Abstract
Purpose
To implement and evaluate combined parallel magnetic resonance imaging (MRI) and partial Fourier acquisition and reconstruction for rapid hyperpolarized carbon-13 (13C) spectroscopic imaging. Short acquisition times mitigate hyperpolarized signal losses that occur due to T1 decay, metabolism, and radiofrequency (RF) saturation. Human applications additionally require rapid imaging to permit breath-holding and to minimize the effects of physiologic motion.
Materials and Methods
Numerical simulations were employed to validate and characterize the reconstruction. In vivo MR spectroscopic images were obtained from a rat following injection of hyperpolarized 13C pyruvate using an 8-channel array of carbon-tuned receive elements.
Results
For small spectroscopic matrix sizes, combined parallel imaging and partial Fourier undersampling resulted primarily in decreased spatial resolution, with relatively less visible spatial aliasing. Parallel reconstruction qualitatively restored lost image detail, although some pixel spectra had persistent numerical error. With this technique, a 30 × 10 × 16 matrix of 4800 3D MR spectroscopy imaging voxels from a whole rat with isotropic 8 mm3 resolution was acquired within 11 seconds.
Conclusion
Parallel MRI and partial Fourier acquisitions can provide the shorter imaging times and wider spatial coverage that will be necessary as hyperpolarized 13C techniques move toward human clinical applications.
Keywords: carbon-13, hyperpolarized, parallel imaging, spectroscopy
Hyperpolarized carbon-13 (13C) techniques exploit dynamic nuclear polarization (1) to increase the nuclear polarization of 13C nuclei by several orders of magnitude compared to the polarization achieved at thermal equilibrium. This increase in polarization permits imaging and spectroscopy of 13C-labeled molecules with high sensitivity and image contrast. Hyperpolarized compounds have been used to study metabolism in several preclinical animal models of malignancy, including (among others) tumors of the prostate, liver, and brain as well as lymphoma (2). Other applications have included the measurement of tissue pH as well as cardiac metabolism (2).
Human clinical applications of hyperpolarized 13C techniques present multiple technical challenges, especially the need for imaging speed. Hyperpolarized magnetization is lost due to T1 decay, metabolism, magnetization saturation, and substrate washout, constraining the nuclear magnetic resonance (NMR) acquisition time. Abdomen, pelvis, and cardiac applications additionally benefit from rapid imaging to allow for breath-holding and to minimize the effects of motion within the heart and bowel.
Because of this special demand for imaging speed, parallel imaging strategies are well suited for hyperpolarized 13C applications. Parallel magnetic resonance imaging (MRI) acquisitions (3–5) and reconstructions permit rapid imaging by exploiting spatial information embedded in coil array sensitivity profiles to reconstruct omitted gradient-encoded κ-space points. A 3-element array of carbon-tuned surface coils has been used in conjunction with sensitivity encoding (SENSE) to reduce scan time by a factor of two (6). In that implementation, coil sensitivity information was obtained using a separate phantom-based reference scan. Using a phantom to measure coil sensitivities restricts the use of flexible coil arrays, which is desirable in many clinical patient studies, particularly for human abdominal imaging.
Self-calibrated parallel MR techniques (4,7,8) overcome the need to obtain an external sensitivity reference by acquiring a small number of extra central k-space lines, effectively embedding a low-resolution reference image within the acquired data. These approaches are particularly appealing for hyperpolarized 13C because subject-based coil sensitivities are nearly impossible to measure due to the low sensitivity and thermal polarization of 13C nuclei. Self-calibrated SENSE has been successfully applied to hyper-polarized 13C spectroscopy and a 4-coil array for imaging a mouse (9). However, for the small matrix sizes typically used in hyperpolarized 13C imaging and spectroscopy, the acquisition of additional lines for coil calibration can represent a large relative burden in terms of imaging time.
In this work a combined parallel and partial Fourier reconstruction for accelerated hyperpolarized 13C spectroscopy was implemented. The addition of the partial Fourier reconstruction allows the omission of additional κ-space lines and partially compensates for the extra imaging time required to acquire extra calibration lines. Although the approach is applicable to both spectroscopy and imaging, this work is focused on spectroscopic acquisitions because phase variations in spectroscopy present the biggest potential challenges for the partial Fourier component of the reconstruction. The basic theory behind combining partial Fourier and parallel MRI is reviewed with the goal of addressing special issues that arise when applying these techniques to spectroscopic imaging applications. Numerical simulations are described in order to evaluate the theoretical accuracy of the reconstruction.
Finally, in vivo MR spectroscopic imaging (MRSI) results from a rat following injection of hyperpolarized 13C pyruvate using an 8-channel array of carbon-tuned surface coils are presented. The data demonstrate the feasibility of acquiring a 30 × 10 × 16 matrix of 4800 3D MRSI voxels with isotropic 8 mm3 resolution within 11 seconds. This matrix size, spatial resolution, and imaging time approach those required for future human abdominal hyperpolarized 13C-MR studies of liver, kidney, metastatic cancers, etc. within a breath-hold.
MATERIALS AND METHODS
Image Reconstruction
Multiple approaches have been described for combining parallel MR reconstructions with partial Fourier, or “phase-constrained,” image reconstructions in the context of proton MRI (10–13). For the present work, the approach used in Ref. (10) was adopted, which employs a phase-constrained version of generalized encoding matrix reconstruction (5,14). In Ref. (10), the low-resolution coil sensitivity reference provided from a self-calibrated dataset also provided the phase information required for a partial Fourier reconstruction. In addition, it was shown that performing the two reconstructions simultaneously was superior to performing them sequentially. Performing a partial Fourier reconstruction on the undersampled coil images prior to the parallel reconstruction leads to phase errors due to the fact that the low-resolution full field of view (FOV) phase reference does not adequately describe the phase variations of the aliased images. Additionally, the extra constraints provided by the partial Fourier reconstruction improve the numerical conditioning of the reconstruction matrix.
In the following section the reconstruction approach described in Ref. (10) is adapted for use in spectroscopy. In this type of reconstruction (which is similar to the generalized form of SENSE), the received signal from a κ-space point κj and coil number l is written in terms of the underlying spin density ρ(xi) at point xi:
| [1] |
The encoding matrix, B, is defined as:
| [2] |
and Cl(xi) is the sensitivity profile of coil l evaluated at point xi. The underlying spin density is recovered from the acquired undersampled data using the Moore-Penrose pseudo-inverse (5,8,15):
| [3] |
In the equation above, Ψ is the noise correlation matrix. In this equation B’ denotes the complex conjugate transpose. It has been shown (10) that applying a partial Fourier or phase constrained reconstruction amounts to requiring the reconstructed spin density, ρ, to be real. Eq. [1] becomes:
| [4] |
Note that Eq. [4] has twice as many constraints as Eq. [1] for the same number of unknowns.
Three modifications of this formalism permit the reconstruction of spectroscopic data. First, s and ρ become functions of both position, x, and frequency, ω. Because the coil sensitivity is independent of frequency, Eq. [3] is modified in a straightforward way, by formally introducing the frequency dependence:
| [5] |
The reconstruction of an entire spectrum is simply comprised of multiple independent reconstructions for each frequency. While a signal threshold could potentially be used to only reconstruct those portions of the data that have signal (shortening the reconstruction time), the present work attempts a reconstruction for every frequency whether there is signal present or not. In practice, the spin density, ρ(x), may not strictly be real. Broadly speaking, phase offsets in MRS come from two sources. First, areas of high magnetic susceptibility, field inhomogeneity, or transmit inhomogeneity may cause a spatially varying phase offset. Second, spectra typically have a frequency-dependent phase offset that results from asymmetry in the acquired echo. This is particularly true for the free induction decay (FID)-based spectroscopic readouts used in this work (see Materials and Methods, below), in which negative timepoints are not acquired. In order to incorporate this into the reconstruction formalism, ρ is rewritten as:
| [6] |
where is the magnitude of the spin density; ϕfield and ϕspect are the phase offsets introduced by magnetic field inhomogeneities and the spectral readout, respectively. ϕspect is expected to predominantly depend on frequency, but there may also be a small spatial dependence where peaks overlap and the signals from multiple metabolites add with different ratios.
The effects of ϕfield and ϕspect are incorporated into the reconstruction along the same lines as in Ref. (10). In a self-calibrated reconstruction (8), the coil sensitivities are generally not “absolute” coil sensitivities, Cl(xi). Instead, the coil sensitivities are measured from the low-resolution fully sampled center of κ-space. Formally, this “in vivo” sensitivity reference data is written as
| [7] |
where σ(x) is the low-resolution representation of the underlying spin density, ρ(x). Further, if the phase offsets, ϕfield and ϕspect, vary slowly with position,
and:
| [8] |
As used in this work, “coil sensitivity” refers to the physical spatial dependence of each receiver coil's reception profile, which is independent of frequency over the bandwidth of the acquisition and also independent of the underlying spin density. “In vivo sensitivity reference data” (Eq. [7]) refers to the frequency-dependent low-resolution reference that is extracted from the central κ-space lines of acquired data.
It has been shown (8,10,15) that when “in vivo” sensitivity reference data are used, then the reconstruction equation (Eq. [5]) yields the magnitude of the underlying spin density, or , modulated by the low-resolution representation of the spin density:
| [9] |
Multiplication by a sum-of-squares combination of the in vivo coil sensitivity yields the reconstructed image:
| [10] |
This result has two important consequences. First, unlike the nonspectroscopic case in Ref. (10), the in vivo sensitivity reference data in Eq. [8] is now dependent on frequency. Therefore, a separate in vivo sensitivity reference dataset must be extracted for each frequency. Second, the reconstructed spectrum is the magnitude spectrum. In exchange for the time savings achieved using the half-Fourier reconstruction, the phase information is lost, and the spectrum can no longer be rephased to remove the dispersive component. This will lead to a broadening of the spectral lines. Note that in the reconstruction equation above, although the in vivo measured sensitivity reference data are a function of frequency, the true spatial coil sensitivity profile that modulates the final reconstructed image is only a function of position.
In spectroscopy it is often necessary to perform intermediate processing steps on individual component images before combining them. For example, spectral apodization, or line broadening, is typically used to filter noise from the spectra. This type of filtering cannot be reliably performed on the magnitude reconstructed data. Individual coil images can be reconstructed by replacing the postmultiplication factor in Eq. [11]:
| [11] |
The component coil images reconstructed using Eq. [11] can be individually processed and then combined. In our case, we applied line broadening in the time domain and then reconstructed the images in a sum-of-squares fashion. Other combinations of coil data are possible. For example, if the absolute coil sensitivity functions are known, then an optimal phased combination (15) can be performed. This would have the advantage of preserving phase information.
Numerical Simulations
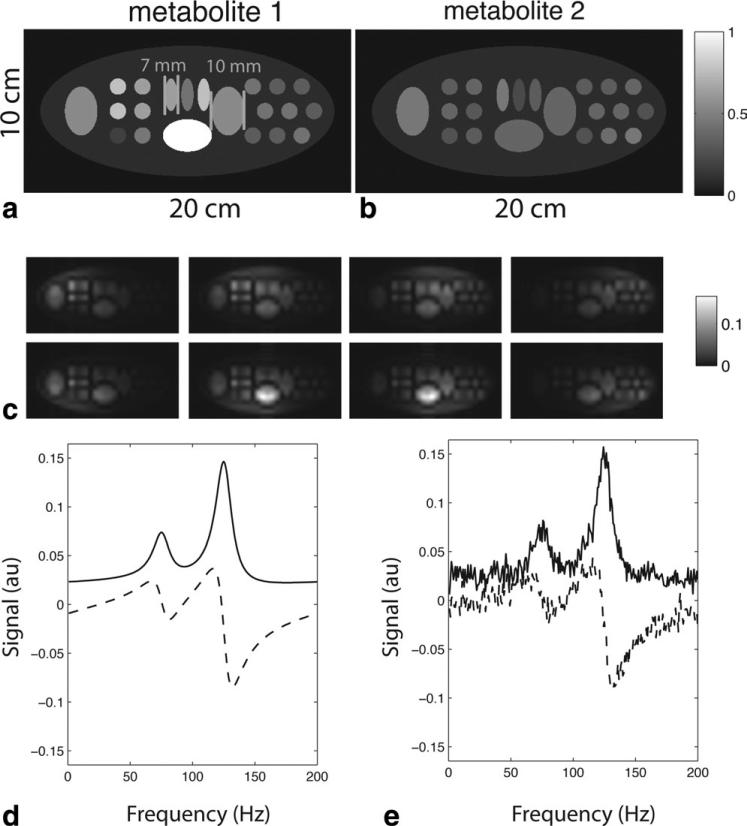
Numerical simulations, image reconstructions, and data display (both simulated and in vivo) were implemented using MatLab (MathWorks, Natick, MA). Simulated reconstructions were performed in order to assess the image reconstruction in a controlled fashion. A numerical phantom with two chemical species, each with a different spatial distribution, was simulated (Fig. 1a,b). Individual component coil images were obtained by modeling an 8-element array of rectangular coils. Four elements were placed above and below the image. Each element was 5.5 × 10.5 cm. Adjacent elements overlapped 1.3 cm and the total length of each 4-element panel was 18 cm. The separation of the two 4-element panels was 11 cm. Coil sensitivities were calculated using the principle of reciprocity and the Biot-Savart law in the quasi-static regime.
Figure 1.
Numerical phantom used for simulations. a,b: Relative distributions of two simulated metabolites. c: Low-resolution component coil images generated from a simulated 8-element array (coil array geometry described in the text). d: Representative spectrum from a single simulated pixel (solid = real, dashed = imaginary). e: Simulated spectrum with noise.
After high-resolution component coil images were generated, simulated images from each component coil with reduced resolution were created by extracting the central 32 × 16 points in frequency domain (Fig. 1c), giving a spatial resolution of ~6 mm2. This low-resolution dataset was used as the reference for our reconstruction. The smallest ellipses in the simulated phantom were 7.6 mm wide, with a spacing of 3.4 mm, which was smaller than the nominal 6 mm spatial resolution of the simulated acquisition. This relationship between object spacing and spatial resolution was chosen to emulate an important aspect of in vivo spectral acquisitions, in which some spatial variation occurs at a higher resolution than the spectral acquisition.
For each pixel a spectrum was generated by simulating a time-domain free induction decay resulting from two chemical species, which were separated by 50 Hz. A total of 256 timepoints were used, with a bandwidth of 200 Hz. T2* exponential decay was assumed to be 20 msec. This combination of T2* decay and spectral separation was chosen to ensure sufficient overlap between adjacent spectral peaks. An example spectrum from one pixel is shown in Fig. 1d. T1 decay and other time-dependent sources of signal loss (metabolism, radiofrequency saturation) were not explicitly included in the simulations. The signal-to-noise ratio (SNR) advantages from reduced imaging time in hyperpolarized parallel acquisition have been investigated previously (9) and we wished to isolate the additional effects of a phased constrained reconstruction on spectral quality.
Reconstructions were performed with or without adding Gaussian noise to each coil channel. The simulated spectrum with added noise is shown in Fig. 1e.
Undersampled datasets were generated by selecting a limited number of κ-space points from the full data. Four sampling patterns were considered. Either 6 × 6 or 10 × 10 κ-space points (central lines, labeled “CL” in the figures) were fully sampled from the center of κ-space and used to form a sensitivity reference. Outside of the fully sampled center, phase encode lines in the positive half of κ-space were undersampled uniformly in one dimension by an “outer reduction factor” (labeled ORF) of either 2 or 3. With the exception of fully sampled sensitivity reference points, no data from the negative portions of κ-space were used. For simulations that included Gaussian noise, a Lorentzian filter with 5 Hz of line broadening was applied to the reconstructed spectra generated from each individual coil:
| [12] |
where κ denotes the amount of line broadening and BW is the bandwidth. These coil spectra were then combined in a sum-of-squares fashion. For select levels of undersampling, reconstructions were also performed on conventionally undersampled full Fourier data, without the added phase constraint of Eq. [4].
The SNR behavior of the reconstruction was investigated through Monte Carlo simulation by generating 64 different noisy datasets, and reconstructing each one independently. Signal and noise were measured by computing the average and standard deviation of the reconstructed images, respectively, on a pixel-by-pixel and frequency-by-frequency basis. A g-factor was computed from the signal-to-noise using the standard formula found in Ref. (5).
Images were generated from the spectroscopic voxels by integrating either over the entire spectrum or each of the peaks individually. For display purposes, images were interpolated by a factor of 4 using bilinear interpolation. For simulated reconstructions the average error of the reconstruction was computed on a pixel-by-pixel basis by evaluating the absolute magnitude percentage deviation of the reconstructed from the original “reference” spectrum for every frequency:
| [13] |
where the sum is carried over all locations and frequencies within the region of interest. For reconstructions in which spectral apodization, or line broadening, was employed, a line-broadened reference was used for comparison. In order to isolate artifacts introduced by the reconstruction from the effects of noise, percentage error was computed based on comparison of the mean signal computed over all Monte Carlo repetitions. Because spectral apodization introduces a nonlinear suppression of noise, signal-to-noise comparisons and g-factor calculations were based on reconstructions performed without spectral apodization.
In Vivo Experiments: Hardware
All scans were performed at a magnetic field strength of 3.0 T using a GE (Milwaukee, WI) MR scanner with multinuclear capability. A photograph and schematic diagram of the surface coils used for in vivo experiments are shown in Fig. 2a,b, respectively. An 8-channel custom coil array with two linear panels was employed; each panel contained a linear arrangement of four surface coils. The general design of the individual coil elements was similar to that described in Ref. (6). Each rectangular coil element was ~5 × 10 cm and tuned to 32.14 MHz. Adjacent elements were overlapped in order to minimize mutual inductance. Carbon-tuned preamplifiers with low input impedance were employed to further reduce inductive coupling. The total length of each panel of surface coils was ~18 cm. In the animal imaging configuration used here, the distance between the center coil elements was ~9 cm and the distance between the edges of the panels was ~6 cm. The coil array was designed for human use and therefore suboptimal for animal imaging, but allowed for an in vivo demonstration of our method and will facilitate rapid translation to clinical studies.
Figure 2.
a: Photograph of animal setup. Top half of the clamshell transmitter is shown along with the top panel of the coil array. The bottom panel of the coil array is covered by a sheet, and the bottom half of the clamshell transmitter is embedded in the patient table. b: Schematic of the eight-element coil array. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
For RF excitation, a custom-built 13C transmitter was used. The detailed design has also been described (6). Briefly, the excitation coil consisted of a capacitively coupled Helmholtz pair with elements anterior and posterior to the imaging volume. Surface coils were decoupled from the transmitter during RF transmit using blocking networks activated by pin diodes. Although the surface coils were decoupled from each other during signal reception as described above, dynamic disable circuitry was not available to decouple the transmitter from the surface coil array during signal reception.
In Vivo Experiments: Hyperpolarization and Animal Setup
Animal studies were carried out under a protocol approved by the local Institutional Animal Care and Use Committee. Normal male Sprague–Dawley rats were placed on a heated pad and anesthetized with isoflurane (1.5%–2%). The rats were positioned supine, resting along the bottom panel of the array. A heating system was placed below the rat and a heated bag of saline was placed on the animal's abdomen.
Samples of 32 μL [1-13C] pyruvic acid (Isotec, Miamisburg, OH) and 15 mM OX63 trityl were polarized and dissolved using a Hypersense DNP polarizer (Oxford Instruments, Carteret, NJ), as described previously (6,16). The samples were carried to the adjacent MRI scanner and 3.0 mL boluses of 100 mM hyperpolarized pyruvic acid were injected through a tail vein catheter over a period of 12 seconds, followed by a 0.5 cm3 bolus of normal saline. The volume of pyruvate was at the upper range for that routinely injected into rats in order to increase overall signal available for this proof-of-concept study. As this was a proof-of-principle study, either two or three experiments were performed during each imaging session, depending on the scanner time available. Injections were separated by ~1 hour, which was the time required for the polarization of the next pyruvate sample.
In Vivo Experiments: Image Acquisition and Reconstruction
A 3D spectroscopic imaging sequence with flyback echo planar readout was used (17). Phase encode directions were chosen superior–inferior and anterior–posterior with respect to the animal (in an axial plane with respect to the main magnetic field). Frequency-encoding and echo planar readout were performed in the right–left direction with respect to the animal (in the direction of the main magnetic field). Imaging commenced at 30 seconds following the start of the injection of hyperpolarized material. A free induction decay echo planar spectroscopic imaging pulse sequence (FID-EPSI) was used with flyback trajectory and a progressive flip angle (18), a 10 cm slab excitation pulse, and concentric phase encoding with TE/TR = 50/130 msec. Each flyback readout yielded 16 spatial points and 59 spectral points, with a spectral bandwidth of 581 Hz. Images were acquired with 0.8 cm isotropic spatial resolution (field of view 24 × 8 cm, voxel size of 0.51 cc). Fully encoded acquisitions were acquired as well as acquisitions with various levels of undersampling. A central region of 6 × 6 fully sampled phase encode points was used as a sensitivity reference. The positive outer portion of κ-space was undersampled by a multiple of either 2 or 3. Negative κ-space points were not acquired, with the exception of the fully sampled sensitivity region. The fully sampled 30 × 10 × 16 acquisition consisted of 300 phase encode points, requiring 39 seconds. Accelerated acquisitions had either 104 or 80 total phase encode points, requiring 13.4 or 10.4 seconds, respectively, yielding acceleration factors of 2.9 or 3.75, respectively.
Images were reconstructed using the strategy outlined above. The noise correlation matrix was measured using two frequency-encoding slices that lay outside of the image. Coil sensitivity data were obtained from the fully sampled centers of κ-space using simple linear interpolation. After reconstructing each coil image spectra, ~5 Hz of spectral apodization (line broadening) was applied. The reconstructed coil spectra were combined in a sum-of-squares fashion to yield the final spectrum.
Anatomic T2-weighted fast spin-echo reference images were obtained using the proton body coil for transmit and receive. Echo train length = 16, TE = 102 msec, TR = 4.5 seconds (sag) TE = 85 msec, TR = 2.1 seconds (cor) TE = 102 msec, TR = 6.7 seconds (axial).
In Vivo Experiments: Estimation of Polarization
In order to quantitatively compare different acquisitions in hyperpolarized experiments, it was necessary to adjust for the shot-to-shot variation in polarization level. Because a separate NMR polarimeter was not available at the time of these experiments, an indirect estimation of the polarization was performed from the raw acquired data. Because of the centric ordering of κ-space, the κ = 0 line was always the first point acquired in each experiment and was always acquired at a fixed time after the pyruvate injection (30 sec). Using a progressive flip angle (18), the magnitude of the signal at this point can be expressed in terms of the polarization, flip angle, and overall magnetization:
| [14] |
where M0 is the average spin density and N is the total number of acquired points and p was the polarization level. The relative polarization was estimated by taking the ratio of the maximum κ-space signal for each acquired coil and adjusting for different initial flip angle, α1 (expressed in terms of the total number of acquired points). Use of this indirect estimate assumed similar decay over the first 30-second delay between injection and imaging.
RESULTS
Numerical Simulations
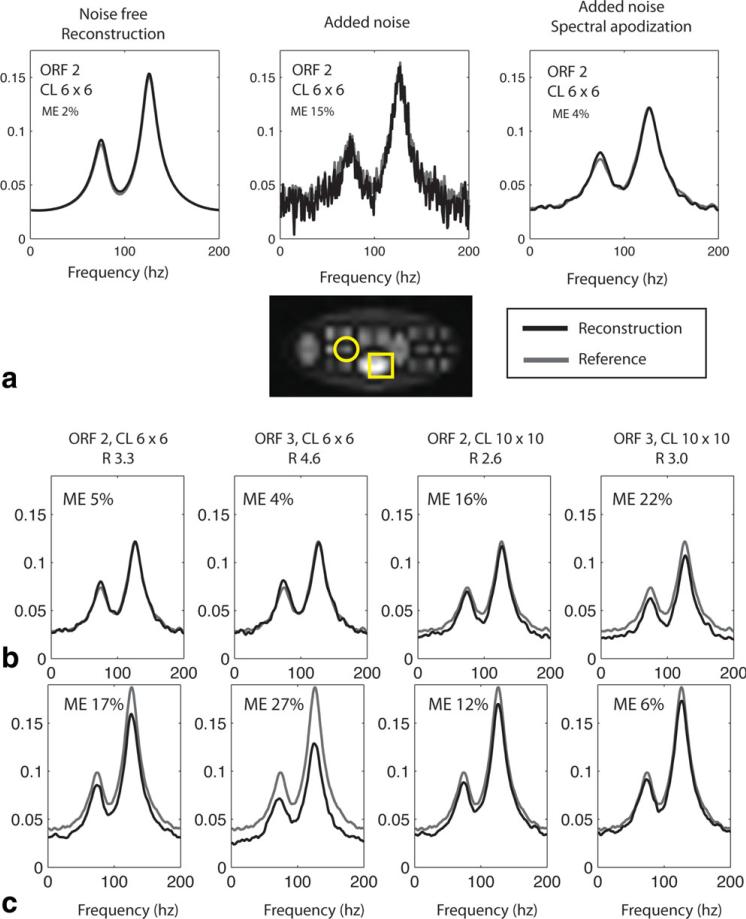
The numerical phantom was described in detail in the Materials and Methods section and is shown in Fig. 1a,b. For a single 6 mm2 voxel, Fig. 3a compares the effects of adding noise to the reconstruction and demonstrates the benefits of spectral apodization, or “line broadening,” on the final spectrum. All results are for a simulated acquisition with 6 × 6 center lines, outer reduction factor of 2, and partial Fourier reconstruction (155 acquired κ-space points out of 512, net acceleration factor of 3.3). The left plot is the reconstructed noise-free data. The middle plot is a reconstruction of noisy data. In the right-most plot, 5 Hz of spectral apodization was added. For this pixel, all three reconstructions (black lines) closely approximated the reference (dark gray lines). The average percentage error for each reconstructed frequency was 2% in the noise-free case, 15% for the case in which noise was added, and 4% for the case in which noise was added but spectral apodization was used. Spectral apodization improved the noise characteristics of the spectrum while continuing to approximate the reference spectrum.
Figure 3.
Reconstructions of simulated spectra. a: Effect of spectral apodization. Reconstructed spectra are shown for a sample voxel (indicated by the circle). Left panel: reconstruction of noise-free data. Center panel: reconstruction of data with added noise. Right image: reconstruction with added noise and 5 Hz of spectral apodization. (black = reconstruction, gray = reference). b: Effects of using different numbers of central reference lines (CL) or different outer reduction factors (ORF) for the reconstruction for voxel shown by circle. c: Same plots as (b), but for voxel shown by square. ME = mean percentage error. R = acceleration factor.
Reconstructed spectra for various undersampling patterns are compared in Fig. 3b. The results for two representative pixels are examined in detail, which illustrate pixels with either relatively small (top row, circle in image) or relatively large (bottom row, square in image) residual deviation from the reference data. Each plot compares the “reference” spectrum (dotted line) with the reconstructed undersampled data (solid line). Noise was added in each case, and each spectrum had spectral apodization applied. Displayed plots and mean error are for the “signal,” or average over all Monte Carlo simulations. For reconstructions using 6 × 6 central lines and outer reduction factors of 2 and 3 (acceleration factors of 3.3 and 4.6), the first pixel had average percentage error 5% and 4%, respectively. For the second pixel, average percentage error was 17% and 27%. Incorporating more sensitivity data and using 10 × 10 center lines (acceleration factors of 2.5 and 3.0) the reconstruction of the first pixel was degraded slightly (average percentage error of 16% and 22%) while reconstruction of the second pixel was improved (average percentage error of 12% and 6%).
The global performance of the reconstruction for simulated data is illustrated by Fig. 4. In Fig. 4a, images were created by computing the area under the magnitude spectra for each pixel. Displayed images were interpolated by a factor of 4 for a final displayed resolution of 128 × 64. All images were displayed according to the same grayscale.
Figure 4.
Reconstructions of simulated images. a: Images generated by reconstructed spectra for different numbers of central lines (CL) or different outer reduction factors. Pixel values correspond to the total area under the spectra. Top row contains zero-filled undersampled images. Bottom row contains reconstructed images. Bottom image is the reference image prior to decimation. b: Line profiles corresponding to the top dashed line in the reference image in (a). Gray plots (both rows): reference image prior to decimation. Black plots, top row: zero-filled data prior to reconstruction. Black plots, bottom row: reconstructed data. c: Line profiles corresponding to the bottom dashed line in the reference image in (a). Y axis has arbitrary image units. ORF = outer reduction factor. CL = central lines. ME = mean percentage error. R = acceleration factor.
The top row of images show images obtained if no image reconstruction was performed, ie, the nonacquired κ-space data were replaced by zeros. The bottom row of images reflects the results of combined partial Fourier and parallel MR image reconstruction. Qualitatively, the undersampled κ-space trajectories predominantly lead to a reduction in spatial resolution. A much smaller relative amount of spatial aliasing was seen at the edges of the field of view. Applying the partial Fourier and parallel MR image reconstruction, much of the spatial detail was restored. Note that the undersampled images are not provided as “reference” reconstructions but rather to illustrate the visual effects of undersampling according to these sampling patterns.
The results are examined quantitatively in Fig. 4b,c. Line profiles through the upper 1/3 and lower 1/3 of the phantom from Fig. 4a are shown. In the plots, dark gray lines represent the “reference” image prior to undersampling and solid lines reflect either the zero-filled undersampled images or the reconstructed images. The undersampled line profile is again seen to represent a smoothed representation of the image that has lost spatial detail compared to the reference. The reconstructed line profiles recovered much of the lost spatial detail, with the average percentage error in the reconstructed images ranging from 10%–27%. As was seen in the single-pixel spectra (Fig. 3b), a larger percentage error of 27% occurred for the line that passed through the large oval in the inferior part of the phantom, for an undersampling pattern with 6 × 6 central lines and an outer reduction factor of 3. While the reconstruction improved at this location when 10 × 10 central lines were used, the change in average error (15% vs. 21%) was out of proportion to the change in net acceleration factor required to obtain the extra central lines (acceleration of 3.0 vs. 4.6).
The results of Monte Carlo simulations are shown in Fig. 5. Profiles are shown for the bottom line of the image shown in Fig. 4b. SNR is plotted in 5a and g-factor is plotted in 5b. The right-most column shows the results for comparison from a nonphase constrained reconstruction with 2D undersampling and outer reduction factors of 3 and 2 in the x- and y-directions, respectively. Net acceleration for this undersampling pattern (4.3) was still less than the net acceleration for the half Fourier acquisition and 1D acceleration with outer reduction factor of 3 (column 2, R = 4.7).
Figure 5.
SNR results of Monte Carlo simulation. All results are shown for the line profile corresponding to bottom white dashed line in Fig. 4a. a: Signal-to-noise ratio. Black lines: accelerated reconstruction. Gray lines: reference profile. b: g-factor. ORF = outer reduction factor. CL = central lines. ME = mean percentage error. R = acceleration factor.
In Vivo Acquisition
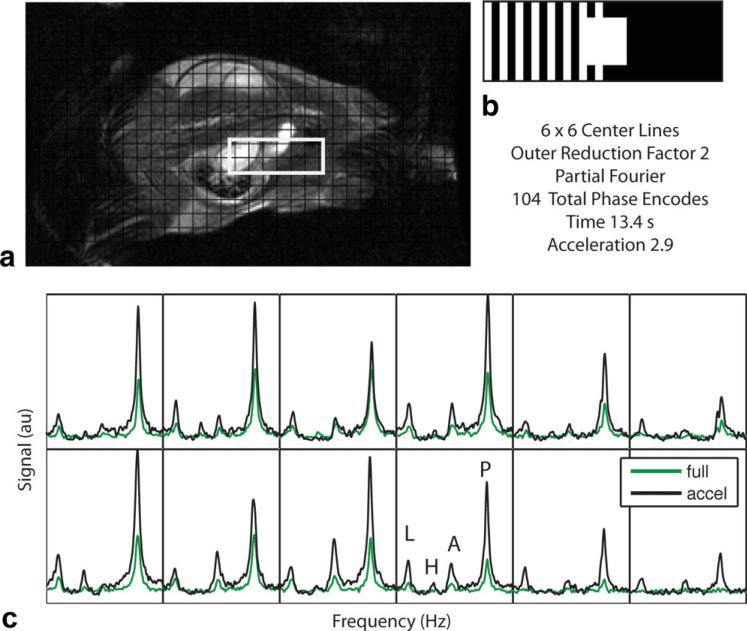
Reconstructed spectra for a coronal slice through the rat kidney are shown in Fig. 6 for acquisitions employing partial Fourier undersampling, an outer reduction factor of 2, and 6 × 6 central sensitivity lines. Detailed spectra through the kidney (outlined by the square) are shown. The reconstructed spectra, which were obtained in 13.4 seconds, are compared with the full 39-second acquisition. The spectra are adjusted for differing overall pyruvate polarization levels, as estimated from the component coil data as described above. In the first set of experiments, the overall polarization from the fully encoded acquisition was ~91% of the accelerated acquisition. In the second set of experiments, in which the outer reduction factor was 3, the polarization of the fully encoded acquisition differed from the accelerated acquisition by a value of 98%. Pyruvate signal was higher in the accelerated spectrum than the full spectrum for all voxels shown. There were some voxels (bottom right in Fig. 6c) where the accelerated acquisition had visible lactate signal but the full acquisition did not.
Figure 6.
Sample coronal slice for the in vivo reconstructed spectra for a combined parallel image and partial Fourier reconstruction with outer reduction factor of 2 and 6 × 6 center lines (acceleration factor 2.9). a: T2-weighted anatomic reference image and grid corresponding to the spectral voxel locations. b: Phase-encode sampling pattern (white corresponds to acquired phase encode points). c: Detailed spectra corresponding to voxels in the kidney (white box in (a)). Black line = reconstruction of under-sampled dataset, green line = sum of squares reconstruction of fully encoded dataset. Metabolite peaks as labeled. P = pyruvate, A = alanine, H = pyruvate hydrate, L = lactate.
Images generated from the reconstructed spectra are shown in Fig. 7. Colored overlays reflect the area under each spectral peak. For visualization purposes, the lactate and alanine signals are multiplied by a factor of 3. Images are also adjusted for differing polarization levels using the adjustment factors described above. Results from the full 39-second reference scan (top row), together with the zero-filled undersampled image (middle row) and image obtained via parallel and partial Fourier reconstruction (bottom row) are shown. The set of nine colored overlays corresponding to each undersampling pattern were displayed using the same color scale. High pyruvate signal was seen within the kidneys, heart, and vasculature. The undersampled images showed very little spatial aliasing. Rather, there was obvious blurring of the signal in the direction of undersampling, which was visible particularly at the kidneys. Following parallel reconstruction, much of this lost spatial resolution was recovered. Increased image heterogeneity, especially for the nonpyruvate metabolites, likely reflects a degradation in signal-to-noise compared to the zero-filled data. This is expected given the overall increased spatial resolution of the reconstructed data as well as SNR losses expected from parallel MR reconstruction.
Figure 7.
Reconstructed metabolite images. Images generated by computing the areas under the spectral peaks. T2-weighted coronal images and acquisition patterns are shown in the left column (white pixels correspond to acquired phase-encode points). For each undersampling pattern, top row: fully encoded images (39 sec). middle row: undersampled acquisition, zero-filled. Bottom row: undersampled acquisition, reconstructed. Within each set of 3 × 3 images, the color scales are the same. Alanine and lactate images have been scaled by a factor of 3 for improved visualization.
DISCUSSION
Hyperpolarization of injectable MR compounds promises to be a powerful tool for the in vivo study of tissue metabolism. Imaging speed is crucial to mitigate the effects of T1 decay, RF saturation, substrate washout, and metabolism. Abdominal and cardiac applications also require sampling within a breath-hold. Parallel MRI and partial Fourier acquisitions can play an important role in achieving the shorter imaging times and wider spatial coverage that will be necessary as hyperpolarized 13C techniques move towards human clinical applications. Using these techniques, together with a human-sized 8-element carbon coil array and a clinical 3T MRI scanner, we have demonstrated the ability to acquire a spectroscopic imaging volume with matrix size 30 × 10 × 16 within 10.4–13.4 seconds. Although our model system (a rat) had a small imaging volume, the 30 × 10 × 16 matrix size approaches that which might eventually be used for clinical abdominal imaging performed at isotropic spatial resolution between 1 and 1.5 cm. The imaging times achieved here can fit comfortably within a breath-hold.
We described numerical simulations and in vivo acquisitions from normal rats. Because the fully sampled acquisition was much longer than the under-sampled acquisition and suffered both resolution and signal loss due to T1 decay, no true reference acquisition was available to assess the accuracy of the in vivo reconstruction. The simulations provided an objective reference with which to compare the reconstructed data. The SNR effects of reduced T1 decay through parallel imaging and hyperpolarized acquisition have been evaluated in prior work (9) and therefore not explicitly included in the simulations. Simulations instead focused on evaluating the combined parallel and partial Fourier reconstruction of low-resolution spectroscopic data. Simulated coil array and sample geometry were chosen to match the in vivo experiment. The simulated line-shape was based on a free induction decay, which introduced frequency-dependent phase variation. Simulated spectral peaks were chosen with different spatial distributions and overlapping spectra. The effects of added noise were also evaluated.
Simulations revealed undersampling of the full data to be manifest chiefly as a blurring of the image, with less prominent spatial aliasing than is usually seen in proton imaging. This effect likely reflected the fact that the fully sampled center of κ-space represented a larger relative proportion of the low-resolution spectroscopic acquisition than it does with higher-resolution proton imaging. Parallel reconstruction recovered much of the lost spatial resolution (evident by recovery of small spatial details in the simulated image). Although the reconstruction recovered most of the qualitative spatial detail that existed in the reference image, there were some simulated voxels that demonstrated larger residual errors than expected following image reconstruction. The source of this residual error is uncertain. Although adding extra center lines for reference data improved the quantitative reconstruction of some pixels, having the extra center lines led to larger errors in other pixels. Part of this error may reflect residual spatial aliasing artifact. It is possible that even 10 × 10 center sensitivity lines did not contain the required spatial information to obtain all of the required sensitivity reference data. Furthermore, it has been shown that parallel imaging reconstructions alter the voxel shape and point spread function of the reconstructed data (14), which may have a significant effect for relatively low resolution spectroscopic images. Finally, the application of alternative reconstruction strategies, including generalized autocalibrating partially parallel acquisitions (GRAPPA) (4), may improve the quantitative reconstruction results. Future work will be devoted to exploring these possibilities.
In vivo results were shown through the kidneys of normal rats. While the in vivo results were more difficult than the numerical simulations to assess quantitatively, they have similar qualitative features. Future experiments will examine qualitative and quantitative performance for pathologic states such as tumors.
The spectroscopic parallel imaging reconstruction described in this work was an extension of the technique described in Ref. (10), where a self-calibrating generalized encoding matrix reconstruction was combined with a partial Fourier acquisition to achieve shorter imaging times than with either technique alone. Several modifications were required in order to apply this technique to spectroscopic imaging. Other methods (11–13) have been described for integrating partial Fourier acquisition with parallel imaging. Potentially, these could be extended to spectroscopy as well and compared to the technique presented here.
In this study a signal-free region at the edge of the image was used to measure the noise correlation matrix, which can potentially have nonzero signal due to Gibbs ringing and other artifacts. A separate noise reference scan (with no RF excitation) can potentially be used for noise measurement to avoid these artifacts.
Spectroscopic acquisitions, particularly half-echo or free induction decay approaches, such as those shown here, generally have a phase offset that varies strongly with spectral frequency and with position. This phase variation might be anticipated to be a concern when performing partial Fourier reconstructions. The theoretical and experimental evidence presented here suggests that this additional phase variation is not a concern as long as each spectral frequency is reconstructed independently, with its own autocalibrated sensitivity reference. Additionally, the combined use of temporal and spatial partial Fourier data is not without sacrifice: the spectrum that is reconstructed is the magnitude spectrum with its associated spectral broadening. It is no longer possible to rephase the spectrum and eliminate the dispersive component. This sacrifice is minimal, however, when imaging carbon, due to the large peak separation when compared to proton imaging.
As mentioned above, this technique uses “in vivo” coil sensitivities that are extracted from autocalibrating lines at the center of κ-space. One sacrifice that is made by using this type of in vivo reference is that the final images remain modulated by the sum-of-squares coil sensitivity profile. This may become a more significant problem in imaging larger samples, where greater spatial variation of coil sensitivities is expected. This limitation will occur with any parallel imaging technique that does not include an explicit measurement of the underlying component coil sensitivities. In proton imaging, the coil sensitivities can be explicitly measured by using an image from the body proton coil to normalize the acquired coil data. This approach is not available with carbon imaging. Measurement of explicit coil sensitivities in the carbon imaging experiment remains a challenge.
It is well known that 2D acceleration along two different phase encode directions can be advantageous when acquiring 3D data, taking advantage of multiple directions with coil sensitivity variation. The experiments described here are restricted to one dimension primarily because the experimental subject (a rat) did not have enough tissue filling the anterior–posterior direction to justify undersampling in that direction. However, this would be applicable for future human studies.
In these experiments the phase encode direction was placed along the long axis of the animal. For applications focused on imaging small animals, such as the rats used in this study, it would be more efficient to place the phase encode directions along the short axes (anterior–posterior and left–right in this case) and place the frequency encode direction along the long axis. However, we mainly intended these experiments as a feasibility study for larger matrix sizes that might be seen in human imaging. Therefore, we chose to keep the phase encode direction along the longer axis of the animal.
Other strategies have been proposed for accelerating the acquisition of hyperpolarized 13C images, including the development of new reconstruction based on compressed sensing (19). The maximum acceleration factor demonstrated here (3.75) is on the same order of magnitude as that found in Ref. (19). At the same time, parallel MRI and compressed sensing have definite synergy, which will almost certainly lead to higher levels of acceleration in the future.
In conclusion, while further development is necessary, the combination of partial Fourier and parallel imaging acquisition will play an important role allowing future human breath-held acquisitions of hyperpolarized 13C spectroscopic imaging.
ACKNOWLEDGMENT
We thank Kristen Scott for technical assistance.
Contract grant sponsor: National Institutes of Health (NIH); Contract grant numbers: P41EB013598, R00-EB012064; Contract grant sponsor: UC Discovery matching grant (ITLbio 178688) with GE Healthcare.
REFERENCES
- 1.Ardenkjaer-Larsen JH, Fridlund B, Gram A, et al. Increase in signal-to-noise ratio of > 10,000 times in liquid-state NMR. Proc Natl Acad Sci U S A. 2003;100:10158–10163. doi: 10.1073/pnas.1733835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kurhanewicz J, Vigneron DB, Brindle K, et al. Analysis of cancer metabolism by imaging hyperpolarized nuclei: prospects for translation to clinical research. Neoplasia. 2011;13:81–97. doi: 10.1593/neo.101102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sodickson DK, Manning WJ. Simultaneous acquisition of spatial harmonics (SMASH): fast imaging with radiofrequency coil arrays. Magn Reson Med. 1997;38:591–603. doi: 10.1002/mrm.1910380414. [DOI] [PubMed] [Google Scholar]
- 4.Griswold MA, Jakob PM, Heidemann RM, et al. Generalized auto-calibrating partially parallel acquisitions (GRAPPA). Magn Reson Med. 2002;47:1202–1210. doi: 10.1002/mrm.10171. [DOI] [PubMed] [Google Scholar]
- 5.Pruessmann KP, Weiger M, Scheidegger MB, et al. SENSE: sensitivity encoding for fast MRI. Magn Reson Med. 1999;42:952–962. [PubMed] [Google Scholar]
- 6.Tropp J, Lupo JM, Chen A, et al. Multi-channel metabolic imaging, with SENSE reconstruction, of hyperpolarized [1-(13)C] pyruvate in a live rat at 3.0 Tesla on a clinical MR scanner. J Magn Reson. 2011;208:171–177. doi: 10.1016/j.jmr.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jakob PM, Griswold MA, Edelman RR, et al. AUTO-SMASH: a self-calibrating technique for SMASH imaging. SiMultaneous Acquisition of Spatial Harmonics. Magma. 1998;7:42–54. doi: 10.1007/BF02592256. [DOI] [PubMed] [Google Scholar]
- 8.McKenzie CA, Yeh EN, Ohliger MA, et al. Self-calibrating parallel imaging with automatic coil sensitivity extraction. Magn Reson Med. 2002;47:529–538. doi: 10.1002/mrm.10087. [DOI] [PubMed] [Google Scholar]
- 9.Arunachalam A, Whitt D, Fish K, et al. Accelerated spectroscopic imaging of hyperpolarized C-13 pyruvate using SENSE parallel imaging. NMR Biomed. 2009;22:867–873. doi: 10.1002/nbm.1401. [DOI] [PubMed] [Google Scholar]
- 10.Willig-Onwuachi JD, Yeh EN, et al. Phase-constrained parallel MR image reconstruction. J Magn Reson. 2005;176:187–198. doi: 10.1016/j.jmr.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 11.Bydder M, Perthen JE, Du J. Optimization of sensitivity encoding with arbitrary k-space trajectories. Magn Reson Imaging. 2007;25:1123–1129. doi: 10.1016/j.mri.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 12.Lew C, Pineda AR, Clayton D, et al. SENSE phase-constrained magnitude reconstruction with iterative phase refinement. Magn Reson Med. 2007;58:910–921. doi: 10.1002/mrm.21284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blaimer M, Gutberlet M, Kellman P, et al. Virtual coil concept for improved parallel MRI employing conjugate symmetric signals. Magn Reson Med. 2009;61:93–102. doi: 10.1002/mrm.21652. [DOI] [PubMed] [Google Scholar]
- 14.Sodickson DK, McKenzie CA. A generalized approach to parallel magnetic resonance imaging. Med Phys. 2001;8:1629–1643. doi: 10.1118/1.1386778. [DOI] [PubMed] [Google Scholar]
- 15.Roemer PB, Edelstein WA, Hayes CE, et al. The NMR phased array. Magn Reson Med. 1990;16:192–225. doi: 10.1002/mrm.1910160203. [DOI] [PubMed] [Google Scholar]
- 16.Hu S, Chen AP, Zierhut ML, et al. In vivo carbon-13 dynamic MRS and MRSI of normal and fasted rat liver with hyperpolarized 13C-pyruvate. Mol Imaging Biol. 2009;11:399–407. doi: 10.1007/s11307-009-0218-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cunningham CH, Vigneron DB, Chen AP, et al. Design of flyback echo-planar readout gradients for magnetic resonance spectroscopic imaging. Magn Reson Med. 2005;54:1286–1289. doi: 10.1002/mrm.20663. [DOI] [PubMed] [Google Scholar]
- 18.Zhao L, Mulkern R, Tseng C, et al. Gradient-echo imaging considerations for hyperpolarized 129Xe MR. J Magn Reson Ser B. 1996;113:179–183. [PubMed] [Google Scholar]
- 19.Hu S, Lustig M, Balakrishnan A, et al. 3D compressed sensing for highly accelerated hyperpolarized (13)C MRSI with in vivo applications to transgenic mouse models of cancer. Magn Reson Med. 2010;63:312–321. doi: 10.1002/mrm.22233. [DOI] [PMC free article] [PubMed] [Google Scholar]