Abstract
Quorum sensing is an intercellular form of communication that bacteria use to coordinate group behaviors such as biofilm formation and the production of antibiotics and virulence factors. The term quorum sensing was originally coined to describe the mechanism underlying the onset of luminescence production in cultures of the marine bacterium Vibrio fischeri. Luminescence and, more generally, quorum sensing are important for V. fischeri to form a mutualistic symbiosis with the Hawaiian bobtail squid, Euprymna scolopes. The symbiosis is established when V. fischeri cells migrate via flagella-based motility from the surrounding seawater into a specialized structure injuvenile squid called the light organ. The cells grow to high cell densities within the light organ where the infection persists over the lifetime of the animal. A hallmark of a successful symbiosis is the luminescence produced by V. fischeri that camouflages the squid at night by eliminating its shadow within the water column. While the regulatory networks governing quorum sensing are critical for properly regulating V. fischeri luminescence within the squid light organ, they also regulate luminescence-independent processes during symbiosis. In this review, we discuss the quorum-sensing network of V. fischeri and highlight its impact at various stages during host colonization.
Keywords: quorum sensing, symbiosis, Vibrio fischeri, Euprymna scolopes, luminescence, motility
1. Introduction
1.1. Quorum Sensing in Vibrio fischeri
Quorum sensing (QS) describes the mechanism of intercellular communication by which bacteria can alter group behavior in accordance with population density [1]. This process depends on the synthesis and diffusion of signaling molecules, called autoinducers, into the surrounding environment. Upon reaching a threshold concentration, autoinducers will trigger cellular responses, typically by altering gene expression across the entire population. QS controls a wide variety of processes in bacteria including bioluminescence production, sporulation, competence, biofilm formation and the synthesis of antibiotics and virulence factors [2,3].
QS was first discovered in the Gram-negative, marine bacterium Vibrio fischeri as the mechanism that controls the induction of luminescence within growing cultures [4]. V. fischeri belongs to the family Vibrionaceae, which is comprised of many bacterial species that are commonly found in fresh and marine water habitats. Vibrio fischeri was originally described as a member of the genus Vibrio. More recently, however, Vibrio fischeri, along with Vibrio logei, Vibrio salmonicida, and Vibrio wodanis, has been placed in a new genus called Aliivibrio, as these species form a monophyletic clade that can be differentiated based on phenotypic traits and biochemical tests from the other members of the genus [5]. Because the majority of studies focusing on QS and host-microbe interactions use the Vibrio nomenclature, we will continue its use in this review.
The proteins required for luminescence in V. fischeri are encoded within the luxICDABEG operon [6,7]. Light is released during the oxidation reaction by the enzyme luciferase, which is comprised of two subunits, α and β, encoded by luxA and luxB, respectively. The enzymatic reaction converts a long-chain fatty acid (RCHO), reduced flavin mononucleotide (FMNH2), and O2 into aliphatic acid (RCOOH) and FMN. A reductase complex composed of the proteins encoded by luxCDE synthesizes the substrate RCHO [8]. LuxG converts FMN to FMNH2 [9].
Multiple QS systems control luminescence in V. fischeri (Figure 1). Directly regulating expression of the luxICDABEG operon is the LuxI-LuxR QS system. LuxI synthesizes the autoinducer N-3-oxohexanoyl-homoserine lactone (3-oxo-C6-HSL) [10] that, at a threshold concentration (100–200 nM), binds and activates the transcription factor LuxR [11]. The LuxR/3-oxo-C6-HSL complex binds as a dimer upstream of the luxICDABEG operon and recruits RNA polymerase to initiate transcription of the operon [12]. Interestingly, while the overall luminescence output of a population of cells increases as the level of 3-oxo-C6-HSL increases, the luminescence levels of individual cells display significant heterogeneity [13]. Whether this cell-cell heterogeneity in luminescence output has a biological role remains unclear at this time.
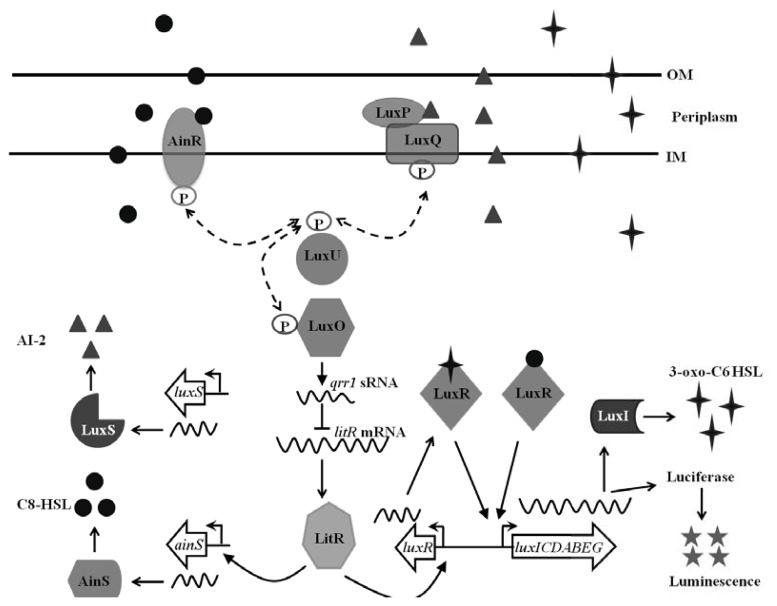
Figure 1.
The QS network of V. fischeri. V. fischeri has three QS systems: LuxI-LuxR, AinS-AinR, and LuxS-LuxP/Q. In the absence of C8-HSL and AI-2 autoinducers, LuxO is phosphorylated by the kinase activities of the histidine kinases AinR and LuxQ. Phosphorylated LuxO activates expression of the sRNA Qrr1, which degrades via Hfq the mRNA of litR, thereby reducing the level of the transcription factor LitR. Accumulation of C8-HSL and AI-2 at high cell density results in decreased phosphorylation of LuxO, which enhances the level of LitR. LitR activates transcription of luxR, which encodes the transcription factor that, when bound by the autoinducer 3-oxo-C6-HSL, directly regulates expression of the luminescence (lux) genes. C8-HSL can also affect luminescence by directly binding to LuxR. The LuxR/C8-HSL complex can activate transcription of the lux genes, although less effectively than the LuxR/3-oxo-C6-HSL complex. In addition to encoding the light-producing enzyme luciferase, the lux operon contains luxI, which encodes the synthase LuxI that synthesizes 3-oxo-C6-HSL. As described in the main text, synthesis of both C8-HSL and 3-oxo-C6-HSL is autoregulated by separate positive feedback loops. OM = outer membrane, IM = inner membrane.
Two additional QS systems, AinS-AinR and LuxS-LuxP/Q, indirectly control luminescence by modulating luxR transcription (Figure 1). AinS synthesizes N-octanoyl-homoserine lactone (C8-HSL), which is an autoinducer detected by the histidine kinase AinR. LuxS synthesizes autoinducer 2 (AI-2), which binds to the periplasmic protein LuxP [14,15]. Based on studies of the homologous system in V. harveyi, LuxP forms a complex with LuxQ, a histidine kinase that dimerizes within the inner membrane [15]. Binding of AI-2 to LuxP induces a rotational shift in the corresponding dimer of LuxQ that inhibits kinase activity. AinR and LuxP/Q act in parallel by controlling the phosphorylation state of LuxU, which serves as an intermediate step within the phosphorelay that dictates the phosphorylation state, and consequently, activity of the response regulator LuxO. Under conditions of low cell density, i.e., in the absence of sufficient levels of autoinducers, the histidine kinases activate the phosphorelay, leading to high levels of phosphorylated LuxO. Phosphorylated LuxO activates transcription of qrr1, which encodes a small regulatory RNA that post-transcriptionally represses the transcription factor LitR via the RNA chaperone Hfq [16]. Under conditions of high cell density, the phosphorelay is reversed, which stabilizes litR transcript. LitR enhances luxR expression, thereby contributing to light production [17].
The QS network of V. fischeri is significantly different than those of other Vibrionaceae members [18]. For instance, the LuxI-LuxR system is exclusively present in the Aliivibrio clade of the Vibrionaceae family, which includes the fish pathogen Aliivibrio salmonicida as well as Vibrio fischeri [19]. In non-Aliivibrio members of Vibrionaceae that are bioluminescent, the luminescence genes are directly regulated by the corresponding LitR homologue instead of a LuxI-LuxR system. As a result, a distinguishing feature of the V. fischeri QS network is that, in contrast to parallel QS networks in V. harveyi and V. cholerae, it is both parallel and hierarchal. In other words, in all Vibrionaceae members, histidine kinases, like AinS-AinR and LuxS-LuxP/Q in V. fischeri, are arranged in parallel within the network and converge on the regulator LitR. The V. fischeri network is also hierarchal because the LuxI-R system acts downstream of LitR, and therefore depends, in part, on the activities of the parallel QS systems. Another significant difference among the QS systems of different Vibrionaceae members is the number of qrr genes within their genomes. In particular, V. fischeri possesses a single qrr gene encoding Qrr1 that is sufficient to repress litR expression [16], whereas multiple Qrr sRNAs regulate the expression of the LitR homologues in V. harveyi (LuxRVH) and V. cholerae (HapR) [20,21].
In V. fischeri, the QS network also appears to have crosstalk among the different autoinducers: namely, C8-HSL, the autoinducer synthesized by AinS, can also directly bind to LuxR [14,22]. However, the LuxR/C8-HSL complex is not as effective as the LuxR/3-oxo-C6-HSL complex in activating transcription of the lux operon [22,23]. As a result, the two autoinducers show competitive binding to LuxR that alters the transcriptional level of the lux operon and resulting luminescence. At high concentration, C8-HSL acts as a competitive inhibitor and suppresses luminescence; however, the suppression can be overcome by sufficiently high concentrations of 3-oxo-C6-HSL [14,22]. Interestingly, the impact of deleting ainS on luminescence depends on the particular strain of V. fischeri, highlighting the complexity by which C8-HSL affects luminescence [14,22]. The crosstalk by autoinducers does not appear to affect the heterogeneity in the luminescence output observed among individual cells [24].
The QS network of V. fischeri also contains feedback loops that ultimately impact its overall dynamics. For instance, the LuxI-derived autoinducer 3-oxo-C6-HSL enhances its own synthesis as the luxI gene is upregulated by the LuxR/3-oxo-C6-HSL. Interestingly, a positive feedback loop is also involved in QS by C8-HSL, as ainS expression is apparently controlled through LitR [25]. How different architectures of various QS networks can impact the magnitudes and temporal profiles of QS-regulated phenotypes is well documented [2,3]. As described throughout this review, the particular intricacies of the QS network of V. fischeri, which experiences both free-living and symbiotic conditions, has evolved to accommodate such disparate life styles.
Notably, the QS network of V. fischeri regulates processes in addition to bioluminescence. Flagella-based motility is controlled, in part, by the LuxO-Qrr1-LitR pathway, so that motility is enhanced under the conditions of low cell density that repress bioluminescence. Phosphorylation of LuxO, or equivalently expression of qrr1, results in enhanced motility. Epistasis experiments have demonstrated that LitR is the transcription factor that mediates the effect on motility by LuxO and Qrr1 [26]. AinS also regulates via LitR the expression of acs, which encodes acetyl-CoA synthetase and, as a result, can control acetate metabolism [27]. More recently, the LuxP/Q complex was shown to impact biofilm formation by V. fischeri, which is a process mediated by an 18-gene, symbiosis polysaccharide (syp) locus [28]. Interestingly, the mechanism may involve direct phosphotransfer between LuxU and the response regulator SypG, although this has yet to be shown.
1.2. The Euprymna scolopes-Vibrio fischeri Symbiosis
Recent studies of the human gut microbiome have highlighted the general importance of beneficial bacteria on animal physiology and development [29]. V. fischeri forms monospecific, beneficial symbioses in many marine animals, including various squid and fish [30,31]. Of these hosts, E. scolopes, a species of bobtail squid found in the offshore waters of the Hawaiian Islands, is the most studied. The symbiosis is highly specific, such that the squid is colonized exclusively by only certain strains of V. fischeri [31,32]. In addition, bacterial transmission is horizontal, i.e., V. fischeri cells are acquired each generation by juveniles from seawater. Because of these features, the symbiosis offers an excellent model to study the establishment, development, and maintenance of a beneficial bacterial infection along an epithelial surface [33]. The nocturnal squid uses bioluminescence emitted by V. fischeri for camouflage by shining light downwards to disrupt its shadow within the water column [34]. To harness this bioluminescence, E. scolopes houses populations of V. fischeri within an organ referred to as the light organ that is located in the middle of the mantle cavity, just inside of the ventral surface of the mantle (Figure 2). The light organ possesses bilateral symmetry and, in juvenile squid, exhibits on each side two appendages that are surrounded by a field of ciliated epithelia [35]. At the base of the appendages on either side of the light organ are three pores, which lead through ciliated ducts to crypt spaces deep within the organ, where bacteria reside extracellularly during the lifetime of the host.
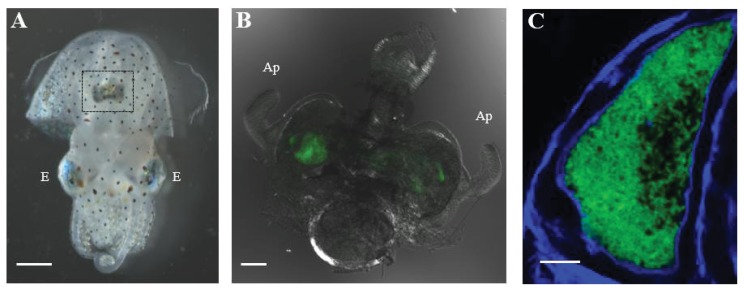
Figure 2.
The light organ of a juvenile E. scolopes harboring V. fischeri. (A) Bright field image showing the ventral side of a juvenile E. scolopes. The dark structure highlighted in the box is the light organ. Scale bar = 1 mm. E = eye; (B) Differential interference contrast (DIC) image of a 48-h p.i. light organ colonized with GFP-labeled V. fischeri cells (green). Scale bar = 100 μm. Ap = appendages; (C) Confocal image of a light organ crypt colonized with GFP-labeled V. fischeri cells (green). Host actin is stained with phalloidin (blue). Scale bar = 10 μm.
The initial process of colonization begins within 2–4 h after juvenile squid hatch from their eggs into seawater containing as little as 1000 V. fischeri CFU/mL [36]. The first event in establishing the symbiosis is the direct interaction of bacterial cells, including V. fischeri, to host cilia that are associated with surface epithelial cells [37,38]. Beating of these cilia collect bacteria in aggregates, which are comprised of up to a few hundred cells within host-derived mucus that is secreted by surface epithelial cells of the light organ [37]. Interestingly, hyper-motile mutants of V. fischeri show a delay in aggregate formation, which highlights the importance of proper regulatory control over motility throughout the colonization process [39]. Although mucus secretion is a general host response to the presence of bacteria [32], the aggregate is dominated by V. fischeri cells [40]. The mechanism by which V. fischeri dominates the aggregate despite its low abundance in seawater is largely unknown. It is unlikely that V. fischeri cells within the aggregate break down the mucus for nutrients more efficiently than others as the cells do not multiply in the aggregate. Chemotaxis towards N-acetylneuraminic acid, a component of squid mucus, suggests that V. fischeri has evolved to respond to host-derived compounds [41]. After 2–4 h within the aggregate, V. fischeri migrates through the mucus towards the pores. Non-motile V. fischeri and bacterial-sized particles do not display this behavior, suggesting that V. fischeri actively participates in the process [37]. Chemotaxis to host-derived compounds does not play a role during these steps, since a cheA transposon-insertion mutant of V. fischeri migrates to the pores in similar fashion to wild-type cells [42].
Once V. fischeri has entered a pore, it travels through ciliated ducts to gain access to deep crypt spaces, where it begins to multiply and establishes a stable association [33]. Within each duct, a gradient of chitin-derived oligosaccharides, which are chemoattractants for V. fischeri, recruit potential symbionts into the crypt spaces [42]. As described above, the effect of this chemotactic gradient seems to be important only for entry into the pores and travelling through the ducts, not for the initial migration through the mucus [42]. Within the deep crypts, V. fischeri grows rapidly for the first 12 h, until the population size reaches about 105 CFU [43]. This growth is concomitant with luminescence, which can be detected as early as 7 h and achieves steady-state levels by 12 h [43].
2. Impact of QS Network on the Euprymna scolopes-Vibrio fischeri Symbiosis
2.1. Impact of LuxI-LuxR Signaling
Luminescence is critical for the Euprymna scolopes-Vibrio fischeri symbiosis to persist. The requirement of bacterial luminescence was first demonstrated using non-luminescent mutants. In particular, mutants containing disruptions in luxI, luxR or luxA, which are as capable as their parental wild-type strain in initiating the symbiosis, have approximately three-fold lower colonization levels (as measured by CFU counts) than wild-type cells at 48 h post-inoculation (p.i.) (Tables 1 and 2) [26,44]. Furthermore, when co-colonized with a wild-type strain at a 1:1 ratio, the luxA mutant was outcompeted both at 24 h and 48 h p.i. (Table 3) [44]. Similar colonization results were obtained using a mutant containing an in-frame deletion of luxCDABEG, which retains an intact LuxR-LuxI signaling system [45]. The inability of the non-luminous V. fischeri strains to persist inside the light organ constitutes a long standing puzzle in the squid-Vibrio symbiosis research. However, recent studies are offering useful insights into the puzzle. Luminescence appears to be a “secret language” of communication between the two partners [46]. Luminescence is the second most important signal, after the presence of the symbiont itself, to induce the host gene expression in the light organ [47]. Furthermore, a recent study showed that luminescence is critical for cyclic expression of a cryptochrome gene that encodes a blue-light receptor, which is known to be involved in circadian clock functions in other animals [48]. As the symbiosis initiates and becomes established, V. fischeri induces several developmental events in the nascent light organ through bacteria-derived signals called microbe associated molecular patterns (MAMPs), which include the lipid A component of LPS and monomers of peptidoglycan (also referred to as tracheal cytotoxin) [49]. Non-luminous strains show defects in inducing many of these developmental events [44,46,50]. Although these studies illustrate the major role of luminescence in host development, the mechanism by which the squid responds to luminescence is not yet known. Recent evidence of the host’s ability to detect the luminescence has led to the speculation that the squid can recognize and specifically eliminate non-luminous cells from the light organ [51]. The down-regulation of a host gene encoding the blood pigment hemocyanin in the light organ when the host is colonized by the luxA mutant [47], suggests that the host may not be able to provide the sufficient oxygen levels for the non-luminous strains that may adversely affect both bacterial growth and light organ development.
Table 1.
Bioluminescence levels emitted by animals colonized by various V. fischeri mutants. The luminescence levels of squid colonized by mutant strains are relative to those of animals colonized by a wild-type V. fischeri strain, which is defined as 100%. The luminescence levels shown in the table for some of the animals are approximate, and we therefore refer readers to the original studies for more details.
| Strain | Luminescence at different stages of symbiosis | References | |||
|---|---|---|---|---|---|
|
| |||||
| 12 h | 24 h | 48 h | 72 h | ||
| ainS | 10%–20% | 10%–20% | 10%–40% | ND | [14,25] |
| luxO | 100% | 100% | 100% | ND | [14] |
| ainS, luxO | 100% | 100% | 100% | ND | [14] |
| luxS | 100% | 100% | ND | ND | [25] |
| luxS, ainS† | 50% | 50% | ND | ND | [25] |
| litR | 100% | 100% | 100% | ND | [17] |
| luxR | BD | BD | ND | ND | [44] |
| luxI | BD | BD | ND | ND | [44] |
| luxA | BD | BD | ND | ND | [44] |
Luminescence is relative to the ainS mutant instead of the wild type strain;
ND = Not Determined; BD = Below Detection.
Table 2.
Bacterial levels in animals colonized by various V. fischeri mutants. The bacterial loads of squid colonized by mutant strains are relative to those of animals colonized by a wild-type V. fischeri strain, which is defined as 100%. The colonization levels shown in the table for some of the animals are approximate, and we therefore refer readers to the original studies for more details.
| Strain | Colonization level at different stages of symbiosis | References | |||
|---|---|---|---|---|---|
|
| |||||
| 12 h | 24 h | 48 h | 72 h | ||
| ainS | 45% | 75% | 40% | 20% | [14,26] |
| luxO | 37% | ND | ND | 100% | [14,26] |
| LuxO(D47E) | 52% | ND | ND | 97% | [26] |
| ainS, luxO | 36% | ND | ND | 100% | [14,26] |
| luxS | 95% | 75% | 90% | ND | [25,26] |
| luxS, ainS | 48% | 50% | 20% | ND | [25,26] |
| litR | 51% | 100% | 100% | ND | [17,26] |
| luxR | 119% | 100% | 25%–35% | ND | [26,44] |
| luxI | 115% | 100% | 25%–35% | ND | [26,44] |
| luxA | ND | 100% | 25%–35% | ND | [44] |
| luxCDABEG | ND | ND | 25%–35% | ND | [45] |
| ainS, luxI | 79% | 75% | 30% | 20% | [14,26] |
ND = Not Determined.
Table 3.
Competition advantage of different V. fischeri mutants in co-colonization experiments.
| Strains in mixed inoculums (1:1 ratio) | Dominant strain at different stages of symbiosis | References | |||
|---|---|---|---|---|---|
|
|
|
||||
| Strain 1 | Strain 2 | 12 h | 24 h | 48 h | |
| ESR1 | luxA | ND | ESR1 | ESR1 | [44] |
| ESR1 | luxR | ND | - | ESR1 | [44] |
| ES114 | litR | ND | ND | litR | [16,17] |
| ES114 | qrr1 | ND | ND | ES114 | [16] |
| ES114 | luxO | ND | ND | ES114 | [16] |
| ES114 | luxO, litR | ND | ND | None | [16] |
| ES114 | qrr1, litR | ND | ND | None | [16] |
| ES114 | ainS | ND | ND | ES114 | [14] |
| ES114 | luxCDABEG | ND | ND | ES114 | [45] |
ND = Not Determined.
While the defects in colonization by the luxA or luxCDABEG mutants are easily attributed to the disruption of the luciferase enzyme, the corresponding colonization defects of the luxI and luxR mutants could be due to luminescence-independent functions, as LuxR also regulates 18 non-lux genes, including several peptidases and proteases [52,53]. Interestingly, QsrP, which is a putative periplasmic protein encoded by a LuxR-regulated gene, is highly abundant in V. fischeri cells associated with adult light organs [54]. Future colonization experiments are required to determine the role of QsrP and other non-lux genes in the context of symbiosis.
2.2. Impact of AinS-AinR Signaling
Luminescence in V. fischeri is thought to be induced sequentially by the multiple QS systems [14]. At intermediate cell density (108–109 cells/mL), the AinS-AinR system induces luminescence 1) by enhancing luxR expression via increased levels of LitR and 2) from the direct binding of C8-HSL to LuxR, which forms a complex that can also activate transcription [55]. Within the light organ, where bacterial cell densities can exceed 1010 cells/mL, luminescence is predominantly induced by LuxI-LuxR signaling. However, the AinS-AinR system does continue to impact light production in situ, as demonstrated by the reduced luminescence the ainS mutant exhibits at 48 h p.i. (Table 1) [14]. Although the reported animal luminescence levels yield large variations, they were routinely less than 40% of those produced by animals colonized by the wild-type strain [14]. Normal animal luminescence levels are restored in a luxO ainS double mutant, showing that the effect of the AinS signal on luminescence in situ occurs through LuxO [14]. Interestingly, these studies revealed that colonization levels of luxI, ainS, and ainS luxI mutants are comparably reduced at 48 h and 72 h relative to wild type (Table 2) suggesting AinS signaling is as important as LuxI signaling for the persistence of symbionts within the light organ [14]. It is tempting to attribute the colonization defect associated with the ainS mutant to the requirement of bacterial luminescence by the host at this stage of symbiosis [44]. However, a luminescence level as low as 10% of that emitted by animals colonized by wild-type cells is sufficient to rescue colonization defects associated with non-luminous strains [44], which suggests AinS also regulates colonization factors that are independent of luminescence. Notably, a litR mutant does not impact colonization levels or animal luminescence at 24 or 48 h p.i., which suggests that these AinS-dependent colonization factors are also independent of LitR [16,17]. It may be possible that the colonization factors are encoded by LuxR-regulated, non-lux genes, and that AinS impacts expression of those genes through the direct binding of C8-HSL to LuxR. However, further investigations are required to discern the luminescence-independent roles of AinS-AinR signaling in persistence.
AinS-AinR signaling also plays a significant role during the initial steps of host colonization that is distinct from its role in persistence (Tables 1 and 2). At 12 h p.i., the colonization levels of the ainS and litR mutants are each two-fold lower than wild-type levels [26]. Beyond 24 h, the litR mutant displays normal colonization levels and, in fact, out-competes wild-type cells, suggesting that the role of AinS-AinR signaling changes as the symbiosis develops [17,26]. Interestingly, deletion of luxO does not restore normal colonization levels in an ainS mutant, which has been observed at 72 p.i. [14,26]. It was hypothesized that an appropriate level of LuxO protein is crucial for the proper functioning of signaling [26]. Alternatively, AinS-AinR signaling may only be important at a certain stage of colonization initiation, and constitutive expression of LitR due to the luxO mutation has a negative impact.
Microarray experiments designed to compare the transcriptome of the ainS mutant with a mutant expressing a constitutively active variant of LuxO (LuxOD47E) revealed multiple genes regulated by the AinS-AinR system that are independent of luminescence [26]. LuxO activates several genes involved in flagella-based motility (e.g., flagellin genes), which is a trait critical for V. fischeri to colonize the light organ [39,56,57]. Non-motile mutants are able to form normal aggregate outside of the light organ, but do not migrate into the light organ [58]. Interestingly, hypermotile mutants exhibit colonization defects during both initiation and persistence phases of colonization [39]. More specifically, hypermotile mutants show a delay in both aggregation and migration behaviors important for colonization initiation. Moreover, they colonize the light organ at 10%–50% of wild-type levels at 12 and 24 p.i., (Table 4) demonstrating that the proper regulation of genes involved in motility remains important after the initial colonization event. This general persistence defect associated with hypermotile strains suggests that proper regulation of motility is critical for symbiosis.
Table 4.
Motility-associated phenotypes, aggregation behaviors, and colonization levels of various V. fischeri mutants.
| Strain | Motility behavior on agar | Flagellation | Aggregation behavior | Colonization level at different stages of symbiosis | Reference | |||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| 0.3%–0.7% | 0.25% | 12 h | 24 h | 48 h | ||||
| ES114 | Motile | Motile | Normal flagella | Normal | 100% | 100% | 100% | [56] |
| N210 | Non-motile | ND | Normal flagella | ND | ND | BD | ND | [56] |
| NF201 | Non-motile | ND | No flagella | ND | ND | BD | ND | [56] |
| NM200 | Non-motile | ND | Abnormal flagella | ND | ND | BD | ND | [56] |
| DM66 | Hypermotile | ND | Hyper flagellation | Delayed | 50% | 60% | 100% | [39] |
| DM73 | Hypermotile | ND | Hyper flagellation | Delayed | ND | 40% | ND | [39] |
| DM61 | Hypermotile | ND | Hyper flagellation | Delayed | 0.1%–10% | 0.1%–10% | 0.1%–10% | [39] |
| flaA | ND | Less motile | Hypo flagellation | Normal | ND | 20%–25% | ND | [58] |
| flrA | Non-motile | ND | No flagella | Normal | ND | BD | ND | [58] |
| ainS | Motile | Hyper-motile | ND | Normal | [26] | |||
| litR | ND | Hyper-motile | ND | ND | [26] | |||
| luxO | ND | Non-motile | ND | ND | See Table 1 | [26] | ||
| luxR | ND | Motile | ND | ND | [26] | |||
| luxI | Motile | Motile | ND | ND | [26] | |||
ND = Not Determined; BD = Below Detection.
The transcriptome analysis described above also revealed that the expression of additional regulatory proteins is controlled by the AinS-AinR system, which highlights the links between the QS network and other regulatory pathways [26]. For instance, the sigma factor RpoQ, which was shown to be repressed by LuxO in the transcriptome analysis, leads to chitinase activity, thereby linking QS to the ability of V. fischeri to break down chitin [59]. Intriguingly, RpoQ also represses both luminescence and motility suggesting that high levels of RpoQ can counteract the ability of LitR to activate luxR expression.
2.3. Impact of the LuxS/LuxP/Q Signaling
In contrast to a mutation in ainS, mutation of luxS alone does not affect colonization levels at 12, 24, or 48 h p.i. (Table 2) [25,26]. In addition, the level of the ainS luxS double mutant at 12 h p.i. is comparable to the ainS mutant [26]. A synergistic effect from the two signaling pathways only manifests at 24 h p.i., as demonstrated by the reduced colonization level of the double mutant compared to the ainS mutant [25]. Interestingly, the luxS mutant colonizes the host normally when inoculated with wild-type cells [25]. It was hypothesized that the signal secreted by wild-type cells rescues any colonization defect associated with the luxS mutant. In contrast, the defects associated with mutations in either ainS and luxI continue to manifest in competition experiments involving the wild-type strain [14,44].
3. Influence of Abiotic Environment Cues on the QS Network
Wild-type V. fischeri ES114 produces a level of luminescence in culture that is three orders of magnitude lower than that achieved within the light organ [60]. This observation has been attributed in part to differences in the cell density between the light organ and culture environments that impact the level of 3-oxo-C6-HSL. The addition of exogenous 3-oxo-C6-HSL induces culture luminescence to a level comparable to that observed in the light organ [44]. The low level of the 3-oxo-C6-HSL in culture is puzzling considering a positive feedback loop amplifies synthesis of the autoinducer. However, an alternative explanation is that environmental factors other than population density impact the synthesis of autoinducers [61]. For instance, recent studies have suggested conditions of low iron in the host may affect luminescence by regulating components of the QS network [62,63]. In particular, low iron conditions lead to increased luminescence by removing Fur-mediated repression of litR expression [63]. Luminescence of wild-type V. fischeri is enhanced under reduced levels of inorganic phosphate (Pi), and this effect is lost in a phoB mutant [64]. The phoB gene encodes a response regulator that is phosphorylated by the sensor PhoR under low Pi conditions. These results suggest that the effect of Pi levels is mediated by the PhoR/PhoB two-component system [65]. Luminescence is also repressed by oxygen as a result of the two-component system ArcA-ArcB [45,66]. In fact, de-repression by the ArcA-ArcB system leads to an increase in luminescence that depends on the presence of the positive feedback loop associated with luxI [66].
4. Concluding Remarks and Future Perspective
Together, the studies described above highlight the complexity of QS in V. fischeri. The LuxI-LuxR, AinS-AinR, and LuxS-LuxP/Q QS systems form an interconnected signaling network that V. fischeri uses to regulate various factors involved in the squid-Vibrio symbiosis (Figure 3). While regulation of light production by QS has received substantial attention, the control by this regulatory network over many other bacterial activities, including motility, chitinase activity, biofilm formation, and acetate metabolism, is also an interesting direction of research. All three QS systems affect bacterial persistence through their ability to regulate luminescence production. In contrast, only AinS-AinR signaling impacts the initial steps of colonization, primarily through the regulation of motility and other uncharacterized factors by LitR. The QS network has also evolved to respond to various environmental conditions, such as the levels of oxygen, iron, and phosphate. The Euprymna scolopes-Vibrio fischeri symbiosis will continue to elucidate the various functions of quorum sensing in the context of host colonization.
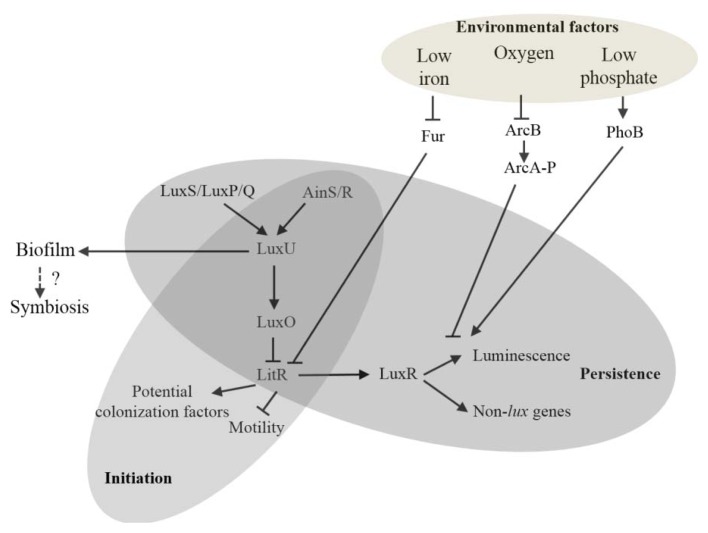
Figure 3.
Role of the QS network of V. fischeri in symbiosis with E. scolopes. The QS network of V. fischeri, which is formed by three interconnected QS systems, regulates the factors that V. fischeri uses to initiate and persist in a mutualistic symbiosis with E. scolopes. Motility and other unknown factors are regulated by the AinS-AinR system and play important roles in the initial steps of host colonization. Luminescence, which is controlled directly by the LuxI-LuxR system and indirectly by the AinS-AinR and LuxS-LuxP/Q systems, is required for bacterial persistence. Various environmental signals integrate into the QS network affecting the luminescence output. The QS network is also linked to other regulatory pathways controlling bacterial activities such as biofilm formation, whose functions are unclear in the symbiosis.
Acknowledgments
We thank the three anonymous reviewers for their constructive criticisms of the manuscript. This work was supported by NIH Grant 4R00GM097032 to T.M.
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Schuster M., Sexton D.J., Diggle S.P., Greenberg E.P. Acyl-homoserine lactone quorum sensing: From evolution to application. Annu. Rev. Microbiol. 2013;67:43–63. doi: 10.1146/annurev-micro-092412-155635. [DOI] [PubMed] [Google Scholar]
- 2.Waters C.M., Bassler B.L. Quorum sensing: Cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 3.Ng W.L., Bassler B.L. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nealson K.H., Platt T., Hastings J.W. Cellular control of the synthesis and activity of the bacterial luminescent system. J. Bacteriol. 1970;104:313–322. doi: 10.1128/jb.104.1.313-322.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Urbanczyk H., Ast J.C., Higgins M.J., Carson J., Dunlap P.V. Reclassification of Vibrio fischeri, Vibrio logei, Vibrio salmonicida and Vibrio wodanis as Aliivibrio fischeri gen. nov., comb. nov., Aliivibrio logei comb. nov., Aliivibrio salmonicida comb. nov. and Aliivibrio wodanis comb. nov. Int. J. Syst. Evol. Microbiol. 2007;57:2823–2829. doi: 10.1099/ijs.0.65081-0. [DOI] [PubMed] [Google Scholar]
- 6.Engebrecht J., Nealson K., Silverman M. Bacterial bioluminescence: Isolation and genetic analysis of functions from Vibrio fischeri. Cell. 1983;32:773–781. doi: 10.1016/0092-8674(83)90063-6. [DOI] [PubMed] [Google Scholar]
- 7.Miyashiro T., Ruby E.G. Shedding light on bioluminescence regulation in Vibrio fischeri. Mol. Microbiol. 2012;84:795–806. doi: 10.1111/j.1365-2958.2012.08065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boylan M., Miyamoto C., Wall L., Graham A., Meighen E. Lux C, D and E genes of the Vibrio fischeri luminescence operon code for the reductase, transferase, and synthetase enzymes involved in aldehyde biosynthesis. Photochem. Photobiol. 1989;49:681–688. doi: 10.1111/j.1751-1097.1989.tb08441.x. [DOI] [PubMed] [Google Scholar]
- 9.Nijvipakul S., Wongratana J., Suadee C., Entsch B., Ballou D.P., Chaiyen P. LuxG is a functioning flavin reductase for bacterial luminescence. J. Bacteriol. 2008;190:1531–1538. doi: 10.1128/JB.01660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schaefer A.L., Val D.L., Hanzelka B.L., Cronan J.E., Jr, Greenberg E.P. Generation of cell-to-cell signals in quorum sensing: Acyl homoserine lactone synthase activity of a purified Vibrio fischeri LuxI protein. Proc. Natl. Acad. Sci. USA. 1996;93:9505–9509. doi: 10.1073/pnas.93.18.9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanzelka B.L., Greenberg E.P. Evidence that the N-terminal region of the Vibrio fischeri LuxR protein constitutes an autoinducer-binding domain. J. Bacteriol. 1995;177:815–817. doi: 10.1128/jb.177.3.815-817.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stevens A.M., Dolan K.M., Greenberg E.P. Synergistic binding of the Vibrio fischeri LuxR transcriptional activator domain and RNA polymerase to the lux promoter region. Proc. Natl. Acad. Sci. USA. 1994;91:12619–12623. doi: 10.1073/pnas.91.26.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perez P.D., Hagen S.J. Heterogeneous response to a quorum-sensing signal in the luminescence of individual Vibrio fischeri. PLoS One. 2010;5:e15473. doi: 10.1371/journal.pone.0015473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lupp C., Urbanowski M., Greenberg E.P., Ruby E.G. The Vibrio fischeri quorum-sensing systems ain and lux sequentially induce luminescence gene expression and are important for persistence in the squid host. Mol. Microbiol. 2003;50:319–331. doi: 10.1046/j.1365-2958.2003.t01-1-03585.x. [DOI] [PubMed] [Google Scholar]
- 15.Neiditch M.B., Federle M.J., Pompeani A. J., Kelly R.C., Swem D.L., Jeffrey P.D., Bassler B.L., Hughson F.M. Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell. 2006;126:1095–1108. doi: 10.1016/j.cell.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyashiro T., Wollenberg M.S., Cao X., Oehlert D., Ruby E.G. A single qrr gene is necessary and sufficient for LuxO-mediated regulation in Vibrio fischeri. Mol. Microbiol. 2010;77:1556–1567. doi: 10.1111/j.1365-2958.2010.07309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fidopiastis P.M., Miyamoto C.M., Jobling M.G., Meighen E.A., Ruby E.G. LitR, a new transcriptional activator in Vibrio fischeri, regulates luminescence and symbiotic light organ colonization. Mol. Microbiol. 2002;45:131–143. doi: 10.1046/j.1365-2958.2002.02996.x. [DOI] [PubMed] [Google Scholar]
- 18.Milton D.L. Quorum sensing in vibrios: Complexity for diversification. Int. J. Med. Microbiol. 2006;296:61–71. doi: 10.1016/j.ijmm.2006.01.044. [DOI] [PubMed] [Google Scholar]
- 19.Nelson E.J., Tunsjo H.S., Fidopiastis P.M., Sorum H., Ruby E.G. A novel lux operon in the cryptically bioluminescent fish pathogen Vibrio salmonicida is associated with virulence. Appl. Environ. Microbiol. 2007;73:1825–1833. doi: 10.1128/AEM.02255-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tu K.C., Bassler B.L. Multiple small RNAs act additively to integrate sensory information and control quorum sensing in Vibrio harveyi. Genes Dev. 2007;21:221–233. doi: 10.1101/gad.1502407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lenz D.H., Mok K.C., Lilley B.N., Kulkarni R.V., Wingreen N.S., Bassler B.L. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 22.Kuo A., Callahan S.M., Dunlap P.V. Modulation of luminescence operon expression by N-octanoyl-L-homoserine lactone in ainS mutants of Vibrio fischeri. J. Bacteriol. 1996;178:971–976. doi: 10.1128/jb.178.4.971-976.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaefer A.L., Hanzelka B.L., Eberhard A., Greenberg E.P. Quorum sensing in Vibrio fischeri: Probing autoinducer-LuxR interactions with autoinducer analogs. J. Bacteriol. 1996;178:2897–2901. doi: 10.1128/jb.178.10.2897-2901.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perez P.D., Weiss J.T., Hagen S.J. Noise and crosstalk in two quorum-sensing inputs of Vibrio fischeri. BMC Syst. Biol. 2011;5 doi: 10.1186/1752-0509-5-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lupp C., Ruby E.G. Vibrio fischeri LuxS and AinS: Comparative study of two signal synthases. J. Bacteriol. 2004;186:3873–3881. doi: 10.1128/JB.186.12.3873-3881.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lupp C., Ruby E.G. Vibrio fischeri uses two quorum-sensing systems for the regulation of early and late colonization factors. J. Bacteriol. 2005;187:3620–3629. doi: 10.1128/JB.187.11.3620-3629.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Studer S.V., Mandel M.J., Ruby E.G. AinS quorum sensing regulates the Vibrio fischeri acetate switch. J. Bacteriol. 2008;190:5915–5923. doi: 10.1128/JB.00148-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ray V.A., Visick K.L. LuxU connects quorum sensing to biofilm formation in Vibrio fischeri. Mol. Microbiol. 2012;86:954–970. doi: 10.1111/mmi.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sommer F., Backhed F. The gut microbiota—Masters of host development and physiology. Nat. Rev. Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 30.Visick K.L., Ruby E.G. Vibrio fischeri and its host: It takes two to tango. Curr. Opin. Microbiol. 2006;9:632–638. doi: 10.1016/j.mib.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 31.Mandel M.J., Wollenberg M.S., Stabb E.V., Visick K.L., Ruby E.G. A single regulatory gene is sufficient to alter bacterial host range. Nature. 2009;458:215–218. doi: 10.1038/nature07660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nyholm S.V., Deplancke B., Gaskins H.R., Apicella M.A., McFall-Ngai M.J. Roles of Vibrio fischeri and nonsymbiotic bacteria in the dynamics of mucus secretion during symbiont colonization of the Euprymna scolopes light organ. Appl. Environ. Microbiol. 2002;68:5113–5122. doi: 10.1128/AEM.68.10.5113-5122.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nyholm S.V., McFall-Ngai M.J. The winnowing: Establishing the squid-vibrio symbiosis. Nat. Rev. Microbiol. 2004;2:632–642. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- 34.Jones B.W., Nishiguchi M.K. Counterillumination in the Hawaiian bobtail squid, Euprymna scolopes Berry (Mollusca: Cephalopoda) Mar. Biol. 2004;144:1151–1155. [Google Scholar]
- 35.Sycuro L.K., Ruby E.G., McFall-Ngai M. Confocal microscopy of the light organ crypts in juvenile Euprymna scolopes reveals their morphological complexity and dynamic function in symbiosis. J. Morphol. 2006;267:555–568. doi: 10.1002/jmor.10422. [DOI] [PubMed] [Google Scholar]
- 36.Lee K.H., Ruby E.G. Detection of the light organ symbiont, Vibrio fischeri, in Hawaiian seawater by using lux gene probes. Appl. Environ. Microbiol. 1992;58:942–947. doi: 10.1128/aem.58.3.942-947.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nyholm S.V., Stabb E.V., Ruby E.G., McFall-Ngai M.J. Establishment of an animal-bacterial association: Recruiting symbiotic vibrios from the environment. Proc. Natl. Acad. Sci. USA. 2000;97:10231–10235. doi: 10.1073/pnas.97.18.10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altura M.A., Heath-Heckman E.A., Gillette A., Kremer N., Krachler A.M., Brennan C., Ruby E.G., Orth K., McFall-Ngai M.J. The first engagement of partners in the Euprymna scolopes-Vibrio fischeri symbiosis is a two-step process initiated by a few environmental symbiont cells. Environ. Microbiol. 2013 doi: 10.1111/1462-2920.12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Millikan D.S., Ruby E.G. Alterations in Vibrio fischeri motility correlate with a delay in symbiosis initiation and are associated with additional symbiotic colonization defects. Appl. Environ. Microbiol. 2002;68:2519–2528. doi: 10.1128/AEM.68.5.2519-2528.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nyholm S.V., McFall-Ngai M.J. Dominance of Vibrio fischeri in secreted mucus outside the light organ of Euprymna scolopes: The first site of symbiont specificity. Appl. Environ. Microbiol. 2003;69:3932–3937. doi: 10.1128/AEM.69.7.3932-3937.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeLoney-Marino C.R., Wolfe A.J., Visick K.L. Chemoattraction of Vibrio fischeri to serine, nucleosides, and N-acetylneuraminic acid, a component of squid light-organ mucus. Appl. Environ. Microbiol. 2003;69:7527–7530. doi: 10.1128/AEM.69.12.7527-7530.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mandel M.J., Schaefer A.L., Brennan C.A., Heath-Heckman E.A., Deloney-Marino C.R., McFall-Ngai M.J., Ruby E.G. Squid-derived chitin oligosaccharides are a chemotactic signal during colonization by Vibrio fischeri. Appl. Environ. Microbiol. 2012;78:4620–4626. doi: 10.1128/AEM.00377-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruby E.G., Asato L.M. Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch. Microbiol. 1993;159:160–167. doi: 10.1007/BF00250277. [DOI] [PubMed] [Google Scholar]
- 44.Visick K.L., Foster J., Doino J., McFall-Ngai M., Ruby E.G. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J. Bacteriol. 2000;182:4578–4586. doi: 10.1128/jb.182.16.4578-4586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bose J.L., Rosenberg C.S., Stabb E.V. Effects of luxCDABEG induction in Vibrio fischeri: Enhancement of symbiotic colonization and conditional attenuation of growth in culture. Arch. Microbiol. 2008;190:169–183. doi: 10.1007/s00203-008-0387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McFall-Ngai M., Heath-Heckman E.A., Gillette A.A., Peyer S.M., Harvie E.A. The secret languages of coevolved symbioses: Insights from the Euprymna scolopes-Vibrio fischeri symbiosis. Semin. Immunol. 2012;24:3–8. doi: 10.1016/j.smim.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chun C.K., Troll J.V., Koroleva I., Brown B., Manzella L., Snir E., Almabrazi H., Scheetz T.E., de Bonaldo M.F., Casavant T.L., et al. Effects of colonization, luminescence, and autoinducer on host transcription during development of the squid-vibrio association. Proc. Natl. Acad. Sci. USA. 2008;105:11323–11328. doi: 10.1073/pnas.0802369105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heath-Heckman E.A., Peyer S.M., Whistler C.A., Apicella M.A., Goldman W.E., McFall-Ngai M.J. Bacterial bioluminescence regulates expression of a host cryptochrome gene in the squid-vibrio symbiosis. MBio. 2013;4 doi: 10.1128/mBio.00167-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koropatnick T.A., Engle J.T., Apicella M.A., Stabb E.V., Goldman W.E., McFall-Ngai M.J. Microbial factor-mediated development in a host-bacterial mutualism. Science. 2004;306:1186–1188. doi: 10.1126/science.1102218. [DOI] [PubMed] [Google Scholar]
- 50.Koropatnick T.A., Kimbell J.R., McFall-Ngai M.J. Responses of host hemocytes during the initiation of the squid-vibrio symbiosis. Biol. Bull. 2007;212:29–39. doi: 10.2307/25066578. [DOI] [PubMed] [Google Scholar]
- 51.Tong D., Rozas N.S., Oakley T.H., Mitchell J., Colley N.J., McFall-Ngai M.J. Evidence for light perception in a bioluminescent organ. Proc. Natl. Acad. Sci. USA. 2009;106:9836–9841. doi: 10.1073/pnas.0904571106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Callahan S.M., Dunlap P.V. LuxR- and acyl-homoserine-lactone-controlled non-lux genes define a quorum-sensing regulon in Vibrio fischeri. J. Bacteriol. 2000;182:2811–2822. doi: 10.1128/jb.182.10.2811-2822.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Antunes L.C., Schaefer A.L., Ferreira R.B., Qin N., Stevens A.M., Ruby E.G., Greenberg E.P. Transcriptome analysis of the Vibrio fischeri LuxR-LuxI regulon. J. Bacteriol. 2007;189:8387–8391. doi: 10.1128/JB.00736-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schleicher T.R., Nyholm S.V. Characterizing the host and symbiont proteomes in the association between the Bobtail squid, Euprymna scolopes, and the bacterium, Vibrio fischeri. PLoS One. 2011;6:e25649. doi: 10.1371/journal.pone.0025649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Egland K.A., Greenberg E.P. Conversion of the Vibrio fischeri transcriptional activator, LuxR, to a repressor. J. Bacteriol. 2000;182:805–811. doi: 10.1128/jb.182.3.805-811.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Graf J., Dunlap P.V., Ruby E.G. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J. Bacteriol. 1994;176:6986–6991. doi: 10.1128/jb.176.22.6986-6991.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Millikan D.S., Ruby E.G. Vibrio fischeri flagellin A is essential for normal motility and for symbiotic competence during initial squid light organ colonization. J. Bacteriol. 2004;186:4315–4325. doi: 10.1128/JB.186.13.4315-4325.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Millikan D.S., Ruby E.G. FlrA, a sigma54-dependent transcriptional activator in Vibrio fischeri, is required for motility and symbiotic light-organ colonization. J. Bacteriol. 2003;185:3547–3557. doi: 10.1128/JB.185.12.3547-3557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cao X., Studer S.V., Wassarman K., Zhang Y., Ruby E.G., Miyashiro T. The novel sigma factor-like regulator RpoQ controls luminescence, chitinase activity, and motility in Vibrio fischeri. MBio. 2012;3 doi: 10.1128/mBio.00285-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boettcher K.J., Ruby E.G. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J. Bacteriol. 1990;172:3701–3706. doi: 10.1128/jb.172.7.3701-3706.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stabb E.V., Schaefer A., Bose J.L., Ruby E.G. Quorum Signaling and Symbiosis in the Marine Luminous Bacterium Vibrio fischeri. In: Winans S.C., Bassler B.L., editors. Chemical Communication among Bacteria. ASM Press; Washington, DC, USA: 2008. [Google Scholar]
- 62.Septer A.N., Lyell N.L., Stabb E.V. The iron-dependent regulator fur controls pheromone signaling systems and luminescence in the squid symbiont Vibrio fischeri ES114. Appl. Environ. Microbiol. 2013;79:1826–1834. doi: 10.1128/AEM.03079-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Septer A.N., Wang Y., Ruby E.G., Stabb E.V., Dunn A.K. The haem-uptake gene cluster in Vibrio fischeri is regulated by Fur and contributes to symbiotic colonization. Environ. Microbiol. 2011;13:2855–2864. doi: 10.1111/j.1462-2920.2011.02558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lyell N.L., Dunn A.K., Bose J.L., Stabb E.V. Bright mutants of Vibrio fischeri ES114 reveal conditions and regulators that control bioluminescence and expression of the lux operon. J. Bacteriol. 2010;192:5103–5114. doi: 10.1128/JB.00524-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Makino K., Shinagawa H., Amemura M., Kawamoto T., Yamada M., Nakata A. Signal transduction in the phosphate regulon of Escherichia coli involves phosphotransfer between PhoR and PhoB proteins. J. Mol. Biol. 1989;210:551–559. doi: 10.1016/0022-2836(89)90131-9. [DOI] [PubMed] [Google Scholar]
- 66.Septer A.N., Stabb E.V. Coordination of the arc regulatory system and pheromone-mediated positive feedback in controlling the Vibrio fischeri lux operon. PLoS One. 2012;7:e49590. doi: 10.1371/journal.pone.0049590. [DOI] [PMC free article] [PubMed] [Google Scholar]