Abstract
The hormone abscisic acid (ABA) is a small molecule involved in pivotal physiological functions in higher plants. Recently, ABA has been also identified as an endogenous hormone in mammals, regulating different cell functions including inflammatory processes, stem cell expansion, insulin release, and glucose uptake. Aptamers are short, single-stranded (ss) oligonucleotidesable to recognize target molecules with high affinity. The small size of the ABA molecule represented a challenge for aptamer development and the aim of this study was to develop specific anti-ABA DNA aptamers. Biotinylated abscisic acid (bio-ABA) was immobilized on streptavidin-coated magnetic beads. DNA aptamers against bio-ABA were selected with 7 iterative rounds of the systematic evolution of ligands by exponential enrichment method (SELEX), each round comprising incubation of the ABA-binding beads with the ssDNA sequences, DNA elution, electrophoresis, and polymerase chain reaction (PCR) amplification. The PCR product was cloned and sequenced. The binding affinity of several clones was determined using bio-ABA immobilized on streptavidin-coated plates. Aptamer 2 and aptamer 9 showed the highest binding affinity, with dissociation constants values of 0.98±0.14 μM and 0.80±0.07 μM, respectively. Aptamers 2 and 9 were also able to bind free, unmodified ABA and to discriminate between different ABA enantiomers and isomers. Our findings indicate that ssDNA aptamers can selectively bind ABA and could be used for the development of ABA quantitation assays.
Introduction
Aptamers are nucleic acid macromolecules of single-stranded (ss) DNA or RNA that can fold into 3-dimensional structures upon interaction with other molecules, providing preferential binding sites for molecular recognition of analytes (Ellington and Szostak, 1990; Tuerk and Gold, 1990). ssDNA aptamers typically have a complex secondary structure, due to many intramolecular base pairing opportunities (Zhang et al., 2001), which imparts increased stability to the folded molecule. This provides a binding affinity in the picomolar to nanomolar range for complexes of aptamers with high molecular weight ligands and in the micromolar range for complexes formed with small molecules (Floege et al., 1999; Hermann and Patel, 2000; Stojanovic et al., 2001; Huang et al., 2005; Baker et al., 2006; Lai et al., 2007). Aptamers have been used successfully for several different in vitro and in vivo applications, including protein quantification and purification and inhibition of receptors or of enzyme activities. Aptamers can be produced by chemical synthesis, ensuring high product reproducibility, and can be easily chemically modified, allowing the insertion of functional groups for their immobilization or detection. Moreover, the procedure used to generate aptamers, systematic evolution of ligands by exponential enrichment (SELEX), may be modified in order to minimize the effect of the matrix on selectivity, affinity, sensitivity, and stability of the aptamers. Unlike antibodies, aptamers can be developed also against toxic molecules or against targets with low or no immunogenicity. The SELEX process has thus provided the means to generate ssDNA or RNA oligonucleotides specific for a given ligand in alternative to monoclonal antibodies. The diversity of structures exhibited by a random DNA oligonucleotide library allows the selection of aptamers binding simple targets, such as a single amino acid (Connell et al., 1993), or complex targets, such as a virus (Pan et al., 1995). Aptamers with high affinity and specificity have been selected for many different ligands and are emerging as a new class of molecular tools that compete with antibodies in affinity enrichment, analysis, imaging, diagnostics, and therapeutics.
Here, we describe the in vitro selection of novel ssDNA aptamers that bind abscisic acid (ABA). ABA is a polyterpenoid (Mw=264.32 g/mol) plant hormone that regulates several physiological functions in plants, including the response to abiotic stress and the regulation of seed dormancy and germination (Nambara and Marion, 2005; Xue-Xuan et al., 2010). Recently, ABA has been shown to behave also as an endogenous mammalian hormone with pleiotropic functions, that is, stimulation of the activation of human and murine innate immune cells (Bruzzone et al., 2007; Bodrato et al., 2009; Magnone et al., 2009), expansion of human mesenchymal stem cells (Scarfì et al., 2008) and human hematopoietic progenitors (Scarfì et al., 2009), stimulation of insulin release from human and rodent pancreatic β cells (Bruzzone et al., 2008) and of glucose uptake by adipocytes and muscle cells (Bruzzone et al., 2012).
Availability of ABA-binding ssDNA aptamers could allow the development of new molecular tools for the detection of this hormone in biological samples and to inhibit its functional effects in human and plant cells.
Materials and Methods
Reagents and materials
Biotinylated abscisic acid (bio-ABA) was synthesized from (±)2-cis, 4-trans abscisic acid by coupling (+)-biotin-ɛ-aminocaproyl hydrazide to the carbonylic group on the ABA carbon ring via an ɛ-aminocaproyl hydrazide linker, as described in (Yamazaki et al., 2003). Analog number 10 (2′α, 3′α-dihydro-2′α, 3′α–epoxy-abscisic acid) was synthesized as described in (Grozio et al., 2011). Synthetic ssDNA oligonucleotides (RND40), containing a central randomized region of 40 nucleotides flanked by two conserved 18-nucleotide regions (GCGGATGAAGACTGGTGT-40N-GCCCTAAATACGAGCAAC) and the specific unlabeled F3 (5′-GCGGATGAAGACTGGTGT-3′) and R3 (5′- GTTGCTCGTATTTAGGGC-3′) primers, were obtained from Metabion (Martinsried, Germany). Digoxigenin-labeled F3 and R3 primers were purchased from TIB Molbiol. Dimethyl sulphoxide (DMSO), bovine serum albumin (BSA), (±)2-cis, 4-trans ABA, its purified enantiomers and (+)-biotin-ɛ-aminocaproyl hydrazide (bio-ABA linker) were purchased from Sigma. (±)2-trans,4-trans abscisic acid was prepared and high-performance liquid chromatography (HPLC) purified as described by Bangerth (BANGERTH, 1982). [3H]-abscisic acid (10 Ci/mmol) was purchased from American Radiolabeled Chemicals, Inc. Anti-digoxigenin horseradish peroxidase (HRP)-coupled antibody and ABTS solution (2,2′-azinobis [3-ethylbenzothiazoline-6-sulfonic acid]-diammonium salt) were purchased from Roche Diagnostics Gmbh and Boehringer–Mannheim, respectively.
Immobilized bio-ABA
A volume of 250 μL of magnetic streptavidin-coated beads (10 mg/mL, binding capacity: 600 pmol biotin/mg resin, FLUKA) was washed twice with phosphate buffered saline containing 1 mM MgCl2 and three times with the selection buffer (20 mM Tris-HCl, pH7.4, 1 mM MgCl2, 150 mM NaCl, 5 mM KCl, 0.2% BSA) and resuspended in selection buffer at 10 mg/mL. In order to saturate all binding sites of streptavidin, 1 μg bio-ABA (1.6 nmol) (bio-ABA beads) or 0.6 μg of bio-ABA linker (1.6 nmol) (bio-linker beads) were incubated with 250 μL of streptavidin-coated beads for 30 minutes at room temperature with mild shaking. Beads were further incubated for 1 hour at 4°C with a 5% (w/v) solution of BSA in selection buffer to saturate unoccupied groups. The beads were finally washed twice with selection buffer, resuspended at 10 mg/mL (in 250 μL) in selection buffer and stored at 4°C. The amount of bio-ABA attached to the beads was calculated as the difference between the respective initial and unbound concentrations, measured by HPLC on a Hewlett-Packard HP1090 instrument. The column was an Atlantis C18 (3.9×150 mm, 5μm, Waters): solvent A was water containing 0.01% (v/v) trifluoroacetic acid pH 3.3; solvent B was solvent A containing 80% (v/v) acetonitrile. The solvent program was a linear gradient (at a flow rate of 0.8 mL/minute) starting at 100% solvent A and increasing to 100% solvent B in 30 minutes. Bio-ABA was detected with an HP1040A diode array spectrophotometric detector set at 260 nm.
In vitro selection procedure
Seven iterative rounds of selection and amplification of ssDNA aptamers were performed, as described in (Morris et al., 1998). The ssDNA library was denatured at 90°C for 10 minutes and then cooled on ice for 10 minutes. In order to remove aptamers with affinity for the matrix, 500 pmol of ssDNA were pre-incubated with 80 μL of streptavidin-coated beads for 30 minutes at room temperature. The beads were removed and the supernatant was incubated with 80 μL of bio-ABA beads (containing 512 pmol of bio-ABA) for 30 minutes at room temperature. At the end of the incubation, the supernatant was removed and the beads were washed twice with 160 μL selection buffer, resuspended in 100 μL distilled water and heated for 3 minutes at 95°C. The supernatant was collected and 10 μL were amplified by polymerase chain reaction (PCR) using the 5′-digoxigenin-labeled F3 primer and the 5′-phosphate-labeled R3 primer. A pilot PCR was performed to determine the optimal number of cycles required for amplification by evaluating the amount, size, and purity of the band on agarose gel. PCR conditions were then chosen as follows: 1 μM of each primer and 200 μM dNTPs in a final volume of 100 μL; an initial denaturation step at 95°C for 2 minutes, 25 amplification cycles comprising 15 seconds at 95°C, 15 seconds at 55°C, and 30 seconds at 72°C, and an extension step of 4 minutes at 72°C. An aliquot of the PCR product was analyzed for size and purity on a 2.5% agarose gel; the rest was precipitated with ethanol. The double-stranded (ds) DNA (250 pmol) was subjected to exonuclease activity to eliminate the 5′-phosphate-labeled chain, and ssDNA aptamers were then denatured at 95°C for 10 minutes, immediately cooled for 10 minutes on ice, and used as the starting DNA pool in the second selection round on bio-ABA beads. After the fourth SELEX round, a counter selection step on bio-linker beads was introduced to eliminate nonspecific dsDNA aptamers: the denatured PCR product was preincubated with 80 μL of bio-linker beads (512 pmol of bio-linker) for 30 minutes at room temperature prior to incubation with the bio-ABA beads.
After the fourth and the seventh round, the aptamer pool was amplified and labeled by PCR using 5′digoxigenin-labeled F3 and R3 primers to evaluate the binding affinity of the selected aptamers for bio-ABA by enzyme-linked oligonucleotide assay (ELONA).
Cloning and sequencing
After the seventh round, the recovered dsDNA was cloned using pGEM®-T Easy Vector Systems (Promega). The ligation products (5 ng) were introduced by electroporation into Escherichia coli JM 109 cells. White colonies were picked and expanded in liquid culture; the plasmid DNA was purified by miniprep kit (ATP Biotech Inc.) and amplified either with 5′digoxigenin-labeled F3 and unlabeled R3 primers or with unlabeled F3 and 5′digoxigenin-labeled R3 primer in order to test the binding capacity of both strands to bio-ABA. The 2 complementary sequences of each clone were 5′digoxigenin-labeled and tested for their individual binding affinity for bioABA by ELONA. The plasmid DNA was sequenced by the Sanger method using the T7 Promoter Primer (Promega). Alignment of the aptamer sequences was performed with the ClustalW multiple sequence alignment. Analysis of the secondary structure of selected aptamers was performed with the free energy minimization algorithm (ZUKER, 2003) using the Internet-tool mfold (http://mfold.bioinfo.rpi.edu) and QGRS Mapper, a web-based server for predicting G-quadruplexes in nucleotide sequences (http://bioinformatics.ramapo.edu/QGRS/analyze.php).
ELONA
Aptamers obtained after the seventh round of selection (SEL 7, 250 ng) and from the different clones (200 ng) were amplified by PCR with 5′digoxigenin-labeled F3 or 5′digoxigenin-labeled R3 primers. Prior to the assay, the DNA was denatured at 95°C for 10 minutes and immediately cooled for 10 minutes on ice.
For ELONA of SEL 7, streptavidin-coated 96-well plates (Streptavidin Immobilizer™ Module, CovaLink) were incubated for 18 hours at room temperature with 200 μL per well of a 1.6 μM solution of bio-ABA or of bio-linker in selection buffer. The plates were washed 3 times with selection buffer and then incubated for 1 hour at 37°C with 200 μL per well of a 5% (v/v) solution of BSA in selection buffer. The plates were washed again in selection buffer and 200 μL per well of heat-denatured digoxigenin-labeled SEL 7 or RND40 library was added at a concentration of 8 nM in selection buffer. The plate was incubated at 37°C for 1 hour, washed 4 times with selection buffer, and 200 μL per well of a 1:1000 dilution of anti-digoxigenin-HRP antibody was added. After incubation for 1 hour at 37°C on a shaking platform, the plates were washed 4 times and developed using the ABTS solution according to the manufacturer's instructions. Absorbance at 405 nm was determined on a Tecan microplate reader.
For ELONA of the different clones, the streptavidin-coated plates were incubated for 18 hours at room temperature with 200 μL per well of a 1.6 μM solution of bio-ABA in selection buffer. After repeated washing with selection buffer, 200 μL per well of a 200 nM solution of digoxigenin-labeled clones, or digoxigenin-labeled RND40 or digoxigenin-labeled SEL 7, was added and the plates were incubated for 1 hour at 37°C and processed for color development as described above. Absorbance due to aptamer binding to BSA-coated wells (background) was subtracted from all values.
Binding assays
All aptamer clones were tested for their binding capacity to bio-ABA immobilized on streptavidin-coated magnetic beads or to streptavidin-coated magnetic beads incubated with DMSO (negative control). Different concentrations of heat-denatured 5′digoxigenin-labeled aptamers (ranging from 0.1 to 8 μM) were dissolved in binding buffer (20 mM Tris-HCl, pH 7.4, 2 mM MgCl2, 100 mM NaCl, 5 mM KCl, 1 mM CaCl2, 0.02% Tween20) and incubated with 100 μL of bio-ABA beads (10 mg/mL beads with 6 μM bio-ABA for each binding reaction) or DMSO for 1 hour at 37°C. Unbound aptamers were removed by washing the beads twice with 1.5 mL binding buffer and then with binding buffer containing 500 mM NaCl. Afterward, 200 μL of a 1:1000 dilution of anti-digoxigenin-HRP antibody in binding buffer was added. Following a 1-hour incubation at 37°C, the beads were washed twice in binding buffer and color was developed using 100 μL of ABTS solution. The absorbance at 405 nm was determined with a Tecan microplate reader after 30 minutes at 37°C. Aptamers that bind bio-ABA cause a colorimetric reaction, due to the anti-digoxigenin-HRP antibody, that is proportional to the amount of aptamer bound to the bio-ABA resin. Saturation curves were calculated by subtracting the absorbance values of negative controls, performed on beads incubated with DMSO (vehicle). Assuming a 1-to-1 molar ratio between bio-ABA, aptamer, and antibody, the dissociation constant Kd was calculated by nonlinear regression analysis of the saturation curves.
In parallel, selected aptamers were tested for their ability to bind free, not immobilized, ABA. Equilibrium binding experiments with [3H]-ABA were performed with the equilibrium filtration method (Jenison et al., 1994). A series of [3H]-ABA concentrations, ranging from 0.05 to 2.5 μM, were incubated with 0.06 μM heat-denatured aptamers in 50 μL of binding buffer without Tween2 at 25°C for 30 min. The incubations were then loaded onto centrifugal filter columns (modified polyethersulfone, 3 kDa molecular weight cutoff, Amicon,) and centrifuged for 30 minutes at 16,000g. Controls included the same amounts of [3H]-ABA incubated with the RND40 library as negative controls or incubated with the aptamers in the presence of excess unlabeled ABA (1 mM; nonspecific binding). The radioactivity of the filters was determined on a Beckman β-counter. Saturation curves were obtained by subtracting the nonspecific binding to each value. The Kd was calculated by nonlinear regression analysis.
Determination of the specificity of individual aptamers by affinity elution assay
The ability of the selected aptamers to bind the individual ABA enantiomers, (+)2-cis, 4-trans abscisic acid and (-)2-cis, 4-trans abscisic acid, the ABA isomer (±) 2,4-trans abscisic acid, the bio-linker, (+)-biotin-ɛ-aminocaproyl hydrazide or the ABA analog 2′-α, 3′-α-dihydro-2′α, 3′α–epoxy-abscisic acid (analog 10) in solution was tested by affinity elution assay.
The 5′-digoxigenin labeled aptamers (2 μM) were incubated with bio-ABA-coated beads (6 μM bio-ABA for each binding reaction) without (control) or with 600 μM of the above competing molecules for 1 hour at 37°C in 100 μL of binding buffer. Each incubation was performed in triplicate. The beads were washed twice with 1.5 mL of binding buffer and once with 1.5 mL of binding buffer containing 500 mM NaCl and then incubated with 200 μL of a 1:1000 dilution of anti-digoxigenin-HRP antibody. After 1-hour incubation at 37°C, the beads were washed twice with 1.5 mL of binding buffer and once with 1.5 mL of binding buffer containing 500 mM NaCl, and color development was obtained after addition of the ABTS solution. Absorbance at 405 nm (A405) was determined with a microplate reader, after 30 minutes of incubation at 37°C. A405 values of aptamer incubations with bio-ABA coated beads. The percentage of aptamer binding to the competing free molecule was calculated as
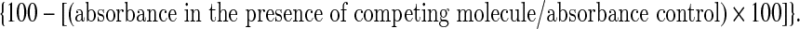 |
Circular dichroism
Circular dichroism (CD) experiments were carried out on clone number 2 and on clone number 9 at a concentration of 10 μM in 20 mM phosphate buffer pH 7.3, containing 100 mM KCl. The ssDNA aptamers were heated to 95°C for 3 minutes and then cooled to room temperature for 4 hours before obtaining the CD spectra. Far ultraviolet (UV) CD spectra were recorded on a Jasco J-500A spectropolarimeter equipped with a Jasco IF-500-2 data processor The spectropolarimeter was calibrated with an aqueous solution of (1S)-(+)-10-camphor sulphonic acid at 290.5 nm, in accordance with the instrument manual. Each spectrum was obtained by averaging three scans from 200 to 350 nm at a rate of 20 nm/minute with a step resolution of 0.2 nm, a time constant of 2 seconds, and a bandwidth of 2.0 nm. A thermostated 0.1-cm path-length cell was used, and all measurements were taken at 20°C. The data were expressed as [Θ], the mean residue ellipticity in units of degrees centimeter squared for decimole of residue. All spectra were corrected by subtracting the appropriate buffer background and normalized to mean residue ellipticity.
Determination of intracellular cAMP levels
Human granulocytes (1.5×107/mL) resuspended in Hank's balanced salt solution with 1.25 mM Ca2+ and 0.8 mM Mg2+ were preincubated for 5 minutes at 25°C with 10 μM cAMP phosphodiesterase inhibitor (Sigma). Duplicate 200-μL aliquots of cells were incubated for 1 minute at 25°C without (control) or with 1 μM ABA, in the absence or presence of 2 or 20 μM of clone number 2 or clone number 9. The intracellular cAMP concentration ([cAMP]i) was determined by radioimmunoassay (Bruzzone. et al., 2007).
Results
Selection of anti-ABA aptamers from a combinatorial ssDNA library
The in vitro selection of ABA-binding aptamers was performed with a modified FluMag-SELEX technology, which uses digoxigenin labels for DNA quantification and magnetic beads for target immobilization (Stoltenburg et al., 2005). Each round of this SELEX process comprised the following steps: (1) binding of ssDNA to the target, immobilized on magnetic beads; (2) washing of the DNA–target complex; (3) heat elution of bound ssDNA from the target; (4) amplification of eluted ssDNA by PCR; and (5) preparation/purification of the relevant ssDNA from the dsDNA PCR products.
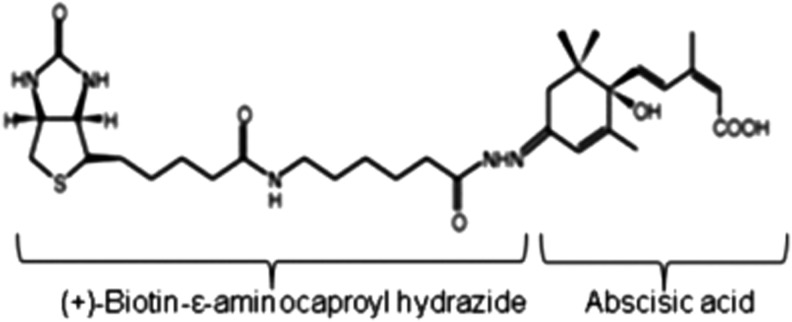
ABA-binding DNA aptamers were selected using biotinylated ABA (BioABA, Fig. 1) immobilized on streptavidin-functionalized magnetic beads and blocked with 5% BSA to avoid nonspecific binding. We also performed a counter selection on magnetic beads covered with the bio-linker alone.
FIG. 1.
Structure of biotinylated abscisic acid (bio-ABA). (+)-Biotin-ɛ-aminocaproyl hydrazide (bio-ABA linker) was linked to the C′4 position of (±)2-cis,4-trans abscisic acid (ABA) and bio-ABA was then immobilized on streptavidin-coated magnetic beads for aptamer selection.
Target immobilization on the surface of magnetic beads allows an easy and efficient handling during the SELEX process. A library of ssDNA with a 40-nucleotide (nt) random region flanked by two 18-nt fixed regions for PCR amplification was screened. The initial library contained 1014–1016 different sequences. Upon mixing with bio-ABA, oligomers from the library may bind to the target. During this process, a bound candidate or candidates of the pool may be specific for the target and can be amplified with primers F3 and R3. The number of PCR cycles was also optimized to avoid overamplification (see methods section). ssDNA sequences bound to the target were eluted from the immobilized bio-ABA by denaturation. The amount of ABA-bound ssDNA recovered after round 8 was similar to that obtained after round 7, possibly because of saturation of the binding sites present on the beads. Thus, no further selection rounds were performed. The aptamer population obtained from round 7, SEL7, was amplified by PCR using digoxigenin-labeled F3 and R3 primers.
ABA binding to the SEL7 aptamer population and to its clones (individual aptamers)
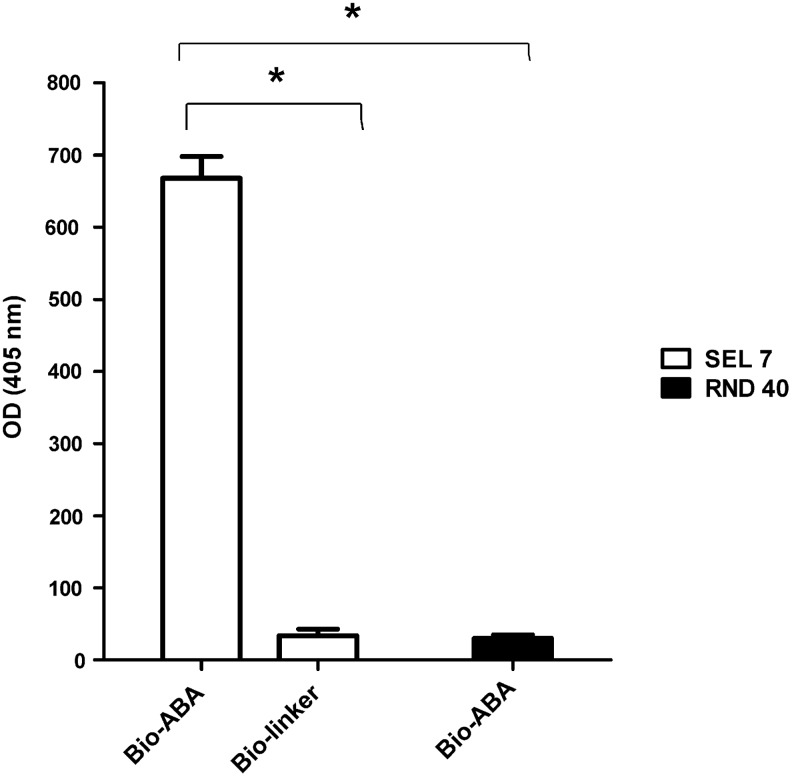
The ABA-binding capacity of the starting ssDNA library (RND40) and of the SEL7 population sequences was compared by ELONA (Fig. 2). The RND40 aptamer population at 8 nM showed no detectable binding to bio-ABA. Conversely, the SEL7 population was able to bind ABA with a higher affinity, as indicated by the intensity of the signal obtained at 8 nM aptamer. Moreover, SEL7 aptamers did not bind to the negative control (bio-linker), indicating specificity towards ABA. These results indicated that the SEL7 population was indeed enriched in ABA-binding sequences compared to RND40.
FIG. 2.
ABA binding to the SEL7 aptamer population (aptamers obtained after the seventh round of selection) compared with the starting ssDNA library. Bio-ABA or the bio-linker were plated at 1.6 nmol/well in 96-well plates and incubated with 8 nM of digoxigenin-labeled aptamers from the SEL 7 population (white bars) or the starting library (RND40). After washing of excess, unbound aptamers, anti-digoxigenin antibodies coupled to horseradish peroxidase (HRP) were added and the reaction was revealed with the 2,2′-azinobis [3-ethylbenzothiazoline-6-sulfonic acid]-diammonium salt (ABTS) solution. Results are the mean of three different experiments, performed in triplicate. *p<0.5, by t-test.
Thus, the SEL7 population was cloned into pGEM®-T Easy Vector, and several clones positive at the blue/white screening were picked and amplified by PCR. The digoxigenin-labeled sequences were then tested for their ABA binding capacity by ELONA.
Among the clones tested, two (clone 2 and clone 9) bound to immobilized ABA with a similar or higher signal compared to the SEL7 aptamer population.
Secondary structure of the anti-ABA aptamers
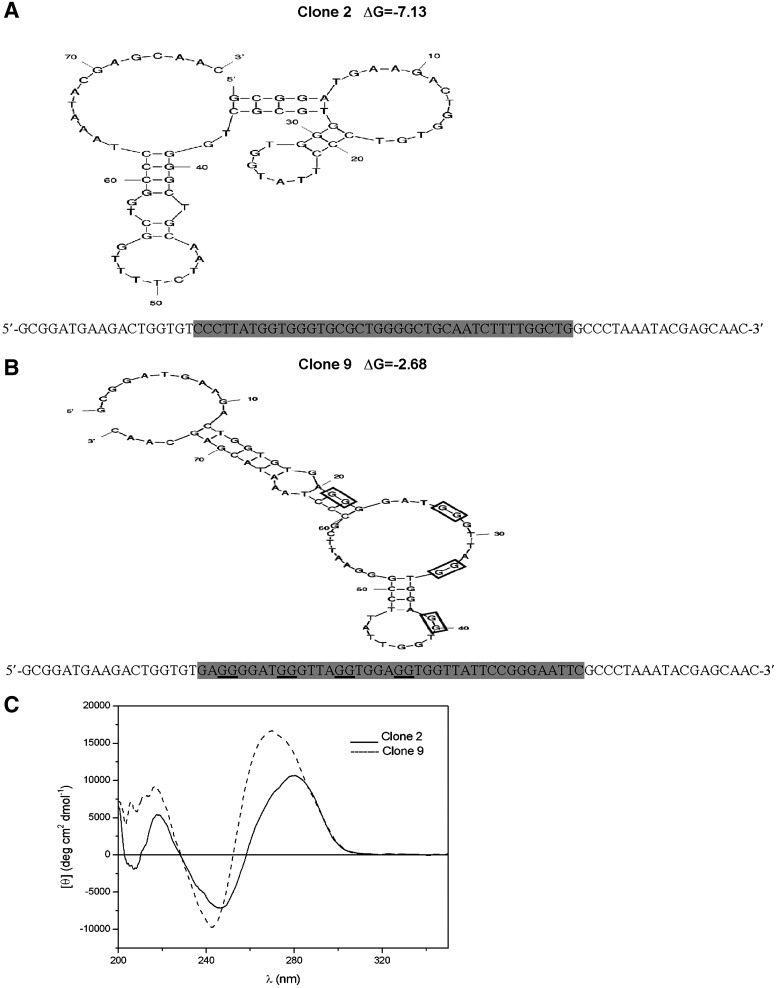
The nucleotide sequence of the positive clones and the respective secondary structures with the lowest free energy (ΔG) of folding (as predicted using mfold) are shown in Fig. 3A, B. These secondary structures were obtained with the free energy minimization algorithm (ZUKER, 2003), which generates several possible structures for each aptamer, with progressively increasing ΔG values (SANTALUCIA, 1998) (i.e., decreasing stability: thus, the structures shown in Fig. 3A, B cannot be predicted as the real ones).
FIG. 3.
Sequence and secondary structure of ABA-binding subclones from the SEL7 population. Predicted structures of clone 2 (A) and clone 9 (B). Structures and free energy values (ΔG, in kcal/mol) were obtained using mfold as described in Materials and Methods. The 40-nucleotide variable region of each aptamer (highlighted in gray) is flanked by two 18-nt constant primer binding regions. The underlined G-doublets in clone number 9 are those with the highest probability (G-score) of participating to the G-quadruplex formation based on the algorithm used by the QGRS Mapper software. (C) The single-stranded (ss) DNA aptamers (10 μM) were thermally denatured and then cooled to 20°C before obtaining the circular dichroism (CD) spectra: shown is the average of three scans from 200 to 350 nm at a rate of 20 nm per minute with a step resolution of 0.2 nm, a time constant of 2 seconds, and a bandwidth of 2.0 nm. Spectra were corrected by subtracting the appropriate buffer background and normalized to mean residue ellipticity.
Sequence alignment with the program ClustalW did not detect conserved motifs among clones 2 and 9. However, the two sequences show a high percentage of guanine nucleotides (clone 2: 37.5%; clone 9: 47.5%), which are spaced in a semiregular pattern as G doublets or quadruplets, particularly in clone 9. CD experiments were performed to investigate the secondary structure of aptamers. It has been reported that the CD spectrum of a typical parallel G-quadruplex structure has a positive peak at 265 nm and at 210 nm and a negative peak near 240 nm, and that of a typical antiparallel G-quadruplex shows a positive peak near 295 nm and a negative peak close to 265 nm (Paramasivan et al., 2007), while that of a B-form DNA structure has a positive peak between 270 and 280 nm and at 220 nm, and a negative peak at 245 nm and at 210 nm (Rajendran and Nair, 2006; Kypr et al., 2009).
The CD spectra of clone 2 and of clone 9 after thermal denaturation and renaturation are shown in Fig. 3C. Clone 9 shows a negative peak at 240 nm and two positive peaks at 220 and 270 nm, consistent with a parallel G-quadruplex [Paramasivan et al., 2007]. The CD spectrum of clone 2 shows two negative peaks at 210 nm and 245 nm and two positive peaks at 220 and 280 nm; this spectrum is consistent with B-form DNA and indicates that clone 2 is unlikely to form extensive G-quadruplexes (Kypr et al., 2009, Lai and DeStefano, 2012). CD spectra were unchanged upon incubation of both clones for 5 days at room temperature (not shown).
Determination of the dissociation constants (Kd) of the cloned aptamers
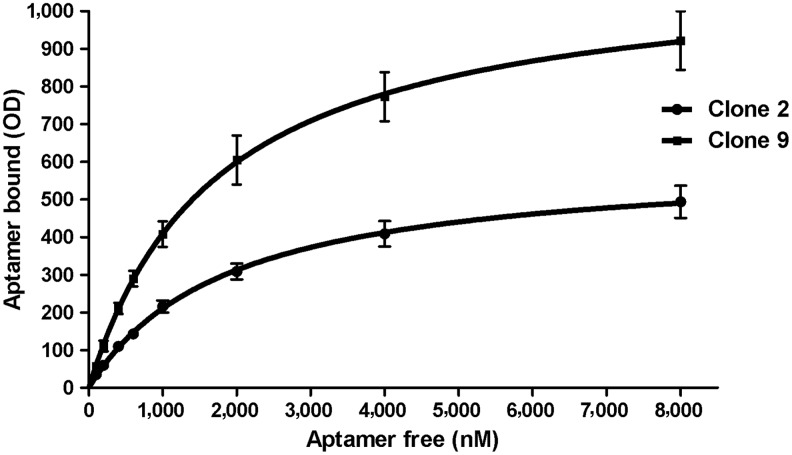
Increasing concentrations (0.1–8 μM) of each digoxigenin-labeled aptamer clone derived from the SEL7 population were incubated with a constant concentration of bio-ABA (6 μM), immobilized on magnetic beads. Fig. 4 shows the nonlinear regression analysis of the results from these binding experiments. The dissociation constants (Kd), as calculated by nonlinear regression analysis with GraphPad v5, were in the low micromolar range: 1.71±0.26 μM and 1.56±0.09 μM for clone 2 and clone 9, respectively.
FIG. 4.
Saturation binding of selected aptamer clones on immobilized bio-ABA. Binding of clone 2 and of clone 9 to bio-ABA was assessed by enzyme-linked oligonucleotide assay (ELONA), as described under Materials and Methods. A fixed concentration (6 μM) of bio-ABA immobilized on streptavidin-coated magnetic beads was incubated with increasing concentrations (0.1–8 μM) of digoxigenin-labeled aptamers. Anti-digoxigenin-HRP antibodies were then added and revealed with ABTS® solution at 405 nm. All experiments were performed in triplicate and each point is the average of three determinations. R2 values were 0.9997 for clone 2 and 0.9999 for clone 9.
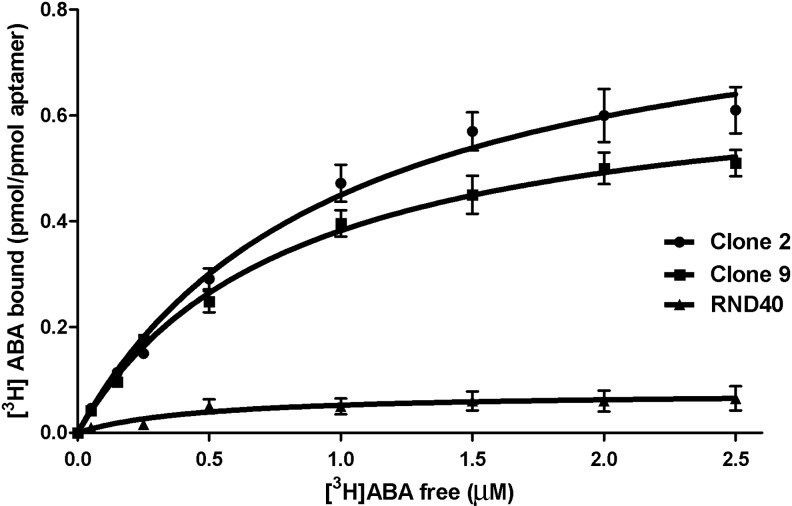
To verify whether the cloned aptamers also bound free, unmodified ABA, clone 2 and clone 9 (0.06 μM) were incubated with unbound 3[H]ABA (0.05–2.5μM). Controls included [3H]-ABA incubated with aptamers in the presence of excess unlabeled ABA (nonspecific binding), or incubated with the RND40 library (negative control). After subtraction of the nonspecific binding, experimental values were interpolated by nonlinear regression analysis (Fig. 5). The Kd values obtained for clone 2 (0.98±0.14 μM) and for clone 9 (0.80±0.07 μM), were similar to, or slightly lower than, respectively, those obtained on immobilized ABA. The RND40 library showed a markedly lower binding capacity than the selected aptamer clones. Thus, although the aptamers were initially selected for their ability to bind immobilized BioABA, they were also able to bind free, unmodified ABA with a similar affinity.
FIG. 5.
Equilibrium binding of ssDNA aptamers to free [3H]-ABA. Equilibrium binding assays were performed by filtration on 3-kDa mol weight cutoff filters, as described in the Methods section. Specific binding was calculated by subtracting the value of the nonspecific binding from each experimental point. Dissociation constants (Kd) were calculated using nonlinear regression analysis. Each point is the mean of 3 determinations. R2 values were 0.9931 for clone 2 and 0.9969 for clone 9.
To verify whether the cloned aptamers were able to bind ABA in a biological specimen, 6 nCi [3H]-ABA was added as a tracer to 200 μL human plasma containing 1 nM ABA, which was then incubated without (control) or with 0.5 μM clone 9 for 1 hour at 22°C. Samples were then loaded onto a silica-membrane-based DNA-binding column and the radioactivity of the flow-through was measured. In the control incubation, the tracer radioactivity was found entirely in the flow-through; conversely, in the presence of clone 9, 40±5% (mean±standard deviation from 4 experiments) of the tracer radioactivity was retained on the column and could be eluted with deionized water.
Specificity of the cloned aptamers for ABA
Bio-ABA used for aptamer selection was synthesized from the racemic mixture of the 2 ABA enantiomers (+)2-cis,4-trans ABA and (–)2-cis,4-trans ABA (Yamazaki et al., 2003). Thus, we investigated whether the cloned aptamers could distinguish between the 2 enantiomers and whether they could bind to structurally related molecules, such as the ABA isomer (±)2-trans,4-trans ABA or the ABA analog, 2′α, 3′α-dihydro-2′α, 3′α–epoxy-abscisic acid (analog 10), which has been recently shown to bind to the human ABA receptor LANCL2 (Grozio et al., 2011; Sturla L. et al., 2011). To this end, affinity elution assays were performed, where the cloned aptamers were first bound to bio-ABA-coated magnetic beads and then challenged with one of the target molecules in solution (600 μM). Upon binding to the target molecule, the aptamer dissociates from the immobilized bio-ABA and is released into the supernatant. The aptamer remaining on the beads is then revealed with the anti-digoxigenin-HRP antibody.
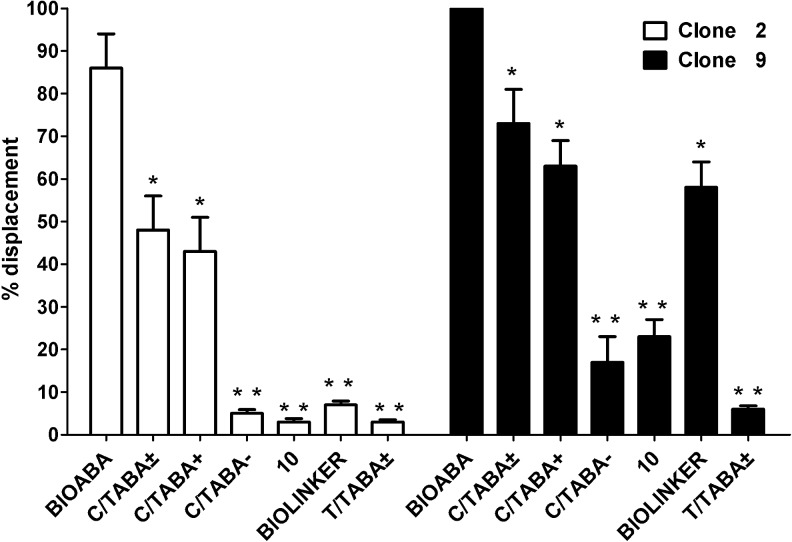
Excess free bio-ABA induced the release of clone 2 (86%) and of clone 9 (100%) from immobilized bio-ABA (Fig. 6). Clone 9 was also dissociated from bio-ABA-coated beads by excess free unconjugated, racemic ABA (73%), by (+)2-cis,4-trans ABA (63%) and by the biolinker (58%); conversely, (–)2-cis, 4-trans ABA, (±)2-trans,4-trans ABA or the ABA analog 10 did not induce a significant dissociation of clone 9 from immobilized bio-ABA (<20%). Clone 2 was released from bio-ABA-coated beads in the presence of excess racemic 2-cis,4-trans ABA (48%) or of the (+) enantiomer (43%), while none of the other compounds could displace the aptamer bound to the beads. Neither clone 2 nor clone 9 were displaced by free biotin (not shown).
FIG. 6.
Affinity elution assays on the cloned aptamers. The indicated cloned, digoxigenin-labeled aptamers were first incubated with bio-ABA immobilized on magnetic beads (0.6 nmol/well), then challenged without (control) or with an excess (60 nmol/well) of the indicated target molecules in solution. The amount of aptamer retained on the beads was determined by immunoenzymatic reaction (see Methods). Results are expressed as the ratio between the absorbance at 405 nm in the presence and in the absence of the target molecules. [bio-ABA: biotinylated abscisic acid; C/TABA±: (±)2-cis,4-trans-ABA; C/TABA+: (+)2-cis,4-trans-ABA; C/TABA-: (–)2-cis,4-trans-ABA; analog 10: 2′α, 3′α-dihydro-2′α, 3′α–epoxy abscisic acid; bio-linker: (+)-biotin-ɛ-aminocaproyl hydrazide; ±T/TABA: (±)2-trans,4-trans ABA]. *p<0.05 compared with bio-ABA; **p<0.005 compared with bio-ABA, by t-test.
These results indicate that both aptamer clones preferentially bind the (+) enantiomer of 2-cis,4-trans ABA and that they do not recognize ABA-related structures [(–)2-cis, 4-trans ABA, (±)2-trans,4-trans ABA or the ABA analog 10]. Clone 9, but not clone 2, apparently recognizes a part of the bio-ABA molecule comprising the biolinker, but not biotin, as both 2-cis,4-trans ABA and the biolinker are similarly effective in competing for immobilized bio-ABA.
Clone numbers 2 and 9 prevent the ABA-induced increase of the [cAMP]i in human granulocytes
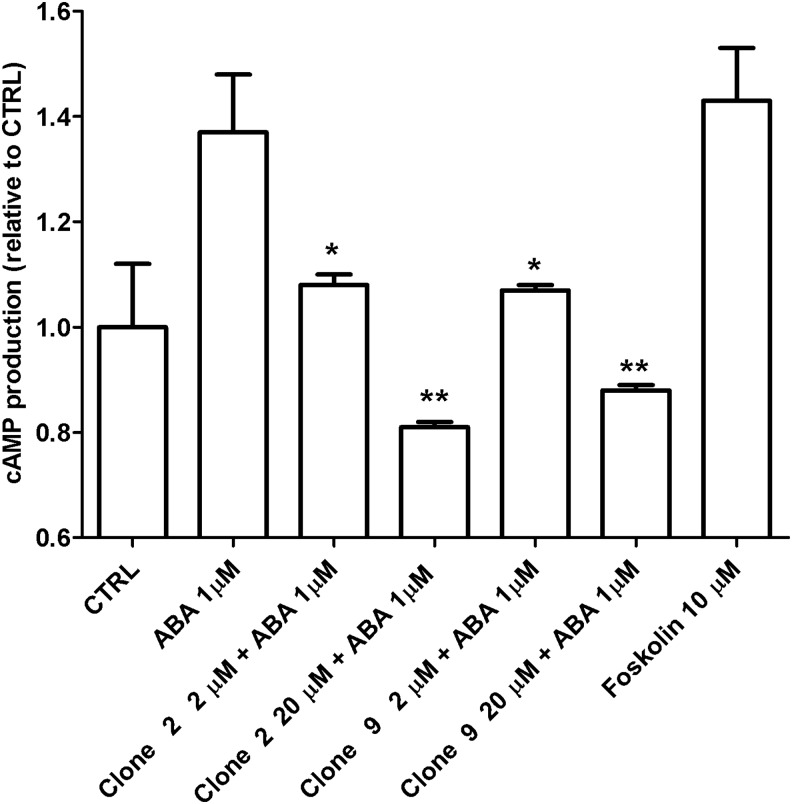
ABA induces an increase of the [cAMP]i in human granulocytes, resulting in their functional activation (Bruzzone S. et al., 2007). To verify whether clones 2 and 9 could prevent exogenous ABA from inducing an increase of the [cAMP]i, granulocytes were incubated in the presence of 1 μM ABA, with or without 2- or 20-μM aptamers. As shown in Fig. 7, both aptamers prevented the ABA-induced increase of the [cAMP]i.
FIG. 7.
Effect of clones 2 and 9 on the intracellular cAMP concentration ([cAMP]i) in human granulocytes. Human granulocytes were incubated for 1 minute without (control) or with 1 μM ABA, without or with clone 2 or clone 9 at 2 or 20 μM. The [cAMP]i rise was calculated as the ratio between the [cAMP]i increase over control values in cells incubated with both ABA and aptamer, and the [cAMP]i increase over control values in cells incubated with ABA alone. Incubation of granulocytes with aptamer alone did not modify the [cAMP]i as compared to control values, at any of the concentrations tested. Forskolin was used as a positive control. Each condition was assayed in duplicate; results shown are the mean±standard deviation from 3 experiments. *p<0.05 compared with ABA 1 μM; **p<0.005 compared with ABA 1 μM, by t-test.
Discussion
Immunoassays (Daie and Wyse, 1982) and analytical methods based on HPLC and HPLC- tandem mass spectrometry have been reported for determination of ABA levels in mammalian cells (Zocchi et al., 2001; Bruzzone et al., 2007). Aptamers are chemically synthesized nucleic acid ligands that bind to their targets with high affinity and specificity. In the present study, an ABA-binding ssDNA aptamer was selected. The binding properties of ssDNA aptamers are similar to those of RNA aptamers, but DNA aptamers are more resistant to nuclease degradation then RNA aptamers, and are therefore stable in biological fluids without modifications, at least for short periods of time (Klussmann, 2006). The SELEX procedure was performed with 1015 molecules from an ssDNA library containing 40 randomized nucleotides and two 18 nucleotides constant regions on each side, for PCR amplification. The SELEX procedure comprised seven iterative rounds of incubation, partitioning, elution, and amplification. To separate the bound and unbound sequences, the target must be immobilized on a solid surface. Several methods for immobilization of the SELEX targets have been reported such as affinity chromatography matrices like agarose and Sephadex, magnetic beads, and affinity capillary electrophoresis columns (GOPINATH, 2007). We chose magnetic beads because of their easy handling and densely coated surface area. Moreover, use of magnetic beads allows the utilization of small sample volumes, reducing procedural costs (Bruno and Kiel, 2002). There are many methods for studying the binding affinity of aptamers, such as surface plasmon resonance, quartz crystal microbalance, and enzyme-linked oligonucleotide assay (ELONA) (Wendy and Yingfu, 2008). Here we used streptavidin-coated 96-well plates and magnetic beads coated with bio-ABA. Anti-ABA aptamers were labeled with 5′-digoxigenin to detect binding.
The selected aptamers did not share a common sequence within the variable part of their structure. However, clone2 and clone 9 showed a high percentage of guanines (clone 2: 37.5%; clone 9: 47.5%), which may fold into a structure called quadruplex (Smith and Feigon, 1992; WILLIAMSON, 1993), as suggested by the mfold and QGRS Mapper softwares. The G-quadruplex can be stabilized by monovalent cations such as, K+ and Na+, both present in the selection and binding buffer used in this study. CD analysis confirmed presence of a parallel G-quadruplex structure in clone 9, but not in clone 2. Thus, the contribution of a G-quadruplex structure to the overall secondary structure of clone 2, if present, is minor.
Depending on the sequence of each aptamer, the constant regions used for PCR amplification may or may not be important for the final structure of the molecule. Although it is known that sometimes the sequences corresponding to the primers can be deleted totally or partially, the analysis of the secondary structures obtained using mfold software shows that the conserved regions of clone 2 and clone 9 probably contribute to stabilize the secondary structure of the aptamers.
The dissociation constants (Kd) of aptamer numbers 2 and 9 with bio-ABA were similar and were 1.7±0.2 μM and 1.6±0.1 μM, respectively. Other aptamers, selected for small molecules, showed a Kd around this value, for example aptamers selected for amino acids such as citrulline (Kd: 0.3μM) (FAMULOK, 1994) and arginine (Kd: 65μM) (Geiger et al., 1996) and those binding ATP (Kd: 6μM) (Huizenga and Szostak, 1995) or xanthine (Kd: 3.3μM) (Kiga et al., 1998). The micromolar range of the Kd of the anti-ABA aptamers is probably due to the paucity of ABA sites required to establish non-covalent bonds with the aptamers and to the fact that ABA is a weak acid (pKa: 4.8), causing the dissociation of the ABA carboxylic group in the selection and in the binding buffer (pH 7.4). The negatively charged ABA may have generated a Coulomb repulsion with the negatively charged phosphate chain of the aptamers. Aptamers 2 and 9 were also able to bind free (unbound) ABA, with Kd values of 0.98±0.14 μM and 0.80±0.07 μM, respectively. These values are similar to those calculated for bio-ABA; therefore, biotinylation of ABA did not affect the capacity of the selected aptamers to bind to the naturally occurring hormone, present in plants and in human plasma. We used affinity elution assays for the determination of aptamer selectivity. Affinity elution assays have been used for simple and fast screening of several organic molecules (Mann et al., 2005; Reinemann et al., 2009). Aptamers 2 and 9 showed a similar specificity profile for the different ABA isomers and enantiomers. Both aptamers were able to bind to (+)2-cis,4-trans-ABA, but not to (–)2-cis,4-trans-ABA, neither (±)2-trans,4-trans ABA or the ABA analog, 2′α, 3′α-dihydro-2′α, 3′α–epoxy abscisic acid. Thus, aptamers 2 and 9 are selective for a single ABA enantiomer, (+)2-cis,4-trans-ABA, the major form of ABA present in plants. Selectivity of aptamers for a specific isomer of the target molecule has been already used for the development of analytic assays, capable of discriminating between isomeric forms of the same molecule (Lin et al., 2009). Aptamer 9 also bound to the biolinker, (+)-biotin-ɛ-aminocaproyl hydrazide, indicating that counter-selection with the linker alone during the SELEX procedure did not remove all the linker-specific sequences. Nevertheless, the capacity of aptamer 9 to bind the linker, but not unmodified biotin, should not affect its utilization for the development of ABA-detecting assays, because (+)-biotin-ɛ-aminocaproyl hydrazide is not a likely contaminant of biological samples. Although there are already commercially available antibodies against ABA, with an affinity constant in the order of 10 nM (Walker-Simmons et al., 1991), the use of aptamers for the development of quantitative detection assays for ABA could be advantageous, reducing costs and production times, preventing variability between different lots and avoiding the use of laboratory animals. Also, aptamers have a higher thermal stability than proteins and could be used over several denaturation and renaturation steps. The recent discovery of ABA in animal cells and of its effects on inflammation and glucose metabolism (Bruzzone et al., 2007; Bruzzone et al., 2008; Magnone et al., 2009; Bruzzone et al., 2012) should increase the demand for rapid and inexpensive assays for measurement of ABA in animal samples. Finally, given the role of ABA as an endogenous activator of innate immune cells, ABA-binding aptamers may have anti-inflammatory properties by interfering with the autocrine and paracrine effects of ABA on inflammatory cells. Indeed, both aptamer clones were able to prevent the ABA-induced increase of the [cAMP]i in human granulocytes (Fig. 7), an essential step of the ABA-signaling pathway in inflammatory cells.
Acknowledgments
This work was supported in part by the Italian Ministry of Education, University, and Scientific Research, the University of Genova, the Fondazione CARIGE, the Compagnia di S. Paolo, and Interuniversity Consortium for Biotechnology (C.I.B).
Author Disclosure Statement
No competing financial interests exist.
References
- BAKER B.R. LAI R.Y. WOOD M.S. DOCTOR E.H. HEEGER A.J. PLAXCO K.W. An electronic, aptamer-based small-molecule sensor for the rapid, label-free detection of cocaine in adulterated samples and biological fluids. J. Am. Chem. Soc. 2006;128:3138–3139. doi: 10.1021/ja056957p. [DOI] [PubMed] [Google Scholar]
- BANGERTH F. Changes in the ratio of cis-trans to trans-trans abscisic acid during ripening of apple fruits. Planta. 1982;155:199–203. doi: 10.1007/BF00392716. [DOI] [PubMed] [Google Scholar]
- BODRATO N. FRANCO L. FRESIA C. GUIDA L. USAI C. SALIS A. MORESCHI I. FERRARIS C. VERDERIO C. BASILE G., et al. Abscisic acid activates the murine microglial cell line N9 through the second messenger cyclic ADP-ribose. J. Biol. Chem. 2009;284:14777–14787. doi: 10.1074/jbc.M802604200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRUNO J.G. KIEL J.L. Use of magnetic beads in selection and detection of biotoxin aptamers by electrochemiluminescence and enzymatic methods. Biotechniques. 2002;32:178–180. doi: 10.2144/02321dd04. [DOI] [PubMed] [Google Scholar]
- BRUZZONE S. AMERI P. BRIATORE L. MANNINO E. BASILE G. ANDRAGHETTI G. GROZIO A. MAGNONE M. GUIDA L. SCARFÌ S., et al. The plant hormone abscisic acid increases in human plasma after hyperglycemia and stimulates glucose consumption by adipocytes and myoblasts. FASEB J. 2012;26:1251–1260. doi: 10.1096/fj.11-190140. [DOI] [PubMed] [Google Scholar]
- BRUZZONE S. BODRATO N. USAI C. GUIDA L. MORESCHI I. NANO R. ANTONIOLI B. FRUSCIONE F. MAGNONE M. SCARFÌ S., et al. Abscisic acid is an endogenous stimulator of insulin release from human pancreatic islets with cyclic ADP-ribose as second messenger. J. Biol. Chem. 2008;283:32188–32197. doi: 10.1074/jbc.M802603200. [DOI] [PubMed] [Google Scholar]
- BRUZZONE S. MORESCHI I. USAI C. GUIDA L. DAMONTE G. SALIS A. SCARFÌ S. MILLO E. DE FLORA A. ZOCCHI E. Abscisic acid is an endogenous cytokine in human granulocytes with cyclic ADP-ribose as second messenger. Proc. Natl. Acad. Sci. U. S. A. 2007;104:5759–5764. doi: 10.1073/pnas.0609379104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CONNELL G.J. ILLANGESEKARE M. YARUS M. Three small ribooligonucleotides with specific arginine sites. Biochemistry. 1993;32:5497–54502. doi: 10.1021/bi00072a002. [DOI] [PubMed] [Google Scholar]
- DAIE J. WYSE R. Adaptation of the enzyme-linked immunosorbent assay (ELISA) to the quantitative analysis of abscisic acid. Anal. Biochem. 1982;119:365–371. doi: 10.1016/0003-2697(82)90599-1. [DOI] [PubMed] [Google Scholar]
- ELLINGTON A.D. SZOSTAK J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- FAMULOK M. Molecular recognition of amino acids by RNA-aptamers – an L-citrulline binding RNA motif and its evolution into an l-arginine binder. J. Am. Chem. Soc. 1994;116:1698–1706. [Google Scholar]
- FLOEGE J. OSTENDORF T. JANSSEN U. BURG M. RADEKE H.H. VARGEESE C. GILL S.C. GREEN L.S. JANJIC’ N. Novel approach to specific growth factor inhibition in vivo: antagonism of platelet-derived growth factor in glomerulonephritis by aptamers. Am. J. Pathol. 1999;154:169–179. doi: 10.1016/S0002-9440(10)65263-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GEIGER A. BURGSTALLER P. VON DER ELTZ H. ROEDER M. FAMULOK A. RNA Aptamers that bind l-arginine with sub-micromolar dissociation constants and high enantioselectivity. Nucleic Acids Res. 1996;24:1029–1036. doi: 10.1093/nar/24.6.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOPINATH S.C. Methods developed for SELEX. Anal. Bioanal. Chem. 2007;387:171–182. doi: 10.1007/s00216-006-0826-2. [DOI] [PubMed] [Google Scholar]
- GROZIO A. MILLO E. GUIDA L. VIGLIAROLO T. BELLOTTI M. SALIS A. FRESIA C. STURLA L. MAGNONE M. GALATINI A., et al. Functional characterization of a synthetic abscisic acid analog with anti-inflammatory activity on human granulocytes and monocytes. Biochem. Biophys. Res. Commun. 2011;415:696–701. doi: 10.1016/j.bbrc.2011.10.143. [DOI] [PubMed] [Google Scholar]
- HERMANN T. PATEL D.J. Adaptive recognition by nucleic acid aptamers. Science. 2000;287:820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- HUANG C.C. HUANG Y.F. CAO Z. TAN W. CHANG H.T. Aptamer-modified gold nanoparticles for colorimetric determination of platelet-derived growth factors and their receptors. Anal. Chem. 2005;77:5735–5741. doi: 10.1021/ac050957q. [DOI] [PubMed] [Google Scholar]
- HUIZENGA D.E. SZOSTAK J.W. A DNA aptamer that binds adenosine and ATP. Biochemistry. 1995;34:656–665. doi: 10.1021/bi00002a033. [DOI] [PubMed] [Google Scholar]
- JENISON R.D. GILL S.C. PARDI A. POLISKY B. High-resolution molecular discrimination by RNA. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- KIGA D. FUTAMURA Y. SAKAMOTO K. YOKOYAMA S. An RNA aptamer to the xanthine/guanine base with a distinctive mode of purine recognition. Nucleic Acids Res. 1998;26:1755–1760. doi: 10.1093/nar/26.7.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KLUSSMANN S. The Aptamer Handbook: Functional Oligonucleotides and their Applications. Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim Germany; 2006. Aptamers to Small Molecules; pp. 94–115. [Google Scholar]
- KYPR J. KEJNOVSKÀ I. RENČIUK D. VORLÌČKOVÁ M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009;37:1713–1725. doi: 10.1093/nar/gkp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAI Y. DESTEFANO J. J. DNA aptamer to human immunodeficiency virus reverse transcriptase selected by a primer-free SELEX method: characterization and comparison with other aptamers. Nucleis Acid Ther. 2012;22:162–176. doi: 10.1089/nat.2011.0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAI R.Y. PLAXCO K.W. HEEGER A. Aptamer-based electrochemical detection of picomolar platelet-derived growth factor directly in blood serum. J. Anal. Chem. 2007;78:229–233. doi: 10.1021/ac061592s. [DOI] [PubMed] [Google Scholar]
- LIN P.H. TONG S.J. LOUIS S.R. CHANG Y. CHEN W.Y. Thermodynamic basis of chiral recognition in a DNA aptamer. Phys Chem. Chem. Phys. 2009;11:9744–9750. doi: 10.1039/b907763d. [DOI] [PubMed] [Google Scholar]
- MAGNONE M. BRUZZONE S. GUIDA L. DAMONTE G. MILLO E. SCARFÌ S. USAI C. STURLA L. PALOMBO D. DE FLORA A. ZOCCHI E. Abscisic acid released by human monocytes activates monocytes and vascular smooth muscle cell responses involved in atherogenesis. J. Biol. Chem. 2009;284:17808–17818. doi: 10.1074/jbc.M809546200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MANN D. REINEMANN C. STOLTENBURG R. STREHLITZ B. In vitro selection of DNA aptamers binding ethanolamine. Biochem. Biophys. Res. Commun. 2005;338:1928–1934. doi: 10.1016/j.bbrc.2005.10.172. [DOI] [PubMed] [Google Scholar]
- MORRIS K.N. JENSEN K.B. JULIN C.M. WEIL M. GOLD L. High affinity ligands from in vitro selection: complex targets. Proc. Natl. Acad. Sci. USA. 1998;95:2902–2907. doi: 10.1073/pnas.95.6.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NAMBARA E. MARION P. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005;56:165–185. doi: 10.1146/annurev.arplant.56.032604.144046. [DOI] [PubMed] [Google Scholar]
- PAN W. CRAVEN R.C. QIU Q. WILSON C.B. WILLS J.W. GOLOVINE S. WANG J.F. Isolation of virus neutralizing RNAs from a large pool of random sequences. Proc. Natl. Acad. Sci. U. S. A. 1995;92:11509–11513. doi: 10.1073/pnas.92.25.11509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PARAMASIVAN S. RUJAN I. BOLTON P.H. Circular dichroism of quadruplex DNAs: applications to structure, cation effect and ligand binding. Methods. 2007;43:324–331. doi: 10.1016/j.ymeth.2007.02.009. [DOI] [PubMed] [Google Scholar]
- RAJENDRAN A. NAIR B.U. Unprecedented dual binding behaviour of acridine group of dye: a combined experimental and theoretical investigation for the development of anticancer chemotherapeutic agents. Biochim. Biophys. Acta. 2006;1760:1794–1801. doi: 10.1016/j.bbagen.2006.08.011. [DOI] [PubMed] [Google Scholar]
- REINEMANN C. STOLTENBURG R. STREHLITZ B. Investigations on the specificity of DNA aptamers binding to ethanolamine. Anal. Chem. 2009;81:3973–3978. doi: 10.1021/ac900305y. [DOI] [PubMed] [Google Scholar]
- SANTALUCIA J., Jr A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. U. S. A. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCARFÌ S. FERRARIS C. FRUSCIONE F. FRESIA C. GUIDA L. BRUZZONE S. USAI C. PARODI A. MILLO E. SALIS A., et al. Cyclic ADP-ribose-mediated expansion and stimulation of human mesenchymal stem cells by the plant hormone abscisic acid. Stem Cells. 2008;26:2855–2864. doi: 10.1634/stemcells.2008-0488. [DOI] [PubMed] [Google Scholar]
- SCARFÌ S. FRESIA C. FERRARIS C. BRUZZONE S. FRUSCIONE F. USAI C. BENVENUTO F. MAGNONE M. PODESTÀ M. STURLA L., et al. The plant hormone abscisic acid stimulates the proliferation of human hemopoietic progenitors through the second messenger cyclic ADP-ribose. Stem Cells. 2009;27:2469–2477. doi: 10.1002/stem.173. [DOI] [PubMed] [Google Scholar]
- SMITH F.W. FEIGON J. Quadruplex structure of Oxytricha telomeric DNA oligonucleotides. Nature. 1992;356:164–168. doi: 10.1038/356164a0. [DOI] [PubMed] [Google Scholar]
- STOJANOVIC M.N. PRADA P.D. LANDRY D.W. Aptamer-based folding fluorescent sensor for cocaine. J. Am. Chem. Soc. 2001;123:4928–4931. doi: 10.1021/ja0038171. [DOI] [PubMed] [Google Scholar]
- STOLTENBURG R. REINEMANN C. STREHLITZ B. FluMag-SELEX as an advantageous method for DNA aptamer selection. Anal. Bioanal. Chem. 2005;383:83–91. doi: 10.1007/s00216-005-3388-9. [DOI] [PubMed] [Google Scholar]
- STURLA L. FRESIA C. GUIDA L. GROZIO A. VIGLIAROLO T. MANNINO E. MILLO E. BAGNASCO L. BRUZZONE S. DE FLORA A. ZOCCHI E. Binding of abscisic acid to human LANCL2. Biochem. Biophys. Res. Commun. 2011;415:390–395. doi: 10.1016/j.bbrc.2011.10.079. [DOI] [PubMed] [Google Scholar]
- TUERK C. GOLD L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- WALKER-SIMMONS M.K. REANEY M.J.T. QUARRIE S.A. PERATA P. VERNIERI P. ABRAMS S.R. Monoclonal antibody recognition of abscisic acid analogs. Plant Physiol. 1991;95:46–51. doi: 10.1104/pp.95.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WENDY M. YINGFU L. Recent progress in nucleic acid aptamer-based biosensors and bioassays. Sensors. 2008;8:7050–7084. doi: 10.3390/s8117050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WILLIAMSON J.R. Guanine quartets. Curr. Opin. Struct. Biol. 1993;3:357. [Google Scholar]
- XUE-XUAN X. HONG-BO S. YUAN-YUAN M. GANG X. JUN-NA S. DONG-GANG G. CHENG-JIANG R. Biotechnological implications from abscisic acid (ABA) roles in cold stress and leaf senescence as an important signal for improving plant sustainable survival under abiotic-stressed conditions. Crit. Rev. Biotechnol. 2010;30:222–230. doi: 10.3109/07388551.2010.487186. [DOI] [PubMed] [Google Scholar]
- YAMAZAKI D. YOSHIDA S. ASAMI T. KUCHITSU K. Visualization of abscisic acid-perception sites on the plasma membrane of stomatal guard cells. Plant J. 2003;35:129–139. doi: 10.1046/j.1365-313x.2003.01782.x. [DOI] [PubMed] [Google Scholar]
- ZHANG Y. ZHOU H. OU-YANG Z.-C. Stretching single-stranded DNA: interplay of electrostatic, base-pairing, and base-pair stacking interactions. Biophys. J. 2001;81:1133–1143. doi: 10.1016/S0006-3495(01)75770-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZOCCHI E. CARPANETO A. CERRANO C. BAVESTRELLO G. GIOVINE M. BRUZZONE S. GUIDA L. FRANCO L. USAI C. The temperature-signaling cascade in sponges involves a heat-gated cation channel, abscisic acid, and cyclic ADP-ribose. Proc. Natl. Acad. Sci. U. S. A. 2001;98:14859–14864. doi: 10.1073/pnas.261448698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZUKER M. Mfold web server for nucleic acid folding and hybridization prediction. Nucl. Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]