. In addition, the findings of decreased MYC expression and reduced TCF7L2 binding upstream of MYC in knockout mice from the same study are also consistent with our findings that CCAT2 regulates MYC transcriptional activity.
CCAT2-enhanced invasion and metastasis appear to occur through the MYC-regulated miRNAs miR-17-5p and miR-20a. The role of MYC in metastasis has been controversial, and a recent publication shows that, in breast cancer, MYC suppresses metastasis by direct transcriptional silencing of integrins (Liu et al. 2012). Of interest, miR-20a was identified as being overexpressed in the transition from colon mucosa to early adenoma, and was predicted to target multiple WNT pathway genes (Bartley et al. 2011). The observation according to which CCAT2-enhanced invasion is mediated by MYC-regulated miRNAs and also possibly by other pro-metastatic targets such as CDC25A (Feng et al. 2011), rather than by MYC itself, suggests a complex regulatory network that needs to be further explored in the future. This is also in agreement with the recent observation by The Cancer Genome Atlas (TCGA) Consortium that MYC-driven transcriptional activation and repression play important roles in colon cancer (The Cancer Genome Atlas Network 2012). It is also possible that the effect of CCAT2 on metastasis is mediated through its general activation of WNT signaling (Nguyen et al. 2009; Yu et al. 2012) or another unknown mechanism.
The TOP-Flash luciferase assay suggests increased WNT activity in CCAT2-overexpressing cells. Therefore, we postulated that the increased MYC transcription could result from increased WNT activity, because MYC itself represents a well-established WNT target (He et al. 1998). Recent studies have identified RNA–protein interaction as one of the main regulatory mechanisms of lncRNAs (Rinn and Chang 2012). Coimmunoprecipitation of CCAT2 (RNA) and TCF7L2 (protein) suggests that CCAT2 may broadly regulate WNT target genes by binding to TCF7L2 and modulating its transcriptional activity. Various mechanisms can be envisaged to explain how the CCAT2/TCF7L2 association can enhance WNT activity. We speculate that CCAT2 may affect TCF7L2-driven gene transcription through modulating association of TCF7L2 with its partners, which include the negative regulators Groucho/TLE and CTBP1 and the activator CTNNB1 (Clevers 2006), or changing TCF7L2 protein function by bridging TCF7L2 with other proteins in the transcription complexes. Alternatively, the association of CCAT2 with TCF7L2 may modify the protein structure and enhance its function. Regarding the mechanism of MYC regulation, it is also possible that CCAT2 participates in the loop formation between the genomic loci of rs6983267 with MYC promoter (Pomerantz et al. 2009) and works together with the enhancer element to activate MYC transcription. Notably, we also show that WNT signaling regulates CCAT2 expression, suggesting a feedback loop between CCAT2 and WNT signaling as previously shown for other targets such as CDH1 (Heuberger and Birchmeier 2010) and AXIN2 (Lustig et al. 2002).
We demonstrated CCAT2's ability to underlie chromosomal instability, manifested by the loss or gain of large portions or whole chromosomes, eventually resulting in aneuploidy, a characteristic trait of MSS CRCs. These data support a scenario in which, together with mutations in coding genes such as APC (Fodde et al. 2001; Kaplan et al. 2001) and BUB1 (Cahill et al. 1998), CCAT2 contributes to CIN in colon cancer. This is likely to occur through CCAT2's effects on downstream mediators such as MYC and/or other WNT target genes. The direct involvement of MYC in CIN has been previously demonstrated by showing that transient excess of MYC activity could elicit genomic instability and carcinogenesis (Felsher and Bishop 1999). Inversely, Myc deletion rescues APC deficiency in the mouse intestine (Sansom et al. 2007). Thus, it is plausible that MYC acts as a mediator of CCAT2-induced CIN phenotype. Furthermore, the MIR17HG miRNAs, known to be MYC regulated, can also affect CIN as evidenced by diffuse large B-cell lymphoma that has high levels of these miRNAs and nonrandom chromosomal abnormalities (Li et al. 2009). Alternatively, the CIN-inducing effect can result from the general activation of WNT signaling by CCAT2. This hypothesis is supported by previous findings that CTNNB1/TCF7L2-mediated transcription (Aoki et al. 2007) or ectopic expression of the WNT target gene conductin (AXIN2) (Hadjihannas et al. 2006) directly induces CIN. However, it should be noted that although CIN can be the underlying mechanism for CCAT2-promoted tumor growth and metastasis, mechanistic connections of these phenotypes are not established in this study.
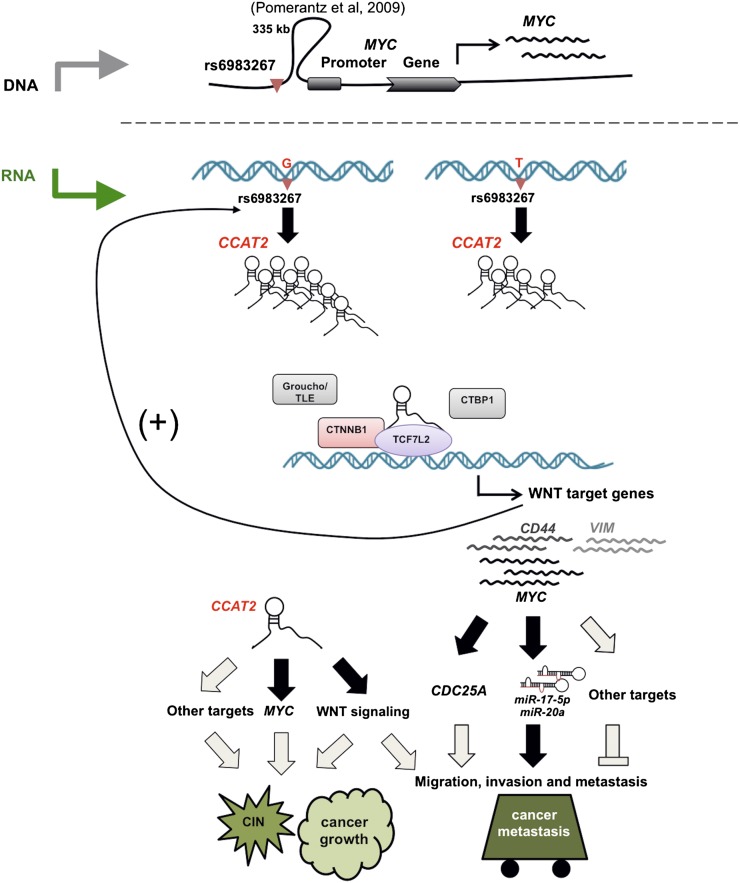
Finally, our Pyrosequencing data on heterogeneous cell line models and CRC samples suggest that rs6983267 status affects the CCAT2 expression, providing an additional mechanism linking the risk allele of rs6983267 with higher CRC risk. Although we demonstrated that WNT signaling regulates CCAT2 expression, it remains to be determined if this accounts for the SNP-caused differential expression of CCAT2. In addition, in CRC patients with GG (but not TT) genotype, CCAT2 shows significant association with MYC expression levels, suggesting that the SNP status may also affect CCAT2 function as an RNA transcript. Although most of the previous studies failed to identify a correlation between this SNP and MYC expression, recent research found higher MYC expression in CRC samples containing GG alleles, probably due to the improved purity of the cancer tissues owing to the use of laser microdissection (Takatsuno et al. 2012). Our findings on SNP-induced differential CCAT2 expression and function, together with the DNA enhancer element (Pomerantz et al. 2009; Tuupanen et al. 2009), provide an explanation on the cancer risk conferred by the rs6983267 risk allele (Fig. 7).
Figure 7.
A model of CCAT2 locus involvement in CRC. (Upper panel) An ∼335-kb DNA loop brings the rs6983267 genomic region close to the MYC locus, and this physical association may contribute to the enhancer function of the SNP-containing region on MYC transcription (Pomerantz et al. 2009). (Lower panel) The enhancer region is transcribed into a long noncoding RNA (CCAT2), and the SNP status affects CCAT2 expression by an as-yet-unknown mechanism. The CCAT2 transcript up-regulates WNT activity and increases expression levels of WNT target genes (including MYC). This regulation by CCAT2, possibly through its physical interaction with TCF7L2, may lead to genomic instability and promote cell growth. We demonstrated that MYC-regulated miR-17-5p and miR-20a participate in the CCAT2-enhanced cell invasion and speculate that other mechanisms, such as MYC-related mechanisms (CDC25A) or enhanced WNT signaling (VIM and CD44), may exist to coordinate the metastatic phenotype elicited by CCAT2. Finally, we demonstrated that CCAT2 expression is regulated by transcriptional factors TCF7L2, indicating a positive feedback loop between CCAT2 and WNT signaling. Our findings provide an additional explanation on the SNP-conferred CRC risk. (Black) Demonstrated; (gray) hypothesized interactions.
The discovery of CCAT2 introduces a novel mechanism for the involvement of ncRNAs in WNT signaling and colon cancer pathogenesis. The recent findings showing that the CIN phenotype is a predictive marker for survival and may be used to select high-risk patients with stages II and III CRC (Watanabe et al. 2012), combined with the specific involvement of CCAT2 in MSS CRCs indicate that CCAT2 has the potential to become a useful diagnostic and/or prognostic marker, although further validation in larger cohorts and by multiple independent studies is necessary.
Methods
Patient samples
One hundred and ninety-one colorectal cancer samples and 70 non-neoplastic mucosa samples were used in this study. The tumor and control samples were obtained from three different sources: an Italian cohort from the University of Ferrara (CRC = 81 and normal colon mucosa = 25), a Japanese cohort from the University of Kyushu (CRC = 45 and paired normal colon mucosa = 45), an Australian cohort from Princess Alexandra Hospital (CRC = 65), and a C-CFR cohort from Northern American colon cancer patients with MSS, CIMP-negative colon cancer at three participating centers of the Colorectal Cancer Family Registry (Mayo Clinic, Rochester, Minnesota, USA; Mount Sinai Hospital, Ontario, Canada; and Cleveland Clinic, Cleveland, Ohio, USA) (CRC = 24 and paired normal colon mucosa = 24). Tissue samples were obtained from fresh surgical specimens frozen in liquid nitrogen and stored at −80°C. All the samples were obtained with the patients' informed consent and were histologically confirmed.
Breast tumors were selected from the tumor bank at the Erasmus Medical Center (Rotterdam, The Netherlands). The frozen tumor samples were collected from female patients with breast cancer who entered the clinic from 1979 to 1998 and for whom detailed clinical follow-up data and tumor RNA for CCAT2 analysis were available (Sieuwerts et al. 2007; Smid et al. 2011). The protocol for studying biological markers associated with disease outcome was approved by the medical ethics committee of the Erasmus Medical Center (MEC 02.953).
The British CRC samples were derived from a set of CRC patients recruited as part of the CORGI study who had known genotypes at the 8q24 tagSNP (Tomlinson et al. 2007).
RNA isolation, cDNA synthesis, and quantification of specific RNA species
RNA was extracted using the TRIzol protocol (Invitrogen) and DNase-digested (Ambion); the quality of the RNA was assessed using agarose gel electrophoresis. cDNA was synthesized using the SuperScript III cDNA kit (Invitrogen), and diluted cDNA was used for RT-PCR analysis using iQ SYBR Green Supermix (Bio-Rad) with the appropriate primers (Supplemental Table S2). The 2−ΔCt method was used to calculate the relative abundance of RNA genes compared with GAPDH expression. qRT-PCR analysis for miRNAs was performed with the TaqMan MicroRNA Assay Kit (Applied Biosystems). Primers and probes for miR-17, miR-20a, miR-146a, let-7a, and RNU6-6P snRNA were purchased from Applied Biosystems. The 2−ΔCt method was used to calculate the relative abundance of miRNA genes compared with RNU6-6P expression.
Cell lines
COLO320DM, HCT116, RKO, and HEK293 cells were obtained from the American Type Culture Collection and validated by the Characterized Cell Line Core at The University of Texas MD Anderson Cancer Center using STR DNA fingerprinting.
Stable cell line generation
Stable HCT116 cells for CCAT2 overexpression experiments were produced by pMX retrovirus or pcDNA3.1 expression vector transfection. For the pcDNA3.1 single clones, we screened CCAT2 expression in the HCT116 cells transfected with CCAT2 construct and randomly chose two clones with high CCAT2 expression (OC1 and OC2), and one clone with basal level of CCAT2 expression (E2). E1 was originated from the HCT116 cells transfected with the empty pcDNA3.1 vector.
COLO320 stable cells for CCAT2 knockdown experiments were generated using pRS-Retro-GFP/neo vector retrovirus (OligoEngine) or the Knockout Single Vector Inducible RNAi System (Clontech).
Virus production
The CCAT2-containing genomic region was amplified with a primer combination (CCAT2 F2 and R2) from HCT116 genomic DNA with Pfu polymerase (Invitrogen). After sequencing of the PCR product, we cloned it into the pMX plasmid. Then we produced CCAT2-containing retrovirus in 293 GP2 cell lines and used virus-containing supernatant to infect HCT116 cells. After infection, HCT116 cells were grown in complete media containing puromycin (1 μg/mL). The shRNAs against CCAT2 (Supplemental Table S2) were inserted into the pRS-Retro-GFP/neo vector according to the manufacturer's protocol (OligoEngine).
Plasmid production
The sequence used for retrovirus production was also cloned into a pcDNA3.1+ vector. The pcDNA CCAT2 plasmid was linearized with BglII and transfected into HCT116 cells using Lipofectamine. Selection with G418 (0.5 mg/mL) was carried out until single colonies were generated. The tet-on inducible shCCAT2 clones (Supplemental Table S2) were generated using the Knockout Single Vector Inducible RNAi System according to the manufacturer's instructions.
CCAT2 in situ hybridization
The frozen tissue slides of two MSS CRCs, two MSI-H CRCs, and two normal mucosae were incubated with a double-DIG-labeled CCAT2 probe or control RNU6-6P snRNA probe (Exiqon) and detected with a polyclonal anti-DIG antibody and alkaline phosphatase–conjugated secondary antibody (Ventana) using NBT-BCIP as the substrate. The signal intensities of CCAT2 and RNU6-6P expression were quantified by using the intensity measurement tools of the Image-Pro Plus software package (Media Cybernetics).
In vivo tumorigenic assay
Swiss nu-nu/Ncr nude mice (spontaneous mutant T-cell-deficient mice) were obtained from the Animal Production Area of the Department of Experimental Radiation Oncology at MD Anderson Cancer Center. The mice were maintained under specific pathogen-free conditions and used in accordance with institutional guidelines. HCT116 cells (1 × 106) were injected subcutaneously into the right flanks of the mice, and tumor size was measured weekly for 4 wk.
In vivo metastasis assay
A midline incision was made on the left side of the flank, and the spleen was exteriorized. HCT116 cells (1 × 106 cells) were injected into the spleen, after which the wound was closed with surgical metal clips. The mice were sacrificed after 8 wk, and their spleens and livers were removed and examined for tumor metastases on the surface of the liver. The liver specimens were formalin fixed and paraffin embedded for histological analysis.
In vitro migration assays
We used two assays: In the first, HCT116 cells (200,000 cells in 200 μL of serum-free media) were seeded into a Transwell insert in a 24-well format (8 μm; BD Biosciences). To the bottom part of the well, 800 μL of medium with serum was added. Twenty-four hours after seeding, cells were fixed with paraformaldehyde and stained with crystal violet/methanol solution. Cells at the top of the Transwell were removed using a cotton swab, and photos of the Transwells were taken using Geldoc (Bio-Rad). The intensity of the crystal violet staining in each Transwell was evaluated using Photoshop software (Adobe Systems) and compared among the different treatments.
The second type of assay, a cell migration assay, was performed according to the protocol previously described (Spannuth et al. 2009). Cells were suspended in serum-free media (65,000 cells/insert) and seeded onto the gelatin-coated inserts. The cells that migrated to the bottom of the wells were fixed, stained, and counted using a microscope. For each well, 10 different fields were counted, and the average number of cells was determined. Both types of assays were performed in triplicate experiments.
SiRNA treatment
Cells were seeded onto 24-well or six-well plates. After reaching 50% confluence, cells were transfected with 50 nM siRNA by Lipofectamine 2000 (Life Technologies) for 48 h before collecting RNA or protein for analysis.
Genomic instability analysis
Cells were exposed to Colcemid (0.04 μg/mL) for 25 min at 37°C and to hypotonic treatment (0.075 M KCl) for 20 min at room temperature, and then were fixed in a methanol and acetic acid mixture (3:1 by volume) for 15 min and washed three times in the fixative. The slides were air-dried, stained in 4% Giemsa, and coded for the blind analysis. Later, the slides were decoded for the evaluation of results. Slides were analyzed for several parameters, including chromosome aberrations (as evidenced by both chromosome- and chromatid-type breaks), fragments, tetraploidy, fusions, and formation of tri-radials.
Spectral karyotyping
Spectral karyotyping was performed according to the manufacturer's protocol using Human Paint probes (Applied Spectral Imaging, ASI). Images were captured using a Nikon 80i microscope equipped with Spectral karyotyping software from ASI.
CREST staining
After fixation, cells were incubated for 1 h at room temperature with human serum from scleroderma patients (kindly provided by Maureen Mayes from the University of Texas–Houston Medical School) that produced, during the course of their disease, anti-centromere antibodies used for staining. Staining was visualized using FITC-conjugated donkey anti-human antibody. Beta-tubulin was detected using Cy3-conjugated antibody (Sigma-Aldrich).
Subcellular fractionation
The separation of nuclear and cytosolic fractions was performed using the PARIS Kit (Life Technologies) according to the manufacturer's instructions.
RNA immunoprecipitation
We used the Magna RIP Kit (Millipore) according to the manufacturer's instructions. Cells were prepared in RIP lysis buffer, and the RNA–protein complexes were immunoprecipitated using anti-TCF7L2 or normal goat IgG (control). RNA was purified using phenol:chloroform:isoamyl alcohol and subjected to reverse transcription-PCR or real-time PCR analysis (Supplemental Table S2). Control amplification was carried out on input RNA before immunoprecipitation.
TOP-Flash WNT reporter
Cells were transfected with 250 ng of the TOP-FLASH or FOP-FLASH reporter constructs together with 25 ng of the Renilla luciferase vector. Luciferase activity was measured by the Dual–Luciferase Reporter Assay System (Promega) 48 h after transfection, and TOP or FOP values were normalized to Renilla values. The TOP/FOP ratios were calculated and used as indicators of the endogenous level of WNT signaling.
Immunofluorescent assay
Cells were seeded on a 96-well plate. After 48 h, the cells were permeabilized and fixed in 4% paraformaldehyde in PBS buffer at room temperature for 15 min. The cells were then incubated with anti-vimentin (V9, Novus) overnight and finally incubated with secondary antibody and DAPI for 1 h. All matched samples were photographed using an immunofluorescence microscope and identical exposure times. Each experiment was performed in triplicate and repeated three times.
Allele quantification by Pyrosequencing
We used the Pyrosequencing technology (QIAGEN) to measure the frequency of the G and T alleles of the CCAT2 transcript. The principles of the method and analysis have been previously reported (Wang and Elbein 2007). In our study, nested PCR reactions were done for each sample. The first PCR was performed using the primers 1-F (TTTAGCAGCTGCATCGCTCCATAG) and 2-R (CTCCCTCCCCCACATAAAAT). The second PCR combined a universal biotinylated primer for subsequent single-strand DNA capture for the Pyrosequencing reaction. One microliter of the first PCR was used for the nested amplification, and a reaction was performed to capture the forward strand using the primers SNP/F1 (GGGACGAATAAACTCTCCTCCTAC), SNP/RU1 (GGGACACCGCTGATCGTTTACTCCCTCCCCCACATAAAAT), and the universal biotinylated primers UBP (Bio-GGGACACCGCTGATCGTTTA). An independent PCR reaction was performed to capture the reverse strand using the primers SNP/FU2 (GGGACACCGCTGATCGTTTAGAGGTGTAGCCAGAGTTAATACCC), SNP/R2 (CTCCCTCCCCCACATAAAAT), and UBP (Bio-GGGACACCGCTGATCGTTTA). After cleanup in a vacuum prep station, the single-strand DNA was combined with sequencing primers to read the forward (SNP/S1, AGCTCAGCAGATGAAA, and the sequence to analyze GG/TCACTG) and reverse (SNP/S2, TCTTTGTACTTTTCTCAGTG, and the sequence to analyze A/CCTTTCATC) strands of the target SNP. Each measurement was performed in duplicate, and the values from the forward and reverse strands was combined to give the frequency of the G and T alleles.
Statistical analysis
For the colorectal cancer samples, the statistical analyses were performed in R (version 2.14.2). The Shapiro-Wilk test was applied to determine whether data followed a normal distribution. Accordingly, the t-test or the nonparametric Mann-Whitney-Wilcoxon test was applied to assess the relationship between CCAT2 expression levels and clinical parameters. Fisher's exact test was used to assess the association between CCAT2 and the number of metastases in mice. For the breast cancer cohorts, data were analyzed using the STATA statistical package, release 9 (STATA Corp.) and SPSS 15.0 (IBM Corporation). We investigated the prognostic value of the clinical and biological variables with metastasis-free survival (MFS) as the end point using Cox univariate and multivariate regression analyses. CCAT2 RNA levels were analyzed as a log-transformed continuous variable and also, for visualization in Kaplan-Meier survival analysis, as a dichotomized variable based on the median level of CCAT2 RNA, with the log-rank test used to evaluate differences. A Spearman's rank-order correlation test was applied to measure the strength of association between the expressions of CCAT2 and MYC in colorectal cancer samples and between CCAT2 RNA and CIN score in breast cancer samples. All tests were two sided, and P < 0.05 was considered statistically significant.
Data access
The CCAT2 RNA sequence reported in this paper has been deposited in GenBank (http://www.ncbi.nlm.nih.gov/nuccore) under accession number GQ911591.
Acknowledgments
G.A.C. is the Alan M. Gewirtz Leukemia & Lymphoma Society Scholar. He is also supported as a Fellow at The University of Texas MD Anderson Research Trust, as a University of Texas System Regents Research Scholar, and by the CLL Global Research Foundation. Work in Dr. Calin's laboratory is supported in part by the NIH/NCI (CA135444); a Department of Defense Breast Cancer Idea Award; Developmental Research Awards in Breast Cancer, Ovarian Cancer, Brain Cancer, Prostate Cancer, Multiple Myeloma, and Leukemia SPOREs; an MDACC Sister Institution Network Fund grant in colorectal cancer; the Laura and John Arnold Foundation; the RGK Foundation; and the Estate of C.G. Johnson, Jr. H.L. is an Odyssey Fellow, and his work is supported in part by the Odyssey Program and the Estate of C.G. Johnson, Jr. at the University of Texas MD Anderson Cancer Center. R.F. and Y.A. were supported by grants from the Dutch Cancer Society (EMCR 2001-2482), the Netherlands Organisation for Scientific Research (NWO/Vici 016.036.636), the BSIK (Kennisinfrastructuur) program of the Dutch Government grant 03038 (http://www.stemcells.nl), the Netherlands Institute for Regenerative Medicine (NIRM; http://www.nirm.nl), and the EU FP6 and FP7 consortia Migrating Cancer Stem Cells program (MCSCs; http://www.mcscs.eu) and TuMIC (integrated concept of tumor metastasis; http://itgmv1.fzk.de/www/tumic/tumic_main.htm). K.M. and M.M. were supported by the Funding Program for Next Generation World-Leading Researchers (LS094) and Core Research for Evolutional Science and Technology, respectively. S.R.H. is the recipient of the Frederick F. Becker Distinguished University Chair in Cancer Research from The University of Texas. Genomic instability analysis was performed in the MD Anderson Cancer Center Core laboratory supported by NCI grant number CA016672. Short tandem repeat DNA fingerprinting was performed by the Cancer Center Support grant-funded Characterized Cell Line Core, NCI number CA16672. Allele quantification was performed in the MD Anderson Cancer Center DNA Methylation Analysis Core. The C-CFR Cohort study was supported by the National Cancer Institute, National Institutes of Health under RFA number CA-95-011 and through cooperative agreements with the University of Hawaii Colorectal Cancer Family Registry (U01 CA074806), Mayo Clinic Cooperative Family Registry for Colon Cancer Studies (U01 CA074800), Ontario Registry for Studies of Familial Colorectal Cancer (U01 CA074783), the University of Southern California Familial Colorectal Neoplasia Collaborative Group (U01 CA074799), and members of the Colon Cancer Family Registry and P.I.s. In addition, this work was partly supported by grant 5U24 CA074806 and an award by Affymetrix Collaborations in Cancer Research Program. We also thank the participants of the Colon Family Registry, who have contributed to a better understanding of the genetic contributions to colon cancer. The CCAT2 expression analysis for C-CFR samples was performed at the Genomics Shared Resource of the University of Hawaii Cancer Center, which is supported by 2P30CA071789-13. We thank Dawn Chalaire from the Department of Scientific Publications at The University of Texas MD Anderson Cancer Center for her help with the editing of this manuscript. We acknowledge Dr. Bryant G. Darnay, who kindly provided the pMX vector and offered assistance with the retrovirus production, and Dr. Maureen Mayes for kindly providing serum from scleroderma patients.
Author contributions: G.A.C. and R.F. designed this study. H.L., R.S., R.F., S.R.H., and G.A.C. wrote the manuscript. R.S., H.L., Y.A., M. Nicoloso, M. Shimizu, R.R., J.S., N.N., E.B., I.D.V., M.C., J.F.G., B.H.B., S.S., M. Shariati, F.H., V.B., A.S.M., J.W., M.A.S., W.Z., and A.M.S. performed most of the wet-lab experiments. R.G., G.A.P., D.C.G., K.Y., I.T., I.B.-N., K.M., M.M., A.M.S., J.W., M.M., I.T., S.G., G.C., S.N.T., L.L.M., M.T, J.A.F., S.R.H, and G.L. provided the patient specimens. X.Z. designed probes and performed the ISH experiments, and S.R.H. assisted with interpretation. H.L., R.S., Y.A., M.F., C.I., X.Z., R.V.D., A.M.S., J.A.F., M. Nicoloso, S.K., R.F., and G.A.C. analyzed the data. All authors read and approved the manuscript's content.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.152942.112.
References
- Ambros V 2001. microRNAs: Tiny regulators with great potential. Cell 107: 823–826 [DOI] [PubMed] [Google Scholar]
- Aoki K, Aoki M, Sugai M, Harada N, Miyoshi H, Tsukamoto T, Mizoshita T, Tatematsu M, Seno H, Chiba T, et al. 2007. Chromosomal instability by β-catenin/TCF transcription in APC or β-catenin mutant cells. Oncogene 26: 3511–3520 [DOI] [PubMed] [Google Scholar]
- Bartley AN, Yao H, Barkoh BA, Ivan C, Mishra BM, Rashid A, Calin GA, Luthra R, Hamilton SR 2011. Complex patterns of altered microRNA expression during the adenoma-adenocarcinoma sequence for microsatellite-stable colorectal cancer. Clin Cancer Res 17: 7283–7293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D 2004. Ultraconserved elements in the human genome. Science 304: 1321–1325 [DOI] [PubMed] [Google Scholar]
- Bertucci F, Lagarde A, Ferrari A, Finetti P, Charafe-Jauffret E, Van Laere S, Adelaide J, Viens P, Thomas G, Birnbaum D, et al. 2012. 8q24 cancer risk allele associated with major metastatic risk in inflammatory breast cancer. PLoS ONE 7: e37943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B 1998. Mutations of mitotic checkpoint genes in human cancers. Nature 392: 300–303 [DOI] [PubMed] [Google Scholar]
- Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE, et al. 2007. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell 12: 215–229 [DOI] [PubMed] [Google Scholar]
- Camps J, Morales C, Prat E, Ribas M, Capella G, Egozcue J, Peinado MA, Miro R 2004. Genetic evolution in colon cancer KM12 cells and metastatic derivates. Int J Cancer 110: 869–874 [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network 2012. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487: 330–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT 2008. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet 40: 43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H 2006. Wnt/β-catenin signaling in development and disease. Cell 127: 469–480 [DOI] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. 2012. Landscape of transcription in human cells. Nature 489: 101–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhrai-Rad H, Pourmand N, Ronaghi M 2002. Pyrosequencing: An accurate detection platform for single nucleotide polymorphisms. Hum Mutat 19: 479–485 [DOI] [PubMed] [Google Scholar]
- Felsher DW, Bishop JM 1999. Transient excess of MYC activity can elicit genomic instability and tumorigenesis. Proc Natl Acad Sci 96: 3940–3944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Wu Z, Wu Y, Hankey W, Prior TW, Li L, Ganju RK, Shen R, Zou X 2011. Cdc25A regulates matrix metalloprotease 1 through Foxo1 and mediates metastasis of breast cancer cells. Mol Cell Biol 31: 3457–3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodde R, Kuipers J, Rosenberg C, Smits R, Kielman M, Gaspar C, van Es JH, Breukel C, Wiegant J, Giles RH, et al. 2001. Mutations in the APC tumour suppressor gene cause chromosomal instability. Nat Cell Biol 3: 433–438 [DOI] [PubMed] [Google Scholar]
- Galaktionov K, Chen X, Beach D 1996. Cdc25 cell-cycle phosphatase as a target of c-myc. Nature 382: 511–517 [DOI] [PubMed] [Google Scholar]
- Ghoussaini M, Song H, Koessler T, Al Olama AA, Kote-Jarai Z, Driver KE, Pooley KA, Ramus SJ, Kjaer SK, Hogdall E, et al. 2008. Multiple loci with different cancer specificities within the 8q24 gene desert. J Natl Cancer Inst 100: 962–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilles C, Polette M, Mestdagt M, Nawrocki-Raby B, Ruggeri P, Birembaut P, Foidart JM 2003. Transactivation of vimentin by β-catenin in human breast cancer cells. Cancer Res 63: 2658–2664 [PubMed] [Google Scholar]
- Hadjihannas MV, Bruckner M, Jerchow B, Birchmeier W, Dietmaier W, Behrens J 2006. Aberrant Wnt/β-catenin signaling can induce chromosomal instability in colon cancer. Proc Natl Acad Sci 103: 10747–10752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiman CA, Le Marchand L, Yamamato J, Stram DO, Sheng X, Kolonel LN, Wu AH, Reich D, Henderson BE 2007. A common genetic risk factor for colorectal and prostate cancer. Nat Genet 39: 954–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW 1998. Identification of c-MYC as a target of the APC pathway. Science 281: 1509–1512 [DOI] [PubMed] [Google Scholar]
- Heuberger J, Birchmeier W 2010. Interplay of cadherin-mediated cell adhesion and canonical Wnt signaling. Cold Spring Harb Perspect Biol 2: a002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jho EH, Zhang T, Domon C, Joo CK, Freund JN, Costantini F 2002. Wnt/β-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol 22: 1172–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan KB, Burds AA, Swedlow JR, Bekir SS, Sorger PK, Nathke IS 2001. A role for the Adenomatous Polyposis Coli protein in chromosome segregation. Nat Cell Biol 3: 429–432 [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B 1997. Genetic instability in colorectal cancers. Nature 386: 623–627 [DOI] [PubMed] [Google Scholar]
- Li C, Kim SW, Rai D, Bolla AR, Adhvaryu S, Kinney MC, Robetorye RS, Aguiar RC 2009. Copy number abnormalities, MYC activity, and the genetic fingerprint of normal B cells mechanistically define the microRNA profile of diffuse large B-cell lymphoma. Blood 113: 6681–6690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MF, Jungreis I, Kellis M 2011. PhyloCSF: A comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 27: i275–i282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Radisky DC, Yang D, Xu R, Radisky ES, Bissell MJ, Bishop JM 2012. MYC suppresses cancer metastasis by direct transcriptional silencing of αv and β3 integrin subunits. Nat Cell Biol 14: 567–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig B, Jerchow B, Sachs M, Weiler S, Pietsch T, Karsten U, van de Wetering M, Clevers H, Schlag PM, Birchmeier W, et al. 2002. Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol Cell Biol 22: 1184–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell KO, Ricci MS, Miyashita T, Dicker DT, Jin Z, Reed JC, El-Deiry WS 2000. Bax is a transcriptional target and mediator of c-myc-induced apoptosis. Cancer Res 60: 6318–6325 [PubMed] [Google Scholar]
- Nguyen DX, Chiang AC, Zhang XH, Kim JY, Kris MG, Ladanyi M, Gerald WL, Massague J 2009. WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell 138: 51–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT 2005. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435: 839–843 [DOI] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. 2010. Long noncoding RNAs with enhancer-like function in human cells. Cell 143: 46–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, Doddapaneni H, Beckwith CA, Chan JA, Hills A, Davis M, et al. 2009. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet 41: 882–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY 2012. Genome regulation by long noncoding RNAs. Annu Rev Biochem 81: 145–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom OJ, Meniel VS, Muncan V, Phesse TJ, Wilkins JA, Reed KR, Vass JK, Athineos D, Clevers H, Clarke AR 2007. Myc deletion rescues Apc deficiency in the small intestine. Nature 446: 676–679 [DOI] [PubMed] [Google Scholar]
- Sieuwerts AM, Usher PA, Meijer-van Gelder ME, Timmermans M, Martens JW, Brunner N, Klijn JG, Offenberg H, Foekens JA 2007. Concentrations of TIMP1 mRNA splice variants and TIMP-1 protein are differentially associated with prognosis in primary breast cancer. Clin Chem 53: 1280–1288 [DOI] [PubMed] [Google Scholar]
- Smid M, Hoes M, Sieuwerts AM, Sleijfer S, Zhang Y, Wang Y, Foekens JA, Martens JW 2011. Patterns and incidence of chromosomal instability and their prognostic relevance in breast cancer subtypes. Breast Cancer Res Treat 128: 23–30 [DOI] [PubMed] [Google Scholar]
- Sotelo J, Esposito D, Duhagon MA, Banfield K, Mehalko J, Liao H, Stephens RM, Harris TJ, Munroe DJ, Wu X 2010. Long-range enhancers on 8q24 regulate c-Myc. Proc Natl Acad Sci 107: 3001–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spannuth WA, Nick AM, Jennings NB, Armaiz-Pena GN, Mangala LS, Danes CG, Lin YG, Merritt WM, Thaker PH, Kamat AA, et al. 2009. Functional significance of VEGFR-2 on ovarian cancer cells. Int J Cancer 124: 1045–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spizzo R, Nicoloso MS, Croce CM, Calin GA 2009. SnapShot: MicroRNAs in Cancer. Cell 137: 586. [DOI] [PubMed] [Google Scholar]
- Sur IK, Hallikas O, Vaharautio A, Yan J, Turunen M, Enge M, Taipale M, Karhu A, Aaltonen LA, Taipale J 2012. Mice lacking a Myc enhancer that includes human SNP rs6983267 are resistant to intestinal tumors. Science 338: 1360–1363 [DOI] [PubMed] [Google Scholar]
- Takatsuno Y, Mimori K, Yamamoto K, Sato T, Niida A, Inoue H, Imoto S, Kawano S, Yamaguchi R, Toh H, et al. 2012. The rs6983267 SNP is associated with MYC transcription efficiency, which promotes progression and worsens prognosis of colorectal cancer. Ann Surg Oncol 20: 1395–1402 [DOI] [PubMed] [Google Scholar]
- Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, Spain S, Penegar S, Chandler I, Gorman M, Wood W, et al. 2007. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet 39: 984–988 [DOI] [PubMed] [Google Scholar]
- Tuupanen S, Turunen M, Lehtonen R, Hallikas O, Vanharanta S, Kivioja T, Bjorklund M, Wei G, Yan J, Niittymaki I, et al. 2009. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet 41: 885–890 [DOI] [PubMed] [Google Scholar]
- Wang H, Elbein SC 2007. Detection of allelic imbalance in gene expression using Pyrosequencing. Methods Mol Biol 373: 157–176 [DOI] [PubMed] [Google Scholar]
- Wang L, Park HJ, Dasari S, Wang S, Kocher JP, Li W 2013. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res 41: e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Kobunai T, Yamamoto Y, Matsuda K, Ishihara S, Nozawa K, Yamada H, Hayama T, Inoue E, Tamura J, et al. 2012. Chromosomal instability (CIN) phenotype, CIN high or CIN low, predicts survival for colorectal cancer. J Clin Oncol 30: 2256–2264 [DOI] [PubMed] [Google Scholar]
- Wielenga VJ, Smits R, Korinek V, Smit L, Kielman M, Fodde R, Clevers H, Pals ST 1999. Expression of CD44 in Apc and Tcf mutant mice implies regulation by the WNT pathway. Am J Pathol 154: 515–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik SE, Rossi S, Shimizu M, Nicoloso MS, Cimmino A, Alder H, Herlea V, Rassenti LZ, Rai KR, Kipps TJ, et al. 2010. Non-coding RNA sequence variations in human chronic lymphocytic leukemia and colorectal cancer. Carcinogenesis 31: 208–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Ting DT, Stott SL, Wittner BS, Ozsolak F, Paul S, Ciciliano JC, Smas ME, Winokur D, Gilman AJ, et al. 2012. RNA sequencing of pancreatic circulating tumour cells implicates WNT signalling in metastasis. Nature 487: 510–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Xiao Z, Lai D, Sun J, He C, Chu Z, Ye H, Chen S, Wang J 2012. miR-21, miR-17 and miR-19a induced by phosphatase of regenerating liver-3 promote the proliferation and metastasis of colon cancer. Br J Cancer 107: 352–359 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ, Roussel MF 1998. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev 12: 2424–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]