Abstract
Induced pluripotent stem (iPS) cells are attractive for cell replacement therapy, because they overcome ethical and immune rejection issues that are associated with embryonic stem cells. iPS cells have been derived from autonomous fibroblasts at low efficiency using multiple ectopic transcription factors. Recent evidence suggests that the epigenome of donor cell sources plays an important role in the reprogramming and differentiation characteristics of iPS cells. Thus, identification of somatic cell types that are easily accessible and are more amenable for cellular reprogramming is critical for regenerative medicine applications. Here, we identify ciliary body epithelial cells (CECs) as a new cell type for iPS cell generation that has higher reprogramming efficiency compared with fibroblasts. The ciliary body is composed of epithelial cells that are located in the anterior portion of the eye at the level of the lens and is readily surgically accessible. CECs also have a reduced reprogramming requirement, as we demonstrate that ectopic Sox2 and c-Myc are dispensable. Enhanced reprogramming efficiency may be due to increased basal levels of Sox2 in CECs. In addition, we are the first to report a cellular reprogramming haploinsufficiency observed when reprogramming with fewer factors (Oct4 and Klf4) in Sox2 hemizygous cells. Taken together, endogenous Sox2 levels are critical for the enhanced efficiency and reduced exogenous requirement that permit facile cellular reprogramming of CECs.
Introduction
Pluripotent cells offer an exciting opportunity to replenish damaged cells and restore organ function. Somatic cells can be reprogrammed into induced pluripotent stem (iPS) cells, nearly indistinguishable from embryonic stem (ES) cells, and can be differentiated into cells of all three germ layers [1,2]. However, fibroblast reprogramming into iPS cells is inefficient and slow [1]. Current studies indicate adult stem cells reprogram more efficiently than terminally differentiated cells [3,4]. For example, neural stem cells and dermal papilla cells can be reprogrammed with higher efficiency and less exogenous input compared with fibroblasts [5–8]. This enhanced reprogramming efficiency is due, in part, to the higher level of expression of some endogenous reprogramming factors compared with that of fibroblasts [5,7]. Thus, the identification of accessible cell types with enhanced reprogramming capabilities is warranted for facile cellular reprogramming [9].
The mammalian ciliary body is a surgically accessible region of the anterior portion of the eye that is anatomically contiguous with the retinal pigment epithelium. The ciliary body region contains nonpigmented and pigmented epithelial cells. Recent studies have demonstrated that epithelial cells in this region express neural stem cell markers in vitro [10]. However, it is unclear whether these epithelial cells are more susceptible to reprogramming. Here, we report that ciliary body epithelial cells (CECs) are an easily accessible cell type and can be efficiently reprogrammed.
Materials and Methods
All procedures on mice were performed in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research and approved by the Institutional Animal Care and Use Committee at the University of North Carolina at Chapel Hill.
CEC and fibroblasts culture
We used 5–6-week-old Sox2EGFP knock-in [11] or wild-type CD-1 strain mice, and 4F2A reprogrammable transgenic mice [12]. Dissection of adult mouse ciliary body was performed as previously described [10,13,14]. Briefly, eyes were enucleated and placed in artificial cerebral spinal fluid. Eyes were halved, and the cornea, iris, lens, and posterior segment were dissected. The remaining ciliary margin was treated with a mixture of protease, and the ciliary epithelium was scraped away from the sclera as described [10]. The ciliary epithelium was then triturated into single cells with fire-polished pipettes. After centrifugation, the cells were resuspended in CECs growth medium consisting of neurobasal medium, 1% fetal bovine serum (FBS), 1×B27 supplement, 1% penicillin/streptomycin, 2 mM l-glutamine, bFGF (20 ng/mL; Peprotech), EGF (20 ng/mL; Peprotech), and heparin (2 μg/mL; Sigma), and plated in 60-mm gelatin-coated tissue culture dishes in a 37°C CO2 incubator [14,15]. Adult tail tip fibroblasts (TTFs) were prepared as described [16]. All cell culture reagents were from Invitrogen unless otherwise noted.
iPS cells generation
CECs at passage 2 (8 days after isolation) were plated at 1×105 cells/well of gelatin-coated six-well plates in CECs growth medium. The next day, for the four-factor transduction, concentrated lentiviruses containing CMV promoter-driven human Oct4, Sox2, Klf4, and c-Myc (Cellomics Technology) were added to the cells at a multiplicity of infection of 10 with 4 μg/mL polybrene (Sigma) in CECs growth medium. For the three-factor transduction, lentiviruses coding human Oct4, Sox2, and Klf4 were added. For the two-factor transduction, lentiviruses coding human Oct4 and Klf4, or Oct4 and Sox2 were used; whereas for the one-factor transduction, only lentivirus coding human Oct4 was added. Twenty-four hours postinfection, the viral infection mix was exchanged for fresh CECs growth medium. The next day, transduced CECs were subcultured onto mitomycin C-treated SNL feeder cells (a mouse fibroblast STO cell line-derived, neomycin-resistant, leukemia inhibitory factor (LIF)-producing cell line Cell Biolabs) in six-well plates at a split ratio of 1:4 in ES cell medium minus LIF containing DMEM supplemented with 15% FBS, 2 mM l-glutamine, 100 μM nonessential amino acids, 100 μM 2-mercaptoethanol, 50 U/mL penicillin, and 50 μg/mL streptomycin [16].
For reprogramming CECs and TTFs from 4F2A mice, cells were seeded at 1×104 cells/well in fibroblast medium on SNL feeder cells in 12-well plates. The next day, ES cell medium minus LIF containing 2 μg/mL doxycycline was added and exchanged every 2 day. After the ES cell-like colonies appeared at day 12–20, cultures were washed twice with DMEM and then grown in ES cell medium plus LIF without doxycycline for 4–5 days. The colonies were then scored by staining for alkaline phosphatase or Nanog.
The efficiency of generating iPS cells was calculated by dividing the number of GFP-positive or alkaline phosphatase (or Nanog)-positive ES cell-like colonies by the seeded cell number.
ES cell-like (or Sox2-GFP-positive) colonies were picked, dissociated with trypsin, and expanded on SNL feeders in KnockOut DMEM supplemented with 15% KSR, 100 μM MEM nonessential amino acids, 100 μM 2-mercaptoethanol, 2 mM l- glutamine, 50 U penicillin, and 50 μg/mL streptomycin [17]. For RNA and genomic DNA extraction, iPS cells were depleted of feeder cells for at least two passages on gelatin-coated plates in serum-free N2B27 medium supplemented with LIF and 2i inhibitors [18], CHIR99021 (3 μM; Stemgent) and PD0325901 (1 μM; Stemgent).
Viral vector integration analysis
Genomic DNA was prepared with the Trizol reagent (Invitrogen) according to the manufacturer's instructions. Genomic PCR was performed with the PCR 2×reaction mix Immomix Red (Bioline) with the following cycling conditions: 95°C for 10 min followed by 35 cycles of amplification (15 s at 94°C, 30 s at 60°C, and 1 min at 72°C) and 10 min at 72°C at the end. Primers are listed in Supplementary Table S1 (Supplementary Data are available online at www.liebertpub.com/scd).
Karyotyping
Karyotyping was performed by KaryoLogic, Inc. (www.karyologic.com/).
Bisulfite genomic sequencing
Genomic DNA was isolated with Quick-gDNA MiniPrep (Zymo Research) and then treated for bisulfite sequencing with EZ DNA Methylation-Lightning Kit (Zymo Research). Bisulfite PCR primers [1] are listed in Supplementary Table S1. Amplified fragments were cloned using the Zero Blunt TOPO PCR Cloning Kits (Invitrogen). Five to eight clones for each gene were sequenced with the SP6 and T7 primers.
RNA isolation, RT-PCR, and real-time PCR
Total RNAs from cultured CECs (passage 2) and iPS cells were extracted using the Trizol reagent according to the manufacturer's instructions, and quantified with the NanoDrop spectrophotometer (Thermo Scientific). Total RNA from mouse ES-D3 cell line was purchased from Cell Biolabs. Total RNAs were treated with amplification-grade DNase I and then reverse transcribed with Superscript III First-Strand Synthesis System (Invitrogen). PCRs were performed with the PCR 2×reaction mix (Bioline) using the GeneAmp PCR system 2700 (Applied Biosystems) with the following conditions: 10 min at 95°C, 15 s at 94°C, 30 s at 60°C, 30 s at 72°C, 30 cycles of 15 s at 94°C, 30 s at 55°C, 30 s at 72°C, and 10 min at 72°C at the end. Real-time PCR was run on a 7500 Real-Time PCR System (Applied Biosystems) with a SYBR Green qPCR Master Mix (Applied Biosystems). Primers are listed in Supplementary Table S1. The mRNA level of each gene was normalized to the mRNA encoding glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Differences between samples and controls were calculated based on 2−ΔΔCT method.
Immunofluorescence staining and alkaline phosphatase staining
Cells were fixed with 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) for 10 min and treated for 30 min with blocking solution (0.1% Triton X-100, 1% normal goat serum, and 1% bovine serum albumin in PBS). Cells were then incubated with primary antibodies overnight at 4°C. After rinsing thrice with PBS, cells were incubated with secondary antibodies, rinsed, and counter-stained with Hoechst 33342. Primary and secondary antibodies were listed in Supplementary Table S2. iPS cells were also stained with an Alkaline Phosphatase Staining Kit (Stemgent) following the manufacturer's instructions.
In vitro differentiation
iPS cells were dissociated with trypsin, and 5×105 cells were cultured in suspension in 100-mm bacterial culture dishes in ES medium minus LIF. After 3 days of culturing to form embryoid bodies (EBs), cell aggregates were seeded onto gelatin-coated tissue culture plates and cultured for an additional 10 days. The cells were then harvested for total RNA extraction. Spontaneous differentiation was examined by RT-PCR for representative lineage-specific markers with the indicated primers (Supplementary Table S1).
Teratoma formation
iPS cells (1.5×106) were mixed with Matrigel (BD Biosciences) and subcutaneously injected into the dorsal flank of NOD-SCID mice. Teratomas were isolated at 4–6 weeks after injection, fixed in 4% PFA. Half of the harvested tissue was embedded in paraffin, and sectioned (5 μm thick) for staining with hematoxylin and eosin. The other half was embedded in Tissue-Tek OCT compound (Sakyra Finetek), and frozen sections (10 μm thick) were prepared for immunofluorescence staining.
Statistics
n=3 in all real-time PCR experiments. P value was calculated using Student's t-test.
Results and Discussion
Generation of iPS cells from mouse CECs
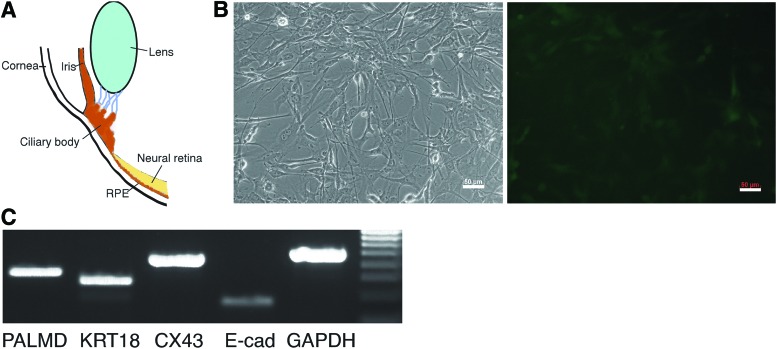
We isolated adult mouse CECs from hemizygote Sox2EGFP knock-in mice, and cultured them as a monolayer in serum-supplemented medium (Fig. 1A, B). Cultured Sox2EGFP CECs (passage 2) displayed a heterogeneous morphology characteristic of ciliary epithelial cells [14] and expressed weak EGFP (Fig. 1B). RT-PCR detected mRNAs of epithelial markers (cytokeratin-18, connexin-43, and E-cadherin) and differentiated ciliary epithelial marker palmdelphin in these cells (Fig. 1C) [13,14].
FIG. 1.
Mouse ciliary body epithelial cell (CEC) culture. (A) Diagram of the anatomical position of the ciliary body. (B) Brightfield (left) and fluorescent (right) images of monolayer of Sox2EGFP CECs. The Sox2EGFP CECs were heterogeneous and some expressed weak EGFP. Scale bar: 50 μm. (C) RT-PCR showed that Sox2EGFP CECs expressed epithelial markers cytokeratin-18, connexin-43, E-cadherin, and palmdelphin. Color images available online at www.liebertpub.com/scd
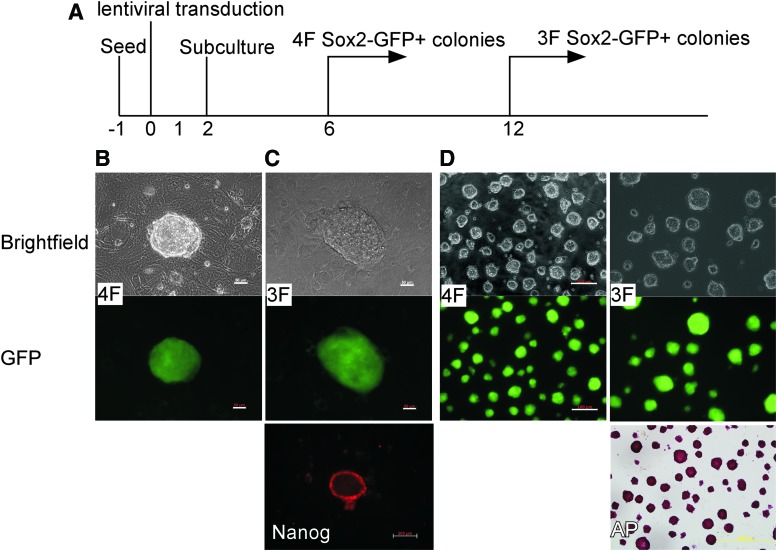
To test whether Sox2EGFP CECs can be reprogrammed into iPS cells, we transduced the Sox2EGFP CECs with lentiviral vectors carrying the human cDNAs encoding four transcription factors: Oct4, Sox2, c-Myc, and Klf4. As detailed in the timeline (Fig. 2A), Sox2EGFP CECs were transduced with either four (4F: Oct4, Sox2, c-Myc, and Klf4) or three (3F: Oct4, Sox2, and Klf4) reprogramming factors. After 4F transduction, several small-cell GFP-positive colonies with a large nuclear to cytoplasmic ratio emerged at day 6 after infection (Fig. 2B). At day 21, more than 100 ES cell-like colonies were observed in each well of transduced CECs, and the nominal reprogramming efficiency was calculated as 0.4%, which is more than seven times higher than with 4F reprogramming of TTFs reported in the literature [1,19]. We then used a doxycycline-inducible transgenic system to confirm this observation. TTFs and CECs were cultured from 6-week-old 4F2A mice carrying two copies of the 4F2A (OSKM) transgene and two copies of rtTA transgene. The cells were then reprogrammed in the presence of doxycycline as described [12]. After doxycycline administration, ES cell-like colonies appeared at day 6 in the CEC culture, and at day 16, in the TTFs. As expected, more AP-positive colonies were observed in the CEC group than in the TTF group (Supplementary Fig. S1). On day 25, the AP-positive (or Nanog-positive) colonies were counted, and we found that the number of colonies in the CEC group was 6 times higher than in the TTF group (46/8, 38/6). Taken together, these data suggest that CECs are more amenable to facile reprogramming into iPS cells.
FIG. 2.
Generation of induced pluripotent stem (iPS) cells from Sox2EGFP CECs with four and three reprogramming factors. (A) Timeline for reprogramming Sox2EGFP CECs into iPS cells. CECs from hemizygote Sox2 mouse were transduced with Oct4, Sox2, Klf4, and c-Myc (4F) or with Oct4, Sox2, and Klf4 (3F). Transduction mix was exchanged for fresh growth medium at day 1. At day 2, transduced CECs were subcultured onto SNL feeder cells (a mouse fibroblast STO cell line-derived, neomycin-resistant, leukemia inhibitory factor (LIF)-producing cell line) in embryonic stem (ES) cell medium minus leukemia inhibitory factor (LIF). (B) Representative 4F Sox2-GFP+ iPS colony. Scale bar, 50 μm. (C) Representative 3F Sox2-GFP+ iPS colony, plus Nanog staining. Scale bar, 50 μm. (D) Representative 4F and 3F Sox2-GFP+ iPS cell lines cultured in N2B27+ 2i/LIF medium on gelatin-coated plate without feeder cells, plus AP staining of 3F Sox2-GFP+ iPS cell line. Scale bar, 100 μm. Color images available online at www.liebertpub.com/scd
Since fibroblasts can be reprogrammed without ectopic c-Myc expression [20,21], we next infected Sox2EGFP CECs with lentiviruses expressing Oct4, Sox2, and Klf4. For the 3F transduction, GFP-positive iPS colonies appeared at day 12 postinfection (Fig. 2C) with considerably fewer (40/100,000) ES cell-like colonies emerging 21 days postinfection. The efficiency of iPS cell formation was 0.04% at day 28, which is ∼10-fold lower than 4F reprogramming. As with other cell types, 3F transduction of Sox2EGFP CECs suffered delayed kinetics and lower efficiency [9,20–22].
We randomly picked several GFP-positive iPS colonies for expansion and characterized their pluripotency features at the molecular level. Most of the iPS colonies (5/6 4F-iPS colonies and 6/6 3F-iPS colonies) were expandable into stable iPS cell lines, displaying proliferation and a morphology characteristic of ES cells (Fig. 2D).
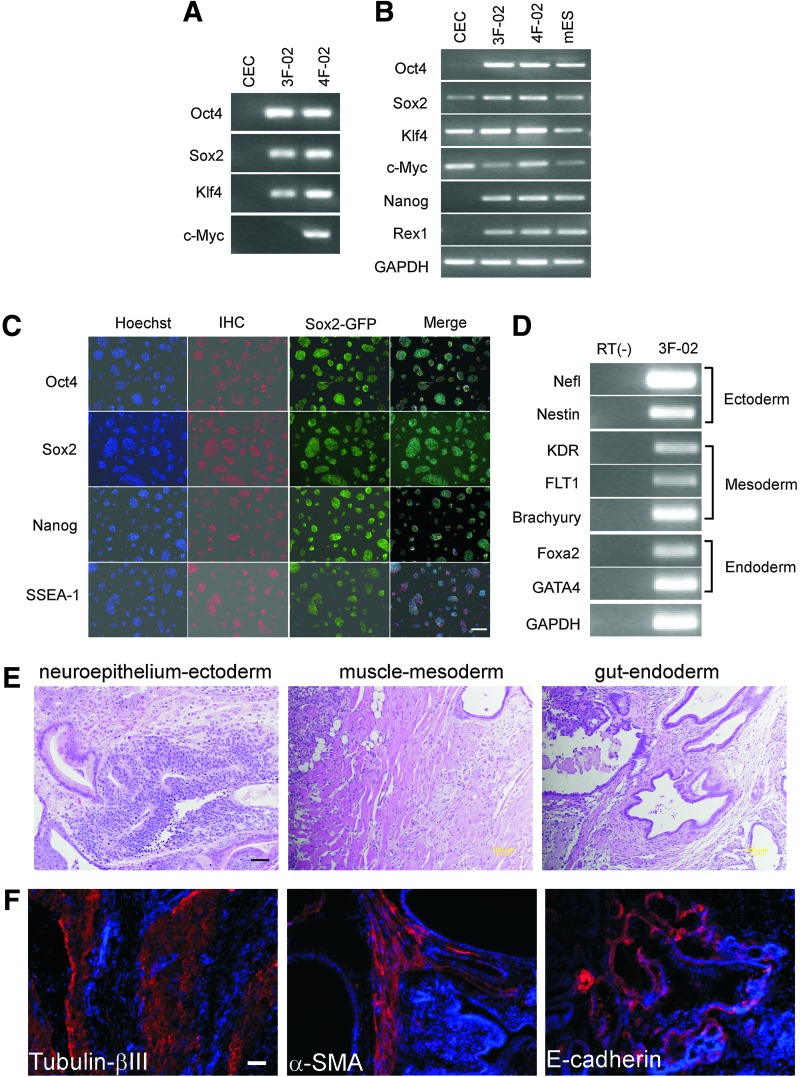
We next examined the integration of the viral transgenes by genotyping PCR. All four viral transgenes were detected in 4F-iPS cells; whereas in 3F-iPS cells, the c-Myc transgene was absent (Fig. 3A), ruling out the possibility that cross-contamination occurred between 4F and 3F clones. RT-PCR showed that all tested endogenous pluripotency-associated transcription factors were expressed in all iPS cell lines (Fig. 3B). Except for high expression of Klf4 and c-Myc, and low expression of Sox2, the other pluripotency genes examined were not expressed in CECs. We confirmed the RT-PCR results with immunofluorescence staining for several transcription markers such as Oct4, Sox2, and Nanog, and for pluripotent cell surface marker SSEA-1 for the 4F (data not shown) and 3F (Fig. 3C) iPS lines.
FIG. 3.
Characteristics of the mouse CEC-derived 4F and 3F Sox2-GFP+ iPS cells. (A) Genotyping of 4F and 3F iPS cells. Genomic PCR showed all four reprogramming factors in 4F iPS cells, and the absence of the c-Myc transgene in 3F iPS cells. Parental CECs served as negative control. (B) RT-PCR of pluripotency-associated genes and GAPDH. 4F and 3F iPS cells express all tested endogenous pluripotency genes. Parental CECs express Sox2, Klf4, and c-Myc. ES cells served as positive control. (C) Immunostaining of pluripotency markers Oct4, Sox2, Nanog, and SSEA-1 in 3F iPS cells. Sox2-GFP shows GFP expression from the Sox2 locus. Scale bar, 100 μm. (D) RT-PCR of representative lineage markers after differentiation of embryoid bodies (EBs) generated from 3F iPS clones. (E) Hematoxylin and eosin staining of teratoma sections derived from 3F iPS clones showing all three embryonic germ layers. Scale bar, 100 μm. (F) Immunostaining of teratoma sections derived from 3F iPS clones showing tubulin-β III positive neural epithelium, α-smooth muscle actin-positive muscle, and E-cadherin-positive endodermal cells. Scale bar, 100 μm. Color images available online at www.liebertpub.com/scd
We next investigated the differentiation potential of 3F-iPS cells in vitro using the EB assay [1,5]. 3F-iPS cells derived from Sox2EGFP CECs effectively formed EBs (data not shown). Ten days after EB transfer onto gelatin-coated plates, the attached cells expressed markers of the three germ layers, including the endoderm markers GATA4 and hepatocyte nuclear factor-3β (Foxa2), the mesoderm markers vascular endothelial growth factor receptors and Brachyury, and the ectoderm marker Nestin as determined by RT-PCR (Fig. 3D). To fully characterize the differentiation potential of CEC-derived iPS cells in vivo, we generated teratomas in immune-deficient NOD-SCID mice after a subcutaneous injection of 4F- and 3F-iPS cells. Histological examinations showed that the teratomas contained derivatives of the three embryonic germ layers, including neural tissues (ectoderm), muscle (mesoderm), and gut-like epithelial tissues (endoderm) (Fig. 3E). We confirmed these morphological findings with immunofluorescence staining: neural epithelial-like tissues were positive for tubulin-β III expression, muscle-like tissues were positive for α-smooth muscle actin, and gut-like epithelium was positive for E-cadherin expression (Fig. 3F).
Generation of 2F iPS cells from mouse CECs
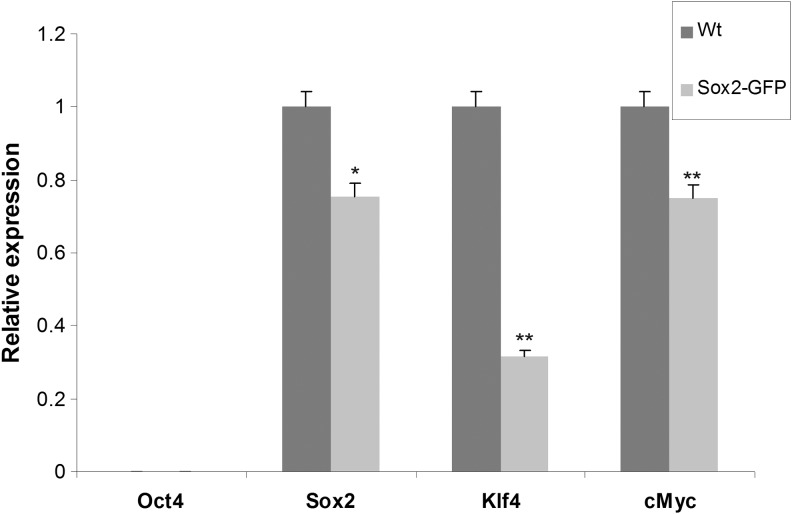
Cells with high levels of endogenous Sox2 have been previously reprogrammed without ectopic Sox2 expression [5,7,19]. Given that Sox2 is expressed in Sox2EGFP CECs (Fig. 3B), we attempted to reprogram Sox2EGFP CECs with Oct4 and Klf4 (2F) only. At a much delayed time point (28 days postinfection), only three GFP-positive colonies (3/100,000) were observed. These three colonies lost GFP expression gradually during expansion, and they were not expandable into stable iPS cell lines. However, when propagated in serum-free N2B27 medium supplemented with 2i/LIF, all three colonies were converted to stable iPS cell lines with stable GFP expression (data not shown). These observations suggest that these three colonies were partially reprogrammed, because with the addition of small molecules modulating signal transduction, they can transition to the completely reprogrammed iPS state as previously demonstrated [22]. A previous report demonstrates that hemizygote Sox2EGFP knock-in mice express less Sox2 than wild-type mice [11]. We confirmed this finding in CECs using quantitative RT-PCR and found 25% less Sox2 compared with wild-type CECs (Fig. 4). Interestingly, except for Oct4, which was undetectable, c-Myc was reduced to 25% and Klf4 was surprisingly reduced much more to 70% in Sox2EGFP CECs (Fig. 4). Given the instrumental role that Sox2 plays in somatic cell reprogramming, and that three reprogramming factors are significantly reduced in Sox2EGFP CECs, we hypothesized that a reprogramming haploinsufficiency exists in hemizygous Sox2EGFP CECs.
FIG. 4.
Endogenous expression of reprogramming factors in wild-type and Sox2EGFP CECs. Quantitative real-time PCR was performed on passage 2 wild-type and hemizygous Sox2EGFP CECs. Neither of the cells expresses Oct4 at appreciable levels. Compared with the wild type, expression levels of Sox2, Klf4, and c-Myc are reduced in hemizygous Sox2EGFP CECs. Data are normalized for GAPDH expression and presented as mean±SEM for three replicates. *P value<0.05; **P value<0.01. Color images available online at Y
To examine the possibility that an iPS reprogramming haploinsufficiency exists in Sox2 hemizygous cells, we infected wild-type CECs to assess their potential to form iPS cells. Wild-type CECs with the 3F reprogramming protocol mentioned earlier produced more iPS colonies than Sox2EGFP CECs. The appearance of ES cell-like colonies occurred 1 day earlier. The efficiency of 3F iPS generation from wild-type CECs was 0.21%, about five times higher than Sox2EGFP CECs. Moreover, 2F experiments revealed iPS colonies after 3 weeks using wild-type CECs with a respectable reprogramming efficiency of 0.046% (46/100,000) at day 28 (Fig. 5A). Analysis of genomic DNA from 2F-iPS cells via genotyping PCR showed transgenes Oct4 and Klf4 and confirmed the absence of viral Sox2 and c-Myc (Fig. 5B). 2F-iPS cells derived from wild-type CECs expressed pluripotency markers, as demonstrated by RT-PCR (Fig. 5C), which were confirmed with immunofluorescence staining (Fig. 5D). Most of them were expandable and maintained ES cell-like morphology after serial passaging (Fig. 5E). Furthermore, the transgenes hOct4 and hKlf4 were significantly attenuated at late passages (Supplementary Fig. S2). These 2F-iPS cells maintained normal karyotypes (Supplementary Fig. S3), acquired a stem cell methylation pattern in Oct4 and Nanog promoters (Supplementary Fig. S4), and could differentiate in vitro using the EB formation, and into endodermal, mesodermal, and ectodermal derivatives in teratomas (Fig. 5F, G). These results suggest that ectopic Sox2 and c-Myc expression is dispensable for reprogramming wild-type CECs into iPS cells. We then attempted reprogramming wild-type CECs with viral Oct4 and Sox2 twice, and reproducibly noticed a morphological change in CECs, but did not observe iPS colonies at day 35.
FIG. 5.
Generation of iPS cells from mouse wild-type CECs with two reprogramming factors. (A) Brightfield and AP staining of representative 2F iPS colony. Scale bar, 50 μm. (B) Genotyping of 2F iPS cells. Genomic PCR showed only Oct4 and Klf4 transgenes in 2F iPS cells as expected. (C) RT-PCR of pluripotency-associated genes and GAPDH. 2F iPS cells express all tested endogenous pluripotency genes, and parental wild-type CECs express Sox2, Klf4 and c-Myc. (D) Immunostaining of pluripotency markers Oct4, Sox2, Nanog, and SSEA-1 in 2F iPS cells. Scale bar, 100 μm. (E) Representative 2F iPS cell line cultured in N2B27+ 2i/LIF medium on gelatin-coated plate without feeder cells. Scale bar, 50 μm. (F) RT-PCR of typical lineage markers after differentiation of EBs derived from 2F iPS clones. (G) Teratoma formation. Hematoxylin and eosin staining of teratoma sections derived from 2F iPS clones showed all three embryonic germ layers. Immunostaining of teratoma sections showed tubulin-β III-positive neural epithelium, α-smooth muscle actin-positive muscle, and E-cadherin-positive endodermal cells. Scale bar, 100 μm. Color images available online at www.liebertpub.com/scd
Since endogenous Sox2 and c-Myc expression were sufficient for 2F reprogramming of wild-type CECs, and wild-type CECs express Klf4 as well, we tried to derive iPS cells with viral Oct4 alone. As with the Oct4 and Sox2 combination, some morphologic changes occurred in the CECs, but ES cell-like colonies did not emerge at day 35, despite three attempts.
Mesenchymal-epithelial transition (MET) is an essential early step for reprogramming fibroblasts [23,24]. Human epithelial cells reprogram with a higher efficiency than fibroblasts [24,25], suggesting that bypassing MET is beneficial for reprogramming. Ciliary epithelium is neuroepithelium, and it shares the same developmental origin as neural retina and retinal pigment epithelium. These specialized epithelial cells express not only typical epithelial cell markers, such as cytokeratin and connexin-43, but also neural progenitor markers such as Pax6 and Sox2 [14]. E-cadherin, an important regulator of epithelial homeostasis, is highly expressed in ES/iPS cells. It is also required for fibroblast reprogramming [26]. Although mRNA of E-cadherin is expressed in CECs in our RT-PCR data (Fig. 1C), the protein was undetectable ([14] and our unpublished data). To determine the relative E-cadherin expression levels between CECs and TTFs, we performed qPCR experiments and found essentially no difference in E-cadherin mRNA between the two cell populations (Supplementary Fig. S5). However, CECs expressed relatively high levels of Sox2 and EpCAM (epithelial cell adhesion molecule) compared with TTFs (Supplementary Fig. S5), EpCAM has been shown to promote somatic cell reprogramming even more profoundly than E-cadherin [27]. It is, thus, tempting to speculate that CEC reprogramming benefits from high basal expression of EpCAM, in addition to high endogenous expression of reprogramming factor Sox2, although further investigations are required to address this hypothesis.
CECs are a new cell source that may have advantages for cellular reprogramming. Specifically, we demonstrate the emergence of ES cell like colonies in a short time frame, increased efficiency, and a reduced exogenous transcription factor requirement. We also discovered that endogenous Sox2 levels are critical for facile cellular CEC reprogramming. This observation may be applicable to other cell types, such as neural stem cells or dermal papilla cells, that have higher endogenous Sox2 expression.
CECs have the advantage of being easily accessible unlike neural stem cells [5]. The ciliary body region is a readily accessible region of the eye using several techniques [28,29]. Access to ciliary body tissue for tumor biopsy is well established [30], and recently, one group described a technique to culture ciliary body cells from human patients using minimal tissue [31].
CECs can be reprogrammed with a reduced genetic input compared with fibroblasts. Fewer transcription factors and retroviruses may reduce the risk of insertional mutagenesis and cancer formation risk. Taken together, these characteristics may be advantageous for clinical therapy.
Supplementary Material
Acknowledgments
S.H.C. would like to acknowledge Hope for Vision, Research to Prevent Blindness Career Development Award for grant support, and NEI K-08 Career Development Award. The authors would like to thank Larysa Pevny for sharing Sox2EGFP mice and data review.
Author Disclosure Statement
The authors declare no conflict of interest.
References
- 1.Takahashi K. Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Lowry WE. Richter L. Yachechko R. Pyle AD. Tchieu J. Sridharan R. Clark AT. Plath K. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci U S A. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eminli S. Foudi A. Stadtfeld M. Maherali N. Ahfeldt T. Mostoslavsky G. Hock H. Hochedlinger K. Differentiation stage determines potential of hematopoietic cells for reprogramming into induced pluripotent stem cells. Nat Genet. 2009;41:968–976. doi: 10.1038/ng.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan KY. Eminli S. Hettmer S. Hochedlinger K. Wagers AJ. Efficient generation of iPS cells from skeletal muscle stem cells. PLoS One. 2011;6:e26406. doi: 10.1371/journal.pone.0026406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim JB. Zaehres H. Wu G. Gentile L. Ko K. Sebastiano V. Arauzo-Bravo MJ. Ruau D. Han DW. Zenke M. Scholer HR. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454:646–650. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- 6.Kim JB. Sebastiano V. Wu G. Arauzo-Bravo MJ. Sasse P. Gentile L. Ko K. Ruau D. Ehrich M, et al. Oct4-induced pluripotency in adult neural stem cells. Cell. 2009;136:411–419. doi: 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 7.Tsai SY. Clavel C. Kim S. Ang YS. Grisanti L. Lee DF. Kelley K. Rendl M. Oct4 and klf4 reprogram dermal papilla cells into induced pluripotent stem cells. Stem Cells. 2010;28:221–228. doi: 10.1002/stem.281. [DOI] [PubMed] [Google Scholar]
- 8.Tsai SY. Bouwman BA. Ang YS. Kim SJ. Lee DF. Lemischka IR. Rendl M. Single transcription factor reprogramming of hair follicle dermal papilla cells to induced pluripotent stem cells. Stem Cells. 2011;29:964–971. doi: 10.1002/stem.649. [DOI] [PubMed] [Google Scholar]
- 9.Yamanaka S. Strategies and new developments in the generation of patient-specific pluripotent stem cells. Cell Stem Cell. 2007;1:39–49. doi: 10.1016/j.stem.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 10.Tropepe V. Coles BL. Chiasson BJ. Horsford DJ. Elia AJ. McInnes RR. van der Kooy D. Retinal stem cells in the adult mammalian eye. Science. 2000;287:2032–2036. doi: 10.1126/science.287.5460.2032. [DOI] [PubMed] [Google Scholar]
- 11.Taranova OV. Magness ST. Fagan BM. Wu Y. Surzenko N. Hutton SR. Pevny LH. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev. 2006;20:1187–1202. doi: 10.1101/gad.1407906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carey BW. Markoulaki S. Beard C. Hanna J. Jaenisch R. Single-gene transgenic mouse strains for reprogramming adult somatic cells. Nat Methods. 2010;7:56–59. doi: 10.1038/nmeth.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cicero SA. Johnson D. Reyntjens S. Frase S. Connell S. Chow LM. Baker SJ. Sorrentino BP. Dyer MA. Cells previously identified as retinal stem cells are pigmented ciliary epithelial cells. Proc Natl Acad Sci U S A. 2009;106:6685–6690. doi: 10.1073/pnas.0901596106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gualdoni S. Baron M. Lakowski J. Decembrini S. Smith AJ. Pearson RA. Ali RR. Sowden JC. Adult ciliary epithelial cells, previously identified as retinal stem cells with potential for retinal repair, fail to differentiate into new rod photoreceptors. Stem Cells. 2010;28:1048–1059. doi: 10.1002/stem.423. [DOI] [PubMed] [Google Scholar]
- 15.MacNeil A. Pearson RA. MacLaren RE. Smith AJ. Sowden JC. Ali RR. Comparative analysis of progenitor cells isolated from the iris, pars plana, and ciliary body of the adult porcine eye. Stem Cells. 2007;25:2430–2438. doi: 10.1634/stemcells.2007-0035. [DOI] [PubMed] [Google Scholar]
- 16.Takahashi K. Okita K. Nakagawa M. Yamanaka S. Induction of pluripotent stem cells from fibroblast cultures. Nat Protoc. 2007;2:3081–3089. doi: 10.1038/nprot.2007.418. [DOI] [PubMed] [Google Scholar]
- 17.Okada M. Oka M. Yoneda Y. Effective culture conditions for the induction of pluripotent stem cells. Biochim Biophys Acta. 2010;1800:956–963. doi: 10.1016/j.bbagen.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 18.Ying QL. Wray J. Nichols J. Batlle-Morera L. Doble B. Woodgett J. Cohen P. Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Utikal J. Maherali N. Kulalert W. Hochedlinger K. Sox2 is dispensable for the reprogramming of melanocytes and melanoma cells into induced pluripotent stem cells. J Cell Sci. 2009;122:3502–3510. doi: 10.1242/jcs.054783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakagawa M. Koyanagi M. Tanabe K. Takahashi K. Ichisaka T. Aoi T. Okita K. Mochiduki Y. Takizawa N. Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 21.Wernig M. Meissner A. Cassady JP. Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell. 2008;2:10–12. doi: 10.1016/j.stem.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 22.Silva J. Barrandon O. Nichols J. Kawaguchi J. Theunissen TW. Smith A. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008;6:e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Samavarchi-Tehrani P. Golipour A. David L. Sung HK. Beyer TA. Datti A. Woltjen K. Nagy A. Wrana JL. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 24.Li R. Liang J. Ni S. Zhou T. Qing X. Li H. He W. Chen J. Li F, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 25.Aasen T. Raya A. Barrero MJ. Garreta E. Consiglio A. Gonzalez F. Vassena R. Bilic J. Pekarik V, et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat Biotechnol. 2008;26:1276–1284. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- 26.Redmer T. Diecke S. Grigoryan T. Quiroga-Negreira A. Birchmeier W. Besser D. E-cadherin is crucial for embryonic stem cell pluripotency and can replace OCT4 during somatic cell reprogramming. EMBO Rep. 2011;12:720–726. doi: 10.1038/embor.2011.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang HP. Chen PH. Yu CY. Chuang CY. Stone L. Hsiao WC. Li CL. Tsai SC. Chen KY, et al. Epithelial cell adhesion molecule (EpCAM) complex proteins promote transcription factor-mediated pluripotency reprogramming. J Biol Chem. 2011;286:33520–33532. doi: 10.1074/jbc.M111.256164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Damato BE. Local resection of uveal melanoma. Dev Ophthalmol. 2012;49:66–80. doi: 10.1159/000328261. [DOI] [PubMed] [Google Scholar]
- 29.Engin G. Yilmazli C. Engin KN. Gulkilik G. Bilgic L. Combined cyclectomy-trabeculectomy procedure for refractory glaucoma. Ophthalmic Surg Lasers Imaging. 2004;35:507–511. [PubMed] [Google Scholar]
- 30.Forrest AW. Keyser RB. Spencer WH. Iridocyclectomy for melanomas of the ciliary body: a follow-up study of pathology and surgical morbidity. Ophthalmology. 1978;85:1237–1249. doi: 10.1016/s0161-6420(78)35560-3. [DOI] [PubMed] [Google Scholar]
- 31.Garcia-Valenzuela E. Stem cell potential of ciliary epithelium. Clinical-Ophthalmology.com Clinical-Ophthalmology.com
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.