Abstract
Treating intracellular pathogens remains a considerable medical challenge because of the inefficient intracellular delivery of antimicrobials and the frequent emergence of bacterial resistance to therapeutic agents deemed the drugs of last resort. We investigated the capability of antisense peptide nucleic acids (PNAs) conjugated to the (KFF)3K cell penetrating peptide to target RNA polymerase α subunit (rpoA) and RNA polymerase sigma 70 (rpoD) in the intracellular pathogen Listeria monocytogenes. The PNAs tested displayed a concentration dependent inhibition of L. monocytogenes growth in pure culture at the micromolar level and significantly reduced intracellular L. monocytogenes in infected cell culture and Caenorhabditis elegans whole animal model. In vitro, the combined PNAs treatment was synergistic resulting in a clearance of L. monocytogenes at 0.5× the individual PNA concentration. This study demonstrates the potential of anti-rpoA PNA as an antibacterial agent and will provide the basis for improving and developing these PNAs to better target intracellular pathogens like Listeria. This study also establishes C. elegans as a potential model for the screening of PNAs.
Introduction
Listeria monocytogenes is a food-borne pathogen with a public health burden of approximately $2.6 billion annually in the United States alone. L. monocytogenes can cross the intestinal epithelial barrier and preferentially infect phagocytes, which spread via lymphatic and blood circulations and disseminate to deeper tissues and cause invasive fatal disease, especially in immunocompromised persons (Buffer et al., 2012; Hoffmann et al., 2012). Although clinical experience suggests that a combination of ampicillin and an aminoglycoside should be used for treatment of invasive listeriosis, effective eradication of the intracellular L. monocytogenes remains challenging and the fatality rate remains high and lifelong suppressive therapy may be necessary for immunocompromised patients in order to prevent relapses (SCHLECH, 2000). In addition, there is associated nephrotoxicity and renal failure observed with aminoglycosides in listeriosis treatment (Mitja et al., 2009). Furthermore, many of the available antimicrobials, although active in vitro, cannot efficiently penetrate the cell membrane. The exploration of a novel antibacterial agent has increased significantly in the last few years, in particular, antisense peptide nucleic acid (PNA) (Nielsen et al., 1991; Hanvey et al., 1992; Good and Nielsen, 1998a, 1998b; Kurupati et al., 2007; Soofi and Seleem, 2012). PNAs are DNA mimics that firmly anneal to complementary strands of DNA and/or RNA and disrupt the expression of targeted genes (Good and Nielsen, 1998a). A number of studies have shown that PNAs can be used for silencing genes critical for bacterial viability, thereby inhibiting bacterial growth (Good and Nielsen, 1998b; Nekhotiaeva et al., 2004; Kulyte et al., 2005).
It has also been shown that PNAs conjugated with a cell penetration peptide (CPP) can be effectively delivered across the macrophage membrane and into the intracellular bacteria (Soofi and Seleem, 2012). In the present study PNAs covalently attached with CPP were used to target two proposed essential genes in L. monocytogenes: RNA polymerase α subunit (rpoA), which is involved in the transcription, and RNA polymerase sigma 70 (sigma D factor) (rpoD), an initiation factor that promotes the attachment of RNA polymerase to specific initiation sites (Yura and Ishihama, 1979). The efficacy of engineered antisense conjugates was explored in vitro, in cell culture and in infected whole animal model (Caenorhabditis elegans). The results showed that targeted PNA-based gene inhibition represents great potency in bacterial inhibition in a sequence-specific and dose dependent manner at micromolar concentrations.
Materials and Methods
Chemicals used
Gentamicin, (GIBCO, Invitrogen); trypticase soy broth (TSB) and trypticase soy agar (TSA) (BD/Difco); and fetal bovine serum (FBS) were purchased from Life Technologies); Dulbecco's modified Eagle's medium (DMEM) and other chemicals were purchased from Sigma-Aldrich Chemicals. PNA-peptide conjugates were purchased from Panagene. They were resuspended in DNase, RNase free water and heated to 50°C for 5 minutes to ensure complete dissolving.
Bacterial and worm strains
L. monocytogenes F4244 was routinely grown at 37°C in TSB or on TSA. C. elegans strain N2 was maintained on nematode growth media (NGM) plates seeded with Escherichia coli OP50 (STIERNAGLE, 2006).
Cell line and culture conditions
The murine macrophage-like cell line J774A.1 was maintained in an atmosphere containing 5% CO2 at 37°C in DMEM supplemented with 10% FBS. The J774A.1 cells were seeded in 96-well tissue culture plates (USA Scientific) to obtain semi-confluent monolayers (2.0×105 cells) and were grown for 18 hours in an atmosphere containing 5% CO2 at 37°C.
PNA design
The specific nucleotide sequence of the PNA construct was chosen to be complementary to a specific region of the critical genes' mRNA including the translation start codon and the 5′ terminal region since this region is accessible for ribosome assembly and consistent success has been experienced targeting this region (Dryselius et al., 2003; Rasmussen et al., 2007). The sequences of L. monocytogenes F4424 rpoA and rpoD genes 5′ terminal region were confirmed by amplification and sequencing using the following primers: rpoA-forward (5′-acaagcagctggtcttgaagtaac-3′) and rpoA-reverse (5′-accttcaattacagaaaactcatg-3′) for rpoA gene and rpoD-forward (5′-agagctagcgactttaaatcgtgaaaatga-3′) and rpoD-reverse (5′-tgcatcgtcagaaacttcaattcc-3′) for ropD gene. PNAs were synthesized and purified by Panagene Inc. The PNAs were conjugated by manual coupling chemistry with the cell penetrating peptides (CPP) (KFF)3K that improves antisense potency and facilitate their uptake through the bacterial cell envelope (Eriksson et al., 2002). The PNA sequences are: anti-rpoA (H-KFFKFFKFFK-o-cgatcattcaaa-NH2), anti-rpoD (H-KFFKFFKFFK-o-tcataactgcc-NH2), and control PNA (H-KFFKFFKFFK-o-catactcttcct-NH2).
In vitro testing
The PNAs were added to ∼5×103 colony forming units (CFU) of L. monocytogenes at a concentration of 5, 10, 15, 30, and 40 μM and incubated at 37°C for 8 hours in triplicates. The combined effect of the PNAs (anti-rpoA and anti-rpoD) was tested at 5, 10, and 15 μM and incubated at 37°C for 8 hours in triplicates. The bacterial cultures were serially diluted in Dulbecco's phosphate buffered saline and plated on TSA. CFU were determined after 18–20 hours incubation at 37°C. Each experiment was repeated at least twice.
Cell culture testing
To test the PNA efficiency in clearing intracellular infection, monolayers of J774A.1 cells were infected with L. monocytogenes at a multiplicity of infection of 10 bacteria per macrophage for 1 hour. The PNAs, at a concentration of 15 μM, were evaluated after 24 hours incubation with the infected macrophages in an atmosphere containing 5% CO2 at 37°C. Cells were lysed and CFU were determined after 20 hours incubation at 37°C and the results were compared with the control (water).
Antimicrobial activity of PNAs in Listeria-infected C. elegans model
One thousand worms at the L1 stage were grown for 6 days at 25°C on 10 cm NGM agar spread with a lawn of E. coli strain OP50. Embryos from the gravid adults were isolated by a previously described hypochlorite method (Powell and Ausubel, 2008) and hatched at 25°C for 24 hours. One thousand of the L1 hatchlings were grown on NGM agar with a lawn of E. coli OP50 at 25 °C for 24 hours to generate young adults. Adult worms were resuspended and washed 5 times with M9 in a 1:10 ratio to remove E. coli. (STIERNAGLE, 2006). Worms were infected with L. monocytogenes for 17 hours. Infected worms were washed 5 times with M9 buffer and 50 worms in triplicates were incubated for 24 hours with 15 and 30 μM PNAs. Untreated C. elegans and Gentamicin at similar concentration were used as a control. Worms incubated with PNA were examined microscopically for morphological changes and viability (live worms are sinusoidal with movement, whereas dead worms are rigid rods). After treatment, infected worms were washed 5 times with M9 buffer and 200 mg of 1.0-mm silicon carbide particles (Biospec Products) were added to each tube. The tubes were vortexed at maximum speed for 1 minute, which disrupts the worms but does not affect bacterial survival, and the resulting suspension was diluted and plated onto TSA containing nalidixic acid to select for L. monocytogenes.
Statistical analysis
Statistical analysis was conducted using Kaleida Graph, version 4.03 (Synergy software). Statistical significance was determined using ANOVA and Fisher's least significant difference comparison test and α=0.05.
Results and Discussion
Bacterial RNA polymerase (RNAP) provides an appealing target for the development of antisense agents. Particularly, rpoA and rpoD genes are evolutionarily conserved, share high similarity between bacterial strains, and are different than the eukaryotic homolog (Bai et al., 2012; Soofi and Seleem, 2012). PNA is a large hydrophilic molecule, and as such, it does not cross cell membranes easily. The CPP, (KFF)3K was conjugated with PNAs because it improves antisense effects by improving PNA uptake properties (Eriksson et al., 2002).
Anti-rpoA and anti-rpoD PNAs inhibited L. monocytogenes in broth culture
The effect of anti-rpoA and anti-rpoD PNAs on the growth of L. monocytogenes cultures were evaluated at different concentrations. Table 1 shows the log CFU reduction in the PNA-treated cultures from that of the untreated control (water). At all concentrations tested, anti-rpoA antisense-PNA showed significant reduction of the growth rate of L. monocytogenes in a dose dependent manner, while anti-rpoD PNA caused a significant reduction in the numbers of L. monocytogenes at concentrations of 10 μM and higher. This difference in reduction can be explained by higher gene stringency requirement in L. monocytogenes for rpoA gene in comparison to rpoD (Goh et al., 2009). Complete clearance of L. monocytogenes was observed at 40 μM for both PNAs. The control PNA (random sequence) showed reduction of L. monocytogenes growth rate at very high concentration (30 and 40 μM). Although the control PNA has a random nucleotide sequence, 10 out of its 12 nucleotides were complementary to a branched chain fatty acid (butyrate) kinase gene (buk) in L. monocytogenes. The possible annealing of this control PNA sequence to the buk gene could explain the lower growth rate that was observed at high concentration. This emphasizes the importance of well-designed PNAs, which would minimize unspecific binding and ensure the silencing of targeted genes. Table 2 shows the log CFU reduction in the combined PNA-treated cultures from that of the untreated control (water). There was a synergistic effect (at 0.5× the individual PNA) when using the combined PNAs (anti-rpoA and anti-rpoD) seen by significant reduction of the growth rate of L. monocytogenes in a dose dependent manner and complete clearance at 10 μM and higher.
Table 1.
Effect of the Anti-rpoA and Anti-rpoD PNAs on Growth of Listeria monocytogenes in Pure Culture
| |
5 μM |
10 μM |
15 μM |
30 μM |
40 μM |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| PNA Name | Log CFU (±SD) | Reduction log from H2O | Log CFU (±SD) | Reduction log from H2O | Log CFU (±SD) | Reduction log from H2O | Log CFU (±SD) | Reduction log from H2O | Log CFU (±SD) | Reduction log from H2O |
| Anti-rpoA | 4.96±(0.31) | 1.31* | 4.30±(0.13) | 1.97* | 3.83±(0.23) | 2.44* | 3.52±(0.29) | 2.68* | Cleared | 6.16* |
| Anti-rpoD | 6.09±(0.06) | 0.19 | 5.79±(0.13) | 0.48* | 5.43±(0.09) | 0.85* | 3.69±(0.32) | 2.50* | Cleared | 6.16* |
| Control PNA |
6.63±(0.12) | −0.36 | 6.53±(0.05) | −0.26 | 6.14±(0.25) | 0.14 | 5.23±(0.26) | 0.97* | 4.44±(0.33) | 1.71* |
| Water | 6.27±(0.59) | 0 | 6.27±(0.59) | 0 | 6.27±(0.59) | 0 | 6.19±(0.22) | 0 | 6.16±(0.87) | 0 |
The peptide nucleic acids (PNAs) were incubated with 5×103 L. monocytogenes at 37°C for 8 hours. Colony forming units (CFU) were counted on trypticase soy agar.
Values found to be significantly different (p≤0.05) from water by statistical analysis (analysis of variance and Fisher's least significant difference comparisons test).
CFU, colony forming units; SD, standard deviation.
Table 2.
Effect of the Combined Anti-rpoA and Anti-rpoD PNAs on Growth of L. monocytogenes in Pure Culture
| |
5/5μM |
10/10μM |
15/15μM |
|||
|---|---|---|---|---|---|---|
| PNA Name | Log CFU (±SD) | Reduction log from H2O | Log CFU (±SD) | Reduction log from H2O | Log CFU (±SD) | Reduction log from H2O |
| Anti-rpoA/Anti-rpoD | 3.58±(0.20) | 1.81* | Cleared | 5.39* | Cleared | 5.39* |
| Control PNA | 5.38±(0.39) | 0.01 | 5.20±(0.20) | 0.18 | 5.02±(0.09) | 0.36 |
| Water | 5.39±(0.36) | 0 | 5.39±(0.36) | 0 | 5.39±(0.36) | 0 |
The PNAs were incubated with 8×103 L. monocytogenes at 37°C for 8 hours. CFU were counted on TSA.
Values found to be significantly different (p≤0.05) from water by statistical analysis (analysis of variance and Fisher's least significant difference comparisons test).
PNA effect on intracellular Listeria using infected cell culture
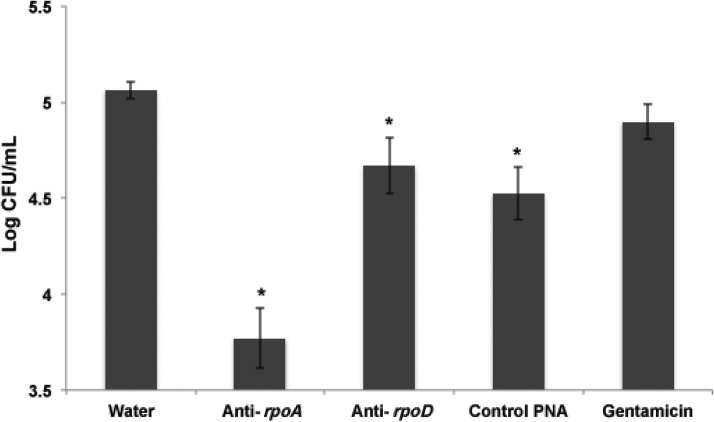
Because L. monocytogenes infect and replicate inside a variety of phagocytic cells, including macrophages, it was important to know whether the PNAs would inhibit growth once L. monocytogenes are in that intracellular location. To test the efficiency of the anti-rpoA and anti-rpoD PNAs in clearing intracellular infection, murine macrophage-like cells J774A.1 were infected with L. monocytogenes and treated with the 2 PNAs at a concentration of 15 μM. As shown in Fig. 1, the number of viable L. monocytogenes cells in the culture was significantly reduced in the presence of antisense peptide-PNA conjugates. Compared with the intracellular L. monocytogenes growth of the untreated cell cultures (water), anti-rpoA and anti-rpoD resulted in a 1.29 and 0.39 log reduction respectively. By contrast, treating the infected cell cultures with gentamicin did not result in statistically significant reduction in the number of viable L. monocytogenes cells. The PNA-dependent intracellular reduction indicated that rpoA and rpoD genes differ in their stringency growth requirement, suggesting that rpoA is a much better target. The relatively high molecular weights, and possibly the polarity, of the PNAs may account for the decreased antibacterial activity that is observed when comparing in vitro and in cell culture activity against Listeria.
FIG. 1.
Effect of the anti-rpoA and anti-rpoD PNA treatment on Listeria infected J774A.1 cells. Intracellular Listeria monocytogenes reduction in infected macrophage cell line J774.A1 after 24 hours incubation with 15 μM PNAs. Bars represent average log of Listeria colony forming units (CFU) with error bars representing 95% confidence interval. *Values found to be significantly different (p≤0.05) from that of water by statistical analysis (analysis of variance and Fisher's least significant difference comparisons test).
In vivo evaluation of the effect of PNA using Listeria-infected C. elegans worms
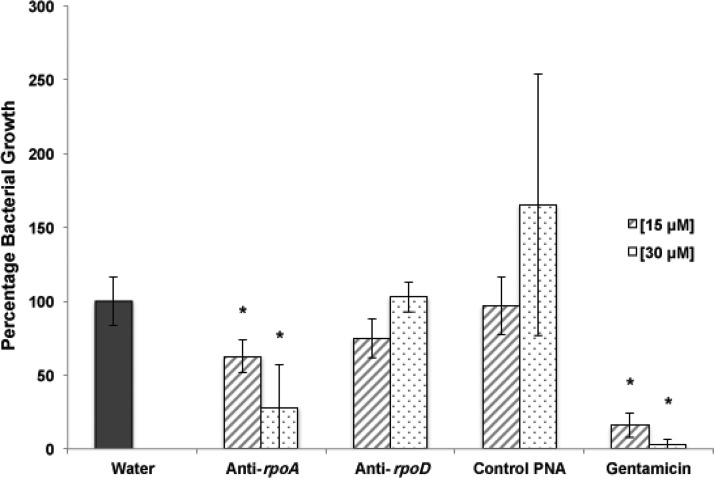
C. elegans has been previously used to screen libraries of chemical compounds for antimicrobial activity against multiple bacterial pathogens (Moy et al., 2009). It has been also established that L. monocytogenes can be used to infect and kill C. elegans (Thomsen et al., 2006; Gottlieb et al., 2008). In this study, C. elegans was used as a unique whole animal model to evaluate the efficiency of the PNA treatments against Listeria in vivo. Two concentrations of the PNAs, 15 and 30 μM, were used to infected C. elegans (Fig. 2). While anti-rpoD PNA was not able to reduce the numbers of L. monocytogenes, significant reduction was observed for anti-rpoA PNA (Fig. 2). When the infected C. elegans were treated with anti-rpoA PNA, 37% and 72% growth reduction were observed for the 15 and 30 μM treatments, respectively. These results suggest that the effect of the anti-rpoA PNA was concentration dependent and that higher PNA concentrations are needed to significantly reduce the number of L. monocytogenes in infected C. elegans. One of the main factors limiting PNA antibacterial activity is bioavailability of the PNA inside of the target bacterium. With complex system like C. elegans there are additional barriers constrained accumulation of the PNA-peptide conjugates inside of the bacterium, thereby reducing antibacterial activity as compared to in vitro models. Also, nonspecific biodistribution and binding of the PNAs, high molecular weights, and possibly the polarity may account for the decreased antibacterial activity that is observed when comparing in vitro and in cell culture activity with C. elegans (Rasmussen et al., 2007; Soofi and Seleem, 2012).
FIG. 2.
Effect of anti-rpoA and anti-rpoD PNA treatments on Listeria infected Caenorhabditis elegans. In vitro C. elegans infection assay. Intracellular L. monocytogenes reduction in infected C. elegans after 24 hours incubation with 15 and 30 μM PNAs. Bars represent percentage of Listeria growth with error bars representing 95% confidence interval. *Values found to be significantly different (p≤0.05) from that for water by statistical analysis (analysis of variance and Fisher's least significant difference comparisons test).
In conclusion, rpoA and rpoD genes were identified as potential targets for inhibition of L. monocytogenes using antisense technology. Micromolar concentration of the peptide-conjugated PNAs were successful inhibiting the growth of pure cultures of Listeria in a concentration dependent fashion and significantly reduced the viable numbers of intracellular L. monocytogenes in infected macrophages and infected C. elegans. Overall, anti-rpoA PNA was more potent in inhibiting L. monocytogenes growth than anti-rpoD PNA. The lower inhibition rate of growth obtained by targeting rpoD gene does not necessary eliminate the potential of this target. The conjugates used in this study have not been optimized and further modifications may allow for greater antibacterial activity. More investigation and optimization are needed to better establish C. elegans as a model for in vivo screening of PNAs.
Increasing number of pathogens that are resistant to traditional therapeutics and the difficulties in discovery of new antimicrobials have prompted consideration of new strategies such as antisense PNA. This study reveals the potential usefulness of antisense PNA constructs as novel therapeutic agents against intracellular L. monocytogenes. Although the preliminary results are very encouraging, several issues must be addressed (safety, pharmacological properties, and cost) before using PNA for further drug development against multidrug-resistant bacteria. The therapeutic potential of these peptides is currently limited due to high production costs. Nonetheless, there is a need for an appropriate carrier system that will deliver PNAs specifically to the infected macrophages and the resident pathogens, which will reduce the therapeutic dose and avoid possible off-target effects.
Acknowledgments
We would like to thank Muhammad Soofi and Danielle McPherson for their helpful comments and discussion. We also would like to thank Haroon Mohammad for reviewing the manuscript and for his suggestions. We would like to thank Dr. Arun K. Bhunia for providing the bacterial strains. We also would like to thank the Caenorhabditis Genetics Center (CGC) funded by National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440) for providing C. elegans and E. coli strains.
Author Disclosure Statement
No competing financial interests exist.
References
- BAI H. YOU Y. YAN H. MENG J. XUE X. HOU Z. ZHOU Y. MA X. SANG G. LUO X. Antisense inhibition of gene expression and growth in gram-negative bacteria by cell-penetrating peptide conjugates of peptide nucleic acids targeted to rpoD gene. Biomaterials. 2012;33:659–667. doi: 10.1016/j.biomaterials.2011.09.075. [DOI] [PubMed] [Google Scholar]
- BUFFER J.L. MEDEIROS L.C. KENDALL P. SCHROEDER M. SOFOS J. Health professionals' knowledge and understanding about L. monocytogenes indicates a need for improved professional training. J. Food Protect. 2012;75:1310–1316. doi: 10.4315/0362-028X.JFP-12-006. [DOI] [PubMed] [Google Scholar]
- DRYSELIUS R. ASWASTI S.K. RAJARAO G.K. NIELSEN P.E. GOOD L. The translation start codon region is sensitive to antisense PNA inhibition in Escherichia coli. Oligonucleotides. 2003;13:427–433. doi: 10.1089/154545703322860753. [DOI] [PubMed] [Google Scholar]
- ERIKSSON M. NIELSEN P.E. GOOD L. Cell permeabilization and uptake of antisense peptide-peptide nucleic acid (PNA) into Escherichia coli. J. Biol. Chem. 2002;277:7144–7147. doi: 10.1074/jbc.M106624200. [DOI] [PubMed] [Google Scholar]
- GOH S. BOBEREK J.M. NAKASHIMA N. STACH J. GOOD L. Concurrent growth rate and transcript analyses reveal essential gene stringency in Escherichia coli. PLoS One. 2009;4:e6061. doi: 10.1371/journal.pone.0006061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOOD L. NIELSEN P.E. Antisense inhibition of gene expression in bacteria by PNA targeted to mRNA. Nat. Biotechnol. 1998a;16:355–358. doi: 10.1038/nbt0498-355. [DOI] [PubMed] [Google Scholar]
- GOOD L. NIELSEN P.E. Inhibition of translation and bacterial growth by peptide nucleic acid targeted to ribosomal RNA. Proc. Natl. Acad. Sci. U. S. A. 1998b;95:2073–2076. doi: 10.1073/pnas.95.5.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOTTLIEB C. THOMSEN L. INGMER H. MYGIND P. KRISTENSEN H.-H. GRAM L. Antimicrobial peptides effectively kill a broad spectrum of Listeria monocytogenes and Staphylococcus aureus strains independently of origin, sub-type, or virulence factor expression. BMC Microbiol. 2008;8:205. doi: 10.1186/1471-2180-8-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HANVEY J.C. PEFFER N.J. BISI J.E. THOMSON S.A. CADILLA R. JOSEY J.A. RICCA D.J. HASSMAN C.F. BONHAM M.A. AU K.G., et al. Antisense and antigene properties of peptide nucleic acids. Science. 1992;258:1481–1485. doi: 10.1126/science.1279811. [DOI] [PubMed] [Google Scholar]
- HOFFMANN S. BATZ M.B. MORRIS J.G., JR Annual cost of illness and quality-adjusted life year losses in the United States due to 14 foodborne pathogens. J. Food Prot. 2012;75:1292–1302. doi: 10.4315/0362-028X.JFP-11-417. [DOI] [PubMed] [Google Scholar]
- KULYTE A. NEKHOTIAEVA N. AWASTHI S.K. GOOD L. Inhibition of Mycobacterium smegmatis gene expression and growth using antisense peptide nucleic acids. J. Mol. Microbiol. Biotechnol. 2005;9:101–109. doi: 10.1159/000088840. [DOI] [PubMed] [Google Scholar]
- KURUPATI P. TAN K.S.W. KUMARASINGHE G. POH C.L. Inhibition of gene expression and growth by antisense peptide nucleic acids in a multiresistant beta-lactamase-producing Klebsiella pneumoniae strain. Antimicrob. Agents Chemother. 2007;51:805–811. doi: 10.1128/AAC.00709-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MITJA O. PIGRAU C. RUIZ I. VIDAL X. ALMIRANTE B. PLANES A.M. MOLINA I. RODRIGUEZ D. PAHISSA A. Predictors of mortality and impact of aminoglycosides on outcome in listeriosis in a retrospective cohort study. J. Antimicrob. Chemother. 2009;64:416–423. doi: 10.1093/jac/dkp180. [DOI] [PubMed] [Google Scholar]
- MOY T.I. CONERY A.L. LARKINS-FORD J. WU G. MAZITSCHEK R. CASADEI G. LEWIS K. CARPENTER A.E. AUSUBEL F.M. High-throughput screen for novel antimicrobials using a whole animal infection model. ACS Chem. Biol. 2009;4:527–533. doi: 10.1021/cb900084v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NEKHOTIAEVA N. AWASTHI S.K. NIELSEN P.E. GOOD L. Inhibition of Staphylococcus aureus gene expression and growth using antisense peptide nucleic acids. Mol. Ther. 2004;10:652–659. doi: 10.1016/j.ymthe.2004.07.006. [DOI] [PubMed] [Google Scholar]
- NIELSEN P.E. EGHOLM M. BERG R.H. BUCHARDT O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- POWELL J.R. AUSUBEL F.M. Models of Caenorhabditis elegans infection by bacterial and fungal pathogens. In: Ewbank J., editor; Vivier E., editor. Methods in Molecular Biology, vol. 415: Innate Immunity. Humana Press, Inc.; Totowa, New Jersey: 2008. pp. 403–427. [DOI] [PubMed] [Google Scholar]
- RASMUSSEN L.C. SPERLING-PETERSEN H.U. MORTENSEN K.K. Hitting bacteria at the heart of the central dogma: sequence-specific inhibition. Microb. Cell Fact. 2007;6:24. doi: 10.1186/1475-2859-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHLECH W.F. Foodborne listeriosis. Clin. Infect. Dis. 2000;31:770–775. doi: 10.1086/314008. [DOI] [PubMed] [Google Scholar]
- SOOFI M.A. SELEEM M.N. Targeting Salmonella essential genes with antisense peptide nucleic acid. Antimicrob. Agents Chemother. 2012;12:6407–6409. doi: 10.1128/AAC.01437-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STIERNAGLE T. Maintenance of C. elegans WormBook and the c. elegans research community. 2006. www.wormbook.org/chapters/www_strainmaintain/strainmaintain.html. www.wormbook.org/chapters/www_strainmaintain/strainmaintain.html [DOI] [PMC free article] [PubMed]
- THOMSEN L.E. SLUTZ S.S. TAN M.-W. INGMER H. Caenorhabditis elegans is a model host for Listeria monocytogenes. Appl. Environ. Microbiol. 2006;72:1700–1701. doi: 10.1128/AEM.72.2.1700-1701.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YURA T. ISHIHAMA A. Genetics of bacterial RNA polymeras. Annu. Rev. Genet. 1979;13:59–97. doi: 10.1146/annurev.ge.13.120179.000423. [DOI] [PubMed] [Google Scholar]