Abstract
Human endogenous retroviruses (HERVs) make up 8% of the human genome. The HERV-K (HML-2) family is the most recent group of these viruses to have inserted into the genome, and we have detected the activation of HERV-K (HML-2) proviruses in the blood of patients with HIV-1 infection. We report that HIV-1 infection activates expression of a novel HERV-K (HML-2) provirus, termed K111, present in multiple copies in the centromeres of chromosomes throughout the human genome yet not annotated in the most recent human genome assembly. Infection with HIV-1 or stimulation with the HIV-1 Tat protein leads to the activation of K111 proviruses. K111 is present as a single copy in the genome of the chimpanzee, yet K111 is not found in the genomes of other primates. Remarkably, K111 proviruses appear in the genomes of the extinct Neanderthal and Denisovan, while modern humans have at least 100 K111 proviruses spread across the centromeres of 15 chromosomes. Our studies suggest that the progenitor K111 integrated before the Homo-Pan divergence and expanded in copy number during the evolution of hominins, perhaps by recombination. The expansion of K111 provides sequence evidence suggesting that recombination between the centromeres of various chromosomes took place during the evolution of humans. K111 proviruses show significant sequence variations in each individual centromere, which may serve as markers in future efforts to annotate human centromere sequences. Further, this work is an example of the potential to discover previously unknown genomic sequences through the analysis of nucleic acids found in the blood of patients.
Human endogenous retroviruses (HERVs) account for 8% of the human genome. These ancient viruses infected the germ cells of mammals and other vertebrates multiple times over millions of years, and as such, their proviruses (viral DNA genomes that are integrated into the host DNA) have been transmitted over the generations in a Mendelian fashion and now remain in the genome (Nelson et al. 2003; Jern and Coffin. 2008; Subramanian et al. 2011). The HERV-K (HML-2) family constitutes the most recent insertion form of these viruses (Barbulescu et al. 1999; Okahara et al. 2004). HERV-K (HML-2) has replicated during the evolution of humans by reinfection (Belshaw et al. 2004) and today accounts for ∼3000 proviral fragments (Paces et al. 2004). At least 91 full-length HERV-K (HML-2) viral elements have been reported to exist in the human genome (Subramanian et al. 2011), most of which have accumulated mutations. The HERV-K (HML-2) family exists in the genome in proviral forms consisting of three retroviral genes (gag, pol, and env), and two accessory genes (rec and np9) flanked by two long terminal repeats (LTRs), with the 5′ LTR serving as the viral transcriptional promoter (Bannert and Kurth 2004). More than 2500 HERV-K (HML-2) elements exist as solitary LTRs (solo LTR), which originated by recombination between the 5′ and 3′ LTRs of full-length proviruses, removing the internal viral genes (Hughes and Coffin 2004). Integration of HERV-K (HML-2) in human DNA produced 5- to 6-bp target site duplication sequences on each side of the provirus. However, target site duplication is not apparent in all HERV-K (HML-2) proviruses, as homologous recombination between certain different HERV-K (HML-2) proviruses created hybrid proviruses with different flanking target site sequences (Hughes and Coffin 2005).
The HERV-K (HML-2) group is made up of human-specific proviruses, of which 11 are polymorphically inserted among humans (Barbulescu et al. 1999; Turner et al. 2001; Subramanian et al. 2011; Contreras-Galindo et al. 2012). Among all of the HERVs, HERV-K (HML-2) appears to be the most transcriptionally active (Tönjes et al. 1996; Seifarth et al. 1998; Johnston et al. 2001; Sugimoto et al. 2001; Wang-Johanning et al. 2001; Yi et al. 2001; Ruda et al. 2004) and has been found to produce virus-like particles (VLPs) in breast cancer, leukemia, melanoma, and teratocarcinoma cell lines, although these particles appear not to be infectious (Löwer et al. 1993; Seifarth et al. 1998; Bieda et al. 2001; Muster et al. 2003; Büscher et al. 2005). We detected activation of HERV-K (HML-2) proviruses in the blood of patients with HIV-1 infection and with certain types of cancers, such as lymphoma and breast cancer, leading to the production of viral RNA, proteins, and VLPs that are found in the blood of patients (Contreras-Galindo et al. 2006, 2008, 2012).
In our previous work, we discovered many HERV-K (HML-2) RNA sequences in the plasma of HIV-1 patients that we could not assign to known human proviruses. These HERV-K (HML-2) sequences were only ∼95% similar to the closest known HERV-K (HML-2) proviruses but were 98% similar to one provirus found in the genome of the chimpanzee. We used the flanking sequence of that locus in the genome of the chimpanzee to amplify the previously unknown human provirus, which we termed K111 (Contreras-Galindo et al. 2012). In the present study, we report the detection of hundreds of K111 variants dispersed throughout the centromeres of at least 15 human chromosomes. Our studies indicate that the progenitor K111 inserted in the genome of the hominid lineage before the split of humans and chimpanzees. Furthermore, as only a few K111 proviruses can be identified in the genome of the extinct Denisovan and Neanderthal, and as multiple copies of K111 proviruses exist in the genome of modern humans, our studies indicate that the expansion of K111 took place during the evolution of hominins. Sequence analysis indicated that K111 proviruses expanded in copy number both within and among several human chromosomes by a mechanism resembling recombination, providing striking evidence for likely homologous recombination between centromeres during the evolution of humans. Finally, we further explored the mechanism for the activation of K111 during HIV-1 infection and found that the HIV-1 Tat protein is responsible, at least in part, for the expression of K111 by inducing loss of heterochromatin in pericentromeric regions. By studying endogenous retroviruses expressed in the blood of living patients, we have thus uncovered new centromeric proviruses hidden in the human genome.
Results
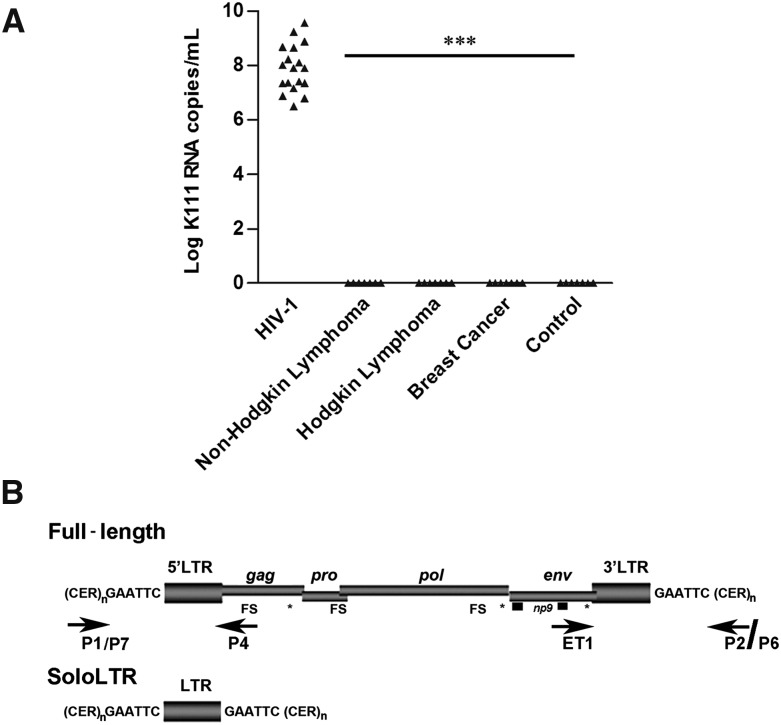
We first found a phylogenetically distinct HERV-K (HML-2) viral RNA, termed K111, in the blood of HIV-1–infected patients and not in breast cancer or lymphoma patients tested (Fig. 1; Supplemental Fig. S1; Contreras-Galindo et al. 2012). K111 sequences are not found in the current version of the annotated human genome (GRCh37/hg19; Feb. 2009); however, a full-length K111 provirus we termed CERV-K111 was found close to the telomere of the q arm of chromosome 7 in the chimpanzee (NCBI Nucleotide database [http://www.ncbi.nlm.nih.gov/nuccore] acc. no. NW_003457191.1). Using this information, we designed primers (P1/P7 and P4; ET1 and P2/P6) (Fig. 1B) that target the flanking regions and the internal viral genes of the K111 provirus to amplify K111 in humans by PCR, and found that while K111 RNA was particularly found in the blood of the HIV-1–infected patients studied, K111 exists at the genomic DNA level in all 189 human samples tested, including those of healthy subjects. We detected two forms of K111, the full-length provirus as well as the solo LTR, in all of these 189 human DNA samples (Fig. 1B; Supplemental Information).
Figure 1.
Identification and genomic organization of K111 proviruses. (A) Quantitation of K111 env titers by qRT-PCR in the plasma of patients with HIV-1 and other diseases. The K111 env titers were measured by qRT-PCR using the probe K111P (see Supplemental Methods) that specifically discriminates the K111 env gene from other HERV-K (HML-2) env sequences due to a 6-bp mutation. K111 titers were detected in the plasma of patients with HIV infection but not in the plasma of healthy individuals or the plasma of patients with lymphoma or breast cancer. (B) Genomic organization of K111 full-length and solo LTR, target site duplication “GAATTC,” and centromeric flanking sequences (CER:D22Z3). Frame shift (FS) and stop codon (asterisks) mutations are indicated. The positions of the primers used to amplify K111 5′ LTR and 3′ LTR insertions are indicated by arrows.
K111 integrated into the centromeric repeat CER:D22Z3, which has been assigned to the centromere of chromosome 22 (Metzdorf et al. 1988; Dunham et al. 1999) and created a characteristic GAATTC target site duplication flanking each side of the provirus after integration (Fig. 1B). We then asked whether other copies of K111 could be found in human sequence databases. BLAST analysis of the flanking CER:D22Z3 and LTR of K111 to the NCBI database revealed four K111-related insertions, two solo LTRs and two full-length proviruses. The K111 solo LTRs were assigned to the centromeres of chromosomes 9 and 22 but have not been annotated in the human genome draft sequence (GRCh37/hg19; Feb. 2009). To verify that these K111 solo LTRs originated by recombination between the LTRs of K111 proviruses and to rule out the possibility that these solo LTRs are derived from other HERV-K (HML-2) proviruses, we performed phylogenetic (Supplemental Fig. S2A) and sequence recombination (Supplemental Fig. S2B,C) analysis, which indicated that K111 solo LTRs arose from recombinational deletion of the 5′ and 3′ LTRs of full-length K111 proviruses as predicted.
The two other K111-related full-length proviruses retrieved in the BLAST analysis, the previously described K105 and one we termed K112, were assigned to the centromere and the pericentromere of chromosome 21, respectively (Supplemental Fig. S3; Barbulescu et al. 1999; Kurdyukov et al. 2001), but have not been annotated in the current human genome draft sequence (GRCh37/hg19; Feb. 2009). As only sequences from the LTRs and the flanking regions of K105 and K112 were obtained in these studies, we used primers that recognize sequences specific to K112 and K105 to confirm the existence of these proviruses in humans and obtained additional K105 (6361 bp, accession number JQ790992) and K112 (2042 bp, accession number JQ790991) sequences (Supplemental Fig. S3). The sequence of the 5′ LTR of the K105 provirus is 100% identical to our full-length K111; the 3′ LTR of K105 and the gag (partial sequence), pol, and env genes are ∼98.5% similar to the K111 provirus, suggesting that K105 is a variant of K111 (Supplemental Fig. S3). The 5′ LTR sequence of provirus K112 is characterized by a 22-bp deletion and the partial sequence of K112 is ∼96.4% similar to K111, suggesting that K112 is another variant of K111 (Supplemental Fig. S3). Therefore, at least four K111 variants are found in the NCBI database, although they were not previously characterized as such.
By studying the expression of HERV-K (HML-2) in the blood of HIV-1–infected patients, we thus uncovered the retroviral insertion K111 that is present in the genomes of all humans. We therefore wished to further characterize the new K111 endogenous retrovirus. Analysis of the K111 RNA env sequences found in HIV-1 patients showed that K111s have accumulated a balanced number of synonymous and nonsynonymous mutations over the years, suggesting that the DNA sequence has been maintained and propagated by a process resembling recombination rather than infection (Contreras-Galindo et al. 2012). As K111 proviruses are integrated into CER:D22Z3 repeats, we used BLAST analysis of human databases to look for CER:D22Z3 sequences, in which we predicted that other K111-related insertions might be found to be integrated. We identified sequences similar to CER:D22Z3 in 10 human centromeres and one telomere (Supplemental Fig. S3B). In order to calculate the time of integration of K111 and test the possibility of whether centromeric K111 expanded by recombination during the hominid evolution, we searched for K111-related 5′ and 3′ insertions in the DNA of New and Old World monkeys and primates, including humans, by PCR (Fig. 1B; Supplemental Results). One K111 insertion was detected in the chimpanzee, but none were found in other primates or monkeys (Fig. 2A). This is consistent with results obtained in previous studies (Barbulescu et al. 1999; Subramanian et al. 2011). K111 solo LTR insertion was detected in human DNA (Fig. 2A) but not in the chimpanzee or other primates, suggesting that full-length K111 has undergone recombinational deletion between the LTRs only during human evolution. We dated the integration time of K111 into the hominid lineage by molecular clock sequence analysis of the LTRs of K111 (see Supplemental Results) and found that K111 integrated ∼2.6–6.3 Myr ago, around the time of the Homo-Pan divergence, an event calculated to have happened <6.3 Myr ago (Patterson et al. 2006).
Figure 2.
Expansion of K111 proviruses in humans took place after the Homo-Pan divergence. (A) Detection of K111 full-length and solo LTR insertions from DNA of New and Old World primates. Full-length K111 proviruses were detected by PCR only in the human and chimpanzee. The 5′ flanking K111 insertions were amplified with the primers P1/P7 and P4, and the 3′ flanking K111 insertions were amplified with the primers ET1 and P6. Solo LTRs were amplified by PCR using the primers P1 and P2 (see Supplemental Methods) and were seen only in humans. Arrows indicate individual insertional polymorphisms. (B) Detection of K111 insertions in human chromosomes. DNA from human–hamster hybrid cell lines, which carry only one specific human chromosome, were analyzed by PCR for the presence of K111 using the set of primers that amplify the 5′ LTR insertion as described. Other bands (e.g., the PCR products detected in chromosomes 3, 6, and 8) were shown by sequencing to be the result of nonspecific PCR amplification.
We sequenced the K111 insertions amplified in human DNA, and the sequence variation in these PCR products showed the existence of multiple copies of K111 in humans. We also detected several K111 5′ and 3′ insertions only in humans (Fig. 2A), further suggesting that the expansion of K111 took place during the evolution of hominins. The CER:D22Z3 elements flanking the K111 insertions are characterized by different patterns of sequence repetition (Supplemental Fig. S4), indicating that K111 proviruses are found at multiple loci and suggesting that recombination may have involved not only the K111 sequence but also flanking CER:D22Z3 elements.
We then studied whether the K111 proviruses amplified in human DNA by PCR arose by recombination or by independent infection and integration using phylogenetic analyses of 5′ and 3′ LTR sequences. At the time of integration, the 5′ and 3′ LTRs of each provirus are identical; thus in a phylogenetic tree, the 5′ and 3′ LTRs for each provirus should appear as sister taxa. Our phylogenetic reconstruction of the many LTRs from K111-related proviruses amplified by PCR in human samples and other HERV-K (HML-2) proviruses shows two major branches (Supplemental Fig. S5). One branch represents multiple proviruses that independently infected humans over millions of years, such as K101, K102, K110, and K113. The 5′ and 3′ LTRs of each of these proviruses cluster as sister taxa, and therefore, the observed results can only be explained by the LTRs being copied via duplication during the replication event that immediately precedes integration. The other major branch depicts an apparently single ancestral retroviral infection with K111 that subsequently expanded into many copies of K111-related proviruses by a mechanism resembling recombination. The ancestral K111 branch diverges into two branches corresponding to the 5′ and 3′ LTRs. As predicted, the oldest LTR sequences found in each one of these branches corresponds to the chimpanzee K111 5′ and 3′ LTRs, which likely represent the ancestral K111 progenitor. Human K111 5′ and 3′ LTR sequences do not cluster as sister taxa, unlike other HERV-K (HML-2), excluding the possibility that K111 expanded in copy number via multiple independent infections. Rather, the 5′ and 3′ LTRs were distinct at the time of integration, suggesting subsequent recombination rather than infectious replication. Therefore, the evolutionary relationships of the K111 5′ LTRs, as well as of the K111 3′LTRs, suggest that K111 expanded by a mechanism resembling recombination, thus preserving the original sequence of the progenitor K111 5′ and 3′ LTR.
As we found CER:D22Z3-like elements in the centromeres (and one telomere) of ∼10 chromosomes in human databases (Supplemental Fig. S3), we wondered whether K111 sequences might be found embedded in multiple CER:D22Z3 elements. Thus, we looked for the presence of K111-related insertions in each human chromosome using DNA from human/rodent cell hybrids (Fig. 2B), each one harboring one human chromosome, by PCR. Interestingly, K111 insertions were detected in human chromosomes 1, 4, 7, 9, 12, 13, 14, 15, 17, 18, 20, 21, 22, X, and Y, but not in the other human chromosomes. K111 solo LTRs were detected only in chromosomes 15 and 22. These DNAs were prepared in an outside laboratory, and the possibility of DNA contamination was further ruled out by amplification of chromosome-specific genes in five of the chromosomes where K111 insertions were amplified (Supplemental Fig. S6).
We next studied whether one or multiple K111 proviruses exist in each human chromosome using phylogenetic analyses of the sequences obtained by PCR in the human/rodent cell hybrids. Phylogenetic reconstruction (Fig. 3), and the differences in nucleotide sequences of individual K111 insertions seen in each chromosome (Supplemental Fig. S7), demonstrated the existence of at least 100 K111 proviruses (Fig. 3). All K111 variants detected in human chromosomes are flanked by CER:D22Z3 repeats and the same GAATTC target site duplication preceding the proviral LTRs. In Figure 3, the color of each sequence in the tree represents the chromosome from which the sequence was amplified. As sequences of the same color cluster near each other in the tree, it is clear from the phylogenetic analysis that the K111 loci found in each chromosome are much more similar to each other than to the K111 loci of other chromosomes. Therefore, the phylogenetic tree groups K111 sequences in separate branches, each one corresponding to the K111 loci present in a specific chromosome. Specific informative nucleotide sequence substitutions are found in the K111 loci of each chromosome (Supplemental Fig. S7). Thus, these studies indicate that K111 loci are not randomly distributed between/among the chromosomes, but rather phylogenetically distinct informative substitutions exist in the K111 loci of a given chromosome. In addition, the existence of phylogenetically informative substitutions of the K111s distinct to each chromosome suggests that sequence errors are not responsible for generating the novel K111 loci. These sequence studies further suggest that K111 loci may have increased in copy number within each centromere by intrachromosomal recombination. As predicted, the K112 and K105 proviruses clustered to K111 insertions amplified in chromosome 21. Several distinct K111 insertions were found in chromosomes 15, 21, and 22, correlating with the greater intensity of the amplification products found in these particular chromosomes (Figs. 2B, 3). Evidence from recombinant sequences (i.e., clones 1A6, 7A6, and 14A4) also suggests that interchromosomal recombination occurred between certain chromosomes.
Figure 3.
Identification of K111 proviruses in individual human chromosomes. Bayesian inference tree of the 5′ LTR and 3′ LTRs, and flanking CER:D22Z3 sequences, of K111 proviruses amplified from human chromosomes. Sequences are colored to indicate from which human chromosome they arise. Note that each color tends to cluster to specific evolutionary branches, indicating that individual K111s often spread within an individual chromosome. Posterior probability values greater than 70 are shown for an unrooted tree. The tree was generated using Bayesian inference with four independent chains run for at least 1,000,000 generations until sufficient trees were sampled to generate >99% credibility. 5′ (black) and 3′ (blue) LTRs and solo LTR (red) lineages are shown along with the chimpanzee LTRs (CERV-K111). Informative nucleotide sequence substitutions were found that are specific for the K111 group of sequences found in each chromosome.
We next investigated whether the K111 sequences obtained by PCR can be detected by another methodology, using bioinformatics approaches and next-generation deep sequencing of human DNA samples. Hence, we isolated DNA from splenic fibroblasts and adjacent malignant lymphocytes from a patient with large B-cell lymphoma. We enriched for HERV-K (HML-2) LTR sequences using a set of probes that span the HERV-K (HML-2) LTR, and performed deep sequencing analysis. By use of this different approach, 90% of the K111 insertions detected by PCR were independently discovered (Supplemental Table S1), confirming the existence of multiple K111 insertions in humans. We are able to assign most of these K111 insertions to chromosomal locations based on PCR studies using monochromosomal cell hybrids (Fig. 2B).
As the genome sequence of two archaic hominins, the Neanderthal and the Denisovan, have been previously obtained from fossil samples (Noonan et al. 2006; Green et al. 2010; Reich et al. 2010; Meyer et al. 2012), we searched for K111 insertions in the genome of these ancient human relatives. First we looked for K111 virus–host junctions by retrieving reads that contain 20 bp of flanking CER:D22Z3 sequence, the GAATTC target site duplication, and 20 bp of the K111 LTR. We found sequence reads that meet these criteria in the Neanderthal genome and the Denisovan genome, showing the presence of K111 in these extinct hominins. We then searched for read sequences identical to the K111 insertions amplified by PCR in human/rodent cell hybrids. Bioinformatic analyses identified reads of seven K111 insertions in the Neanderthal and four K111 insertions in the Denisovan (Supplemental Table S2). Many of the K111 insertions were identified in independent sequence reads, confirming the existence of K111s in the genomes of these hominins. It is likely that several other K111 insertions were not identified because of the difficulties in sequencing ancient DNA as well as the depth of genome coverage obtained in these studies (Green et al. 2010; Reich et al. 2010). A recent report identified several more HERV-K (HML-2) proviruses in the genome of the Denisovan than of the Neanderthal (Agoni et al. 2012). K111 was not identified in this study, likely due to the filtering out of repetitive elements not assembled in the human genome reference sequence (GRCh37/hg19; Feb. 2009). Taken together, our studies indicate that the expansion of K111 occurred after the panini and hominini separation, continued to some degree in the Neanderthals and Denisovans, and expanded extensively during the evolution of the Homo sapiens to exist as at least 100 K111 proviruses in the genome of modern humans.
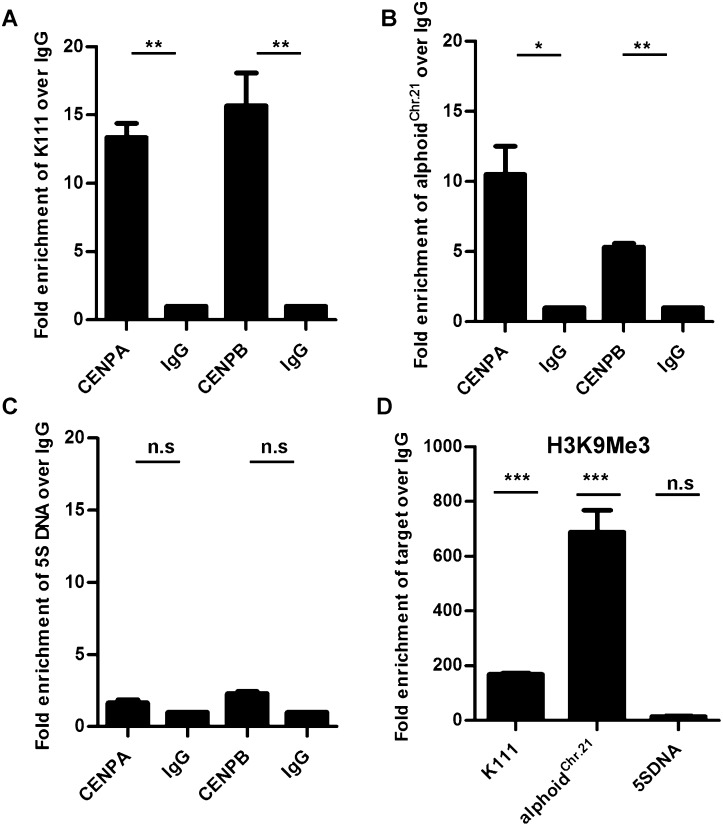
We next performed chromatin immunoprecipitation (ChIP) assays to further confirm that K111 sequences localize to centromeric chromatin. The histone 3 variants, centromere protein A (CENPA) (Verdaasdonk and Bloom 2011) and the centromere protein B (CENPB) (Masumoto et al. 2004), are both DNA-binding proteins specific to the centromere; the histone post-translational modification mark H3K9me3 is one of the hallmarks of pericentromeric heterochromatin (Mosch et al. 2011). We immunoprecipitated CENPA, CENPB, and the H3K9me3 mark in chromatin extracts from HeLa cells using specific antibodies, and the K111 DNA linked to these centromere proteins or chromatin marks was then quantitated by qPCR. Our studies show a ∼15-fold enrichment of K111 associated with CENPA and CENPB when ChIP was performed with specific antibodies compared with ChIP performed with control IgG (Fig. 4A). Immunoprecipitation using antibodies to CENPA and CENPB also yielded an enrichment of the endogenous 11-mer alphoid repeat of chromosome 21 (alphoidChr.21) as previously reported (Nakano et al. 2008), and the enrichment was comparable to that seen with K111 (Fig. 4B). In contrast, antibodies specific to CENPA and CENPB did not enrich the 5S ribosomal DNA gene, which is found in the q arm of chromosome 1 (Fig. 4C). Immunoprecipitation of the H3K9me3 histone mark, which is found abundantly in pericentromeric regions (Mosch et al. 2011), yielded a marked enrichment of K111 DNA (∼170-fold change) and endogenous alphoidChr.21 repeat DNA (∼650-fold change), but did not significantly enrich the 5s ribosomal DNA (Fig. 4D). These results confirm that K111 sequences reside in the centromeric and pericentromeric domains of the centromeres. So, although we cannot rule out the possibility that some K111 are found in telomeres, it appears that the vast majority of them are in centromeres.
Figure 4.
ChIP analysis shows that K111 proviruses are found in centromeric and pericentromeric regions. Quantitative PCR of K111 DNA (A), and the centromeric 11-mer alphoid repeat of chromosome 21 (alphoidChr.21) DNA (B), immunoprecipitated by antibodies to CENPA and CENPB or control IgG. K111 is enriched ∼15-fold in the CENP protein fractions compared with control IgG (A), while the alphoidChr.21 is enriched approximately eightfold (B). Quantitative PCR of 5S ribosomal DNA, present in the q arm of chromosome 1, shows no significant enrichment with antibodies to CENPA and CENPB (C). Quantitative PCR of K111, alphoidChr.21, and 5S ribosomal DNA in H3K9me3-associated fractions shows that K111 is enriched ∼170-fold. The 11-mer alphoidChr.21 repeat is enriched ∼650-fold in the H3K9me3-associated fractions, while 5S ribosomal DNA shows no significant enrichment (D). Graphs show the relative enrichment normalized to control IgG-precipitated fractions. Asterisks indicate statistical significance: (***) P < 0.001, (**) P < 0.01, (*) P < 0.05, (n.s.) not significant.
Lastly, we asked why K111, which is present in the genomes of all subjects tested, is particularly seen in the blood of HIV-1–infected patients. Thus, we tested whether the activation of K111 expression seen in HIV-1 patients is the direct result of HIV-1 infection or indirect pathology associated with the infection. We quantitated the K111 RNA levels by qRT-PCR in both HIV-1–infected cell lines (Fig. 5A) and peripheral blood lymphocytes (PBLs) freshly infected with HIV-1 (Fig. 5B). Infection of human cell lines and PBLs with HIV-1 induced the expression of K111 RNA, which is otherwise silenced in most uninfected cells. Further, as we had previously observed that the HIV-1 transactivator (Tat) protein activates the HERV-K (HML-2) promoter (Gonzalez-Hernandez et al. 2012), we tested whether stimulation of human cells with Tat was sufficient to induce K111 expression by qRT-PCR. The concentration of the HIV-1 Tat protein that we used in these experiments correlates well with the levels of Tat found in the blood of HIV-1–infected patients (Gonzalez-Hernandez et al. 2012). Indeed, addition of recombinant Tat to PBLs or overexpression of Tat in cell lines led to a marked increase in K111 RNA (Fig. 5C). As expression of K111 proviruses is likely to be repressed at baseline due to the condensed structure of chromatin at the centromere, and as other groups have shown that Tat drives a transition from heterochromatin to euchromatin by activating histone acetylases (Easley et al. 2010), we tested whether expression of Tat leads to a more open chromatin structure over K111 proviruses. We examined the effect of Tat on the chromatin structure associated with K111 by ChIP assays using antibodies that detect the heterochromatic histone marks H3K9me3, which associates with K111 DNA as shown above, and H4K20me3. We immunoprecipitated histones with the H3K9me3 or H4K20me3 marks using modification-specific antibodies on chromatin extracts from control HeLa and Tat-expressing HeLa cells and quantitating the K111 DNA linked to the histone proteins bearing these marks by qPCR (Fig. 5D). These studies demonstrated a marked loss of heterochromatic marks, and hence heterochromatin, over K111 in Tat-expressing cell lines compared with the parental cell line. In contrast, Tat did not induce loss of the same heterochromatin marks over the RPL30 and MYOD1 genes, which are epigenetically regulated under other circumstances (Supplemental Fig. S8). Therefore, Tat appears to activate K111 proviruses by inducing loss of heterochromatin in pericentromeric regions, opening up the chromatin structure and allowing for active transcription. Modulation of the transcription factors NFkappaB and NF-AT may further contribute to the activation of the K111 promoter (Gonzalez-Hernandez et al. 2012). However, as K111 is found in highly heterochromatic regions (centromeres), it appears that the opening of chromatin by Tat would be a crucial step in activating expression of K111 proviruses. Reconstruction of K111 env sequences found in the blood of HIV-1 patients and K111-related DNA proviruses shows that HIV-1 induces activation of several distinct K111 proviruses (Supplemental Fig. S9).
Figure 5.
HIV-1 infection, and HIV-1 Tat protein, activate K111 provirus expression in part by inducing loss of heterochromatin. HIV-1 infection of cell lines (A) or human peripheral blood lymphocytes (PBLs; B) activates the expression of K111 as detected by qRT-PCR of K111 env RNA using the K111-specific probe K111P. K111 RNA expression was not detected, or was detected only at low titer, in uninfected cells. (C) Activation of K111 in cell lines constitutively expressing Tat, or in human PBLs by addition of exogenous HIV-1-Tat. K111 RNA levels measured by qRT-PCR were essentially undetectable in cells without Tat but were found at titers ranging from 104–106 in Tat-stimulated cells. (D) ChIP assays show that HIV-1-Tat reduces the heterochromatic marks H3K9me3 and H4K20me3 at K111 proviral loci. Histones containing the H3K9me3 or H4K20me3 modification were specifically immunoprecipitated from the nuclear extracts of control HeLa and HeLa Tat expressing cells, and the K111 DNA bound to the histones containing these heterochromatin marks was measured by qPCR using the K111-specific probe K111P. Tat expressing cell lines: Jurkat Tat, Hltat, and HeLaTatIII. (***) P < 0.0001, (**) P < 0.001.
Discussion
In this study, we report that in addition to the 91 full-length HERV-K (HML-2) proviruses previously found in the human genome draft sequence (GRCh37/hg19; Feb. 2009; Subramanian et al. 2011), hundreds more HERV-K (HML-2) proviruses not yet published or deposited in the NCBI database exist in the human genome. These proviruses are related to a common provirus insertion, termed K111, which is found in our closest relative, the chimpanzee, as one single copy. The repetitive nature of these proviruses and their flanking sequences, their location in the centromere, and the existence of similar sequences in several chromosomes that resemble segmental duplications may have made these sequences previously impossible to assemble in the human genome sequence. Strikingly, K111 is not found in monkeys and lower primates, is found in a single copy in chimpanzees, in a small number of copies in Neanderthals and Denisovans, but in at least 100 copies in modern humans.
K111 proviruses are members of the HERV-K (HML-2) group that has entered the human genome on multiple separate occasions and is now represented by insertions in multiple locations throughout the genome. In contrast to other HERV-K (HML-2), the K111 insertions can be recognized by the GAATTC target site nucleotide sequence duplication on each side of the provirus. Other HERV-K (HML-2) have different target site duplication sequences, indicating that they entered the genome in multiple, independent events, unlike K111, which appears to have inserted only once but then spread to other genomic locations by recombination. In addition, unlike the case with other HERV-K (HML-2), several CER:D22Z3 elements are found flanking each side of the K111 proviral variants. These findings suggest that K111s arose from one ancestral infection and expanded by recombination into the centromeres of several chromosomes during human evolution. Premature stop codon mutations present in all of the viral genes of the K111 genome (except np9) suggest that K111 is not replication competent (Fig. 1B; Contreras-Galindo et al. 2012). However, evidence for recombination between K111 and other HERV-K (HML-2) RNAs found in the plasma of HIV patients suggests that K111 could complement other HERV-K (HML-2) loci in trans, but only under the circumstance of active expression of K111, as seen in HIV-1 infection (Contreras-Galindo et al. 2012).
Copying of other HERV-K (HML-2) loci by the process of segmental duplication has been reported (Reus et al. 2001). However, the present report is the first sequence-based study to discover and show copying of a HERV lineage and its progeny within multiple human centromeres, and suggests that centromeres from different chromosomes exchanged genetic material by recombination during hominid evolution. Interestingly, an endogenous retrovirus in the kangaroo (KERV) has recently been shown to have expanded its progeny into areas restricted to centromeric and pericentromeric domains (Carone 2008; Ferreri et al. 2011).
As CER:D22Z3 elements are also found in a telomere, some of the K111 sequences may also be from these chromosomal regions. However, our assignment of K111 sequences primarily to centromeres is based on six observations: (1) In situ hybridization analysis of CER:D22Z3 elements, which flank the K111s, demonstrated that these elements are found in the centromeres of chromosomes 21 and 22 (Metzdorf et al. 1988; Müllenbach et al. 1992); (2) mapping of certain K111 variants, referred to by us and others as K105 and K112, using hybridization analysis demonstrated that these proviruses are present in the centromere and pericentromere domains of chromosome 21, respectively (Kurdyukov et al. 2001); (3) BLAST analysis of CER:D22Z3 elements to unpublished centromeric sequences revealed sequences >95% similar to CER:D22Z3 elements in several human centromeres; (4) centromeric regions make up part of the 5% of the human genome that has not yet been sequenced and assembled into the human genome draft sequence (Eichler et al. 2004; Zeitlin. 2010), making this a logical place to find previously unreported repeated elements; (5) although BLAST analysis of CER:D22Z3 elements retrieved one single match to a telomeric sequence in chromosome 4 (Supplemental Fig. S3), no other evidence suggests that K111 sequences are present in telomeres; and (6) we confirmed the existence of K111 sequences in centromeres using ChIP assays (Fig. 4).
This study provides evidence in support of the expansion of specific repetitive elements in centromeres by recombination, a topic about which little has previously been known (Warburton et al. 1993; Jaco et al. 2008). Centromeres are the last frontier of eukaryotic genomes, consisting of highly repetitive sequences that are recalcitrant to subcloning, sequencing, and assembly, and therefore constitute the largest missing pieces of the Human Genome Project (∼5%) (Eichler et al. 2004; Nagaki et al. 2004; Zeitlin 2010). Centromeric sequences remain unassembled to this date, in part due to the high concentration of segmental duplications found in those areas (defined as fragments of genomic sequence with high sequence identity [>90% sequence identity in fragments >1 kb in length]). It is not surprising, then, that segments containing K111 variants may fall under this category and therefore would be missed in the current assembly of the human genome. Centromeres have previously been defined as regions of infrequent crossovers, and as such, recombination in the centromere has been assumed to be inefficient (Copenhaver et al. 1999; Yan et al. 2005). Nevertheless, an earlier study revealed evidence for recombination, at least between sister centromeres (Jaco et al. 2008). Other more recent studies in maize inferred by sequence analysis that recombination through gene conversion is common within centromeres and may play a role in determining the distribution of centromeric repeats (Shi et al. 2010). Our observations provide what appear to be the first sequence-based data to suggest that recombination takes place within and among the centromeres of humans. The sites of the actual recombination events that result in K111 expansion may be the K111 sequences themselves, the highly repetitive CER:D22Z3 elements, other repetitive elements outside the CER:D22Z3 repeat, or a combination of these types of recombination events. Whatever the site(s) of the recombination events in these repetitive elements, it is known that tandem repeats undergo extensive growth and shrinking due to unequal recombination events (Warburton et al. 1993). This could explain why K111, which exists as only one peri-telomeric insertion in the chimpanzee, would have spread so effectively within and among chromosomes after its initial jump to the centromere of a human chromosome.
Using the human/rodent chromosomal hybrids, we have identified unique genomic sequence differences in the K111 proviruses present in each centromere. These sequences provide informative nucleotide substitutions distinct enough to assign a K111 provirus to a specific chromosomal centromere. K111 proviruses thus appear to have unique sequences in different chromosomes, and their nucleotide substitutions can perhaps provide new geographic points to help better annotate centromere sequences and understand the biology of human centromeres. In effect, K111 sequences could serve as “barcodes” for specific chromosomal centromeres. Clearly, with broader sequencing of the entire K111 proviral genome in each centromere, we might be able to find even more detailed and informative patterns of nucleotide substitutions, which would serve as better and more distinct barcodes for each individual centromere. These K111 proviruses are therefore islands of endogenous retroviruses in a sea of repetitive elements that might serve as reference points in the future to begin to better order the human centromere.
As HIV infection and Tat modulate the expression of the K111 proviruses, whether or not this process plays a role in the pathogenesis of HIV must be further explored. In any case, it is striking that by studying HIV-1–infected individuals, we have been able to uncover a previously undiscovered, large centromeric lineage of HERVs with the potential to guide future studies of the DNA sequence and function of centromeres. The present study therefore suggests that the human genome can be further annotated by studying nucleic acid sequences found in the blood of patients with specific diseases.
Methods
Real-time qPCR specific for K111
Titers of K111 cellular DNA, plasma RNA, supernatant RNA, and cellular RNA were measured by qPCR or qRT-PCR using a probe that specifically discriminates the K111 env gene from other HERV-K (HML-2) env sequences due to a 6-bp mutation (underlined in the probe sequence K111P). The qPCR was performed according to the method previously described (Contreras-Galindo et al. 2008, 2012) using the primers K111F and K111R and the FAM-labeled probe K111P. For additional information, see Supplemental Material.
Chromatin immunoprecipitation
ChIP assays to assess the association of centromeric proteins and heterochromatic marks with K111 were performed using antibodies to the centromere proteins CENPA and CENPB and the heterochromatic histone mark H3K9me3 or with nonspecific IgG antibodies, while assays to assess the effect of Tat on chromatin were performed with antibodies to the heterochromatic marks H3K9me3 and H4K20me3 (for additional information, see Supplemental Material).
PCR for 5′ and 3′ K111 LTR insertions
The K111-related variants inserted in New and Old World monkeys, humans, and human/rodent chromosomal somatic hybrid DNA were amplified by PCR using the Expand Long Range dNTPack PCR kit (Roche Applied Science). For additional information, see Supplemental Material.
Details regarding study subjects, viral RNA and cellular RNA extraction, RT-PCR of the HERV-K (HML-2) env gene, PCR amplification of K111-related proviruses, HIV-1 infection/Tat stimulation of PBLs and cell lines, sequence and bioinformatics analyses, and statistical analysis are in Supplemental Material.
Data access
All the sequences reported in this article have been deposited in the NCBI Nucleotide database (http://www.ncbi.nlm.nih.gov/nuccore), under accession numbers GU476554–GU476555, and JQ790968–JQ790992.
Acknowledgments
We thank Jen Lewis for manuscript preparation. The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: HIV-1 BAL, ADA, 89.6, NL4.3, and HIV Tat86. We thank Martin Kircher for access to the Neanderthal and Denisovan sequence reads. This work was supported by a generous grant (05-5089) to M.H.K. from the Concerned Parents for AIDS Research (CPFA) and by grants RO1 AI062248 and RO1 CA144043 to D.M.M. from the National Institutes of Health. R.C-G. was supported by a Research Supplement to Promote Diversity in Health-Related Research grant 3R01CA144043-03S1 from the National Institutes of Health. M.J.G-H. was supported by a Rackham Merit Fellowship and by the NIH Ruth L. Kirschstein NRSA Individual Predoctoral Fellowship to Promote Diversity in Health-Related Research grant 1F31CA150523-01. D.D. and M.J.G-H. were supported by the Molecular Mechanisms of Microbial Pathogenesis Training Grant from the University of Michigan (5T32AI007528-13). A.M.C. and D.R. were supported by the Howard Hughes Medical Institute. G.S.O. acknowledges support from NIH grants RM-08-029, P30U54ES017885, U54DA021519, and UL1RR24986. D.M.M. was the recipient of a Burroughs Wellcome Fund Clinical Scientist Award in Translational Research.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.144303.112.
Freely available online through the Genome Research Open Access option.
References
- Agoni L, Golden A, Guha C, Lenz J 2012. Neandertal and Denisovan retroviruses. Curr Biol 22: R437–R438 [DOI] [PubMed] [Google Scholar]
- Bannert N, Kurth R 2004. Retroelements and the human genome: New perspectives on an old relation. Proc Natl Acad Sci 101: 14572–14579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbulescu M, Turner G, Su M, Kim R, Jensen-Seaman MI, Deinard AS, Kidd KK, Lenz J 1999. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr Biol 9: 861–868 [DOI] [PubMed] [Google Scholar]
- Belshaw R, Pereira V, Katzourakis A, Talbot G, Paces J, Burt A, Tristem M 2004. Long-term reinfection of the human genome by endogenous retroviruses. Proc Natl Acad Sci 101: 4894–4899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieda K, Hoffmann A, Boller K 2001. Phenotypic heterogeneity of human endogenous retrovirus particles produced by teratocarcinoma cell lines. J Gen Virol 82: 591–596 [DOI] [PubMed] [Google Scholar]
- Büscher K, Trefzer U, Hofmann M, Sterry W, Kurth R, Denner J 2005. Expression of human endogenous retrovirus K in melanomas and melanoma cell lines. Cancer Res 65: 4172–4180 [DOI] [PubMed] [Google Scholar]
- Carone DM. 2008. Marsupial centromere structure. In The role of retroviruses and RNA in mammalian centromere competency, pp. 39–45. ProQuest, Ann Arbor, MI. [Google Scholar]
- Contreras-Galindo R, Kaplan MH, Markovitz DM, Lorenzo E, Yamamura Y 2006. Detection of HERV-K(HML-2) viral RNA in plasma of HIV type 1-infected individuals. AIDS Res Hum Retroviruses 22: 979–984 [DOI] [PubMed] [Google Scholar]
- Contreras-Galindo R, Kaplan MH, Leissner P, Verjat T, Ferlenghi I, Bagnoli F, Giusti F, Dosik MH, Hayes DF, Gitlin SD, et al. 2008. Human endogenous retrovirus K (HML-2) elements in the plasma of people with lymphoma and breast cancer. J Virol 82: 9329–9336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras-Galindo R, Kaplan MH, Contreras-Galindo AC, Gonzalez-Hernandez MJ, Ferlenghi I, Giusti F, Lorenzo E, Gitlin SD, Dosik MH, Yamamura Y, et al. 2012. Characterization of human endogenous retroviral elements in the blood of HIV-1-infected individuals. J Virol 86: 262–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copenhaver GP, Nickel K, Kuromori T, Benito MI, Kaul S, Lin X, Bevan M, Murphy G, Harris B, Parnell LD, et al. 1999. Genetic definition and sequence analysis of Arabidopsis centromeres. Science 286: 2468–2474 [DOI] [PubMed] [Google Scholar]
- Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, Beare DM, Clamp M, Smink LJ, et al. 1999. The DNA sequence of human chromosome 22. Nature 402: 489–495 [DOI] [PubMed] [Google Scholar]
- Easley R, Van Duyne R, Coley W, Guendel I, Dadgar S, Kehn-Hall K, Kashanchi F 2010. Chromatin dynamics associated with HIV-1 Tat-activated transcription. Biochim Biophys Acta 1799: 275–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE, Clark RA, She X 2004. An assessment of the sequence gaps: Unfinished business in a finished human genome. Nat Rev Genet 5: 345–354 [DOI] [PubMed] [Google Scholar]
- Ferreri GC, Brown JD, Obergfell C, Jue N, Finn CE, O'Neill MJ, O'Neill RJ 2011. Recent amplification of the kangaroo endogenous retrovirus, KERV, limited to the centromere. J Virol 85: 4761–4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Hernandez MJ, Swanson MD, Contreras-Galindo R, Cookinham S, King SR, Noel RJ Jr, Kaplan MH, Markovitz DM 2012. Expression of Human Endogenous Retrovirus Type-K (HML-2) is activated by the Tat protein of HIV-1. J Virol 86: 7790–7805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH, et al. 2010. A draft sequence of the Neandertal genome. Science 328: 710–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JF, Coffin JM 2004. Human endogenous retrovirus K solo-LTR formation and insertional polymorphisms: Implications for human and viral evolution. Proc Natl Acad Sci 101: 1668–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JF, Coffin JM 2005. Human endogenous retroviral elements as indicators of ectopic recombination events in the primate genome. Genetics 171: 1183–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaco I, Canela A, Vera E, Blasco MA 2008. Centromere mitotic recombination in mammalian cells. J Cell Biol 181: 885–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jern P, Coffin JM 2008. Effects of retroviruses on host genome function. Annu Rev Genet 42: 709–732 [DOI] [PubMed] [Google Scholar]
- Johnston JB, Silva C, Holden J, Warren KG, Clark AW, Power C 2001. Monocyte activation and differentiation augment human endogenous retrovirus expression: Implications for inflammatory brain diseases. Ann Neurol 50: 434–442 [DOI] [PubMed] [Google Scholar]
- Kurdyukov SG, Lebedev YB, Artamonova II, Gorodentseva TN, Batrak AV, Mamedov IZ, Azhikina TL, Legchilina SP, Efimenko IG, Gardiner K, et al. 2001. Full-sized HERV-K (HML-2) human endogenous retroviral LTR sequences on human chromosome 21: Map locations and evolutionary history. Gene 273: 51–61 [DOI] [PubMed] [Google Scholar]
- Löwer R, Boller K, Hasenmaier B, Korbmacher C, Müller-Lantzsch N, Löwer J, Kurth R 1993. Identification of human endogenous retroviruses with complex mRNA expression and particle formation. Proc Natl Acad Sci 90: 4480–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H, Nakano M, Ohzeki J 2004. The role of CENP-B and α-satellite DNA: de novo assembly and epigenetic maintenance of human centromeres. Chromosome Res 12: 543–556 [DOI] [PubMed] [Google Scholar]
- Metzdorf R, Göttert E, Blin NA 1988. A novel centromeric repetitive DNA from human chromosome 22. Chromosoma 97: 154–158 [DOI] [PubMed] [Google Scholar]
- Meyer M, Kircher M, Gansauge MT, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prüfer K, de Filippo C, et al. 2012. A high-coverage genome sequence from an archaic Denisovan individual. Science 338: 222–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosch K, Franz H, Soeroes S, Singh PB, Fischle W 2011. HP1 recruits activity-dependent neuroprotective protein to H3K9me3 marked pericentromeric heterochromatin for silencing of major satellite repeats. PLoS ONE 6: e15894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müllenbach R, Lutz S, Holzmann K, Dooley S, Blin N 1992. A non-alphoid repetitive DNA sequence from human chromosome 21. Hum Genet 89: 519–523 [DOI] [PubMed] [Google Scholar]
- Muster T, Waltenberger A, Grassauer A, Hirschl S, Caucig P, Romirer I, Födinger D, Seppele H, Schanab O, Magin-Lachmann C, et al. 2003. An endogenous retrovirus derived from human melanoma cells. Cancer Res 63: 8735–8741 [PubMed] [Google Scholar]
- Nagaki K, Cheng Z, Ouyang S, Talbert PB, Kim M, Jones KM, Henikoff S, Buell CR, Jiang J 2004. Sequencing of a rice centromere uncovers active genes. Nat Genet 36: 138–145 [DOI] [PubMed] [Google Scholar]
- Nakano M, Cardinale S, Noskov VN, Gassmann R, Vagnarelli P, Kandels-Lewis S, Larionov V, Earnshaw WC, Masumoto H 2008. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev Cell 14: 507–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PN, Carnegie PR, Martin J, Davari Ejtehadi H, Hooley P, Roden D, Rowland-Jones S, Warren P, Astley J, Murray PG 2003. Demystified: Human endogenous retroviruses. Mol Pathol 56: 11–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan JP, Coop G, Kudaravalli S, Smith D, Krause J, Alessi J, Chen F, Platt D, Pääbo S, Pritchard JK, et al. 2006. Sequencing and analysis of Neanderthal genomic DNA. Science 314: 1113–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okahara G, Matsubara S, Oda T, Sugimoto J, Jinno Y, Kanaya F 2004. Expression analyses of human endogenous retroviruses (HERVs): Tissue-specific and developmental stage-dependent expression of HERVs. Genomics 84: 982–990 [DOI] [PubMed] [Google Scholar]
- Paces J, Pavlícek A, Zika R, Kapitonov VV, Jurka J, Paces V 2004. HERVd: The Human Endogenous RetroViruses Database: Update. Nucleic Acids Res 32: D50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Richter DJ, Gnerre S, Lander ES, Reich D 2006. Genetic evidence for complex speciation of humans and chimpanzees. Nature 441: 1103–1108 [DOI] [PubMed] [Google Scholar]
- Reich D, Green RE, Kircher M, Krause J, Patterson N, Durand EY, Viola B, Briggs AW, Stenzel U, Johnson PL, et al. 2010. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468: 1053–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reus K, Mayer J, Sauter M, Scherer D, Müller-Lantzsch N, Meese E 2001. Genomic organization of the human endogenous retrovirus HERV-K(HML-2.HOM) (ERVK6) on chromosome 7. Genomics 72: 314–320 [DOI] [PubMed] [Google Scholar]
- Ruda VM, Akopov SB, Trubetskoy DO, Manuylov NL, Vetchinova AS, Zavalova LL, Nikolaev LG, Sverdlov ED 2004. Tissue specificity of enhancer and promoter activities of a HERV-K(HML-2) LTR. Virus Res 104: 11–16 [DOI] [PubMed] [Google Scholar]
- Seifarth W, Baust C, Murr A, Skladny H, Krieg-Schneider F, Blusch J, Werner T, Hehlmann R, Leib-Mösch C 1998. Proviral structure, chromosomal location, and expression of HERV-K-T47D, a novel human endogenous retrovirus derived from T47D particles. J Virol 72: 8384–8391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wolf SE, Burke JM, Presting GG, Ross-Ibarra J, Dawe RK 2010. Widespread gene conversion in centromere cores. PLoS Biol 8: e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian RP, Wildschutte JH, Russo C, Coffin JM 2011. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology 8: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto J, Matsuura N, Kinjo Y, Takasu N, Oda T, Jinno Y 2001. Transcriptionally active HERV-K genes: Identification, isolation, and chromosomal mapping. Genomics 72: 137–144 [DOI] [PubMed] [Google Scholar]
- Tönjes RR, Löwer R, Boller K, Denner J, Hasenmaier B, Kirsch H, König H, Korbmacher C, Limbach C, Lugert R, et al. 1996. HERV-K: The biologically most active human endogenous retrovirus family. J Acquir Immune Defic Syndr Hum Retrovirol 1: S261–S267 [DOI] [PubMed] [Google Scholar]
- Turner G, Barbulescu M, Su M, Jensen-Seaman MI, Kidd KK, Lenz J 2001. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr Biol 11: 1531–1535 [DOI] [PubMed] [Google Scholar]
- Verdaasdonk JS, Bloom K 2011. Centromeres: Unique chromatin structures that drive chromosome segregation. Nat Rev Mol Cell Biol 12: 320–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang-Johanning F, Frost AR, Johanning GL, Khazaeli MB, LoBuglio AF, Shaw DR, Strong TV 2001. Expression of human endogenous retrovirus k envelope transcripts in human breast cancer. Clin Cancer Res 7: 1553–1560 [PubMed] [Google Scholar]
- Warburton PE, Waye JS, Willard HF 1993. Nonrandom localization of recombination events in human alpha satellite repeat unit variants: Implications for higher-order structural characteristics within centromeric heterochromatin. Mol Cell Biol 13: 6520–6529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Jin W, Nagaki K, Tian S, Ouyang S, Buell CR, Talbert PB, Henikoff S, Jiang J 2005. Transcription and histone modifications in the recombination-free region spanning a rice centromere. Plant Cell 17: 3227–3238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi JM, Kim HM, Kim HS 2001. Molecular cloning and phylogenetic analysis of the human endogenous retrovirus HERV-K long terminal repeat elements in various cancer cells. Mol Cells 12: 137–141 [PubMed] [Google Scholar]
- Zeitlin SG 2010. Centromeres: The wild west of the post-genomic age. Epigenetics 5: 34–40 [DOI] [PubMed] [Google Scholar]