Abstract
The LRRK2 (leucine-rich repeat protein kinase-2) is mutated in a significant number of Parkinson’s disease patients, but little is known about its regulation and function. A common mutation changing Gly2019 to serine enhances catalytic activity, suggesting that small-molecule inhibitors might have utility in treating Parkinson’s disease. We employed various approaches to explore the substrate-specificity requirements of LRRK2 and elaborated a peptide substrate termed Nictide, that had 20-fold lower Km and nearly 2-fold higher Vmax than the widely deployed LRRKtide substrate. We demonstrate that LRRK2 has marked preference for phosphorylating threonine over serine. We also observed that several ROCK (Rho kinase) inhibitors such as Y-27632 and H-1152, suppressed LRRK2 with similar potency to which they inhibited ROCK2. In contrast, GSK429286A, a selective ROCK inhibitor, did not significantly inhibit LRRK2. We also identified a mutant LRRK2[A2016T] that was normally active, but resistant to H-1152 and Y-27632, as well as sunitinib, a structurally unrelated multikinase inhibitor that, in contrast with other compounds, suppresses LRRK2, but not ROCK. We have also developed the first sensitive antibody that enables measurement of endogenous LRRK2 protein levels and kinase activity as well as shRNA (short hairpin RNA) methods to reduce LRRK2 expression. Finally, we describe a pharmacological approach to validate whether substrates are phosphorylated by LRRK2 and use this to provide evidence that LRRK2 may not be rate-limiting for the phosphorylation of the proposed substrate moesin. The findings of the present study will aid with the investigation of LRRK2.
Keywords: leucine-rich repeat protein kinase-2 (LRRK2), moesin, Parkinson’s disease, phosphorylation, Rho kinase (ROCK)
INTRODUCTION
Autosomal dominant point mutations within the gene encoding LRRK2 (leucine-rich repeat protein kinase-2) predispose humans to PD (Parkinson’s disease) [1,2]. Patients with LRRK2 mutations generally develop PD at the normal age of 60–70 years, with clinical appearance and symptoms indistinguishable from idiopathic PD [3]. Mutations in LRRK2 account for 4 % of familial PD and are observed in 1 % of sporadic PD patients [3]. Little is known about how LRRK2 is regulated, what its substrates are and how mutations cause PD.
LRRK2 is a large multi-domain protein kinase of 2527 residues, consisting of leucine-rich repeats (residues 1010–1287), a GTPase domain (residues 1335–1504), a COR [C-terminal of Roc (Ras in complex proteins)] domain (residues 1517–1843), a serine/threonine protein kinase domain (residues 1875–2132) and a WD40 repeat (residues 2231–2276) [4]. Over 40 mutations have thus far been reported which mainly comprise amino acid substitutions [5]. The most frequent mutation comprises an amino acid substitution of the highly conserved Gly2019 located within the subdomain VII DFG (Asp-Phe-Gly) motif of the kinase domain to a serine residue [5]. Several studies have reported that this mutation enhances the protein kinase activity of LRRK2 2–3-fold, suggesting that LRRK2 inhibitors may have utility for the treatment of PD [6]. Other than the non-specific/multi-kinase protein kinase inhibitors staurosporine (IC50 2 nM), K252 (IC50 4 nM) and Su-11248/sunitinib (IC50 15 nM) [7,8], no selective LRRK2 inhibitors have been reported thus far.
We previously undertook a KESTREL (kinase substrate tracking and elucidation) screen in rat brain extracts to identify proteins phosphorylated by the activated PD LRRK2[G2019S] mutant. This led to the observation that moesin, a member of the ERM (ezrin/radixin/moesin) proteins that anchors the actin cytoskeleton to the plasma membrane, is efficiently phosphorylated by LRRK2 at Thr558, a previously identified in vivo phosphorylation site that regulates the ability of moesin to bind actin [9]. LRRK2 also phosphorylated the other ERM proteins, ezrin and radixin, that are related to moesin at the residue equivalent to Thr558, as well as a peptide encompassing Thr558 (LRRKtide) [9]. Previous work had suggested that ROCK (Rho kinase) could also phosphorylate ERM proteins at the residue equivalent to Thr558 of moesin both in vitro and when overexpressed in cells [10–12]. No evidence has been published to demonstrate that LRRK2 phosphorylates ERM proteins in cells.
To aid the functional characterization of LRRK2, we have analysed the substrate specificities of LRRK2 and elaborated the peptide substrate Nictide that has a 20-fold lower Km and nearly 2-fold higher Vmax than the widely deployed LRRKtide substrate [9]. We also observed that some previously reported ROCK inhibitors also inhibited LRRK2 with a similar potency to the inhibition of ROCK. Moreover, we demonstrate that sunitinib can be deployed as a control compound that inhibits LRRK2, but not ROCK, and the selective ROCK inhibitor GSK429286A [13] can be deployed as a compound that inhibits ROCK, but not LRRK2. Modelling studies enabled us to generate an inhibitor-resistant mutant of LRRK2 that is normally active, but 20-fold less sensitive to inhibition by the LRRK2 inhibitors. We also develop the first robust assay that allows the protein kinase activity of endogenous LRRK2 to be quantified and present a pharmacological strategy that can be deployed to validate LRRK2 substrates. The findings of the present study will help with dissecting the regulation and function of LRRK2.
MATERIALS AND METHODS
Antibodies
A GST (glutathione transferase)-fusion protein of amino acids was expressed in bacteria and purified by glutathione–Sepharose chromatography. Following cleavage of the GST tag, LRRK2-(100–500) was used as an immunogen to raise a polyclonal antibody (S348C). Antibodies were affinity-purified from antisera using the LRRK2-(100–500) protein immunogen. Antibody (S374C) against LRRK2 was raised against a peptide immunogen similar to what was described previously [14], encompassing amino acids 2498–2514 [CINLPHEVQNLEKHIEVR with an additional N-terminal cysteine residue for coupling to KLH (keyhole-limpet haemocyanin)]. Antibodies were affinity-purified against the peptide. Anti-moesin (S135C) and anti-ezrin antibodies (S245C) were generated by injection of purified full-length protein into sheep, followed by affinity-purification of the antibody against the antigen. Antibody production in animals was carried out in accordance with U.K. Home Office regulations. Pan-phospho-ERM antibody (S296C) was generated by injection of the KLH-conjugated phosphopeptide CDKYKTpLRQI into sheep and was affinity-purified by positive and negative selection against the phospho- and de-phospho-peptides respectively. Sheep polyclonal antibody (S662B) was raised against MBP (maltose-binding protein)–MYPT (myosin phosphatase target) from chicken (amino acids 714–1004). Rabbit polyclonal antibody against MYPT pThr850 was from Upstate (# 36-003). Anti-GFP (green fluorescent protein) antibody (S268B) was raised against recombinant GFP protein and affinity-purified against the antigen. Antibody (S221B) against ERK (extracellular-signal-regulated kinase) 1/2 was raised against GST–ERK1 protein. Anti-FLAG M2 antibody and affinity matrix were from Sigma (A2220).
LRRK2 and ROCK kinase assays
Peptide kinase assays were set up in a total volume of 40 μl with either recombinant GST–LRRK2-(1326–2527) (0.5 μg, which is ~ 10 % pure [9], thus corresponding to 8 nM LRRK2 in the final kinase assay) or immunoprecipitated LRRK2 or recombinant His6–ROCK2-(2–543) (80 ng, corresponding to 30 nM ROCK2 in the final kinase assay) in 50 mM Tris/HCl (pH 7.5), 0.1 mM EGTA, 10 mM MgCl2 and 0.1 mM [γ-32P]ATP (~ 500–1000 c.p.m./pmol) in the presence of the indicated concentration of peptide substrate. In reactions where kinase inhibitors were assayed, inhibitors were dissolved in DMSO at 0.1 % of the reaction volume. After incubation for 15 min at 30 °C, reactions were terminated by applying 35 μl of the reaction mixture on to P81 phosphocellulose paper and immersion in 50 mM phosphoric acid. After extensive washing, reaction products were quantified by Cerenkov counting. One unit of LRRK2 activity was defined as the amount of enzyme that catalysed the incorporation of 1 nmol of 32P into LRRKtide. Km and Vmax parameters were determined by performing the assay described above using various concentrations of LRRKtide or Nictide. The Km and Vmax parameters were calculated using the GraphPad Prism program. Km and Vmax values are rounded in Figure 1(C) to reflect the need to estimate values due to the nature of the peptide assay, wherein high concentrations of peptide became inhibitory to the kinase. IC50 values were calculated using non-linear regression analysis using GraphPad Prism.
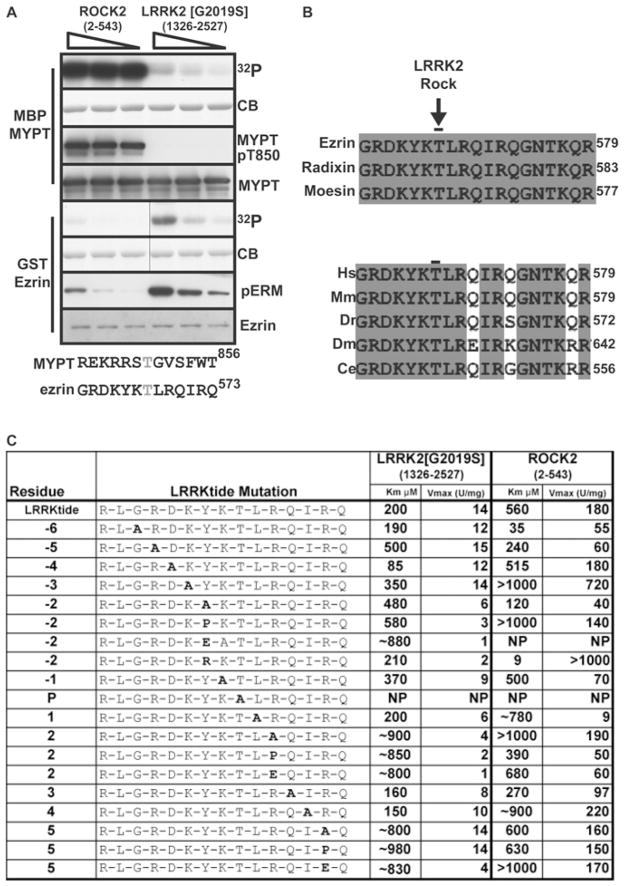
Figure 1. Comparison of ROCK substrates as substrates for LRRK2.
(A) Upper panels: His6–ROCK2-(2–543) and GST–LRRK2[G2019S]-(1326–2527) were diluted to a concentration where they possessed identical activity towards LRRKtide and then incubated with 2 μM MBP–MYPT-(714–1004) or heat-treated GST–ezrin (full-length) [9] in the presence of Mg2+-[γ-32P]ATP. Enzyme was decreased successively 2-fold, as indicated by the decreasing slope of the triangle. Reactions were terminated after 15 min by addition of sample buffer and products were subjected to SDS/PAGE. Gels were analysed by staining with Colloidal Blue (CB) and phosphorylation was monitored by autoradiography. Immunoblotting analysis was also undertaken with the indicated antibodies. Lower panels: comparison of the amino acid sequences surrounding Thr850-MYPT and Thr567-ezrin showing little similarity between peptides. (B) Sequence alignments of the indicated species of ERM proteins surrounding the ROCK/LRRK2 phosphorylation site. Identical residues are shaded. The sequences used were human moesin (GenBank® accession number NP_002435), human ezrin (GenBank® accession number NP_003370) and human radixin (GenBank® accession number NP_002897). Human ezrin (Hs) was also aligned with Mus musculus (Mm) moesin (GenBank® accession number NP_034963), Danio rerio (Dr) radixin (GenBank® accession number NP_001004296), Drosophila melanogaster (Dm) moesin (GenBank® accession number NP_727290) and Caenorhabditis elegans (Ce) ERM-1A (GenBank® accession number NP_491560). (C) Analysis of substrate-recognition determinants in LRRKtide. Residues from −6 to +5 of the LRRKtide substrate (RLGRDKYKTLRQIRQ) were mutated to the residue indicated in bold. These peptides were analysed for their ability to be phosphorylated by GST–LRRK2[G2019S]-(1326–2527) purified from HEK-293 cells or ROCK2-(2–543) purified from baculovirus. NP denotes that the peptide was phosphorylated poorly and that kinetic analysis was not feasible. For Km values above 500 μM, a ~ sign is added to stress that these values were inferred from kinetic analysis undertaken at peptide concentrations of lower than 1 mM. Similar results were obtained in at least two experiments.
For assays using recombinant proteins as substrates, the assays were set up in a total volume of 25 μl with recombinant kinase [12 nM GST–LRRK2-(1326–2527) or 5 nM His6–ROCK2-(2–543)] in 50 mM Tris/HCl (pH 7.5), 0.1 mM EGTA, 10 mM MgCl2 and 0.1 mM [γ-32P]ATP (~ 500 c.p.m./pmol), with substrate at 2 μM. After incubation for 15 min at 30 °C, the reactions were stopped by the addition of Laemmli sample buffer. Reaction products were resolved by electrophoresis on SDS/polyacrylamide gels. The incorporation of phosphate into protein substrates was determined by autoradiography and/or immunoblotting with phosphospecific antibodies.
Immunological procedures
Cell lysates (10–30 μg) were resolved by electrophoresis on SDS/polyacrylamide gels or Novex 4–12 % gradient gels, and electroblotted on to nitrocellulose membranes. Membranes were blocked with 5 % (w/v) dried non-fat skimmed milk powder in TBST [Tris-buffered saline with Tween 20: 50 mM Tris/HCl (pH 7.5), 0.15 M NaCl and 0.1 % Tween 20]. For the anti-phospho-MYPT antibody, primary antibody was used at a concentration of 1 μg/ml, diluted in 5 % BSA in TBST. Anti-phospho-ERM antibody was used at 1 μg/ml in the presence of 10 μg/ml LRRKtide, diluted in 5 % (w/v) dried non-fat skimmed milk powder in TBST. All other antibodies were used at 1 μg/ml in 5 % (w/v) dried non-fat skimmed milk powder in TBST. Detection of immune complexes was performed using horseradish-peroxidase-conjugated secondary antibodies (Pierce) and an enhanced chemiluminescence reagent. For immunoprecipitations, antibody was non-covalently coupled to Protein G–Sepharose at a ratio of 1 μg antibody/μl of beads, or anti-FLAG M2–agarose was utilized. The indicated amount of cell lysate was incubated with a 5 μl bed volume of coupled antibody for 1 h. Immune complexes were washed twice with lysis buffer supplemented with 0.5 M NaCl and twice with buffer A. Precipitates were either used as a source of kinase or analysed immediately by immunoblotting.
Lentivirus shRNA (short hairpin RNA) production and transduction
For knockdown of endogenous mouse LRRK2, MISSION™ shRNA constructs in the lentiviral expression vector pLKO.1-Puro were purchased from Sigma. Clones TRCN0000022655 termed shRNA LRRK2 #1 (5′-CCGGCGCAGCTTTCAGCGATTCTAACTCGAGTTAGAATCGCTGAAAGCTGCGTTTTT-3′) and TRNC0000022654 termed shRNA LRRK2 #2 (5′-CCGGGCTTACTACTTCACGATATTTCTCGAGAAATATCGTGAAGTAGTAAGCTTTTT-3′) were used for experiments. A non-targeting scrambled shRNA construct (5′-CCGGCCTAAGGTTAAGTCGCCCTCGCTCTAGCGAGGGCGACTTAACCTTAGGTTTTT-3′) also in pLKO.1-Puro was used as a control. For the production of lentiviral transduction particles, HEK (human embryonic kidney)-293T cells growing in 10-cm-diameter dishes were transfected with 11 μg of shRNA encoding plasmid along with 7 μg of the packaging plasmid pCMV delta R8.2 and 4 μg of the envelope plasmid pCMV-VSV-G using the polyethyleneimine method. At 24 and 48 h post-transfection, virus-containing medium was collected. Medium from both time points was pooled, centrifuged at 524 g for 5 min, filtered through a 0.2 μm filter, divided into aliquots of 1.5 ml and stored at −80 °C. For lentiviral transduction of RAW 264.7 cells (mouse leukaemic monocyte macrophage cell line), 1 ml of viral supernatant and 5 μg/ml polybrene was added to cells at 50 % confluence in six-well dishes. Virus-containing medium was removed after 24 h and successfully infected cells were selected with the addition of 5 μg/ml puromycin to the medium. Cells were maintained in medium with 5 μg/ml puromycin until being harvested for experiments.
Other methods
The Supplementary Online Data section at http://www.BiochemJ.org/bj/424/bj4240047add.htm contains further detailed information on the materials, general methods, buffers, cell culture, purification of recombinant proteins, the specificity kinase panel, MS methodology, computer analysis and alternative procedure to verify binding of phosphorylated peptides to P81 paper that were employed in the present study.
RESULTS
Comparison of the substrate specificities of LRRK2 and ROCK
We first compared the rates at which recombinant ROCK2-(2–543) and LRRK2-(1326–2527) lacking the N-terminal non-catalytic leucine-rich repeats, phosphorylated ezrin and MYPT (a well-characterized ROCK substrate [15]). Under conditions in which equimolar MYPT and ezrin were present, ROCK2 phosphorylated MYPT, but barely ezrin (Figure 1A). In contrast, LRRK2 phosphorylated ezrin, but did not phosphorylate MYPT (Figure 1A). Comparison of residues surrounding Thr850 (the major ROCK phosphorylation site on MYPT [16]) and Thr567 (LRRK2 phosphorylation site on ezrin [9]) revealed that, overall, these peptides were dissimilar with only the −3 position (arginine in MYPT/lysine in ezrin) possessing homology (Figure 1A, lower panel).
The human sequence surrounding the LRRK2 phosphorylation site of ezrin is identical in moesin and radixin, and also strikingly conserved in Drosophila and Caenorhabditis elegans ERM homologues (Figure 1B, lower panel). To investigate the substrate-specificity determinants of LRRK2, we tested how mutation of different residues affected the kinetics of LRRK2 phosphorylation of the LRRKtide peptide that encompasses the Thr567 ERM phosphorylation motif (Figure 1C). The wild-type LRRKtide peptide was phosphorylated by LRRK2 with a Km of 200 μM and Vmax of 14 units/mg. The following mutations to alanine suppressed phosphorylation by increasing the Km value: Arg−5 (2.5-fold), Tyr−2 (2.4-fold), Arg+2 (4.5-fold) and Arg+5 (4-fold). Mutation of Tyr−2 to glutamic acid increased the Km 4.4-fold, whereas its mutation to arginine slightly decreased the Km, suggesting that an aromatic residue at this position is not essential (Figure 1C). Mutation of the + 2 and + 5 residues of the peptide to proline or glutamic acid increased the Km similarly to the alanine mutation. Only mutation of Asp−4 to alanine moderately enhanced peptide phosphorylation by decreasing the Km 2.3-fold (Figure 1C). Several mutations also markedly decreased Vmax values, which included Tyr−2 (2–10-fold), Leu+1 (2-fold), Arg+2 (4–28-fold) and Arg+5 (to glutamic acid, 4-fold). Binding of phosphopeptides to P81 paper was used as our assay to isolate and quantify phosphorylation of peptides (see the Materials and methods section). In order to verify that substitution of residues in LRRKtide did not affect binding to P81 paper, seven different phosphorylated peptides were purified by HPLC and demonstrated to interact with similarly high efficiency to P81 paper (Supplementary Figure S1 at http://www.BiochemJ.org/bj/424/bj4240047add.htm).
We also investigated how mutations in LRRKtide affected phosphorylation by ROCK2. In contrast with LRRK2, we observed that several mutations substantially improved peptide phosphorylation by decreasing the Km value. The most dramatic change involved mutation of Tyr−2 to an arginine residue that is found in most ROCK substrates. This decreased the Km value over 60-fold and increased the Vmax over 5-fold (Figure 1C). Mutation of Leu+1 to alanine abolished phosphorylation by ROCK2, but had no effect on LRRK2 phosphorylation (Figure 1C).
Elaboration of an optimal LRRK2 peptide substrate
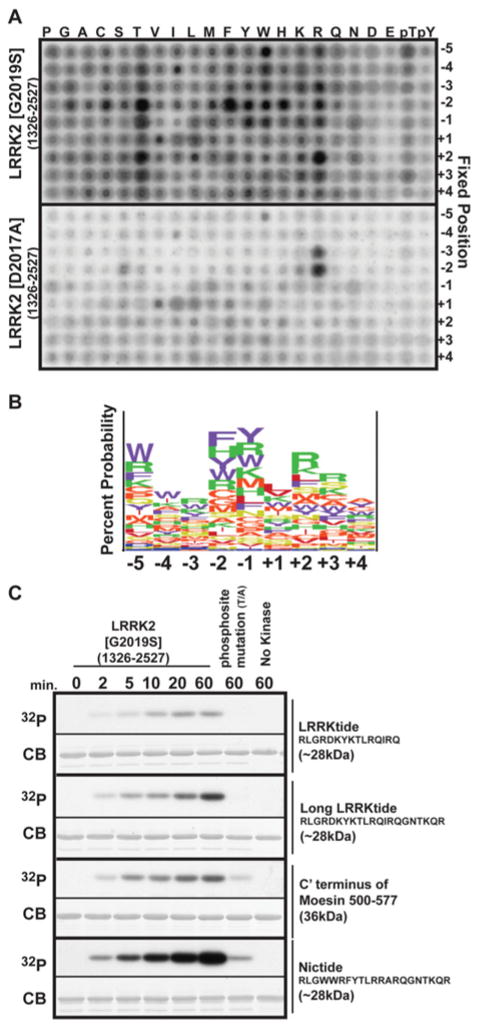
To investigate further and improve the optimal phosphorylation motif for LRRK2, we utilized a positional scanning peptide library approach [17,18]. This assay utilizes 198 biotinylated peptide libraries. Each library contains a 1:1 mixture of serine and threonine at the central position and one additional position fixed to one of the 20 amino acids, phosphothreonine or phosphotyrosine. Phosphothreonine and phosphotyrosine were included to allow identification of kinases that possess a requirement for priming phosphorylation events. All other positions contain an equimolar degenerate mixture of natural amino acids (except serine, threonine and cysteine). Recombinant LRRK2[G2019S] or kinase-inactive LRRK2[D2017A] was used to phosphorylate all 198 peptide libraries simultaneously in solution using [γ-32P]ATP, and biotinylated peptides were captured on a streptavidin-coated membrane. The relative preference for each amino acid at each position was determined by quantifying 32P-radioactivity incorporation following phosphoimaging (Figure 2A). The quantitative results of the LRRK2-[G2019S] screen were also input as a matrix into the enoLOGOS program [19], and the relative preferences for each amino acid is displayed in Figure 2(B). We found that LRRK2 exhibited preferred sequence specificity at multiple positions relative to the phosphorylation site, with strong preferences for −5 (tryptophan and arginine), −2 (phenylalanine, tyrosine, histidine and threonine), −1 (tyrosine, arginine and tryptophan), +2 (arginine and threonine) and +3 (arginine) positions. This is consistent with the kinetic studies shown in Figure 1, demonstrating that mutation of these residues increased the Km and, in some cases, also decreased the Vmax values. An aspartic acid or glutamic acid residue at any position with the peptide reduced LRRK2 phosphorylation (Figure 2A). For experiments undertaken with kinase-inactive LRRK2[D2017A], vastly lower overall levels of phosphorylation were observed, but nevertheless some contaminant kinase activity with preference for arginine residues at the −3 and −2 positions was still found. Similar results were also reported in previous studies employing recombinant kinase-inactive GST–IκB (inhibitor of nuclear factor κB) kinase-β derived from HEK-293 cells [20]. This trace level of protein kinase activity probably results from protein kinases that contaminate the GST-purified kinase from HEK-293 cell extracts.
Figure 2. Determination of the preferred substrate-phosphorylation sequence for LRRK2.
(A) Recombinant HEK-293-purified GST–LRRK2[G2019S]-(1326–2527) and catalytically inactive GST–LRRK2[D2017A]-(1326–2527) was used to screen a positional scanning peptide library consisting of 189 biotinylated peptide libraries in individual kinase assays. Reaction products were bound to streptavidin-coated membrane and, after washing, phosphorylation was visualized by phosphoimaging. (B) Logo plot of the LRRK2 phosphorylation site was derived from empirical data from (A) inputted into enoLOGOS. The height of the stack of single amino acid letters indicates the entropy of the site, and the size of each letter indicates its preference at the position relative to the phosphorylation site between −5 and +4. The largest letters at each position in the logo were chosen to substitute for residues in a longer version of the LRRKtide substrate peptide to derive Nictide, shown below the logo. (C) GST-fusion proteins with the indicated peptide sequences of LRRKtide, the longer LRRKtide, the C-terminus of moesin-(500–577) and the Nictide substrates were subjected to phosphorylation by HEK-293-purified LRRK2[G2019S]-(1326–2527). Reactions were stopped by the addition of sample buffer, and products were subjected to SDS/PAGE. Gels were analysed by staining with Colloidal Blue (CB), and phosphorylation was monitored by autoradiography (32P). Similar results were obtained in replicate experiments.
Elaboration of Nictide LRRK2 substrate
The data from the positional scanning peptide library indicated that the optimal LRRK2 phosphorylation motif between −5 and +4 positions is WWRFYTLRRA. In order to generate an improved substrate for LRRK2, we substituted this motif into the moesin sequence, from which the LRRK2tide peptide was derived. Since sequences as distant as the +5 residues affected kinetics of LRRKtide phosphorylation (Figure 1C) and the LRRKtide peptide terminated at the +6 position, we decided to incorporate the WWRFYTLRRA motif into a longer variant of the LRRKtide peptide encompassing a further six residues of moesin. The resulting sequence, RLGWWRFYTLRRARQGNTKQR, was termed Nictide (reflecting the names of the first two authors of this study). We first compared the phosphorylation by LRRK2[G2019S] of GST fused to the original LRRKtide sequence, the longer version of LRRKtide, the entire C-terminus of moesin (residues 500–577) as well as Nictide. This revealed that GST–Nictide was phosphorylated to a significantly greater extent by LRRK2 than the other GST-fusion proteins (Figure 2C). Mutation of the threonine residue predicted to comprise the LRRK2 phosphorylation site, virtually abolished phosphorylation of the GST-fusion proteins. Our results also demonstrate that the expanded LRRKtide sequence was more efficiently phosphorylated by LRRK2 than the original shorter variant (Figure 2C).
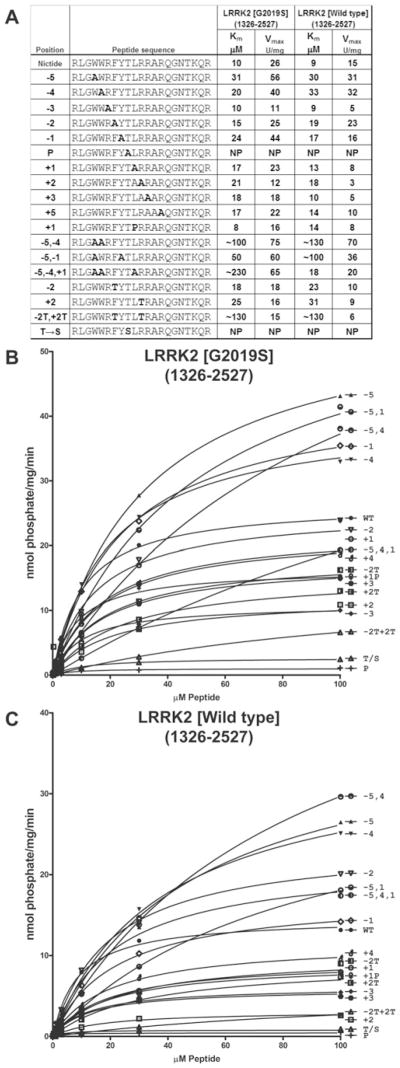
We next generated the synthetic Nictide peptide and found that it was phosphorylated by LRRK2[G2019S] with a Km of 10 μM (20-fold lower than LRRKtide) and a Vmax of 26 units/mg (1.7-fold higher than LRRKtide). Nictide was phosphorylated by wild-type LRRK2 with a similar Km, but lower Vmax, consistent with previous work showing that the G2019S mutation stimulates LRRK2 activity [6]. We also studied the effects of mutating individual residues of Nictide on phosphorylation by LRRK2[G2019S] as well as wild-type LRRK2 (Figure 3). The mutations affected wild-type LRRK2 and LRRK2[G2019S] similarly. Most mutations moderately affected Km values, with the largest effect being the Trp−5 to alanine mutation increasing the Km value 3-fold. Other mutations decreased the Vmax value of phosphorylation 2–5-fold (Trp−4 to alanine and Arg+2 to alanine) (Figure 3). Several mutations (Trp−5 to alanine, Trp−4 to alanine and Tyr−1 to alanine) increased Km values 2–3-fold, but enhanced Vmax of LRRK2 phosphorylation ~ 2-fold. We also combined the −5, −4 and −1 mutations that enhanced Vmax and found that, although high Vmax values were maintained, the Km values were substantially increased 5–20-fold compared with Nictide. Interestingly, mutation of the threonine residue phosphorylated by LRRK2 to serine abolished phosphorylation of the peptide by LRRK2 (Figure 3). Surprisingly, mutation of Leu+1 to a proline residue, which would inhibit phosphorylation of most substrates by non-CMGC proline-directed kinases, only decreased the Vmax of LRRK2 phosphorylation under 2-fold without affecting the Km. This suggests that LRRK2, despite not belonging to the CMGC kinase family does have the potential to phosphorylate serine/threonine residues followed by a proline residue. The positional scanning peptide library data also indicated that there could be a preference for a threonine residue at the −2 and +2 positions (Figure 2A). As similar preference for threonine at −2 and +2 positions has also been observed in other kinase scanning peptide library screens [21], we decided to introduce a threonine residue at either −2 or +2 positions and found that this moderately increased the Km and reduced the Vmax value (Figure 3). Introduction of threonine residues at both the −2 and +2 positions increased the Km value over 10-fold, suggesting that threonine at these positions is not well tolerated. To verify that substitution of residues in Nictide did not affect binding to P81 paper, we purified five different phosphorylated peptides by HPLC and demonstrated that they all interacted with similar high efficiency to P81 paper (see Supplementary Figure S1).
Figure 3. Kinetic analysis of the Nictide substrate.
(A) Residues from −5 to +5 of the Nictide substrate (RLGWWRFYTLRRARQGNTKQR) were mutated to the residue indicated in bold. These peptides were analysed for their ability to be phosphorylated by GST–LRRK2[G2019S]-(1326–2527) or GST–LRRK2[wild-type]-(1326–2527) purified from HEK-293 cells, and Km and Vmax values were derived by non-linear regression analysis as described in the Materials and methods section. NP denotes that the peptide was phosphorylated poorly and that kinetic analysis was not feasible. For Km values above 500 μM, a ~ sign is added to stress that these values were inferred from kinetic analysis undertaken at peptide concentrations of lower than 1 mM. Similar results were obtained in at least three experiments. (B) Average values from a representative experiment from which data in (A) were derived for GST–LRRK2[G2019S]-(1326–2527). (C) As in (B) except with GST–LRRK2[wild-type]-(1326–2527).
We also compared how other LRRK2 PD-associated mutations phosphorylated Nictide compared with LRRKtide. Using both peptides, we found similar results to those we reported previously [9], namely 1.5–2-fold elevated activity for LRRK2[G2019S] and 2–4-fold decreased activity for LRRK2[I2012T], LRRK2[I2020T] and LRRK2[G2385R] (see Supplementary Figure S2 at http://www.BiochemJ.org/bj/424/bj4240047add.htm).
Identification of selective small-molecule tool inhibitors of LRRK2
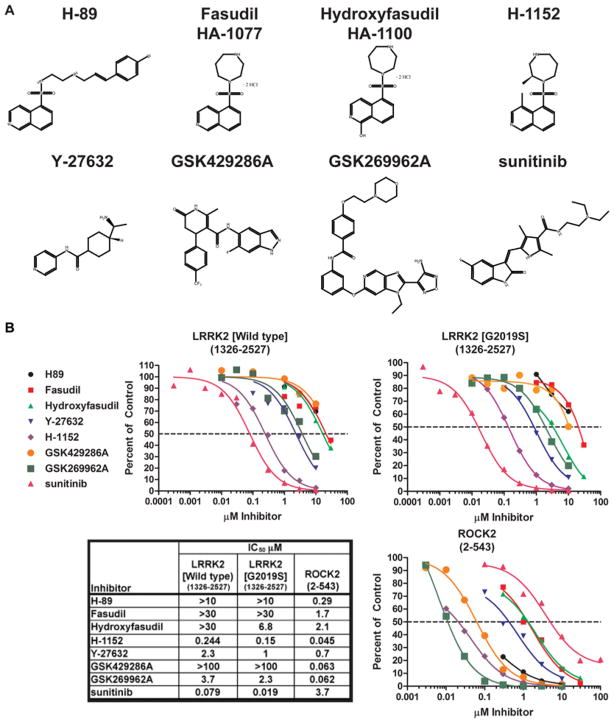
In the course of comparing the substrate specificity of LRRK2[G2019S] and ROCK2, we observed that several commonly deployed ROCK inhibitors (Y-27632 [22], hydroxyfasudil [23] and H-1152 [24]) also inhibited LRRK2[G2019S] (Figure 4). Y-27632 inhibited LRRK2[G2019S] (IC50 1 μM) with a similar potency to ROCK2 (IC50 0.7 μM). LRRK2[G2019S] was inhibited by hydroxyfasudil (IC50 6.8 μM) and H-1152 (IC50 0.15 μM) ~ 3-fold more weakly than ROCK (Figure 4B). H-1152 and hydroxyfasudil belong to a well-studied series of isoquinolinesulfonamides [25], whereas Y-27632 is structurally unrelated (Figure 4A). Other isoquinolinesulfonamides reported to inhibit ROCK, namely H-89 [26] and fasudil [27], only inhibited LRRK2[G2019S] weakly (Figure 4B). In contrast, the dihydropyridone indazole amide ROCK inhibitor GSK429286A [13] and the aminofurazan ROCK inhibitor GSK269962A [28] were observed to suppress ROCK (IC50 ~ 60 nM), but not LRRK2 (IC50 > 1000 nM). We confirmed that the structurally unrelated drug sunitinib inhibited LRRK2 with a similar potency (IC50 ~ 19 nM) to previous reports [7,8], but, in contrast with Y-27632 and H-1152, sunitinib inhibited ROCK only weakly (IC50 ~ 3700 nM). Comparing the potency of H-1152, Y-27632 and sunitinib for LRRK2[G2019S] and wild-type LRRK2, we observed that wild-type LRRK2 was moderately less sensitive to these compounds than the activated mutant. The IC50 of inhibition of wild-type LRRK2 was increased 2-fold for H-1152 and Y-27632 and 4-fold for sunitinib (Figure 4B).
Figure 4. Characterization of ROCK inhibitors as LRRK2 inhibitors.
(A) The chemical structures of the inhibitors utilized in the present study. (B) GST–LRRK2[wild-type]-(1326–2527) or GST–LRRK2[G2019S]-(1326–2527) or His6–ROCK2-(2–543) were assayed in the presence or absence of the indicated concentration of the indicated inhibitor, in the presence of 100 μM ATP. The results are presented as percentage of kinase activity relative to the control measured in the presence of DMSO. Results are the average for at least duplicate reactions where similar results were observed in at least one other experiment. The inhibitors are represented as follows: H-89 by a black line and circles; fasudil by a red line and squares; hydroxyfasudil by a bright green line and triangles; Y-27632 by a blue line and inverted triangles; H-1152 by a purple line and diamonds; GSK429286A by an orange line and circles; GSK269962A by a dark green line and squares; and sunitinib by a pink line and triangles. The table shows the IC50 values (in μM) derived from the graphs.
LRRK2 tool compound selectivity profiles
To compare the relative selectivity profiles of Y-27632, H-1152 and fasudil (HA-1077) with GSK429286A, GSK269962A and sunitinib, we profiled these inhibitors against a panel of 85 protein kinases at ATP concentrations which approximate the Km constant for ATP (see Table 1, where definitions for kinases mentioned in this section can be found). This revealed that GSK429286A is a very selective ROCK2 inhibitor and, at 1 μM, it reduced ROCK2 activity over 20-fold, under conditions in which the only other kinase on the panel that was significantly inhibited was MSK1 whose activity was reduced ~ 5-fold. Y-27632 is also selective, and at 10 μM in addition to inhibiting ROCK2 only suppressed activity of PRK2 and MNK1 over 5-fold. H-1152 (1 μM) in addition to ROCK, inhibited Aurora B kinase and BRSK2 over 5-fold. Fasudil, hydroxyfasudil, sunitinib and GSK269962A were less selective. Fasudil (10 μM) inhibited RSK1, S6K1, PRK2, MSK1, MNK1 and MELK over 5-fold. Hydroxyfasudil (10 μM) inhibited RSK1, S6K1, PRK2 and MSK1. Sunitinib (1 μM) and GSK269962A significantly inhibited 12 and seven kinases respectively out of the 85 kinases profiled (Table 1).
Table 1. Kinase profiling of LRRK2 and ROCK inhibitors.
Results are presented as the percentage of kinase activity compared with control incubations in which inhibitor was omitted. Protein kinases were assayed as described in the Materials and methods section.
| Kinase | HA-1100 (10 μM) | HA-1077 (10 μM) | H-1152 (1 μM) | Y-27632 (10 μM) | GSK429286A (1 μM) | GSK269962A (1 μM) | Sunitinib (0.1 μM) | Sunitinib (1 μM) |
|---|---|---|---|---|---|---|---|---|
| LRRK2 wild-type | 72 ± 11 | 77 ± 13 | 20 ± 3 | 22 ± 2 | 98 ± 22 | 71 ± 0 | 42 ± 2 | 9 ± 0 |
| LRRK2[G2019S] | 26 ± 1 | 67 ± 3 | 13 ± 1 | 11 ± 1 | 97 ± 5 | 66 ± 1 | 14 ± 1 | 3 ± 0 |
| ROCK2 | 7 ± 2 | 3 ± 1 | 3 ± 0 | 4 ± 1 | 3 ± 1 | 1 ± 1 | 92 ± 2 | 67 ± 5 |
| MKK1 | 63 ± 3 | 41 ± 2 | 69 ± 15 | 71 ± 17 | 61 ± 13 | 54 ± 2 | 37 ± 2 | 16 ± 1 |
| ERK1 | 109 ± 6 | 104 ± 2 | 105 ± 8 | 92 ± 1 | 90 ± 1 | 106 ± 2 | 108 ± 2 | 104 ± 7 |
| ERK2 | 102 ± 8 | 98 ± 2 | 94 ± 4 | 94 ± 2 | 94 ± 9 | 105 ± 4 | 101 ± 1 | 103 ± 10 |
| JNK1 | 105 ± 9 | 115 ± 8 | 96 ± 5 | 106 ± 6 | 111 ± 3 | 105 ± 9 | 123 ± 16 | 104 ± 5 |
| NK2 | 90 ± 5 | 94 ± 4 | 91 ± 6 | 96 ± 9 | 101 ± 0 | 101 ± 12 | 101 ± 8 | 88 ± 1 |
| p38α MAPK | 98 ± 1 | 108 ± 13 | 109 ± 11 | 103 ± 4 | 103 ± 6 | 108 ± 6 | 111 ± 2 | 103 ± 0 |
| p38β MAPK | 90 ± 6 | 111 ± 2 | 107 ± 2 | 101 ± 10 | 97 ± 8 | 113 ± 2 | 116 ± 0 | 113 ± 4 |
| p38γ MAPK | 97 ± 9 | 99 ± 5 | 101 ± 18 | 96 ± 13 | 112 ± 13 | 72 ± 14 | 109 ± 21 | 98 ± 18 |
| p38δ MAPK | 100 ± 4 | 77 ± 1 | 93 ± 4 | 86 ± 1 | 94 ± 7 | 86 ± 7 | 110 ± 2 | 107 ± 1 |
| ERK8 | 76 ± 1 | 68 ± 44 | 91 ± 11 | 83 ± 6 | 95 ± 0 | 27 ± 1 | 85 ± 1 | 59 ± 4 |
| RSK1 | 11 ± 15 | 14 ± 5 | 57 ± 1 | 26 ± 3 | 20 ± 25 | 12 ± 1 | 38 ± 53 | 78 ± 9 |
| RSK2 | 41 ± 6 | 31 ± 1 | 76 ± 1 | 42 ± 5 | 48 ± 14 | 24 ± 4 | 60 ± 23 | 41 ± 5 |
| PDK1 | 112 ± 4 | 97 ± 1 | 74 ± 5 | 120 ± 9 | 120 ± 5 | 117 ± 0 | 124 ± 2 | 95 ± 1 |
| PKBα | 27 ± 7 | 27 ± 1 | 64 ± 8 | 56 ± 14 | 72 ± 16 | 26 ± 0 | 87 ± 10 | 73 ± 6 |
| PKBβ | 100 ± 3 | 77 ± 11 | 112 ± 9 | 105 ± 1 | 104 ± 8 | 64 ± 1 | 101 ± 4 | 88 ± 2 |
| SGK1 | 55 ± 13 | 69 ± 21 | 92 ± 8 | 50 ± 8 | 131 ± 0 | 24 ± 9 | 79 ± 26 | 26 ± 5 |
| S6K1 | 11 ± 1 | 13 ± 0 | 72 ± 2 | 70 ± 11 | 24 ± 4 | 2 ± 0 | 82 ± 4 | 41 ± 3 |
| PKA | 89 ± 10 | 21 ± 0 | 76 ± 5 | 106 ± 16 | 88 ± 24 | 22 ± 3 | 104 ± 3 | 97 ± 3 |
| PRK2 | 12 ± 0 | 6 ± 0 | 70 ± 69 | 7 ± 3 | 40 ± 3 | 2 ± 0 | 98 ± 3 | 81 ± 17 |
| PKCα | 64 ± 5 | 39 ± 5 | 70 ± 11 | 52 ± 1 | 82 ± 4 | 32 ± 0 | 93 ± 13 | 77 ± 9 |
| PKCζ | 94 ± 8 | 66 ± 2 | 86 ± 6 | 69 ± 2 | 81 ± 4 | 5 ± 1 | 110 ± 1 | 94 ± 6 |
| PKD1 | 60 ± 3 | 25 ± 1 | 79 ± 16 | 89 ± 0 | 85 ± 3 | 85 ± 6 | 79 ± 7 | 34 ± 5 |
| MSK1 | 17 ± 0 | 14 ± 1 | 39 ± 4 | 41 ± 8 | 20 ± 1 | 1 ± 0 | 94 ± 12 | 60 ± 8 |
| MNK1 | 76 ± 8 | 20 ± 3 | 50 ± 6 | 20 ± 10 | 91 ± 6 | 86 ± 2 | 99 ± 10 | 83 ± 5 |
| MNK2 | 77 ± 4 | 27 ± 6 | 49 ± 9 | 44 ± 2 | 88 ± 6 | 51 ± 2 | 83 ± 17 | 80 ± 7 |
| MAPKAPK2 | 96 ± 9 | 91 ± 9 | 93 ± 0 | 85 ± 4 | 91 ± 1 | 88 ± 1 | 91 ± 17 | 95 ± 5 |
| PRAK | 88 ± 16 | 93 ± 12 | 87 ± 17 | 88 ± 14 | 92 ± 17 | 86 ± 13 | 102 ± 1 | 75 ± 15 |
| CaMKKβ | 93 ± 3 | 95 ± 5 | 81 ± 6 | 102 ± 8 | 100 ± 1 | 98 ± 7 | 87 ± 12 | 39 ± 6 |
| CaMK1 | 108 ± 14 | 109 ± 4 | 105 ± 2 | 85 ± 22 | 102 ± 18 | 122 ± 31 | 113 ± 19 | 66 ± 2 |
| SmMLCK | 66 ± 3 | 66 ± 6 | 96 ± 5 | 70 ± 1 | 73 ± 3 | 65 ± 3 | 49 ± 2 | 26 ± 3 |
| PHK | 89 ± 4 | 46 ± 6 | 26 ± 2 | 79 ± 1 | 97 ± 8 | 92 ± 3 | 11 ± 1 | 2 ± 0 |
| CHK1 | 87 ± 1 | 103 ± 5 | 89 ± 14 | 96 ± 9 | 111 ± 6 | 88 ± 2 | 76 ± 0 | 33 ± 3 |
| CHK2 | 89 ± 13 | 34 ± 2 | 44 ± 0 | 93 ± 7 | 102 ± 6 | 52 ± 1 | 23 ± 1 | 5 ± 1 |
| GSK3β | 91 ± 11 | 91 ± 5 | 72 ± 28 | 92 ± 3 | 93 ± 13 | 12 ± 0 | 108 ± 3 | 93 ± 11 |
| CDK2/cyclin A | 98 ± 5 | 82 ± 6 | 74 ± 7 | 72 ± 4 | 49 ± 34 | 50 ± 3 | 98 ± 2 | 87 ± 5 |
| PLK1 | 101 ± 9 | 111 ± 14 | 105 ± 1 | 108 ± 16 | 102 ± 5 | 105 ± 6 | 103 ± 9 | 109 ± 10 |
| Aurora B kinase | 59 ± 1 | 51 ± 1 | 11 ± 2 | 79 ± 1 | 59 ± 5 | 36 ± 0 | 72 ± 5 | 29 ± 4 |
| AMPK | 72 ± 2 | 60 ± 1 | 45 ± 4 | 77 ± 4 | 78 ± 15 | 87 ± 0 | 51 ± 3 | 15 ± 4 |
| MARK3 | 84 ± 2 | 79 ± 2 | 56 ± 3 | 105 ± 5 | 98 ± 7 | 67 ± 0 | 77 ± 4 | 36 ± 4 |
| BRSK2 | 65 ± 3 | 38 ± 2 | 17 ± 4 | 43 ± 4 | 91 ± 10 | 95 ± 7 | 94 ± 0 | 40 ± 6 |
| MELK | 76 ± 2 | 19 ± 1 | 70 ± 4 | 79 ± 2 | 83 ± 0 | 44 ± 3 | 55 ± 6 | 17 ± 1 |
| CK1 | 100 ± 0 | 77 ± 1 | 112 ± 9 | 107 ± 12 | 106 ± 3 | 102 ± 2 | 50 ± 2 | 10 ± 0 |
| CK2 | 84 ± 4 | 81 ± 9 | 84 ± 17 | 85 ± 2 | 82 ± 0 | 88 ± 2 | 91 ± 2 | 72 ± 1 |
| DYRK1A | 96 ± 16 | 102 ± 3 | 96 ± 6 | 96 ± 3 | 98 ± 7 | 95 ± 7 | 103 ± 6 | 83 ± 8 |
| DYRK2 | 89 ± 1 | 80 ± 2 | 84 ± 2 | 92 ± 5 | 92 ± 7 | 64 ± 2 | 92 ± 6 | 76 ± 9 |
| DYRK3 | 99 ± 2 | 39 ± 5 | 94 ± 0 | 100 ± 6 | 94 ± 6 | 77 ± 9 | 104 ± 5 | 84 ± 3 |
| NEK2a | 103 ± 2 | 98 ± 9 | 99 ± 3 | 104 ± 0 | 106 ± 5 | 94 ± 5 | 103 ± 12 | 87 ± 6 |
| NEK6 | 96 ± 8 | 107 ± 28 | 99 ± 16 | 80 ± 18 | 87 ± 24 | 82 ± 19 | 95 ± 26 | 83 ± 15 |
| IKKβ | 86 ± 6 | 79 ± 4 | 100 ± 8 | 79 ± 4 | 97 ± 7 | 83 ± 3 | 90 ± 9 | 80 ± 4 |
| PIM1 | 90 ± 14 | 84 ± 14 | 90 ± 5 | 94 ± 12 | 89 ± 8 | 31 ± 6 | 95 ± 5 | 86 ± 14 |
| PIM2 | 104 ± 1 | 111 ± 3 | 91 ± 7 | 102 ± 3 | 106 ± 9 | 89 ± 5 | 111 ± 7 | 104 ± 2 |
| PIM3 | 75 ± 1 | 81 ± 1 | 96 ± 1 | 87 ± 2 | 89 ± 1 | 35 ± 4 | 84 ± 1 | 50 ± 8 |
| SRPK1 | 90 ± 12 | 92 ± 10 | 125 ± 1 | 65 ± 35 | 71 ± 8 | 90 ± 13 | 80 ± 0 | 86 ± 4 |
| MST2 | 44 ± 4 | 51 ± 2 | 41 ± 2 | 61 ± 8 | 108 ± 3 | 98 ± 2 | 59 ± 7 | 16 ± 2 |
| EF2K | 103 ± 14 | 120 ± 4 | 99 ± 24 | 81 ± 2 | 99 ± 4 | 113 ± 30 | 97 ± 15 | 106 ± 26 |
| HIPK2 | 109 ± 3 | 127 ± 6 | 10 ± 0 | 104 ± 1 | 104 ± 3 | 17 ± 0 | 79 ± 2 | 30 ± 1 |
| PAK4 | 104 ± 4 | 99 ± 17 | 99 ± 7 | 101 ± 23 | 95 ± 1 | 95 ± 3 | 99 ± 9 | 72 ± 5 |
| PAK5 | 113 ± 7 | 112 ± 16 | 100 ± 1 | 101 ± 11 | 107 ± 5 | 102 ± 4 | 121 ± 4 | 99 ± 3 |
| PAK6 | 103 ± 5 | 110 ± 8 | 103 ± 7 | 96 ± 7 | 109 ± 7 | 106 ± 2 | 115 ± 8 | 102 ± 10 |
| MST4 | 82 ± 2 | 79 ± 8 | 90 ± 7 | 67 ± 7 | 70 ± 5 | 86 ± 9 | 87 ± 5 | 80 ± 6 |
| TBK1 | 98 ± 5 | 110 ± 3 | 98 ± 7 | 69 ± 9 | 101 ± 0 | 77 ± 2 | 106 ± 9 | 54 ± 1 |
| IKKε | 99 ± 2 | 95 ± 8 | 80 ± 4 | 76 ± 8 | 109 ± 15 | 66 ± 21 | 101 ± 1 | 63 ± 3 |
| GCK | 73 ± 7 | 55 ± 4 | 56 ± 1 | 89 ± 4 | 97 ± 7 | 78 ± 9 | 45 ± 2 | 11 ± 1 |
| IRAK4 | 87 ± 4 | 69 ± 7 | 102 ± 7 | 101 ± 8 | 94 ± 1 | 78 ± 2 | 76 ± 19 | 23 ± 4 |
| NUAK1 | 36 ± 11 | 21 ± 2 | 51 ± 10 | 106 ± 1 | 51 ± 1 | 39 ± 4 | 30 ± 1 | 15 ± 3 |
| MLK1 | 75 ± 16 | 56 ± 5 | 82 ± 18 | 90 ± 19 | 81 ± 14 | 77 ± 7 | 74 ± 2 | 40 ± 0 |
| MINK1 | 68 ± 21 | 54 ± 6 | 104 ± 5 | 81 ± 0 | 75 ± 9 | 70 ± 4 | 46 ± 1 | 8 ± 0 |
| MLK3 | 45 ± 60 | 72 ± 8 | 85 ± 9 | 110 ± 3 | 116 ± 23 | 97 ± 1 | 72 ± 9 | 26 ± 1 |
| LKB1 | 50 ± 0 | 33 ± 0 | 50 ± 3 | 52 ± 12 | 58 ± 1 | 58 ± 4 | 59 ± 1 | 52 ± 2 |
| HER4 | 97 ± 5 | 129 ± 4 | 107 ± 2 | 69 ± 9 | 50 ± 2 | 93 ± 3 | 126 ± 14 | 103 ± 9 |
| TTK | 82 ± 2 | 73 ± 5 | 93 ± 9 | 73 ± 2 | 83 ± 0 | 69 ± 0 | 99 ± 5 | 72 ± 6 |
| Src | 111 ± 2 | 120 ± 6 | 81 ± 0 | 90 ± 4 | 85 ± 4 | 105 ± 0 | 98 ± 7 | 52 ± 1 |
| Lck | 79 ± 29 | 109 ± 7 | 74 ± 1 | 83 ± 32 | 82 ± 7 | 102 ± 4 | 51 ± 4 | 11 ± 2 |
| CSK | 95 ± 7 | 104 ± 12 | 101 ± 4 | 104 ± 14 | 106 ± 19 | 98 ± 15 | 98 ± 10 | 89 ± 2 |
| FGFR1 | 83 ± 2 | 65 ± 5 | 35 ± 3 | 95 ± 9 | 95 ± 3 | 96 ± 18 | 77 ± 2 | 29 ± 0 |
| IRR | 89 ± 1 | 86 ± 4 | 88 ± 2 | 91 ± 1 | 106 ± 1 | 61 ± 7 | 90 ± 1 | 77 ± 24 |
| EPH A2 | 95 ± 8 | 94 ± 6 | 49 ± 5 | 106 ± 4 | 105 ± 12 | 108 ± 4 | 104 ± 3 | 91 ± 9 |
| SYK | 112 ± 11 | 104 ± 1 | 104 ± 2 | 94 ± 8 | 78 ± 9 | 73 ± 5 | 107 ± 13 | 94 ± 10 |
| YES1 | 111 ± 4 | 136 ± 6 | 76 ± 8 | 96 ± 5 | 58 ± 5 | 114 ± 11 | 30 ± 3 | 7 ± 1 |
| IGF-1R | 91 ± 9 | 92 ± 1 | 113 ± 2 | 99 ± 5 | 97 ± 1 | 104 ± 7 | 84 ± 1 | 33 ± 6 |
| VEGFR | 115 ± 1 | 70 ± 4 | 82 ± 20 | 105 ± 4 | 117 ± 12 | 91 ± 7 | 33 ± 0 | 9 ± 0 |
| BTK | 108 ± 20 | 112 ± 21 | 82 ± 9 | 85 ± 17 | 66 ± 3 | 107 ± 4 | 90 ± 1 | 52 ± 12 |
| IR-HIS | 100 ± 4 | 105 ± 1 | 108 ± 6 | 113 ± 1 | 107 ± 13 | 93 ± 1 | 104 ± 7 | 65 ± 5 |
| EPH-B3 | 111 ± 2 | 65 ± 9 | 69 ± 11 | 83 ± 12 | 87 ± 9 | 92 ± 10 | 107 ± 9 | 103 ± 0 |
The results are means ± S.D. of triplicate determinations. Bold and underlined values indicates inhibition of 80 % or more. Abbreviations not defined in main text: AMPK, AMP-activated-protein kinase; BRSK, brain-specific kinase; BTK, Bruton’s tyrosine kinase; CaMK1, calmodulin-dependent kinase; CaMKK, CaMK kinase; CDK, cyclin-dependent kinase; CHK, checkpoint kinase; CK1, casein kinase 1; CSK, C-terminal Src kinase; DYRK, dual-specificity tyrosine-phosphorylated and regulated kinase; EF2K, elongation-factor-2 kinase; EPH, ephrin; FGFR, fibroblast growth factor receptor; GCK, germinal centre kinase; GSK3, glycogen synthase kinase 3; HIPK, homeodomain-interacting protein kinase; HER4, V-erb a erythroblastic leukaemia viral oncogene homologue 1; IRAK, interleukin-1 receptor-associated kinase 4; IGF1R, insulin-like growth factor 1 receptor; IKK, IκB kinase; IR, insulin receptor; IRR, insulin-related receptor; JNK, c-Jun N-terminal kinase; Lck, lymphocyte cell-specific protein tyrosine kinase; MAPK, mitogen-activated protein kinase; MAPKAPK, MAPK-activated protein kinase; MARK, microtubule-affinity-regulating kinase; MELK, maternal embryonic leucine-zipper kinase; MKK1, MAPK kinase-1; MLCK, smooth muscle myosin light-chain kinase; MNK, MAPK-integrating protein kinase; MLK, mixed lineage kinase; MINK, misshapen-like kinase; MSK, mitogen- and stress-activated protein kinase; MST, mammalian homologue Ste20-like kinase; NEK, NIMA (never in mitosis in Aspergillus nidulans)-related kinase; NUAK1, SNF1-like kinase1; PAK, p21-activated protein kinase; PHK, phosphorylase kinase; PIM, provirus integration site for Moloney murine leukaemia virus; PDK1, 3-phosphoinositide-dependent protein kinase-1; PKB, protein kinase B; PKC, protein kinase C; PKD, protein kinase D; PLK, Polo-like kinase; PRAK, p38-regulated activated kinase; PRK, PKC-related kinase; RSK, ribosomal S6 kinase; S6K, p70 ribosomal S6 kinase; SGK1, serum- and glucocorticoid-induced protein kinase 1; SRPK, serine/arginine protein kinase; SYK, spleen tyrosine kinase; TBK1, TANK [TRAF (tumour-necrosis-factor-receptor-associated factor)-associated nuclear factor κB activator]-binding kinase 1; TTK, tau tubulin kinase; VEGFR, vascular endothelial growth factor receptor; YES1, Yamaguchi sarcoma viral oncogene homologue 1.
LRRK2 homology model and development of inhibitor-resistant mutants
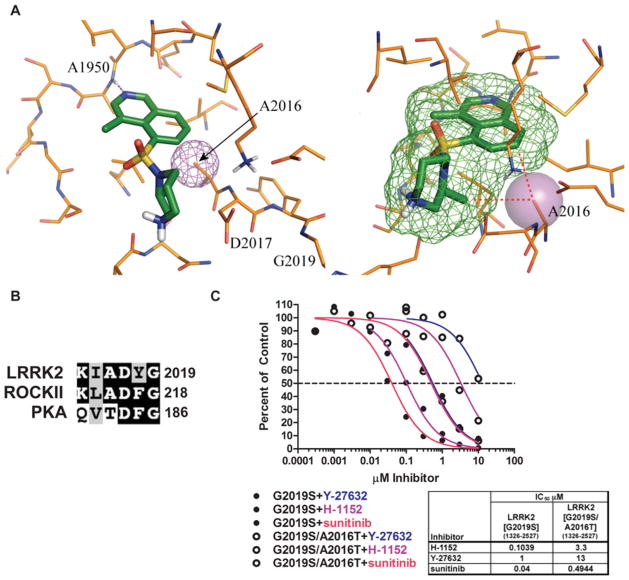
In the absence of an available crystal structure of the LRRK2 kinase domain, we utilized the observation that some ROCK1 inhibitors displayed similar activity against LRRK2, to use ROCK1 as a template from which to construct a homology model of the LRRK2 kinase domain. An unpublished structure of ROCK1 complexed with fasudil, similar to the published structure of ROCK2 [29], was used as the template for the model because of its slightly higher resolution. A binding model of H-1152 in LRRK2 was obtained by superimposing the protein Cα atoms of the LRRK2 model with the reported ROCK1–H-1152 complex [30] (Figure 5A). The binding mode of H-1152 to LRRK2 is predicted to be similar to the binding mode to ROCK1 [30]. In the complex with ROCK1, the isoquinoline nitrogen of H-1152 accepts the hinge hydrogen bond from the backbone NH atom of Met156, and the same interaction is predicted to take place with the backbone NH atom of the equivalent LRRK2 residue, Ala1950. The two methyl groups of H-1152 help to restrict the conformational freedom of the inhibitor, in addition to making lipophilic contacts with the ATP site [30]. These lipophilic contacts are also well conserved in LRRK2.
Figure 5. Design of an LRRK2 inhibitor-desensitized mutant.
(A) Structural model of the LRRK2 kinase domain in complex with H-1152 based on a ROCK crystal structure. The predicted positions of Ala1950, Ala2016 and Asp2017 are indicated. The right-hand panel is rotated slightly. (B) Sequence alignment of the amino acids surrounding the aspartic acid residue of subdomain VII for LRRK2, PKA and ROCK2. Identical amino acids are shaded in black, and similar amino acids are shaded in grey. Terminal amino acid residues are numbered. (C) GST–LRRK2[G2019S]-(1326–2527) and GST–LRRK2[G2019S/A2016T]-(1326–2527) were assayed in the presence or absence of the indicated concentration of the inhibitor, in the presence of 100 μM ATP and expressed as a percentage of the control reactions performed in the presence of vehicle alone. GST–LRRK2[G2019S]-(1326–2527) reactions are represented by filled circles and GST–LRRK2[G2019S/A2016T]-(1326–2527) reactions are represented by open circles. The colour scheme is the same as in Figure 4(B).
The model also highlights the position of Ala2016 in which the side-chain Cβ atom of Ala2016 is located close to H-1152, 4.3 Å (1 Å = 0.1 nm) away from the homopiperazine methyl group. The C-7 atom of the isoquinoline ring is even closer, only 3.7 Å away. In the case of ROCK2, the residue equivalent to Ala2016 is Ala215 (Figure 5B). This residue in both LRRK2 and ROCK1 lies just before the subdomain VII DFG motif. Previous work has shown that Ala215 on ROCK plays an important role in controlling the specificity of the interaction with H-1152 by forming two van der Waals interactions with H-1152 [30]. PKA (protein kinase A) is inhibited ~ 50-fold more weakly by H-1152, and has Thr183 in the equivalent position. Mutation of Thr183 to alanine in PKA did not affect basal activity, but enhanced its inhibition by H-1152 4-fold [31]. We therefore mutated Ala2016 in LRRK2 to a threonine residue and found that this did not inhibit LRRK2[G2019S] basal activity, but increased the IC50 of inhibition by H-1152 ~ 30-fold (Figure 5D). We also observed that the LRRK2[A2016T] mutant was 13- and 12-fold more resistant to inhibition by Y-27632 and sunitinib respectively (Figure 5D). In the LRRK2[A2016T] mutant the Thr2016 side chain would clash with these atoms of H-1152, probably forcing it to bind in a rotated and less favourable orientation in the ATP site. This is the likely explanation of its reduced activity against the mutant compared with the wild-type LRRK2. We also mutated Ala2016 to other residues, but found that these mutations markedly inhibited intrinsic LRRK2 activity (results not shown).
Evaluation of ROCK and LRRK2 inhibitors in cells
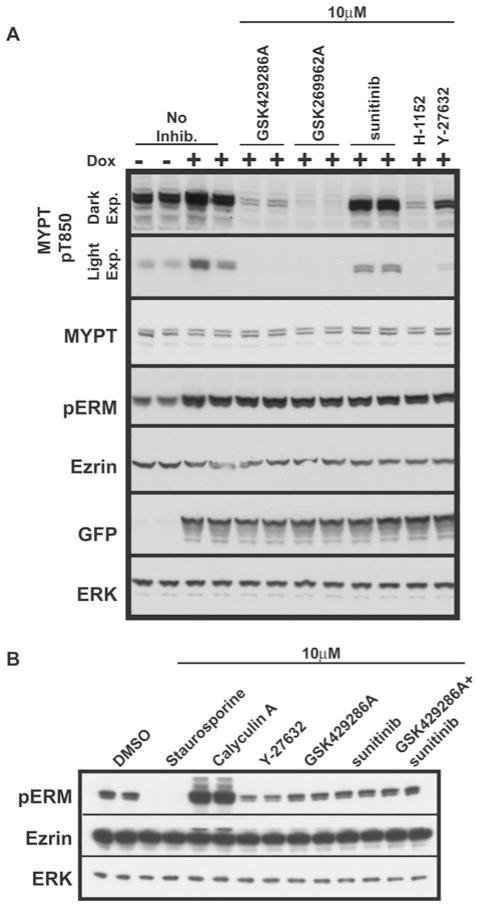
We next investigated the effect that GSK429286A, GSK269962A, sunitinib, H-1152 and Y-27632 had on phosphorylation of MYPT and ERM proteins in HEK-293 cells. We observed significant basal phosphorylation of MYPT at Thr850 and ERM proteins at site(s) equivalent to Thr567 on ezrin (Figure 6A). In order to activate ROCK, we induced stable expression of a constitutively activated G14V-Rho mutant which increased phosphorylation of both MYPT and ezrin ~ 3-fold (Figure 6A). Treatment of cells with 10 μM GSK429286A or GSK269962A, but not sunitinib, ablated phosphorylation of MYPT at Thr850 to a similar extent as H-1152 and Y-27632, consistent with ROCK mediating this phosphorylation. However, in the same extracts, GSK429286A, GSK269962A, sunitinib, H-1152 or Y-27632 did not inhibit ERM protein phosphorylation either in the presence or absence of G14V-Rho (Figure 6A). We also found that adding both GSK429286A and sunitinib together to inhibit both LRRK2 and ROCK did not affect basal ERM phosphorylation (Figure 6B).
Figure 6. Testing the efficacy of LRRK2 and ROCK inhibitors in vivo.
(A) Flp-in T-REx cells that harbour GFP-tagged constitutively active G14V-Rho were either left uninduced or induced by the inclusion of 1 μg/ml doxycycline (Dox) in the culture medium. At 7 h after induction, cells were treated with 10 μM GSK429286A, GSK269962A, H-1152 or Y-27632 for 1 h. Cells were lysed in direct SDS lysis buffer and resolved on 4–12 % Novex gels and subjected to immunoblot analysis with the indicated antibodies. (B) As in (A), except that HEK-293 cells were used and GSK429286A was used in conjunction with sunitinib.
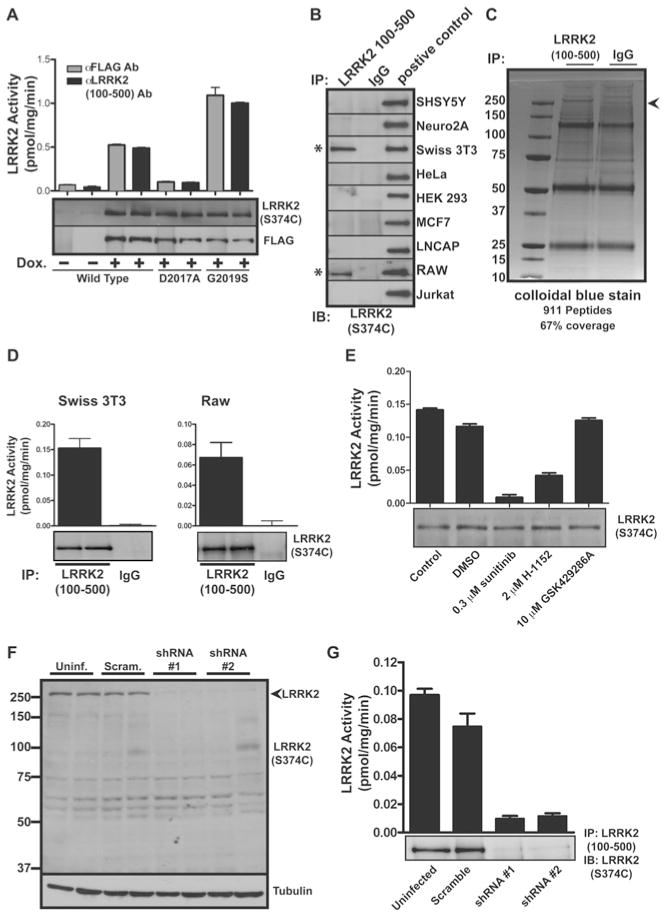
Immunoprecipitation and assay of endogenous LRRK2
In order to measure protein kinase activity of endogenous LRRK2 (which to our knowledge has not been achieved previously), we generated numerous anti-LRRK2 antibodies and evaluated their ability to immunoprecipitate and immunoblot recombinant full-length FLAG–LRRK2. This revealed that the antibody raised against a fragment of LRRK2 encompassing amino acids 100–500 immunoprecipitated and immunoblotted FLAG–LRRK2 with a similar efficiency to that of anti-FLAG antibody (Figure 7A). Moreover, overexpressed FLAG–LRRK2 was immunoprecipitated with the anti-LRRK2-(100–500) antibody and possessed similar activity to enzyme immunoprecipitated with anti-FLAG antibody, indicating that the anti-LRRK2-(100–500) antibody is not inhibiting LRRK2 protein kinase activity (Figure 7A). We next attempted to immunoprecipitate endogenous LRRK2 from extracts derived from a panel of cell lines using the anti-LRRK2-(100–500) antibody. This revealed that Swiss-3T3 fibroblasts and RAW 264.7 macrophages express detectable levels of endogenous LRRK2 protein (Figure 7B). The LRRK2 immunoprecipitates derived from Swiss-3T3 cells were subjected to electrophoresis on a polyacrylamide gel. Staining with Colloidal Blue revealed a protein migrating with a molecular mass of ~ 280 kDa which was confirmed to comprise LRRK2 by MS analysis (Figure 7C). We next subjected LRRK2 and control immunoprecipitates derived from Swiss-3T3 and RAW 264.7 cells to protein kinase assays employing Nictide as a substrate. This revealed significant protein kinase activity with LRRK2 immunoprecipitates, but not with the control immunoprecipitate (Figure 7D). Moreover, the protein kinase activity detected in the LRRK2 immunoprecipitate was suppressed by H-1152 and sunitinib, but not by the selective GSK429286A ROCK inhibitor (Figure 7E). This emphasizes that these compounds are also capable of suppressing the activity of full-length LRRK2 containing leucine-rich repeats.
Figure 7. Analysis of endogenous LRRK2.
(A) Doxycycline (Dox.)-inducible HEK-293 cells overexpressing full-length FLAG-tagged human LRRK2 and LRRK2 containing D2017A and G2019S mutations were generated as described in the Materials and methods section. Anti-LRRK2-(100–500) antibody (Ab) S348C was used to retrieve recombinant protein. Precipitates were assayed for kinase activity using LRRKtide and were immunoblotted with anti-FLAG and S374C anti-LRRK2-(2498–2514) antibodies. (B) Extracts of the indicated cell lines were screened for the presence of LRRK2 protein following immunoprecipitation with the indicated antibodies and immunoblotting with the anti-LRRK2-(2498–2514) antibody S374C. (C) Anti-LRRK2-(100–500) antibody S348C and control IgG were used in immunoprecipitations of 60 mg of Swiss-3T3 lysate. Specific bands corresponding to the predicted molecular mass of LRRK2 were excised and tryptic peptides were identified by MS. Molecular masses are indicated in kDa. (D) Endogenous LRRK2 kinase activity from S348C anti-LRRK2-(100–500) antibody was measured against the Nictide substrate in Swiss-3T3 and RAW 264.7 cells following immunoprecipitation. (E) Immunoprecipitates from Swiss-3T3 cell lines were assayed against Nictide substrate in the presence of the indicated concentrations of the indicated inhibitors. Kinase assays of immune complexes were carried out in triplicate and are representative of at least two separate experiments. Results are means+S.E.M. (F) Lysates of RAW 264.7 cells, stably transduced with the indicated shRNA-expressing lentivirus, were resolved by SDS/PAGE (8 % gels) and immunoblotted with the anti-LRRK2-(2498–2514) antibody S374C. Anti-tubulin was used as a loading control. Uninf., uninfected (i.e. no shRNA); Scram., scrambled shRNA. Molecular masses are indicated in kDa. (G) Endogenous LRRK2 kinase activity against Nictide was assayed as in (E), from RAW 264.7 cells stably transduced with the indicated shRNA-expressing lentivirus. IB, immunoblot; IP, immunoprecipitation.
shRNA-mediated knockdown of endogenous LRRK2
To establish further that we were indeed observing endogenous LRRK2, we generated RAW 264.7 macrophage cell lines that were stably infected with lentiviral vectors expressing scrambled shRNA as well as two distinct shRNAs targeting LRRK2. This revealed that both shRNA vectors markedly reduced expression of LRRK2 observed in total cell extracts (Figure 7F), as well as immunoprecipitates (Figure 7G) compared with non-infected cells or cells infected with a scrambled shRNA. We also observed a concomitant reduction in LRRK2 activity associated with the immunoprecipitates derived from cell lines expressing LRRK2 targeting shRNA (Figure 7G).
DISCUSSION
Our data reveal that LRRK2 tolerates a wider range of amino acids in its substrates compared with some other protein kinases that have strong requirements for specific amino acids within the substrates that they phosphorylate (Figures 1 and 2). Significant substrate specificity preferences are the −5 (tryptophan and arginine), −2 (phenylalanine, tyrosine and histidine), −1 (tyrosine, arginine and tryptophan), +2 (arginine) and +3 (arginine) positions. Importantly, our data suggest that LRRK2 has a strong preference for phosphorylating threonine, as mutation of the phosphorylated threonine residue in Nictide to serine abolished phosphorylation of the peptide by LRRK2 (Figure 3). Positional scanning peptide library analysis also suggested that LRRK2 poorly tolerated acidic glutamic acid or aspartic acid residues at all positions surrounding the phosphorylation site (Figure 2A). LRRKtide only possesses a single acidic residue (Asp−4), and the only mutation we tested that improved LRRK2 phosphorylation, was mutation of this residue to alanine (Figure 1C). It is possible that this knowledge of substrate specificity of LRRK2 may aid in the identification of LRRK2 substrates and/or potential phosphorylation sites within identified substrates. This analysis has also enabled us to generate the Nictide peptide, a much improved substrate compared with LRRKtide peptide that is widely deployed to assay LRRK2. A key advantage of Nictide is that it can be used at much lower concentrations in kinase assays. We have been able to assay the activity of endogenous LRRK2 using Nictide, with virtually no background activity observed in the control immunoprecipitate (Figure 7E). To our knowledge, this is the first time that activity of endogenous LRRK2 has been assessed. When trying to assay activity of endogenous LRRK2 employing LRRKtide at concentrations of 300 μM (Km value), we observed significant background activity in the pre-immune immunoprecipitation (N. Dzamko, unpublished work). Assessment of activity of endogenous LRRK2 is important, as it paves the way to study LRRK2 activity in cells/tissues derived from PD patients. It will enable evaluation of whether LRRK2 protein kinase activity is controlled by extracellular agonists and may also help in the screening for inhibitors for LRRK2. We also observed that fusing Nictide to GST, yielded a highly expressed protein in Escherichia coli (4 mg/l) which was efficiently phosphorylated by LRRK2 in vitro at a greater initial rate than GST–ezrin-(505–586) or GST–LRRKtide (Figure 2C). GST–Nictide would serve as a good positive control when evaluating efficiency of phosphorylation of LRRK2 substrates that are identified in future studies.
Our analysis reveals that the substrate specificity of LRRK2 is quite distinct from that of ROCK2. LRRK2 does not phosphorylate MYPT, and ROCK2 poorly phosphorylates ezrin. Moreover, mutations in LRRKtide affected phosphorylation by LRRK2 and ROCK2 in different ways. For example, mutation of the +1 position of the LRRKtide peptide from a leucine residue to alanine abolished ROCK phosphorylation, without affecting LRRK2 phosphorylation. Many LRRKtide mutations enhanced phosphorylation by ROCK, but inhibited phosphorylation by LRRK2. Consistent with ROCK2 phosphorylating ezrin poorly in vitro, we also found that in vivo various ROCK inhibitors failed to inhibit ERM phosphorylation under conditions which they suppressed MYPT phosphorylation. This is consistent with other studies where the Y-27632 ROCK inhibitor was found not to suppress ERM phosphorylation [32,33]. Taken together, these data cast doubt on earlier suggestions that ERM proteins are physiologically phosphorylated by ROCK isoforms.
The finding that the H-1152, Y-27632 and sunitinib failed to suppress ERM phosphorylation indicates that either LRRK2 does not phosphorylate ERM in HEK-293 cells or that LRRK2 is not the sole kinase that phosphorylates ERM proteins. As we were unable to detect significant levels of endogenous LRRK2 in HEK-293 cells (Figure 7), we overexpressed LRRK2 and LRRK2[G2019S] in HEK-293 cells, but this also failed to induce phosphorylation of ERM proteins (R.J. Nichols, unpublished work). Taken together, this suggests that, although ERM proteins are efficiently phosphorylated by LRRK2 in vitro, there is no strong evidence that ERM proteins comprise physiological substrates for LRRK2. Recent studies in Drosophila [34,35] and primary mouse lymphocytes [36] have suggested that the SLK/LOK STE20 protein kinase might be a key player in controlling ERM phosphorylation. Consistent with this, ERM phosphorylation is reduced but not abolished in lymphocytes derived from SLK/LOK-knockout mice [36]. It was shown in 1994 that SLK has an optimal motif of R-R/K-F-G-S/T-L-R-R-F/I [37], resembling the ERM site D-K-Y-K-T-L-R-Q-I and is also remarkably similar to the optimal substrate specificity of LRRK2, W-R-F-Y-T-L-R-R-A. It would also be interesting to test whether residual ERM phosphorylation observed in the SLK/LOK-knockout cells was reduced further by treatment with sunitinib and Y-27632 LRRK2 inhibitors. Another recent study has indicated that another STE20 family kinase termed Mst4 kinase can phosphorylate ezrin in polarized epithelial cells in a pathway controlled by the LKB1 tumour suppressor [38]. MST4 was not inhibited by any compound used in the present study (Table 1).
The finding that the widely utilized ROCK inhibitor Y-27632 (used in > 1400 papers) as well as H-1152 and hydroxyfasudil inhibit recombinant as well as endogenous LRRK2 with a similar potency to that which they target ROCK2 was unexpected, as LRRK2 and ROCK2 are not closely related kinases. LRRK2 lies within the tyrosine-like kinases of the human kinome, whereas ROCK2 belongs to the distinct AGC branch [39]. It is therefore possible that some of the physiological effects observed with these ROCK inhibitors could result from inhibition of LRRK2 rather than ROCK isoforms. Identification of GSK429286A is significant, in that not only is it more selective than Y-27632 as assessed on our kinase-specificity panel, but also it does not significantly inhibit LRRK2 even at doses as high as 30 μM (500-fold higher than IC50 of inhibition of ROCK2). GSK429286A will be a useful reagent to use in addition to Y-27632 to assess cellular roles of ROCK isoforms. The finding that the LRRK2[G2019S] mutant was 2–4-fold more sensitive H-1152, Y-27632 and sunitinib than the wild-type LRRK2 also indicates that it may be possible to develop compounds that have greater potency towards the PD mutant. It has also been reported that the LRRK2[G2019S] and LRRK2[I202T] mutants that possess elevated activity were also moderately more sensitive to a panel of non-selective kinase inhibitors [40]. If compounds that specifically inhibited PD mutant forms of LRRK2 could be elaborated, they might have lower side effects and not suppress the normal functions of wild-type LRRK2. In drug discovery screens being undertaken to identify LRRK2 inhibitors, it could be beneficial to screen compounds against both mutant and wild-type forms of LRRK2.
Molecular modelling of the kinase domain of LRRK2 and comparing it with the structures of other kinases revealed a model of how LRRK2 might interact with H-1152. Several residues in the active site of ROCK that are key for binding to H-1152 are also conserved in LRRK2. These include Ala2016, the equivalent of Ala215 in ROCK2 that plays an important role in mediating binding to the inhibitor [30]. Mutation of Ala2016 in LRRK2 to a threonine residue, equivalent to Thr182 in PKA that is weakly inhibited by H-1152, did not affect the basal LRRK2 activity, but markedly suppressed inhibition of LRRK2 by H-1152 and other ROCK inhibitors. The inhibitor-resistant LRRK2[A2016T] mutant might aid in exploring the physiological roles of LRRK2. The wild-type and the LRRK2[A2016T] mutant could be overexpressed in cells and phosphorylation of any target should be less sensitive to inhibition by H-1152, Y-27632 or sunitinib in the cells overexpressing the drug-resistant mutant.
Our findings also provide a pharmacological strategy in which phosphorylation of identified LRRK2 substrates could be validated. We suggest that phosphorylation of a LRRK2 substrate should be suppressed by Y-27632 and H-1152 (dual ROCK and LRRK2 inhibitors) as well as sunitinib (inhibits LRRK2, but not ROCK), but not be affected by GSK429286A (inhibits ROCK, but not LRRK2). In contrast, ROCK-mediated processes should be sensitive to GSK429286A in addition to Y-27632 and H-1152, but should not be inhibited by sunitinib. Consistent with this, sunitinib does not inhibit the phosphorylation of MYPT at Thr850 under conditions where this phosphorylation is inhibited by GSK429286A, Y-27632 and H-1152 (Figure 6).
In conclusion, we have undertaken basic analysis of the LRRK2 substrate specificity and developed improved assays to isolate and assess its activity. This will aid in assessing how LRRK2 is regulated and might also facilitate identification of LRRK2 inhibitors that might have potential for treatment of PD. We have also developed a strategy making use of Y-276332 or H-1152, sunitinib and GSK429286A to explore the roles of the LRRK2 kinase. Finally, we recommend that effects prescribed to ROCK based mainly on the use of Y-276332 and/or H-1152 be re-evaluated with the more selective GSK429286A inhibitor to ensure that these are indeed mediated by ROCK and not LRRK2.
Supplementary Material
Acknowledgments
We thank David Campbell for undertaking MS analysis as well as the Sequencing Service (School of Life Sciences, University of Dundee, Scotland) for DNA sequencing, the Post Genomics and Molecular Interactions Centre for Mass Spectrometry facilities (School of Life Sciences, University of Dundee, Scotland) and the protein production and antibody purification teams [Division of Signal Transduction Therapy (DSTT), University of Dundee] co-ordinated by Hilary McLauchlan and James Hastie for expression and purification of antibodies and Christian Peifer for the inhibitor structures. We also thank Miratul Muqit, Andrew Eatherton, David Livermore, and Dennis Lee for helpful discussions.
FUNDING
We thank the Medical Research Council, The Michael J. Fox Foundation and the pharmaceutical companies supporting the Division of Signal Transduction Therapy Unit (AstraZeneca, Boehringer-Ingelheim, GlaxoSmithKline, Merck & Co. Inc., Merck KgaA and Pfizer) for financial support. N. D. is supported by a Medical Research Council Technology (MRCT) Industry collaborative award. L. C. was supported by the National Institutes of Health [grant number GM56203].
Abbreviations used
- ERK
extracellular-signal-regulated kinase
- ERM
ezrin/radixin/moesin
- GFP
green fluorescent protein
- GST
glutathione transferase
- HEK
human embryonic kidney
- KLH
keyhole-limpet haemocyanin
- LRRK2
leucine-rich repeat protein kinase-2
- MBP
maltose-binding protein
- MYPT
myosin phosphatase target
- PD
Parkinson’s disease
- PKA
protein kinase A
- ROCK
Rho kinase
- shRNA
short hairpin RNA
- TBST
Tris-buffered saline with Tween 20
Footnotes
AUTHOR CONTRIBUTION
Jeremy Nichols, Nicolas Dzamko, Alastair Reith and Dario Alessi planned the experiments and analysed the experimental data. Jeremy Nichols undertook most of the experimentation shown in Figures 1–6, while Nicolas Dzamko did most of the work for Figure 7. Jessica Hutti and Lewis Cantley undertook the positional scanning peptide library analysis, Jennifer Moran undertook kinase profiling, Paul Bamborough did the molecular modelling for Figure 5(A), Paul Bamborough and Alastair Reith provided GSK429286A and GSK269962A, and Jeremy Nichols and Dario Alessi wrote the paper.
References
- 1.Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, Kachergus J, Hulihan M, Uitti RJ, Calne DB, et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 2.Paisan-Ruiz C, Jain S, Evans EW, Gilks WP, Simon J, van der Brug M, Lopez de Munain A, Aparicio S, Gil AM, Khan N, et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 3.Healy DG, Falchi M, O’Sullivan SS, Bonifati V, Durr A, Bressman S, Brice A, Aasly J, Zabetian CP, Goldwurm S, et al. Phenotype, genotype, and worldwide genetic penetrance of LRRK2-associated Parkinson’s disease: a case-control study. Lancet Neurol. 2008;7:583–590. doi: 10.1016/S1474-4422(08)70117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mata IF, Wedemeyer WJ, Farrer MJ, Taylor JP, Gallo KA. LRRK2 in Parkinson’s disease: protein domains and functional insights. Trends Neurosci. 2006;29:286–293. doi: 10.1016/j.tins.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 5.Biskup S, West AB. Zeroing in on LRRK2-linked pathogenic mechanisms in Parkinson’s disease. Biochim Biophys Acta. 2009;1792:625–633. doi: 10.1016/j.bbadis.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greggio E, Cookson MR. Leucine-rich repeat kinase 2 mutations and Parkinson’s disease: three questions. ASN NEURO. 2009;1(1):art:e00002. doi: 10.1042/AN20090007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Covy JP, Giasson BI. Identification of compounds that inhibit the kinase activity of leucine-rich repeat kinase 2. Biochem Biophys Res Commun. 2009;378:473–477. doi: 10.1016/j.bbrc.2008.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anand VS, Reichling LJ, Lipinski K, Stochaj W, Duan W, Kelleher K, Pungaliya P, Brown EL, Reinhart PH, Somberg R, et al. Investigation of leucine-rich repeat kinase 2: enzymological properties and novel assays. FEBS J. 2009;276:466–478. doi: 10.1111/j.1742-4658.2008.06789.x. [DOI] [PubMed] [Google Scholar]
- 9.Jaleel M, Nichols RJ, Deak M, Campbell DG, Gillardon F, Knebel A, Alessi DR. LRRK2 phosphorylates moesin at threonine-558: characterization of how Parkinson’s disease mutants affect kinase activity. Biochem J. 2007;405:307–317. doi: 10.1042/BJ20070209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsui T, Maeda M, Doi Y, Yonemura S, Amano M, Kaibuchi K, Tsukita S, Tsukita S. Rho-kinase phosphorylates COOH-terminal threonines of ezrin/radixin/moesin (ERM) proteins and regulates their head-to-tail association. J Cell Biol. 1998;140:647–657. doi: 10.1083/jcb.140.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oshiro N, Fukata Y, Kaibuchi K. Phosphorylation of moesin by rho-associated kinase (Rho-kinase) plays a crucial role in the formation of microvilli-like structures. J Biol Chem. 1998;273:34663–34666. doi: 10.1074/jbc.273.52.34663. [DOI] [PubMed] [Google Scholar]
- 12.Tran Quang C, Gautreau A, Arpin M, Treisman R. Ezrin function is required for ROCK-mediated fibroblast transformation by the Net and Dbl oncogenes. EMBO J. 2000;19:4565–4576. doi: 10.1093/emboj/19.17.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodman KB, Cui H, Dowdell SE, Gaitanopoulos DE, Ivy RL, Sehon CA, Stavenger RA, Wang GZ, Viet AQ, Xu W, et al. Development of dihydropyridone indazole amides as selective Rho-kinase inhibitors. J Med Chem. 2007;50:6–9. doi: 10.1021/jm0609014. [DOI] [PubMed] [Google Scholar]
- 14.Biskup S, Moore DJ, Rea A, Lorenz-Deperieux B, Coombes CE, Dawson VL, Dawson TM, West AB. Dynamic and redundant regulation of LRRK2 and LRRK1 expression. BMC Neurosci. 2007;8:102. doi: 10.1186/1471-2202-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sward K, Dreja K, Susnjar M, Hellstrand P, Hartshorne DJ, Walsh MP. Inhibition of Rho-associated kinase blocks agonist-induced Ca2+ sensitization of myosin phosphorylation and force in guinea-pig ileum. J Physiol. 2000;522:33–49. doi: 10.1111/j.1469-7793.2000.0033m.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng J, Ito M, Ichikawa K, Isaka N, Nishikawa M, Hartshorne DJ, Nakano T. Inhibitory phosphorylation site for Rho-associated kinase on smooth muscle myosin phosphatase. J Biol Chem. 1999;274:37385–37390. doi: 10.1074/jbc.274.52.37385. [DOI] [PubMed] [Google Scholar]
- 17.Hutti JE, Jarrell ET, Chang JD, Abbott DW, Storz P, Toker A, Cantley LC, Turk BE. A rapid method for determining protein kinase phosphorylation specificity. Nat Methods. 2004;1:27–29. doi: 10.1038/nmeth708. [DOI] [PubMed] [Google Scholar]
- 18.Turk BE, Hutti JE, Cantley LC. Determining protein kinase substrate specificity by parallel solution-phase assay of large numbers of peptide substrates. Nat Protoc. 2006;1:375–379. doi: 10.1038/nprot.2006.57. [DOI] [PubMed] [Google Scholar]
- 19.Workman CT, Yin Y, Corcoran DL, Ideker T, Stormo GD, Benos PV. enoLOGOS: a versatile web tool for energy normalized sequence logos. Nucleic Acids Res. 2005;33:W389–W392. doi: 10.1093/nar/gki439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hutti JE, Turk BE, Asara JM, Ma A, Cantley LC, Abbott DW. IκB kinase β phosphorylates the K63 deubiquitinase A20 to cause feedback inhibition of the NF-κB pathway. Mol Cell Biol. 2007;27:7451–7461. doi: 10.1128/MCB.01101-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller ML, Jensen LJ, Diella F, Jorgensen C, Tinti M, Li L, Hsiung M, Parker SA, Bordeaux J, Sicheritz-Ponten T, et al. Linear motif atlas for phosphorylation-dependent signaling. Sci Signaling. 2008;1:ra2. doi: 10.1126/scisignal.1159433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishizaki T, Uehata M, Tamechika I, Keel J, Nonomura K, Maekawa M, Narumiya S. Pharmacological properties of Y-27632, a specific inhibitor of rho-associated kinases. Mol Pharmacol. 2000;57:976–983. [PubMed] [Google Scholar]
- 23.Shimokawa H, Seto M, Katsumata N, Amano M, Kozai T, Yamawaki T, Kuwata K, Kandabashi T, Egashira K, Ikegaki I, et al. Rho-kinase-mediated pathway induces enhanced myosin light chain phosphorylations in a swine model of coronary artery spasm. Cardiovasc Res. 1999;43:1029–1039. doi: 10.1016/s0008-6363(99)00144-3. [DOI] [PubMed] [Google Scholar]
- 24.Sasaki Y, Suzuki M, Hidaka H. The novel and specific Rho-kinase inhibitor (S)-(+)-2-methyl-1-[(4-methyl-5-isoquinoline)sulfonyl]-homopiperazine as a probing molecule for Rho-kinase-involved pathway. Pharmacol Ther. 2002;93:225–232. doi: 10.1016/s0163-7258(02)00191-2. [DOI] [PubMed] [Google Scholar]
- 25.Tamura M, Nakao H, Yoshizaki H, Shiratsuchi M, Shigyo H, Yamada H, Ozawa T, Totsuka J, Hidaka H. Development of specific Rho-kinase inhibitors and their clinical application. Biochim Biophys Acta. 2005;1754:245–252. doi: 10.1016/j.bbapap.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 26.Chijiwa T, Mishima A, Hagiwara M, Sano M, Hayashi K, Inoue T, Naito K, Toshioka T, Hidaka H. Inhibition of forskolin-induced neurite outgrowth and protein phosphorylation by a newly synthesized selective inhibitor of cyclic AMP-dependent protein kinase, N-[2-(p-bromocinnamylamino)ethyl]-5-isoquinolinesulfonamide (H-89), of PC12D pheochromocytoma cells. J Biol Chem. 1990;265:5267–5272. [PubMed] [Google Scholar]
- 27.Shibuya M, Suzuki Y, Sugita K, Saito I, Sasaki T, Takakura K, Nagata I, Kikuchi H, Takemae T, Hidaka H, et al. Effect of AT877 on cerebral vasospasm after aneurysmal subarachnoid hemorrhage: results of a prospective placebo-controlled double-blind trial. J Neurosurg. 1992;76:571–577. doi: 10.3171/jns.1992.76.4.0571. [DOI] [PubMed] [Google Scholar]
- 28.Doe C, Bentley R, Behm DJ, Lafferty R, Stavenger R, Jung D, Bamford M, Panchal T, Grygielko E, Wright LL, et al. Novel Rho kinase inhibitors with anti-inflammatory and vasodilatory activities. J Pharmacol Exp Ther. 2007;320:89–98. doi: 10.1124/jpet.106.110635. [DOI] [PubMed] [Google Scholar]
- 29.Yamaguchi H, Kasa M, Amano M, Kaibuchi K, Hakoshima T. Molecular mechanism for the regulation of rho-kinase by dimerization and its inhibition by fasudil. Structure. 2006;14:589–600. doi: 10.1016/j.str.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 30.Jacobs M, Hayakawa K, Swenson L, Bellon S, Fleming M, Taslimi P, Doran J. The structure of dimeric ROCK I reveals the mechanism for ligand selectivity. J Biol Chem. 2006;281:260–268. doi: 10.1074/jbc.M508847200. [DOI] [PubMed] [Google Scholar]
- 31.Bonn S, Herrero S, Breitenlechner CB, Erlbruch A, Lehmann W, Engh RA, Gassel M, Bossemeyer D. Structural analysis of protein kinase A mutants with Rho-kinase inhibitor specificity. J Biol Chem. 2006;281:24818–24830. doi: 10.1074/jbc.M512374200. [DOI] [PubMed] [Google Scholar]
- 32.Matsui T, Yonemura S, Tsukita S, Tsukita S. Activation of ERM proteins in vivo by Rho involves phosphatidyl-inositol 4-phosphate 5-kinase and not ROCK kinases. Curr Biol. 1999;9:1259–1262. doi: 10.1016/s0960-9822(99)80508-9. [DOI] [PubMed] [Google Scholar]
- 33.Lee JH, Katakai T, Hara T, Gonda H, Sugai M, Shimizu A. Roles of p-ERM and Rho-ROCK signaling in lymphocyte polarity and uropod formation. J Cell Biol. 2004;167:327–337. doi: 10.1083/jcb.200403091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carreno S, Kouranti I, Glusman ES, Fuller MT, Echard A, Payre F. Moesin and its activating kinase Slik are required for cortical stability and microtubule organization in mitotic cells. J Cell Biol. 2008;180:739–746. doi: 10.1083/jcb.200709161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kunda P, Pelling AE, Liu T, Baum B. Moesin controls cortical rigidity, cell rounding, and spindle morphogenesis during mitosis. Curr Biol. 2008;18:91–101. doi: 10.1016/j.cub.2007.12.051. [DOI] [PubMed] [Google Scholar]
- 36.Belkina NV, Liu Y, Hao JJ, Karasuyama H, Shaw S. LOK is a major ERM kinase in resting lymphocytes and regulates cytoskeletal rearrangement through ERM phosphorylation. Proc Natl Acad Sci USA. 2009;106:4707–4712. doi: 10.1073/pnas.0805963106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Songyang Z, Blechner S, Hoagland N, Hoekstra MF, Piwnica-Worms H, Cantley LC. Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr Biol. 1994;4:973–982. doi: 10.1016/s0960-9822(00)00221-9. [DOI] [PubMed] [Google Scholar]
- 38.ten Klooster JP, Jansen M, Yuan J, Oorschot V, Begthel H, Di Giacomo V, Colland F, de Koning J, Maurice MM, Hornbeck P, Clevers H. Mst4 and Ezrin induce brush borders downstream of the Lkb1/Strad/Mo25 polarization complex. Dev Cell. 2009;16:551–562. doi: 10.1016/j.devcel.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 39.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 40.Reichling LJ, Riddle SM. Leucine-rich repeat kinase 2 mutants I2020T and G2019S exhibit altered kinase inhibitor sensitivity. Biochem Biophys Res Commun. 2009;384:255–258. doi: 10.1016/j.bbrc.2009.04.098. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.