Abstract
RNA affinity tags (aptamers) have emerged as useful tools for the isolation of RNAs and ribonucleoprotein complexes from cell extracts. The streptavidin binding RNA aptamer binds with high affinity and is quickly and cleanly eluted with biotin under mild conditions that retain intact complexes. We describe the use of the streptavidin binding aptamer as a tool for purification and discuss strategies towards the design and production of tagged RNAs with a focus on structured target RNAs. The aptamer site can be further exploited as a unique region for the hybridization of oligonucleotide probes and localization by fluorescent in situ hybridization (FISH). The aptamer insertion will allow the localization of a population of RNA species (such as mutants) to be viewed specifically, while in the presence of the wild type RNA. We describe the production of labeled oligonucleotide probes and the preparation of yeast cells for the localization of RNAs by FISH.
Keywords: RNA, RNP isolation, Aptamer, FISH, Ribonucleoprotein, Streptavidin
1. Introduction
Aptamers are nucleic acid sequences that have been selected from a large pool of random sequences to exhibit a particular desired property, usually tight binding to a specific target molecule. The desired properties of each aptamer are obtained through multiple rounds of selection by utilizing the systematic evolution of ligands by exponential enrichment (SELEX) method (1, 2). Both DNA and RNA aptamers have been isolated with many uses including a wide range of therapeutics, biosensors, chiral reagents, and affinity tags (3).
The more recently developed RNA affinity tags complement the more widely used protein based affinity tags (4) and these aptamers have become useful research tools allowing the specific purification of RNA targets and their associated complexes from cellular extracts.
1.1. Affinity Purification of RNAs and RNA–Protein Complexes
A number of RNA aptamers have been used for the purification of RNAs and RNA–protein complexes from cellular extracts, these include aptamers that bind to immobilized streptavidin, tobramycin, streptomycin, or sephadex (5–8). We are most experienced with the streptavidin binding aptamer (S1 aptamer) and focus on this example throughout this chapter. The streptavidin binding aptamer has many desirable properties; it binds to immobilized streptavidin with high affinity (Kd ~ 70 nM), the binding is stable to high salt conditions (400 mM NaCl), and the aptamer is quickly and cleanly eluted from the affinity resin by the binding of the small molecule D-biotin to streptavidin. Initial experiments utilized the full SELEX derived sequence to purify RNase P from yeast cells (5) (see Figs. 1 and 2). However, only the minimal streptavidin binding aptamer is needed for full binding activity and the extraneous sequence can be completely omitted (Fig. 1) (9). The streptavidin binding aptamer has been used to purify the multi subunit ribonucleoprotein RNase P from both yeast (5) and human cells (10), mutant ribosomes from bacteria (11), and also telomerase from yeast cells (12).
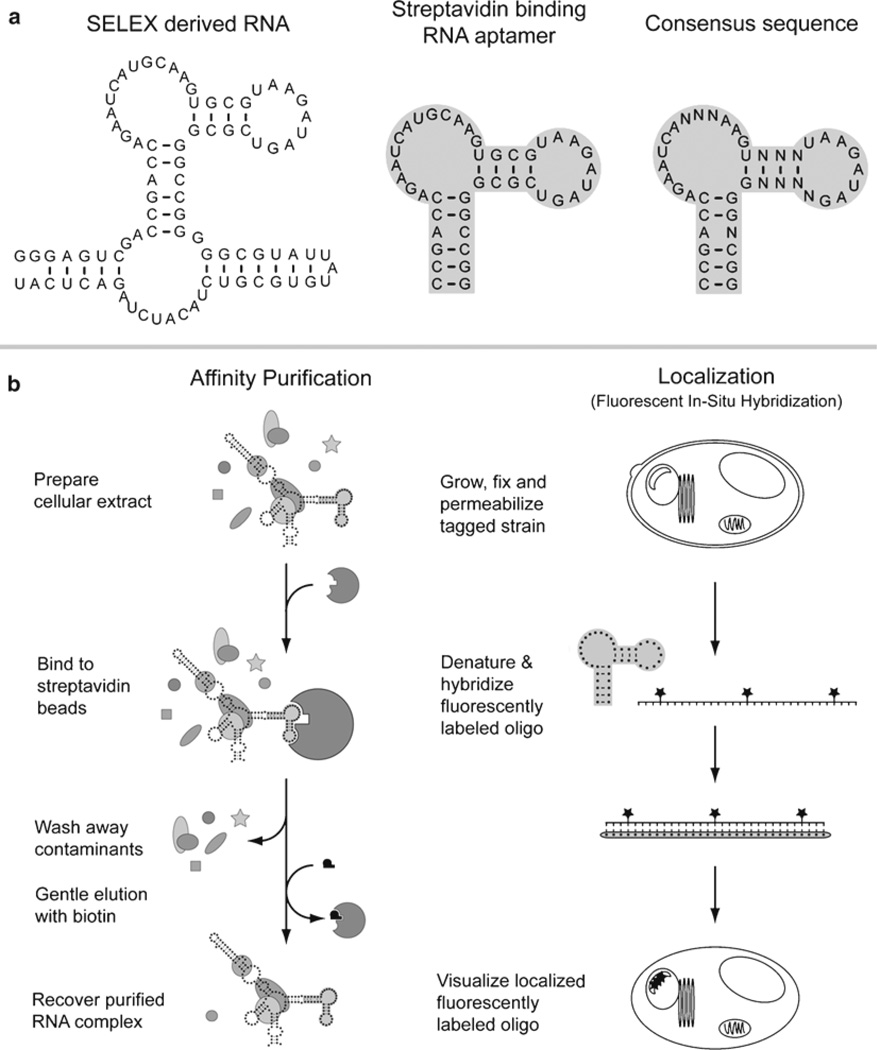
Fig. 1.
The streptavidin binding RNA aptamer. (a) The original SELEX derived streptavidin binding sequence is shown along with the minimal streptavidin binding aptamer (shaded). The aptamer was derived from a population of species and the consensus sequence from multiple isolated clones is shown. (b) The streptavidin binding aptamer has been used for affinity purifications of ribonucleoprotein complexes (left) and the sequence has also been targeted as a unique hybridization region for fluorescent in situ hybridizations to localize ribonucleoprotein complexes (right).
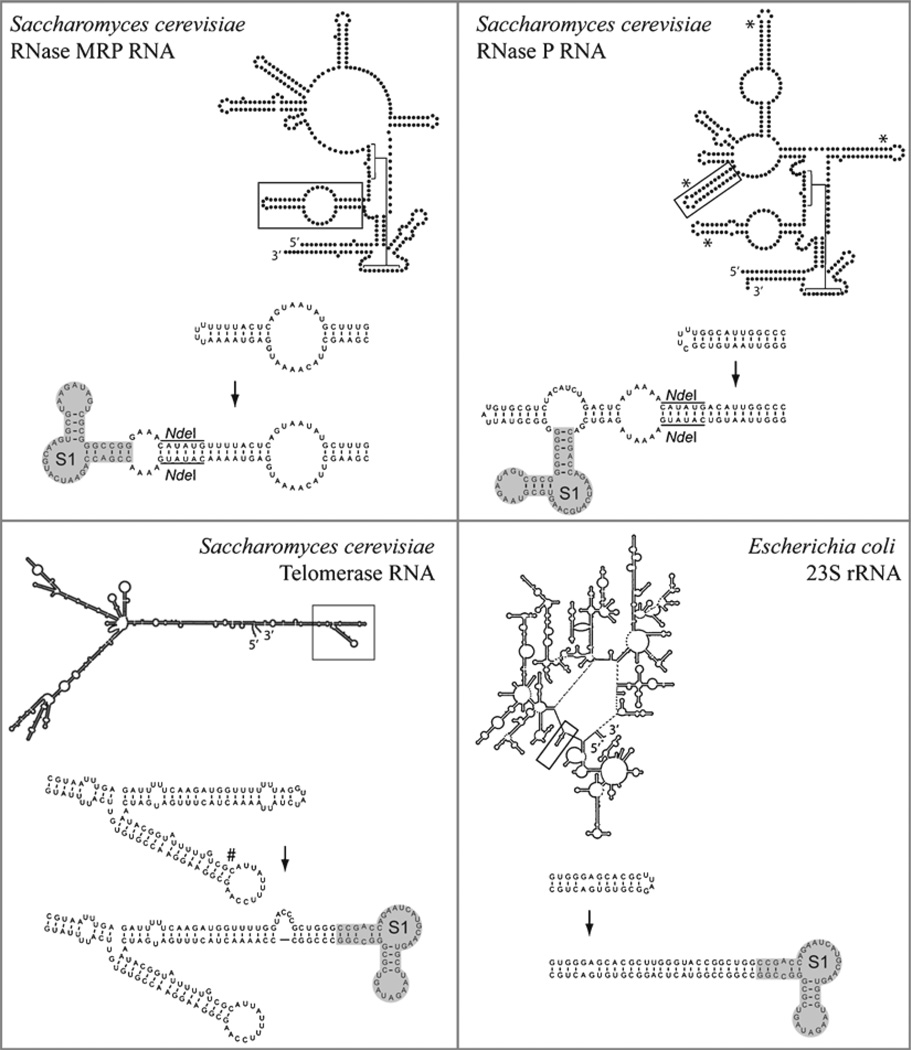
Fig. 2.
Insertion of the streptavidin binding aptamer into ribonucleoprotein complexes. (Yeast RNase P and RNase MRP) Aptamer insertions into the yeast RNase MRP and RNase P holoenzymes have been used to purify these ribonucleoprotein complexes. The yeast RNase P has been extensively tagged, initially with the full SELEX derived sequence (shown) and subsequently with the minimal streptavidin binding aptamer sequence. The yeast RNase P can be tagged in four different locations that are indicated (*). In each case, these positions are known to be phylogenetically variable and solvent exposed and the aptamer insertion does not alter the growth or pre-tRNA processing significantly. (Yeast telomerase) The yeast telomerase was tagged at two positions, the successful aptamer insertion is shown and the unsuccessful position is also indicated (#). The purified telomerase was subsequently shown to be active in a telomerase assay. (Bacterial ribosomes) The streptavidin aptamer was used to specifically isolate ribosomal subunits containing a known lethal mutation. Coexpression of the lethal mutation alongside the wildtype sequence was necessary to sustain bacterial growth.
It is important to note that in each of these published studies the aptamer was placed internally by insertion into a solvent-accessible stem loop within the structured target RNA (see Fig. 2). Early attempts to use the streptavidin binding aptamer as a flanking tag (i.e., placed either 5′ or 3′ to the target RNA) were not successful (unpublished communications). These studies targeted unstructured RNAs (mRNAs and pre-mRNAs) and the tagged constructs were found to be functional in vitro, but failed to perform in vivo, in a manner consistent with the aptamer becoming unwound or otherwise obscured by protein binding and RNP formation in the cellular environment. In contrast, the tobramycin aptamer has been used as a flanking tag to successfully purify pre-mRNA complexes (13). Comparison of the two approaches suggests that the use of a G-C rich stabilizing stem at the base of the tobramycin aptamer was instrumental for success. In this chapter, we will only describe the use of aptamers tags placed internally within a structured target RNA. Although these constructs are more difficult to design and test, we have had greater success when employing the aptamer tag internally.
1.2. Localization of RNAs and RNA–Protein Complexes
Fluorescence in situ hybridization (FISH) is an efficient mechanism for localizing RNAs and their associated complexes (14). FISH uses a fluorescently labeled oligonucleotide probe complementary to the sequence of interest. Although any RNA can be visualized with an appropriate antisense probe, there can be distinct advantages to exploiting an existing aptamer insertion into a target RNA as a unique target region for FISH. Cells with the aptamer tagged complex are first fixed with formaldehyde and permeabilized on a microscope slide. The probe is then hybridized to the aptamer, and the complex is directly detectable using fluorescent microscopy (see Note 1). The localization can be further extended by the simultaneous hybridization of multiple fluorescent probes. This multicolor FISH permits one to see the location of an aptamer tagged RNA relative to that of other labeled features, such as the nucleus or nucleolus.
The aptamer sequence can serve as a unique site for the identification of only a single population of RNA species. When a functional tagged RNA is coexpressed alongside the wild type RNA it becomes possible to further manipulate the tagged RNA and examine the effects of various mutations on the localization of the tagged RNA. In this situation, the strain is kept viable by the presence of the wild type RNA and the localization of only the mutated RNA population can be detected using the unique tag. Similar approaches have been used to examine nucleo–cytoplasmic shuttling of snRNAs in heterokaryons by detecting the unique sequence region of a functional hybrid U6 snRNA in the presence of the wild type U6 snRNA (15). We have used the streptavidin binding aptamer sequence to successfully localize the RNase P RNA and have also observed mis-localization of a number of RNA mutants (Fig. 3) (16).
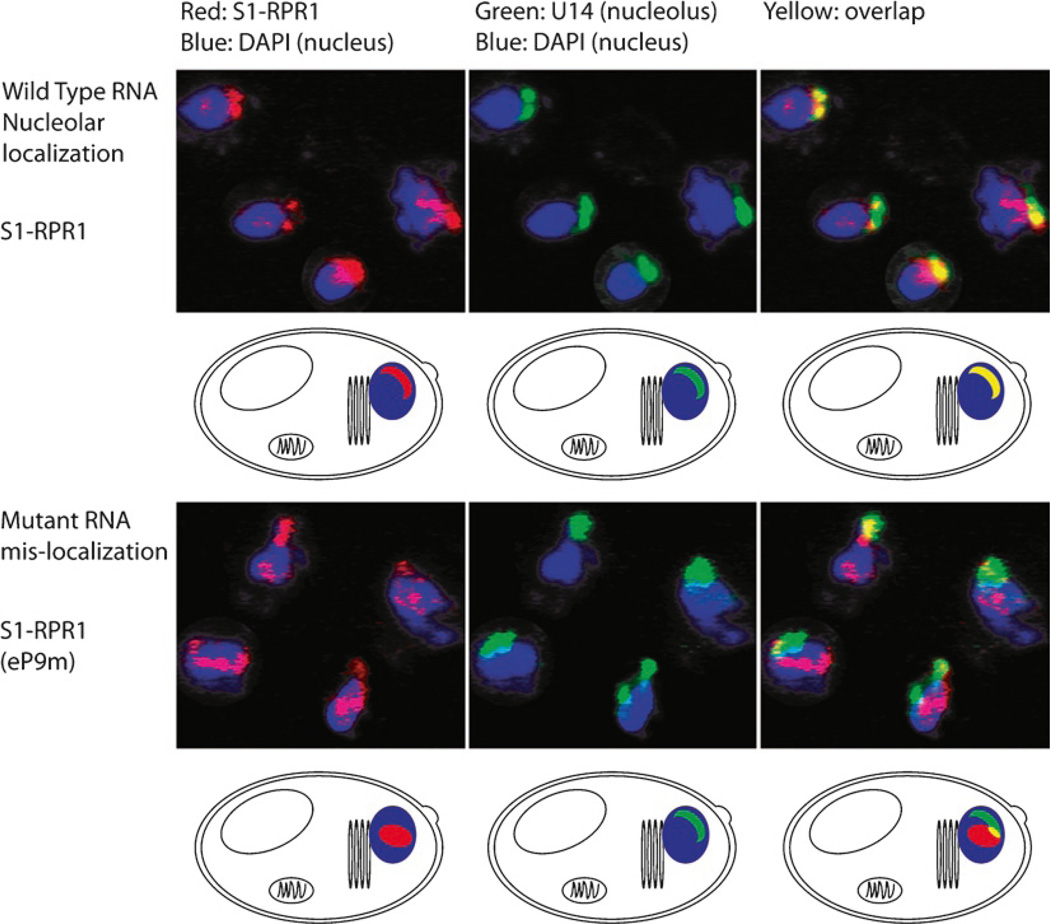
Fig. 3.
Localization of yeast RNase P. The RNA subunit of the RNase P ribonucleoprotein (RPR1) is localized using a fluorophore labeled antisense oligo (red) targeting the streptavidin binding aptamer insertion into the RNA subunit (S1-RPR1). The U14 RNA is also visualized using an antisense oligo (green) and serves as a nucleolar marker. The overlap (yellow) shows that the wild type RNA is predominantly nucleolar. The complex becomes mislocalized to the nucleoplasm (blue) when an RNA mutation within the eP9 stem loop is introduced into the RNA subunit [S1-RNA (eP9m)]. Adapted from ref. 16 with permission of Cold Spring Harbor Press.
There are also other practical reasons for utilizing an existing aptamer insertion as this helps to ensure that the target region is solvent exposed, aiding effective strand invasion and hybridization of the fluorescently tagged oligonucleotide to the target RNA. This approach also allows the same fluorescent oligonucleotide probe to be used for the examination of different aptamer tagged target RNA species. Developing unique antisense probes for each target RNA has drawbacks since each fluorescent probe is costly to produce and must be tested to ensure effective strand invasion and hybridization to the target.
2. Materials
2.1. Design of Tagged RNAs
Software allowing secondary structure prediction. Downloadable programs can be found for both the PC (RNAstructure: http://rna.urmc.rochester.edu/rnastructure.html) and Mac (Mulfold: http://iubio.bio.indiana.edu/soft/molbio/mac/). A second program for the PC is (RNAdraw: http://www.rnadraw.com) and links to web-based RNA folding servers can also be found at this address.
2.2. Construction of the Tagged RNA Sequence
2.2.1. Two Step PCR
Genomic DNA or plasmid bearing the target RNA sequence.
Oligonucleotides (Integrated DNA Technologies).
DNA Polymerase, supplied with commercial buffer (Pfu – Stratagene) (Taq – Roche).
Agarose gel electrophoresis equipment.
Gel purification mini-prep kit (Promega).
Restriction enzymes, supplied with commercial buffer (New England Biolabs).
T4 DNA ligase, supplied with commercial buffer (New England Biolabs).
Competent E. coli cells DH5α or XL1-Blue.
Access to an automated DNA sequencing facility.
2.2.2. Site-Directed Mutagenesis
Suitable vector bearing the target RNA sequence.
QuikChange mutagenesis Kit (Stratagene).
Oligonucleotides (Integrated DNA Technologies).
Restriction enzymes, supplied with commercial buffer (New England Biolabs).
T4 DNA ligase, supplied with commercial buffer (New England Biolabs).
Competent E. coli cells DH5α or XL1-Blue.
Access to an automated DNA sequencing facility.
2.3. Preparation of Yeast Extracts
Yeast strain carrying the appropriate tagged RNA.
Growth media (YPD) 10 g/l Bacto Yeast extract, 20 g/l Bacto peptone, and 20 g/l Dextrose. Combine and autoclave.
1× Lysis Buffer: 50 mM Hepes at pH 7.4, 10 mM MgCl2, 100 mM NaCl, 1 mM DTT, 0.1% Triton-X100, 10% glycerol, and Complete® protease inhibitors (Roche).
Acid washed glass beads 425–600 µm.
Protein content assay, Micro Bicinchoninic acid assay (Pierce).
2.4. Affinity Purification
Streptavidin agarose (Sigma #S1638).
Avidin from egg white (Sigma).
1× Lysis buffer, as above but without Complete®.
Ultra-Free MC centrifugal filter device (Millipore).
D-biotin (Sigma).
2.5. Localization of Tagged RNAs via Fluorescent In Situ Hybridization
2.5.1. Preparation of Fluorescently Labeled Oligonucleotide Probes
Oligonucleotides, FPLC purified bearing appropriate amino modifications for labeling (Integrated DNA Technologies) (see Note 2).
Fluorescent Dyes, purchased as an N-hydroxysuccinimide (NHS) activated ester (Invitrogen or Amersham).
Anhydrous dimethyl sulfoxide (DMSO).
Labeling buffer: 0.1 M sodium tetraborate at pH 8.5.
Micro Bio-Spin 30 columns (Bio-Rad).
2.5.2. Fixation and Permeabilization of Yeast Cells
40% Paraformaldehyde.
Sorbitol buffer: 1.2 M sorbitol and 0.1 M potassium phosphate at pH 7.5.
Spheroplast buffer: 1.2 M Sorbitol, 0.1 M Potassium Phosphate at pH 7.5, 1× Vanadyl Ribonucleoside Inhibitor (New England Biolabs), 28 mM β-mercaptoethanol, and 0.06 mg/ml PMSF at pH 7.5, store frozen.
Zymolyase 20T (Seikagaku Biobusiness Corp, via amsbio.com).
Microscope slides: eight-well not poly-lysine coated (MP Biomedicals LLC).
2.5.3. Hybridization of Fluorescent Oligonucleotide Probe
20× SSC: 300 mM NaCl and 30 mM Citrate at pH 7.0.
10× SSCP: 1.5 M NaCl, 0.15 M NaCitrate, and 0.2 M NaHPO4 at pH 6.0.
Denaturating solution: 70% Formamide, 2× SSC. Make fresh for each hybridization.
Hybridization mix: 50% Formamide, 2× SSCP, 10% Dextran Sulfate, and 0.4 mg/ml ssDNA. Prewarm an aliquot of 50% (w/v) Dextran Sulfate for the hybridization mix to 70–72°C.
2.5.4. DAPI Staining and Slide Mounting
Post-hybridization wash: 50% Formamide, 2× SSC.
DAPI: 4′,6-diamidino-2-phenylindole (Roche).
Prolong Antifade Kit: Mounting Solution Components A & B (Molecular Probes/Invitrogen).
2.5.5. Visualization of Aptamer Tagged RNA
Fluorescent microscope with deconvolution technology or a confocal microscope. We use a Nikon Microscope with deconvolution.
CCD (charge couple device) cameras. We use a cooled CCD camera which allows the detection of low signals.
Imaging software: We use Esee (ISIS) for microscope imaging to take down background and superimpose images from different channels. We also use a combination of Isee (ISIS) and Photoshop (Adobe) for manipulation of the images.
3. Methods
3.1. Guidelines for Designing Aptamer Tagged RNAs
When introducing the aptamer sequence into a target RNA it is vital that both the aptamer and the target RNA maintain their correct structures. The S1 aptamer has been most successfully applied when used internally, within structured target RNAs (5, 10–12) and examples are shown in Fig. 2. In each of these cases, the target RNA had a predicted secondary structure that was used to aid the design of the successful aptamer tagged RNA.
3.1.1. Identifying Sites for Aptamer Insertion into a Target RNA
It is essential to start with some knowledge of the structure of the target RNA. The S1 aptamer has been used most effectively when it has been placed at the end of a solvent exposed stem structure. It is equally important that the stem-loop does not have a conserved function so that it can be freely manipulated without affecting the function of the target RNA. In most cases, the three dimensional structure is not available, however a secondary structure can be predicted and in many cases this data is available for a particular target RNA across several species (17–19). A phylogenetic analysis of RNA structure can be extremely useful to identify stem structures that are variable between species. Variations in the length of a stem and, most importantly, in the sequence of the stem-loop are often good indicators that the stem structure does not have an essential conserved function within the target RNA. These phylogenetically variable stems are more likely to tolerate manipulations and are therefore good targets for the insertion of the aptamer sequence. Although the identification of variable stems and stem-loops provides a good starting point these stems also need to be solvent exposed in order to provide accessibility of the aptamer tag to the affinity resin and allow purification. A complementary approach is to use chemical or enzymatic footprinting data to identify RNA structures that are accessible to these probes and therefore exposed to solvent. The combination of both phylogenetic and footprinting analyses will allow a solvent accessible and variable stem-loop structure(s) to be identified that have a greater likelihood of being successfully tagged.
In the case of the yeast RNase P RNA, both phylogenetic studies (20) and footprinting data (21) are available and there are five solvent exposed stem-loops that also do not have significant sequence conservation (16). To date, we have successfully introduced the S1 aptamer at each of four different positions within the RNase P RNA (Fig. 2). In each case, the aptamer insertion allows the purification of the enzyme and a thorough analysis of the growth phenotype and pre-tRNA processing profiles establishes that these aptamer insertions have no detectible effect on enzyme function ((5, 16) and unpublished data, S.C. Walker).
When phylogenetic or footprinting data is not available the best approach is to use trial and error in combination with best judgment based on what is known about the particular target RNA under study. In all cases it is still possible that the insertion of the aptamer sequence might have an adverse affect upon the folding of the target RNA. The potential effects of the aptamer sequence on the folding of the target RNA can be tested by folding the proposed sequence in silico using secondary structure prediction software. If the target RNA is large (>200 nt) it can often be more reliable to truncate the sequence and predict the folding of only the aptamer tagged stem and the surrounding sequence (approx. ±100 nt).
Ultimately, the only true test of correct aptamer folding and function within a target RNA is to produce and test the tagged target RNA in vivo. We strongly recommend the creation of a minimum of two constructs tagged at different positions. The tagged RNAs should be designed using all available knowledge (secondary structure, phylogenetic analysis, footprinting, or three dimensional structure) to guide the choice of stem structures that will be tagged.
3.1.2. Designing the Hybrid RNA
Once appropriate sites have been identified within the target RNA it is useful to draw out the final designs for the stem structure bearing the aptamer sequence. Different linker designs have been successfully used to fuse the aptamer to the target RNA including both rigid and flexible linkers of various lengths, examples are shown in Fig. 2. A long and flexible linker may provide better access to the affinity resin leading to a successful purification. However, it is also likely that a longer linker will have a greater potential to interfere with the correct folding of the target RNA and large flexible regions can be very sensitive sites for nuclease digestion. We have used both rigid and flexible linker designs and have not observed any significant differences when these have been used at the same position within the yeast RNase P RNA (S. C. Walker, unpublished data). However, since each target RNA will behave differently it is important to recognize that in certain contexts, flexible linkers may be sensitive to nucleases or that rigid linkers may not allow for a productive interaction with the affinity resin. In such cases, the investigator can respond by redesigning the linker or repositioning the aptamer tag.
3.2. Constructing the Hybrid Aptamer Target RNA Sequence
We describe two different approaches to facilitate the introduction of the aptamer sequence into an appropriate position within the target RNA. In each case we assume that the target RNA is being expressed from a plasmid bearing appropriate promoter and terminator signals.
3.2.1. Construction of the Aptamer Tagged RNA Sequence Using Two-Step PCR
After the aptamer insertion has been designed it is useful to split the sequence into upstream and downstream fragments (Fig. 4). Amplification of the downstream fragment will require two oligonucleotides (Primer 1 and Primer 2). The first oligonucleotide (Primer1) must have sufficient complementarity to the 5′ end of the fragment sequence (20–25 nt) and the aptamer sequence, which does not anneal to the template, is added to the 5′ end of this oligonucleotide. The second oligonucleotide (Primer 2) must have sufficient complementarity to the 3′ end of the fragment sequence (20–25 nt) and a suitable restriction site can be added at the 5′ end of the oligonucleotide to facilitate cloning.
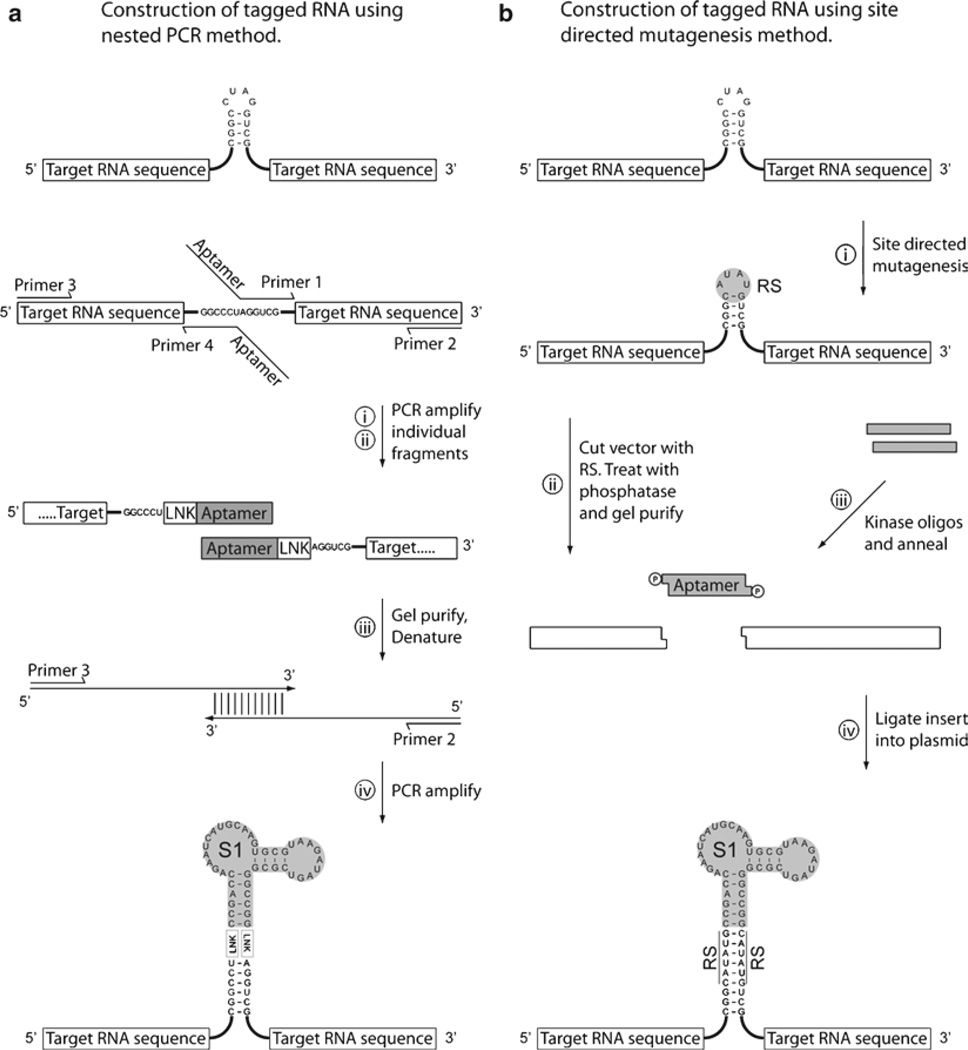
Fig. 4.
Strategies for the construction of aptamer-tagged RNA sequences. (a) The insertion of the aptamer at the required position using a PCR based strategy. Individual PCR fragments are produced, (i) and (ii), where the aptamer sequence is introduced at the desired location by incorporation into the 5′ regions of the primers. The two aptamer-tagged sequence fragments are isolated and used in trace amounts as template for a further round of PCR. The outlying primers (2 and 3) can be used to amplify the entire region and produce the final product bearing an accurately inserted aptamer within the target RNA sequence. (b) The insertion of an aptamer at a required position using a cloning based strategy. Firstly, sitedirected mutagenesis is used to introduce a suitable restriction site into the target RNA sequence (plasmid borne). The restriction site is used to facilitate the introduction of a synthetic insert bearing the required aptamer sequence. The presence of a correctly orientated insert can be performed by PCR screening colonies prior to sequencing.
To produce the upstream fragment a similar method is used where the aptamer is added at the 3′ end of the fragment (Primer 4) and the restriction site, or other required sequence (see Note 3), at the 5′ end of the fragment (Primer 3). The two fragments are then used as template in a third PCR where they are joined together via the introduced aptamer sequence and amplified by the outlying primers (Primer 2 and Primer 3) as outlined below.
The downstream fragment (Fig. 4a (i)) is produced by PCR using primers 1 and 2 using a suitable template DNA encoding the target RNA sequence.
The upstream fragment (Fig. 4a (ii)) is produced by PCR using primers 3 and 4 using a suitable template DNA encoding the target RNA sequence.
The PCR inserts are checked on an agarose gel next to size markers and gel-purified.
The two purified PCR fragments are mixed in equimolar amounts and used as template (1–5 ng total) in PCR with primers 2 and 3 to amplify the entire fragment bearing the tagged RNA sequence and introduced 5′ and 3′ restriction sites. The first few rounds of the PCR create the full length gene fragment due to the overlapping half gene fragments which will be extended by the DNA polymerase.
The correct size of the PCR fragment is checked on an agarose gel next to size markers and the DNA is gel isolated.
The fragment is digested with the appropriate restriction enzymes at the termini and ligated into a suitable plasmid at the correct restriction sites.
The ligation reaction is transformed in to competent bacterial host cells (e.g., E. coli DH5α or XL1-Blue cells) and plated onto selective media.
Plasmid is isolated from individual colonies and the PCR insert is verified by appropriate restriction digests and DNA sequencing.
3.2.2. Insertion of the Aptamer Sequence via a User Created Restriction Site
A second cloning strategy can provide some advantages in certain situations. The target stem-loop structure is mutated to a single hexameric restriction site and the aptamer sequence is purchased as two complementary oligonucleotides and inserted into this restriction site. After insertion into the target RNA, the introduced aptamer sequence is flanked by two complementary copies of the initial restriction site. The overall cloning strategy is represented in Fig. 4b. This sequential approach allows the effects of the mutated stem-loop upon target RNA function to be tested in vivo and is a good indicator of the likely success of the aptamer insertion at that position. A second advantage is that the resulting restriction site(s) can be used to facilitate further manipulations of the target RNA at this site. When using a single restriction site the direction of the insert must be screened by PCR or by sequencing. In some cases the sequential introduction of two restrictions sites, to facilitate the insertion of the aptamer sequence, may be necessary.
For the specific introduction of restriction sites into a particular sequence, we routinely use the Quikchange® kit (Stratagene™). This kit uses two user designed oligonucleotides bearing the mutated sequence to generate two mutant strands from a plasmid template. The parental (methylated) DNA is digested, using DpnI, and the resulting annealed double stranded nicked cDNA molecules are transformed into E. coli where the nicked cDNA is repaired to generate a plasmid bearing the required mutations. For more exact details of the technique, including oligonucleotide design, we refer the reader to the product manual.
Restriction site(s) are introduced into the plasmid sequence by site-directed mutagenesis using the Quikchange® kit (Stratagene™).
The plasmid is cut with the appropriate restriction enzyme(s). After digestion, the cut vector is treated with alkaline phosphatase to remove terminal phosphates and prevent re-ligation to itself. The DNA is then purified from an agarose gel in preparation for ligation with the insert.
The aptamer sequence including any linker is purchased as two individual DNA oligonucleotides bearing the correct overhangs for ligation. In order to facilitate ligation these oligonucleotides are individually phosphorylated with polynucleotide kinase (300 pmol in a 50 µl volume). The enzyme is removed by phenol/chloroform extraction and the two oligonucleotides are annealed in equal concentration, by heating to 65°C and cooling on ice, to create a double stranded insert.
The insert is ligated into the vector at the restriction site.
The ligation reaction is transformed in to competent bacterial (E. coli DH5α or XL1-Blue) cells and plated onto selective media.
Plasmid is isolated from individual colonies and the presence and correct orientation of the inserted aptamer is verified by appropriate restriction digests and automated DNA sequencing.
3.3. Preparation of Tagged Yeast Strains
Once a plasmid has been constructed that will allow the expression of the aptamer tagged target RNA, this can be used to prepare a yeast strain for study. In the yeast system it is possible to use a knockout strain such that the only copy of the target RNA in the final yeast strain is the tagged RNA on the introduced plasmid (see Note 4). In the resulting yeast strains, the aptamer tagged RNA is the only source of the RNA. When the target RNA is essential, the viability of these yeast strains is the direct indicator that the particular aptamer insertion is tolerated and does not interfere with the essential function of the target RNA. When the target RNA is not essential or when co-expression is the only option, then efforts to assess the integrity of the isolated complexes through an appropriate activity assay are required. An example is the successful processing of pre-tRNA substrates in vitro that was shown by the human RNase P holoenzyme, isolated using the streptavidin aptamer (10). In contrast, co-transcription of the tagged RNA alongside the chromosomal wild type sequence has been exploited in order to specifically isolate known lethal mutants of the 23S rRNA sequence for further study (11).
3.3.1. Preparation of Yeast Extracts for Binding
The particular method used to generate crude extracts will vary depending upon the system in which the tagged RNA is being expressed. In general, any protocol for the preparation of crude extracts should focus upon firstly maintaining the temperature at 4°C throughout and secondly producing the extract and binding it to beads as quickly as possible to minimize ribonuclease (and protease) degradation of the RNP complexes. For the preparation of extracts from yeast cultures we use the following protocol.
A single colony of the appropriate yeast strain is grown to saturation in YPD media at 30°C and used to inoculate 1 l of YPD media.
The culture is grown at 30°C until an OD600 of around1–2 and the cells are harvested by centrifugation at 4,000×g for 15 min at 4°C.
The pellet is washed and resuspended in 10 pellet volumes of ice-cold sterile water, then re-pelleted by centrifugation at 4,000 × g for 5 min at 4°C. This washing procedure is carried out three to four times.
The pellet is resuspended in 1 ml of lysis buffer per 2 g of wet cell paste, containing Complete® protease inhibitors (Roche).
-
The cells are lysed by vortexing with of 1/3 volume (~1 ml) of acid washed beads, 425–600 µm (Sigma), for 20–30 min. Efforts to keep the mixture cool during the lysis procedure are strongly recommended. Periodic cooling on crushed dry ice can be used to keep the sample cool, taking care to avoid freezing the sample.
For larger scale preparations (>20 l culture), we have carried out lysis by passing the resuspended cells (step 4) through a microfluidiser eight to ten times (model #110Y, Microfluidics Corp.). Again, efforts to keep the mixture well cooled throughout the lysis process are strongly recommended.
The extract is cleared by centrifugation at 14,000 × g for 20 min at 4°C. A second ultracentrifugation step is optional at 142,000 × g for 1 h at 4°C.
The protein content in the cleared extract is determined using a Micro BCA assay (Pierce).
3.4. Isolation of Aptamer-Tagged RNA Complexes from Extracts Using Streptavidin
In the case of the streptavidin aptamer, a background of free biotin and biotinylated cellular material can be blocked by the addition of egg white avidin (Sigma) to the extract prior to binding to the streptavidin affinity resin (the RNA affinity tag does not bind to egg white avidin, only streptavidin). For optimal yields from different extracts, it can be useful to experimentally determine the correct amount of avidin required to block biotin in the extract. In our hands, extracts produced from yeast cultures can be sufficiently blocked by the addition of 10 µg avidin per milligram of protein in the extract. The streptavidin aptamer can stably bind to the affinity resin at up to 400 mM NaCl allowing purification under different salt conditions to be attempted if desired. A control pull down can be performed by pre-blocking the streptavidin affinity resin with D-biotin prior to incubation with the cell extract.
Block the extract with 5–20 µg of avidin per milligram of protein in the extract. Incubate at 4°C for 10 min prior to binding to the affinity resin.
Incubate the extract with 10–20 µl of streptavidin beads per milligram of protein in the extract. Binding should be carried out at 4°C for 1 h in 1× lysis buffer at an appropriate volume (five to ten times the bed volume). The beads are separated by centrifugation 4,000 × g for 5 min at 4°C.
Wash the beads with 20 bed volumes of 1× lysis buffer five times for 5 min each at 4°C.
Transfer the beads to an Ultrafree-MC centrifugal filter unit (0.45 µm pore size Mllipore) and wash twice with 5 bed volumes of 1× lysis buffer at 4°C.
Elute the beads by incubating for 0.5–1 h at 4°C with 2 bed volumes of 1× lysis buffer containing 5 mM D-biotin.
3.5. Localization of Tagged RNAs via Fluorescent In Situ Hybridization
3.5.1. Preparation of Fluorescently Labeled Oligonucleotide Probes
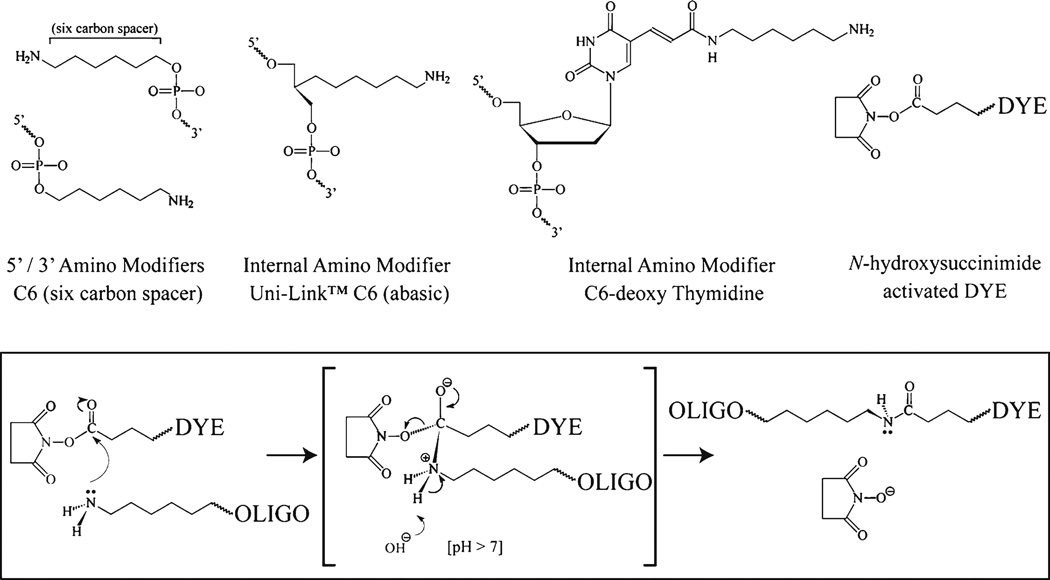
The aptamer sequence is localized by hybridizing a fluorescently labeled antisense oligonucleotide and detecting its fluorescence via microscopy. The oligonucleotide is synthesized with reactive amine group modifications that will facilitate chemical coupling of the fluorescent dye. There are a number of reactive amine modifications available and a subset of these are shown in Fig. 5. The positioning of the reactive amine groups within the oligonucleotide is important as the signal from the fluorescent dye can be quenched if the dye molecules are too close to each other (<10 nucleotides) or placed adjacent to a guanine. The following antisense probe is suitable for the detection of the streptavidin binding aptamer sequence and is modified to accept up to three fluorescent dye molecules.
Anti-S1 probe 5′-GAC(T)ATCTTACGCAC(T)TGCATGATTC(T)GGTCGGT-3′
(T) = internal amino modifier (C6-deoxy Thymidine)
Fig. 5.
Labeling amine modified oligonucleotides using activated ester chemistry. A number of amino modifications are available for oligonucleotide synthesis; common amino modifiers are shown including end modifications (5′ and 3′) and internal modifications. The fluorophore (DYE) is purchased as an activated ester form to facilitate coupling to the oligonucleotide via the amine modification. The reactive chemistry is summarized; the amine reacts with the ester group resulting in the formation of a stable carboxamide bond between the oligonucleotide and the fluorescent dye. The reaction is facilitated by the enhanced stability of the N-hydroxysuccinimide leaving group.
The amine groups can be coupled to a fluorescent dye by using amine reactive fluorescent dyes. The reaction is facilitated by activated ester chemistry where the oligonucleotide amine reacts with an activated ester group linked to the fluorescent dye resulting in the formation of a stable carboxamide bond between the oligonucleotide and the fluorescent dye (see Fig. 5). The chemistry is facilitated by the presence of a stable leaving group on the ester linkage of the fluorescent dye. Fluorescent dyes are available with a variety of leaving groups that react with amines in similar ways, including succinimidyl esters, sulfosuccinimidyl esters, tetrafluorophenyl esters and sulfodichlorophenol esters. Activated ester fluorescent dyes are unstable and have a limited shelf life (6–12 months) when stored cold (< −20°C) in the lyophilized form. The dyes must be dissolved in anhydrous solvent (DMSO) immediately prior to their use.
The reaction is set up with an excess of the amine reactive fluorescent dye and it is essential to remove this prior to purification of the labeled oligonucleotide. Generally, the amine reactive chemistry does not proceed to completion and the unreacted oligonucleotide must be separated from the labeled oligonucleotide species before these can be used for FISH. It is necessary to first remove the excess unreacted dye before attempting a high resolution purification of the labeled species. The final purification can be carried out by either preparative denaturing PAGE or reverse phase HPLC, we describe the use of preparative denaturing PAGE. When using oligonucleotides with multiple reactive amine modifications, the reaction products will be a mixture of mono-, di- and tri-labeled species. Each conjugated dye molecule will retard the migration of the oligonucleotide (by approx. 1–2 nt) and will allow the separation of the labeled species from the unlabeled oligo.
Dissolve the amine modified oligonucleotide to a final concentration of 3 mM (approx. 42 µg/µl for the anti-S1 probe) using deionized water (see Note 2). This stock solution can be stored at −20°C.
Prepare a fresh solution of 0.1 M sodium tetraborate buffer and adjust to pH 8.5 with HCl.
- Prepare the amine reactive fluorescent dye by dissolving it in fresh, anhydrous DMSO to a final concentration of 15 mM. Fluorescent dyes are light sensitive and it is best to use a foil wrapped or amber colored Eppendorf tube. For each labeling reaction prepare 15 µl of the dye solution, any remaining unused solution is unstable and it is preferable to prepare fresh solution for each set of labeling reactions.
Cy3 Mono NHS Ester (766 Da) 15 mM ~11.5 mg/ml Cy5 Mono NHS Ester (792 Da) 15 mM ~11.9 mg/ml Oregon Green 488-X1 NHS Ester (623 Da) 15 mM ~ 9.4 mg/ml -
Combine the following in a foil wrapped or amber colored eppendorf. The final molar ratio of fluorescent dye to reactive amine is (12:1), in this case the concentration of the anti-S1 probe is adjusted from 18 to 6 nmol as it bears three reactive amines.
-
–14 µl of fluorescent dye solution (210 nmol)
-
–2 µl of DNA oligonucleotide (6 nmol oligo/18 nmol reactive amine)
-
–75 µl of labeling buffer
-
–4 µl of deionized water.
Incubate the tube in the dark at room temperature using a rocker/shaker to gently mix the reaction, allow the reaction to proceed overnight.
-
–
An initial ethanol precipitation step will remove a significant amount of unreacted or degraded fluorescent dye. Precipitate the oligonucleotide by adding 10 µl of sodium acetate (3 M NaOAc pH 5.2) followed by 500 µl of ice cold ethanol. Invert the tube to mix and incubate at −80°C for at least 1 h (preferably overnight). Pellet the DNA by centrifugation at ≥14,000 × g for 30 min. Carefully remove the supernatant and wash the pellet twice with 70% ethanol to help remove excess unreacted fluorescent dye. Resuspend the pellet in 25–50 µl of buffer (10 mM Na-Phosphate pH 7.4).
Remaining unreacted dye is removed by spin column gel filtration chromatography. Carefully load the resuspended oligo (25–50 µl) onto the center of a prespun Bio-Spin 30 column and spin to recover the labeled probe according to the manufacturer’s protocol.
Add an equal volume of formamide to the oligonucleotide and load onto a 15% denaturing polyacrylamide gel (approx. 1–1.5 mm thickness). Load additional lanes as tracking markers, with bromophenol blue (BPB) and any leftover unused amine reactive fluorescent dye respectively. Run the gel until the BPB is approximately three-quarters of the way down the gel (the BPB migrates at approx. 15 nt). Transfer the gel onto saran wrap and locate the bands using a hand-held UV lamp and an X-ray film intensifying screen. Unreacted oligonucleotide will run faster than the labeled oligonucleotides and will form a dark UV shadow. The conjugated oligonucleotides will run with the dye color and can also be located by their UV shadow, although this may be less intense due to the fluorescence of the dye. Cut out the labeled oligonucleotides and elute from the gel slices overnight using the crush and soak method.
Precipitate the labeled oligonucleotides and resuspend in deionized water. Read the concentration of oligonucleotide in a spectrophotometer at 260 nm and adjust the final concentration to 2 µM.
3.5.2. Fixation and Permeabilization of Yeast Cells
Grow 50 ml of cells in synthetic minimal medium to midlog phase (OD = 0.2–0.4 at A600).
Crosslink cells by adding 5 ml of 40% paraformaldehyde (3.6% final concentration) to each culture and incubate for 30 min at room temperature, while shaking (see Note 5).
Harvest the fixed cells by centrifugation 4,000 × g for 5 min. Wash the cells by resuspending in 10 ml of ice-cold sorbitol buffer and repeat centrifugation to pellet. Repeat this wash step.
Digest the cell wall by first resuspending the cell pellet in 1 ml of spheroplast buffer and then adding Zymolyase up to 0.2 mg/ml final concentration. Mix gently and incubate at 37°C for 30–45 min.
Harvest the spheroplasted cells by centrifugation 4,000 × g for 2 min at 4°C. Wash the spheroplasts by resuspending in 1 ml of ice-cold sorbitol buffer and repeat centrifugation to pellet. Resuspend in a final volume of 500–800 µl ice-cold sorbitol buffer.
Spot 20 µl of spheroplasted cells onto each well of several slides. Incubate at 4°C for 30–60 min, and then aspirate excess liquid from each well. Wash each well with a droplet of ice cold sorbitol buffer and carefully aspirate. Store in slide jar filled with 70% ethanol at −20°C for at least 16 h before use.
3.5.3. Hybridization of Fluorescent Oligonucleotide Probe
Prepare the fluorescent oligonucleotide probe(s) by adding 30 ng (approximately 1 µl of a 2 µM solution) of each probe to 20 µl of hybridization mix; this is enough for one well on a slide (see Notes 6 and 7).
Denature slides by incubating for 10 min in prewarmed (72°C) denaturing solution.
Wash the slides in a series of cold (−20°C) ethanol washes: 70, 80, 90, and 100% ethanol for 1 min each. Allow slides to air dry at room temperature and immediately add denatured probe (step 4) once dry.
Denature probe by incubating at 72°C for 10 min. Immediately load 20 µl probe/hybridization mix to each spot.
Incubate at 36°C in dark, in a slide warmer/humid chamber for 24–48 h.
3.5.4. DAPI Staining and Slide Mounting
Incubate slides in post-hybridization wash buffer for 20–30 min.
Wash slides twice in 1× PBS for 5 min.
Dry slides on paper towel in the dark room, take care not to dry completely. To speed up drying, you can aspirate liquid surrounding wells.
Spot ~15 µl of 1 mg/ml of DAPI onto each well. Incubate for 3 min.
Wash slides twice in a jar of 1× PBS for 2 min.
Dry slides on a paper towel in the dark room, take care not to dry completely.
Combine mounting solution components A & B when ready to put on cover slip. Prewarming component B to 50°C aids pipetting. Add one drop of mounting solution to each well and gently press on the cover slip. Dry overnight in the dark on a flat surface. Once dry the slides can be viewed directly or sealed and stored upright in a covered slidebox at −20°C.
3.5.5. Visualization of Aptamer Tagged RNA
High resolution imaging will be required for fine discrimination of molecular compartments. Current methods generally employ either laser confocal or computer assisted deconvolution of Z-stacks acquired on a widefield fluorescence microscope. The user will be limited by the tools available and each approach has its own advantages, we refer the reader to the following review (22).
Acknowledgments
This work was supported by a National Institute of Health grant (R01GM082875) to DRE.
Footnotes
The sensitivity of FISH is such that the threshold level of detection is in the region of 10–20 copies of a localized target RNA per cell. For low copy targets, the sensitivity can be increased by utilizing oligonucleotides with multiple attached fluorophores.
It is important that the modified oligonucleotide is free from any contaminating amine compounds that would also react with the amine reactive fluorescent dye (including Tris buffers). We recommend purchasing the oligonucleotide with FPLC purification and resuspending in purified water. Additional chloroform extractions (3×) followed by ethanol precipitation can be performed if contamination is suspected.
For the rapid production of templates to be used for in vitro transcription of the target RNA, the T7 RNA polymerase promoter sequence (5′-TAATACGACTCACTATAGG-3′) can be encoded by the outlying primer and thus incorporated into the final PCR product. In vitro RNA transcripts produced from such templates can be used to test the binding of the tagged RNA to the affinity matrix in the absence of cellular components.
When the knockout strain involves a target RNA that is essential for yeast growth it is necessary to first supply the gene on a “rescue” plasmid in the absence of the chromosomal copy. When the rescue plasmid is marked with the URA3 auxotrophic marker this can be counterselected using 5-fluoroorotic acid (5-FOA).
The cells must be fixed with formaldehyde under normal growth conditions before centrifugation or any other processing steps are performed. If the cells are manipulated prior to fixation it is possible that fixation will capture a stress response which can significantly impact the final observations and the interpretation of localization data.
Multiple labeled oligonucleotide probes can be used with different fluorophores to highlight known locations within the yeast cell. For example, we have routinely employed an antisense probe against the yeast U14 RNA as a marker for the nucleolus.
When performing an in situ hybridization experiment, proper controls are necessary to ensure that observable signal is specific. Specificity can be determined with competition studies using both labeled and excess unlabeled probe. Excess unlabeled probe can displace the specific binding of the labeled probe, but will not affect nonspecific binding of labeled probe.
References
- Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Wilson DS, Szostak JW. In vitro selection of functional nucleic acids. Annu Rev Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- Nimjee SM, Rusconi CP, Sullenger BA. Aptamers: an emerging class of therapeutics. Annu Rev Med. 2005;56:555–583. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- Nygren PA, Stahl S, Uhlen M. Engineering proteins to facilitate bioprocessing. Trends Biotechnol. 1994;12:184–188. doi: 10.1016/0167-7799(94)90080-9. [DOI] [PubMed] [Google Scholar]
- Srisawat C, Engelke DR. Streptavidin aptamers: affinity tags for the study of RNAs and ribonucleoproteins. RNA. 2001;7:632–641. doi: 10.1017/s135583820100245x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srisawat C, Goldstein IJ, Engelke DR. Sephadex-binding RNA ligands: rapid affinity purification of RNA from complex RNA mixtures. Nucleic Acids Res. 2001;29:E4. doi: 10.1093/nar/29.2.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmuth K, Urlaub H, Vornlocher HP, Will CL, Gentzel M, Wilm M, Luhrmann R. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc Natl Acad Sci U S A. 2002;99:16719–16724. doi: 10.1073/pnas.262483899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachler M, Schroeder R, vonAhsen U. StreptoTag: a novel method for the isolation of RNA-binding proteins. RNA. 1999;5:1509–1516. doi: 10.1017/s1355838299991574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srisawat C, Engelke DR. RNA affinity tags for purification of RNAs and ribo-nucleoprotein complexes. Methods. 2002;26:156–161. doi: 10.1016/S1046-2023(02)00018-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Altman S. Partial reconstitution of human RNase P in HeLa cells between its RNA subunit with an affinity tag and the intact protein components. Nucleic Acids Res. 2002;30:3706–3711. doi: 10.1093/nar/gkf499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonov AA, Sergiev PV, Bogdanov AA, Brimacombe R, Dontsova OA. Affinity purification of ribosomes with a lethal G2655C mutation in 23 S rRNA that affects the translocation. J Biol Chem. 2003;278:25664–25670. doi: 10.1074/jbc.M302873200. [DOI] [PubMed] [Google Scholar]
- Shcherbakova DM, Sokolov KA, Zvereva MI, Dontsova OA. Telomerase from yeast Saccharomyces cerevisiae is active in vitro as a monomer. Biochemistry (Mosc) 2009;74:749–755. doi: 10.1134/s0006297909070074. [DOI] [PubMed] [Google Scholar]
- Hartmuth K, Vornlocher HP, Luhrmann R. Tobramycin affinity tag purification of spliceosomes. Methods Mol Biol. 2004;257:47–64. doi: 10.1385/1-59259-750-5:047. [DOI] [PubMed] [Google Scholar]
- Long RM, Elliott DJ, Stutz F, Rosbash M, Singer RH. Spatial consequences of defective processing of specific yeast mRNAs revealed by fluorescent in situ hybridization. RNA. 1995;1:1071–1078. [PMC free article] [PubMed] [Google Scholar]
- Olson BL, Siliciano PG. A diverse set of nuclear RNAs transfer between nuclei of yeast heterokaryons. Yeast. 2003;20:893–903. doi: 10.1002/yea.1015. [DOI] [PubMed] [Google Scholar]
- Xiao S, Day-Storms JJ, Srisawat C, Fierke CA, Engelke DR. Characterization of conserved sequence elements in eukaryotic RNase P RNA reveals roles in holoenzyme assembly and tRNA processing. RNA. 2005;11:885–896. doi: 10.1261/rna.7282205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL. RNA secondary structure analysis using the Vienna RNA package. Curr Protoc Bioinformatics. 2004;Chapter 12(Unit 12):2. doi: 10.1002/0471250953.bi1202s04. [DOI] [PubMed] [Google Scholar]
- Hofacker IL. RNA consensus structure prediction with RNAalifold. Methods Mol Biol. 2007;395:527–544. doi: 10.1007/978-1-59745-514-5_33. [DOI] [PubMed] [Google Scholar]
- Tranguch AJ, Engelke DR. Comparative structural analysis of nuclear RNase P RNAs from yeast. J Biol Chem. 1993;268:14045–14055. [PubMed] [Google Scholar]
- Tranguch AJ, Kindelberger DW, Rohlman CE, Lee JY, Engelke DR. Structure-sensitive RNA footprinting of yeast nuclear ribonuclease P. Biochemistry. 1994;33:1778–1787. doi: 10.1021/bi00173a022. [DOI] [PubMed] [Google Scholar]
- Lichtman JW, Conchello JA. Fluorescence microscopy. Nat Methods. 2005;2:910–919. doi: 10.1038/nmeth817. [DOI] [PubMed] [Google Scholar]