Abstract
Recent studies have reported associations of sirtuin 1 (SIRT1) single nucleotide polymorphisms (SNPs) to both obesity and BMI. This study was designed to investigate association between SIRT1 SNPs, SIRT1 gene expression and obesity. Case-control analyses were performed using 1,533 obese subjects (896 adults, BMI >40 kg/m2 and 637 children, BMI >97th percentile for age and sex) and 1,237 nonobese controls, all French Caucasians. Two SNPs (in high linkage disequilibrium (LD), r2 = 0.96) were significantly associated with adult obesity, rs33957861 (P value = 0.003, odds ratio (OR) = 0.75, confidence interval (CI) = 0.61–0.92) and rs11599176 (P value: 0.006, OR = 0.74, CI = 0.61–0.90). Expression of SIRT1 mRNA was measured in BMI-discordant siblings from 154 Swedish families. Transcript expression was significantly correlated to BMI in the lean siblings (r2 = 0.13, P value = 3.36 × 10−7) and lower SIRT1 expression was associated with obesity (P value = 1.56 × 10−35). There was also an association between four SNPs (rs11599176, rs12413112, rs33957861, and rs35689145) and BMI (P values: 4 × 10−4, 6 × 10−4, 4 × 10−4, and 2 × 10−3) with the rare allele associated with a lower BMI. However, no SNP was associated with SIRT1 transcript expression level. In summary, both SNPs and SIRT1 gene expression are associated with severe obesity.
INTRODUCTION
The sirtuin 1 (SIRT1) gene belongs to a family of NAD+-dependent protein deacetylases known as sirtuins and is expressed in a wide range of tissues including the brain, adipose, kidney, muscle, and liver (1,2). Its yeast ortholog, Sir2, is a crucial protein in the pathway that controls the positive effects of caloric restriction on life span in lower organisms and it is thought to mediate the same process in mammals (3,4). In humans, the SIRT1 protein deacetylates a number of transcription factors and histones that are involved in energy regulation (2). SIRT1 activity is under the control of NAD+ levels, which was recently demonstrated to be mediated by AMP-activated protein kinase (5,6). AMP-activated protein kinase is an enzyme important in the regulation of mitochondrial biogenesis in response to energy deprivation (7). As such SIRT1 functions as an energy sensing molecule that controls transcriptional regulation in response to the energy status of the cell (6).
Levels of the transcription factor peroxisome proliferator-activated receptor γ (PPARγ) and its gene targets are inversely proportional to SIRT1 expression in adipocytes and since PPARγ upregulates genes affecting lipogenesis and insulin-dependent glucose transport, SIRT1 activity results in fat mobilization (8). It has also been demonstrated to reduce adipocyte differentiation through PPARγ inhibition in mice (9). SIRT1 also upregulates the transcriptional coactivator PPARγ-coactivator 1 α (PGC1-α) which leads to an increase in fatty acid oxidation and gluconeogenesis and a decrease in glycolysis (10–12).
SIRT1 has also been found to regulate the expression of adiponectin, although these two studies reported opposite effects (13,14). Adiponectin is secreted by adipocytes and leads to an increased sensitivity to insulin in liver and muscle (15). Reduced levels of adiponectin in the blood are linked to obesity and related insulin resistance (16) and variants in its gene have been reported to associate with obesity and with type 2 diabetes (17–19). SIRT1 activators have been shown to protect against obesity and insulin resistance in mice fed a high-fat diet through the activation of PGC1-α. SIRT1 activators are the subjects of research into therapeutics for the treatment of type 2 diabetes due to their positive effects on fat oxidation (10,20–22).
Three single nucleotide polymorphisms (SNPs) within the SIRT1 gene, rs3740051, rs2236319, and rs2272773 have been associated with rates of energy expenditure in Finnish subjects (10) and it is plausible that a reduced rate of energy expenditure caused by variants in SIRT1 could predispose an individual to obesity. Three recent studies have reported association of SIRT1 SNPs with obesity. One SNP, rs7069102, was found to be associated with obesity in a Belgian study (n = 1,068 cases and 313 controls, P = 0.007) (23). Another, rs2273773 was found to be associated with BMI in a Dutch study (n = 3,575, P = 0.001) (24). A third study investigated three SNPs within two Dutch populations (n = 6,251 from a population-based study and n = 2,347 from a family-based study) and found two SNPs rs7895833 and rs1467568 to be associated with BMI (P = 0.02 and 0.008) and obesity (P = 0.007 and 0.0009)(25). None of these genetic studies investigated SIRT1 expression, however a recent study in Danish women reported a significantly higher rate of SIRT1 transcription in lean compared to obese (n = 24, P < 0.02) (26).
Given its role in energy regulation and the associations already reported SIRT1 is a plausible candidate gene for polygenic obesity and this study was designed to explore the possibility of genetic association between variants in the SIRT1 gene, SIRT1 gene expression levels and common severe obesity.
METHODS AND PROCEDURES
Case-control subjects
Two groups of cases were included in this study: (i) 896 unrelated morbidly obese (BMI over 40 kg/m2) adults (689 females and 207 males; mean BMI = 47.5 kg/m2, s.d. = 7.5 kg/m2; mean age = 44.3 years, s.d. = 11.9 years) (ii) 637 unrelated severely obese children (age < 18 years), with a BMI >97th percentile for age and sex (341 females and 296 males; mean BMI = 29.6 kg/m2, s.d. = 6.5 kg/m2; mean zBMI = 4.27, s.d. = 1.2; mean age = 11 years, s.d. = 3.2 years) (27). These were all French Caucasians recruited through a multimedia campaign run by the Centre National de la Recherche Scientifique (CNRS), Hotel Dieu Hospital, the Pasteur Institute, Lille and the Department of Paediatric Endocrinology of Jeanne de Flandres Hospital. Control subjects were unrelated adult nonobese and nondiabetic French Caucasians composed of 532 individuals (243 males, 289 females; mean BMI = 21.3 kg/m2, s.d. = 2.0 kg/m2; mean age = 22.7 years, s.d. = 3.5 years) from the Haguenau cohort (28) together with 705 subjects (282 males, 423 females; mean BMI = 23.3 kg/m2, s.d. = 1.8 kg/m2; mean age = 53.9 years, s.d. = 5.6 years) that were selected among participants of the Data from the Epidemiology Study on the Insulin Resistance (D.E.S.I.R) study (29). BMI was calculated as weight (in kilograms) divided by the square of height (in metres). Height and weight were measured by trained professionals. Control participants were pooled for case control analyses and χ2 tests did not show significant difference in genotype or allele frequencies between both control groups (0.204 ≤ P ≤ 1). Both adult and child obesity case groups were analyzed with the same set of controls. This genetic study was approved by the ethical committee of Hôtel-Dieu in Paris and Centre Hospitalier Régional Universitaire in Lille.
Families
The study cohort comprises 154 Swedish nuclear families (732 subjects) ascertained via an extremely BMI-discordant sibpair (at least 10 units). The average family size was 4.75. Blood samples for DNA extraction as well as a subcutaneous adipose tissue biopsy have been obtained from all participants. BMI was measured for all subjects. The median (1st–3rd quartiles) was 27.5 (24.0–33.2) ranging between 16.9 and 57.8. Informed written consent was obtained from all participants. This study was approved by the ethics committee at Gothenburg University (30).
Genotyping
Tagging SNPs were selected from the HapMap database (http://www.hapmap.org/) with r2 ≥ 0.8 and a minor allele frequency ≥0.05 using Haploview 4.0 software (31). This gave three SNPs capturing all 21 SNPs within SIRT1 in HapMap, with a mean r2 of 0.975. An additional 24 SNPs were selected from the NCBI database, each with a minor allele frequency ≥0.05, to provide overall coverage of approximately one SNP per kb in the SIRT1 gene (with the three tag SNPs accounting for 21 other SNPs).
Genotyping was carried out on the Sequenom MassArray platform (Sequenom, San Diego, CA) (32). PCR and extension primers were designed using MassARRAY Assay Design 3.1 software (Sequenom). For a multiplex PCR, 2 μl (2.5 ng/μl) of each DNA sample were mixed with 2.18 μl H2O, 0.5 μl Qiagen Hot Star Buffer, 0.2 μl MgCl2 (25 mmol/l), 0.02 μl Qiagen Hot Star Taq (5 units/μl), 0.1 μl of 10 mmol/l dNTP mix and 0.5 μl of 1 μmol/l primer mix. PCR was then carried out at 95 °C for 15 min, then five cycles of 95 °C for 20 s, 65 °C for 30 s, 72 °C for 1 min, followed by five cycles of 95 °C for 20 s, 58 °C for 30 s, 72 °C for 1 min, followed by 38 cycles of 95 °C for 20 s, 53 °C for 30 s, 72 °C for 1 min, and ending with 72 °C for 3 min. Shrimp alkaline phosphatase and iPLEX Gold extension reactions were carried out according to the manufacturer’s instructions. For each reaction, 15–25 nl of sample was dispensed onto a SpectroCHIP and the chip was then analyzed using the Sequenom MALDI-TOF mass spectrometer. Mass Array Typer 3.4 software was then used to call the genotypes based on the calculated mass of the extension products.
Genotyping was considered satisfactory if the success rate for the SNP was ≥85% and the genotype distribution did not depart significantly from Hardy–Weinberg equilibrium (HWE) (P > 0.05 for χ2 test between expected and observed values) in the control groups. Hardy–Weinberg test was evaluated using the online Finetti HWE calculator (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl). In the family data, Mendelian errors were detected using Pedstats while genotypes that resulted in tight double recombinants were identified with Merlin and treated as missing data in the analyses (33).
RNA isolation
Subcutaneous adipose tissue biopsies were immediately frozen in liquid nitrogen and stored at −80 °C until analysis as described previously (30). RNA was extracted using Qiagen Lipid Tissue (Qiagen, Hilden, Germany) according to the manufacturers’ recommendations. The RNA concentration was measured spectrophotometrically using NanoDrop (Thermo Fisher Scientific, Waltham, MA) and the quality assessed by agarose gel electrophoresis.
Gene expression: measurement
Sufficient amount of high quality RNA for gene expression analysis was available from 376 siblings. Gene expression was measured using the Affymetrix Human Genome U133 Plus 2.0 gene expression arrays (Affymetrix, Santa Clara, CA) according to the manufacturers recommendations. In brief, RNA from subcutaneous tissue was reverse transcribed into cDNA and biotin-labelled cRNA was prepared by in vitro transcription (Enzo Diagnostics, Farmingdale, NY). After hybridization (according to the Minimum Information About a Microarray Experiment guideline (34)), the arrays were scanned using an Affymetrix confocal laser scanner (GeneArray scanner GCS3000; Affymetrix) and visualised using GeneChip Operating Software (GCOS; Affymetrix). Gene expression levels were normalized using the Robust Multiarray Average method (35).
Gene expression: replication
Technical replication of the gene expression results from the Affymetrix microarray was achieved using a TaqMan assay (Applied Biosystems, Carlsbad, CA). The replication cohort consisted of 71 unrelated individuals from the SibPair cohort described above for whom sufficient RNA remained after the microarray experiment (obese males: n = 12, lean males: n = 13, obese females: n = 24, lean females: n = 22). All groups were age and BMI-matched. The standard curve was generated using cDNA generated from a pool of RNA from 18 of the samples. These were representative for the cohort in terms of age, BMI, and gender distribution.
The cDNA synthesis was carried out using the High Capacity cDNA Reverse Transcription Kit from Applied Biosystems. 1 μg of RNA was used in a 100μl reaction. A total of 2 μg RNA (110 ng of each sample) was used in a 100 μl reaction for the RNA pool. For each sample 3.5 μl of cDNA was diluted with 14 μl of H2O and 5 μl of diluted cDNA was used in each well. Each sample was run in triplicate. TaqMan reagents (from Applied Biosystems) were: TaqMan Gene expression mastermix (2×) and AssayIDs Hs01009006_m1 (SIRT1) and Hs00204094_m1 (LRP10).
Statistical analysis
Case-control association analysis was performed using PLINK v1.07 (http://pngu.mgh.harvard.edu/purcell/plink/) (36). Association to obesity was analyzed by comparing allele frequencies using the assoc command. One million permutations were used to compensate for the effects of multiple testing as this method is considered the most rigorous (37). Odds ratios (OR) are reported with a 95% confidence interval (CI). Haploview 4.0 was used to calculate and display linkage disequilibrium values in the region. Linkage disequilibrium blocks were identified using the CI method described by Gabriel et al (38). Haplotypes were constructed and analyzed for association using PLINK v1.07 using linear regression and one hundred thousand permutations to correct for multiple testing. Quantitative analysis of age- and sex-corrected BMI within the controls was performed using the Kruskal–Wallis Test as implemented in SPSS v15 (SPSS, Chicago, IL). The family data was analyzed for association with BMI using the within-families model of association implemented in quantitative transmission disequilibrium test (39).
To analyze the microarray transcription data, an independent samples t-test was used to compare obese to nonobese siblings using SPSS v15 (SPSS). R2 and P values were calculated by linear regression using R project (http://www.r-project.org/). To correct for relatedness between sib-pairs, regressions were evaluated considering a clustering option within pairs. For the TaqMan replication, the expression of the SIRT1 gene was corrected using the expression of the LRP10 gene as a baseline and lean and obese expression were compared using an unpaired, two-sided, t-test.
RESULTS
Of the 27 SNPs genotyped within the French cases and controls, 22 achieved a success rate of ≥85% with an average success rate of 90.2%. Six of these were found to be monomorphic and a further five had minor allele frequencies below 1% and so were not taken forward for analysis (See Supplementary Table S1 online). We did not observe significant departure from HWE (P < 0.05). Of the remaining eleven SNPs, five were significantly associated with obesity (See Table 1).
Table 1.
Association analysis of SNP genotypes in the French Case-Control cohorts with obesity using PLINK
| Genotype numbers and minor allele frequencies (MAF) |
Results of association of SNP genotypes with obesity |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Controls |
Morbidly obese adults |
Severely obese children |
Morbidly obese adults |
Severely obese children |
||||||||||||||
| SNP | 1,1a | 1,2 | 2,2 | MAF | 1,1 | 1,2 | 2,2 | MAF | 1,1 | 1,2 | 2,2 | MAF | Empirical P value |
Corrected P value |
Odds ratio (95% CI) |
Empirical P value |
Corrected P value |
Odds ratio (95% CI) |
| rs33957861 | 874 | 254 | 21 | 12.9% | 663 | 133 | 14 | 9.9% | 482 | 121 | 8 | 11.2% | 0.006 | 0.034 b | 0.75 (0.61–0.91) |
0.153 | 0.635 | 0.85 (0.69–1.06) |
| rs737477 | 1051 | 132 | 6 | 6.1% | 449 | 56 | 1 | 5.7% | 504 | 69 | 5 | 6.8% | 0.716 | 1.000 | 0.94 (0.69–1.29) |
0.382 | 0.945 | 1.14 (0.86–1.51) |
| rs2236318 | 319 | 568 | 223 | 45.7% | 249 | 388 | 146 | 43.4% | 176 | 282 | 124 | 45.5% | 0.166 | 0.684 | 0.91 (0.8–1.04) |
0.936 | 1.000 | 0.99 (0.86–1.15) |
| rs10823103 | 503 | 475 | 111 | 32.0% | 378 | 311 | 66 | 29.3% | 286 | 223 | 62 | 30.4% | 0.086 | 0.430 | 0.88 (0.76–1.02) |
0.348 | 0.924 | 0.93 (0.79–1.08) |
| rs12413112 | 876 | 238 | 22 | 12.4% | 671 | 149 | 7 | 9.9% | 457 | 119 | 7 | 11.4% | 0.013 | 0.085 | 0.77 (0.63–0.95) |
0.396 | 0.955 | 0.91 (0.73–1.13) |
| rs11599176 | 886 | 261 | 25 | 13.3% | 684 | 156 | 8 | 10.1% | 463 | 122 | 9 | 11.8% | 0.003 | 0.019 b | 0.74 (0.61–0.9) |
0.217 | 0.767 | 0.87 (0.71–1.08) |
| rs10997865 | 480 | 511 | 129 | 34.3% | 354 | 323 | 81 | 32.0% | 249 | 264 | 71 | 34.8% | 0.135 | 0.597 | 0.9 (0.78–1.03) |
0.802 | 1.000 | 1.02 (0.88–1.18) |
| rs11596401 | 468 | 520 | 123 | 34.5% | 339 | 329 | 70 | 31.8% | 254 | 255 | 72 | 34.3% | 0.083 | 0.443 | 0.89 (0.77–1.02) |
0.936 | 1.000 | 0.99 (0.86–1.15) |
| rs35689145 | 948 | 187 | 9 | 9.0% | 693 | 107 | 1 | 6.8% | 526 | 80 | 4 | 7.2% | 0.015 | 0.101 | 0.74 (0.58–0.94) |
0.076 | 0.390 | 0.79 (0.61–1.03) |
| rs33955981 | 518 | 456 | 115 | 31.5% | 389 | 333 | 76 | 30.4% | 280 | 243 | 60 | 31.1% | 0.474 | 0.984 | 0.95 (0.83–1.09) |
0.830 | 1.000 | 0.98 (0.84–1.15) |
| rs2234975 | 952 | 220 | 7 | 9.9% | 720 | 132 | 9 | 8.7% | 499 | 75 | 6 | 7.5% | 0.188 | 0.728 | 0.87 (0.7–1.07) |
0.018 | 0.123 | 0.74 (0.57–0.95) |
P values < 0.05 are shown in boldface.
Corrected P values result from one million permutations.
1 denotes the common allele and 2 denotes the rare allele.
Statistically significant P values.
CI, confidence interval.
One SNP, rs2234975 was nominally associated with obesity in the severely obese children (P = 0.018, OR = 0.74, CI = 0.57–0.95) although this did not withstand permutation correction (P = 0.123). No association was found with this SNP in the morbidly obese adults (P = 0.188). Four SNPs, rs33957861 (P = 0.006, OR = 0.75, CI = 0.61–0.92), rs12413112 (P = 0.013, OR = 0.77, CI = 0.63–0.95), rs11599176 (P = 0.003, OR = 0.74, CI = 0.61–0.90) and rs35689145 (P = 0.015, OR = 0.74, CI = 0.58–0.95) were found to be nominally associated with obesity in the morbidly obese adults and two of these SNPs survive correction using one million permutations (rs33957861, P = 0.034 and rs11599176, P = 0.019). These SNPs were all within high linkage disequilibrium (LD) (r2 values range from 0.63 to 0.96) (see Figure 1). None of these SNPs were found to be associated with obesity in the severely obese children after correction (see Table 1). All associated SNPs had a greater frequency of the minor allele in the controls compared to the cases, suggesting a protective effect. Quantitative trait analysis revealed no association between any of the SIRT1 SNPs and age and sex-corrected BMI in either French control group (data not shown).
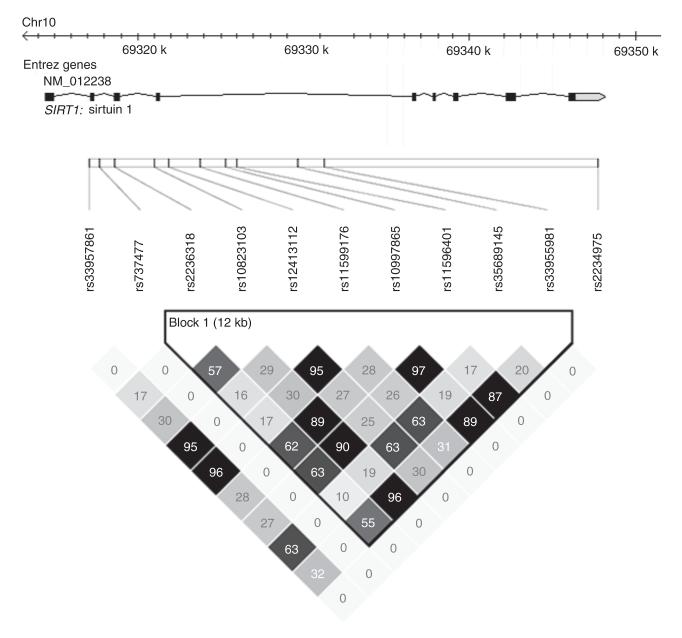
Figure 1.
Linkage disequilibrium (r2) plot of sirtuin 1 (SIRT1) SNPs genotyped in this study using genotype data from control samples only. Figures within the squares are the r2 value expressed as a percentage.
Linkage disequilibrium analysis using r2 within the control samples revealed strong linkage disequilibrium throughout the gene with one block including three of the SNPs associated with adult obesity (rs35689145, rs12413112, and rs11599176) (see Figure 1). Haplotype association analysis carried out on the four nominally associated SNPs within the adult case-control cohort revealed a significant association to obesity (P = 0.028, see Table 2).
Table 2.
Haplotype association analysis to obesity in French morbidly obese adult case-controls using the four SNPs found to be nominally associated with adult obesity
| SNP 1 | SNP 2 | SNP 3 | SNP 4 | Haplotypes testedb | Empirical P value | Corrected P value |
|---|---|---|---|---|---|---|
| rs33957861 | rs12413112 | rs11599176 | rs35689145 | CATA (AGGG) | 0.0079 | 0.0275a |
Corrected P values result from 100,000 permutations.
Statistically significant P values.
Haplotypes displayed are wild-type (variant).
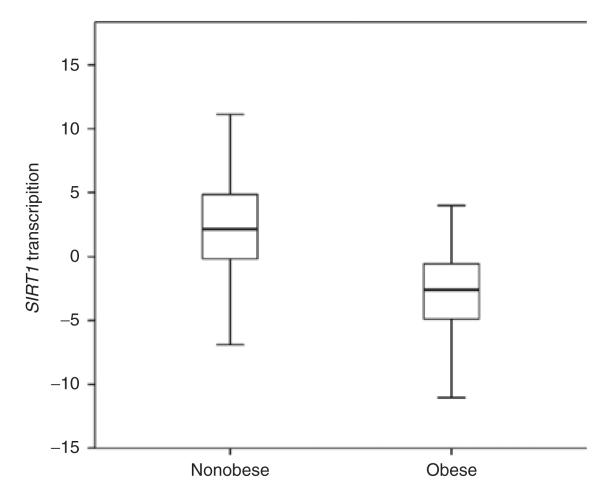
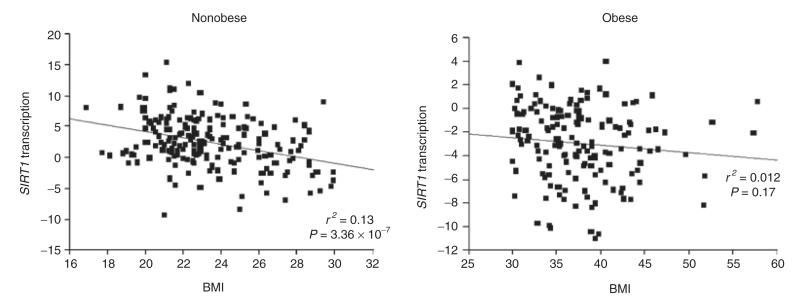
In the Swedish BMI-discordant siblings, transcript levels of SIRT1 were found to be significantly higher in lean compared to obese siblings (P = 1.56 × 10−35) (see Figure 2). BMI was found to be negatively correlated to SIRT1 expression levels in lean siblings (r2 = 0.13, P = 3.36 × 10−7) but not in obese siblings (r2 = 0.012, P = 0.17) (see Figure 3). The transcript levels of SIRT1 were confirmed as higher in lean subjects compared to obese in the subset of unrelated lean and obese subjects, using a TaqMan assay for SIRT1 expression (P = 0.016) (data not shown).
Figure 2.
Transcription levels of sirtuin 1 (SIRT1) in nonobese and obese siblings. SIRT1 transcription was measured as the DNA microarray signal value corrected for age, sex, and relatedness. Mean difference in SIRT1 transcription (95% CI) was 5.19 (4.46–5.92), P = 1.56 × 10−35. CI, confidence interval.
Figure 3.
SIRT1 transcription levels plotted against BMI in nonobese (n = 196) and obese (n = 156) siblings. SIRT1 transcription was the DNA microarray signal value corrected for age, sex, and relatedness.
The five SNPs found to be nominally associated in the French case-control groups were genotyped in the Swedish families. Four SNPs, rs11599176, rs12413112, rs33957861, and rs35689145 were found to be significantly associated with BMI (P = 4 × 10−4, 6 × 10−4, 4 × 10−4, and 0.020 respectively) (see Table 3). For each SNP, the minor allele was found to be associated with reduced BMI with a mean BMI difference of up to 2.8 kg/m2 between the homozygous common allele and homozygous minor allele (see Table 3). This again suggests a protective effect. No SNPs were found to be significantly associated with levels of SIRT1 transcript (data not shown).
Table 3.
Analysis of SIRT1 SNPs and BMI in Swedish families
| Genotype numbers |
Mean BMI ± 95% CI (kg/m2) |
||||||
|---|---|---|---|---|---|---|---|
| SNP | 1,1a | 1,2 | 2,2 | 1,1 | 1,2 | 2,2 | P value |
| rs33957861 | 535 | 153 | 9 | 29.2 ± 0.55 | 28.4 ± 1.2 | 26.4 ± 3.0 | 0.0004b |
| rs12413112 | 527 | 149 | 8 | 29.3 ± 0.56 | 28.4 ± 1.2 | 26.8 ± 3.3 | 0.0006b |
| rs11599176 | 527 | 149 | 8 | 29.3 ± 0.56 | 28.4 ± 1.2 | 26.8 ± 3.0 | 0.0004b |
| rs35689145 | 594 | 91 | 2 | 29.1 ± 0.54 | 28.3 ± 1.5 | 31.6 ± 8.9 | 0.0197b |
| rs2234975 | 579 | 99 | — | 29.2 ± 0.57 | 28.8 ± 1.1 | — | 0.9226 |
Analysis was performed using the within-families model of association in quantitative transmission disequilibrium test.
1 denotes major allele, 2 denotes minor allele.
Statistically significant P values.
DISCUSSION
These results demonstrate an association between SNPs in the SIRT1 gene and severe obesity using both unrelated cases and controls and families. For each of these SNPs, the minor allele was present at a higher frequency in the controls, indicating a protective association. The same four SNPs that were found to be nominally associated with obesity in the adult case-control study, were also found to be associated with BMI in the Swedish families. Strong linkage disequilibrium was identified throughout the gene, with one LD block, defined using Gabriel’s CI method (36), containing the two HapMap SNPs genotyped in this study, rs12413112 and rs11596401. This is consistent with the HapMap data in which there is also a large degree of linkage disequilibrium throughout the gene and one block is identified containing these two SNPs. The lack of association between SNPs and BMI in the French control groups may be due to the relative lack of variance (mean BMI ± 2.0 kg/m2) in these cohorts.
For each SNP the rare homozygote status was linked to a lower BMI, up to 2.8 kg/m2 lower than the common homozygote. Although the low numbers of subjects with the rare homozygote mean that this difference may not be accurate it is clear that the minor allele is associated with a reduced BMI and this again indicates a protective association. The correlation of SIRT1 expression to BMI in the lean group along with the association observed between SIRT1 expression and obesity in siblings further supports a contribution of SIRT1 to obesity. The replication of this result using the TaqMan assay is significant but unsurprisingly not as significant as the original result as it is a simple case-control analysis using a small subset of the cohort. The lack of association between any SIRT1 SNPs and transcription levels of the gene may be due to not genotyping the causative variant. An alternative explanation is that SIRT1 variants are causatively associated with obesity risk, but not transcription level, and it is being obese that modifies the expression of SIRT1 through other mechanisms.
Our results corroborate and expand on two recent genetic studies. The first reported a protective association between the SIRT1 SNP rs7069102 (tagged by rs11596401 in our study with r2 = 0.956 in HapMap) and obesity (P = 0.007) (23). The second reported a protective association of SIRT1 SNPs rs1467568 (P = 0.0009) and rs7895833 (P = 0.007) and obesity (25). Rs1467568 is also tagged by rs11596401 in our study (r2 = 1.0 in HapMap) but rs7895833 is located 21 kb upstream of SIRT1 and was not tagged by any SNP we investigated, though this would be a good target to investigate for association with the transcript expression level. While rs11596401 was not found to be associated with obesity in our study (P = 0.073 in adults and P = 1.000 in children), it is clear that in general terms, genetic markers in this gene are associated with obesity. As the causative variant has not been identified and because the association is not very strong (P > 10−4), it is not surprising that these associations are observed to different markers, this is probably just due to the variability introduced by different recruitment strategies or to differences in LD structure between the different populations. The finding that SIRT1 expression levels were significantly higher in nonobese compared to obese corroborates with a Danish study in which SIRT1 transcription was found to be significantly higher in lean compared to obese women (26).
The absence of SIRT1 association in any genome wide association study may be due to the stringent multiple testing corrections required and this highlights the continuing importance of candidate gene studies. It is notable that nominally-associated SNPs located near to SIRT1 (P ≤ 0.05) are present in our own obesity GWAS (genome-wide association study) data, with rs730821 (17 kb upstream of SIRT1) being associated with obesity in both French children (P = 0.013) (40) and in adult type 2 diabetics (P = 0.011) (41). No significant association is present within the Wellcome Trust Case-Control Consortium GWAS data (P < 0.05) (42). Although these are clearly not statistically significant after genome wide multiple testing correction, these nominal associations indicate that the raw GWAS association results support our findings.
All SNPs that were found to be associated are located within introns, with the exception of rs2234975, which is located in the 3′ untranslated region and so none of them directly affect the amino acid sequence of the protein. Therefore, it is likely that these SNPs are tagging another variant that is causative. The fact that the haplotype produced a lower P value than the individual SNPs may be evidence that these SNPs are tagging a more significantly associated variant, although we have not calculated whether the difference in P values is significant. With some associations reported upstream of SIRT1 (e.g., rs7895833 and rs730821) it seems likely that this untyped variant is within the region 20 kb upstream of the transcription start site, possibly within the promoter region where it could directly affect the expression levels of the SIRT1 protein. We cannot rule out the possibility that individuals with the protective minor alleles may have a higher transcription level of SIRT1 in adipose or other tissues, even though we could not demonstrate an association between the SNPs and SIRT1 transcript expression levels in subcutaneous adipose tissue. It is possible that the SIRT1 protein influences susceptibility to obesity through its effects on the expression of genes important in lipid and glucose metabolism, including PPARGC1A (8), PPARG (10–12), and ADIPOQ (13–19). Alternatively, the associated SNPs may affect splicing of the messenger RNA either directly or through LD with another variant that does.
In conclusion, our results indicate that the SIRT1 gene has a role in the development of polygenic obesity and this will require both further investigations of other populations as well as resequencing and functional work to locate the causative variant underlying this genetic association.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to acknowledge the contribution of the patients and other individuals involved in the recruitment of subjects to this study. This work was supported by grants from the Wellcome Trust (grant ref 079534/z/06/z), Swedish Research Council, the Swedish foundation for Strategic Research to Sahlgrenska Center for Cardiovascular and Metabolic Research, the Swedish Diabetes foundation, the Swedish federal government under the LUA/ALF agreement. This work was partly supported by the French Government “Agence Nationale de la Recherche”, and the charities: “Association Française des Diabétiques” and “Programme national de recherche sur le diabète”. We thank Marianne Deweider and Frederic Allegaert for the DNA bank management. We are indebted to all subjects who participated to this study. The D.E.S.I.R. study has been supported by INSERM contracts with CNAMTS, Lilly, Novartis Pharma and Sanofi-Aventis; by INSERM (Réseaux en Santé Publique, Interactions entre les déterminants de la santé), Cohortes Santé TGIR, the Association Diabète Risque Vasculaire, the Fédération Française de Cardiologie, La Fondation de France, ALFEDIAM, ONIVINS, Ardix Medical, Bayer Diagnostics, Becton Dickinson, Cardionics, Merck Santé, Novo Nordisk, Pierre Fabre, Roche, Topcon. Members of the DESIR Study Group: INSERM U1018: B.B., MA Charles. Ducimetière, E Eschwège; INSERM. U367: F Alhenc-Gelas; CHU D’Angers: Y Gallois, A Girault; Bichat. Hospital: F Fumeron, M.M.; CHU Rennes: F Bonnet; CNRS UMR8090. LILLE: P.F.; Centres d’Examens de Santé: Alençon, Angers, Caen, Chateauroux, Cholet, Le Mans, Tours; Institute de Recherche Médecine. Générale: J. Cogneau; General practitioners of the region; Institut Inter-régional pour la Santé: C Born, E Caces, M Cailleau, O.L., JG Moreau, F Rakotozafy, J Tichet, S Vol.
Footnotes
SUPPLEMENTARY MATERIAL Supplementary material is linked to the online version of the paper at http://www.nature.com/oby
DISCLOSURE The authors declared no conflict of interest.
REFERENCES
- 1.Cohen HY, Miller C, Bitterman KJ, et al. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 2.Yang T, Fu M, Pestell R, Sauve AA. SIRT1 and endocrine signaling. Trends Endocrinol Metab. 2006;17:186–191. doi: 10.1016/j.tem.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 3.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Picard F, Guarente L. Molecular links between aging and adipose tissue. Int J Obes (Lond) 2005;29(Suppl 1):S36–S39. doi: 10.1038/sj.ijo.0802912. [DOI] [PubMed] [Google Scholar]
- 5.Cantó C, Gerhart-Hines Z, Feige JN, et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freyssenet D. Energy sensing and regulation of gene expression in skeletal muscle. J Appl Physiol. 2007;102:529–540. doi: 10.1152/japplphysiol.01126.2005. [DOI] [PubMed] [Google Scholar]
- 7.Zong H, Ren JM, Young LH, et al. AMP kinase is required for mitochondrial biogenesis in skeletal muscle in response to chronic energy deprivation. Proc Natl Acad Sci USA. 2002;99:15983–15987. doi: 10.1073/pnas.252625599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Picard F, Kurtev M, Chung N, et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bäckesjö CM, Li Y, Lindgren U, Haldosén LA. Activation of Sirt1 decreases adipocyte formation during osteoblast differentiation of mesenchymal stem cells. J Bone Miner Res. 2006;21:993–1002. doi: 10.1359/jbmr.060415. [DOI] [PubMed] [Google Scholar]
- 10.Lagouge M, Argmann C, Gerhart-Hines Z, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 11.Gerhart-Hines Z, Rodgers JT, Bare O, et al. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci USA. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiao L, Shao J. SIRT1 regulates adiponectin gene expression through Foxo1-C/enhancer-binding protein alpha transcriptional complex. J Biol Chem. 2006;281:39915–39924. doi: 10.1074/jbc.M607215200. [DOI] [PubMed] [Google Scholar]
- 14.Qiang L, Wang H, Farmer SR. Adiponectin secretion is regulated by SIRT1 and the endoplasmic reticulum oxidoreductase Ero1-L alpha. Mol Cell Biol. 2007;27:4698–4707. doi: 10.1128/MCB.02279-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trujillo ME, Scherer PE. Adiponectin–journey from an adipocyte secretory protein to biomarker of the metabolic syndrome. J Intern Med. 2005;257:167–175. doi: 10.1111/j.1365-2796.2004.01426.x. [DOI] [PubMed] [Google Scholar]
- 16.Arita Y, Kihara S, Ouchi N, et al. Paradoxical decrease of an adipose-specific protein, adiponectin, in obesity. Biochem Biophys Res Commun. 1999;257:79–83. doi: 10.1006/bbrc.1999.0255. [DOI] [PubMed] [Google Scholar]
- 17.Bouatia-Naji N, Meyre D, Lobbens S, et al. ACDC/adiponectin polymorphisms are associated with severe childhood and adult obesity. Diabetes. 2006;55:545–550. doi: 10.2337/diabetes.55.02.06.db05-0971. [DOI] [PubMed] [Google Scholar]
- 18.Stumvoll M, Tschritter O, Fritsche A, et al. Association of the T-G polymorphism in adiponectin (exon 2) with obesity and insulin sensitivity: interaction with family history of type 2 diabetes. Diabetes. 2002;51:37–41. doi: 10.2337/diabetes.51.1.37. [DOI] [PubMed] [Google Scholar]
- 19.Menzaghi C, Ercolino T, Di Paola R, et al. A haplotype at the adiponectin locus is associated with obesity and other features of the insulin resistance syndrome. Diabetes. 2002;51:2306–2312. doi: 10.2337/diabetes.51.7.2306. [DOI] [PubMed] [Google Scholar]
- 20.Milne JC, Lambert PD, Schenk S, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baur JA, Pearson KJ, Price NL, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feige JN, Lagouge M, Canto C, et al. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008;8:347–358. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 23.Peeters AV, Beckers S, Verrijken A, et al. Association of SIRT1 gene variation with visceral obesity. Hum Genet. 2008;124:431–436. doi: 10.1007/s00439-008-0567-8. [DOI] [PubMed] [Google Scholar]
- 24.van den Berg SW, Dollé ME, Imholz S, et al. Genetic variations in regulatory pathways of fatty acid and glucose metabolism are associated with obesity phenotypes: a population-based cohort study. Int J Obes (Lond) 2009;33:1143–1152. doi: 10.1038/ijo.2009.152. [DOI] [PubMed] [Google Scholar]
- 25.Zillikens MC, van Meurs JB, Rivadeneira F, et al. SIRT1 genetic variation is related to BMI and risk of obesity. Diabetes. 2009;58:2828–2834. doi: 10.2337/db09-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pedersen SB, Ølholm J, Paulsen SK, Bennetzen MF, Richelsen B. Low Sirt1 expression, which is upregulated by fasting, in human adipose tissue from obese women. Int J Obes (Lond) 2008;32:1250–1255. doi: 10.1038/ijo.2008.78. [DOI] [PubMed] [Google Scholar]
- 27.Rolland-Cachera MF, Cole TJ, Sempé M, et al. Body Mass Index variations: centiles from birth to 87 years. Eur J Clin Nutr. 1991;45:13–21. [PubMed] [Google Scholar]
- 28.Jaquet D, Collin D, Lévy-Marchal C, Czernichow P. Adult height distribution in subjects born small for gestational age. Horm Res. 2004;62:92–96. doi: 10.1159/000079709. [DOI] [PubMed] [Google Scholar]
- 29.Jaziri R, Lobbens S, Aubert R, et al. DESIR Study Group. The PPARG Pro12Ala polymorphism is associated with a decreased risk of developing hyperglycemia over 6 years and combines with the effect of the APM1 G-11391A single nucleotide polymorphism: the Data From an Epidemiological Study on the Insulin Resistance Syndrome (DESIR) study. Diabetes. 2006;55:1157–1162. doi: 10.2337/diabetes.55.04.06.db05-0676. [DOI] [PubMed] [Google Scholar]
- 30.Jernås M, Olsson B, Arner P, et al. Regulation of carboxylesterase 1 (CES1) in human adipose tissue. Biochem Biophys Res Commun. 2009;383:63–67. doi: 10.1016/j.bbrc.2009.03.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 32.Sauer S, Gut IG. Genotyping single-nucleotide polymorphisms by matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;782:73–87. doi: 10.1016/s1570-0232(02)00692-x. [DOI] [PubMed] [Google Scholar]
- 33.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 34.Brazma A, Hingamp P, Quackenbush J, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 35.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 36.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salyakina D, Seaman SR, Browning BL, Dudbridge F, Muller-Myhsok B. Evaluation of Nyholt’s procedure for multiple testing correction. Hum Hered. 2005;60:19–25. doi: 10.1159/000087540. discussion 61. [DOI] [PubMed] [Google Scholar]
- 38.Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 39.Abecasis GR, Cardon LR, Cookson WO. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyre D, Delplanque J, Chèvre JC, et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet. 2009;41:157–159. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- 41.Sladek R, Rocheleau G, Rung J, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 42.Saxena R, Voight BF, Lyssenko V, et al. Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.