Summary
Elevated Aurora kinase-A expression is correlated with abrogation of DNA damage induced apoptotic response and mitotic spindle assembly checkpoint (SAC) override in human tumor cells. We report that Aurora-A phosphorylation of p73 at serine235 abrogates its transactivation function and causes cytoplasmic sequestration in a complex with the chaperon protein mortalin. Aurora-A phosphorylated p73 also facilitates inactivation of SAC through dissociation of the MAD2-CDC20 complex in cells undergoing mitosis. Cells expressing phosphor-mimetic mutant (S235D) of p73 manifest altered growth properties, resistance to cisplatin induced apoptosis, as well as premature dissociation of the MAD2-CDC20 complex, and accelerated mitotic exit with SAC override in the presence of spindle damage. Elevated cytoplasmic p73 in Aurora-A overexpressing primary human tumors corroborates the experimental findings.
Introduction
Aurora kinase-A (hereafter referred to as Aurora-A), a key regulator of the mitotic cell division cycle, is overexpressed in many human tumors and is associated with abrogation of DNA damage induced apoptotic response and spindle assembly checkpoint (SAC) override in cancer cells. Aurora-A, a cancer susceptibility gene (Ewart-Toland et al., 2003), plays essential roles in the commitment of proliferating cells to G2/M progression, centrosome maturation-separation, bipolar spindle formation, and spindle damage recovery (Marumoto et al., 2005, Katayama et al., 2008, Macurek et al., 2008, Seki et al., 2008). We and others have previously identified functional inactivation of p53 tumor suppressor protein after Aurora-A phosphorylation at serine 315 and serine 215 residues; the former facilitates Mdm2 mediated degradation, and the latter causes loss of DNA binding ability in human cells (Katayama et al., 2004, Liu et al., 2004). Aurora-A phosphorylation of BRCA1 at serine 308 is correlated with silencing of DNA damage induced G2/M checkpoint (Ouchi et al., 2004). Furthermore, overexpression of Aurora-A makes HeLa cells resistant to taxol induced cell death due to mitotic SAC override (Anand et al., 2003). A recent study found that treatment of p53-deficient cells with Aurora-A small molecule inhibitors activates p73 trans-activation function with up-regulation of its down-stream target genes during induction of cell death (Dar et al., 2008). However, the molecular mechanism(s) underlying the observed effects have not been elucidated.
The role of p73 in tumorigenesis has been debated because loss of function mutations in the gene is rare. However, recently developed transactivation competent (TA) p73 specific gene-knockout mice have a high incidence of spontaneous and carcinogen induced tumors (Tomasini et al., 2008). In addition, oocytes and cells lacking TAp73 exhibit abnormal spindle structure and mitotic slippage with spindle poisons, indicating participation of TAp73 in the SAC pathway (Tomasini et al., 2009). More recent studies have demonstrated that TAp73 interacts with SAC proteins Bub1, Bub3, and BubR1. TAp73 deficient or knock down cells reveal mis-localization of Bub1 and BubR1 at the kinetochore and reduced BubR1 kinase activity, associated with aneuploidy and chromosome instability (Tomasini et al., 2009, Vernole et al., 2009). Together with pro-apoptotic function of TAp73 in response to genotoxic stress, these results suggest that p73 is directly involved in maintaining genomic stability and regulating SAC pathway.
In view of Aurora-A over expression reported to induce resistance to DNA damage mediated apoptosis response and SAC over ride, we investigated the possible role of Aurora-A functional interaction with p73 and the underlying molecular mechanisms involved in the development of these phenotypes.
Results
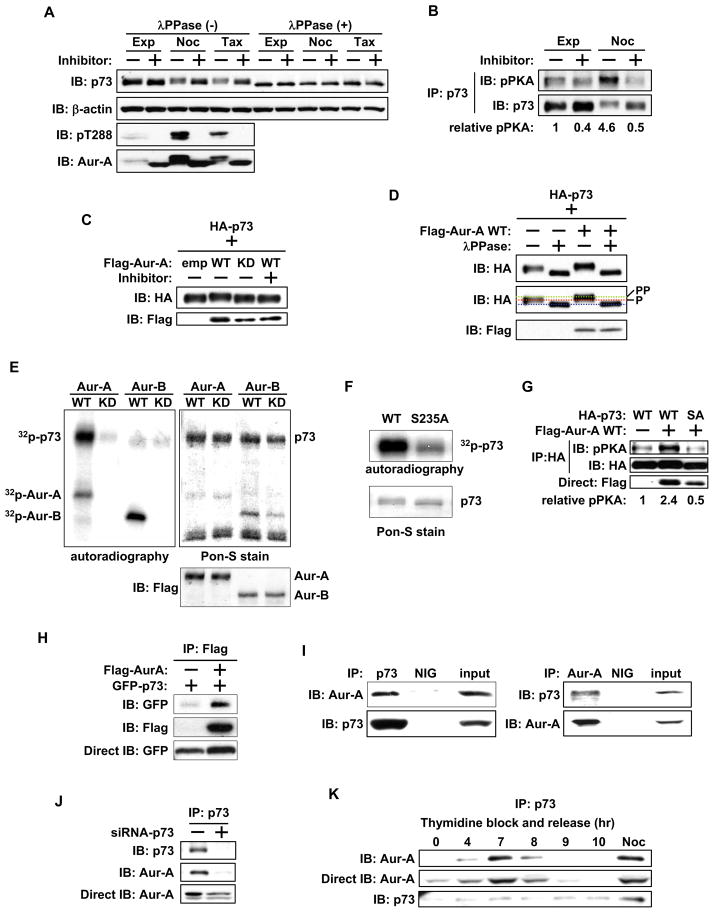
Aurora-A phosphorylates p73
We hypothesized that direct phosphorylation of p73 by Aurora-A negatively regulates p73 transactivation function and consequential activation of apoptosis response. Because p73 is reported to be phosphorylated in mitosis (Fulco et al., 2003), we treated nocodazole and taxol arrested mitotic Cos-1 cells with Aurora-A specific inhibitor MLN8054 and proteasome inhibitor MG132 to detect Aurora-A specific post-translational p73 modification. p73 from inhibitor treated mitotic cells migrated faster than that in untreated cells, whereas p73 from exponentially growing cells had intermediate mobility (Figure 1A). The slower migrating form was seen in cells with active Aurora-A, detected with anti-phosphor T288 antibody. To determine whether slower mobility of p73 was due to phosphorylation and whether Aurora-A is directly involved in p73 phosphorylation, we treated cell extracts with λPPase, with or without Aurora-A inhibitor. While inhibitor treatment alone resulted in minimal increase in mobility, λPPase treatment, both with or without of Aurora-A inhibitor led to similar yet markedly faster migration in p73. These results indicate that slower mobility was due to multiple phosphorylations, possibly catalyzed by several kinases, including Aurora-A. Aurora-A inhibition alone resulted in a minor downward shift in gel mobility due to selective interference with Aurora-A phosphorylation, but the more rapidly migrating form was due to complete de-phosphorylation with λPPase (Figure 1A). To determine direct involvement of Aurora-A in p73 phosphorylation in vivo, we performed p73 immunoprecipitation, followed by immunoblotting with the anti-phosphor-PKA substrate antibody, which recognizes the Aurora-A consensus phosphorylation motif in substrate proteins (Katayama et al., 2007, Plotnikova and Golemis 2011). We observed clear phosphor-PKA signal in immunoprecipitated p73 from nocodazole treated mitotic cells, which was diminished in inhibitor treated samples. In exponentially growing cells, the phosphor-PKA signal changed little after treatment (Figure 1B). These findings further verified the involvement of Aurora-A in p73 phosphorylation in vivo (Figure 1C and 1D).
Figure 1. Aurora A phosphorylates and interacts with p73.
(A) Cos-1 cells, grown with or without nocodazole or taxol for 20 hr, were cultured with or without Aurora-A inhibitor MLN8054 in the presence of MG132 for 4 hr. Whole cell extracts (WCE) were incubated with (+) or without (−) λPPase for 30 min. Samples were analyzed by immunoblotting with the indicated antibodies.
(B) WCE prepared in (A) were immunoprecipitated with anti-p73 antibody and subjected to immunoblotting with anti-pPKA substrate (top) and anti-p73 (bottom) antibodies. The pPKA signal was normalized by the amount of precipitated HA-p73, and the relative pPKA signal was quantified.
(C) HA-p73α was co-transfected with empty vector, Flag-wild type Aurora-A (WT), or Flag-kinase dead Aurora-A (KD) into Cos-1 cells. 24 hr later, cells were cultured with (+) or without (−) Aurora-A inhibitor for 4 hr. WCE were subjected to immunoblotting with anti-HA (top) and anti-Flag (bottom) antibodies.
(D) WCE prepared in (C) were treated without (−) or with (+) λPPase for 30 min and analyzed as in (C). The middle panel shows the differential migration of HA-p73α.
(E) Anti-Flag M2 antibody immunoprecipitates from nocodazole treated mitotic 293T cells transfected with Flag-Aurora-A-WT, Flag-Aurora-A-KD, Flag -Aurora-B-WT, or Flag -Aurora-B-KD were incubated with GST-p73α in the presence of [γ32p]ATP. GST-p73α was resolved by SDS-PAGE and visualized by autoradiography (left) and Pon-S staining (right). Immunoprecipitated Aurora-A and Aurora-B were detected with anti-Flag M2 antibody (bottom right).
(F) Immunoprecipitates with anti-Aurora-A antibody from nocodazole treated mitotic 293T cells were incubated with GST-p73α-WT or GST-p73α-S235A (SA) in the presence of [γ32p]ATP. GST-p73 protein was resolved and visualized as in (E).
(G) HA-p73α-WT or -SA were co-transfected with either empty vector or Flag-Aurora-A-WT into Cos-1 cells. 24 hr later, WCE were immunoprecipitated with anti-HA antibody, followed by immunoblotting with anti-pPKA substrate (top) and anti-HA (middle) antibodies. WCE were directly immunoblotted with anti-Flag antibody (bottom). The pPKA signal was normalized by the amount of precipitated HA-p73, and the relative pPKA signal was quantified.
(H) GFP-p73α was co-transfected with empty vector or Flag-Aurora-A in 293T cells. 24 hr later, cells were immunoprecipitated with anti-Flag antibody, followed by immunoblotting with indicated antibodies (top and middle). Aliquots of the same cell lysates were directly immunoblotted with anti-GFP antibody (bottom).
(I) Cos-1 cells were immunoprecipitated with anti-p73 (right) or anti-Aurora-A (left) antibodies or normal IgG (NIG). Immunoprecipitates were subjected to immunoblotting with anti-p73 and anti-Aurora-A antibodies. Input indicates the immunoblot of WCE.
(J) Cos-1 cells were transfected with control siRNA (−) or p73 siRNA (+) for 48 hr and subjected to immunoprecipitation with anti-p73 antibody, followed by immunoblotting with anti-p73 (top) and anti-Aurora-A (middle) antibodies, respectively. WCE were directly immunoblotted with anti-Aurora-A antibody (bottom).
(K) Cos-1 cells synchronized by double thymidine block and release were immunoprecipitated with anti-p73 antibody (left), anti-Aurora-A antibody (right), or normal IgG (NIG), followed by immunoblotting with the indicated antibodies.
See also Figure S1.
We next performed an in vitro kinase assay of p73, with or without wild-type (WT) or kinase-dead (KD) Aurora-A, with the closely related paralog Aurora-B as a control. Aurora-A-WT phosphorylated p73, but Aurora-A-KD did not (Figure 1E). Complete absence of phosphorylation signal on p73 with Aurora-B further validated Aurora-A as the bona fide kinase of p73. We next identified the specific Aurora-A phosphorylated amino acid residue in p73 using site directed mutants in Aurora kinase consensus phosphorylation motifs and subjecting them to in vitro kinase assays. The serine 235 to alanine (S235A) mutant of p73 had reduced phosphorylation than did p73-WT, indicating that S235 is phosphorylated by Aurora-A (Figure 1F). We further confirmed this phosphorylation using an anti-phosphor-PKA substrate specific antibody. p73-WT phosphorylation was evident in cells co-expressing Aurora-A but not those expressing the empty vector. Phosphorylation was significantly diminished in cells expressing the S235A mutant, demonstrating that serine 235 in p73 is phosphorylated by Aurora-A (Figure 1G). Intriguingly, transactivation defective ΔNp73 showed minimal loss of phosphorylation in the SA mutant of the conserved motif and appeared to bind WT and phosphor-mimetic (S235D) mutant of p73 with similar efficiency (Figure S1A and S1B).
We determined in vivo interaction between Aurora-A and p73 by immunoprecipitation of 293T cells co-transfected with Flag-Aurora-A and GFP-p73. Anti-Flag antibody revealed a specific interaction between p73 and Aurora-A (Figure 1H). Anti-Flag antibody immunoprecipitations also detected enriched presence of p73-S235D mutant in the immune complex compared with S235A mutant (Figure S1C). To determine the interaction between endogenous Aurora-A and p73, we used synchronized mitotic cells for reciprocal immunoprecipitation experiments, which revealed p73 and Aurora-A in the same complex (Figure 1I) that was absent in the p73 knockdown cells (Fig. 1J). This interaction was also detected in human non-tumorigenic MCF-10A mammary epithelial cells and p53-deficient H1299 lung carcinoma cells (Figure S1D and S1E). Cell cycle dependence of this interaction was analyzed in synchronized cells after double thymidine block and release. Consistent with published data (Fulco et al., 2003), p73 expression was uniform through the cell cycle. The amount of Aurora-A bound to p73 progressively increased, peaking at mitosis, also evident in nocodazole treated cells (Figure 1K).
Aurora-A phosphorylated p73 loses DNA binding and transactivation activity
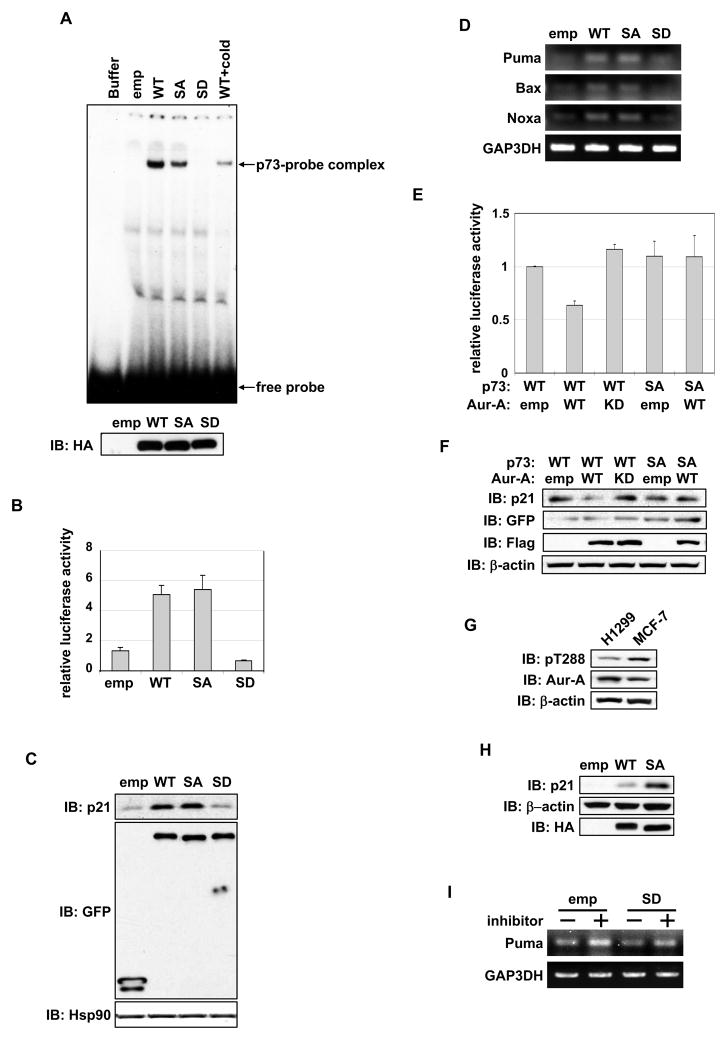
Because the Aurora-A phosphorylation site is located in the DNA binding domain, we determined the effect of Aurora-A phosphorylation on DNA binding and transactivation activity of p73. Electrophoretic mobility shift assay (EMSA) revealed that DNA binding of S235D mutant was markedly inhibited, whereas S235A mutant had weaker DNA binding ability compared with WT (Figure 2A). We next, evaluated the transactivation function of p73 phosphor mutants using a p21 promoter-driven luciferase assay in H1299 cells. S235D mutant had minimal transactivation of the p21 promoter, whereas S235A mutant had similar activity to that of WT (Figure 2B). Endogenous p21 protein levels in cells expressing p73-WT and phosphor mutants were consistent with the p73 transcriptional activity detected by luciferase assay. p21 levels were low in S235D mutant cells compared with WT and S235A mutant cells (Figure 2C). Similarly, p73-S235D mutant cells demonstrated diminished expression of p73 target genes Puma, Bax, and Noxa compared with p73-WT and S235A mutant cells (Figure 2D).
Figure 2. Aurora-A inhibits DNA binding and transactivation activity of p73.
(A) DNA binding activities of in vitro transcribed and translated proteins of HA-p73α wild type (WT), S235A (SA), S235D (SD) and empty vector (emp) were analyzed by EMSA with a [γ32p]ATP-labeled p53/p73 binding consensus oligonucleotide probe. A competition assay was performed with 50-fold molar excess amounts of unlabeled probe (cold). Buffer was used as a negative control. The reaction was separated by a native polyacrylamide gel (top). The amount of proteins used was analyzed by immunoblotting with anti-HA antibody (bottom).
(B) H1299 cells were co-transfected GFP empty or GFP-p73α-WT or mutants with luciferase reporter construct containing p21 promoter and Renilla luciferase reporter internal control plasmid. 24 hr later, cells were subjected to a reporter assay. Data represent mean values with the standard deviation (± SD) from three independent experiments.
(C) Protein expression of p21 in cells transfected with GFP-empty and GFP-p73α constructs used in (B). Proteins were detected by immunoblotting with the indicated antibodies.
(D) mRNA expression of p73 target genes in cells used in (B), analyzed by semi-quantitative RT-PCR.
(E) GFP-p73α-WT or -SA mutant was co-transfected with empty vector, Flag-Aurora-A-WT, and Flag-Aurora-A-KD in H1299 cells and analyzed as in (B). Data represent mean values with ± SD from three independent experiments.
(F) Protein expression of p21 and transfected GFP-p73α and Flag-Aurora-A in cells described in (E), analyzed by immunoblotting with the indicated antibodies.
(G) Expression analysis of active-Aurora-A (pT288) and total Aurora-A in H1299 and MCF-7 cells.
(H) MCF-7 cells transfected with empty vector, HA-p73α-WT, or -SA mutant for 24 hr were analyzed by immunoblotting with the indicated antibodies.
(I) H1299 cells transfected with empty vector or HA-p73α, cultured without (−) or with (+) Aurora-A inhibitor for 24 hr, and analyzed for mRNA expression of Puma by semi-quantitative RT-PCR.
We determined whether p73 activity depends on Aurora-A kinase activity and whether S235A mutant is insensitive to this activity. Luciferase assay revealed that p73-WT activity was inhibited by Aurora-A-WT but not by the KD mutant, whereas S235A mutant was not inhibited by Aurora-A (Figure 2E). Endogenous p21 expression levels in these cells were consistent with the results of luciferase assay (Figure 2F). Similar transactivation activity and endogenous target gene levels in the WT and S235A mutant cells appear to be the result of Aurora-A’s inhibitory phosphorylation interfering with p73-WT’s transactivation function in vivo. To investigate this, we transfected p73-WT and S235A mutant in MCF7 cells, which naturally express high levels of active Aurora-A (Figure 2G). The results revealed distinctly elevated p21 protein levels in cells expressing S235A mutant compared with that in cells expressing WT (Figure 2H). Aurora-A inhibitor treatment of empty vector transfected H1299 cells revealed up-regulation of p73 target PUMA mRNA, whereas S235D interfered with transactivation in a dominant-negative manner (Figure 2I). These results demonstrate that Aurora-A phosphorylation of p73 at serine 235 negatively regulates p73 transactivation.
Aurora-A regulates p73 subcellular localization
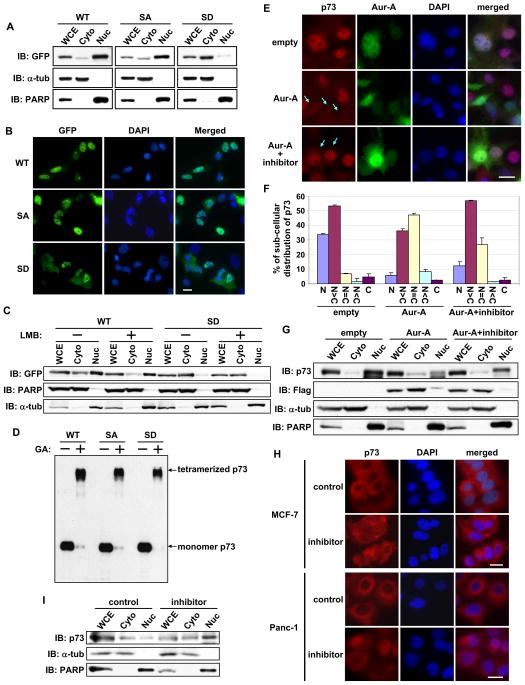
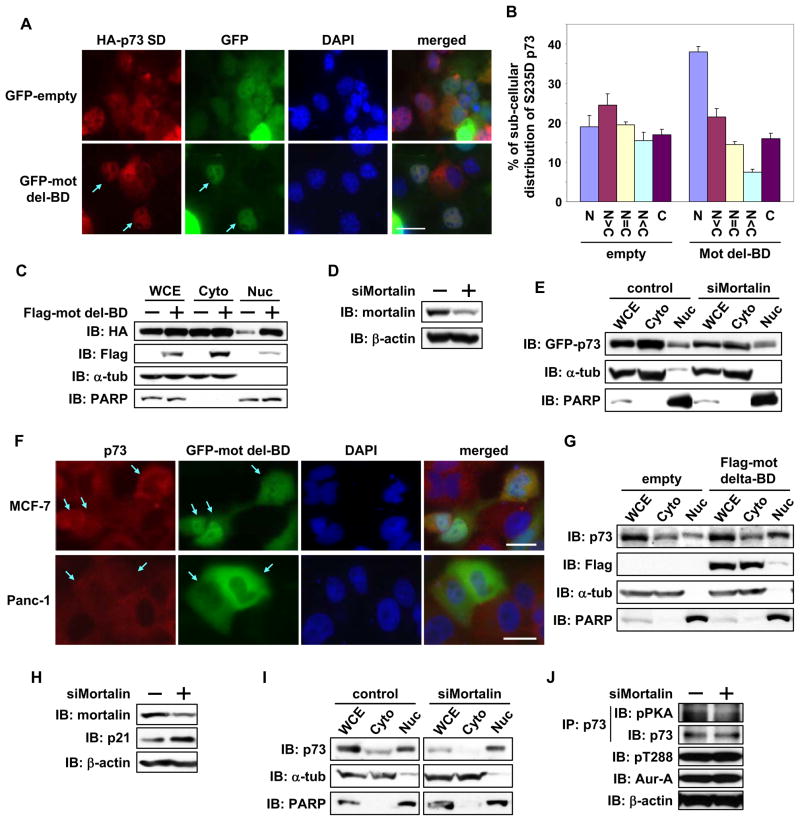
Protein fractionation experiments revealed marked accumulation of S235D mutant in the cytoplasmic fraction, whereas accumulation was predominantly nuclear in the WT and S235A mutant cells (Figure 3A). Similar results were found on immunofluorescence microscopy (Figure 3B) and in different cell lines, such as HeLa, H1299, and MCF7 (data not shown). We next analyzed whether the cytoplasmic distribution of S235D mutant was due to its accelerated export from the nucleus or interference with its nuclear translocation by treating cells with leptomycin B, an inhibitor of nuclear export of proteins. Protein fractionations revealed cytoplasmic localization of S235D mutant regardless of leptomycin B treatment, and more nuclear accumulation of WT (Figure 3C), indicating that phosphorylated p73 at serine 235 is tethered in the cytoplasm. Similar results were observed for S215D mutant of p53 (Figure S2A). Enrichment of the phosphor-mimetic mutant of p73 in the cytoplasmic fraction was also observed in nocadazole arrested mitotic cells with high Aurora-A activity, possibly coinciding with nuclear envelope breakdown (Figure S2B and S2C). Because proteins with aberrant conformations are preferentially transported to the cytoplasm to be degraded, we determined whether cytoplasmic distribution of S235D mutant reflected a conformational change using a glutaraldehyde-based protein cross-linking assay. Since p73 is a tetramer in its natural state and if S235 phosphorylation does not affect monomeric p73 structure, a slower migrating p73 tetramer would still be detectable on SDS-PAGE. High molecular weight S235D and S235A mutants migrated near the tetrameric form of p73-WT (Figure 3D), indicating that p73 phosphorylation status at serine 235 does not cause conformational changes.
Figure 3. Aurora-A regulates subcellular localization of p73.
(A) H1299 cells transfected with GFP-p73α expression plasmids for 24 hr were fractionated into cytosolic (Cyto) and nuclear (Nuc) fractions and analyzed by immunoblotting with anti-GFP antibody. The purity of cytosolic and nuclear fractions was confirmed by immunoblotting with anti-α-tubulin and anti-PARP antibodies, respectively. WCE, whole cell extracts.
(B) H1299 cells transfected with GFP-p73α expression plasmids were analyzed for subcellular localization of GFP fusion p73 by immunofluorescence microscopy and counterstained for DNA with DAPI. Scale bar corresponds to 20μm.
(C) GFP-p73α-WT or -S235D mutant was transfected into H1299 cells. 24 hr later, cells were further cultured with (+) or without (−) leptomycin B for 6 hr and subjected to subcellular fractionation as in (A).
(D) WCE prepared as in (A) were incubated with (+) or without (−) 0.01% glutaraldehyde for 15 min, the products were resolved in SDS-PAGE, followed by immunoblotting with anti-GFP antibody.
(E) Cos-1 cells were transfected with GFP-empty vector or with GFP-Aurora-A WT. 24 hr later, cells were grown for an additional 6 hr with or without MLN8054 and immunostained with anti-p73 antibody (red). DNA was counterstained with DAPI (blue). Arrow indicates GFP-positive cells. Scale bar corresponds to 20μm.
(F) Subcellular distribution of p73 in GFP-positive cells in (E) was analyzed and quantified for nuclear and cytoplasmic localization. Mean values with the standard deviation (± SD) are from two independent experiments (n=100).
(G) Distribution of the proteins in cellular fractions from samples prepared as in (E), analyzed by immunoblotting with the indicated antibodies.
(H) MCF-7 and Panc-1 cells were incubated with or without MLN8054 for 6 hr and subjected to immunostaining as described in (E). Scale bar corresponds to 20μm.
(I) Protein distribution in cellular fractions from samples prepared in (H), analyzed by immunoblotting with the indicated antibodies. See also Figure S2.
To determine whether endogenous p73 is distributed in the cytoplasm with Aurora-A, we performed immunofluorescence microscopy with anti-p73 antibody. Cells overexpressing Aurora-A showed evenly diffused endogenous p73 staining in the cytoplasm and nucleus, which were reversed with Aurora-A inhibitor (Figure 3E and 3F). Protein fractionation experiments further confirmed these findings (Figure 3G). p73 is localized in the cytoplasm of MCF-7 breast cancer cell line and Panc-1 pancreatic cancer cell line, both express elevated Aurora-A levels (Sen et al., 1997, Li et al., 2003). Inhibitor treatment of these cell lines resulted in p73 nuclear localization (Figure 3H), confirming that cytoplasmic distribution of p73 is influenced by Aurora-A kinase activity. Protein fractionation experiments in Panc-1 cells also supported this observation (Figure 3I). Similar results were observed in Aurora-A inhibitor treated MCF-7 cells (data not shown). These results validated that Aurora-A phosphorylation of p73 negatively regulates its nuclear localization.
Mortalin tethers phosphor-p73 in the cytoplasm
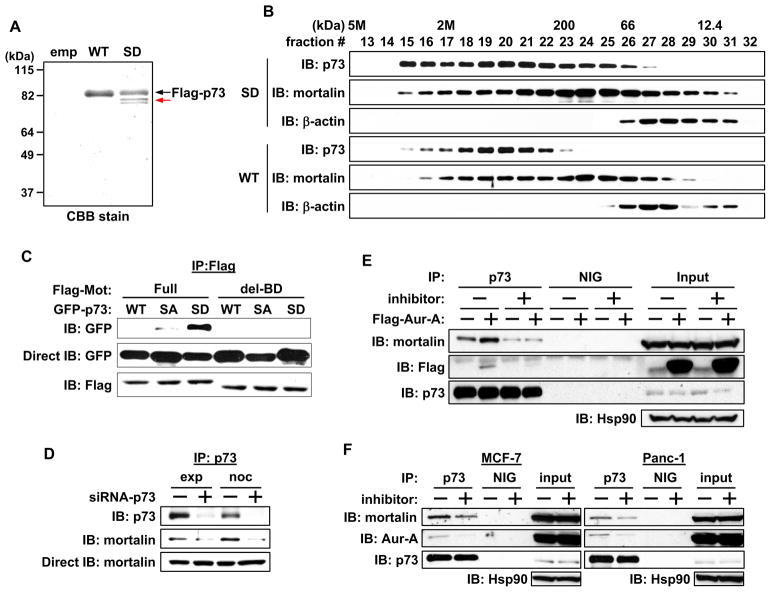
To identify the proteins bound to phosphor-p73, we immunoprecipitated protein complexes with WT and S235D mutant of p73. A protein band of approximately 80 kD molecular weight was detected only in the immune complex of the S235D mutant but not the WT (Figure 4A). Mass spectrometry identified this protein as mortalin, a member of the hsp70 family that is implicated in immortalization and tumorigenesis (Deocaris et al., 2007). Gel filtration column chromatography revealed that p73 and mortalin existed in high molecular weight complexes, distributed over a wide size range. Interestingly, the S235D mutant and mortalin containing complexes were significantly more enriched at greater than 2 mega-Dalton size fractions than were the p73-WT and mortalin complexes (Fig. 4B). Enrichment of S235D mutant and mortalin in the higher molecular complex was also evident in cell extracts resolved on native gels immunoblotted with anti-p73 and mortalin antibodies (Figure S3A). We co-transfected WT or deletion mutant of mortalin (mot del-BD) lacking the p53 binding domain (a.a. 253–282), described earlier (Ma et al., 2006), with WT or phosphor mutants of p73 to determine whether mortalin interaction with the S235D mutant, tethered in the cytoplasm, was mediated through the same domain involved in p53 binding. WT and mutant p73 did not interact with the mortalin deletion mutant, but full-length mortalin’s interaction was enhanced with S235D mutant compared with WT and S235A mutant (Figure 4C). Similar results were seen in p53 co-immunoprecipitation experiments (Figure S3B). These results demonstrate that Aurora-A phosphorylation of p73 and p53 positively regulates their interactions with mortalin, mediated through the same binding domain.
Figure 4. Mortalin interaction with Aurora-A phosphorylated p73.
(A) Flag-empty vector (emp), Flag-p73α-WT, or –SD mutant was transfected into HeLa cells. 24 hr later, cells were immunoprecipitated with anti-Flag antibody conjugated protein-A sepharose and immunoprecipitates were eluted with Flag peptide. Eluted proteins were subjected to SDS-PAGE, and the gel was stained with Coomassie Brilliant Blue (CBB). Arrow in red shows mortalin, identified as the p73-interacting protein from this experiment.
(B) Whole cell extracts (WCE) prepared as in (A) were subjected to gel filtration column chromatography, and an equal volume of eluted proteins in each fraction was analyzed by immunoblotting with the indicated antibodies.
(C) Full-length Flag-mortalin (Full) or p53-binding domain deletion mutant of Flag-mortalin (del-BD) were co-transfected with GFP-p73α–WT, -SA, or -SD in Hela cells. 24 hr later, cells were subjected to immunoprecipitation with anti-Flag antibody, followed by immunoblotting with the indicated antibodies (top and bottom). WCE were directly immunoblotted with anti-GFP antibody (middle).
(D) Cos-1 cells were transfected with control (−) or p73 siRNA (+) for 48 hr and grown with or without nocodazole for 16 hr. Cells were subjected to immunoprecipitation with anti-p73 antibody, followed by immunoblotting with the indicated antibodies (top and middle). WCE were directly immunoblotted with anti-mortalin antibody (bottom).
(E) Cos-1 cells were transfected with Flag-empty vector (−) or with Flag-Aurora-A-WT (+). 24 hr later, cells were cultured for 6 hr with (+) or without (−) MLN8054. Cells were then subjected to immunoprecipitation with anti-p73 antibody or normal IgG (NIG), followed by immunoblotting with the indicated antibodies. Input shows immunoblotting of WCE.
(F) MCF-7 and Panc-1 cells were cultured for 6 hr with (+) or without (−) MLN8054, subjected to immunoprecipitation, and analyzed as in (E).
See also Figure S3.
Immunoprecipitation experiments revealed enhanced interaction of p73 with mortalin in nocodazole treated mitotic cell extracts compared with extracts from exponentially growing cells, indicating the importance of p73 phosphorylation in mitosis for mortalin binding. The specificity of this interaction was verified by immunoprecipitating the extracts from p73 knockdown cells (Figure 4D). The interaction between Aurora-A and p73 was not affected by mortalin deletion mutant (Figure S3C).
To further validate the role of Aurora-A phosphorylation in regulating p73 binding to mortalin co-immunoprecipitation of the two proteins was performed with or without Aurora-A inhibitor treated cells transfected with empty vector or Aurora-A expression vector. Less mortalin bound to p73 in treated cells than in untreated cells. A similar effect was seen in empty vector transfected cells, reflecting the effects of endogenous Aurora-A kinase activity on the binding of p73 to mortalin (Figure 4E). This finding was corroborated in MCF-7 and Panc-1 cells (Figure 4F). Ectopic expression of Aurora-A-KD mutant demonstrated that mortalin protein stability is not affected by Aurora-A kinase activity (Figure S3D). Decreased binding of ectopically expressed and endogenous Aurora-A to p73 in inhibitor treated cells verified that the interaction between Aurora-A and p73 is kinase activity dependent (Figure 4E and 4F).
To determine the effect of mortalin binding on subcellular localization of phosphor-mimetic p73, S235D mutant was cotransfected with the mortalin deletion mutant or an empty vector in Cos-1 cells. In cells with mutant mortalin, the p73 S235D mutant translocated into the nucleus more than in the empty vector transfected cells (Figure 5A and 5B). Protein fractionation experiments also revealed enhanced nuclear accumulation of S235D mutant in mortalin deletion mutant cells than in control cells (Figure 5C). To determine whether loss of mortalin expression had a similar effect on p73 localization, S235D mutant was expressed in cells transfected with control or mortalin targeting siRNAs. Protein fractionation revealed that the nuclear:cytoplasmic ratio was relatively higher in mortalin siRNA transfected cells than in control cells, indicating mortalin involvement in cytoplasmic sequestration of p73 (Figure 5D and 5E). We next analyzed endogenous cytoplasmic p73 in MCF7 and Panc-1 cells after ectopic expression of mortalin deletion mutant. Nuclear staining was detected in 36% of mortalin mutant MCF-7 and Panc-1 cells (n=100) versus 2% of empty vector cells (Figure 5F). p73 was also enriched in the nuclear fraction in mortalin mutant cells, whereas it was localized in the cytoplasm in empty vector cells (Figure 5G). Aurora-A was also distributed in the nucleus in mortalin mutant cells, but its nuclear accumulation was lower than p73 (Figure S4). The microscopy and fractionation experiments demonstrated a positive correlation between nuclear p73 localization and mutant mortalin expression. Moreover, mortalin siRNA transfected Panc-1 cells revealed reduced cytoplasmic localization (Figure 5H and 5I) and phosphorylation of p73 along with increased p21 expression (Figure 5H and 5J), suggesting that mortalin regulates Aurora-A phosphorylation of p73 and its transactivation function. Immunoprecipitation of p73 from empty vector transfected cells demonstrated interaction between p73 and mortalin. This interaction was weakened in the presence of Aurora-A inhibitor, which correlated with positive nuclear p73 staining and loss of Aurora-A interaction with p73. These results point towards an important role for mortalin in cytoplasmic sequestration of p73 following phosphorylation by Aurora-A.
Figure 5. Mortalin interaction regulates cytoplasmic distribution of p73.
(A) HA-p73α-SD was co-transfected with GFP-empty vector or GFP-mortalin-del-BD into Cos-1 cells. 24 hr later, cells were immunostained with anti-HA antibody (red) and counterstained for DNA with DAPI (blue). Arrow shows GFP-positive cells with nuclear staining of HAp73α-SD. Scale bar corresponds to 20μm.
(B) Subcellular distribution of HAp73α SD in GFP-positive Cos-1 cells prepared as in (A) was analyzed and quantified. Mean values with the standard deviation (±SD) are from three independent experiments (n=100).
(C) Cos-1 cells prepared as in (A) were fractionated into cytosolic (Cyto) and nuclear (Nuc) fractions and analyzed by immunoblotting with anti-HA and anti-Flag antibodies. The purity of cytosolic and nuclear fractions was confirmed by immunoblotting for anti-α-tubulin and anti-PARP antibodies respectively.
(D) Cos-1 cells were transfected with control (−) or mortalin siRNA (+) for 48 hr and subjected to immunoblotting with anti-mortalin (top) and anti-β-actin (bottom) antibodies, respectively.
(E) Cos-1 cells transfected with control or mortalin siRNA for 24 hr were transfected with GFP-p73α-SD. 24 hr later, cells were subjected to fractionations, followed by immunoblotting, as in (C).
(F) MCF-7 and Panc-1 cells were transfected with GFP-mortalin-del-BD for 24 hr, immunostained with anti-p73 antibody (red), and counterstained for DNA with DAPI (blue). Arrow indicates GFP-positive cells. Scale bar corresponds to 20μm.
(G) Panc-1 cells were transfected with Flag-empty vector or Flag-mortalin-del-BD. 24 hr later, cells were subjected to fractionation, followed by immunoblotting as in (C).
(H) Panc-1 cells were transfected with control (−) or mortalin siRNA (+) for 48 hr and subjected to immunoblotting with anti-mortalin (top) and anti-β-actin (bottom) antibodies, respectively.
(I) Panc-1 cells prepared in (H) were subjected to fractionations, followed by immunoblotting as in (C).
(J) WCE prepared in (H) were immunoprecipitated with anti-p73 antibody, followed by immunoblotting with the indicated antibodies (top and second). WCE were directly immunoblotted with the indicated antibodies also (third and bottom).
See also Figure S4.
Aurora-A phosphorylation of p73 abrogates cell growth inhibition and DNA damage induced cell death response
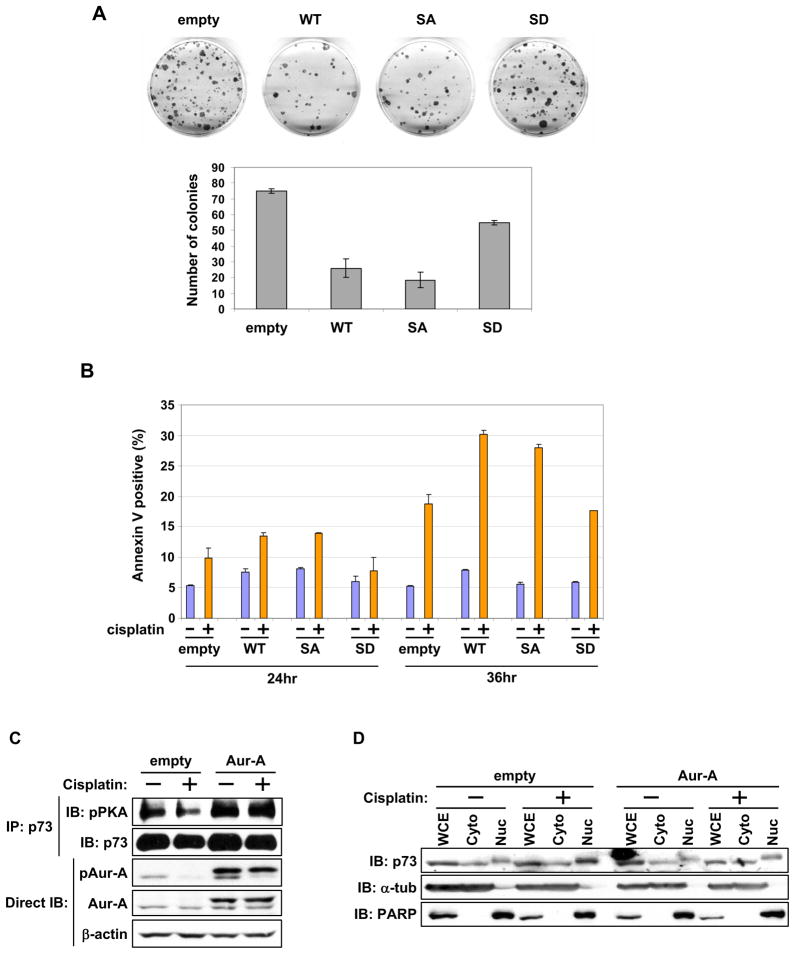
We determined the physiological effects of Aurora-A phosphorylated p73 on cell growth and DNA damage induced cell death response in p53-null Saos-2 and H1299 cells. WT and S235A mutant significantly inhibited colony formation compared with S235D mutant (Figure 6A). Because p73 is a critical regulator of the DNA damage induced cell death pathway, we determined whether p73’s phosphorylation status in H1299 cells influenced cisplatin induced cell death. Consistent with the expected induction of pro-apoptotic genes by p73, cells expressing wild type and S235A mutant showed higher apoptosis than did the vector transfected cells while S235D mutant made cells least sensitive to cisplatin induced cell death (Figure 6B). These results demonstrate that Aurora-A phosphorylation compromises the p73 mediated DNA damage induced cell death response. Next, we determined the plausible differential activation of Aurora-A, p73 phosphorylation and its nuclear-cytoplasmic distribution, with or without DNA damage. DNA damage inducing cisplatin treatment resulted in loss of Aurora-A activation and reduced p73 phosphorylation in empty vector transfected cells, but in the presence of ectopic Aurora-A overexpression, minimal differences in Aurora-A activation, p73 phosphorylation and nuclear cytoplasmic distribution were found between untreated and treated cells (Figure 6C). Empty vector cells showed elevated nuclear distribution of p73 after treatment (Figure 6D).
Figure 6. Role of phosphor-p73 mutant in cell growth and response to DNA damage induced cell death.
(A) Colony formation assay in SAOS-2 cells grown for 3 weeks under G418 selection after transfection with the indicated p73 expression constructs. Representative photos of culture plates from one experiment are shown (top). Mean colony numbers with the standard deviation (± SD) from three independent experiments are shown in the graphs (bottom).
(B) H1299 cells were transfected with the indicated expression constructs. 24 hr later, untreated cells (−) and cells treated with cisplatin (+) for 24 or 36 hr were subjected to annexin-V assay. The graph represents the mean annexin-V-positive apoptotic cells with ± SD from two independent experiments.
(C) Flag-empty vector or Flag-Aurora-A transfected cells for 24 hr were treated with cisplatin (50 uM) for 6 hr. Cells were subjected to immunoprecipitation with anti-p73 antibody, followed by immunoblotting with the indicated antibodies (top and second). Whole cell extracts were directly immunoblotted with antibodies, as shown.
(D) Cells prepared as in (C) were subjected to fractionations, followed by immunoblotting with the indicated antibodies.
Aurora-A phosphorylation inactivates mitotic SAC function of p73
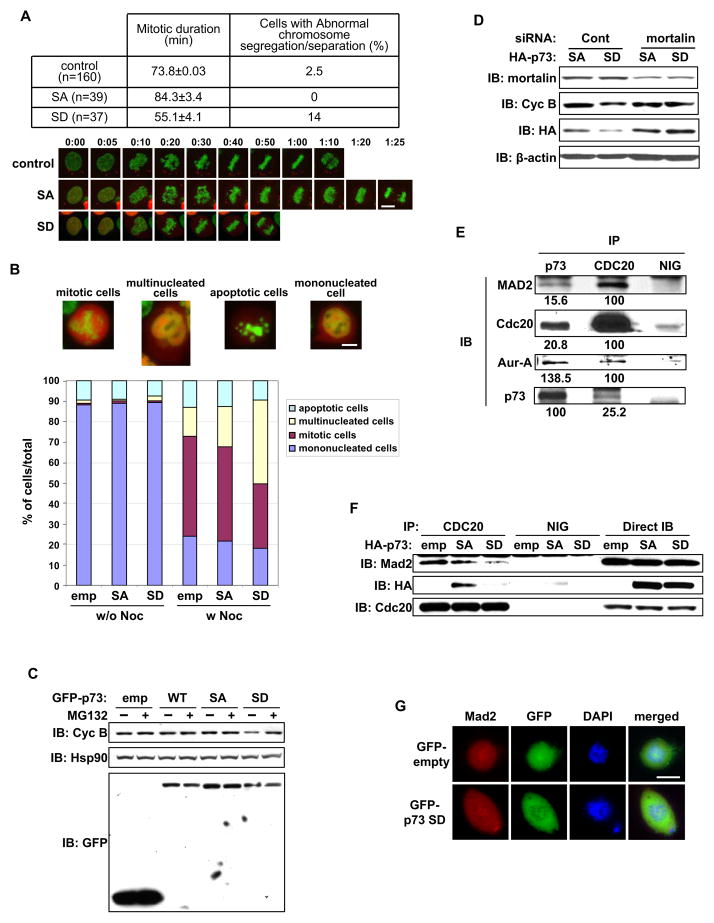
SAC is impaired without p73, thus, we investigated whether Aurora-A phosphorylation of p73 affects SAC response. We ectopically expressed mCherry fusion construct of p73 phosphor mutants in Hela cells in which the chromatin was labeled with stably expressing GFP-tagged histone H2B protein. Time-lapse microscopy revealed that the duration from nuclear envelope breakdown to anaphase was shorter in S235D mutant cells than in controls and S235A mutant cells. S235A mutant cells took the longer to transition into anaphase (Figure 7A). S235D mutant cells had no abnormal chromosome alignment but had frequent chromosome bridges in anaphase-telophase cells, reflecting defects in the chromosome segregation process. To determine whether this resulted from aberrant SAC function, cells expressing p73 phosphor-mutants were grown with or without nocodazole, and quantified in terms of mono- and multi-nucleation, presence in mitosis, or apoptosis induction (Figure 7B). Nocodazole treatment of empty vector and S235A mutant cells had similar effects, with 48.8±1.9% and 46.2±0.4% in mitosis and 14±2.4% and 19.4±1.4% displaying multi-nucleation, respectively. In contrast, nocodazole treatment resulted in fewer S235D mutant cells in mitosis (31.3±1.3%) and more multi-nucleation (41±2.2%). Increased multi-nucleation was also seen in untreated S235D mutant cells than in untreated empty vector and S235A mutant cells (Figure 7B), indicating that Aurora-A phosphorylation of p73 has a role in inactivating the SAC response. Further, p73 phosphor cells were treated with nocodazole, with or without MG132, a proteasome inhibitor that blocks E3 ubiquitin ligase anaphasepromoting complex/cyclosome (APC/C) involved in cyclin B1 degradation. Cyclin B1 levels in S235D mutant cells were lower than in empty vector and S235A mutant cells without MG132 but with MG132, cyclin B1 levels were similar in these cells demonstrating that S235D mutant expression impairs nocodazole induced mitotic arrest (Figure 7C). Nocodazole treated p73-knockdown cells, however, had reduced cyclin B1 levels compared with that in control cells (Figure S5A).
Figure 7. Effects of Aurora-A phosphorylated p73 on mitosis and SAC complex.
(A) HeLa/GFP-H2B cells were transfected with mCherry-p73α-SA or -SD mutant. 24 hr later, cells were monitored by time-lapse microscopy using a low level of light intensity to determine the duration from nuclear envelope breakdown to anaphase. Mock-transfected cells were used as the control. The table represents mean mitotic durations with the standard deviation (± SD) and percentages of cells with defects in chromosome segregation or separation (top). Selected still frames from time-lapse microscopy of transfected cells (bottom) at time intervals of minutes and seconds. Scale bar corresponds to 10μm.
(B) HeLa/GFP-H2B cells transfected with mCherry-empty vector, p73α-SA, or -SD mutant for 24 hr were cultured with (w Noc) or without (w/o Noc) nocodazole. 18 hr later, cells were fixed, and the fate of mCherry-positive cells was analyzed by fluorescence microscopy and quantified. The percentage values of cells in mitosis with the indicated nuclear phenotypes from three independent experiments (n=200), are shown. Scale bar corresponds to 10μm.
(C) 293T cells transfected for 24 hr with the above expression constructs were cultured with nocodazole. 16 hr later, cells were grown with (+) or without (−) MG132 for 4 hr and subjected to immunoblotting with the indicated antibodies.
(D) HeLa/GFP-H2B cells transfected with control or mortalin siRNA for 24 hr were subsequently transfected with HA-p73α-SA or -SD mutant expression constructs. 12 hr later, cells were cultured with nocodazole for 18 hr and subjected to immunoblotting with the indicated antibodies.
(E) Whole cell extracts (WCE) from nocodazole treated 293T cells were immunoprecipitated with normal IgG (NIG), or anti-p73 or anti-CDC20 antibody. Immunoprecipitates were subjected to immunoblotting with the indicated antibodies.
(F) 293T cells transfected with the indicated expression constructs for 24 hr were incubated with nocodazole for 20 hr and then MG132 for 4 hr. Cells were subjected to immunoprecipitation with normal IgG (NIG) or anti-CDC20 antibody, followed by immunoblotting with the indicated antibodies.
(G) Saos-2 cells were transfected with the GFP-empty vector or GFP-p73α-SD mutant construct. 24 hr later, cells were immunostained with anti-Mad2 antibody (red) and counterstained for DNA with DAPI (blue). Scale bar corresponds to 10μm.
See also Figure S5.
We next investigated whether Aurora-A phosphorylation of p73 is a normal physiological event in cells with basal Aurora-A expression or is an unnatural event in Aurora-A overexpressing tumor cells. For the purpose, Aurora-A phosphorylation of p73 was evaluated in synchronized MCF-10A and Cos-1 at prophase; metaphase and anaphase stages. MG132. Western blotting of immunoprecipitated p73 with anti-phosphor PKA substrate antibody revealed that p73 phosphorylation progressively peaked at metaphase but was barely detectable in anaphase, when both amount and activity of Aurora-A were significantly reduced (Figure S5B). These findings indicate that Aurora-A phosphorylation of p73 has a role in regulating SAC during normal mitosis in cells with basal Aurora-A expression. It is conceivable that elevated Aurora-A expression weakens the SAC due to precocious phosphorylation of p73 in tumor cells. Interestingly, co-transfection of S235D mutant with mortalin siRNA failed to override mitotic arrest, as evident from the similar expression levels of cyclin B1 in control and mortalin siRNA transfected cells (Figure 7D), suggesting that silencing of mortalin can rescue phosphor-p73 mediated SAC inactivation.
Co-immunoprecipitation using anti-p73 and anti-CDC20 antibodies revealed complex formation of p73 with Mad2, CDC20, and Aurora-A (Figure 7E). Thus, we determined the effect of p73-S235D mutant expression on these protein-protein interactions in cells treated with nocodazole and MG132. Co-immunoprecipitation experiments with anti-CDC20 antibody revealed a marked reduction in the interaction of both S235D mutant and MAD2 with CDC20 compared with that in empty vector and S235A mutant cells, whereas BubR1’s interaction with CDC20 was not affected in S235D mutant cells (Figure 7F and data not shown). Immunoprecipitation with BubR1 and MAD2 antibodies did not reveal the two proteins in the same complex from nocodazole treated cell extracts (data not shown) indicating that the two checkpoint proteins form independent complexes with CDC20, as reported earlier (Fang, 2002). Immunofluorescence microscopy revealed that kinetochore localized Mad2 is not affected by ectopic expression of S235D mutant (Figure 7G). These results demonstrate that p73 is involved in the formation of a cytoplasmic ternary complex with MAD2 and CDC20. Aurora-A phosphorylation of p73 in this complex releases p73 and the inhibitory complex between MAD2 and CDC20, with the released CDC20 expected to facilitate activation of APC/C, leading to mitotic exit.
Aurora-A overexpressing primary pancreatic cancer shows high cytoplasmic p73 distribution
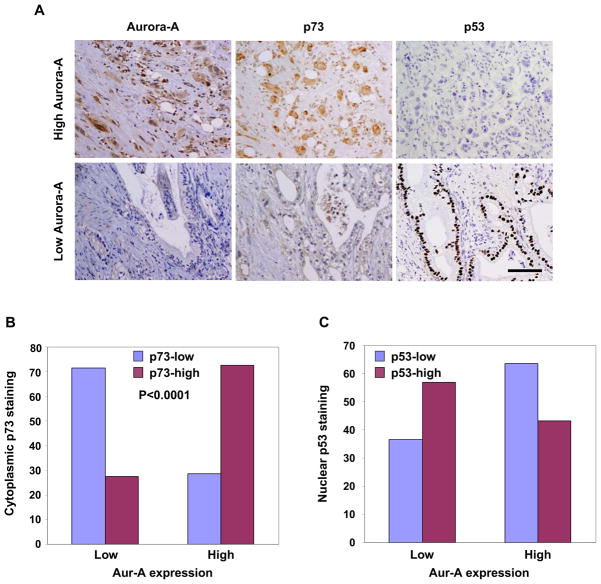
To determine whether cytoplasmic sequestration of p73, consequent to Aurora-A phosphorylation, is reflected in cytoplasmic p73 distribution in Aurora-A overexpressing tumors, we performed immunohistochemical analyses of p73 and Aurora-A in two sets of primary human pancreatic cancer tissues (114 pancreatic ductal adenocarcinomas (PDAC) samples from MD Anderson and 20 from the University of Alabama at Birmingham (UABCC). p53 localization was also determined because Aurora-A phosphor mimetic p53-S215D mutant demonstrated cytoplasmic localization and preferential interaction with mortalin (Figures S2 and S3B). 51 (44.7%) PDAC samples showed high Aurora-A expression. Cytoplasmic p73 staining was clearly detected, but positive cytoplasmic p53 staining was almost undetectable. Among 51 tumors, 37 (72.5%) had high cytoplasmic staining of p73, and 22 (43%) had nuclear staining of p53. Among the remaining 63 Aurora-A low tumors, only 18 (28.6%) had strong cytoplasmic p73 staining, and 40 (63%) had nuclear p53 staining (Figure 8). These results reveal a relationship between Aurora-A expression and cytoplasmic p73 localization and between Aurora-A expression and nuclear p53 localization in primary PDAC tissue. A similar trend between Aurora-A expression and p73 distribution was also found in the UABCC tissue set (data not shown). Nuclear localized mutant p53 is reported in 50%–75% of PDAC, thus, the predominant p53 nuclear distribution was not unexpected. The relationship between high Aurora-A expression and low p53 nuclear staining suggests that Aurora-A overexpression is correlated with p53 gene mutations in PDAC, while p53-WT remains undetectable in the cytoplasm possibly because of enhanced protein degradation after Aurora-A phosphorylation, as previously described (Katayama et al., 2004, Morton et al., 2010).
Figure 8. Correlation of Aurora-A expression levels with p73 and p53 localization in human PDAC.
(A) Representative micrographs of PDAC tissues with varying levels of Aurora-A, p73, and p53 expression. Scale bar corresponds to 50μm.
(B) Bar graph representation of relative cytoplasmic p73 expression and nuclear p53 expression in relation to high or low Aurora-A expression in the TMA containing 114 PDAC tissue samples. Aurora-A expression was correlated significantly with cytoplasmic p73 expression (p<0.0001).
Discussion
Aurora-A overexpression is detected in various tumor types and confers resistance to chemotherapeutic drugs and irradiation (Zhou et al., 1998, Marumoto et al., 2002, Yang et al., 2006). We present evidence that the p73 tumor suppressor protein is a direct downstream target of Aurora-A, which influences cell fate after chemotherapeutic drug induced DNA and spindle damage in tumor cells. Aurora-A phosphorylation of p73 at serine 235 is critical in Aurora-A overexpression mediated abrogation of apoptotic response and mitotic checkpoint override.
Aurora-A inhibits p73 and p53 transactivation functions through a common molecular mechanism
We and the others have reported that Aurora-A phosphorylation of p53 compromises its apoptosis response function induced after cisplatin and irradiation treatment, whereas Aurora-A knockdown sensitizes cells to DNA damage induced p53 dependent apoptosis (Katayama et al., 2004, Liu et al., 2004). The current findings reveal that Aurora-A phosphorylations abrogate DNA damage response functions of both p53 and p73 consequent to their interactions with mortalin and cytoplasmic sequestration. It also appears that with progressively increasing Aurora-A kinase activity during mitosis, p53 and p73 remain localized in the cytoplasm coincidentally with nuclear envelope breakdown. Phosphorylation mediated binding to mortalin, promoting nuclear exclusion of p53 and p73, may be common in tumor cells (Walker et al., 2006, Walker and Böttger, 2008) and consistent with the earlier observations that p53 binding domain on mortalin (Kaul et al., 2001) negatively regulates transcriptional activity, inhibits nuclear translocation of p53, and abolishes p53 dependent suppression of centrosome duplication (Wadhwa et al., 1998, Walker et al., 2006, Ma., et al., 2006). Because the mortalin binding domain of p53 at its C-terminus (Kaul et al., 2001, Wadhwa et al., 2002) is not conserved in p73, it is worth investigating whether Aurora-A phosphorylation of p53 and p73 creates a mortalin binding site or recruits a mortalin interaction factor in a phosphorylation dependent manner. Complex formation between mortalin and p53 has been detected in the mitochondria during p53 induced apoptosis, with and without DNA damage (Marchenko et al., 2000), implicating involvement of mortalin-p53 complex in the transactivation independent apoptotic signaling pathway. However, the molecular mechanisms regulating activation of this pathway remains to be elucidated. WWOX, a putative tumor suppressor protein interacts with p53 and p73 regulating their sub-cellular distribution and apoptosis response functions elicited in mitochondria (Chang et al., 2003, Chang et al., 2005, Aqeilan et al., 2004). Based on the current findings its may be suggested that Aurora-A phosphorylation induced mortalin binding influences interactions of p53 and p73 with WWOX and/or pro-apoptotic mitochondria proteins. Further investigation is needed to understand these pathways.
Molecular mechanism of Aurora-A mediated inactivation of mitotic SAC function of p73
Aurora-A overexpression has been shown to override mitotic SAC and induce aberrant chromosome segregation, resulting in aneuploidy (Anand et al., 2003). However, the underlying molecular mechanism of this effect has remained unclear. We found that p73 was involved in the inhibitory mitotic checkpoint complex of Mad2 and CDC20, preventing activation of the E3 ubiquitin ligase APC/C, and Aurora-A phosphorylation of p73 caused dissociation of the Mad2-CDC20 complex, facilitating mitotic exit. Because p73 is detected in large macromolecular complexes including mortalin, the further studies are needed to determine their functional significance in the regulation of the Mad2-CDC20 containing SAC complex.
We observed no specific localization of wild-type or phosphor-mimetic p73 mutants at the mitotic apparatuses or an effect of phosphor-mimetic mutant on Mad2 mis-localizations at the kinetochore. However, immunostaining with anti-p73 antibody revealed cytoplasmic and mitotic spindle p73 localization. Mitotic SAC generates a diffusible wait signal at microtubule unattached kinetochores that inhibits CDC20 mediated APC activation. MAD2 and BubR1 are the two most critical proteins of this signal (Shah et al., 2004, Peters, 2006), which form separate inactive complexes with CDC20 (Fang, 2002). Although evidence suggests that the soluble MAD2-CDC20 complex acts as a transient precursor to the BubR1-CDC20 inhibitory complex (Kulukian et al, 2009), the exact mechanism is still not well understood. Failure of BubR1 to rescue SAC dysfunction in cells expressing a mutant CDC20 allele that does not bind MAD2 (Li et al., 2009) clearly illustrates critical non-redundant role of Mad2 in SAC activation. Aurora-A phosphorylation of p73 dissociated the MAD2-CDC20 complex, providing evidence that Aurora-A negatively regulates a critical step in the SAC activation pathway. Unlike its effect on Mad2-CDC20 interaction, phosphor-mimetic mutant p73 did not affect the interaction of BubR1 with CDC20. Progressively increasing Aurora-A phosphorylation of p73 from prophase through metaphase, followed by a sharp decline at anaphase and telophase in synchronized non-tumorigenic MCF-10A cells, with basal Aurora-A expression, suggests that this phosphorylation has a role in inactivating SAC during the metaphase-anaphase transition of normal mitosis. Constitutively phosphorylated p73 expressing cells underwent an early transition to anaphase and overrode the mitotic checkpoint, indicating that Aurora-A overexpressing cells are predisposed to abrogate the checkpoint response because of precocious p73 phosphorylation. Our findings do not reveal how this phosphorylation is temporally regulated to coincide with SAC inactivation after chromosome bi-orientation in normal mitosis. Structural studies have revealed that an open conformation of MAD2 (O-MAD2) prevents association with MAD1 or CDC20 (Musacchio and Hardwick, 2002). Thus, it will be interesting to determine whether Mad2-bound p73 phosphorylation induces open conformation changes in the latter, leading to its dissociation from CDC20. Our findings indicate that p73 is a critical regulator of the cytoplasmic MAD2-CDC20 checkpoint protein complex. Additional studies are required to unravel the details of these molecular interactions.
p73 deficient mice have a high incidence of spontaneous tumors (Tomasini et al., 2008) and loss of function is correlated with induction of chromosomal instability (Talos et al., 2007). Evidence supports a role for p73 in mitosis (Fulco et al., 2003, Merlo et al. 2005), including SAC regulation (Tomasini et al., 2009). Thus, p73 plays an important role in faithful chromosome segregation and maintenance of genomic stability. p73 is up-regulated during the transformation process in response to aberrant Rb pathway expression, and a genetic alteration with a dominant negative effect is required to block tumor suppressor function of p73. Published data indicate that overexpression of the dominant negative p73 protein ΔNp73 compromises tumor suppressor function of p73 in pre-malignant stages (Oswald and Stiewe, 2008). ΔNp73 overexpression may disrupt the stochastic balance of Aurora-A mediated p73 SAC function because the two isoforms, despite forming a heterotetramer, do not share the predominant site of Aurora-A phosphorylation in p73.
Our findings demonstrate that elevated Aurora-A expression, a common oncogenic event in human cancers, has the dominant negative effect of inactivating p73 function through increased phosphorylation of the protein sequestered in the cytoplasm. The positive correlation between Aurora-A overexpression and cytoplasmic p73 localization in human pancreatic cancer tissue corroborate the experimental findings and indicates that these tumors have weakened or inactivated DNA and spindle damage induced apoptosis and SAC pathways, making them refractory to conventional radiation and chemotherapeutic regimens. Detailed analyses of p73 phosphorylation profiles of these tumors together with chemosensitivities and radiosensitivities would help resolve the issue and future design of appropriately targeted therapies.
In conclusion, we uncovered a signaling pathway of Aurora-A-p73 axis in which Aurora-A phosphorylation inactivates p73 function in both DNA damage induced cell death and mitotic SAC pathways. Further in-depth studies of Aurora-A involvement in both signaling pathways will improve our understanding of oncogenic function of Aurora-A in cancer biology and help us develop more effective strategies for cancer prevention and treatment.
Experimental Procedures
Detailed experimental procedures are available in the Supplemental Experimental Procedures.
Transfection, Luciferase assay, and siRNA
For transfection, Fugene 6 transfection reagent (Roche), oligofectamine, and lipoectamine 2000 (Invitrogen) were used according to manufacturer’s instructions. For luciferase assays, H1299 or Saos-2 cells were co-transfected with the same amount of wild type (WT) or mutant pEGFPp73α (100–200 ng), luciferase reporter construct (100 ng), and internal control Renilla luciferase expression plasmid (10 ng), with or without increasing amounts of Flag-Aurora-A WT or kinase dead expression plasmids. The total amount of plasmid DNA was kept constant (1 ug) at pcDNA3. Luciferase activities were measured 24 hr after transfection using a dual-luciferase reporter assay kit (Promega). The siRNA sequences were: p73; 5′-CGGAUUCCAGCAUGGACGUdTdT-3′ (Basu et al., 2003), mortalin; 5′-CCGUCUCGUGGGCGCCGCAdTdT-3′ (Ma et al 2006). Additional siRNAs for both genes from Santa-Cruz Biotechnology (cat# sc-35520 for mortalin, cat# sc-36167 for p73) were also used.
Cell fractionation, cross linking assay, in vitro kinase assay, semi-quantitative RT-PCR, immunoprecipitation, western blotting, and immunofluorescence microscopy
Biochemical protein fractionation of cells was performed according to the manufacturer’s protocol (Pierce). Whole cell extracts were prepared in RIPA buffer. All other experimental procedures, conditions, and primer sequences for semi-quantitative RT-PCR have been described (Kawai et al., 2007, Katayama et al., 2007 and 2008, Li et al., 2007).
EMSA
p73 proteins, produced by an in vitro transcription and translation kit (Promega), were incubated with 32P-labeled p21 probe containing the p53 DNA binding site (5′-TACAGAACATGTCTAAGCATGCTGGGG-3′) in the binding buffer (20 mM Hepes pH 7.9, 1 mM MgCl2, 10 uM ZnCl2, 1 mM DTT, 5% glycerol, 670 ng/ul BSA, and 0.5 ug/ul poly(dI-dC) at R.T. for 20 min. For the competition assay, 1 ug of unlabeled probe was added to the reaction. The protein-DNA complexes were resolved by electrophoresis through 4.5% polyacrylamide gel at 4 °C.
λPPase treatment
Whole cell extracts were incubated with 20 units of λPPase (New England Biolabs) or glycerol (solvent) in the supplemented buffer for 30 min at 30°C. The reaction was terminated by adding SDS sample buffer and subjected to SDS-PAGE.
Gel filtration column chromatography
Gel filtration column chromatography was carried out as described previously (Samanta et al., 2010). In brief, 3 mg of whole cell extracts prepared in column elution buffer (30 mM HEPES, pH 7.4, containing 150 mM NaCl and 10% glycerol, and 0.5% NP-40, 1 μM microcystin LR, protease inhibitor cocktail (Roche)) were loaded on the column packed with Superose 6 prep grade gel (GE Healthcare), and 500 μl of elution was collected in each fraction. Equal volumes of eluted fractions were subjected to immunoblotting. The mixture of protein markers containing keyhole limpet hemocyanin (KLH; MW 8.5 million Da), blue dextran (2 million Da), β-amylase (200 kDa), BSA (66 kDa), and cytochrome C (12.4 kDa) were used as the molecular weight standard.
Time-lapse microscopy
Time-lapse microscopy was performed using a Perkin Elmer UltraVIEW ERS spinning disc confocal microscope equipped with an environmental control chamber that maintained the cells at 37°C in a humidified stream of 5% CO2. Individually tagged image format files were imported into Photoshop (Adobe) for analysis.
Colony formation assay and drug treatment
Saos-2 cells, transfected with pEGFP-empty vector, WT and mutants of pEGFP-p73α, were cultured in media containing 200 μg/ml of G418 for 3 weeks and stained with crystal violet. Colonies of 1 mm diameter were counted. H1299 cells transfected for 24 hr were treated with cisplatin at 50 μM for 24 hr and 36 hr. Annexin V-FITC assay was performed according to the manufacturer’s protocol (BD Pharmingen). Nocodazole was used at 50 ng/ml for GFP-H2B HeLa, 350 ng/ml for 293T, 500 ng/ml for MCF-7 and Panc-1, and 1 ug/ml for Cos-1 cells. Aurora-A inhibitor MLN8054 was used at 0.5 μM with or without 20 μM of MG132 for 4–6 hr.
Supplementary Material
Highlights.
Aurora-A abrogates p73 functions in DNA damage response and SAC pathways
Mortalin tethers Aurora-A phosphorylated p73 in the cytoplasm
Aurora-A phosphorylation of p73 regulates Mad2-Cdc20 SAC complex
Cytoplasmic p73 correlates with Aurora-A expression levels in primary human tumors
Significance.
Resistance to DNA and spindle damaging chemotherapeutic drugs is a major challenge to effective therapeutic interventions in human cancer. Elevated Aurora kinase-A expression is correlated with chemotherapy resistance, and p73 is a major determinant of chemosensitivity in tumor cells. Our findings demonstrate that p73 phosphorylation by Aurora-A results in inactivation of its DNA damage and spindle assembly checkpoint response functions. Aurora-A phosphorylated p73 loses chromatin binding affinity, is sequestered in the cytoplasm, and facilitates dissociation of the MAD2-CDC20 spindle assembly checkpoint complex. Elucidation of the molecular mechanisms underlying the development of resistance to DNA and spindle targeted therapeutic agents in Aurora-A overexpressing tumors should help us design more effective therapeutic regimens.
Acknowledgments
The authors thank Dr. Zhi-Min Yuan for the GFP-tagged p73 expression construct and luciferase reporter constructs, Dr. Elsa Flores for the HA-tagged p73 expression constructs, and Dr. Kenji Fukasawa for the mortalin expression constructs. We acknowledge the technical assistance of Ms. Yvette Gonzales and Ms. Aimee LeBlanc. Editorial help of Ms. Amy Sutton from the Department of Scientific Publications is acknowledged. This study was supported by grants from the National Institutes of Health (R01CA089716 and NCI/EDRN UO1CA111302), University Cancer Foundation, MD Anderson to S.S. The DNA analysis facility used in the study is supported by Cancer Center Support grant CA16672.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anand S, Penrhyn-Lowe S, Venkitaraman AR. AURORA-A amplification overrides the mitotic spindle assembly checkpoint, inducing resistance to Taxol. Cancer Cell. 2003;3:51–62. doi: 10.1016/s1535-6108(02)00235-0. [DOI] [PubMed] [Google Scholar]
- Aqeilan RI, Pekarsky Y, Herrero JJ, Palamarchuk A, Letofsky J, Druck T, Trapasso F, Han SY, Melino G, Huebner K, et al. Functional association between Wwox tumor suppressor protein and p73, a p53 homolog. Proc Natl Acad Sci USA. 2004;101:401–406. doi: 10.1073/pnas.0400805101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Totty NF, Irwin MS, Sudol M, Downward J. Akt Phosphorylates the Yes-Associated Protein, YAP, to Induce Interaction with 14-3-3 and Attenuation of p73-Mediated Apoptosis. Mol Cell. 2003;11:11–23. doi: 10.1016/s1097-2765(02)00776-1. [DOI] [PubMed] [Google Scholar]
- Chang NS, Doherty J, Ensign A, Lewis J, Heath J, Schultz L, Chen ST, Oppermann U. Molecular mechanisms underlying WOX1 activation during apoptotic and stress responses. Biochem Pharmacol. 2003;66:1347–1354. doi: 10.1016/s0006-2952(03)00484-2. [DOI] [PubMed] [Google Scholar]
- Chang NS, Doherty J, Ensign A, Schultz L, Hsu LJ, Hong Q. WOX1 is essential for tumor necrosis factor-, UV light-, staurosporine-, and p53-mediated cell death, and its tyrosine 33- phosphorylated form binds and stabilizes serine 46-phosphorylated p53. J Biol Chem. 2005;280:43100–43108. doi: 10.1074/jbc.M505590200. [DOI] [PubMed] [Google Scholar]
- Dar AA, Belkhiri A, Ecsedy J, Zaika A, El-Rifai W. Aurora kinase A inhibition leads to p73-dependent apoptosis in p53-deficient cancer cells. Cancer Res. 2008;68:8998–9004. doi: 10.1158/0008-5472.CAN-08-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deocaris CC, Widodo N, Ishii T, Kaul SC, Wadhwa R. Functional significance of minor structural and expression changes in stress chaperone mortalin. Ann N Y Acad Sci. 2007;1119:165–175. doi: 10.1196/annals.1404.007. [DOI] [PubMed] [Google Scholar]
- Ewart-Toland A, Briassouli P, de Koning JP, Mao JH, Chan F, MacCarthy-Morrogh L, Ponder BA, Nagase H, Burn J, Ball S, Almeida M, Linardopoulos S, Balmain A. Identification of Stk6/STK15 as a candidate low penetrance tumor-susceptibility gene in mouse and human. Nat Genet. 2003;34(4):403–412. doi: 10.1038/ng1220. [DOI] [PubMed] [Google Scholar]
- Fang G. Checkpoint protein BubR1 acts synergistically with Mad2 to inhibit anaphase-promoting complex. Mol Biol Cell. 2002;13:755–766. doi: 10.1091/mbc.01-09-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulco M, Costanzo A, Merlo P, Mangiacasale R, Strano S, Blandino G, Balsano C, Lavia P, Levrero M. p73 is regulated by phosphorylation at the G2/M transition. J Biol Chem. 2003;278:49196–49202. doi: 10.1074/jbc.M304921200. [DOI] [PubMed] [Google Scholar]
- Katayama H, Sasai K, Kawai H, Yuan ZM, Bondaruk J, Suzuki F, Fujii S, Arlinghaus RB, Czerniak BA, Sen S. Phosphorylation by aurora kinase A induces Mdm2-mediated destabilization and inhibition of p53. Nat Genet. 2004;36:55–62. doi: 10.1038/ng1279. [DOI] [PubMed] [Google Scholar]
- Katayama H, Sasai K, Czerniak BA, Carter JL, Sen S. Aurora-A kinase phosphorylation of Aurora-A kinase interacting protein (AIP) and stabilization of the enzyme-substrate complex. J Cell Biochem. 2007;102:1318–1331. doi: 10.1002/jcb.21421. [DOI] [PubMed] [Google Scholar]
- Katayama H, Sasai K, Kloc M, Brinkley BR, Sen S. Aurora kinase-A regulates kinetochore/chromatin associated microtubule assembly in human cells. Cell Cycle. 2008;7:2691–2704. doi: 10.4161/cc.7.17.6460. [DOI] [PubMed] [Google Scholar]
- Kaul SC, Reddel RR, Mitsui Y, Wadhwa R. An N-terminal region of mot-2 binds to p53 in vitro. Neoplasia. 2001;3:110–114. doi: 10.1038/sj.neo.7900139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai H, Lopez-Pajares V, Kim MM, Wiederschain D, Yuan ZM. RING domainmediated interaction is a requirement for MDM2’s E3 ligase activity. Cancer Res. 2007;67:6026–6030. doi: 10.1158/0008-5472.CAN-07-1313. [DOI] [PubMed] [Google Scholar]
- Kulukian A, Han JS, Cleveland DW. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Dev Cell. 2009;16:105–117. doi: 10.1016/j.devcel.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Zhu J, Firozi PF, Abbruzzese JL, Evans DB, Cleary K, Friess H, Sen S. Overexpression of oncogenic STK15/BTAK/Aurora A kinase in human pancreatic cancer. Clin Cancer Res. 2003;9:991–997. [PubMed] [Google Scholar]
- Li M, Fang X, Wei Z, York JP, Zhang P. Loss of spindle assembly checkpoint-mediated inhibition of Cdc20 promotes tumorigenesis in mice. J Cell Biol. 2009;185:983–994. doi: 10.1083/jcb.200904020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Lee YK, Jeng JC, Yen Y, Schultz DC, Shih HM, Ann DK. Role for KAP1 serine 824 phosphorylation and sumoylation/desumoylation switch in regulating KAP1-mediated transcriptional repression. J Biol Chem. 2007;282:36177–36189. doi: 10.1074/jbc.M706912200. [DOI] [PubMed] [Google Scholar]
- Liu Q, Kaneko S, Yang L, Feldman RI, Nicosia SV, Chen J, Cheng JQ. Aurora-A abrogation of p53 DNA binding and transactivation activity by phosphorylation of serine 215. J Biol Chem. 2004;279:52175–52182. doi: 10.1074/jbc.M406802200. [DOI] [PubMed] [Google Scholar]
- Ma Z, Izumi H, Kanai M, Kabuyama Y, Ahn NG, Fukasawa K. Mortalin controls centrosome duplication via modulating centrosomal localization of p53. Oncogene. 2006;25:5377–5390. doi: 10.1038/sj.onc.1209543. [DOI] [PubMed] [Google Scholar]
- Macurek L, Lindqvist A, Lim D, Lampson MA, Klompmaker R, Freire R, Clouin C, Taylor SS, Yaffe MB, Medema RH. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature. 2008;455:119–123. doi: 10.1038/nature07185. [DOI] [PubMed] [Google Scholar]
- Marchenko ND, Zaika A, Moll UM. Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J Biol Chem. 2000;275:16202–16212. doi: 10.1074/jbc.275.21.16202. [DOI] [PubMed] [Google Scholar]
- Marumoto T, Hirota T, Morisaki T, Kunitoku N, Zhang D, Ichikawa Y, Sasayama T, Kuninaka S, Mimori T, Tamaki N, et al. Roles of aurora-A kinase in mitotic entry and G2 checkpoint in mammalian cells. Genes Cells. 2002;7:1173–1182. doi: 10.1046/j.1365-2443.2002.00592.x. [DOI] [PubMed] [Google Scholar]
- Marumoto T, Zhang D, Saya H. Aurora-A - a guardian of poles. Nat Rev Cancer. 2005;5:42–50. doi: 10.1038/nrc1526. [DOI] [PubMed] [Google Scholar]
- Merlo P, Fulco M, Costanzo A, Mangiacasale R, Strano S, Blandino G, Taya Y, Lavia P, Levrero M. A role of p73 in mitotic exit. J Biol Chem. 2005;280:30354–30360. doi: 10.1074/jbc.M500635200. [DOI] [PubMed] [Google Scholar]
- Morton JP, Timpson P, Karim SA, Ridgway RA, Athineos D, Doyle B, Jamieson NB, Oien KA, Lowy AM, Brunton VG, et al. Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer. Proc Natl Acad Sci U S A. 2010;107:246–251. doi: 10.1073/pnas.0908428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, Hardwick KG. The spindle checkpoint: structural insights into dynamic signalling. Nat Rev Mol Cell Biol. 2002;3:731–741. doi: 10.1038/nrm929. [DOI] [PubMed] [Google Scholar]
- Oswald C, Stiewe T. In good times and bad: p73 in cancer. Cell Cycle. 2008;7:1726–1731. doi: 10.4161/cc.7.12.6148. [DOI] [PubMed] [Google Scholar]
- Ouchi M, Fujiuchi N, Sasai K, Katayama H, Minamishima YA, Ongusaha PP, Deng C, Sen S, Lee SW, Ouchi T. BRCA1 phosphorylation by Aurora-A in the regulation of G2 to M transition. J Biol Chem. 2004;279:19643–19648. doi: 10.1074/jbc.M311780200. [DOI] [PubMed] [Google Scholar]
- Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- Plotnikova OV, Golemis EA. Aurora A kinase activity influences calcium signaling in kidney cells. J Cell Biol. 2011;193:1021–1032. doi: 10.1083/jcb.201012061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta AK, Chakraborty SN, Wang Y, Schlette E, Reddy EP, Arlinghaus RB. Destabilization of Bcr-Abl/Jak2 Network by a Jak2/Abl Kinase Inhibitor ON044580 Overcomes Drug Resistance in Blast Crisis Chronic Myelogenous Leukemia (CML) Genes Cancer. 2010;1:346–359. doi: 10.1177/1947601910372232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki A, Coppinger JA, Jang CY, Yates JR, Fang G. Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science. 2008;320:1655–1658. doi: 10.1126/science.1157425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen S, Zhou H, White RA. A putative serine/threonine kinase encoding gene BTAK on chromosome 20q13 is amplified and overexpressed in human breast cancer cell lines. Oncogene. 1997;14:2195–2200. doi: 10.1038/sj.onc.1201065. [DOI] [PubMed] [Google Scholar]
- Shah JV, Botvinick E, Bonday Z, Furnari F, Berns M, Cleveland DW. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr Biol. 2004;14:942–952. doi: 10.1016/j.cub.2004.05.046. [DOI] [PubMed] [Google Scholar]
- Tomasini R, Tsuchihara K, Wilhelm M, Fujitani M, Rufini A, Cheung CC, Khan F, Itie-Youten A, Wakeham A, Tsao MS, et al. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Dev. 2008;22:2677–2691. doi: 10.1101/gad.1695308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasini R, Tsuchihara K, Tsuda C, Lau SK, Wilhelm M, Ruffini A, Tsao MS, Iovanna JL, Jurisicova A, Melino G, et al. TAp73 regulates the spindle assembly checkpoint by modulating BubR1 activity. Proc Natl Acad Sci USA. 2009;106:797–802. doi: 10.1073/pnas.0812096106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernole P, Neale MH, Barcaroli D, Munarriz E, Knight RA, Tomasini R, Mak TW, Melino G, De Laurenzi V. TAp73alpha binds the kinetochore proteins Bub1 and Bub3 resulting in polyploidy. Cell Cycle. 2009;8:421–429. doi: 10.4161/cc.8.3.7623. [DOI] [PubMed] [Google Scholar]
- Wadhwa R, Takano S, Robert M, Yoshida A, Nomura H, Reddel RR, Mitsui Y, Kaul SC. Inactivation of tumor suppressor p53 by mot-2, a hsp70 family member. J Biol Chem. 1998;273:29586–29591. doi: 10.1074/jbc.273.45.29586. [DOI] [PubMed] [Google Scholar]
- Wadhwa R, Yaguchi T, Hasan MK, Mitsui Y, Reddel RR, Kaul SC. Hsp70 family member, mot-2/mthsp70/GRP75, binds to the cytoplasmic sequestration domain of the p53 protein. Exp Cell Res. 2002;274:246–253. doi: 10.1006/excr.2002.5468. [DOI] [PubMed] [Google Scholar]
- Walker C, Böttger S, Low B. Mortalin-based cytoplasmic sequestration of p53 in a nonmammalian cancer model. Am J Pathol. 2006;168:1526–1530. doi: 10.2353/ajpath.2006.050603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker CW, Böttger SA. A naturally occurring cancer with molecular connectivity to human diseases. Cell Cycle. 2008;7:2286–2289. doi: 10.4161/cc.6366. [DOI] [PubMed] [Google Scholar]
- Yang H, He L, Kruk P, Nicosia SV, Cheng JQ. Aurora-A induces cell survival and chemoresistance by activation of Akt through a p53-dependent manner in ovarian cancer cells. Int J Cancer. 2006;119:2304–2312. doi: 10.1002/ijc.22154. [DOI] [PubMed] [Google Scholar]
- Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW, Sahin A, Brinkley BR, Sen S. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.