Abstract
Excessive bone marrow adipocytes (BMAs) formation is tightly associated with development of osteoporosis. Considering the high heterogeneity of bone marrow stromal cells (BMSCs), identification of bone marrow adipocyte progenitors (BMAPs) within heterogeneous BMSCs may provide better cellular models for research regarding osteoporosis development and therapy. However, currently there is no efficient method or specific surface makers that are available for BMAPs isolation. In the current study, we developed a novel BMAPs isolation method based on silica microbeads incubation and subsequent centrifugation in ficoll-paque. The “Sca-1+CD73−CD90−CD105+” subpopulation selected by this method exhibited significantly stronger adipogenic potential than nonselected BMSCs in vitro and could homogeneously differentiate into mature adipocytes within 4 days. Moreover, these cells also highly expressed a series of adipogenesis-related genes even before differentiation. After long-term culture, however, BMAPs would gradually lose high adipogenic ability, but sorting CD105+ cells from BMAPs in later passages was able to retrieve the highly adipogenic subpopulation. In conclusion, this study demonstrated that BMAPs subpopulation could be effectively isolated from heterogeneous BMSCs by a special silica microbeads incubation method and re-enriched by sorting CD105+ cells. These findings offer convenient and repeatable approaches to obtain pure BMAPs for research regarding pathogenic mechanisms and therapeutics development of osteoporosis.
Introduction
Increased bone marrow adiposity is a common phenomenon observed in osteoporosis [1–3]. Even the exact roles of bone marrow adipocytes (BMAs) in osteoporosis development have not been fully revealed [4], more recent studies are supporting the notion that excessive BMAs formation will accelerate the progression of osteoporosis. For example, increased bone marrow adiposity induced by treatment of adipogenic drugs or feeding a high fat diet would lead to reduced bone mineral density [5,6]. Moreover, recent studies also discovered that BMAs were able to suppress new bone formation by inducing osteoblast trans-differentiation to adipocytes or to enhance old bone resorption by promoting osteoclast formation [7–10]. Therefore, these findings supported the detrimental effects of excessive BMAs formation and highlighted the importance of suppressing bone marrow adipogenesis for osteoporosis therapy.
To achieve this goal, previous studies have extensively investigated the molecular mechanisms controlling adipogenic differentiation of bone marrow stromal cells (BMSCs) based on cell lines or primary BMSCs [4,11,12]. However, cellular models based on primary BMSCs or immortalized cell lines are confronted with certain limitations. For example, one drawback for primary BMSCs is their high heterogeneity, especially for adipogenic potentials [13–15]. One recent study found that a significant portion of human primary BMSCs were unable to differentiate into adipocytes in vitro. Moreover, even within the adipogenic capable cells, the adipogenic potentials for different subpopulations also displayed high variations [13]. Therefore, studies based on heterogeneous BMSCs might only reflect the general features of all BMSCs subpopulations rather than the specific features of the highly adipogenic subpopulation. On the other hand, immortalized cell lines, especially preadipocyte cell lines [16], may offer more stable and homogenous models for adipogenesis research. However, these cells have been immortalized and have undergone several gene mutations [17], which may raise the concern about their similarities to the real BMSCs in vivo. Moreover, studies based on cell lines also cannot monitor the real-time cellular changes in the animal models. Due to these limitations, it will be more preferable to directly isolate the bone marrow adipocyte progenitors (BMAPs) for studies regarding bone marrow adipogenesis, as they can truly represent the highly adipogenic subpopulation within BMSCs. By studying the specific features of this subpopulation, researchers may identify more specific molecules or pathways that endow BMSCs with high adipogenic ability and discover more potential targets for suppressing bone marrow adipogenesis.
Nevertheless, previous studies on BMAPs isolation are limited. Even previous studies have demonstrated the existence of BMSC subpopulations that can only differentiate into adipocytes, but their specific markers and whether they possess high adipogenic potential are still unclear [18,19]. Moreover, current strategies to isolate BMSC subpopulations generally required seeding primary BMSCs in low density and subsequent screening of the differentiation abilities from different colonies [13], which may be time consuming and difficult to repeat. Hence, there is a need of an alternative technique that can be used to efficiently and reproducibly isolate specific BMAPs subpopulation.
In this study, we attempted to utilize a special silica microbeads incubation method to isolate the BMAPs subpopulation from mixed BMSCs. This isolation method is based on our previous finding that different subpopulations of BMSCs might possess a different endocytosis ability when cultured in low serum medium [20]. When inert silica microbeads were added, different BMSC subpopulations may engulf different amounts of silica microbeads and, thus, could be isolated based on different cellular densities. The aim of the following study is to evaluate the effectiveness of applying this silica microbeads incubation method to BMAPs isolation.
Materials and Methods
Unless specifically indicated, all chemicals used in the experiments were purchased from Sigma Aldrich.
Animals
Eight-week-old C57BL/6J mice were selected as the donor of BMSCs. Mice were purchased from the NUS laboratory animal center and housed in the animal holding unit of the National University of Singapore. All experiments involving animals complied with the protocol approved by the institutional animal care and use committee (IACUC) of the National University of Singapore.
BMAPs isolation and cell culture
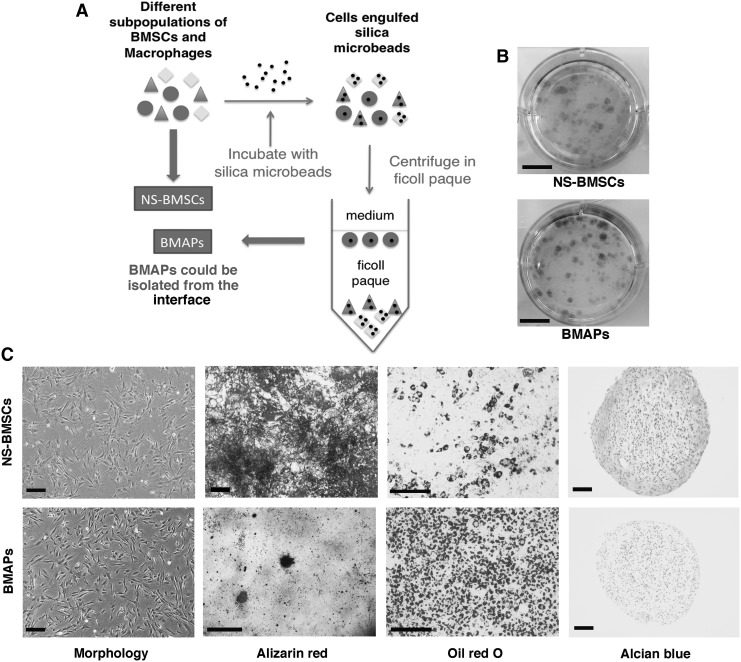
Total BMSCs were obtained by flushing the bone marrow cavities in femur and tibia. BMSCs from one donor (n=4) were then transferred to one T25 flask and cultured in complete MSC medium (Mesencult mouse MSC expansion kit; Stemcell Technologies) at 37°C, 5% CO2, and in a 95% humidity incubator. Nonadherent cells were then removed; adherent cells were cultured until confluence after 3–5 days; and confluent cells were then dethatched by 0.05% Trypsin/EDTA (Life technologies) for 2 min and passaged at a spill ratio of 1:2. For BMSCs without selection [nonselected BMSCs (NS-BMSCs)], attached cells were continually passaged until passage 3 (P3) for hematopoetic cell depletion. For isolation of BMAPs (n=4, repeated in BMSCs from 4 mice), 1×106/mL of BMSCs at P2 were suspended in RPMI-1640 medium with 5% fetal bovine serum (FBS; HyClone). About 100 μg/mL silica microbeads (Bangs Laboratories, Inc.; SS04N, 1–2.49 μm) were then added into the medium. Cells were incubated with the microbeads for 4.5 h at 37°C with manual resuspension every 20 min. After incubation, cells were centrifuged at 300 g for 3 min and then resuspended in 4 mL of phosphate buffer solution (PBS; 1st Bast). For isolation, 4 mL cells in PBS were slowly layered on 3 mL ficoll-paque premium (1.073 g/mL) (Stemcell Technologies) and then centrifuged at 300 g for 30 min. BMAPs could finally be isolated from the interlayer after centrifugation (Fig. 1A). Isolated cells were seeded at 1×104 cell/cm2 in complete MSC medium for expansion. At P3, the left hematopoietic cells in both BMAPs and NS-BMSC were further depleted by Lineage cell depletion kit (Miltenyi Biotec). The negative selected cells were then seeded at a density of 5,000 cells/cm2 in complete MSC medium and kept passaging with the same seeding density.
FIG. 1.
Isolation and characterization of BMAPs. (A) Schematic illustration for isolating BMAPs from heterogeneous BMSCs by the silica microbeads selection method. (B) Colony-forming units assay for NS-BMSCs (top) and BMAPs (bottom) at P6, cells were seeded at 50 cells/cm2 and stained by crystal violet after 10 days. (C) Morphologies and tri-lineages differentiation abilities of NS-BMSCs (top) and BMAPs (bottom) at P6 (From left to right): morphologies before differentiation, alizarin red staining after 3 weeks' osteogenic differentiation, oil red O staining after 7 days' adipogenic differentiation, and alcian blue staining after 28 days' chondrogenic differentiation. Scale bars: (B)=1 cm; (C)=100 μm, except the scale bar dizarin red staining on BMAPs equals 1 mm. BMAPs, bone marrow adipocyte progenitors; BMSCs, bone marrow stromal cells; NS-BMSCs, nonselected BMSCs.
Colony-forming unit assay
After hematopoietic depletion, NS-BMSCs and BMAPs at P6 were seeded in six-well plates at the density of 50 cells/cm2 in triplicate. Cells were cultured for 10 days in complete MSC medium and then stained with 0.25% crystal violet for 10 min.
Osteogenic differentiation
Osteogenic differentiation was induced by adding 50 μg/mL 2-phosphate ascorbic acid, 10−8 M dexamethasone, and 10 mM β-glycerophosphate in complete MSC medium when cells nearly reached confluence. Cells were differentiated for 3 weeks; differentiated cells were stained with 2% alizarin red for 1 min. For quantification, alizarin red stained dye was extracted by adding 10% Cetylpyridinium chloride in PBS (13). One milliliter Cetylpyridinium chloride was added into one well of a 12-well plate, and the reaction was allowed at 37°C for 15 min. One hundred microliter of the reacted solution was then transferred to 1 well of 96-well plates, and absorbance was detected at 560 nm with a fluorescence microplate reader (Tecan Infinite M200; Tecan Group Ltd., AG). Tests were performed in triplicate.
Adipogenic differentiation and quantification
Adipogenic differentiation was induced by supplementing 60 μM indomethacin, 0.5 mM 3-Isobutyl-1-methylxanthine, 5 μg/mL insulin, 1×10−6 M dexamethasone, and 1 μM Rosiglitazone (Cayman Chemical) in complete MSC medium when cells nearly reached confluence. Cells were differentiated for 4–7 days with the medium changed every 3 days. For lipid content detection, differentiated cells were stained by 0.36% Oil Red O in 60% isopropanol for 1 h. For quantification, 1 mL isopropanol was added to 1 well of 12-well plates to extract the staining dye. Extraction absorbance was detected at 550 nm with a fluorescence microplate reader. Experiments were performed in triplicate. For free fatty acids (FFAs)-induced adipogenesis, cells were incubated with 1 μg/mL of green fluorescent FFA BODIPY® FL C12 (Life Technologies) in complete MSC medium for 4 days. After incubation, cells were washed twice with culture medium and observed under a fluorescence microscope (Olympus IX70).
Chondrogenic differentiation
Chondrogenic differentiation was induced by supplementing MSC complete culture medium with 1% insulin-transferrin-selenium (ITS) Premix (BD Biosciences), 10 ng/mL transforming growth factor α (TGF-α; R&D Systems), 50 μg/mL ascorbate-2-phosphate, 10−7 M dexamethasone, 40 μg/mL L-proline, 1% sodium pyruvate, 1% nonessential amino acids (Life Technologies), 1% Glutamax (Life Technologies), and 1% PS at 2.5×105 cells/tube in a 3D pellet culture for 28 days. Differentiation was detected by alcian blue staining on paraffin-embedded slides.
Total RNA extraction, conventional and real-time PCR
Total RNA was extracted by Qiagen RNeasy Mini Kit (Qiagen). cDNA was then synthesized by iScript™ cDNA Synthesis Kit (Bio-Rad). For conventional PCR, newly synthesized cDNA was mixed with PCR Master Mix (2×) (Fermentas) and primers for PCR reactions. For real-time PCR, 2×SYBR® Green PCR Master Mix (Applied Biosystem) was mixed with cDNA templates and primers for real-time PCR analysis in Stepone plus real-time PCR system (Applied Biosystem). Complete reactions conditions of PCR were as follows: Holding stage: 95°C for 20 s; Cycling stage (40 cycles): Step 1, 95°C for 3 s; and Step 2, 60°C for 30 s. Nonreverse transcript controls were used as negative controls for each batch of experiment to confirm the validity of gene expression. Relative gene expression was calculated by ΔΔCt method and normalized to endogenous Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) expression level. qPRC experiments were performed in triplicate. Sequences and sources of the primers used in conventional or real-time PCR are listed in Supplementary Table S1 (Supplementary Data are available online at www.liebertpub.com/scd).
Immunostaining
For immunostaining, cells were first fixed with 4% paraformaldehyde for 20 min and then permeabilized by 0.2% Triton X-100 in PBS for 10 min. Samples were blocked with 2% bovine serum albumin (BSA) in PBS for 1 h. After blocking, primary antibodies (Anti perilipin A; Abcam) were added in 1:200 dilution (5 μg/mL) and incubated at 4°C overnight with rotation. Labeled cells were then washed twice with 0.05% tween20 in PBS and subsequently incubated with secondary antibodies (1:200 dilution, Alexa Fluor® 594 Goat Anti-Rabbit IgG; Life Technologies) in PBS containing 2% BSA for 2 h. Finally, samples were washed thrice with 0.05% tween 20 in PBS. For visualization of nucleus, cells were counter stained with 300 nM DAPI (Life Technologies).
Flow cytometry analysis and cell sorting
For flow cytometry analysis, cells were first detached with 0.05% trysin-EDTA (Life Technologies) and subsequently washed twice with PBS containing 2% FBS. Fluorochrome-conjugated antibodies were then mixed with the cells and incubated at 4°C for 30 min. Labeled cells were washed and then filtered to generate single-cell suspensions. Surface markers expression was analyzed by Cyan flow cytometer (Beckman Coulter). For cell sorting, experiments were performed under sterile conditions. After staining and washing, cells were resuspended in complete MSC medium and then sorted with MoFlo™ XDP cell sorter (Beckman Coulter). Sorted cells were collected with 100% FBS and then seeded at a density of 20,000 cells/cm2. Antibodies used in flow cytometry analysis and cell sorting are listed in Supplementary Table S2.
Statistical analysis
Quantitative results were presented as mean with an error bar (±standard deviation from experiments in triplicate). If quantitative data followed normal distribution, comparisons of data were analyzed by one-way analysis of variance, followed by Tukey's post hoc test for multiple comparisons. Non-normal distributed data were analyzed by Mann–Whitney U test. P<0.05 was considered significant difference. For determining correlation, the Pearson correlation coefficient (R) was calculated for two sets of data, and |R|>0.9 was considered tight correlation. All statistical analyses were performed in IBM SPSS Statistics 20.0.
Results
Colonies-forming ability and tri-lineages differentiation
BMAPs subpopulation could be effectively isolated by silica microbeads incubation method as indicated in (Fig. 1A). In colonies-forming assay, both BMAPs and NS-BMSCs were able to form colonies when seeded in low densities (Fig. 1B). When cultured on plastic plates, the selected BMAPs displayed a homogeneous fibroblast-like morphology, while the NS-BMSCs displayed more heterogeneous morphologies (Fig. 1C). When subjected to tri-lineages differentiation, NS-BMSCs were able to differentiate into osteoblasts, adipocytes, and chondrocytes, which were respectively confirmed by alizarin red, oil red O, and alcian blue staining (Fig. 1C, top). However, different from NS-BMSCs, BMAPs exhibited particularly high adipogenic ability in vitro, but could only weakly differentiate into osteoblasts and nearly could not differentiate into chondrocytes (Fig. 1C, bottom). These findings indicated that the selected BMAPs subpopulation possess distinct differentiation preferences from the NS-BMSCs.
Comparison of surface markers expression
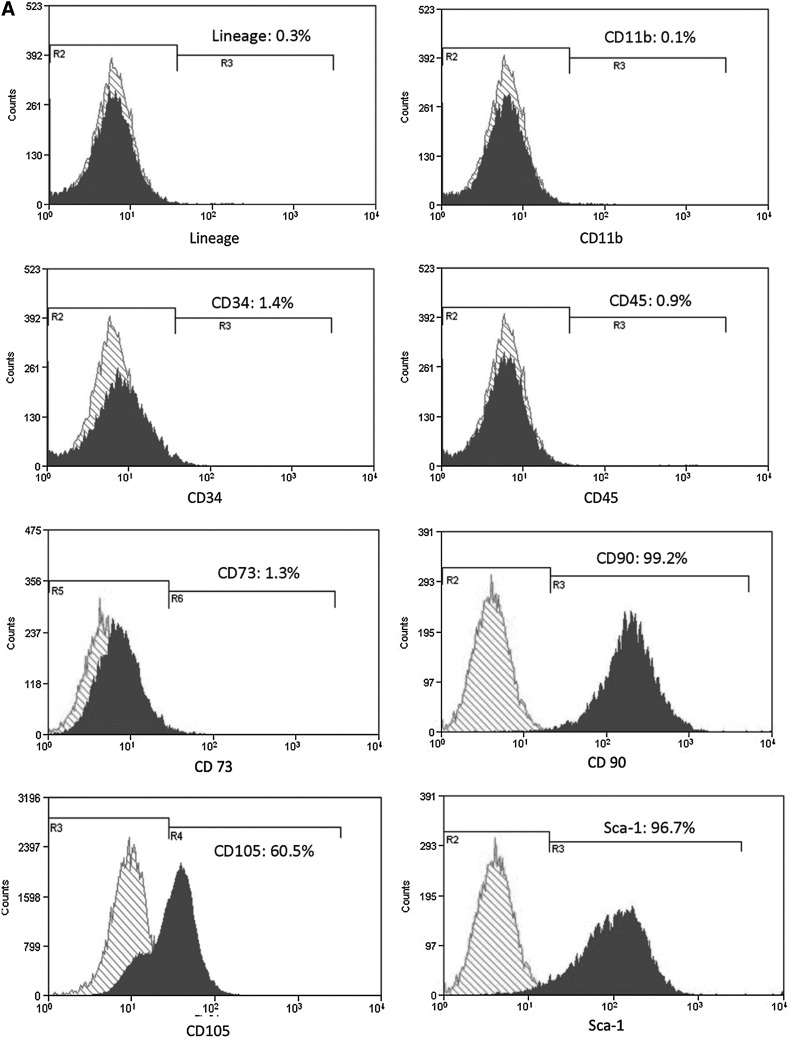
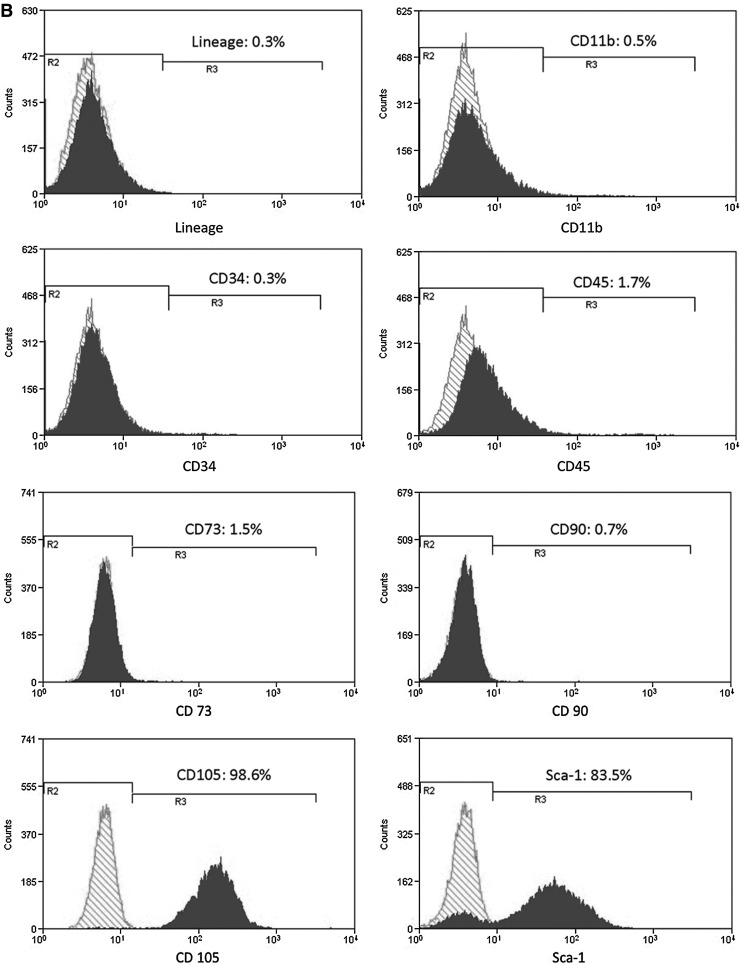
In flow cytometry analyses, NS-BMSCs highly expressed several BMSCs related markers, such as Sca-1 (96.7%), CD90 (99.2%), and CD105 (60.5%), but were nearly negative for CD73 (1.3%) (Fig. 2A); on the other hand, BMAPs only highly expressed CD105 (98.6%) and Sca-1 (83.5%) but were almost negative for CD73 (1.3%) and CD90 (0.7%) expression (Fig. 2B). After negative selection to remove hematopoietic cells, both NS-BMSCs and BMAPs were mostly negative (>98%) for the hematopoietic cell-related markers, such as CD11b (for macrophages), CD31, and CD45 (Fig. 2).
FIG. 2.
Surface markers expression. (A, B) Flow cytometry analyses of surface markers expression on P6 NS-BMSCs (A) and BMAPs (B). Surface markers related to hematopoietic cells (lineage markers, CD34, and CD45), macrophages (CD11b), and BMSCs (CD73, CD90, CD105, and Sca-1) were, respectively, analyzed.
Rapid and strong adipogenic differentiation of BMAPs
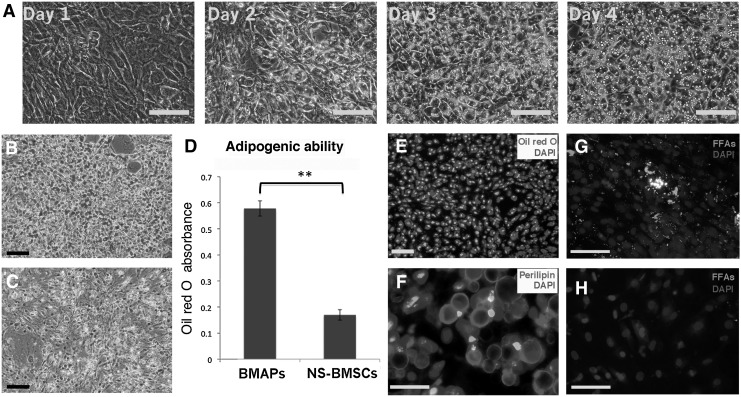
To further investigate the high adipogenic potential of BMAPs, both BMAPs and NS-BMSCs were induced for adipogenic differentiation for 4 days. Interestingly, BMAPs exhibited particularly high sensitivity to adipogenic stimulation and could rapidly differentiate into large lipid-droplets-laden adipocytes within 4 days (Fig. 3A). Oil red O-stained cells under phase contrast (Fig. 3B) and fluorescent view (Fig. 3E) indicated that the differentiation occurred homogeneously in the whole cell population. On the contrary, only a small portion of NS-BMSCs could be induced to lipid-droplets-laden adipocytes after 4 days (Fig. 3C). Absorbance comparison of the extracted oil red O from both cells further confirmed that BMAPs accumulated a significantly higher amount of lipids than NS-BMSCs after adipogenic differentiation (Fig. 3D, P<0.01). Moreover, the differentiated BMAPs were also positive for perilipin staining, which further confirm the maturity of the differentiated cells. To further assess the sensitivity to FFAs stimulation, both types of cells were cultured along with fluorescent FFAs without supplementing differentiation cocktail. After 4 days' incubation, BMAPs but not NS-BMSCs displayed significant lipid accumulation as indicated by the presence of numerous lipid droplets containing green fluorescent FFAs (Fig. 3G, H).
FIG. 3.
Potent adipogenic differentiation potential of BMAPs. (A) Morphology changes of BMAPs (P8) under adipogenic differentiation from day 1 to day 4; images indicated the rapid and homogenous differentiation of BMAPs. (B, C) Oil red O staining of P8 BMAPs (B) and NS-BMSCs (C) after 4 days' adipogenic differentiation. (D) Absorbance comparison of extracted oil red O stains from the 4 days' differentiated BMAPs and NS-BMSCs. (E) Fluorescent view of the 4 days' differentiated BMAP, oil red O emitted red fluorescence under 500 nm fluorescent excitation. (F) Immunostaining of Perilipin on the 4 days' differentiated BMAP. (G, H) incubation of BMAPs (G) and NS-BMSCs (H) with green fluorescent FFAs for 4 days, auto-differentiated cells absorbed green fluorescent FFAs. Nuclei were counter-stained with DAPI. **P<0.01. Scale bars: (A, B, E, G, H)=100 μm; (F)=25 μm. FFAs, free fatty acids.
Gene expression analyses
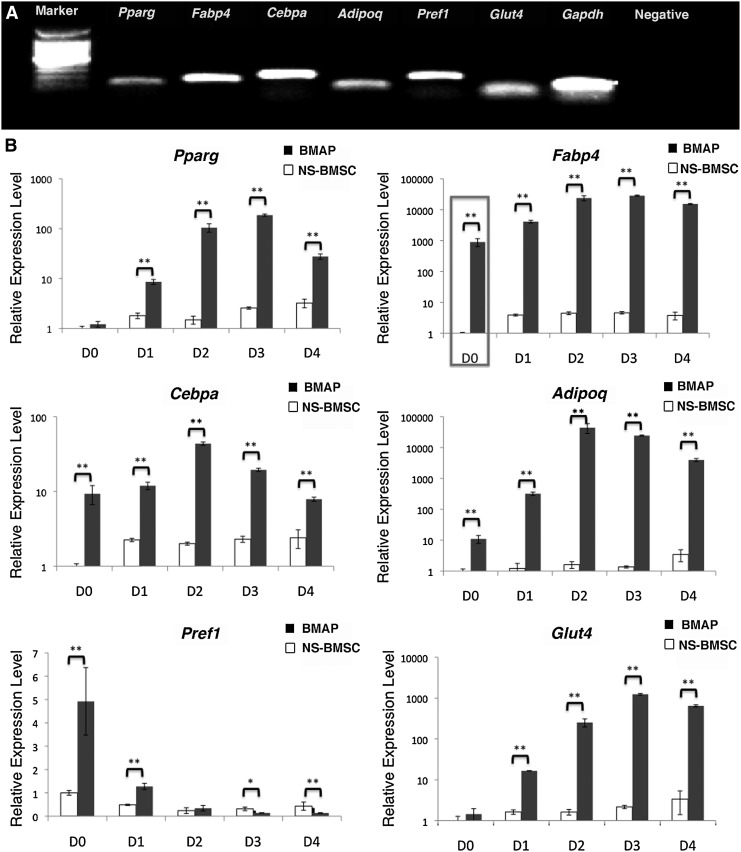
To further confirm the adipogenic differentiation of BMAPs, 4 days' differentiated BMAPs and NS-BMSCs were assessed by genes expression analyses. On day 4, differentiated BMAPs had already highly expressed all tested adipogenic genes as indicated by conventional PCR (Fig. 4A). When compared with the NS-BMSCs in real-time PCR, BMAPs expressed a significantly higher level of all tested adipogenic genes during 4 days' differentiation (Fig. 4B, P<0.01). Moreover, before differentiation (D0), BMAPs had already expressed a significantly higher level of adipogenic genes such as fatty acid-binding protein 4 (Fabp4), CCAAT/enhancer-binding protein α (Cebpa), adiponectin, and preadipocye factor 1 (Pref1) than NS-BMSCs (Fig. 4B, P<0.01). Notably, a particularly high basal expression level of Fabp4 could be observed in the undifferentiated BMAPs (>1,000 times higher than that of NS-BMSCs at D0) (Fig. 4B).
FIG. 4.
Gene expression of the adipogenic differentiated BMAPs. (A) Adipogenic genes' expression on 4 days' differentiated BAMPs (P8). Adipogenic genes involved PPAR-γ, FABP4, C/EBPα, adiponectin, Pref-1, and Glut-4; GADPH was used as the positive control. (B) Real-time PCR analysis of adipogenic genes in P8 BMAPs (black) and NS-BMSCs (white) before and during 4 days' adipogenic differentiation (day0–day4). Gene expression levels were normalized to the expression of GADPH. Box in (B) indicated the significant difference of basal FABP4 gene expression levels. **P<0.01. Pparg, peroxisome proliferator-activated γ; Fabp4, fatty acid-binding protein 4; Cebpa, CCAAT enhancer-binding protein alpha; Adipog, adiponetin; Pref1, preadipocyte factor 1; Glut4, glucose transporter 4; Gapdh, glyceraldehyde 3-phosphate dehydrogenase.
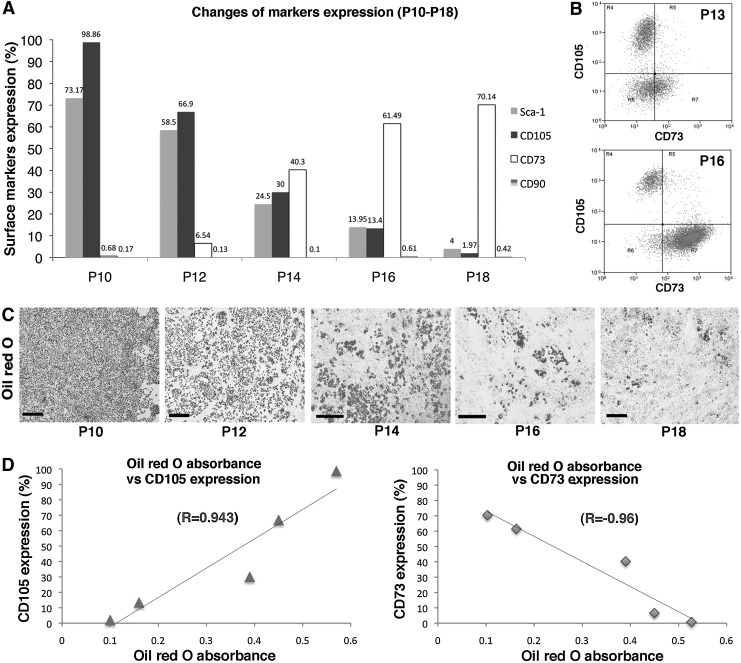
Correlation between CD105 expression and adipogenic potential of BMAPs
At P10, most BMAPs still maintained the “Sca-1+CD90−CD73−CD105+” surface markers expression and high adipogenic differentiation ability (Fig. 5A,C). However, after P10, surface marker expression of BMAPs gradually switched to “Sca-1−CD90−CD73+CD105−,” co-staining of CD105 and CD73 on P13 and P16 BMAPs revealed that CD105+CD73− cells gradually changed to CD105−CD73+ cells during the conversion (Fig. 5B). Accompanied by changes in marker expression, the adipogenic abilities of BMAPs also started to decrease (Fig. 5C). To further determine the correlations among CD105, CD73, and the adipogenic potential of BMAPs, we tested the absorbance of extracted oil red O stains from differentiated cells during P10–P18 (every two passages) and calculated their correlations with the surface markers expression at that passage. Results showed that the CD105's expression was positively correlated to the absorbance of extracted oil red O (R=0.943), while the expression of CD73 expression displayed inverse correlation to the absorbance of oil red O (R=−0.96) (Fig. 5D).
FIG. 5.
Changes in surface markers expression and adipogenic differentiation potential in long-term passaged BMAPs: (A) Alternation of surface markers expression of BMAPs from P10 to P18. (B) Expression of CD105 and CD73 on P13 (top) and P16 (bottom) BMAPs. (C) Oil red O staining for BMAPs at different passages (P10–P18) after 4 days' adipogenic differentiation. (D) Correlations between surface markers expression (left, CD105; right, CD73) and average absorbance of extracted oil red O from differentiated BMAPs (P10–P18). Data used for calculating correlations were averages from three experiments. Scale bars: (C)=500 μm.
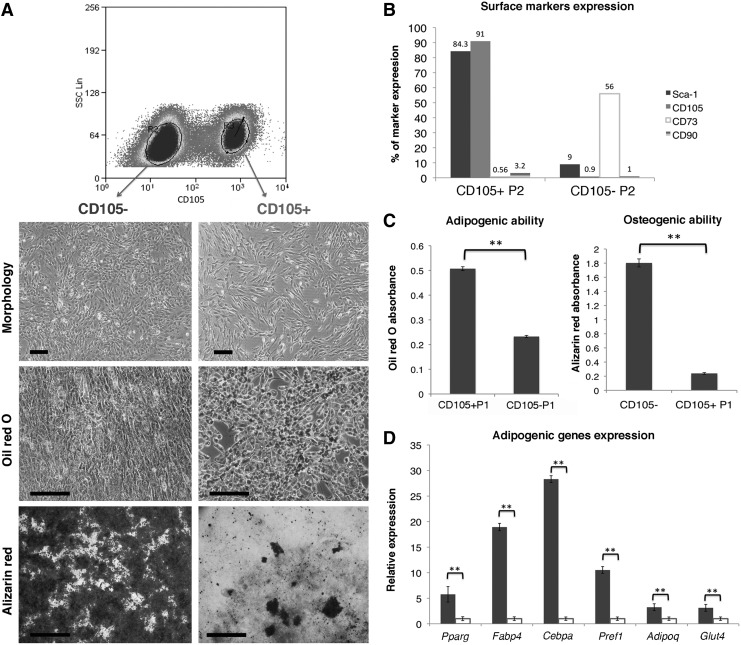
Re-enrichment of BMAPs by sorting CD105+ cells
Based on the tight correlation between CD105 and the adipogenic potential of converting BMAPs, we further tested whether sorting the CD105+ cells could repopulate the BMAPs subpopulation (Fig. 6A). Importantly, the CD105+ sorted cells displayed high adipogenic ability but low osteogenic ability in vitro (Fig. 6A, C). Moreover, their surface markers expression was also “Sca-1+CD90−CD73−CD105+” (Fig. 6B). On the contrary, the sorted CD105− cells displayed high osteogenic but low adipogenic abilities (Fig. 6A, C) in vitro and expressed surface markers similar to BMAPs in later passages (Fig. 6B). In addition, real-time PCR analysis for CD105+ and CD105− subpopulations further confirmed that CD105+ cells expressed a significantly higher basal level of adipogenic genes than the CD105− cells, (Fig. 6D).
FIG. 6.
Re-enrichemnt of BMAPs by sorting CD105+ cells. (A) Sorting of CD105+ and CD105− cells from P14 BMAPs. From top to bottom, morphology, oil red O staining (4 days' differentiaton), and alizarin red staining (3 weeks' differentiation) of the CD105− (left) and CD105+ sorted cells. (B) Surface markers expression of CD105+ sorted cells and CD105− sorted cells at P2 after sorting. (C) Absorbance comparison of the extracted oil red O (left) and alizarin red (right) stains from adipogenic or osteogenic differentiated CD105+ and CD105− sorted cells. (D) Comparison of basal adipogenic genes expression between sorted CD105+ (black bars) and CD105− (white bars) cells. **P<0.01. Scale bars: (A)=200 μm for morphologies and oil red O staining,=1.6 mm for alizarin red staining.
Discussion
In the current study, we applied a special isolation method that involved silica microbeads incubation to isolate BMAPs. Different from previous methods [13,15,18,21], this method did not require colonies selection or specific markers for cell sorting; rather, it utilized the different particle-engulfing abilities of different BMSC subpopulations for cell separation. At first, this method was designed to remove the abundant contaminating macrophages in the primary mouse BMSCs due to their high particle-engulfing abilities. However, unexpectedly, we observed that subpopulation of BMSCs could also be effectively isolated after incubation with silica microbeads. These findings suggested that different subpopulation of BMSCs might possess distinct particle-engulfing ability, and the subpopulations which engulfed more particles would be settled down earlier during centrifugation in ficoll paque due to the increased cellular density. According to this hypothesis, the BMAPs might be the subpopulation that possessed low-particle-engulfing ability, as they were left in the interface after centrifugation. However, further studies are still required to confirm the exact separation mechanisms of this selection method.
Through selection with silica microbeads, a homogenous “Sca-1+CD73−CD90−CD105+” BMSC subpopulation could be effectively isolated. The most important characteristic of this subpopulation was when they were maintained in good condition (cells homogeneously expressed “Sca-1+CD73−CD90−CD105+” surface markers), the whole population was able to homogenously differentiate to mature adipocytes within 4 days in vitro, which was even shorter than the differentiation time line of preadipocyte cell line 3T3-L1 [22]. Concurrently, in contrast to the high adipogenic ability, these cells only displayed low osteogenic and chondrogenic abilities in vitro. Based on their high propensity for adipogenic differentiation, the selected cells could potentially be regarded as the adipocyte progenitors subpopulation within BMSCs.
To further confirm their adipocyte progenitor identities, gene expression analyses were performed, which discovered that the potent adipogenic ability of BMAPs was probably due to their high basal expression levels of adipogenic genes, such as Cebpa, Fabp4, and adiponectin, which, respectively, encode the transcription factor, ligand transporter, and adipokine that stimulate adipogenesis [23,24]. Therefore, the higher basal expression of these genes suggested that BMAPs were much more committed to adipogenic differentiation than NS-BMSCs. Notably, BMAPs had a particularly higher basal expression of Fabp4 than NS-BMSCs, which implied that there existed larger pools of fatty acid-binding proteins inside BMAPs, which allowed them to efficiently transport lipid ligands such as FFAs to the nucleus for inducing adipogenesis [25,26]. Consistent with this hypothesis, BMAPs, but not NS-BMSCs, exhibited significant lipid accumulation when incubated with FFAs alone. This FFA-induced adipogenesis has previously been observed in preadipocytes cell lines [27], and its presence in BMAPs further confirmed the preadipocyte-like characteristic of this subpopulation.
In addition, similar to other primary BMSCs [15], long-term cultured BMAPs would gradually lose high adipogenic differentiation ability. During this change, the overall trend was the conversion from highly adipogenic CD105+CD73− BMAPs to the lower adipogenic CD105−CD73+ cells. Interestingly, both CD105 (positive correlation) and CD73 (negative correlation) expressions exhibited tight correlations with the adpogenic abilities of the converting cells. Based on these findings, the highly adipogenic subpopulation could be successfully repopulated by sorting the CD105+ progenies from later passages. These selected CD105+ cells possessed similar adipogenic ability and surface marker expression as BMAPs in early passages, which suggested that they were the unchanged BMAPs subpopulation within the long-term passaged “BMAPs” that were composed of different altered subpopulations. Selection based on this specific marker not only offered a convenient approach to re-enrich sufficient number of BMAPs for various applications but also enabled effective elimination of the altered low adipogenic cells for maintaining the purity of BMAPs subpopulation. However, due to the broad expression of CD105 on bone marrow hematopoietic and stromal cells, cell sorting based on CD105 alone could hardly isolate a pure BMAPs subpopulation from heterogeneous BMSCs. Therefore, isolation of a pure BMAPs subpopulation may still require silica microbeads selection or sorting with a combination of different markers.
Based on the results just cited, the silica microbeads selection method had exhibited high effectiveness for selecting specific subpopulations from primary BMSCs. Even this method had only been tested in mouse BMSCs, it could possibly be applied to isolate subpopulations from human primary BMSCs or even other mixed cell populations as long as the particle-engulfing abilities were different among subpopulations. Notably, despite the effectiveness, the long-term influences of this method on BMSCs are still unclear. Even we have observed that incubation with silica microbeads would not significantly alter the surface markers expression and differentiation abilities of BMSCs (Supplementary Fig. S1), and the engulfed silica microbeads would be gradually diluted after cell replication and, finally, disappear; more detailed studies are required to investigate the long-term influences of this method on the selected stem cells in future studies.
To summarize, in this study, we developed an efficient and repeatable approach based on silica microbeads incubation for isolating BMAPs subpopulation from heterogeneous BMSCs. To our knowledge, this is the first reported specific method for BMAPs isolation. With the convenient isolation method, BMAPs could potentially be developed as a more specific cellular model for various studies related to bone marrow adipogenesis.
Supplementary Material
Acknowledgments
This work was partially supported by grants from Academic Research Fund, Singapore Ministry of Education: R221000023112 and research grant, National University Health System: R221000053515.
Author Disclosure Statement
All authors indicate no potential conflicts of interest.
References
- 1.Rosen CJ. Ackert-Bicknell C. Rodriguez JP. Pino AM. Marrow fat and the bone microenvironment: developmental, functional, and pathological implications. Crit Rev Eukaryot Gene Expr. 2009;19:109–124. doi: 10.1615/critreveukargeneexpr.v19.i2.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duque G. Bone and fat connection in aging bone. Curr Opin Rheumatol. 2008;20:429–434. doi: 10.1097/BOR.0b013e3283025e9c. [DOI] [PubMed] [Google Scholar]
- 3.Chan GK. Duque G. Age-related bone loss: old bone, new facts. Gerontology. 2002;48:62–71. doi: 10.1159/000048929. [DOI] [PubMed] [Google Scholar]
- 4.Kawai M. Devlin MJ. Rosen CJ. Fat targets for skeletal health. Nat Rev Rheumatol. 2009;5:365–372. doi: 10.1038/nrrheum.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grey A. Bolland M. Gamble G. Wattie D. Horne A. Davidson J. Reid IR. The peroxisome proliferator-activated receptor-gamma agonist rosiglitazone decreases bone formation and bone mineral density in healthy postmenopausal women: a randomized, controlled trial. J Clin Endocrinol Metab. 2007;92:1305–1310. doi: 10.1210/jc.2006-2646. [DOI] [PubMed] [Google Scholar]
- 6.Halade GV. Rahman MM. Williams PJ. Fernandes G. High fat diet-induced animal model of age-associated obesity and osteoporosis. J Nutr Biochem. 2010;21:1162–1169. doi: 10.1016/j.jnutbio.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clabaut A. Delplace S. Chauveau C. Hardouin P. Broux O. Human osteoblasts derived from mesenchymal stem cells express adipogenic markers upon coculture with bone marrow adipocytes. Differentiation. 2010;80:40–45. doi: 10.1016/j.diff.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Goto H. Hozumi A. Osaki M. Fukushima T. Sakamoto K. Yonekura A. Tomita M. Furukawa K. Shindo H. Baba H. Primary human bone marrow adipocytes support TNF-alpha-induced osteoclast differentiation and function through RANKL expression. Cytokine. 2011;56:662–668. doi: 10.1016/j.cyto.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 9.Hozumi A. Osaki M. Goto H. Sakamoto K. Inokuchi S. Shindo H. Bone marrow adipocytes support dexamethasone-induced osteoclast differentiation. Biochem Biophys Res Commun. 2009;382:780–784. doi: 10.1016/j.bbrc.2009.03.111. [DOI] [PubMed] [Google Scholar]
- 10.Maurin AC. Chavassieux PM. Frappart L. Delmas PD. Serre CM. Meunier PJ. Influence of mature adipocytes on osteoblast proliferation in human primary cocultures. Bone. 2000;26:485–489. doi: 10.1016/S8756-3282(00)00252-0. [DOI] [PubMed] [Google Scholar]
- 11.Takada I. Kouzmenko AP. Kato S. Wnt and PPARgamma signaling in osteoblastogenesis and adipogenesis. Nat Rev Rheumatol. 2009;5:442–447. doi: 10.1038/nrrheum.2009.137. [DOI] [PubMed] [Google Scholar]
- 12.Beresford JN. Bennett JH. Devlin C. Leboy PS. Owen ME. Evidence for an inverse relationship between the differentiation of adipocytic and osteogenic cells in rat marrow stromal cell cultures. J Cell Sci. 1992;102(Pt 2):341–351. doi: 10.1242/jcs.102.2.341. [DOI] [PubMed] [Google Scholar]
- 13.Russell KC. Phinney DG. Lacey MR. Barrilleaux BL. Meyertholen KE. O'Connor KC. In vitro high-capacity assay to quantify the clonal heterogeneity in trilineage potential of mesenchymal stem cells reveals a complex hierarchy of lineage commitment. Stem Cells. 2010;28:788–798. doi: 10.1002/stem.312. [DOI] [PubMed] [Google Scholar]
- 14.Pevsner-Fischer M. Levin S. Zipori D. The origins of mesenchymal stromal cell heterogeneity. Stem Cell Rev. 2011;7:560–568. doi: 10.1007/s12015-011-9229-7. [DOI] [PubMed] [Google Scholar]
- 15.Muraglia A. Cancedda R. Quarto R. Clonal mesenchymal progenitors from human bone marrow differentiate in vitro according to a hierarchical model. J Cell Sci. 2000;113(Pt 7):1161–1166. doi: 10.1242/jcs.113.7.1161. [DOI] [PubMed] [Google Scholar]
- 16.Poulos SP. Dodson MV. Hausman GJ. Cell line models for differentiation: preadipocytes and adipocytes. Exp Biol Med (Maywood) 2010;235:1185–1193. doi: 10.1258/ebm.2010.010063. [DOI] [PubMed] [Google Scholar]
- 17.Obinata M. The immortalized cell lines with differentiation potentials: their establishment and possible application. Cancer Sci. 2007;98:275–283. doi: 10.1111/j.1349-7006.2007.00399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Post S. Abdallah BM. Bentzon JF. Kassem M. Demonstration of the presence of independent pre-osteoblastic and pre-adipocytic cell populations in bone marrow-derived mesenchymal stem cells. Bone. 2008;43:32–39. doi: 10.1016/j.bone.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 19.Abdallah BM. Kassem M. New factors controlling the balance between osteoblastogenesis and adipogenesis. Bone. 2012;50:540–545. doi: 10.1016/j.bone.2011.06.030. [DOI] [PubMed] [Google Scholar]
- 20.Liu H. Toh WS. Lu K. MacAry PA. Kemeny DM. Cao T. A subpopulation of mesenchymal stromal cells with high osteogenic potential. J Cell Mol Med. 2009;13:2436–2447. doi: 10.1111/j.1582-4934.2009.00793.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bakondi B. Shimada IS. Perry A. Munoz JR. Ylostalo J. Howard AB. Gregory CA. Spees JL. CD133 identifies a human bone marrow stem/progenitor cell sub-population with a repertoire of secreted factors that protect against stroke. Mol Ther. 2009;17:1938–1947. doi: 10.1038/mt.2009.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zebisch K. Voigt V. Wabitsch M. Brandsch M. Protocol for effective differentiation of 3T3-L1 cells to adipocytes. Anal Biochem. 2012;425:88–90. doi: 10.1016/j.ab.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 23.Rosen ED. Spiegelman BM. Molecular regulation of adipogenesis. Ann Rev Cell Dev Biol. 2000;16:145–171. doi: 10.1146/annurev.cellbio.16.1.145. [DOI] [PubMed] [Google Scholar]
- 24.Rosen ED. MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol. 2006;7:885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- 25.Maeda K. Cao H. Kono K. Gorgun CZ. Furuhashi M. Uysal KT. Cao Q. Atsumi G. Malone H, et al. Adipocyte/macrophage fatty acid binding proteins control integrated metabolic responses in obesity and diabetes. Cell Metab. 2005;1:107–119. doi: 10.1016/j.cmet.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 26.Tan NS. Shaw NS. Vinckenbosch N. Liu P. Yasmin R. Desvergne B. Wahli W. Noy N. Selective cooperation between fatty acid binding proteins and peroxisome proliferator-activated receptors in regulating transcription. Mol Cell Biol. 2002;22:5114–5127. doi: 10.1128/MCB.22.14.5114-5127.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang JY. Della-Fera MA. Rayalam S. Park HJ. Ambati S. Hausman DB. Hartzell DL. Baile CA. Regulation of adipogenesis by medium-chain fatty acids in the absence of hormonal cocktail. J Nutr Biochem. 2009;20:537–543. doi: 10.1016/j.jnutbio.2008.05.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.