Abstract
The human noncoding RNA gene RGM249 has been shown to regulate the degree of cancer cell differentiation. In this study, we investigated the effects of 3 microRNA-like molecules digested from RGM249 on the loss of malignant properties in cancer cells in immunodeficient KSN/Slc mice. We utilized small interfering RNAs (siRNAs) alone or in combination with a cationized drug delivery system (DDS) consisting of atelocollagen or gelatin hydrogel microspheres. The results demonstrated growth inhibition and apoptosis and the inhibition of both neovascularization and metastasis, indicating that the DDSs effectively infiltrated the majority of tumor cells in vivo. Systemic administration of the 3 siRNAs inhibited the metastatic ability of malignant cells. Cotransfection of these siRNAs exerted a regulatory effect upon the genes involved in differentiation, pluripotency, and proliferation in cancer cells. These results suggest that RGM249-derived oligonucleotides may be involved in the regulation of metastasis, proliferation, and differentiation in vivo, and that the tested siRNAs may therefore represent a new anticancer therapeutic approach.
Introduction
Novel mechanisms underlying gene regulation have been identified since the initial discovery of microRNAs (miRNAs) (Lau et al., 2001). miRNAs are endogeneous, small, noncoding RNA (ncRNA) molecules 18–25 nucleotides in length that are transcribed by RNA polymerase 2 and function as post-transcriptional gene regulators (Lagos-Quintana et al., 2001; MELTZER, 2005). Mammalian miRNAs are involved in mediating cellular differentiation and reprogramming, and are considered to play a crucial role in the initiation and progression of human cancer (Chen et al., 2004; Thum et al., 2007; Viswanathan et al., 2008). The alteration of miRNA expression levels influences tumor growth by modulating the functional expression of the targeted genes involved in the regulation of tumor cell apoptosis or proliferation (MELTZER, 2005). miRNAs can act as oncogenes (oncomiR) and/or tumor suppressor genes during tumorigenesis. For example, the Bcl2 oncogene (B-cell CLL/lymphoma 2) is targeted by miR-15a and miR-16; the PTEN (phosphatase and tensin homolog) tumor suppressor by miR-21; HOXD10 (homeobox D10) by miR-10b; Pak [p21 protein (Cdc42/Rac)-activated kinase] by miR-7; MYOD1 (myogenic differentiation 1); estrogen receptor genes by miR-206; and HER2 (ERBB2 gene) by miR-125a (Cimmino et al., 2005). OncomiRs have been investigated in different tumor types, including melanomas, hepatomas, brain tumors, and breast tumors (Chan et al., 2005; Ma et al., 2007; Meng et al., 2007; Satzger et al., 2010).
We previously identified an RNA gene (RGM249) that may indirectly regulate hTERT (human telomerase reverse transcriptase) expression via the induction of double-stranded (ds), small interfering (si), and short hairpin (sh) RNAs. The regulation of this gene may be involved in cell development and differentiation, anti-inflammatory effects that maintain telomere length prior to DNA damage, and cell growth in undifferentiated cancers. RGM249 is strongly expressed in poorly differentiated or undifferentiated malignant tumor cell lines and may play a role in carcinogenesis or the maintenance of low differentiation levels (www.ncbi.nlm.nih.gov/nuccore/EF433558) (Miura et al., 2009). We have previously used siRNAs to study the roles of 3 small RNAs derived from the ncRNA in vivo and investigated whether the prevention of RGM249-derived small RNA binding to the 3′ untranslated regions of their target genes affected their metastatic or proliferative abilities during in vivo tumor growth (Doench et al., 2003; Doench and Sharp, 2004; Liu et al., 2005). We have also investigated the safety, efficacy, and specificity of drug delivery systems (DDSs) by utilizing gelatin hydrogel microspheres or atelocollagen for subcutaneous (s.c.) injection and spermine-pullulan or atelocollagen for intravenous (i.v.) administration (Jo et al., 2006; Kushibiki et al., 2006; Mu et al., 2009; Kimura and Tabata, 2010; Takeshita et al., 2010).
Since 2004, when the first human clinical trials were conducted using a direct intraocular injection of synthetic siRNAs on patients with blinding choroidal neovascularization, other clinical trials have been initiated (Castanotto and Rossi, 2009), and early clinical data have been reported (DeVincenzo et al., 2008; Leachman et al., 2010). However, previous animal studies supported the hypothesis that siRNA functions via a mechanism involving RNA interference (RNAi) (Alvarez et al., 2009). We therefore aimed to begin to elucidate the physiological functions of miRNA-like molecules generated from RGM249 and their roles in carcinogenesis, differentiation, and pluripotency and investigate their potential utility for antitumor therapy or regenerative medicine in vivo.
Materials and Methods
Identification of oligonucleotides digested by Dicer and RNase 3
RGM249 was ligated into the pRNAT-U6.1/neo vector (GenScript USA Inc.). RGM249 mRNA generated by T7 RNA polymerase was digested using the Dicer enzyme (Genlantis) and/or RNase3 (NEB). miRNA was fractionated using the mirVANA miRNA isolation kit (Ambion Japan) and purified with the miRNA isolation kit with or without human anti-Ago2 beads (WAKO). Digested small RNAs were cloned using the miRCAT-microRNA cloning kit (Integrated DNA Technologies) and sequenced using the TOPO vector (Invitrogen). Secondary structures were predicted (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi), and the sequence homologies of the small RNAs were investigated using miRBase (www.mirbase.org).
Cell lines and immunodeficient mice
A human malignant melanoma cell line (HMV-1) and poorly differentiated hepatoma cells (HLF) with strong RGM249 mRNA expressions were obtained from Tohoku University and ATCC, respectively. A total of 1×107 cells were harvested and inoculated into mice either subcutaneously or intravenously. Six-week-old immunodeficient mice (CAnN Cg-Foxn1 BALB/c-nu) were inoculated for in vivo study by RGM249 transfection. Immunodeficient KSN/Slc mice were used to examine the anti-metastatic effects of the 3 small RNAs. Athymic mice were anesthetized by an intraperitoneal injection of 100 mg/kg Nembutal. All animals were housed and fed at the Japanese Association for Accreditation for Laboratory Animal Care–approved facilities, and animal research and handling were performed in strict conformance with federal Institutional Animal Care and Use Committee guidelines.
siRNA preparation and gene expression constructs
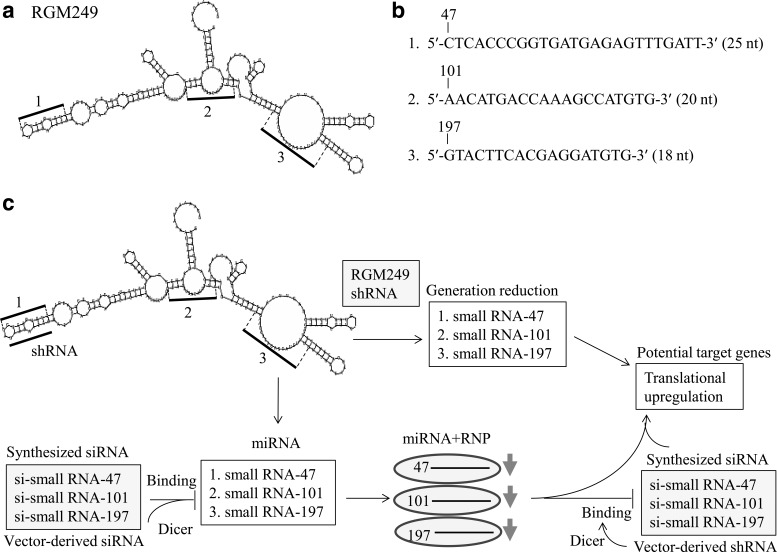
A total of 50 nM of synthesized siRNAs (corresponding to small RNA-47, RNA-101, and RNA-197) were transfected into HMV-1 cells, and the effects were assessed using the FuGENE HD transfection reagent (Roche Diagnostics). For the RGM249 shRNA-generating construct, a specific target sequence (small RNA-47) against RGM249 was chosen using the Stealth RNAi designer, and the shRNA-generating vector was created using BLOCK-it Inducible H1 RNAi Entry Vector (Invitrogen). A vector with 2 base deletions (mtRGM249 shRNA: m1 vector) was generated as a sequence control, and Lac shRNA was used as a mock control. The RGM249 shRNA sequence was as follows: 5′-caccgcagaataaggctagacaaagcgaactttgtctagccttattc tgc-3′. The nucleotides deleted in the m1 sequence are underlined. The sequences corresponding with the 3 siRNAs are shown in Fig. 1b. (47 siRNA-sense: 5′-cucacccggugaugagaguuugauu-3′; 101 siRNA-sense: 5′-aacaugaccaaagccauguguu-3′; and 197 siRNA-sense: 5′-guacuucacgaggauguguu-3′). Scrambled sequences as a negative control of RNAi were as follows: 5′-guugugauuaaugugcgcgaacucc-3′ for small RNA-47, 5′-guacaacgagucuaacacga-3′ for -101, 5′-aggugcguuauaugccga-3′ for -197 (https://www.genscript.com/ssl-bin/app/scramble). shRNA-generating constructs against the small RNAs derived from RGM249 mRNA were produced using pRNATin-H1.4/Lenti. siRNAs were synthesized using Invitrogen's siRNA designer. To obtain RGM249 mRNA, RGM249 was ligated into the pRNAT-U6.1/neo vector, and RGM249 mRNA was digested with Dicer. Digested small RNAs were cloned using the miRCAT-microRNA cloning kit. The sequence homologies of small RNAs were investigated using miRBase. A total of 50 nM of synthesized siRNAs (small RNA-47, RNA-101, and RNA-197) were transfected into HMV-1 cells, and their effects were assessed using the FuGENE HD transfection reagent. The secondary structures and sequences of the small RNAs are shown in Fig. 1a, b. Sufficient siRNAs were generated to achieve an inhibitory effect on miRNA processing (Fig. 1c).
FIG. 1.
RGM249, RGM249-derived small RNA, RGM249 shRNA, and siRNAs corresponding with the 3 small RNAs and the schematic of the strategy used in this study. (a) Secondary structure of the RNA gene RGM249 and the 3 internal sites (bold bars) at which the small RNAs were generated. (b) Sequences of the 3 small RNAs generated from RGM249. (c) Schematic of the small-RNA-silencing method used in this study. Short hairpin RNA (shRNA) with functional sequences against RGM249 and siRNAs against the 3 small RNAs derived from RGM249 led to similar translational upregulation. Small interfering RNAs (siRNAs) corresponding with the small RNAs were expected to function in 2 manners during microRNA (miRNA) processing. Arrows indicate expressive reduction.
Gene transfer procedures in vitro and in vivo
To determine the anti-cancer effects of RGM249-silencing molecules, 50 nM siRNA corresponding with each small RNA or a mixture of all 3 was transfected into HMV-1 cells. Transfectants were collected 24 hours after transfection. RNA was extracted, and its suppressive effect on gene expression levels was evaluated. The migration-inhibition effect (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/nat) of transfection with siRNA for small RNA-197 was confirmed using exCELLigence (Roche) (Meng et al., 2007). We then examined its effects on HLF and HMV-1 cells in vivo. Gelatin hydrogel microspheres were used as carriers for the RGM249 shRNA plasmid, and HLF cells were s.c. inoculated into the right flank of the mice. Spermine-pullulan was used as a liver-targeting DDS for injection into the tail vein. Both DDSs were provided by Prof. Yasuhiko Tabata of Kyoto University (Kushibiki et al., 2006; Kimura and Tabata, 2010). Inoculation into 6-week-old immunodeficient mice (CAnN Cg-Foxn1 BALB/c-nu) was performed every 5 days for 40 days. The animals were sacrificed after 56 days and examined for gross tumor formation and metastasis. Spermine-pullulan was cationized by carbonyldiimidazole activation, and mice subsequently received an i.v. injection every week starting 1 week after the first injection of HLF cells. The injection volume was 200 μL and comprised 1×107 cells. Animals were sacrificed after 28 days and examined for intrahepatic tumors and intraperitoneal dissemination. To determine the anticancer effects of a mixture of siRNAs on the same cell lines, siRNAs against all 3 small RNAs were transfected using atelocollagen (AteloGene™) (Jo et al., 2006; Takeshita et al., 2010), gelatin hydrogel microspheres via s.c. inoculation (n=5), and spermine-pullulan via i.v. injection in immunodeficient KSN/Slc mice (n=5).
Evaluation of gene transfer in vitro and in vivo
MicroRNA expression in tumors was evaluated after categorizing the parts of the tumor as shown in Fig. 2a. RNA quantification was confirmed by sequencing with high reproducibility (Goodarzi et al., 2009). Cells and tissues were subjected to miRNA extraction using a mirVana miRNA Isolation kit, and miRNA expression was examined using a Mir-X™ miRNA qRT-PCR (quantitative reverse transcription-polymerase chain reaction) SYBR® kit to confirm suppression by siRNA and evaluate changes in miRNA expression using the 2–ΔΔCt method according to the manufacturer's recommendations (Supplementary Fig. S2) (see Supplementary Methods). The genes were chosen from those with altered expressions in response to RGM249 overexpression and silencing (data not shown). The primer sequences used for mRNA or miRNA quantification are shown in Fig. 1b and Supplementary Fig. S3. Western blot analyses were performed using the i-Blot gel transfer system (Supplementary Methods). The antibodies (anti-hTERT, -p53, -c-Myc, -Oct4, -AID, -DICER, and -PROM1) were diluted 1:500, and the control anti-β-actin antibody was diluted 1:1,000 (Supplementary Methods). Chemiluminescent signals were detected within 1 minute using LAS-1000 (Supplementary Methods). Palpable tumors were confirmed on day 7 following inoculation, and 10 mice were randomized into 2 groups to receive mixed siRNAs against small RNA-47, RNA-101, and RNA-197 (100 μM) (n=5) or an equivalent volume of vehicle (n=5). Mixed transfected cells only suppressed cell growth in vitro. Animals were sacrificed after 5 weeks for tumor analysis. Volume estimations were determined using the following formula: volume=(π/6×width×length×height). Furthermore, HMV-1 cells (1×107) were injected into the caudal vein of athymic mice 1 week before treatment with siRNAs (400 μM) or vehicle. siRNAs or siRNA mixtures were administered every week, and tumor volume and metastasis were assessed after 5 weeks. Levels of gene transcription and translation were examined in vivo in same manner as in the in vitro study (see Supplementary Methods). This study was approved by the ethics committee of Tottori University (#10-Y-36). The effects of small-RNA-targeted siRNAs and the overexpression of miRNAs in normal human (293FT) cells were examined in transfectants within 24–48 hours following transfection, and changes in gene expression levels and phenotype were investigated.
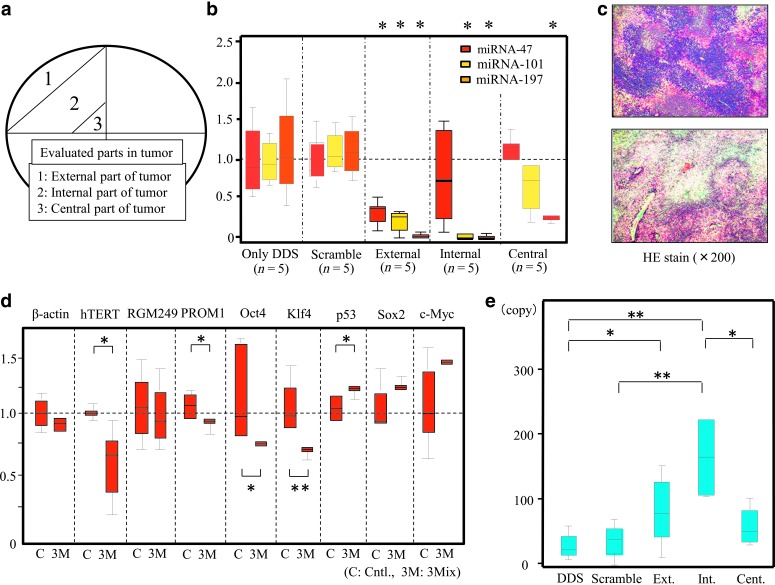
FIG. 2.
Evaluation of gene expression in tumors treated with siRNAs. (a) Tumor classified into 3 parts to examine the delivery by drug delivery system (DDS)+siRNAs: 1, external; 2, internal; and 3, central. The divisions were occasionally approximate because the tumors were fragile. (b) The suppressive effects of a mixture of siRNAs on the respective small RNAs were examined. Data were analyzed using the Kruskal-Wallis test (n=5). * indicates a significant difference in small RNA expression levels compared with levels in tumors treated with DDS alone or a scramble (p<0.01). (c) Microscopic findings [hematoxylin and eosin (HE) stain; 400×] in the control (top) and siRNA-treated tumors (bottom). There was a marked decrease in human malignant melanoma cell line (HMV-I) cells in siRNA-treated tumors, and tumor necrosis was followed by fibrosis. (d) Relative expression levels of tumor-, differentiation-, or pluripotency-related genes were examined at the transcriptional level; this indicated that Oct4, Klf4, p53, hTERT, and PROM1 may be involved in pluripotency, oncogenesis, and cancer stemness. Messenger RNA (mRNA) expression levels were compared with β-actin mRNA expression using the 2–ΔΔCt method. Cntl, control (DDS alone); scramble; 3M, DDS+mixture of small RNA-47, RNA-101, and RNA-197. *p<0.05, **p<0.01. (e) RGM249 mRNA levels were evaluated quantitatively in siRNAs-treated (n=4) and DDS-treated tumors (n=8). The copy number is shown, and significant upregulation was induced in both the external and internal parts of the tumor, which is similar to the in vitro results. *p<0.05; **p<0.01.
Effect of siRNA for three small RNAs derived from RGM249 in normal human cells
To investigate the effects of 3 small RNAs-targeting siRNAs and the miRNAs upregulated by RGM249 shRNA, 293FT cells were transfected with siRNAs for small RNA-47, RNA-101, and RNA-197. The inductive effects of the small RNA-targeted siRNAs were evaluated at the transcriptional and translational levels. Immunohistochemical examination was performed using the pluripotent markers Oct4 and Nanog.
Statistical analysis
Comparisons between 2 groups were analyzed using the Mann-Whitney test with 1 observed variable; p<0.05 was considered to indicate a significant result.
Results
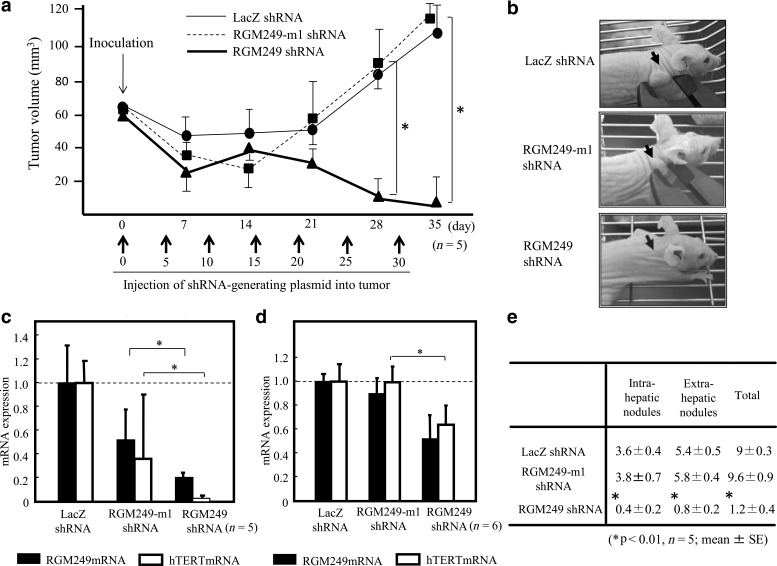
The mature form (secondary structure) of RGM249 and the sequences of the small RNAs are shown in Fig. 1a, b. The sequences were determined following digestion by Dicer and/or RNase 3, and identical sequences were produced using either enzyme. Approximately 50% of the 3 products (Fig. 1a, b) corresponding to RGM249 small RNA-47 were bound to Ago2 and purified using anti-Ago2 beads. However, we were unable to identify any proteins bound to small RNA-101 or 197 (data not shown) (also see miRNA expression in tumor tissues from the animal model experiments). Mice subjected to an s.c. injection of RGM249 shRNA-generating plasmid+cationized DDS (gelatin hydrogel) inhibited the proliferation of HLF hepatoma cells compared with the mice injected with LacZ shRNA-generating plasmid+DDS or RGM249 with 2 base deletions (RGM249-m1) in the shRNA-generating plasmid. The tumor suppressive effect was observed 3 weeks after the first injection (Fig. 3a, b). Four weeks after the first injection, the RGM249 shRNA-generating plasmid+DDS group exhibited a significant suppression of proliferation compared with the RGM249-m1 shRNA-generating and LacZ shRNA-generating plasmid+DDS groups (p=0.034 and p=0.021, respectively). The RGM249-m1 shRNA-generating plasmid+DDS group demonstrated tumor suppression of less than 50% of the tumor volume. Intratumoral expression levels of RGM249 and hTERT mRNA in the RGM249 shRNA-generating plasmid+DDS group were significantly suppressed compared with those of control groups (p=0.036 and p=0.025 for RGM249 mRNA and hTERT mRNA, respectively). RGM249 mRNA in liver tissue was not significantly suppressed in the RGM249-m1 shRNA group; however, hTERT mRNA was significantly suppressed (p=0.028) (Fig. 3c, d).
FIG. 3.
Effect of RGM249 shRNA on the suppression of RGM249-derived small RNAs. (a) Tumor-suppressive effects of RGM249 shRNA in hepatoma (HLF) cells in athymic mice. A significant suppressive effect of RGM249 shRNA was detected at 21 days post-injection. Tumor volume [mean±stadard error (SE)] is depicted, and the data (n=5) were analyzed using the Mann-Whitney test. *p<0.01. (b) Representative photographs showing the macroscopic findings regarding tumor volume in each group. RGM249 shRNA-generating plasmids had an 80% suppressive effect on tumor proliferation after subcutaneous (s.c.) injection on average. These photographs were taken a month after shRNA injection. (c) Suppressive effects of RGM249 shRNA-generating plasmids on gene expression in tumors. Plasmids were induced into inoculated athymic mice by s.c. injection. Closed and open squares show RGM249 and hTERT (human telomerase reverse transcriptase) mRNA expression, respectively. There were significant differences between RGM249-m1 shRNA and RGM249 shRNA and between LacZ shRNA and RGM249 shRNA in terms of both RGM249 and hTERT mRNA levels. The data were analyzed using the Mann-Whitney test. *p<0.05. (d) The preventive effects of the RGM249 shRNA-generating plasmid were evaluated by examining gene expression in metastatic liver nodules after intravenous (i.v.) injection using spermine-pullulan as a liver-targeting DDS. Closed and open squares show RGM249 and hTERT mRNA expression, respectively. There were significant differences between RGM249-m1 shRNA and RGM249 shRNA and between LacZ shRNA and RGM249 shRNA in terms of both RGM249 and hTERT mRNA levels. The data were analyzed using the Mann-Whitney test. *p<0.05. (e) Macroscopic observations of representative intra- and extra-hepatic nodules showing the tumor-preventive effects of RGM249 shRNA-generating plasmids (n=5: mean±SE). Nodule formation was suppressed in the RGM249 shRNA group compared with the other groups. *p<0.01 by Mann-Whitney test.
Macroscopic cancer nodules and microscopic metastatic foci were observed in the livers or lungs of all control mice injected with HLF cells alone and those in the LacZ shRNA-generating plasmid group, and metastasis occurred in the left kidney of 1 mouse. However, only 1 mouse in the group that received the RGM249 shRNA-generating plasmid+DDS exhibited a nodule in the liver, while another mouse exhibited a nodule in the kidney, indicating that RGM249 shRNA suppressed tumorigenicity and metastasis compared with the control groups (Fig. 3e). These findings indicate that RGM249 liver-targeting spermine-pullulan suppressed intraperitoneal carcinogenesis when administered via i.v. injection. Because this precursor gene is specific to humans, we examined the expression levels of RGM249 and hTERT in mouse livers. The expression levels of RGM249 and hTERT mRNAs in the RGM249 shRNA-generating group were both significantly suppressed compared with the LacZ shRNA-generating group (p=0.049 and p=0.046, respectively).
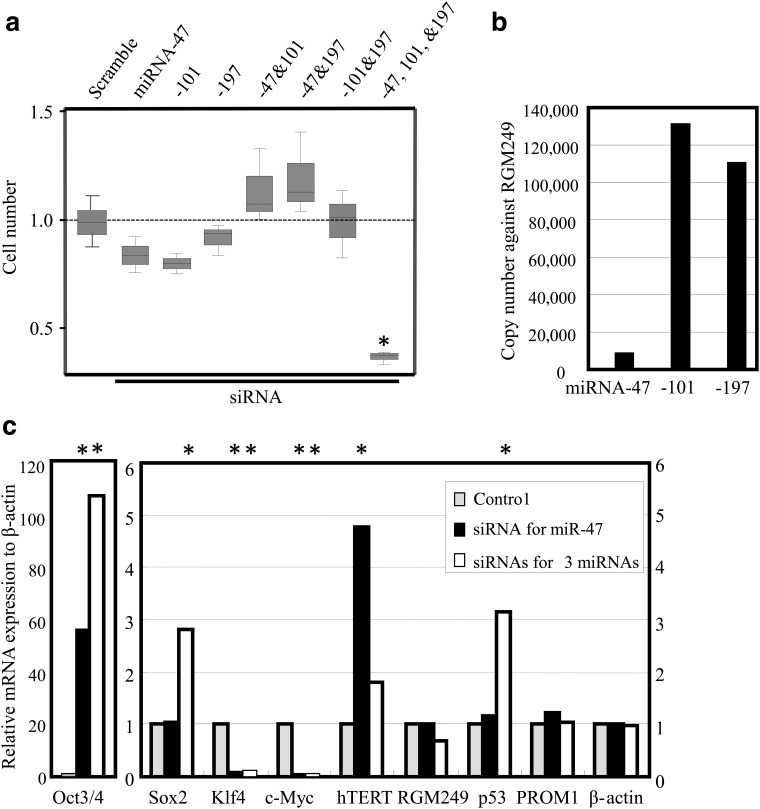
We also investigated the expression levels in the other cell lines that exhibited high RGM249 using the small RNAs previously generated. HMV-1 cells, which express high levels of RGM249, were transfected with siRNAs against 3 small RNAs (-47, -101, and -197) generated from RGM249, and the individual and synergistic effects of the 3 small RNAs were investigated using the siRNA method (see Supplementary Methods). Compared with DDS-treated cells, each siRNA suppressed RGM249 mRNA, and a mixture of all 3 siRNAs demonstrated a marked suppressive effect on cell growth. In contrast, mixtures of only 2 siRNAs did not suppress the cell number at all (Fig. 4a). Baseline expression levels of each small RNA in HMV-1 cells were confirmed, and the expression levels of each small RNA relative to the RGM249 expression level were determined in HMV-1 cells using qRT-PCR (2–ΔΔCt method) (Fig. 4b, Supplementary Fig. S2). Transcriptional expression profiling in transfectants was assessed according to cancer-related (hTERT, c-Myc, p53), pluripotency-related (Oct4, Sox2, Klf4), and stemness-related (PROM1; prominin 1, CD133) genes. Representative results for induction by the siRNA for small RNA-47 and a mixture of siRNAs for the 3 small RNAs are shown because the sequence of the siRNA for small RNA-47 is the same as the sequence used in RGM249 shRNA, as shown in Fig. 1c. The siRNAs for small RNA-101 and RNA-197 did not upregulate the pluripotent marker genes in HMV-1 cells more strongly than siRNA for small RNA-47 or the 3 combined siRNAs (data not shown). Oct4, Sox2, hTERT, and p53 were upregulated following treatment with the 3 short siRNAs, whereas c-Myc and Klf4 were downregulated (Fig. 4c). PROM1 was unaffected.
FIG. 4.
Suppressive effect of siRNAs corresponding with the 3 small RNAs on tumor growth. Synergistic suppressive effects of siRNAs against small RNA-47, -101, and -197 in HMV-I malignant melanoma cells in vitro were evaluated using a DDS to determine their effects (a) against cell proliferation, and (b) relative to expression of each small RNA corresponding with precursor gene (RGM249) expression. Differences between the expression levels of the 3 miRNAs were large, indicating that differences in processing efficiency may be involved; even low levels of small RNA can effectively bind to the target genes, depending on target type or function. (c) The relative expression levels of tumor-, differentiation-, and pluripotency-related genes examined at the transcriptional level indicated that small RNA-197 may be involved in pluripotency and oncogenesis but not in cancer stemness. hTERT may be regulated by small RNA-101 or RNA-197 because their siRNAs strongly suppressed its expression, while small RNA-47 siRNA significantly upregulated hTERT expression. mRNA expression levels were compared with β-actin mRNA. RGM249 mRNA expression gradually recovered to original levels 1 week after transfection. * denotes corresponding significant differences relative to β-actin based on the 2–ΔΔCt method.
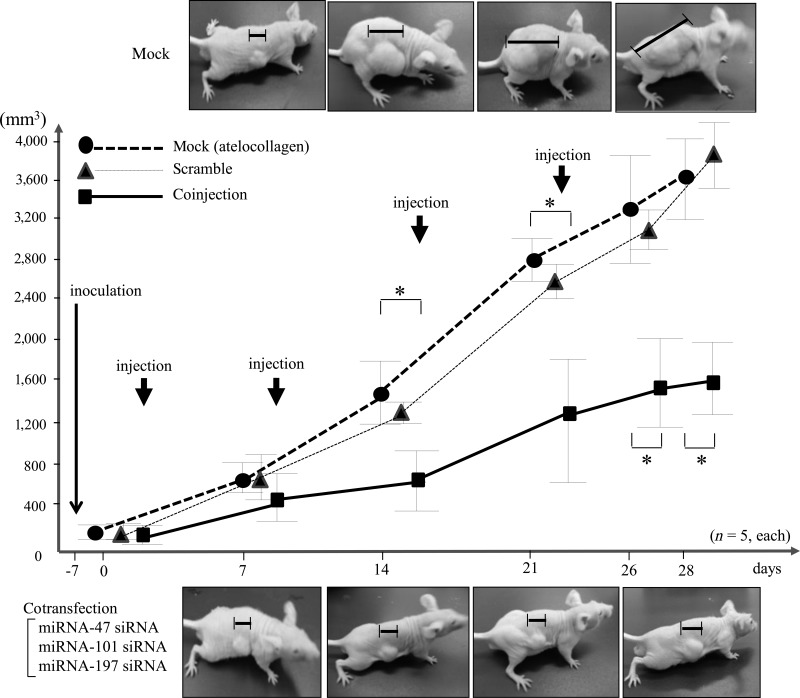
In animal model experiments, the s.c. injection of the 3 siRNAs+DDS (atelocollagen) inhibited the proliferation of HMV-1 cells compared with control injections with DDS alone. Representative photographs of a DDS-alone case (top) or a 3-siRNAs case (bottom) are depicted (p<0.01 by Kruskal-Wallis test, n=5) (Fig. 5). Athymic mice co-injected with siRNAs were sacrificed, and their subcutaneous tumorigenicity and metastasis were examined. Control mice developed multiple nodules in the lungs (mean, 15.8±1.9 nodules) and intraperitoneum (mean, 0.8±0.6 nodules). Several metastatic foci were observed in the peritoneum and retroperitoneum, and invasion was observed (Table 1).
FIG. 5.
HMV-I suppressive effect of the 3 small RNAs derived from RGM249 in athymic mice. Representative photographs (top and bottom) show mice s.c. injected with atelocollagen without nucleotides (mock), scramble, and mice co-injected with the same DDS plus a mixture of siRNAs corresponding with small RNA-47, RNA-101, and RNA-197. DDS alone and scramble failed to suppress tumor growth, but the co-injected mixture significantly suppressed tumor growth. The 2 groups are shown with different start points on the graph to avoid overlapping dots and enable the difference between the 2 groups to be visualized more easily. Data were analyzed using the Kruskal-Wallis test (n=6). * indicates a significant difference (p<0.01) between no RNA and siRNA.
Table 1.
Metastasis-Suppressive Effect of Small Interfering RNAs in Mice Inoculated with HMV-I Human Malignant Melanoma Cells
| Metastasis | Mock-transfection mean±SE (range) | Transfection with 3 miRNAs (s.c.) mean±SE (range) | p |
|---|---|---|---|
| Lung | 15.8±1.9 (10–20) | 2.0±0.4 (0–3) | 0.008 |
| Intraperitoneum | 0.8±0.6 (0–5) | 0 | N.S. |
| Retroperitoneum | 1.2±1.0 (0–1) | 0 | N.S. |
| Liver | 0 | 0 | N.S. |
| Subcutaneous invasion | 0.2±0.2 (1–3) | 0 | 0.005 |
N.S., not significant; s.c., subcutaneous; SE, standard error.
Intratumor expression (see Fig. 2a) of RGM249 small RNAs indicated that the DDS permitted the general invasion of the tumor tissue, as shown in Fig. 2b, with increased small RNA expression in tumors with siRNA compared with tumors with DDS alone. Histological comparison of siRNA-induced suppression of growth and proliferation demonstrated that transfection with the mixture of 3 siRNAs had a necrotic effect on the tumors, which led to fibrosis after apoptosis, compared with the control group (Fig. 2c; hematoxylin and eosinstain×200; top: DDS alone, bottom: combined injection by 3 siRNAs). A comparison of changes in gene expression between growth-suppressed and nonsuppressed tumors demonstrated that siRNAs for RGM249 small RNAs induced the upregulation of p53 mRNA and downregulation of Klf4, which is in accordance with the in vitro results. Conversely, hTERT, Oct4 (POU class 5 homeobox 1), and PROM1 were downregulated compared with the in vitro results (Fig. 2d). Although the 3 small RNA-like molecules targeted by the mixture of siRNAs were suppressed in each tumor section (Supplementary Fig. S4b), the RGM249 mRNA level in the tumors was upregulated by the 3 siRNAs (Fig. 2e).
We further confirmed the suppressive effect of the siRNAs by systemic injection. The intravenous injection of the 3 siRNAs+atelocollagen significantly induced the anti-metastatic ability of HMV-1 cells (p<0.05 for lung, liver, and intraperitoneum) (Table 2); no liver or lung metastasis was observed, and only 1 mouse developed intraperitoneal metastasis.
Table 2.
Metastasis-Suppressive Effect of Small Interfering RNAs in Mice Following Systemic Delivery of HMV-I Cells
| Metastasis | No RNAs mean±SE (range) | Three siRNAs for miRNAs (i.v.) mean±SE (range) | p |
|---|---|---|---|
| Lung | 2.0±0.5 (0–3) | 0±0 (0 for all) | 0.018 |
| Intraperitoneum | 3.8±1.5 (0–8) | 0.2±0.2 (0–1) | 0.044 |
| Liver | 5.2±2.2 (0–13) | 0±0 (0 for all) | 0.005 |
i.v., intravenous.
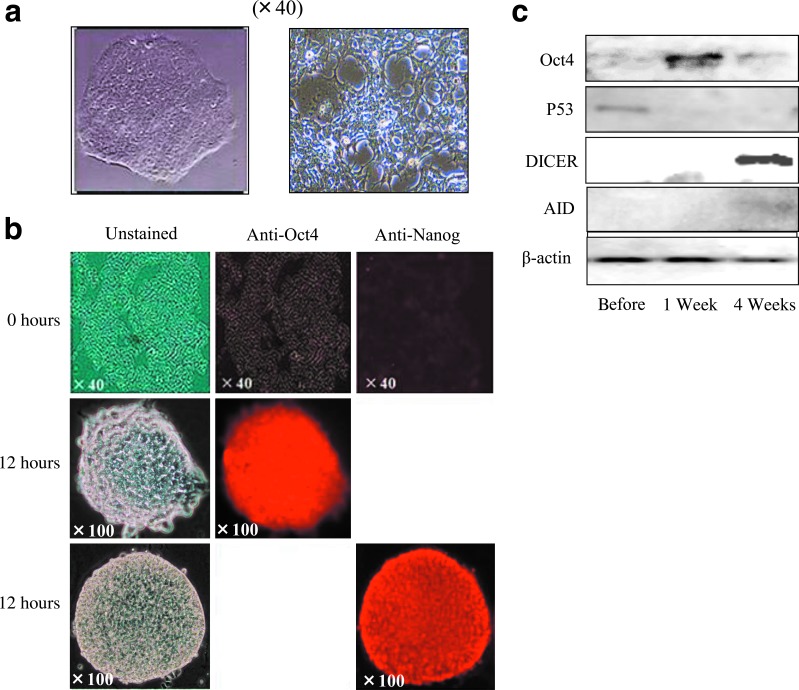
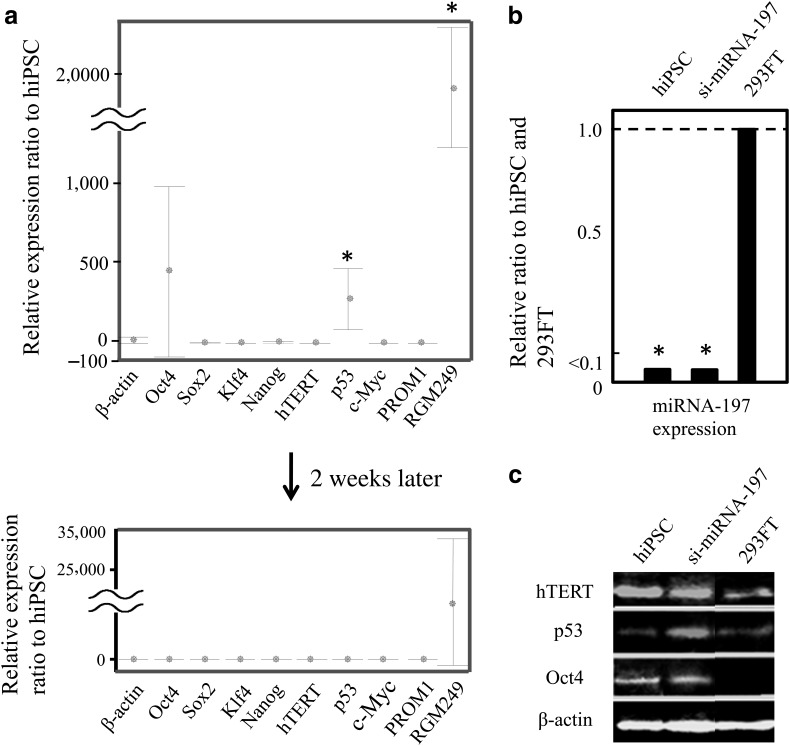
Of the RGM249-derived oligonucleotides and several upregulated small RNAs,11 the siRNA-197 and siRNA-47/101/197 mixture induced the conversion of 293FT cells (Fig. 6b; unstained 0-hour photograph) to undifferentiated cells in addition to the upregulation of Oct4, hTERT, and p53 (Figs. 2d, 6a). Immunocytochemical examination revealed strong expression of Oct4 and Nanog in spherical cells (Fig. 6b). A western blot analysis of p53, Oct4, and cytidine deaminase (AID, also known as AICDA) over a 1-week period demonstrated the transformation of 293FT cells into pluripotent cells (Fig. 6c). The predicted target of each siRNA is depicted in Supplementary Figs. S5, S6. siRNA against small RNA-197 was the most effective for producing pluripotent cells (approximately 1,000 cells in a 10-cm dish). Although neither Oct4 nor Nanog were direct targets according to this prediction, we performed qualitative comparisons with human-induced pluripotent stem cells (hiPSCs) because of the upregulation of pluripotent markers. Representative transcriptional (Fig. 7a), miRNA (Fig. 7b), and translational (Fig. 7c) expression levels in pluripotent cells generated from 293FT cells are shown. Cells transfected with small RNA-197 demonstrated similar expression levels to those in hiPSC at 2 weeks after transfection (Fig. 7a, bottom). The estimation of the methylation-related genes suggested that the transcription of AID and DNA (cytosine-5-)-methyltransferase was upregulated and downregulated, respectively (Supplementary Fig. S7). The cells expressing pluripotent markers exhibited similar expression profiles to those observed to be previously established in hiPSC cells (Okita et al., 2010). These findings indicated a significant upregulation of p53 in our hiPSCs (p<0.05, Mann-Whitney test).
FIG. 6.
siRNAs induce the expression of pluripotent markers in normal human (293FT) cells. (a) Middle: siRNA-treated 293FT cells 3 days after transfection. (b) Microscopic and immunohistochemical findings in 293FT cells transfected with siRNA for small RNA-197. Top: microscopic findings (magnification: 40×) in 293FT cells without transfection (0 hours) (left panel, unstained; center panel, rhodamine staining with anti-Oct4 antibody; right panel, rhodamine staining with anti-Nanog antibody). Middle: undifferentiated status 12 hours after transfection with siRNA against small RNA-197. Microscopic (left panel) and immunocytochemical findings showed strong Oct4 expression (center panel) (magnification: 100×). Bottom: microscopic (left) and immunocytochemical findings showed strong Nanog (right) expression (magnification: 100×). The 0-hour time point indicates the background of staining, and the 12-hour time point shows a formed spherical cell with many nuclei expressing Oct4 or Nanog. Further, 293FT cells maintained their undifferentiated status for up to 7 days in culture medium. (c) Western blot showing translational changes induced by transfection of siRNA-197 in 293FT cells. P53 and Oct4 were downregulated and upregulated, respectively. The activation of Dicer indicates the induction of miRNA biogenesis, causing pluripotency. Cytidine deaminase (AID) was weakly upregulated at 4 weeks. The downregulation of p53 to human-induced pluripotent stem cells (iPSC) levels occurred 1 week after transfection in embryonic stem cell (ES) culture medium.
FIG. 7.
A comparison of gene expression between induced 293FT cells and hiPSCs. (a) Relative levels of transcriptional of each gene in 293FT cells transfected with siRNA for small RNA-197 (si-small RNA-197) were evaluated and compared with expression levels in hiPSCs using 1-step real-time qRT-PCR. Top: 48 hours after transfection. Bottom: cells were maintained in ES medium for 2 weeks after transfection. Although RGM249 was expressed at high levels even 2 weeks after transfection in 293FT cells treated with si-small RNA-197; other genes were expressed at similar levels as in hiPSCs. (b) Relative small RNA-197 expression levels were determined in hiPSC, small-RNA-197-targeted siRNA-induced, and 293FT cells by 2-step real-time qRT-PCR (n=5). The expression levels in transfectants were similar to those in hiPSCs. Effective small RNA-197 silencing was confirmed in transfectants. (c) Translation of hTERT, p53, and Oct4 was examined in hiPSCs and si-small RNA-197-induced 293FT and 293FT 24 hours after transfection. p53 expression was upregulated in treated cells, and Oct4 was expressed at similar levels in all cells, except for 293FT cells. hTERT was not regulated in cells with siRNA or small RNA in a fixed manner.
Discussion
MicroRNAs have been implicated in numerous biological processes (Ghildiyal and Zamore, 2009). A single miRNA can target and potentially silence more than 100 genes, and several miRNAs can target a single gene, with a single miRNA typically exerting only a modest effect on repression (Persengiev et al., 2004; BEHLKE, 2006). These short RNAs modulate cellular gene expression and play a critical role in the development of some diseases through partial or full complementarities with their targets (Bernstein et al., 2003; SUH, 2004; Kanellopoulou et al., 2005). This effect is evident in embryonic stem (ES) cells, which include miR-290-295 and -302 clusters. The most abundant ES cell miRNAs are driven by the pluripotency genes Oct4, Nanog, and Sox2 [SRY (sex determining region Y)-box 2] and target these genes in feed-forward loops (GUNARATNE, 2009). Thus, miRNAs are coexpressed and positively correlated with the targets they repress, suggesting that one of their primary roles is to fine-tune gene expression rather than act simply as an on/off switch. They exert marked changes similar to those induced by certain transcription factor genes (PERSENGIEV, 2004). However, it is possible that exogenous sequences integrated into the human genome are expressed as short transcripts and activate caspase 9/Sp1-p53 signaling.
Let-7 family members (tumor suppressors) have received considerable attention as key molecules in lung cancer (Takamizawa et al., 2004). They exhibit low expression levels in ES cells in a manner similar to a recently discovered paradigm in which miRNAs regulate the self-renewal and differentiation pathways in ES cells by forming an integral biological network together with transcription factors (Iliopoulos et al., 2009; Koh et al., 2010). Recently, Oct4/Sox2-regulated miR-302 has been shown to belong to a handful of factors that are necessary and sufficient for converting differentiated cells to stemness (Iliopoulos et al., 2009; Nagata et al., 2009; Utikal et al., 2009). In contrast, perturbations of miRNA-mRNA networks in ES cells are usually considered to be characteristic of cancer stem cells (Carette et al., 2010; Patru et al., 2010; Tabu et al., 2010). We performed this study to investigate the potential involvement of RGM249 or the possible derivative molecules in stemness, carcinogenesis, pluripotency and/or malignancy.
In this study, we employed a xenograft model to investigate the potential of both shRNA against RGM249 and siRNAs against RGM249-derived small RNAs to suppress malignant properties and regulate cell differentiation. We used 2 different types of materials with high biocompatibilities, and cancer cells in 2 types of athymic mice to determine the direct effects of local injection, the preventive effects of systemic siRNA injection, and the changes in the expression levels of pluripotent markers and cancer-related markers. An in vivo study was performed because both the overexpression and suppression of RGM249 limit tumor growth in vitro. Our findings demonstrated that an s.c. injection of RGM249 shRNA in a xenograft model inhibited the differentiation of poorly-differentiated HLF hepatoma cells. An intravenous injection of RGM249 shRNA also suppressed hepatocarcinogenesis in an orthotopic xenograft model. Although the evaluation of miRNA function based on expression levels has proven challenging (Pekarsky and Croce, 2010), we hypothesized that the positive feedback towards RGM249 might have been induced, as the expression of RGM249-derived small RNAs was prevented by their respective siRNAs. Upregulation of target genes by RGM249-derived small RNAs may presumably orchestrate upstream RGM249. Also, RGM249 mRNA levels in HMV-1 cells were different from those in tumors generated with the cells as shown in Fig. 2e and Supplementary Fig. S4a. This difference may be caused by the transfer efficiency of DDS carrying the molecules into tumors. However, the mechanism including the feedback towards RGM249 mRNA expression remains unknown.
Our findings also suggested that these siRNAs were able to prominently suppress the metastatic abilities of highly malignant cells. Using these antagomiR, we confirmed these results in another cell line (HMV-1) expressing RGM249 at a high level. Our findings indicated that these siRNAs were able to prominently suppress the metastatic abilities of highly malignant cells. Furthermore, we confirmed that the biomaterial (atelocollagen) used in this study infiltrated into the majority of tumor cells and induced necrotic changes as a result of successful intra-tumoral gene regulation. The s.c. injection of these antagomiRs strongly inhibited the proliferative ability of HMV-1 cells in a xenograft model. Similarly, the systemic injection of 3 targeted siRNAs remarkably suppressed the metastatic ability of HMV-1 cells. Cotransfection with 3 siRNAs by either local or systemic administration exhibited a regulatory effect on malignant properties by changing the expression levels of genes involved in differentiation, proliferation, and pluripotency.
Our in vivo experiments suggest that RGM249 may induce a cancer stemness–like status via the transcriptional upregulation of hTERT, PROM1, Oct4, and Klf4 and downregulation of p53 (GUNARATNE, 2009). The administration of 3 small RNAs-targeting siRNAs might induce the translational upregulation of the targeted genes, followed by the induction of anticancerous alterations due to downregulated levels of hTERT, PROM1, Oct4, and Klf4 and upregulated levels of p53. The mechanism underlying cancer regulation found in this study remains unclear. However, antagomiR of small RNA-197 showed a strong inductive effect on spheroid conversion of 293FT cells, suggesting that this molecule may also have a crucial function in cancer regulation, although it remains unclear whether this small RNA-197 is physiologically generated from RGM249. We predicted the target genes of the siRNA corresponding with small RNA-197, as shown in Supplementary Fig. S5, S6 (www.ebi.ac.uk/enright-srv/microcosm; Hokkaido System Science) (Altuvia et al., 2005; Landgraf et al., 2007). For example, Sp1 (CRSP7), which is predicted to be targeted by miRNA-197, potentially induces cancer cell apoptosis via p53 upregulation (Sp1-p53 heterocomplex) (JEANG, 2010). This indicates that small RNA-197 siRNA may potentially bind several key genes in an independent manner.
AntagomiRs for RGM249-derived small RNAs exhibited antimetastatic effects on tumors with both local and systemic administrations in mice. However, we examined the effects of these siRNAs, not in cancer cells, but in human mesangial cells (293FT), and undifferentiated cells with high levels of Oct4 expression were induced and grew more rapidly than those with low Oct4 expression. In particular, siRNA against small RNA-197 effectively induced an undifferentiated state in transfectants. The Oct4 and Nanog gene expressions induced by activation-induced AID are known to be upregulated to a greater extent in transfectants compared with 293FT cells (p=0.05 by Mann-Whitney test; Supplementary Fig. S7) (Bhutani et al., 2010), indicating that demethylation and methylation in these cells might be activated and suppressed, respectively, to maintain the reprogramming level. We previously demonstrated that siRNA against RGM249 mRNA upregulated the expression of several small RNAs (Meng et al., 2007). Furthermore, we found that some cancer cells with miR-520d overexpression converted into iPS-like cells with Oct4 and Nanog positivity (data not shown) (Nakashima et al., 2004; Miyoshi et al., 2010). Specially, the siRNA corresponding with small RNA-197 appears to be implicated in both metastatic and pluripotent statuses (Liao et al., 2008; Ren et al., 2009). A possible interpretation of these results is the occurrence of an event triggered by small RNA-197 silencing; the expression levels of the targeted molecule in induced cells was nearly identical to that in hiPSCs. Thus, silencing of RGM249 mRNA temporally induces a benign or hiPSC-like status in cancer and normal cells, indicating that small RNA-197 might function upstream of pluripotent markers to play a crucial role in cell differentiation or development and be a key molecule in understanding how these si/shRNAs function (on RGM249 or the derived small RNAs, as well as which target proteins' expressions are regulated by si/shRNAs), although further work is needed to draw any definitive conclusions.
Taken together, the sequences corresponding with small RNAs may play complex, intricate, and regulatory roles in cellular processes, such as in differentiation and oncogenesis. The perturbation of their expression levels may thus lead to anti-tumorigenesis (Ghildiyal and Zamore, 2009). The elucidation of the mechanisms underlying the function of the RGM249 gene and its 3 small RNAs, including information regarding its genetic targets, could suggest new approaches to targeted therapy, cell therapy, and regenerative medicine (Davis et al., 2010).
Supplementary Material
Acknowledgments
This study does not affect our adherence to all Nucleic Acid Therapeutics policies on sharing data and materials. The cell lines used in this study were provided by the Cell Resource Center for Biomedical Research, Institute of Development, Aging and Cancer, Tohoku University, Japan, and the Japanese Collection of Research Bioresources (JCRB). All the PCR primers were designed by INTEC Web and Genome Informatics, Corporation (Tokyo, Japan).
Author Disclosure Statement
No competing financial interests exist.
References
- ALTUVIA Y. LANDGRAF P. LITHWICK G. ELEFANT N. PFEFFER S. ARAVIN A. BROWNSTEIN M.J. TUSCHL T. MARGALIT H. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;33:2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALVAREZ R. ELBASHIR S. BORLAND T. TOUDJARSKA I. HADWIGER P. JOHN M. ROEHL I. MORSKAYA S.S. MARTINELLO R. KAHN J., et al. RNA interference-mediated silencing of the respiratory syncytial virus nucleocapsid defines a potent antiviral strategy. Antimicrob. Agents Chemother. 2009;253:3952–3962. doi: 10.1128/AAC.00014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BEHLKE M.A. Progress towards in vivo use of siRNAs. Mol. Ther. 2006;13:644–670. doi: 10.1016/j.ymthe.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BERNSTEIN E. KIM S.Y. CARMELL M.A. MURCHISON E.P. ALCORN H. LI M.Z. MILLS A.A. ELLEDGE S.J. ANDERSON K.V. HANNON G.J. Dicer is essential for mouse development. Nat. Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- BHUTANI N. BRADY J.J. DAMIAN M. SACCO A. CORBEL S.Y. BLAU H.M. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CARETTE J.E. PRUSZAK J. VARADARAJAN M. BLOMEN V.A. GOKHALE S. CAMARGO F.D. WERNIG M. JAENISCH R. BRUMMELKAMP T.R. Generation of iPSCs from cultured human malignant cells. Blood. 2010;115:4039–4042. doi: 10.1182/blood-2009-07-231845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CASTANOTTO D. ROSSI J.J. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHAN J.A. KRICHEVSKY A.M. KOSIK K.S. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- CHEN C.Z. LI L. LODISH H.F. BARTEL D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- CIMMINO A. CALIN G.A. FABBRI M. IORIO M.V. FERRACIN M. SHIMIZU M. WOJCIK S.E. AQEILAN R.I. ZUPO S. DONO M., et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVIS M.E. ZUCKERMAN J.E. CHOI C.H. SELIGSON D. TOLCHER A. ALABI C.A. YEN Y. HEIDEL J.D. RIBAS A. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DEVINCENZO J. CEHELSKY J.E. ALVAREZ R. ELBASHIR S. HARBORTH J. TOUDJARSKA I. NECHEV L. MURUGAIAH V. VAN VLIET A. VAISHNAW A.K., et al. Evaluation of the safety, tolerability and pharmacokinetics of ALN-RSV01, a novel RNAi antiviral therapeutic directed against respiratory syncytial virus (RSV) Antiviral Res. 2008;77:225–231. doi: 10.1016/j.antiviral.2007.11.009. [DOI] [PubMed] [Google Scholar]
- DOENCH J.G. PETERSEN C.P. SHARP P.A. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOENCH J.G. SHARP P.A. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GHILDIYAL M. ZAMORE P.D. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOODARZI H. ELEMENTO O. TAVAZOIE S. Revealing global regulatory perturbations across human cancers. Mol. Cell. 2009;36:900–911. doi: 10.1016/j.molcel.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GUNARATNE P.H. Embryonic stem cell microRNAs: defining factors in induced pluripotent (iPS) and cancer (CSC) stem cells? Curr. Stem Cell Res. Ther. 2009;4:168–177. doi: 10.2174/157488809789057400. [DOI] [PubMed] [Google Scholar]
- ILIOPOULOS D. HIRSCH H.A. STRUHL K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JEANG K.T. Human T cell leukemia virus type 1 (HTLV-1) and oncogene or oncomiR addiction? Oncotarget. 2010;1:453–456. doi: 10.18632/oncotarget.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JO J. YAMAMOTO M. MATSUMOTO K. NAKAMURA T. TABATA Y. Liver targeting of plasmid DNA with a cationized pullulan for tumor suppression. J. Nanosci. Nanotechnol. 2006;6:2853–2859. doi: 10.1166/jnn.2006.466. [DOI] [PubMed] [Google Scholar]
- KANELLOPOULOU C. MULJO S.A. KUNG A.L. GANESAN S. DRAPKIN R. JENUWEIN T. LIVINGSTON D.M. RAJEWSKY K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIMURA Y. TABATA Y. Controlled release of stromal-cell-derived factor-1 from gelatin hydrogels enhances angiogenesis. J. Biomater. Sci. Polym. Ed. 2010;21:37–51. doi: 10.1163/156856209X410193. [DOI] [PubMed] [Google Scholar]
- KOH W. SHENG C.T. TAN B. LEE Q.Y. KUZNETSOV V. KIANG L.S. TANAVDE V. Analysis of deep sequencing microRNA expression profile from human embryonic stem cells derived mesenchymal stem cells reveals possible role of let-7 microRNA family in downstream targeting of hepatic nuclear factor 4 alpha. BMC Genomics. 2010;11(Suppl 1):S6. doi: 10.1186/1471-2164-11-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KUSHIBIKI T. NAGATA-NAKAJIMA N. SUGAI M. SHIMIZU A. TABATA Y. Enhanced anti-fibrotic activity of plasmid DNA expressing small interference RNA for TGF-beta type 2 receptor for a mouse model of obstructive nephropathy by cationized gelatin prepared from different amine compounds. J. Control. Release. 2006;110:610–617. doi: 10.1016/j.jconrel.2005.11.007. [DOI] [PubMed] [Google Scholar]
- LAGOS-QUINTANA M. RAUHUT R. LENDECKEL W. TUSCHL T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- LANDGRAF P. RUSU M. SHERIDAN R. SEWER A. IOVINO N. ARAVIN A. PFEFFER S. RICE A. KAMPHORST A.O. LANDTHALER M., et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAU N.C. LIM L.P. WEINSTEIN E.G. BARTEL D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- LEACHMAN S.A. HICKERSON R.P. SCHWARTZ M.E. BULLOUGH E.E. HUTCHERSON S.L. BOUCHER K.M. HANSEN C.D. ELIASON M.J. SRIVATSA G.S. KORNBRUST D.J., et al. First-in-human mutation-targeted siRNA phase 1b trial of an inherited skin disorder. Mol. Ther. 2010;18:442–446. doi: 10.1038/mt.2009.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIAO R. SUN J. ZHANG L. LOU G. CHEN M. ZHOU D. CHEN Z. ZHANG S. MicroRNAs play a role in the development of human hematopoietic stem cells. J. Cell Biochem. 2008;104:805–817. doi: 10.1002/jcb.21668. [DOI] [PubMed] [Google Scholar]
- LIU J. VALENCIA-SANCHEZ M.A. HANNON G.J. PARKER R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MA L. TERUYA-FELDSTEIN J. WEINBERG R.A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- MELTZER P.S. Cancer genomics: small RNAs with big impacts. Nature. 2005;435:745–746. doi: 10.1038/435745a. [DOI] [PubMed] [Google Scholar]
- MENG F. HENSON R. WEHBE-JANEK H. GHOSHAL K. JACOB S.T. PATEL T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MIURA N. NAKAMURA H. SATO R. TSUKAMOTO T. HADARA T. TAKAHASHI S. ADACHI Y. SHOMORI K. SANO A. KISHIMOTO Y., et al. Clinical usefulness of serum telomerase reverse transcriptase (hTERT) mRNA and epidermal growth factor receptor (EGFR) mRNA as a novel tumor marker for lung cancer. Cancer Sci. 2006;97:1366–1373. doi: 10.1111/j.1349-7006.2006.00342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MIURA N. SATO R. TSUKAMOTO T. SHIMIZU M. KABASHIMA H. TAKEDA M. TAKAHASHI S. HARADA T. WEST J.E. DRABKIN H., et al. A noncoding RNA gene on chromosome 10p15.3 may function upstream of hTERT. BMC Mol. Biol. 2009;10:5. doi: 10.1186/1471-2199-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MIYOSHI N. ISHII H. NAGAI K. HOSHINO H. MIMORI K. TANAKA F. NAGANO H. SEKIMOTO M. DOKI Y. MORI M. Defined factors induce reprogramming of gastrointestinal cancer cells. Proc. Natl. Acad. Sci. U. S. A. 2010;107:40–45. doi: 10.1073/pnas.0912407107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MU P. NAGAHARA S. MAKITA N. TARUMI Y. KADOMATSU K. TAKEI Y. Systemic delivery of siRNA specific to tumor mediated by atelocollagen: combined therapy using siRNA targeting Bcl-xL and cisplatin against prostate cancer. Int. J. Cancer. 2009;125:2978–2990. doi: 10.1002/ijc.24382. [DOI] [PubMed] [Google Scholar]
- NAGATA S. TOYODA M. YAMAGUCHI S. HIRANO K. MAKINO H. NISHINO K. MIYAGAWA Y. OKITA H. KIYOKAWA N. NAKAGAWA M., et al. Efficient reprogramming of human and mouse primary extra-embryonic cells to pluripotent stem cells. Genes Cells. 2009;14:1395–1404. doi: 10.1111/j.1365-2443.2009.01356.x. [DOI] [PubMed] [Google Scholar]
- NAKASHIMA K. COLAMARINO S. GAGE F.H. Embryonic stem cells: staying plastic on plastic. Nat. Med. 2004;10:23–24. doi: 10.1038/nm0104-23. [DOI] [PubMed] [Google Scholar]
- OKITA K. HONG H. TAKAHASHI K. YAMANAKA S. Generation of mouse-induced pluripotent stem cells with plasmid vectors. Nat. Protoc. 2010;5:418–428. doi: 10.1038/nprot.2009.231. [DOI] [PubMed] [Google Scholar]
- PATRU C. ROMAO L. VARLET P. COULOMBEL L. RAPONI E. CADUSSEAU J. RENAULT-MIHARA F. THIRANT C. LEONARD N. BERHNEIM A., et al. CD133, CD15/SSEA-1, CD34, or side populations do not resume tumor-initiating properties of long-term cultured cancer stem cells from human malignant glio-neuronal tumors. BMC Cancer. 2010;10:66. doi: 10.1186/1471-2407-10-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PEKARSKY Y. CROCE C.M. Is miR-29 an oncogene or tumor suppressor in CLL? Oncotarget. 2010;1:224–227. doi: 10.18632/oncotarget.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PERSENGIEV S.P. ZHU X. GREEN M.R. Nonspecific, concentration-dependent stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs) RNA. 2004;10:12–18. doi: 10.1261/rna5160904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- REN J. JIN P. WANG E. MARINCOLA F.M. STRONCEK D.F. MicroRNA and gene expression patterns in the differentiation of human embryonic stem cells. J. Transl. Med. 2009;7:20. doi: 10.1186/1479-5876-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SATZGER I. MATTERN A. KUETTLER U. WEINSPACH D. VOELKER B. KAPP A. GUTZMER R. MicroRNA-15b represents an independent prognostic parameter and is correlated with tumor cell proliferation and apoptosis in malignant melanoma. Int. J. Cancer. 2010;126:2553–2562. doi: 10.1002/ijc.24960. [DOI] [PubMed] [Google Scholar]
- SUH M.R. LEE Y. KIM J.Y. KIM S.K. MOON S.H. LEE J.Y. CHA K.Y. CHUNG H.M. YOON H.S. MOON S.Y., et al. Human embryonic stem cells express a unique set of microRNAs. Dev. Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- TABU K. KIMURA T. SASAI K. WANG L. BIZEN N. NISHIHARA H. TAGA T. TANAKA S. Analysis of an alternative human CD133 promoter reveals the implication of Ras/ERK pathway in tumor stem-like hallmarks. Mol. Cancer. 2010;9:39. doi: 10.1186/1476-4598-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAKAMIZAWA J. KONISHI H. YANAGISAWA K. TOMIDA S. OSADA H. ENDOH H. HARANO T. YATABE Y. NAGINO M. NIMURA Y., et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- TAKESHITA F. PATRAWALA L. OSAKI M. TAKAHASHI R.U. YAMAMOTO Y. KOSAKA N. KAWAMATA M. KELNAR K. BADER A.G. BROWN D., et al. Systemic delivery of synthetic microRNA-16 inhibits the growth of metastatic prostate tumors via downregulation of multiple cell-cycle genes. Mol. Ther. 2010;18:181–187. doi: 10.1038/mt.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- THUM T. GALUPPO P. WOLF C. FIEDLER J. KNEITZ S. VAN LAAKE L.W. DOEVENDANS P.A. MUMMERY C.L. BORLAK J. HAVERICH A., et al. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation. 2007;116:258–267. doi: 10.1161/CIRCULATIONAHA.107.687947. [DOI] [PubMed] [Google Scholar]
- UTIKAL J. MAHERALI N. KULALERT W. HOCHEDLINGER K. Sox2 is dispensable for the reprogramming of melanocytes and melanoma cells into induced pluripotent stem cells. J. Cell Sci. 2009;122:3502–3510. doi: 10.1242/jcs.054783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VISWANATHAN S.R. DALEY G.Q. GREGORY R.I. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.