Abstract
Prolonged antibiotic treatment can lead to detrimental side effects in patients, including ototoxicity, nephrotoxicity, and tendinopathy, yet the mechanisms underlying the effects of antibiotics in mammalian systems remain unclear. It has been suggested that bactericidal antibiotics induce the formation of toxic reactive oxygen species (ROS) in bacteria. We show that clinically relevant doses of bactericidal antibiotics—quinolones, aminoglycosides, and β-lactams—cause mitochondrial dysfunction and ROS overproduction in mammalian cells. We demonstrate that these bactericidal antibiotic–induced effects lead to oxidative damage to DNA, proteins, and membrane lipids. Mice treated with bactericidal antibiotics exhibited elevated oxidative stress markers in the blood, oxidative tissue damage, and up-regulated expression of key genes involved in antioxidant defense mechanisms, which points to the potential physiological relevance of these antibiotic effects. The deleterious effects of bactericidal antibiotics were alleviated in cell culture and in mice by the administration of the antioxidant N-acetyl-L-cysteine or prevented by preferential use of bacteriostatic antibiotics. This work highlights the role of antibiotics in the production of oxidative tissue damage in mammalian cells and presents strategies to mitigate or prevent the resulting damage, with the goal of improving the safety of antibiotic treatment in people.
INTRODUCTION
Antibiotics have led to an extraordinary decrease in morbidity and mortality associated with bacterial infections. Yet, despite the great benefits, antibiotic use has been linked to various adverse side effects, including ototoxicity (1), nephrotoxicity (2), and tendinopathy (3). Although antibiotic targets and modes of action have been widely studied and well characterized in bacteria, the mechanistic effects of commonly prescribed antibiotics on mammalian cells remain unclear. Recently, it has been demonstrated that major classes of bactericidal antibiotics, irrespective of their drug-target interactions, induce a common oxidative damage cellular death pathway in bacteria, leading to the production of lethal reactive oxygen species (ROS) (4–12) via disruption of the tricarboxylic acid (TCA) cycle and electron transport chain (ETC) (4, 6). The role of ROS in antibiotic-induced bacterial killing is currently a matter of debate (13, 14) and the subject of intense experimental investigation in our laboratory and other laboratories; however, the techniques critiqued in (13, 14) were not used in the present study, which focuses on mammalian systems.
Bactericidal and bacteriostatic antibiotics have been shown to target mitochondrial components (15–20). In mammalian cells, the mitochondrial ETC is a major source of ROS during normal metabolism because of leakage of electrons (21). Given the proposed bacterial origin of mitochondria (22), we hypothesized that bactericidal antibiotics commonly disrupt mitochondrial function in mammalian cells, leading to oxidative stress and oxidative damage. Previous work has shown that mammalian cells can be damaged by antibiotic treatment, but these results were shown at concentrations considerably higher than those applied clinically. At these high concentrations, select antibiotics inhibited cell growth and metabolic activity, in addition to impairing mitochondrial function in vitro (23, 24).
Here, we focused on characterizing the mechanistic effects of clinically relevant levels of bactericidal antibiotics on mammalian cells, both in vitro and in vivo. We showed that bactericidal antibiotics— quinolones, aminoglycosides, and β-lactams—caused mitochondrial dysfunction and ROS overproduction in mammalian cells, ultimately leading to the accumulation of oxidative tissue damage. We found that these deleterious effects could be alleviated by administration of the Food and Drug Administration (FDA)–approved antioxidant, N-acetyl- L-cysteine (NAC), or prevented by preferential use of bacteriostatic antibiotics. These results reflect two therapeutic strategies to combat the adverse side effects of long-term antibiotic treatment.
RESULTS
Bactericidal antibiotics induce oxidative stress and damage in mammalian cells
We first examined whether clinically relevant doses of antibiotics induce the formation of ROS in mammalian cells. Here, clinically relevant doses are defined by peak serum levels (25). We exposed a human mammary epithelial cell line, MCF10A, to representative bactericidal antibiotics from three different classes: ciprofloxacin (a fluoroquinolone), ampicillin (a β-lactam), and kanamycin (an aminoglycoside). All three bactericidal antibiotics induced a dose- and time-dependent increase in intracellular ROS production (Fig. 1A and fig. S1), but a bacteriostatic antibiotic (tetracycline) did not lead to a significant increase in ROS production (Fig. 1A). Notably, bactericidal antibiotic–induced ROS was not elevated in mammary epithelial cells as a result of an increased presence of dead or dying cells in vitro (fig. S2). To establish that the observed bactericidal antibiotic–induced oxidative stress was not cell line–specific, we tested several additional cell types, including primary human aortic endothelial cells (PAEC), primary human mammary epithelial cells (HMEC), human gut epithelial cells (CACO-2), and normal human diploid skin fibroblasts (NHDF). In all instances, we found the same pattern of significantly elevated ROS levels induced by bactericidal antibiotics (fig. S3), with little effect seen after bacteriostatic antibiotic treatment.
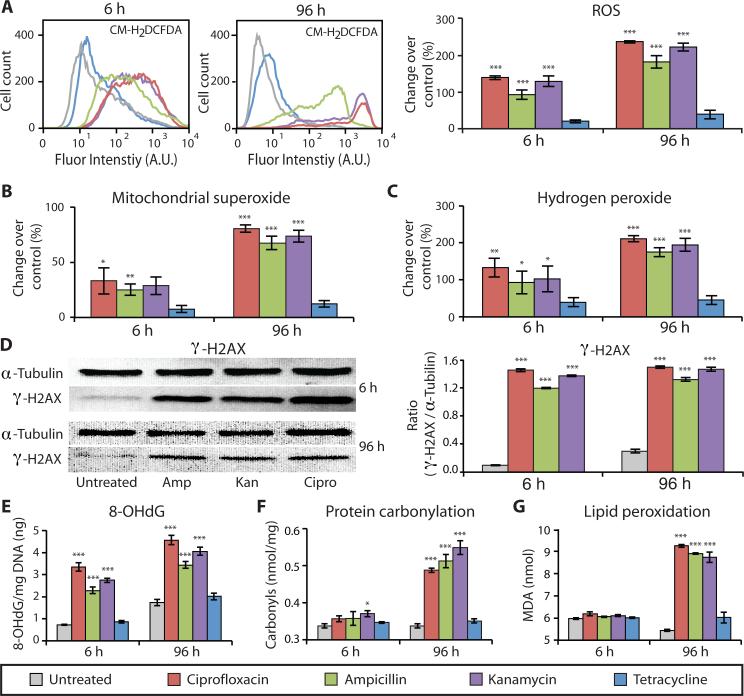
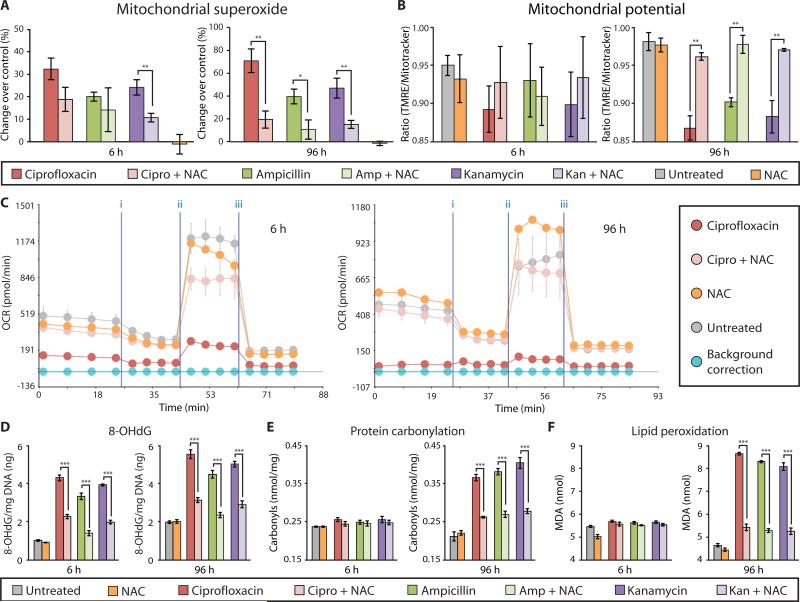
Figure 1. Bactericidal antibiotics cause oxidative damage to mammalian cells.
ROS and oxidative damage were measured in human mammary epithelial cells (MCF10A) after 6 and 96 h of treatment with bactericidal [ciprofloxacin (10 μg/ml), ampicillin (20 μg/ml), kanamycin (25 μg/ml)] or bacteriostatic [tetracycline (10 μg/ml)] antibiotics, compared to untreated cells. (A) ROS was quantified using CM-H2DCFDA by flow cytometry (histograms on left) and a microplate spectrophotometer (bar graphs on right). (B) Mitochondrial superoxide was measured using MitoSox Red. (C) Hydrogen peroxide release was quantified by measuring Amplex Red fluorescence. (D) Antibiotic-induced DNA damage was evaluated by Western blot analysis to measure the abundance of the phosphorylated histone protein, γ-H2AX, compared to α-tubulin serving as the loading control (quantification in the bar graph). (E) Additionally, oxidative DNA damage was assessed by measuring the abundance of 8-hydroxy-2‘-deoxyguanosine (8-OHdG). (F) Oxidative protein damage, protein carbonylation, was detected using an enzyme-linked immunosorbent assay (ELISA). (G) Lipid peroxidation was evaluated by measuring malondialdehyde (MDA). All bar graphs display means ± S.E.M. (n ≥ 3). Comparisons between treatments and untreated controls were made using a Student's t test (*p < 0.5, **p < 0.01, ***p < 0.001).
Superoxide, a reactive oxygen byproduct generated by leakage of electrons from the ETC to oxygen, is a precursor to many other forms of ROS, one instance being the enzymatic conversion into hydrogen peroxide (H2O2) through superoxide dismutase (26). We measured mitochondrial superoxide production and extracellular H2O2 release in mammalian (human) MCF10A cells and found that all three bactericidal antibiotics induced a dose- and time-dependent increase in both mitochondrial superoxide (Fig. 1B and fig. S4) and H2O2 (Fig. 1C and fig. S5). Bacteriostatic antibiotic treatment did not lead to a significant increase in superoxide (Fig. 1B) or H2O2 production (Fig. 1C).
ROS can directly interact with cellular components resulting in DNA, protein, and lipid damage. To characterize DNA damage induced by bactericidal antibiotics in mammalian cells, we used indirect immunofluorescence and Western blot analysis to measure γ-H2AX, a core histone protein that is phosphorylated in response to DNA damage. We observed a persistent and significant increase in γ-H2AX in MCF10A cells exposed to bactericidal antibiotics after 6 and 96 hours of treatment compared with untreated cells (Fig. 1D and fig. S6). Additionally, we quantified the presence of 8-hydroxy-2’-deoxyguanosine (8-OHdG), an oxidized DNA byproduct. Similar to γ-H2AX, we found significantly elevated levels of 8-OHdG after 6 and 96 hours of bactericidal antibiotic treatment (Fig. 1E) but negligible change in response to tetracycline compared with untreated controls.
To further investigate the accumulation of cellular oxidative damage, we measured levels of protein carbonyls, a modification of proteins resulting from oxidative damage, and levels of malondialdehyde (MDA), an end product of lipid peroxidation. We found significantly elevated levels of protein carbonylation (Fig. 1F) and lipid peroxidation (Fig. 1G) in MCF10A cells after 96 hours of bactericidal antibiotic treatment. Consistent with the general ROS, mitochondrial superoxide, and H2O2 results, bacteriostatic antibiotics had little effect on protein carbonylation or lipid peroxidation (Fig. 1, F and G).
Bactericidal antibiotics induce mitochondrial dysfunction
In bacteria, the common mechanism of killing by bactericidal antibiotics advances the finding that toxic ROS are generated via the disruption of the TCA cycle and ETC (4). Bacteriostatic antibiotics do not stimulate ROS production in bacteria, suggesting that bactericidal antibiotics uniquely affect major sources of ROS. In mammalian cells, mitochondria are major sources of intracellular ROS; therefore, we tested the hypothesis that bactericidal antibiotics uniquely disrupt the function of the mitochondrial ETC, leading to the observed overproduction of ROS and oxidative damage (Fig. 1).
We measured the inhibitory effects of bactericidal antibiotics on individual, immunocaptured ETC complexes I to V. Bactericidal antibiotics inhibited complex I activity by 16 to 25% and complex III activity by 30 to 40% compared to untreated samples (Fig. 2A), whereas bacteriostatic antibiotics and negative controls exhibited only 5 to 10% inhibition (fig. S7). In addition, we observed a varied degree of inhibition of complexes II and IV across the tested bactericidal antibiotics, with complex V showing less change in activity compared to complexes I to IV. These data indicate that bactericidal antibiotics inhibit mitochondrial ETC complexes, in particular complexes I and III, which have been identified as major sources of ROS formation (27).
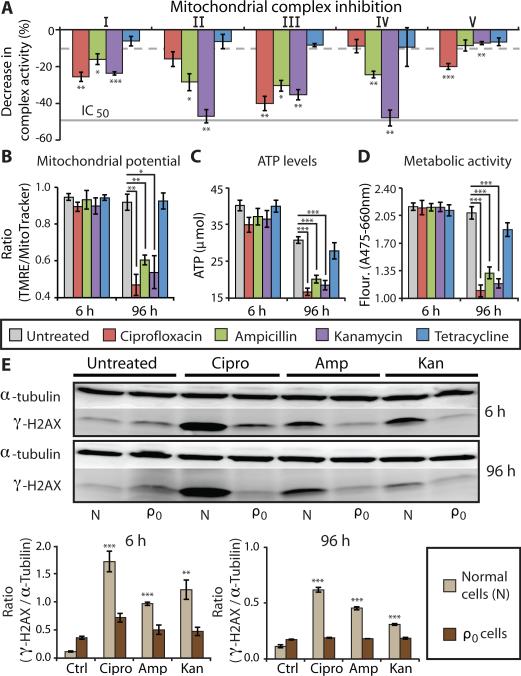
Figure 2. Bactericidal antibiotics induce mitochondrial dysfunction.
(A) The effects of bactericidal [ciprofloxacin (10 μg/ml), ampicillin (20 μg/ml), kanamycin (25 μg/ml)] or bacteriostatic [tetracycline, (10 μg/ml)] antibiotics on the function of individual, isolated ETC protein complexes was measured. Bars represent the change in activity of individual antibiotic-treated complexes compared to untreated complexes. The dashed grey line represents the maximum inhibition (10%) seen across several, independent negative controls. The solid grey line represents the IC50 of positive control drugs shown to inhibit specific target complexes. (See fig. S7 for all positive and negative control results.) (B) Mitochondrial membrane potential was quantified using TMRE and MitoTracker Green. (C) ATP levels were measured using a luciferin/luciferase assay. (D) Metabolic activity was measured using a colorimetric tetrazolium dye (XTT). (E) DNA damage in normal (N) and mtDNA-depleted (ρ0) MCF10A cells was measured by Western blot to evaluate the ratio of γ-H2AX compared to the loading control, α-tubulin. Quantification is shown below the blots. All bar graphs display means ± S.E.M. (n ≥ 3). Comparisons between treatments and untreated controls were made using a Student's t test (*p < 0.5, **p < 0.01, ***p < 0.001).
Because mitochondrial energy metabolism is tightly linked to organelle function, disruption of the ETC should lead to a decrease in mitochondrial membrane potential (ΔΨm), adenosine triphosphate (ATP) levels, and overall metabolic activity. Indeed, bactericidal antibiotic treatment led to a corresponding and significant decrease in all three metabolic functions after 96 hours of treatment (Fig. 2, B to D), further supporting the hypothesis that bactericidal antibiotics induce mitochondrial dysfunction.
If bactericidal antibiotics induce ROS by impairing the function of the ETC, then cells devoid of a functional ETC should show little ROS formation after antibiotic treatment. Mammalian cells devoid of mitochondrial DNA (mtDNA), but retaining their nuclear genomes, lack functional ETC complexes; these cells are referred to as ρ0 cells. By selectively eliminating mtDNA and providing nutrient supplementation, we generated an MCF10A ρ0 cell line (fig. S8A). Bactericidal antibiotic treatments of ρ0 cells showed no difference in ROS production when compared to untreated cells (fig. S8B). This was in sharp contrast to the large, significant increase in ROS production stimulated by bactericidal antibiotic treatment in normal MCF10A cells (Fig. 1 and fig. S8B). Furthermore, we found relatively high levels of nuclear DNA damage (γ-H2AX) in normal cells after bactericidal antibiotic treatments, whereas the level of DNA damage in ρ0 cells was similar to untreated controls (Fig. 2E). These results suggest that the mitochondrial ETC is a major source of bactericidal antibiotic–induced intracellular ROS.
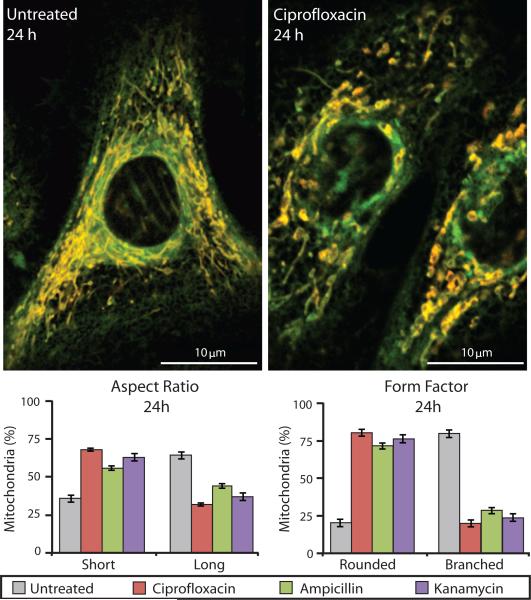
The homeostatic balance between mitochondrial fission and fusion can be altered in response to oxidative stress (28). Disruption of this balance can lead to changes in mitochondrial morphology and function, resulting in the generation of ROS (28). Using live-cell imaging of primary human mammary epithelial cells, we measured the morphological changes in mitochondria and found short, swollen, fragmented mitochondria (smaller aspect ratio) with highly reduced branching (smaller form factor) in bactericidal antibiotic–treated cells compared to long, tubular (larger aspect ratio), and extensively branched (larger form factor) mitochondria in untreated cells (Fig. 3). These results suggest that bactericidal antibiotics shift the balance toward a profission state. Consistent with our data in Fig. 2, a profission state can lead to a loss of membrane potential, loss of metabolic activity, and an overall increase in oxidative stress (28).
Figure 3. Mitochondria in bactericidal antibiotic–treated cells show an abnormal, profission state.
Mitochondrial morphology was measured in primary human mammary epithelial cells using TMRE and MitoTracker Green. Untreated samples (left image) show mitochondria with normal morphology, which are long and highly branched. Ciprofloxacin-treated cells (10 μg/ml) (right image) had abnormally short and truncated mitochondria. The percentages of mitochondria with a short versus long aspect ratio and rounded versus branched form factor were measured for untreated and bactericidal antibiotic–treated cells. Data are means ± S.E.M. (n ≥ 5).
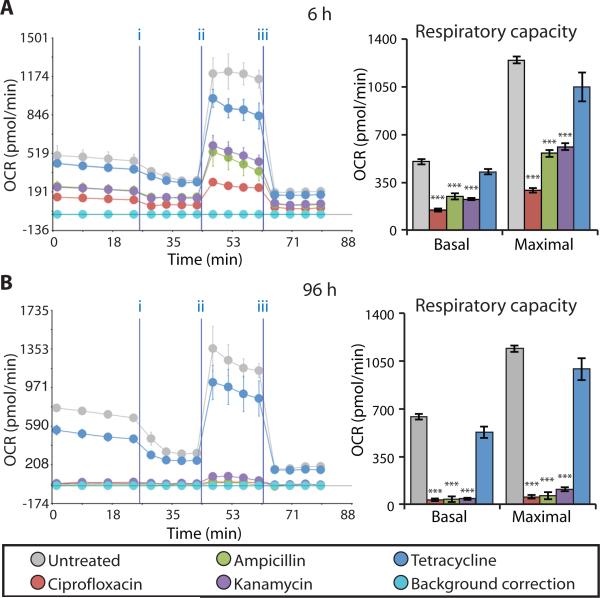
Thus far, we have characterized isolated processes involved in mitochondrial respiration. To directly test the mitochondrial respiratory capacity of antibiotic-treated cells, we measured changes in the oxygen consumption rate (OCR) of intact MCF10A cells using the Seahorse XF24 flux analyzer. Compared to untreated cells, bactericidal antibiotic–treated cells exhibited a significant reduction in both basal respiration and maximal respiratory capacity after 6 hours of treatment (Fig. 4A), which was further reduced after 96 hours of treatment (Fig. 4B). In contrast, cells treated for 6 and 96 hours with tetracycline showed little change in respiratory capacity compared to untreated cells (Fig. 4). These data demonstrate that bactericidal, but not bacteriostatic, antibiotics impair the function of mitochondria and, consequently, the overall respiratory capacity of the cell.
Figure 4. Bactericidal antibiotics decrease mitochondrial basal respiration and maximal respiratory capacity.
(A and B) Oxygen consumption rate (OCR) was measured in MCF10A cells after 6 h (A) and 96 h (B) of bactericidal [ciprofloxacin (10 μg/ml), ampicillin (20 μg/ml), kanamycin (25 μg/ml)] or bacteriostatic [tetracycline, (10 μg/ml)] antibiotic treatment. Cells were treated with antibiotics followed by the Seahorse OCR protocol including treatment with three mitochondrial ETC complex inhibitors: (i) oligomycin, (ii) FCCP, and (iii) antimycin A. OCR measured before (i) represents basal respiration, while OCR measured between (ii) and (iii) represents the maximal respiratory capacity. Representative Seahorse OCR plots are shown and the bar graphs are means ± S.D. for n = 3. Comparisons between treatments and untreated controls were made using a Student's t test (***p < 0.001).
Antibiotic-induced oxidative damage is rescued by an antioxidant
Characterization of bactericidal antibiotic effects on mammalian cells points to widespread cellular oxidative damage. Therefore, we explored the possibility of using an antioxidant to alleviate these deleterious effects. We chose to test the ability of NAC to alleviate bactericidal antibiotic–induced oxidative damage in mammalian cells. NAC was selected because it is an FDA-approved antioxidant that is well tolerated by patients and commonly used to buffer extraneous intracellular ROS in mammalian systems (29, 30). MCF10A cells were pretreated with NAC for 2 hours, followed by bactericidal antibiotic treatment for 6 and 96 hours. NAC pretreatment reduced bactericidal antibiotic–induced ROS levels (Fig. 5A and fig. S9) and restored mitochondrial membrane potential to levels seen in untreated cells after 96 hours of antibiotic treatment (Fig. 5B). Furthermore, NAC restored basal respiration and maximal respiratory capacity of bactericidal antibiotic–treated cells to near-normal levels (Fig. 5C and figs. S10 and S11) and alleviated bactericidal antibiotic–induced DNA damage (γ-H2AX) (fig. S12), 8-OHdG formation (Fig. 5D), protein carbonylation (Fig. 5E), and lipid peroxidation (Fig. 5F) after only 6 hours of treatment. Cells treated with NAC alone showed no significant effects from untreated cells (Student's t test).
Figure 5. NAC rescues bactericidal antibiotic–induced oxidative damage in vitro.
MCF10A cells were incubated with and without NAC (10 mM) for 2 h, followed by treatment with ciprofloxacin (10 μg/ml), ampicillin (20 μg/ml), kanamycin (25 μg/ml), or tetracycline (10 μg/ml) for 6 and 96 h. (A) Mitochondrial superoxide was measured using MitoSox Red. (B) Mitochondrial membrane potential was quantified using TMRE and MitoTracker Green. (C) OCR was measured using the Seahorse XF24 flux analyzer, and a representative diagram testing the ciprofloxacin + NAC is shown. (See fig. S10 and S11 for ampicillin and kanamycin). (D) Oxidative DNA damage was assessed by quantification of 8-OHdG. (E) Protein carbonyls were quantified to evaluate oxidative protein damage. (F) Lipid peroxidation was measured by quantifying malondialdehyde (MDA). Data are means ± S.E.M. (n ≥ 3). Comparisons between treatment + NAC and treatments were made using a Student's t test (*p < 0.5, **p < 0.01, ***p < 0.001).
Combining an antioxidant with a bactericidal antibiotic carries the potential risk of reducing the bacterial killing efficacy of the antibiotic, given that bactericidal antibiotic–induced ROS formation facilitates bacterial killing (4). Tested in bacterial cultures, we found that the concentration of NAC used to rescue antibiotic-induced oxidative damage in mammalian cells did not decrease the bacterial killing efficacy of bactericidal antibiotics (fig. S13). We extended this study to test the antimicrobial efficacy of the antibiotic-NAC combination in a mouse urinary tract infection (UTI) model. Escherichia coli were transurethrally introduced into the bladder of mice. Twenty-four hours after establishing infection, mice were treated with ciprofloxacin (50 μg/ml), NAC (10 mM), and ciprofloxacin + NAC or untreated (vehicle only). Ciprofloxacin + NAC and ciprofloxacin-only treatments showed similar levels of bacterial killing (fig. S14), suggesting that NAC does not interfere with the antimicrobial activity of clinically relevant doses of bactericidal antibiotics. Given that other antioxidants, such as glutathione, have been shown to reduce the bacterial killing efficacy of bactericidal antibiotics (8), further work is needed to characterize the effects of NAC on both Gram-negative and Gram-positive bacteria.
Oxidative damage in vivo caused by bactericidal antibiotics can be rescued
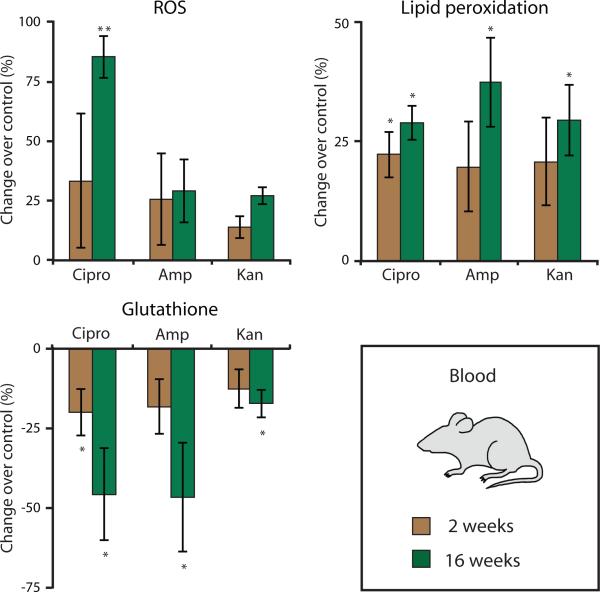
To determine the in vivo relevance of bactericidal antibiotic–induced ROS production and mitochondrial dysfunction, we investigated oxidative stress markers and accumulation of oxidative damage in tissues of mice treated with antibiotics. Mice received clinically relevant doses of antibiotics [ciprofloxacin (12.5 mg/kg per day), ampicillin (28.5 mg/kg per day), and kanamycin (15 mg/kg per day)] in their drinking water (n ≥ 3 animals per treatment group). At 2 and 16 weeks, peripheral blood was drawn from the same animals to measure intracellular ROS, lipid peroxidation, and glutathione levels. A reduction in glutathione is a proxy measure for ROS production because it is an intracellular scavenger of ROS and a key component of the enzymatic antioxidant system (31).
Bactericidal antibiotic–treated mice showed elevated lipid peroxidation in their peripheral blood; these increases were statistically significant after 16 weeks of treatment (Fig. 6). The treated mice also exhibited reduced glutathione levels after 2 weeks of antibiotic treatment, which was significantly decreased after 16 weeks of treatment (Fig. 6). However, only ciprofloxacin led to a significant increase in ROS in treated animals after 16 weeks.
Figure 6. Bactericidal antibiotics induce oxidative damage in mice.
Oxidative stress markers were measured in blood drawn from wild-type mice treated with ciprofloxacin (12.5 mg/kg/day), ampicillin (28.5 mg/kg/day), or kanamycin (15 mg/kg/day) for 2 or for 16 weeks. Data are means ± S.E.M. (n ≥ 3 animals per treatment group). Comparisons between treatments and untreated controls were made using a Student's t test (*p < 0.5, **p < 0.01).
To further characterize the in vivo oxidative stress response, we evaluated the transcriptional changes in key genes involved in antioxidant defense mechanisms. Mammary glands were extracted from mice treated with bactericidal antibiotics in the presence and absence of NAC or the bacteriostatic antibiotic tetracycline. From these tissues, the expression of the antioxidant defense genes—Sod1, Sod2, Gpx1, and Foxo3a—was measured by quantitative polymerase chain reaction (qPCR). After 2 weeks of treatment, these genes showed more than a 2-fold increase in expression in tissues extracted from bactericidal antibiotic–treated mice, which were further elevated to ~10-fold change after 16 weeks of treatment (fig. S15A). These genes were not up-regulated in the tissues of tetracycline-treated mice. Tissues from mice treated with a combination of bactericidal antibiotic + NAC or NAC alone showed decreased expression of the tested genes compared to antibiotic treatment (fig. S15B). These data support our in vitro results, demonstrating that NAC rescued bactericidal antibiotic–induced oxidative damage in MCF10A human epithelial cells (Fig. 5).
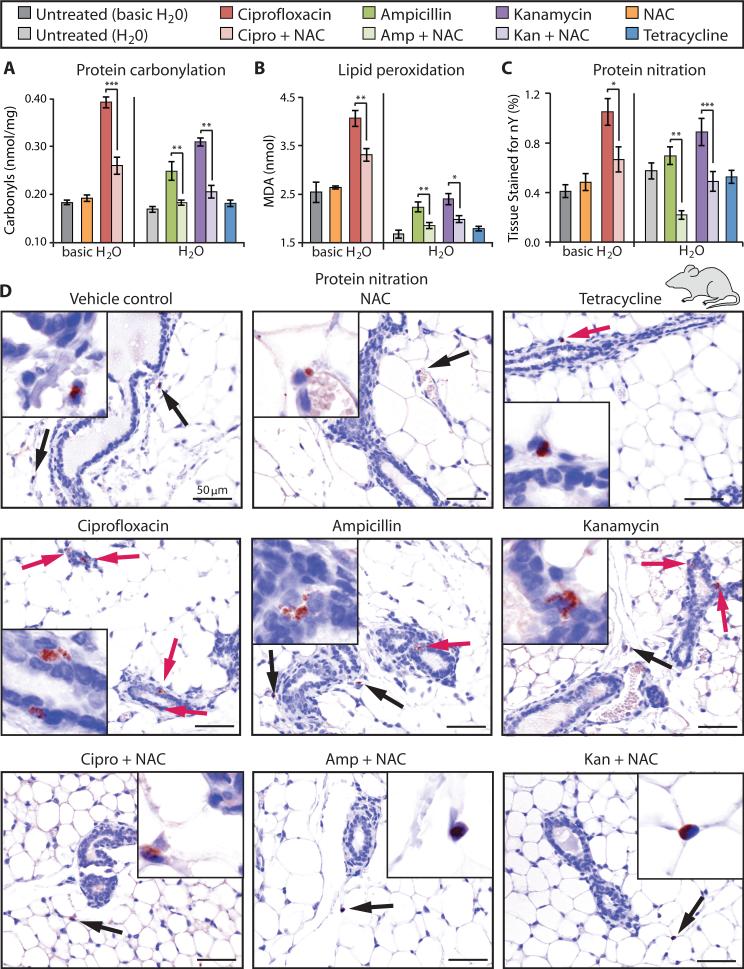
To evaluate bactericidal antibiotic–induced oxidative stress at the tissue level, we measured the accumulation of oxidative tissue damage directly in mammary glands. Quantification of protein carbonylation (Fig. 7A) and lipid peroxidation (Fig. 7B) revealed that bactericidal antibiotic–induced oxidative damage was significantly mitigated by co-administration of NAC. We also found no significant difference between tetracycline-treated and untreated mice. To further characterize these effects, we measured oxidative damage of proteins (nitration) caused by peroxynitrite, the product of the reaction between superoxide and nitric oxide. After 16 weeks of bactericidal antibiotic treatment, with and without NAC, as well as the bacteriostatic antibiotic tetracycline, we evaluated the number and size of anti-nitrotyrosine–stained foci in mouse mammary glands. Animals treated with bactericidal antibiotics showed increased protein nitration compared to untreated mice, whereas mice treated with tetracycline showed no change (Fig. 7C). The observed increase in protein damage caused by bactericidal antibiotic–treated mice was rescued by co-administration of NAC. Immunohistochemical analysis revealed that the damaged proteins in bactericidal antibiotic– treated mice tended to localize to the cytoplasm of mammary ductal epithelial cells (Fig. 7D). However, damaged proteins in NAC-treated mice, in the presence or absence of bactericidal antibiotics, were more likely to be found in the connective tissue, localizing to the nuclei of adipocytes and stromal cells (Fig. 7D).
Figure 7. Oxidative damage induced in mouse mammary gland tissue by bactericidal antibiotics is rescued by an antioxidant.
Mouse mammary glands were harvested 16 weeks after being treated with bactericidal antibiotics [ciprofloxacin (12.5 mg/kg/day), ampicillin (28.5 mg/kg/day), kanamycin (15 mg/kg/day)], with and without NAC (1.5 g/kg/day), or a bacteriostatic antibiotic [tetracycline (27 mg/kg/day)]. Ciprofloxacin requires basic water (pH 8.0) to dissolve, thus the antibiotic treatments in each plot have been grouped according to their control treatments: basic H2O (pH = 8) for ciprofloxacin, and H2O for ampicillin, kanamycin, and tetracycline. (A and B) Protein carbonylation and lipid peroxidation were measured in mouse mammary gland tissue collected from treated mice. (C) Mammary tissue was stained with an anti-nitrotyrosine antibody to measure oxidative protein damage (nitration). Quantification of the protein nitration was defined as the percent of total tissue area that was stained with the anti-nitrotyrosine antibody. Data are means ± S.E.M. (n ≥ 3 animals per treatment group). Comparisons between treatment + NAC and treatments were made using a Student's t test (*p < 0.5, **p < 0.01, ***p < 0.001). (D) Representative IHC images that were quantified in (C). Red arrows indicate protein damage foci (nitration) to ductal epithelial cells, black arrows point to protein damage to connective tissue cells (adipocytes and stromal cells).
DISCUSSION
The emergence of drug-resistant bacterial strains has been one unintended consequence of the ubiquitous and frequent use of antibiotics in medicine and food production. With the belief that antibiotics specifically target bacteria, the consequences of how they interact with mammalian cells have largely been overlooked, despite instances of known adverse effects, including ototoxicity (1), nephrotoxicity (2), and tendinopathy (3). Gaining a deeper mechanistic understanding of the effects of commonly prescribed antibiotics in mammalian systems, particularly for extended periods of treatment, is critical for achieving a clear safety profile for these drugs.
According to the endosymbiotic theory, mitochondria originated from free-living, aerobic bacteria (22). It is likely then that antibiotics target mitochondria and mitochondrial components, similar to their action in bacteria. Indeed, previous studies in mammalian systems have revealed parallel antibiotic-target interactions in mitochondria (15–19, 32). It has been shown, for instance, that aminoglycosides target both bacterial (33) and mitochondrial ribosomes (16), quinolones target bacterial gyrases (34) and mtDNA topoisomerases (15), and β-lactams inhibit bacterial cell wall synthesis (35) and mitochondrial carnitine/acylcarnitine transporters (19). The work presented here advances our understanding of antibiotic action in mammalian systems in two distinct and important ways. First, we show that, regardless of their molecular targets, three major classes of bactericidal antibiotics— quinolones, aminoglycosides, and β-lactams—induce ROS production in mammalian cells, leading to DNA, protein, and lipid damage. Second, we demonstrate that these deleterious effects are produced by clinically relevant doses of bactericidal antibiotics, both in cell culture and in mice. These findings are analogous to our previous work in bacteria, in which we showed that clinically relevant doses of bactericidal antibiotics induce a common oxidative damage pathway (4).
Identifying bactericidal antibiotics as a cause of ROS overproduction and mitochondrial dysfunction in mammalian cells provides a basis for developing therapeutic strategies that could help alleviate adverse side effects associated with antibiotics. For instance, by co-administering an intracellular antioxidant, in this case NAC, we showed that ROS levels and oxidative damage induced by bactericidal antibiotics could be abrogated while having little effect on the bacterial killing efficacy of the antibiotics. Additionally, we showed that bacteriostatic antibiotics, such as tetracycline, did not contribute to the overproduction of harmful ROS in mammalian cells. When appropriate for patient health, substituting a bacteriostatic antibiotic for a bactericidal antibiotic could be a simple treatment strategy aimed at preventing cellular oxidative damage.
To establish the link between bactericidal antibiotics and ROS production, we have limited this study to antibiotic-treated human cell lines and mice. It will be important to extend our investigation to human subjects to confirm our findings, prove their relevance to humans, and maximize the translational value of our work. Epidemiologic studies could provide valuable insight into the clinical implications of antibiotic-induced oxidative damage and define some of the risks associated with bactericidal antibiotic exposure. Another limitation to consider is the set of antibiotics tested. Although we present results on three major classes of bactericidal antibiotics, interrogation across a broader range of antibiotic classes is warranted before conclusions can be drawn about the general category of bactericidal antibiotics. Last, it has been shown that both bactericidal and bacteriostatic antibiotics target mitochondrial components (17–19, 23, 24), yet we observed a marked differential effect between bactericidal and bacteriostatic antibiotic treatments on cellular oxidative damage. Disentangling these differences in antibiotic action will be essential to gain a full understanding of how antibiotics interact with mammalian systems.
In humans, it is likely that the oxidative stress and related oxidative cellular damage induced by bactericidal antibiotics underlie many of the adverse side effects associated with these antibiotics (1–3). In particular, patients with compromised antioxidant defense systems or those genetically disposed to developing a mitochondrial dysfunction disease (36) might be at greater risk from bactericidal antibiotic treatments. Thus, rapid detection (for example, via measurements from peripheral blood) and mitigation of these adverse effects could have important implications for patient care. Our data suggest that oxidative damage markers can be measured in the blood, but further work is needed to determine the efficacy of such a test in humans. Once detected, mitigation strategies such as co-administration with an antioxidant or treatment with bacteriostatic antibiotics could be used. It will be intriguing to explore these possibilities with appropriate clinical trials, with the goal of developing effective antibacterial therapies with minimal adverse side effects.
MATERIALS AND METHODS
Study Design
The objective of our work was to investigate the effects of clinically relevant doses of bactericidal and bacteriostatic antibiotics on mammalian systems in vitro and in vivo. A human mammary epithelial cell line (MCF10A) served as our in vitro model, while 8-week-old female wild-type mice (FVB) were used to study the effect of antibiotics in vivo. To understand the putative role of antibiotic-induced ROS in mammalian cells and accumulation of oxidative damage, an antioxidant (NAC) was used alone or in combination with the bactericidal antibiotics. In vitro, ROS was measured using fluorescent indicators (CM-H2DCFDA, Amplex Red and horseradish peroxidase, MitoSox Red), and oxidative damage to DNA (8-OHdG), proteins (carbonylation) and lipids (peroxidation) was quantified while the functional implications were assessed by evaluation of mitochondrial respiration (oxygen consumption). In vivo, oxidative stress was evaluated in the peripheral blood of treated mice using fluorescent indicators (H2DCFDA, fluor-DHPE, DCF) while oxidative damage to mammary gland tissue (protein carbonylation, lipid peroxidation, protein nitration) was assessed by immunohistochemistry and qPCR quantification of key anti-oxidant genes. Mice were purchased from the vendor and randomly assigned to treatment groups. Antibiotics were prepared and administered in a non-blinded fashion, both in vitro and in vivo. Experiments included three (Figs. 1C, 1D, 1F, 1G, 2A, 2B, 2E, 4, 5B, 6, S3, S5, S6, S7, S9, S10, S11, S13, S15), four (Figs. 1A, 1B, 5A, 5D, 5E, 5F, 8A, 8B, 8C, S1, S2, S4, S14), five (Figs. 1E, 3), six (Figs. 2C, 2D), or eight (Fig. S8) replicates per group.
Cell Culture and Antibiotic Treatments
Human mammary epithelial cells, MCF10A (ATCC), were maintained in DMEM and Ham's F12 50/50 Mix (Cellgro, Mediatech) supplemented with 5% horse serum, epidermal growth factor (100 μg/ml), hydrocortisone (1 mg/ml), cholera toxin (1 mg/ml), insulin (10 mg/ml) and 5 ml of 10,000 units/ml penicillin and 10,000 μg/ml streptomycin. Hereafter, this media will be referred to as ‘complete media’. For antibiotic treatments, complete media was used without the addition of penicillin/streptomycin. We refer to this media as ‘treatment media’. All supplements were purchased from Sigma-Aldrich.
Prior to antibiotic treatments, cells were washed with phosphate-buffered saline (PBS), detached with 0.25% trypsin-EDTA (Life Technologies), and seeded in 6-, 12-, 24- or 96-well plates (Corning) in treatment medium. After 24 h, peak serum levels (25) of antibiotics were added – 10 μg/ml ciprofloxacin, 20 μg/ml ampicillin, 25 μg/ml kanamycin, 10 μg/ml tetracycline, and 100 μg/ml spectinomycin (Fisher Scientific). Cells were plated at roughly equal numbers (near confluence) at each measurement time-point. NAC (10 mM; Sigma-Aldrich) was used as an intracellular ROS scavenger. For antibiotic treatments in conjunction with NAC, cells were incubated in treatment media plus NAC (pH adjusted to 7.4) for 2 h, and then washed with PBS prior to the addition of antibiotics. The growth conditions for the additional cell lines tested are described in the Supplemental Methods.
MCF10A ρ0 Cell Line
MCF10A rho-zero (ρ0) cells are MCF10A cells that have been cultured to eliminate mitochondrial DNA (mtDNA). Following the protocol from King and Attardi (37), MCF10A cells were incubated in complete media supplemented with 4.5 g/l D-glucose, 50 ng/ml ethidium bromide, 50 μg/ml uridine and 1 mM pyruvate for 6 weeks. After complete elimination of mtDNA (as verified through PCR described below), cells were maintained in complete media supplemented with 4.5 g/l D-glucose, 50 μg/ml uridine and 1 mM pyruvate. A set of MCF10A control cells were grown alongside the MCF10A ρ0 cells maintained in complete media supplemented with 4.5 g/l D-glucose, 50 μg/ml uridine and 1 mM pyruvate. The verification of ρ0 is described in the Supplemental Methods.
ROS, H2O2, and Mitochondrial Superoxide
To detect ROS, cells plated in 6- or 96-well tissue culture plates were washed twice with pre-warmed PBS, then incubated with 10 μM CM-H2DCFDA (Life Technologies) at 37°C for 45 min. Cells were washed with PBS to remove excess dye and allowed to recover in treatment media at 37°C for 15 min in the dark. Imaging was carried out on a fluorescence-enabled inverted microscope and quantification was done using a BD FACSAria II flow cytometer (Becton Dickinson) or Spectramax M5 microplate reader (Molecular Devices) at an ex/em of 495/525 nm. This protocol was repeated for all other tested cell lines.
To detect extracellular hydrogen peroxide, we adapted a protocol from Panopoulos et al. (38). Cells plated in 96-well tissue culture plates were washed once with Krebs-Ringer phosphate buffer (KRPG). KRPG containing 0.1% horse serum (KRPG 0.1%) and clinical levels of antibiotics were added to the cells in a total volume of 50 μl. Next, 50 μl of pre-warmed KRPG containing 100 μM Amplex Red reagent and 0.2 units/ml horseradish peroxide (reaction mixture) was added to each well. Absorbance was measured at 560 nm after the addition of reaction buffer using a Spectramax M5 precision microplate reader (Molecular Devices).
To detect mitochondrial superoxide, cells plated in 6- or 96-well tissue culture plates were washed twice with pre-warmed PBS, and then incubated with 5 μM MitoSox Red at 37°C for 10 min in the dark. After incubation, cells were washed and re-suspended in pre-warmed HBSS (Fisher Scientific). Quantification was completed using a BD FACSAria II flow cytometer or a Spectramax M5 microplate reader at an Ex/Em of 510/580 nm.
Oxidative DNA, Protein, and Lipid Damage
8-OHdG levels were quantified using the OxiSelectTM Oxidative DNA Damage ELISA Kit (Cell Biolabs), protein carbonylation was measured using the Protein Carbonyl ELISA Kit (Enzo LifeSciences), and lipid peroxidation was measured using the Lipid Peroxidation MDA Assay Kit (Abcam). All assays were performed according to the manufacturer's protocol and are described in the Supplemental Methods.
Mitochondrial ETC Complex Activity
Direct inhibition of the activity for each of the five electron transport chain complexes was measured using the Mitotox Complete OXPHOS Activity Assay Panel (Abcam), following the manufacturer's protocol. Each of the five complexes was captured from isolated bovine heart mitochondria in their functionally active state using highly specific monoclonal antibodies attached to 96-well microplates. IC50 for known inhibitors of each of the five complexes were used as positive controls – rotenone (Complex I, 17.3 nM), thenoyltrifluoroacetone (TTFA, Complex II, 30 μM), antimycin A (Complex III, 22 nM), potassium cyanide (Complex IV, 3.2 μM) and oligomycin (Complex V, 8 nM). The same positive controls were tested on each complex and used as negative controls for off-target complexes (e.g., rotenone was a negative control for complexes II-V). For each of the complexes treated with antibiotics (10 μg/ml ciprofloxacin, 20 μg/ml ampicillin, 25 μg/ml kanamycin, 10 μg/ml tetracycline, or 100 μg/ml spectinomycin), activity was determined by measuring the decrease in absorbance in mOD/min at room temperature and at specified wavelengths [340 nm (I and V), 600 nm (II) and 550 nm (III and IV)] in kinetic mode [every min for 2 h (I), 1 h (II, IV and V) and every 20 s for 5 min (III)] using a Spectramax M5 microplate reader.
Metabolic Activity, Mitochondrial Potential, and ATP
Metabolic activity was assayed using an XTT cell proliferation kit (ATCC), cellular ATP was measured using the ATPlite Luminescence Assay kit (Perkin Elmer), and mitochondrial potential was calculated using the ratio of TMRE to MitoTracker Green (Life Technologies). All assays were performed according to the manufacturer's protocol and are described in the Supplemental Methods.
Mitochondrial Morphometry
Primary human mammary epithelial cells (HMECs) were plated in glass-bottom confocal dishes in treatment media 24 h before addition of antibiotics. Cells were washed once with PBS and treated with antibiotics for 24 or 96 h. After treatment, cells were washed with 1X PBS and incubated in 7 nM tetramethylrhodamine ester (TMRE) and 10 μM MitoTracker green for 30 min. After incubation, cells were returned to fresh treatment medium with only TMRE (7 nM) and immediately imaged on a Zeiss LSM710 confocal microscope.
Mitochondrial morphometry was carried out using the particle analysis function in the image processing software, ImageJ (NIH) (39), as described in the Supplemental Methods.
Mitochondrial Oxygen Consumption
Oxygen consumption rates (OCR) were measured at 37°C using an XF24 extracellular analyzer (Seahorse Bioscience). MCF10A cells were seeded at a density of 40,000 cells/well on XF24 tissue culture plates overnight and then treated with antibiotics for 6 and 96 h. Measurements of OCR were done according to Seahorse Bioscience protocols as described in the Supplemental Methods.
Bacterial Killing
The growth and survival of untreated exponential-phase wild-type E. coli (MG1655) was compared to cultures treated with bactericidal antibiotics alone (5 μg/ml ciprofloxacin, 5 μg/ml ampicillin or 5 μg/ml kanamycin), 10 mM NAC alone, or a combination of bactericidal antibiotics supplemented with NAC for 3 h. Cells were grown overnight at 37°C and 300 RPM in a light insulated shaker, then diluted 1:500 in 25ml LB in a 250ml flask. Cultures were grown to an optical density (OD 600) of approximately 0.3, as measured using the SPECTRAFluor Plus spectrophotometer (Tecan). Antibiotics were added at this point. For bacterial count measurements (CFU/ml), 100 μl of culture was collected 30 min, 1, 2 and 3 h after addition of antibiotics, washed twice with filtered 1x PBS, pH 7.2 (Fisher Scientific), then serially diluted in 1x PBS. 10μl of each dilution was plated onto 120 mm square dishes (BD Biosciences) containing LB-Agar (Fisher Scientific), and plates were incubated at 37°C overnight. Colonies were counted, and CFU/ml values were calculated using the formula: .
Animal Studies and Tissue Collection
Six- to 8-week old female FVB/NJ mice (Jackson Laboratory) were administered one of the following treatments: ciprofloxacin (12.5 mg/kg/day), ampicillin (28.5 mg/kg/day), kanamycin (15 mg/kg/day), with and without NAC (1.5 g/kg/day), tetracycline (13.5 mg/kg/day), vehicle only (basic water, pH= 8.0, or deionized water). Solutions were made fresh every three days and administered in the drinking water for mice to feed ad libitum. Doses were achieved based on an average mouse weight of 20 g and an approximate intake of drinking water of 4 ml per day. After 2 or 16 weeks, the animals were bled and euthanized. Mammary tissue was collected from each mouse and preserved in RNALater (Ambion, Life Technologies) for quantitative real-time PCR, 10% buffered formalin for immunohistochemistry, and flash frozen for protein extraction. The Institutional Animal Care and Use Committee (IACUC) approved all mouse experiments. Quantitative RT-PCR of oxidative stress genes is described in the Supplemental Methods.
Mouse Blood Oxidative Stress
To measure general oxidative stress, 1×107 peripheral blood cells were incubated with 2’,7’-dichlorodihydrofluorescein diacetate (H2DCFDA) (Life Technologies), dissolved in DMSO (Sigma-Aldrich), at a final concentration of 0.4 mM at 37°C for 15 min at 5% CO2. Reduced glutathione levels were measured by labeling 2 × 107 blood cells with mercury orange (Sigma-Aldrich), dissolved in DMSO, at a final concentration of 40 μM for 3 min at room temperature. To measure lipid peroxidation, 2×107 blood cells were washed with PBS and labeled with N-(fluorescein-5-thiocarbamoyl)-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine, triethyl-ammonium salt (fluor-DHPE) (Life Technologies), dissolved in ethanol, at a final concentration of 50 μM for 1h at 37°C in a 5% CO2 incubator with continuous agitation. After the incubation period with each dye, blood cells were washed twice with PBS to remove unbound label, re-suspended in PBS, and analyzed by flow cytometry (FACSfortessa, Becton Dickinson). A 488 nm argon laser beam was used for excitation. Blood cells labeled with DCF and fluor-DHPE were detected by FL-1 PMT using linear amplification, while mercury orange-labeled red blood cells were detected by FL-2 PMT using log amplification. For each assay, treated and untreated unstained cells were used as controls. Instrument calibration and settings were performed using CaliBRITE-3 beads (Becton Dickinson). The mean fluorescence channel (MFC) of the entire population was calculated for DCF, glutathione and lipid peroxidation by the FACS-equipped BD FACSDiva software.
Urinary Tract Infection Mouse Model
8-week old C57BL/6 female mice were inoculated with 50 μl of 8% (w/v) mucin solution in sterile saline containing 2×109 E. coli (MG1655) cells, via transurethral catheterization into their bladders, as described previously (40). Briefly, mice were anesthetized using 2-4% isoflurane. Urinary catheters (30G × ½ inch hypodermic needle aseptically covered with polyethylene tubing) were coated in medical-grade, sterile lubricating jelly. The bladder of the mouse was gently massaged to expel urine. The lubricated catheter was inserted into the urethral opening. It was then pushed into the urethra until the base of the needle reached the urethral opening. Once fully inserted, 50 μl of the inoculum (containing 2×109 E. coli cells) was injected directly into the bladder.
Infected animals received ciprofloxacin (50 μg/ml), NAC (10 mM), ciprofloxacin and NAC, or vehicle (PBS) only, via intraperitoneal (I.P.) delivery 24 h post-inoculation. Following treatment, animals were observed for an additional 24 or 48 h. At the end of the experiment, animals were euthanized by CO2 asphyxiation followed by cervical dislocation. Bladders were collected in 1 ml of PBS and homogenized for 30 sec for subsequent quantification of bacterial load. For the CFU/bladder measurements, the homogenized bladder was serially diluted in PBS (pH 7.2). A 200 μl portion of each dilution was plated in LB-agar plates and incubated overnight at 37°C. The colonies were counted and CFU/bladder were calculated using the following formula:
Histology and Immunohistochemistry
Six- to 8-week old female FVB/NJ mice (Jackson Laboratory) were administered ciprofloxacin (12.5 mg/kg/day), ampicillin (28.5 mg/kg/day), kanamycin (15 mg/kg/day), with and without NAC (1.5 g/kg/day), tetracycline (13.5 mg/kg/day) or vehicle only [basic or deionized H2O]. Solutions were made fresh every three days and administered in the drinking water for 16 weeks. Mice were euthanized and mammary glands were collected and stored in 10% buffered formalin overnight at 4°C, transferred to 70% ethanol and stored at room temperature until sectioning. Tissues were embedded in paraffin and sectioned for subsequent staining. Mammary gland sections were stained with anti-nitrotyrosine polyclonal antibody (Millipore), followed by anti-rabbit IgG conjugated to HRP (Vector Laboratories). Slides were counterstained with hematoxylin. Image capture and analysis are described in the Supplemental Methods.
Statistical analysis
A one-tailed Student's t test was performed on measurements of oxidative stress in the blood, urinary tract infection model, and for comparisons between antibiotic-treated groups and groups treated with antibiotic & NAC. For all other statistical analyses, a two-tailed Student's t test was performed to compare treatment groups to untreated (control) groups. We assumed equal variance for all groups.
Supplementary Material
One-Sentence Summary.
Mitochondrial dysfunction and oxidative damage induced by bactericidal antibiotics in mammalian cells may be alleviated by an antioxidant or prevented by preferential use of bacteriostatic antibiotics.
Acknowledgments
We thank E. Cameron and P. Belenky for their help with experimental design and troubleshooting, A. Saccone for his assistance with experiments, and T. Ferrante for his assistance with imaging. Funding: This work was supported by the NIH Director's Pioneer Award Program and the Howard Hughes Medical Institute. M.L. was the recipient of a post-doctoral fellowship from Fundacion Ramon Areces and an Evans Center Fellow Award.
Footnotes
Author contributions: All authors designed the study, analyzed results and wrote the manuscript. In vitro experiments were performed by S.K. and S.S. In vivo experiments were performed by S.K. C.S., and J.R.M. Seahorse experiments were completed by S.K. and M.L.
Competing interests: The authors declare no competing financial interests.
REFERENCES AND NOTES
- 1.Brummett RE, Fox KE. Aminoglycoside-induced hearing loss in humans. Antimicrob Agents Chemother. 1989;33:797. doi: 10.1128/aac.33.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mingeot-Leclercq MP, Tulkens PM. Aminoglycosides: nephrotoxicity. Antimicrob Agents Chemother. 1999;43:1003. doi: 10.1128/aac.43.5.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khaliq Y, Zhanel G. Fluoroquinolone-associated tendinopathy: a critical review of the literature. Clin Infect Dis. 2003;36:1404. doi: 10.1086/375078. [DOI] [PubMed] [Google Scholar]
- 4.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 5.Dwyer DJ, Kohanski MA, Hayete B, Collins JJ. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol Syst Biol. 2007;3:91. doi: 10.1038/msb4100135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohanski MA, Dwyer DJ, Wierzbowski J, Cottarel G, Collins JJ. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell. 2008;135:679. doi: 10.1016/j.cell.2008.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foti JJ, Devadoss B, Winkler JA, Collins JJ, Walker GC. Oxidation of the guanine nucleotide pool underlies cell death by bactericidal antibiotics. Science. 2012;336:315. doi: 10.1126/science.1219192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grant SS, Kaufmann BB, Chand NS, Haseley N, Hung DT. Eradication of bacterial persisters with antibiotic-generated hydroxyl radicals. Proc Natl Acad Sci U S A. 2012;109:12147. doi: 10.1073/pnas.1203735109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y, et al. Inhibitors of reactive oxygen species accumulation delay and/or reduce the lethality of several antistaphylococcal agents. Antimicrob Agents Chemother. 2012;56:6048. doi: 10.1128/AAC.00754-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shatalin K, Shatalina E, Mironov A, Nudler E. H2S: a universal defense against antibiotics in bacteria. Science. 2011;334:986. doi: 10.1126/science.1209855. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Zhao X. Contribution of oxidative damage to antimicrobial lethality. Antimicrob Agents Chemother. 2009;53:1395. doi: 10.1128/AAC.01087-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Zhao X, Malik M, Drlica K. Contribution of reactive oxygen species to pathways of quinolone-mediated bacterial cell death. J Antimicrob Chemother. 2010;65:520. doi: 10.1093/jac/dkp486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keren I, Wu Y, Inocencio J, Mulcahy LR, Lewis K. Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science. 2013;339:1213. doi: 10.1126/science.1232688. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Imlay JA. Cell death from antibiotics without the involvement of reactive oxygen species. Science. 2013;339:1210. doi: 10.1126/science.1232751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gootz TD, Barrett JF, Sutcliffe JA. Inhibitory effects of quinolone antibacterial agents on eucaryotic topoisomerases and related test systems. Antimicrob Agents Chemother. 1990;34:8. doi: 10.1128/aac.34.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutchin T, Cortopassi G. Proposed molecular and cellular mechanism for aminoglycoside ototoxicity. Antimicrob Agents Chemother. 1994;38:2517. doi: 10.1128/aac.38.11.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lowes DA, Wallace C, Murphy MP, Webster NR, Galley HF. The mitochondria targeted antioxidant MitoQ protects against fluoroquinolone-induced oxidative stress and mitochondrial membrane damage in human Achilles tendon cells. Free Radic Res. 2009;43:323. doi: 10.1080/10715760902736275. [DOI] [PubMed] [Google Scholar]
- 18.McKee EE, Ferguson M, Bentley AT, Marks TA. Inhibition of Mammalian Mitochondrial Protein Synthesis by Oxazolidinones. Antimicrobial Agents and Chemotherapy. 2006;50:2042. doi: 10.1128/AAC.01411-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pochini L, et al. Interaction of beta-lactam antibiotics with the mitochondrial carnitine/acylcarnitine transporter. Chem Biol Interact. 2008;173:187. doi: 10.1016/j.cbi.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 20.Tune BM. Nephrotoxicity of beta-lactam antibiotics: mechanisms and strategies for prevention. Pediatr Nephrol. 1997 Dec;11:768. doi: 10.1007/s004670050386. [DOI] [PubMed] [Google Scholar]
- 21.Lesnefsky EJ, Moghaddas S, Tandler B, Kerner J, Hoppel CL. Mitochondrial dysfunction in cardiac disease: ischemia--reperfusion, aging, and heart failure. J Mol Cell Cardiol. 2001;33:1065. doi: 10.1006/jmcc.2001.1378. [DOI] [PubMed] [Google Scholar]
- 22.Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 23.Lawrence JW, Claire DC, Weissig V, Rowe TC. Delayed cytotoxicity and cleavage of mitochondrial DNA in ciprofloxacin-treated mammalian cells. Mol Pharmacol. 1996;50:1178. [PubMed] [Google Scholar]
- 24.Duewelhenke N, Krut O, Eysel P. Influence on mitochondria and cytotoxicity of different antibiotics administered in high concentrations on primary human osteoblasts and cell lines. Antimicrob Agents Chemother. 2007;51:54. doi: 10.1128/AAC.00729-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bryskier A. Antimicrobial agents: antibacterials and antifungals. ASM Press; 2005. [Google Scholar]
- 26.Turrens JF. Mitochondrial formation of reactive oxygen species. The Journal of Physiology. 2003;552:335. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dröse S, Brandt U. The Mechanism of Mitochondrial Superoxide Production by the Cytochrome bc1 Complex. Journal of Biological Chemistry. 2008;283:21649. doi: 10.1074/jbc.M803236200. [DOI] [PubMed] [Google Scholar]
- 28.Seo AY, et al. New insights into the role of mitochondria in aging: mitochondrial dynamics and more. J Cell Sci. 2010;123:2533. doi: 10.1242/jcs.070490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Vries N, De Flora S. N-acetyl-l-cysteine. Journal of Cellular Biochemistry. 1993;53:270. doi: 10.1002/jcb.240531040. [DOI] [PubMed] [Google Scholar]
- 30.Aruoma OI, Halliwell B, Hoey BM, Butler J. The antioxidant action of N-acetylcysteine: its reaction with hydrogen peroxide, hydroxyl radical, superoxide, and hypochlorous acid. Free radical biology & medicine. 1989;6:593. doi: 10.1016/0891-5849(89)90066-x. [DOI] [PubMed] [Google Scholar]
- 31.Meister A, Anderson ME. Glutathione. Annual review of biochemistry. 1983;52:711. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- 32.Hobbie SN, et al. Genetic analysis of interactions with eukaryotic rRNA identify the mitoribosome as target in aminoglycoside ototoxicity. Proc Natl Acad Sci U S A. 2008;105:20888. doi: 10.1073/pnas.0811258106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis BD. Mechanism of bactericidal action of aminoglycosides. Microbiological reviews. 1987;51:341. doi: 10.1128/mr.51.3.341-350.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolfson JS, Hooper DC. The fluoroquinolones: structures, mechanisms of action and resistance, and spectra of activity in vitro. Antimicrob Agents Chemother. 1985;28:581. doi: 10.1128/aac.28.4.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomasz A. The mechanism of the irreversible antimicrobial effects of penicillins: how the beta-lactam antibiotics kill and lyse bacteria. Annual review of microbiology. 1979;33:113. doi: 10.1146/annurev.mi.33.100179.000553. [DOI] [PubMed] [Google Scholar]
- 36.Schaefer AM, et al. Prevalence of mitochondrial DNA disease in adults. Annals of Neurology. 2008;63:35. doi: 10.1002/ana.21217. [DOI] [PubMed] [Google Scholar]
- 37.King MP, Attardi G. Isolation of human cell lines lacking mitochondrial DNA. Methods Enzymol. 1996;264:304. doi: 10.1016/s0076-6879(96)64029-4. [DOI] [PubMed] [Google Scholar]
- 38.Panopoulos A, Harraz M, Engelhardt JF, Zandi E. Iron-mediated H2O2 Production as a Mechanism for Cell Type-specific Inhibition of Tumor Necrosis Factor α-Induced but Not Interleukin-1β-induced IκB Kinase Complex/Nuclear Factor-κB Activation. Journal of Biological Chemistry. 2005;280:2912. doi: 10.1074/jbc.M409524200. [DOI] [PubMed] [Google Scholar]
- 39.Abramoff MD, Magalhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11:36. [Google Scholar]
- 40.Hung CS, Dodson KW, Hultgren SJ. A murine model of urinary tract infection. Nat Protoc. 2009;4:1230. doi: 10.1038/nprot.2009.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.