Summary
The evolution of sociality and altruism is enigmatic because cooperators are constantly threatened by cheaters who benefit from cooperation without incurring its full cost [1, 2]. Kin recognition is the ability to recognize and cooperate with genetically close relatives. It has also been proposed as a potential mechanism that limits cheating [3, 4], but there has been no direct experimental support for that possibility. Here we show that kin recognition protects cooperators against cheaters. The social amoebae Dictyostelium discoideum cooperate by forming multicellular aggregates that develop into fruiting bodies of viable spores and dead stalk cells. Cheaters preferentially differentiate into spores while their victims die as stalk cells in chimeric aggregates. We engineered syngeneic cheaters and victims that differed only in their kin-recognition genes, tgrB1 and tgrC1, and in a single cheater allele, and found that the victims escaped exploitation by different types of non-kin cheaters. This protection depends on kin-recognition-mediated segregation, because it is compromised when we disrupt strain segregation. These findings provide direct evidence for the role of kin recognition in cheater control, and suggest a mechanism for the maintenance of stable cooperative systems.
Results
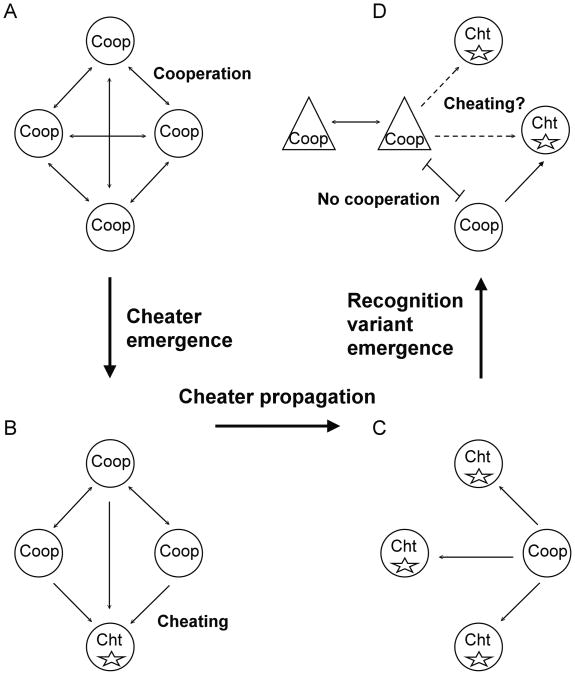
Social cheaters are individuals that reap the benefits of cooperation without fully paying the associated costs [1, 2, 5]. Cheaters arise constantly among cooperators [1, 6, 7]and may overtake a population and collapse a social system if not controlled [3, 8, 9]. Multiple mechanisms may limit cheating, including pleiotropy, high relatedness, and the evolution of cheater-resistance [10-12]. Another possible mechanism is kin recognition, by which altruists preferentially cooperate with genetically related individuals [3, 4]. Figure 1 illustrates how kin recognition may limit the spread of cheaters. A cheater mutation arises in a population of cooperators that recognize one another as kin (Figure 1, A-B). The cheater propagates (Figure 1C), but new mutations that alter kin recognition arise as well (Figure 1D). Individuals that carry a new kin-recognition signal cooperate among themselves while being protected from the cheaters that carry the old kin-recognition signal.
Figure 1. Evolutionary relationships between cheating and kin recognition.
A. Cooperation in a social group is manifested as reciprocal benefits (bi-directional arrows) between cooperating group members (Coop) that share a common kin-recognition signal (circles). B. Mutations (star symbols) can lead to the emergence of cheaters (Cht) within the population. The cheaters carry the common kin-recognition signal and they benefit from the cooperators without paying their fair share of the cost (uni-directional arrows). C. The cheaters propagate in the population and begin to outnumber the other cooperators because of the benefit they receive at a reduced cost. D. Other mutations can lead to the evolution of kin-recognition variants (triangles) among the cooperators. These new variants can cooperate among themselves (bi-directional arrows), but not with the original cooperators (barred line). We propose to test whether the new cooperators would be exploited by the old cheater (dashed arrows, null hypothesis) or whether they would be protected.
Many studies have documented the emergence of cheaters among cooperators and the presence of kin recognition in the same social systems [13-15]. However, there has been no direct demonstration that kin recognition can protect against exploitation by cheaters. We used the social amoeba D. discoideum to address this problem.
D. discoideum are solitary soil amoebae which exhibit social behavior upon starvation [16]. Genetically distinct cells co-aggregate and develop into multicellular structures of approximately 50,000 cells, in which they differentiate into spores and stalk cells. Only the spores survive, whereas 20% of the cells die altruistically as stalks, which are thought to facilitate spore dispersal and therefore increase inclusive fitness [16, 17]. Cheaters in D. discoideum are strains that make more spores than their fair share, where fair share is the ratio between the strains at the onset of development [18]. Wild isolates of D. discoideum can exploit one another [17, 19, 20] and the genetic potential for cheating is considerable, as a single mutation in any one of a large number of genes can lead to cheating [21].
Wild isolates of D. discoideum segregate from genetically distant strains after initial co-aggregation, and develop into mostly clonal fruiting bodies. We have described a kin-recognition mechanism in D. discoideum, in which cells cooperate with genetically related kin during multicellular development [22]. This mechanism is mediated by two transmembrane proteins, TgrB1 and TgrC1 [23, 24]. The tgrB1 and tgrC1 genes are highly polymorphic in natural populations [23], and a matching pair of TgrB1 and TgrC1 is not only necessary and sufficient for cooperation between strains, but also the only component determining kin recognition in D. discoideum [24]. Syngeneic strains that express different pairs of matching tgrB1 and tgrC1 alleles initially co-aggregate upon starvation. After 6-8 hours of development, these mixed strains segregate into distinct sub-aggregates and continue to develop into fruiting bodies that are mostly clonal [24]. The existence of cheating behavior and kin recognition in D. discoideum allows a critical examination of the role of kin recognition in stabilizing social systems.
Cheating and kin recognition have been documented between different wild isolates in D. discoideum, but a recent study found that the genomes of these strains differ by about 40,000 single nucleotide variations (E. Ostrowski, personal communication). It would therefore be difficult to examine the relationship between kin recognition and cheating using wild isolates because the results might be affected by uncharacterized genetic variations. We therefore took a reductionist approach to testing the interplay between kin recognition and cheating in a genetically defined system. D. discoideum genes can be precisely mutated or replaced at their natural chromosomal loci [16]. We used that property to generate strains that differed only in one cheating gene and/or in the tgrB1/tgrC1 kin-recognition locus. These strains recognize each other as non-kin if they carry different sets of tgrB1/tgrC1 alleles and cooperate as kin if they share the same sets of tgrB1/tgrC1 alleles. These otherwise isogenic strains enabled us to test whether kin recognition protects against cheating without the potential confounding effects of other genetic differences. We also designed a cheater-protection assay to measure the cost imposed by cheaters on kin versus non-kin. We mixed cheater mutants with kin and non-kin victims in the test cases and replaced the cheaters with wild-type cells in the controls (Figure 2A). Victim strains are genetically identical to each other and to the wild type except for the designed differences in tgrB1/tgrC1, so each strain should sporulate according to its input proportions in the control, where no cheater is present. On the other hand, victim strains would exhibit reduced spore production in the experiments, where cheaters are present. We used the difference in spore production between the experiment and the control to evaluate the cost to the victims. Higher cost means the victim produced fewer spores when co-developed with a cheater instead of the wild type. The hypothesis that kin recognition protects from cheating would be refuted if the costs were indistinguishable between kin and non-kin and supported if the victims incurred a lower cost when they recognized the cheaters as non-kin.
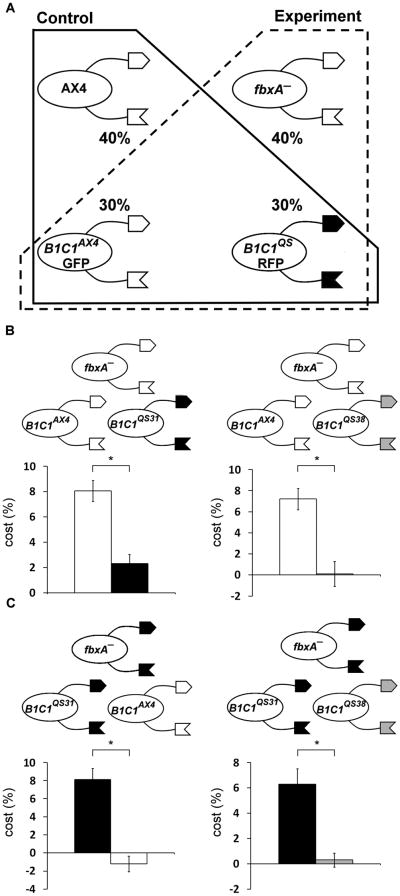
Figure 2. Kin-recognition protects against fbxA− cheating.
Cells were co-developed and spores counted to determine cost. A, Illustration of the assay system. Cells are drawn as ellipses with relevant genotypes within: AX4 – laboratory wild-type strain, fbxA− – cheater strain, B1C1genotype – tgrB1/tgrC1 double-gene replacement strain where ‘genotype’ indicate the allele origin, QS – wild-isolate, GFP and RFP – Green- and Red-Fluorescence Protein labels, respectively. Extracellular arms indicate TgrB1-C1 proteins; the shading represents different alleles. Proportions (%) indicate the initial mixing ratios. Solid frame – control, dashed frame – experiment. B, Assays using fbxA− in the AX4 background. Bars indicate the cost to the compatible strain tgrB1AX4tgrC1AX4–GFP (white) and the incompatible strains tgrB1QS31tgrC1QS31–RFP (black, left panel) and tgrB1QS38tgrC1QS38–RFP (grey, right panel). C, Assays using fbxA− in the tgrB1QS31tgrC1QS31 background. Bars indicate the cost to the compatible strain tgrB1QS31tgrC1QS31–GFP (black) and the incompatible strains tgrB1AX4tgrC1AX4–RFP (white, left panel) and tgrB1QS38tgrC1QS38–RFP (grey, right panel). Data are means +/− s.e.m., n=4 per group, student's t-test *p<0.01.
Kin recognition protects against the obligatory cheater fbxA−
We first tested the hypothesis with the fbxA− mutation, which confers one of the strongest cheater phenotypes [6]. We labeled the victims with Green- or Red- Fluorescent Proteins (GFP and RFP, respectively) to facilitate spore quantification. We mixed the fbxA− strain, which was made in the AX4 background, with the compatible strain tgrB1AX4tgrC1AX4-GFP and the incompatible strains tgrB1QS31tgrC1QS31-RFP, or tgrB1QS38tgrC1QS38-RFP. We expected fbxA− to cheat on the compatible strain (AX4), as published [6], so this experiment tested whether the strain with the switched recognition cues was protected from fbxA−. The results in Figure 2B show that both incompatible strains incurred lower costs than the compatible tgrB1AX4tgrC1AX4- GFP, indicating that the incompatible alleles protected the would-be victims from cheating by fbxA−.
To test for potential confounding interactions between the fbxA− mutation and the tgrB1AX4tgrC1AX4 alleles, we generated the fbxA− mutation in the tgrB1QS31tgrC1QS31 background and repeated the experiment. Figure 2C shows that the incompatible strains tgrB1AX4tgrC1AX4 and tgrB1QS38tgrC1QS38 incurred a lower cost than the compatible strain tgrB1QS31tgrC1QS31. These results further support the finding that kin recognition can protect cooperators from cheating. They also indicate that the fbxA− mutation confers cheating in the tgrB1QS31tgrC1QS31 background and that the tgrB1QS31tgrC1QS31 strain is susceptible to cheating if the cheater carries compatible tgrB1-tgrC1 alleles. We conclude that the tgrB1-tgrC1 alleles, which mediate kin recognition, confer resistance to cheating by fbxA−.
We also examined the protective effect with different cheater frequencies in the populations. We found that the incompatible victims were protected at cheater-victim ratios of either 8:1:1 or 2:9:9 (Figure S1), suggesting that kin recognition protects cooperators against cheaters and the protection is independent of the genetic background and the cheater frequencies.
A potential complication is that the cheater-compatible victim might somehow distract the cheater from interacting with the incompatible victim. To test this possibility, we mixed the cheaters with two incompatible victims, excluding the compatible victim from the assay system. Figure S2 shows that both incompatible strains were protected from the cheater, suggesting that the protection is cell autonomous and independent of the other mixing partners.
A merodiploid strain bridges between incompatible cells
When cells with incompatible tgrB1/tgrC1 allele pairs develop in chimerae, they segregate from one another during the aggregation stage and eventually form clonal fruiting bodies [24]. To test if this segregation is required for protection from cheaters we used a condition that prevents segregation of incompatible strains. tgrB1/tgrC1 merodipoids are strains that carry two pairs of tgrB1/tgrC1 alleles. These strains cooperate with haploid strains that have either one of the matching tgrB1/tgrC1 pairs [24]. Figure 3A shows that merodiploid cells can bridge between incompatible haploid strains and thus prevent segregation during aggregation. When the incompatible haploid strains tgrB1AX4tgrC1AX4 and tgrB1QS31tgrC1QS31 were mixed with the merodiploid strain tgrB1+/QS31tgrC1+/QS31 at equal proportions, the incompatible strains cooperated with one another such that the three strains were equally distributed throughout the aggregate (Figure 3A, left). We propose that the merodiploid cells bridge between the incompatible haploid cells, because decreasing the proportion of the merodiploids in the mix resulted in partial segregation of the haploid cells (Figure 3A, middle). Bridging is also allele-specific because including the merodiploid strain tgrB1+/QS4tgrC1+/QS4, which is compatible with only one of the haploid strains (AX4), resulted in segregation of the two haploid strains (Figure 3A right).
Figure 3. Protection from cheaters depends on segregation.
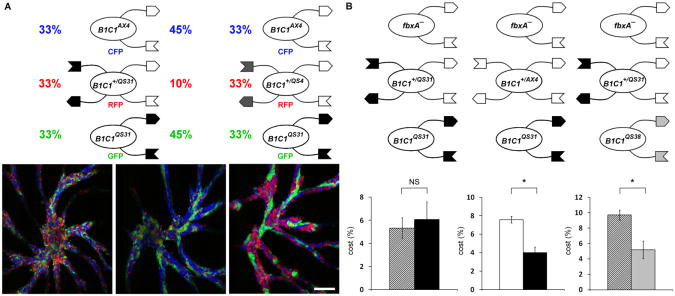
Cells were mixed at the indicated proportions (%) (A) or as described in Figure 2 (B) and allowed to develop. A, Fluorescence micrographs of mixed strains with different tgrB1-C1 alleles during aggregation. Left and middle, incompatible haploids tgrB1AX4tgrC1AX4–CFP and tgrB1QS31tgrC1QS31–GFP, and a merodiploid tgrB1+/QS31tgrC1+/QS31–RFP, which is compatible with both haploids; right, incompatible haploidstgrB1AX4tgrC1AX4–CFP and tgrB1QS31tgrC1QS31– GFP, and a merodiploid tgrB1+/QS4tgrC1+/QS4–RFP, which is compatible with the former haploid. Size bar = 125μm. B, fbxA−–cheater-protection assays in the presence of merodiploid strains. Bars indicate the cost to the merodiploid strains tgrB1+/QS31tgrC1+/QS31–GFP (slashed) and tgrB1+/AX4tgrC1+/Ax4–GFP (white) and to the haploid strains tgrB1QS31tgrC1QS31–RFP (black) and tgrB1QS38tgrC1QS38–RFP (grey). Data are means +/− s.e.m., n=3-7 per group, student's t-test, *p<0.03, NS p=0.57.
Kin-recognition-based protection depends on strain segregation
We examined the effect of merodiploid-bridging on cheater protection. We mixed fbxA− with the incompatible haploid tgrB1QS31tgrC1QS31 and the bridging merodiploid tgrB1+/QS31tgrC1+/QS31, tested the cost to the two victim strains and found that the costs were indistinguishable (Figure 3B, left). In the controls, where the merodiploid could not disrupt the segregation of the incompatible strains from the cheater, the cheater-compatible merodiploids tgrB1+/AX4tgrC1+/AX4 incurred a higher cost than the incompatible tgrB1QS31tgrC1QS31 haploids (Figure 3B, middle), as did the compatible merodiploid tgrB1+/QS31tgrC1+/QS31 compared with the incompatible haploid tgrB1QS38tgrC1QS38 (Figure 3B, right). These results suggest that strain segregation is required for kin-recognition-mediated protection against cheaters.
Kin recognition protects against different facultative cheaters
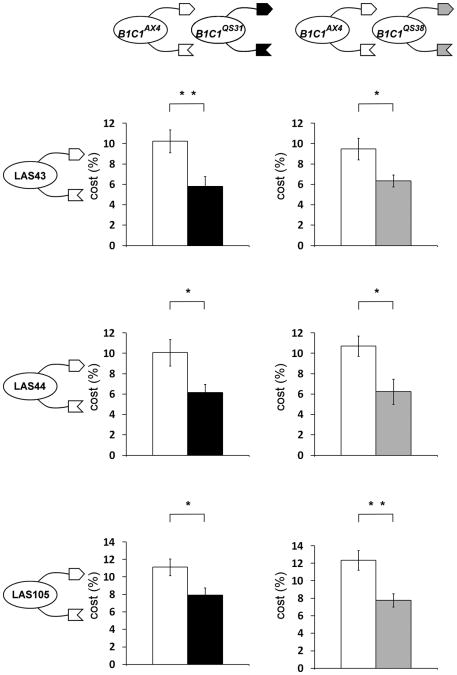
We tested whether kin recognition could protect against different types of cheaters. The fbxA− cells are obligatory cheaters, which depend on the presence of a victim for sporulation [6]. We therefore tested facultative cheaters that can sporulate well in pure populations. Strains LAS43, LAS44, and LAS105 were generated in the AX4 background and are distinct in their cheater mutations and phenotypes [21]. We tested them in the cheater-protection assay with the incompatible strains tgrB1QS31tgrC1QS31 and tgrB1QS38tgrC1QS38. Figure 4 shows that the incompatible strains incurred a lower cost than the compatible tgrB1AX4tgrC1AX4 in all three cases, suggesting that kin recognition provides general protection against cheaters.
Figure 4. Kin-recognition protects against various cheaters.
Cells were developed in chimerae and spores counted to determine cost. The facultative cheaters LAS43, LAS44, and LAS105, made in the AX4 background, were used in independent assays as indicated on the left. Bars represent cost to the compatible strain tgrB1AX4tgrC1AX4–GFP (white) and the incompatible strains tgrB1QS31tgrC1QS31–RFP (black) and tgrB1QS38tgrC1QS38–RFP (grey). Data are means +/− s.e.m., n=4-5 per group, student's t-test, *p<0.05, ** p<0.03.
Discussion
Our results support the hypothesis that kin recognition protects against cheaters because victims that were incompatible with the cheaters incurred a lower cost compared with cheater-compatible victims. However, that protection was not absolute as non-kin victims incurred some cost in many cases. Kin-recognition systems are inherently imperfect [3, 25], so imperfect protection from non-kin cheaters could be the result of incomplete segregation between the incompatible strains. Indeed, segregation in D. discoideum is also not completely exclusive even between the most incompatible wild isolates [22]. Moreover, the merodiploid-cheater mixing experiment (Figure 3B) shows that segregation is a key factor in protection, so non-kin victims probably incur costs from the cheaters because of imperfect segregation. In addition, facultative cheaters can exploit victims by either “self-promotion” or “coercion” [19]. Segregation-mediated protection may be less effective against cheating through the “self-promotion” strategy. Finally, some cheaters could inflict damage prior to segregation [18], but we could not test them because relevant mutations are not known.
D. discoideum cheaters are abundant in nature and are found in close proximity with their victims [17, 20]. Our results suggest that kin recognition is a general defense mechanism that protects against various cheaters. This recognition system limits cheaters to their kin whereas non-kin individuals are protected. However, kin-victims are not completely defenseless from an evolutionary standpoint. The presence of cheaters in a population might select for new tgrB1/tgrC1 allotypes (Figure 1), which could lead to an evolutionary ‘arms-race’ between cheating and kin recognition. This possibility is consistent with the observation that tgrB1 and tgrC1 are highly polymorphic in nature [23]. Other mechanisms also likely play a role in controlling D. discoideum cheaters that are limited to specific cheating mechanisms [10, 12].
Kin recognition has evolved independently in different organisms [26-31] and our results provide direct evidence that it may function in protection from cheaters. Polymorphism is a key element in recognition systems, and the prevalence of cheaters may be an evolutionary pressure that maintains such diversity, along with other selective pressures such as host-parasite interactions [32, 33]. Such evolutionary pressures have been demonstrated in numerous interspecific host-parasite and host-pathogen relationships [3], but not in intraspecific systems such as the one described here.
Experimental Procedures
Cell growth and development
Cells were cultured as described [23]. Media for ura− strains were supplemented with 20 μg/ml uracil and drugs (10 μg/ml G418, 5 μg/ml Blasticidin S) were added as necessary but removed 48 hours before development. For development, cells were washed once with water, resuspended in PDF buffer (20.1 mM KCl, 1 mM CaCl2, 2.5mM MgSO4, 9.2 mM K2HPO4, 13.2 mM KH2PO4, pH = 6.4), deposited on nitrocellulose filters at 1×106 cells/cm2 and incubated in dark humid chambers for 48-72 hours.
Cheater-protection assay
Each set of experiments consisted of two 3-way mixes. In the control, wild-type cells were mixed with GFP- and RFP-labeled strains at a 4:3:3 ratio. The fluorescently-labeled cells differed in their tgrB1-C1 alleles, whereas one of them was compatible with the wild type. In the experiment, the wild type was replaced with a cheater mutant that carried the wild-type tgrB1-C1 alleles. Controls and experiments were performed side-by-side and developed under the same conditions. In each instance, we also developed pure populations of the fluorescent strains and measured the proportion of fluorescent spores produced by each strain. We only considered experiments in which the proportion of fluorescent spores was ≥90% and we used the particular proportion to scale the results of the experiments.
Following development, we harvested the spores in 0.1% NP40 made in KK2 buffer. The total sporulation efficiencies of control and experiment were usually indistinguishable from each other. We measured the proportion of fluorescent spores by flow cytometry (LSRFortessa), counting at least 10,000 spores each time. Cost was calculated by subtracting the percentage of green- or red- fluorescent spores in the experiment from the respective control.
Where GSP is the GFP spore percentage, RSP is the RFP spore percentage, c is the control mix and e is the experiment mix.
For instance, if the GFP- and RFP- spore percentages in the control were 30% each, and in the experiment the GFP- and RFP- spore percentages were 23% each when mixed with a certain cheater, then the cost incurred by the GFP- and RFP- strains would be 30% – 23% = 7%. Each experiment was performed at least in triplicates, done on different days. We evaluated the significance of the differences between the costs to the two fluorescently-labeled strains using a two-tailed student's t-test.
To examine the potential effects of differential fluorescence labeling, we mixed fbxA− with a mix of compatible isogenic GFP- and RFP- labeled tgrB1AX4tgrC1AX4 cells (Figure S3A). We also repeated the cheater-protection assay, which was described in Figure 2B, left panel, with strains in which we switched the GFP- and RFP- labels of the compatible and incompatible strains (Figure S3B). The fluorescent labels did not affect the results in either case.
Segregation assay
We grew the fluorescently-labeled cells in pure populations, washed the cells, mixed them at the indicated proportions at a density of 1×107 cells/ml in PDF buffer, deposited them in 40μl drops on a 5 cm agar plate (2% Noble Agar in KK2 buffer), incubated them in a dark humid chamber and photographed them at the streaming stage (8-12 hours) with fluorescence microscopy.
Supplementary Material
Highlights.
Kin recognition protects D. discoideum against cheaters
The protection depends on kin discriminatory segregation
Kin-recognition protects against different types of cheaters
Acknowledgments
We thank Richard L. Kelley for fruitful suggestions, Elizabeth A. Ostrowski for unpublished genome sequence results and Elizabeth A. Ostrowski, Michelle J. Rubin and Patrick G. Mitchell for discussions and critical reading of the manuscript. This work was supported by grants R01 GM084992 and R01 GM098276 from the National Institutes of Health. H. Ho was a Howard Hughes Medical Institute International Student Research fellow. This project was supported by the Cytometry and Cell Sorting Core at Baylor College of Medicine with funding from the NIH (AI036211, CA125123, and RR024574) and the expert assistance of Joel M. Sederstrom.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Travisano M, Velicer GJ. Strategies of microbial cheater control. Trends in microbiology. 2004;12:72–78. doi: 10.1016/j.tim.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 2.West SA, Griffin AS, Gardner A. Evolutionary explanations for cooperation. Current biology: CB. 2007;17:R661–672. doi: 10.1016/j.cub.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Bourke AFG. Principles of social evolution. Oxford; New York: Oxford University Press; 2011. [Google Scholar]
- 4.Hamilton WD. The genetical evolution of social behaviour. II. Journal of theoretical biology. 1964;7:17–52. doi: 10.1016/0022-5193(64)90039-6. [DOI] [PubMed] [Google Scholar]
- 5.Kessin RH. Evolutionary biology. Cooperation can be dangerous. Nature. 2000;408:917–919. doi: 10.1038/35050184. [DOI] [PubMed] [Google Scholar]
- 6.Ennis HL, Dao DN, Pukatzki SU, Kessin RH. Dictyostelium amoebae lacking an F-box protein form spores rather than stalk in chimeras with wild type. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:3292–3297. doi: 10.1073/pnas.050005097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Velicer GJ, Kroos L, Lenski RE. Developmental cheating in the social bacterium Myxococcus xanthus. Nature. 2000;404:598–601. doi: 10.1038/35007066. [DOI] [PubMed] [Google Scholar]
- 8.Diggle SP, Griffin AS, Campbell GS, West SA. Cooperation and conflict in quorum-sensing bacterial populations. Nature. 2007;450:411–414. doi: 10.1038/nature06279. [DOI] [PubMed] [Google Scholar]
- 9.Hardin G. The Tragedy of the Commons. Science (New York, NY) 1968;162:1243–1248. [PubMed] [Google Scholar]
- 10.Foster KR, Shaulsky G, Strassmann JE, Queller DC, Thompson CR. Pleiotropy as a mechanism to stabilize cooperation. Nature. 2004;431:693–696. doi: 10.1038/nature02894. [DOI] [PubMed] [Google Scholar]
- 11.Gilbert OM, Foster KR, Mehdiabadi NJ, Strassmann JE, Queller DC. High relatedness maintains multicellular cooperation in a social amoeba by controlling cheater mutants. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8913–8917. doi: 10.1073/pnas.0702723104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khare A, Santorelli LA, Strassmann JE, Queller DC, Kuspa A, Shaulsky G. Cheater-resistance is not futile. Nature. 2009;461:980–982. doi: 10.1038/nature08472. [DOI] [PubMed] [Google Scholar]
- 13.Buss LW. Somatic cell parasitism and the evolution of somatic tissue compatibility. Proceedings of the National Academy of Sciences of the United States of America. 1982;79:5337–5341. doi: 10.1073/pnas.79.17.5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grosberg RK, Quinn JF. The genetic control and consequences of kin recognition by the larvae of a colonial marine invertebrate. Nature. 1986;322:456–459. [Google Scholar]
- 15.Stoner DS, Weissman IL. Somatic and germ cell parasitism in a colonial ascidian: possible role for a highly polymorphic allorecognition system. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:15254–15259. doi: 10.1073/pnas.93.26.15254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kessin RH, Franke J. Dictyostelium : evolution, cell biology, and the development of multicellularity. Cambridge, UK ; New York, NY, USA: Cambridge University Press; 2001. [Google Scholar]
- 17.Strassmann JE, Zhu Y, Queller DC. Altruism and social cheating in the social amoeba Dictyostelium discoideum. Nature. 2000;408:965–967. doi: 10.1038/35050087. [DOI] [PubMed] [Google Scholar]
- 18.Shaulsky G, Kessin RH. The cold war of the social amoebae. Curr Biol. 2007;17:R684–692. doi: 10.1016/j.cub.2007.06.024. [DOI] [PubMed] [Google Scholar]
- 19.Buttery NJ, Rozen DE, Wolf JB, Thompson CR. Quantification of social behavior in D. discoideum reveals complex fixed and facultative strategies. Current biology : CB. 2009;19:1373–1377. doi: 10.1016/j.cub.2009.06.058. [DOI] [PubMed] [Google Scholar]
- 20.Fortunato A, Strassmann JE, Santorelli L, Queller DC. Co-occurrence in nature of different clones of the social amoeba, Dictyostelium discoideum. Molecular ecology. 2003;12:1031–1038. doi: 10.1046/j.1365-294x.2003.01792.x. [DOI] [PubMed] [Google Scholar]
- 21.Santorelli LA, Thompson CR, Villegas E, Svetz J, Dinh C, Parikh A, Sucgang R, Kuspa A, Strassmann JE, Queller DC, et al. Facultative cheater mutants reveal the genetic complexity of cooperation in social amoebae. Nature. 2008;451:1107–1110. doi: 10.1038/nature06558. [DOI] [PubMed] [Google Scholar]
- 22.Ostrowski EA, Katoh M, Shaulsky G, Queller DC, Strassmann JE. Kin discrimination increases with genetic distance in a social amoeba. PLoS biology. 2008;6:e287. doi: 10.1371/journal.pbio.0060287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benabentos R, Hirose S, Sucgang R, Curk T, Katoh M, Ostrowski EA, Strassmann JE, Queller DC, Zupan B, Shaulsky G, et al. Polymorphic members of the lag gene family mediate kin discrimination in Dictyostelium. Current biology : CB. 2009;19:567–572. doi: 10.1016/j.cub.2009.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirose S, Benabentos R, Ho HI, Kuspa A, Shaulsky G. Self-recognition in social amoebae is mediated by allelic pairs of tiger genes. Science (New York, NY) 2011;333:467–470. doi: 10.1126/science.1203903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reeve HK. The Evolution of Conspecific Acceptance Thresholds. The American Naturalist. 1989;133:407–435. [Google Scholar]
- 26.De Tomaso AW, Nyholm SV, Palmeri KJ, Ishizuka KJ, Ludington WB, Mitchel K, Weissman IL. Isolation and characterization of a protochordate histocompatibility locus. Nature. 2005;438:454–459. doi: 10.1038/nature04150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Griffin AS, West SA. Kin discrimination and the benefit of helping in cooperatively breeding vertebrates. Science (New York, NY) 2003;302:634–636. doi: 10.1126/science.1089402. [DOI] [PubMed] [Google Scholar]
- 28.Hughes AL, Nei M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature. 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- 29.Mehdiabadi NJ, Jack CN, Farnham TT, Platt TG, Kalla SE, Shaulsky G, Queller DC, Strassmann JE. Social evolution: kin preference in a social microbe. Nature. 2006;442:881–882. doi: 10.1038/442881a. [DOI] [PubMed] [Google Scholar]
- 30.Pfennig DW, Collins JP, Ziemba RE. A test of alternative hypotheses for kin recognition in cannibalistic tiger salamanders. Behavioral Ecology. 1999;10:436–443. [Google Scholar]
- 31.Rosa SF, Powell AE, Rosengarten RD, Nicotra ML, Moreno MA, Grimwood J, Lakkis FG, Dellaporta SL, Buss LW. Hydractinia allodeterminant alr1 resides in an immunoglobulin superfamily-like gene complex. Current biology : CB. 2010;20:1122–1127. doi: 10.1016/j.cub.2010.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rousset F, Roze D. Constraints on the origin and maintenance of genetic kin recognition. Evolution; international journal of organic evolution. 2007;61:2320–2330. doi: 10.1111/j.1558-5646.2007.00191.x. [DOI] [PubMed] [Google Scholar]
- 33.Giron D, Strand MR. Host resistance and the evolution of kin recognition in polyembryonic wasps. Proceedings. Biological sciences/The Royal Society. 2004;271(6):S395–398. doi: 10.1098/rsbl.2004.0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.