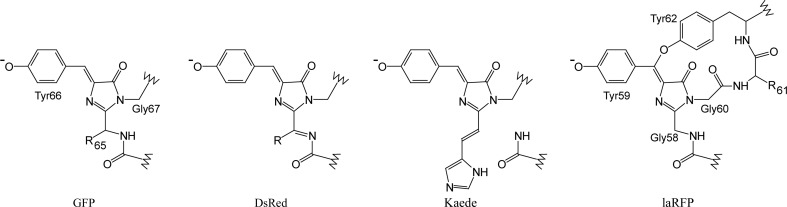
The crystal structure of the novel red emitting fluorescent protein from lancelet Branchiostoma lanceolatum (Chordata) revealed an unusual five residues cyclic unit comprising Gly58-Tyr59-Gly60 chromophore, the following Phe61 and Tyr62 covalently bound to chromophore Tyr59.
Keywords: red fluorescent proteins, GYG chromophore, Branchiostoma lanceolatum, lancelets
Abstract
A key property of proteins of the green fluorescent protein (GFP) family is their ability to form a chromophore group by post-translational modifications of internal amino acids, e.g. Ser65-Tyr66-Gly67 in GFP from the jellyfish Aequorea victoria (Cnidaria). Numerous structural studies have demonstrated that the green GFP-like chromophore represents the ‘core’ structure, which can be extended in red-shifted proteins owing to modifications of the protein backbone at the first chromophore-forming position. Here, the three-dimensional structures of green laGFP (λex/λem = 502/511 nm) and red laRFP (λex/λem ≃ 521/592 nm), which are fluorescent proteins (FPs) from the lancelet Branchiostoma lanceolatum (Chordata), were determined together with the structure of a red variant laRFP-ΔS83 (deletion of Ser83) with improved folding. Lancelet FPs are evolutionarily distant and share only ∼20% sequence identity with cnidarian FPs, which have been extensively characterized and widely used as genetically encoded probes. The structure of red-emitting laRFP revealed three exceptional features that have not been observed in wild-type fluorescent proteins from Cnidaria reported to date: (i) an unusual chromophore-forming sequence Gly58-Tyr59-Gly60, (ii) the presence of Gln211 at the position of the conserved catalytic Glu (Glu222 in Aequorea GFP), which proved to be crucial for chromophore formation, and (iii) the absence of modifications typical of known red chromophores and the presence of an extremely unusual covalent bond between the Tyr59 Cβ atom and the hydroxyl of the proximal Tyr62. The impact of this covalent bond on the red emission and the large Stokes shift (∼70 nm) of laRFP was verified by extensive structure-based site-directed mutagenesis.
1. Introduction
In the last decade, green fluorescent protein (GFP) and related fluorescent proteins (FPs) have established themselves as efficient non-invasive molecular instruments in cell biology and biomedicine, and are used for the visualization and monitoring of internal processes within cells and whole organisms (Chudakov et al., 2010 ▶; Lam et al., 2012 ▶; Wu et al., 2011 ▶). The design of new advanced biomarkers has been strongly supported by X-ray studies, which provide valuable information on the structure–property relationship. Such studies have revealed significant chemical and structural diversity of the chromophores that are positioned within the β-barrel frame of the protein and that play a key role in the manifestation of the FP spectral properties.
To date, GFP-like proteins have been found in several distant groups of marine animals: in two classes [the hydroid jellyfishes Hydrozoa (Prasher et al., 1992 ▶) and the coral polyps Anthozoa (Matz et al., 1999 ▶)] of the phylum Cnidaria; in one species from the comb jelly phlyum Ctenophora (Haddock et al., 2010 ▶); in several species of copepods (phylum Arthropoda; Shagin et al., 2004 ▶) and in lancelets (phylum Chordata; Deheyn et al., 2007 ▶). The greatest spectral diversity has been demonstrated for Anthozoa GFP-like proteins. In addition to the most widespread green FPs, coral species often express cyan, yellow and red FPs and nonfluorescent chromoproteins of various hues from orange to blue.
GFP-like proteins share the same β-barrel fold consisting of 11 β-strands with a distorted α-helix that resides inside the barrel. In contrast to other natural pigments, GFP-like proteins form their chromogenic groups by themselves. A series of post-translational modifications results in a chromophore formed by three internal amino acids within the β-barrel. For example, in A. victoria GFP the Ser65-Tyr66-Gly67 triad undergoes cyclization–dehydration of the backbone and dehydrogenation of the Tyr66 Cα—Cβ bond. These reactions lead to the creation of a green-emitting chromophore formed by two rings (a newly formed five-membered imidazolinone heterocycle and the Tyr66 side chain; Fig. 1 ▶). This GFP-like green chromophore was found to be further modified in red-shifted fluorescent proteins. Thus, the red chromophore of the DsRed type possesses a GFP chromophore core extended by an acylimine group formed by dehydrogenation of the Cα—N bond at position 65 (GFP numbering; Fig. 1 ▶). In some proteins, such as asFP595, zFP538, Kusabira Orange and mOrange, the DsRed-like chromophore is modified owing to the high chemical reactivity of the acylimine group. Another type of red chromophore is the so-called Kaede-like chromophore, which is formed by light-induced cleavage of the protein backbone at the His65 Cα—N bond with concomitant formation of a His65 Cα=Cβ double bond (Fig. 1 ▶). These examples demonstrate the diversity of chemical reactions that take place in the course of chromophore maturation. The immediate amino-acid environment of the chromophore plays a key role in this process, providing the required stereochemical support.
Figure 1.
Chemical structures of the main types of chromophores in fluorescent proteins together with the newly characterized chromophore in lancelet red fluorescent protein (laRFP).
Phylogenetic studies have revealed multiple color diversification events in GFP-like proteins (Labas et al., 2002 ▶; Shagin et al., 2004 ▶). Experimental reconstruction of estimated ancestral genes demonstrated that green fluorescence represents an ancestral state, whereas red-shifted proteins originated independently in different animal groups (Alieva et al., 2008 ▶; Field & Matz, 2010 ▶). The independent origin of the evolutionarily distant red FPs has in some cases resulted in the appearance of red chromophores with different chemical structures.
The recently discovered FPs from lancelets (Deheyn et al., 2007 ▶) remain quite poorly characterized. It was found that individual lancelet genomes contain many (up to 16) genes for GFP homologues (Baumann et al., 2008 ▶; Bomati et al., 2009 ▶). Heterologous expression of some of these genes showed that most GFP-like proteins in lancelets are green-fluorescent. In addition, red FPs from the lancelet Branchiostoma lanceolatum were described in a patent (no peer-reviewed publication is available; Israelsson, 2006 ▶). Notably, this is the only known red FP outside the Cnidaria phylum.
At present, most GFP-like FPs for which three-dimensional structures have been determined are proteins from the Cnidaria phylum. The only exceptions are the structures of the green-fluorescent proteins copGFP (Pontellina plumata; λex/λem = 482/502 nm) and CpYGFP (Chiridius poppei; λex/λem = 509/517 nm) from marine copepods (copFPs; Evdokimov et al., 2006 ▶; Suto et al., 2009 ▶; Wilmann et al., 2006 ▶). They belong to the bilaterian clade and are evolutionarily very distant from cnidarian FPs. Cephalochordates are the modern survivors of an ancient chordate lineage which evolved together with urochordates and vertebrates from a common ancestor over 520 million years ago (Holland et al., 2008 ▶; Putnam et al., 2008 ▶). The amino-acid sequences of the cephalochordate FPs are 35 ± 5% identical to the sequences of bilaterian copepod FPs and have only ∼20% sequence identity to the well studied cnidarian FPs (Baumann et al., 2008 ▶; Bomati et al., 2009 ▶; Deheyn et al., 2007 ▶). Similar to copFPs, a peculiarity of laFPs is an unusual chromophore-forming sequence Gly-Tyr-Gly with Gly as the first residue. The characteristic features of the red-emitting laRFP are a large Stokes shift of ∼70 nm and the presence of the unusual Gln211 instead of the conventional catalytic Glu which was found to be strictly conserved in all wild-type GFP-like proteins. The catalytic Glu coupled with the chromophore-proximal catalytic Arg, another highly conserved residue in the chromophore area, were shown to catalyze chromophore maturation by facilitating oxidation in the course of post-translational modifications (Barondeau et al., 2003 ▶, 2006 ▶; Pouwels et al., 2008 ▶; Sniegowski et al., 2005 ▶; Wood et al., 2005 ▶).
We reasoned that the lancelet red FPs are likely to carry a chromophore with a novel chemical structure. Indeed, lancelets (phylum Chordata) are very distant from corals and jellyfish (phylum Cnidaria). As different red chromophores have evolved in different species even within the class Anthozoa, the appearance of a new structure may be all the more expected in lancelets. The unusual spectral properties of the lancelet red FPs (see below) further support this hypothesis. Here, we present an X-ray crystallographic study of wild-type FPs from the cephalochordate group (Branchiostoma lanceolatum): green laGFP and red laRFP. As a result, we discovered a new red chromophore structure that has not been encountered before (Fig. 1 ▶). Analysis of the three-dimensional structure was supported by rational structure-based site-directed mutagenesis with emphasis on spectral characteristics. We also designed and studied laRFP-ΔS83, a genetically engineered variant of laRFP (through the deletion of Ser83) with a dramatically improved folding and maturation rate.
2. Materials and methods
2.1. Expression, purification, crystallization and mutagenesis
From the proteins described in the patent (Israelsson, 2006 ▶), we chose the red fluorescent protein blFP-R5, with the most red-shifted emission peak at ∼600 nm, for crystal structure determination. The closely related (78% identity) blFP-Y1 was taken as a representative lancelet green FP. Here, these fluorescent proteins are designated laRFP and laGFP, respectively. The cDNAs for these genes were synthesized by the PCR-based oligonucleotide method (Saiki et al., 1985 ▶) according to the GenBank sequences (accession Nos. ACA48230 and ACA48242, respectively) using mammalian-optimized codon usage. For bacterial expression, PCR-amplified BamHI/HindIII fragments encoding the target proteins were cloned into pQE30 vector (Qiagen). Proteins fused to an N-terminal six-histidine tag were expressed in Escherichia coli XL1 Blue strain (Invitrogen). The bacterial cultures were grown overnight at 310 K and were incubated for an additional 12 h at 298 K. No induction by IPTG was applied since promoter leakage was sufficient for effective expression. The cultures were centrifuged and the cell pellets were resuspended in 20 mM Tris–HCl, 100 mM NaCl pH 8.0 buffer and lysed by sonication. The recombinant proteins were purified using TALON metal-affinity resin (Clontech) followed by gel filtration on a Superdex 200 (16/60) size-exclusion column (Amersham). Absorption spectra were recorded with a Varian Cary 100 spectrophotometer, and a Varian Cary Eclipse fluorescence spectrophotometer was used to measure the excitation/emission spectra.
For all proteins, crystals were grown at 293 K by the hanging-drop vapor-diffusion method. Each drop consisted of 2 µl of either ∼13 mg ml−1 laGFP, 9 mg ml−1 laRFP or 36 mg ml−1 laRFP-ΔS83 in 20 mM Tris pH 8.0, 200 mM NaCl, 5 mM EDTA mixed with an equal amount of reservoir solution: 0.2 M MgCl2, 0.1 M bis-tris pH 5.5, 25% PEG 3350 for laGFP, 1 M sodium citrate, 0.1 M Tris pH 7.0 for laRFP and 0.2 M Na2HPO4, 20% PEG 3350 for laRFP-ΔS83. Crystals suitable for X-ray data collection reached their final size in two weeks.
Site-directed mutagenesis was performed by PCR using the overlap extension method with primers containing appropriate target substitutions (Ho et al., 1989 ▶).
2.2. X-ray data collection, structure solution and crystallographic refinement
X-ray diffraction data for all proteins were collected on SER-CAT beamlines 22-ID and 22-BM at the Advanced Photon Source, Argonne National Laboratory, Argonne, Illinois, USA. Prior to data acquisition, the crystals were transferred into a cryoprotectant solution consisting of 20% glycerol and 80% of the corresponding reservoir solution and were flash-cooled in a 100 K nitrogen stream. X-ray data for laGFP were collected in a conventional way. In the case of laRFP, to preserve the extremely unstable covalent bond between the chromophore and Tyr62, a complete data set was obtained from 17 crystals each providing 3° of data. The helical data-collection technique, in which rotation around the ω axis of the goniometer was coupled to gradual translation of the crystal along its longest direction, was used to minimize radiation damage to the laRFP-ΔS83 crystals. The experimental data corresponding to the structure of laRFP with a destroyed covalent bond between the chromophore and Tyr62 (laRFPdam) comprised of the last 30° of data from a 180° data set collected from a single laRFP crystal. All diffraction images were processed with HKL-2000 (Otwinowski & Minor, 1997 ▶). The crystal structure of laRFP was solved by the molecular-replacement method with MOLREP (Vagin & Teplyakov, 2010 ▶; Winn et al., 2011 ▶) using the coordinates of the copGFP (Pontellidae plumata) monomer (34% sequence identity; PDB entry 2g3o; Wilmann et al., 2006 ▶) as a search model. The structures of laGFP, laRFP-ΔS83 and laRFPdam were solved using the coordinates of laRFP. Crystallographic refinement of all four structures was performed with REFMAC5 (Murshudov et al., 2011 ▶) alternating with manual revision of the model using Coot (Emsley & Cowtan, 2004 ▶). Location of water molecules and structure validation were performed with Coot. Crystallographic data and refinement statistics are given in Table 1 ▶. The coordinates and structure factors of laRFP, laRFP-ΔS83, laRFPdam and laGFP have been deposited in the Protein Data Bank as entries 4jf9, 4jge, 4jeo and 4hvf, respectively.
Table 1. Crystallographic data and refinement statistics.
Values in parentheses are for the highest resolution shell. laRFP-ΔS83, laRFP mutant with Ser83 deletion. laRFPdam, structure exposed to prolonged X-ray irradiation.
| Protein | laGFP (PDB code 4hvf) | laRFP (PDB code 4jf9) | laRFP-ΔS83 (PDB code 4jge) | laRFPdam (PDB code 4jeo) |
|---|---|---|---|---|
| Crystallographic data | ||||
| Space group | P212121 | P6422 | P6422 | P6422 |
| Unit-cell parameters (Å) | a = 72.2, b = 105.8, c = 121.1 | a = b = 133.6, c = 156.1 | a = b = 133.8, c = 157.0 | a = b = 134.3, c = 156.9 |
| Z/Z′ | 16/4 | 24/2 | 24/2 | 24/2 |
| Estimated solvent content (%) | 47 | 70 | 70 | 70 |
| Temperature (K) | 100 | 100 | 100 | 100 |
| Wavelength (Å) | 1.00 | 1.00 | 1.00 | 1.00 |
| Resolution range (Å) | 26.56–1.71 (1.77–1.71) | 23.24–2.34 (2.42–2.34) | 22.76–1.94 (2.01–1.94) | 29.83–2.34 (2.42–2.34) |
| Total observations | 742799 | 168256 | 234508 | 197594 |
| Unique reflections observed | 101261 | 34207 | 60055 | 35065 |
| Multiplicity | 7.3 (6.4) | 4.9 (3.9) | 4.0 (3.9) | 5.6 (5.7) |
| 〈I/σ(I)〉 | 22.2 (4.1) | 9.4 (2.3) | 14.2 (3.2) | 13.4 (2.9) |
| R merge | 0.081 (0.415) | 0.175 (0.630) | 0.109 (0.425) | 0.131 (0.627) |
| Completeness (%) | 99.5 (95.3) | 96.3 (95.0) | 97.2 (89.1) | 98.7 (99.0) |
| Refinement statistics | ||||
| Non-H atoms in model | ||||
| Protein | 7345 [4 × residues 1–220] | 3522 [2 × residues 1–220] | 3514 [2 × residues 1–219] | 3522 [2 × residues 1–220] |
| Water | 613 | 328 | 496 | 325 |
| Glycerol | 12 | 6 | 6 | |
| R work † | 0.177 [99.0%] | 0.156 [95.9%] | 0.155 [98.0%] | 0.150 [96%] |
| R free † | 0.213 [1.0%] | 0.198 [4.1%] | 0.194 [2.0%] | 0.197 [4.0%] |
| Mean B factors (Å2) | ||||
| Protein atoms | ||||
| Main chain | 16.3 | 14.4 | 17.7 | 16.4 |
| Side chain | 18.6 | 16.2 | 20.5 | 19.2 |
| Chromophore | 14.0 | 10.4 | 15.1 | 13.6 |
| R.m.s.d.s from ideal values | ||||
| Bond lengths (Å) | 0.030 | 0.024 | 0.033 | 0.027 |
| Bond angles (°) | 2.1 | 2.0 | 2.3 | 2.0 |
| Torsion angles (period 3) (°) | 15 | 16 | 14 | 16 |
| Chirality (Å3) | 0.18 | 0.15 | 0.24 | 0.16 |
| General planes (Å) | 0.017 | 0.011 | 0.015 | 0.013 |
| Ramachandran statistics (%) | ||||
| Preferred | 96.8 | 98.4 | 98.4 | 98.3 |
| Allowed | 2.3 | 1.6 | 1.4 | 1.7 |
| Outliers | 0.9 | 0.0 | 0.2 | 0.0 |
The values in square brackets are the percentages of the data in the working (R work) and free (R free) sets.
3. Results
3.1. Spectral characteristics
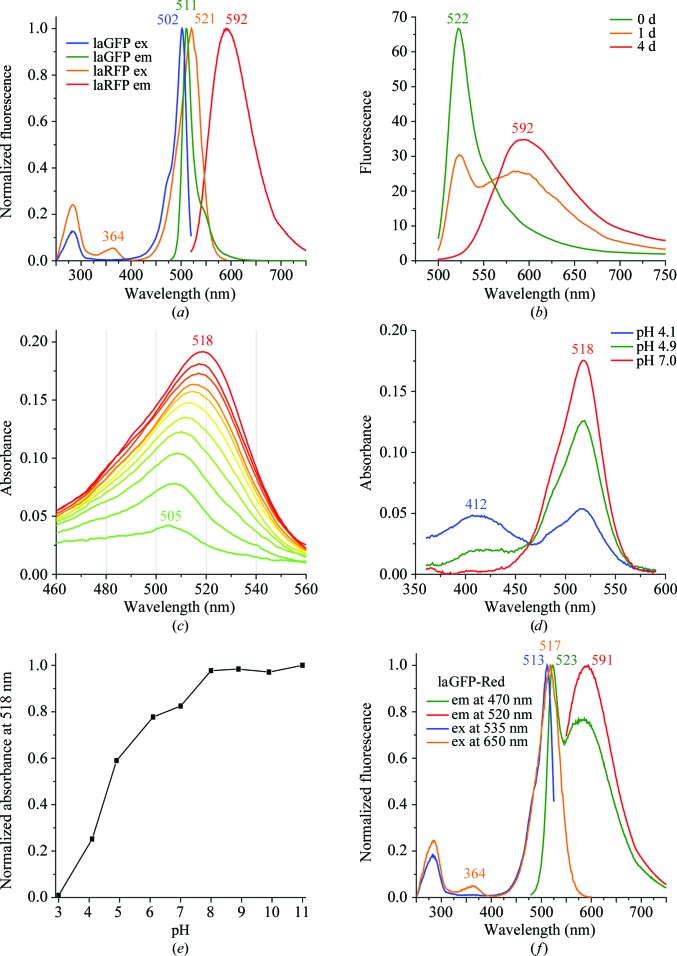
laGFP showed narrow excitation and emission peaks at 502 and 511 nm, respectively (Fig. 2 ▶ a), which are typical of green FPs. In contrast, the laRFP spectra were very unusual. The fully matured protein demonstrated red fluorescence with a major excitation peak at 521 nm and a minor peak at 364 nm and an emission maximum at 592 nm (Fig. 2 ▶ a). The emission peak was very broad, with a full-width at half-maximum (FWHM) of 95 nm (for comparison, the DsRed emission curve has an FWHM of 50 nm). The laRFP fluorescence brightness was found to be rather low, with an extinction coefficient of 71 000 M −1 cm−1 and a quantum yield of 0.1.
Figure 2.
Spectral properties of lancelet FPs. (a) Normalized excitation and emission spectra of laGFP and mature laRFP. (b) The change in the laRFP emission spectrum during the course of its maturation. (c) The change in the laRFP absorption spectrum during the course of its maturation from the green (green line) to the red (red line) state. Spectra were measured at 8 h intervals except for the final curve, which was measured on the fifth day. (d, e) The pH-dependence of the laRFP absorption spectrum. (f) Normalized excitation and emission spectra of the mutant protein laGFP-Red.
Remarkably, laRFP underwent a clearly observable maturation from the green-fluorescent to the red-fluorescent state. Freshly purified protein demonstrated green fluorescence (excitation and emission at 506 and 522 nm, respectively) and slowly converted over several days into the mature red state (Figs. 2 ▶ b and 2 ▶ c). During this conversion, the laRFP absorption maximum shifted from 505 to 518 nm (Fig. 2 ▶ c). This gradual shift apparently represents an overlay of peaks at 505 and 518 nm, which are situated too closely to be resolved as separate interconverting spectral forms. If freshly purified laRFP was placed in low-oxygen conditions (an argon-purged solution in a closed cuvette), its maturation slowed dramatically, so that green emission dominated over red emission even after 5 d of incubation (data not shown). Thus, we concluded that the green-to-red conversion of laRFP requires O2.
pH titration of mature laRFP showed that the native 518 nm peak is stable in alkaline and neutral solutions and converts into a peak at 412 nm upon acidification, with an apparent pK a of 4.7 (Figs. 2 ▶ d and 2 ▶ e). We concluded that the laRFP chromophore exists in a deprotonated (anionic) state owing to ionization of the OH group of Tyr59, a process that is typical for the chromophores of fluorescent proteins.
The deletion of Ser83 in the loop in the β-barrel cap did not change the spectra of laRFP-ΔS83 compared with the parental wild-type laRFP, but dramatically improved the efficiency of its folding, decreasing its maturation time from several days to overnight.
3.2. Overall structure
The asymmetric unit of the laGFP crystal contains one GFP-like tetramer (222 point symmetry) and those of laRFP/laRFPdam and laRFP-ΔS83 contain one dimer. Crystallographic symmetry operations create tetramers out of dimers. The observed electron density allowed the unambiguous fitting of residues 1–220 for all subunits. The principal fold of the monomeric subunit, which is shared with all FP members from the Cnidaria phylum, is an 11-stranded β-barrel closed from both sides by loop caps, with a chromophore embedded in the middle of an internal distorted helix that is wound along the β-barrel axis.
3.3. Structural features of the chromophore area
3.3.1. Red laRFP
No significant differences were found between the structures of laRFP and its improved variant laRFP-ΔS83. Both red-emitting laFPs have a chromophore structure with a basic core typical of the evolutionarily distant green-emitting cnidarian FPs as well as the green-emitting bilaterian laGFP, copGFP (Wilmann et al., 2006 ▶) and CpYGFP (Suto et al., 2009 ▶). The post-translational modification of the chromophore-forming sequence Gly58-Tyr59-Gly60 in laRFP results in a nearly coplanar two-ring chromophore consisting of a five-membered imidazolinone heterocycle with a p-hydroxybenzylidene substituent. The cis orientation of the Tyr59 phenolic ring relative to the Cα—N bond is characterized by torsion angles of ∼4° and ∼19° around the corresponding Cα—Cβ and Cβ—Cγ bonds. The peptide group between the first chromophore residue Gly58 and the preceding Ile57 in laRFP adopts a trans conformation with a single-bond character of the N—Cα (Gly58) bond, which does not lie in the chromophore plane.
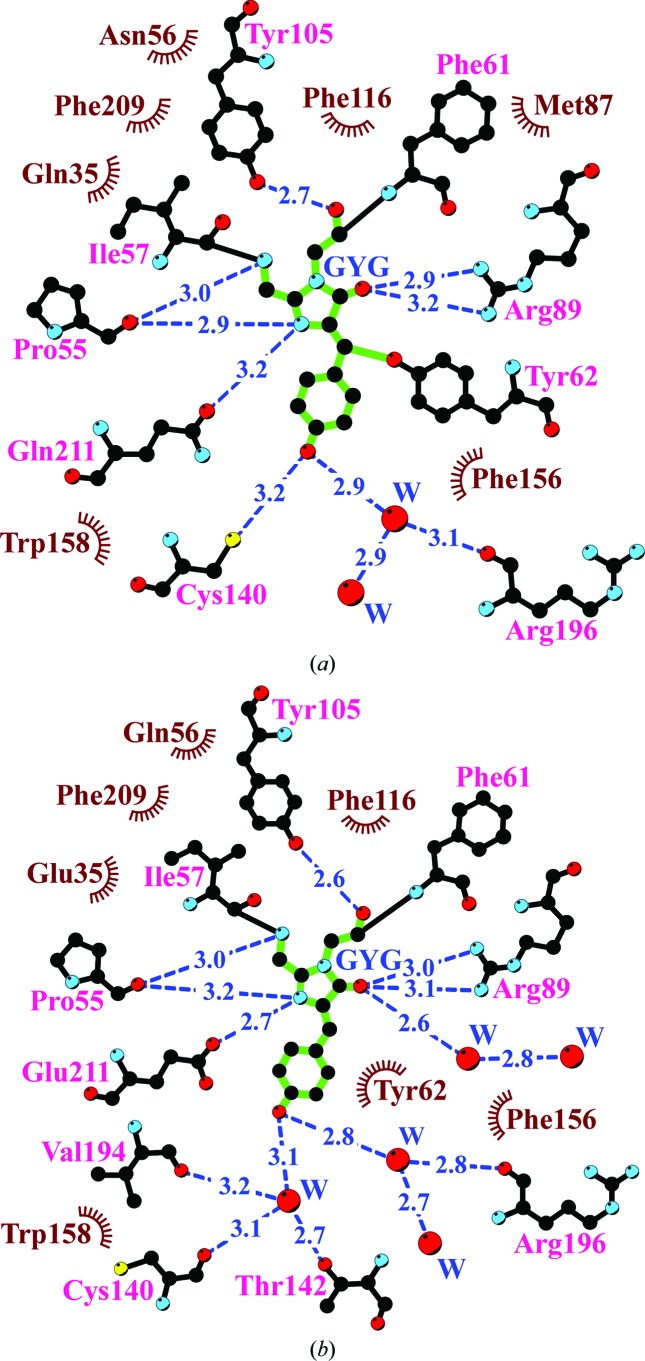
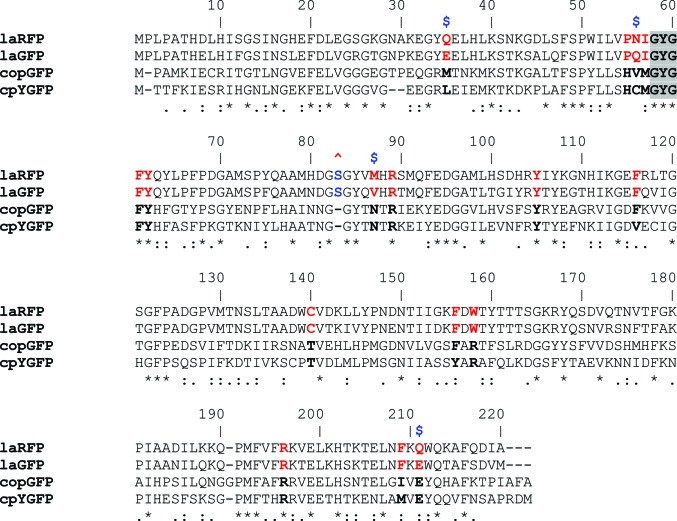
In laRFP, the shell within 3.9 Å distance of the chromophore is composed of 16 residues and two water molecules. The position of the chromophore is stabilized by 17 van der Waals hydrophobic contacts and eight hydrogen bonds to its immediate environment (Fig. 3 ▶ a). Remarkably, nearly half of the residues (Phe61, Tyr62, Tyr105, Phe116, Pe156, Trp158 and Phe209) are aromatic. The nearest shell of laRFP differs from that of laGFP by four amino-acid residues: Gln35, Asn56, Met87 and Gln211 (Fig. 4 ▶). The most remarkable feature of the laRFP environment is the presence of Gln at position 211. In all naturally occurring fluorescent proteins reported to date this position is occupied by a strictly conserved catalytic Glu211 that is considered to be essential for chromophore maturation. This makes laRFP the first wild-type fluorescent protein without a catalytic glutamate.
Figure 3.
The nearest amino-acid environment of the chromophore in the structures of laRFP (a) and laGFP (b). Hydrogen bonds (≤3.3 Å) are shown as blue dashed lines, water molecules (W) as red spheres and van der Waals contacts (≤3.9 Å) as black ‘eyelashes’ (this figure was prepared with LIGPLOT/HBPLUS; Wallace et al., 1995 ▶).
Figure 4.
Alignment of the amino-acid sequences (overall identity of ∼24%). Residues in the nearest environment to the chromophore in the laFPs and the copFPs are shown in bold red and black, respectively; differences between laRFP and laGFP are marked with blue $ symbols. The insertion Ser83 is marked with a red ^ symbol. The pairwise identity between the laFPs is ∼77%, that between the copFPs is ∼50% and that between the laFPs and the copFPs is ∼32%.
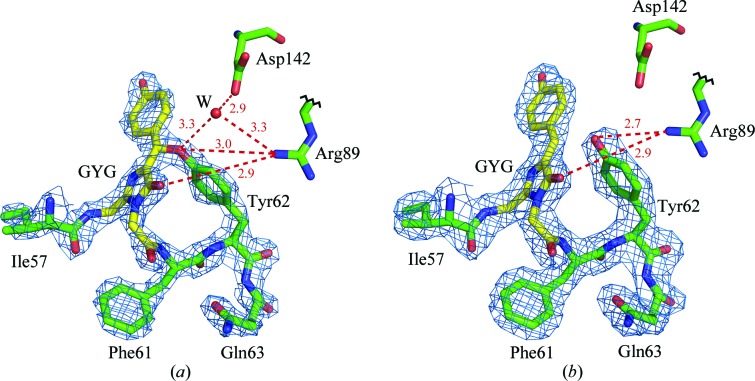
Another dramatic finding that is evident in the laRFP/laRFP-ΔS83 crystal structures is a covalent bond between the chromophore Tyr59 Cβ atom and the phenolic O atom of Tyr62 (Fig. 5 ▶ a). An ∼1.4 Å distance between the corresponding atoms and a high level of electron density unambiguously indicate the presence of a covalent bond. The latter is nearly coplanar with the chromophore and closes a larger cycle comprised of the GYG chromophore, Phe61 and Tyr62. In the course of X-ray experiments, it was found that the Tyr59 Cβ—O Tyr62 covalent bond is highly susceptible to radiation damage. The decomposition of this bond in a crystal exposed to prolonged X-ray irradiation (see §2 and Table 1 ▶) was observed in the laRFPdam structure (Fig. 5 ▶ b) as an increase in the corresponding distance from ∼1.4 to ∼2.7 Å, which was interpreted as a superposition of the covalently bound red form and the unlinked green form. In addition to the covalent bond to the chromophore, the Tyr62 O atom forms a direct hydrogen bond to the proximal catalytic Arg89 and a water-mediated hydrogen bond to Asp142.
Figure 5.
The Gly58-Tyr59-Gly60 chromophore (GYG; shown in yellow) in 2F o − F c electron density: (a) laRFP (density cutoff ρ = 2.0σ) showing the presence of the covalent bond between the chromophore (Tyr59) Cβ and the proximal (Tyr62) O. The potential proton acceptor Asp142 forms a hydrogen bond (shown as a dashed red line), mediated by a water molecule (W), to the Tyr62 hydroxyl. The ligands of water W form a distorted tetrahedron and include another water molecule (not shown). Arg89 forms a hydrogen bond to the Tyr62 hydroxyl that presumably may facilitate proton transfer to Asp142. (b) laRFPdam (density cutoff ρ = 1.0σ) showing the absence of a covalent bond between the chromophore and Tyr62. No water molecule corresponding to W in LaRFP is present, most likely because of a 1 Å shift of the hydroxyl of Tyr62 towards Asp142. This figure was created with PyMOL (DeLano, 2002 ▶).
Understanding the unique spectral behavior of laRFP requires paying particular attention to structural features in the area of the chromophore. The mutually hydrogen-bonded Gln211 and Gln35 interact with the chromophore cyclic unit, forming a direct hydrogen bond to the imidazolinone N atom (Tyr59) and a water-mediated hydrogen bond to the backbone of Phe61, respectively. The side chain of Cys140 is positioned at a hydrogen-bonding distance (3.2 Å) from the hydroxyl of Tyr59. However, it has an unfavorable angular orientation relative to the corresponding C—OH (Tyr59), preventing the formation of an effective hydrogen bond (Fig. 3 ▶ a).
Superposition of the structures of laFPs and copFPs on the basis of their Cα atoms revealed a relative ∼1 Å shift of their chromophores perpendicular to their planes along the axis of the β-barrel. This shift most probably originates from trans–cis isomerization of the peptide bond preceding Pro55, a residue that is close to the chromophore. In copFPs this position is occupied by His, favoring a trans configuration of the corresponding bond.
3.3.2. Green laGFP
LaGFP, with the conserved catalytic Glu211, adopts a conventional bicyclic chromophore structure typical of GFP-like green-emitting FPs and similar to the basic structure of the chromophore core (without the Tyr62 substituent) in laRFP. 17 residues and five water molecules within the nearest shell form nine direct and six water-mediated hydrogen bonds to the chromophore; nine of them are similar to those in laRFP (Fig. 3 ▶ b). In contrast to laRFP, the distance between the chromophore Tyr59 Cβ atom and the proximal Tyr62 hydroxyl in laGFP is 3.45 Å, indicating a noncovalent character of their interaction. The hydroxyl group of Tyr62 forms two direct hydrogen bonds to the catalytic Arg89 and Asp154. It should be noted that the hydrogen bond connecting Gln211 and Gln35 in laRFP is not present in laGFP, in which both these positions are occupied by Glu.
3.4. Structure-based site-directed mutagenesis and photophysical characterization
To identify the key residues responsible for biosynthesis of the unique chromophore structure and, as a consequence, the observed spectral properties of laRFP, we performed careful site mapping around the chromophore both in laRFP and laGFP. The role of the potentially important residues was tested by site-directed mutagenesis with an emphasis on laRFP sites that differed from those in laGFP and copFPs (Table 2 ▶). The most challenging exercise undertaken was stepwise mutagenesis aimed at the conversion of the green-emitting laGFP to a red-emitting variant by the introduction of a minimal number of mutations in the chromophore area.
Table 2. Spectral characteristics.
n/d, nondetectable.
| λex (nm) | λem (nm) | |
|---|---|---|
| laRFP mutants | ||
| WT laRFP | 521 | 592 |
| ΔSer83 | 514 | 594 |
| Tyr62Ser | 514 | 524 |
| Tyr62Gln | 514 | 526 |
| Tyr62Phe | 511 | 526 |
| Gln35Met/Leu | 504 | 523 |
| Cys140Ser | 500 | 518 |
| Cys140Asn | 488 | 523 |
| Gln211Glu | 512 | 521 |
| Pro55His | 494 | 511 |
| Pro55Ala | 490 | 520 |
| laGFP mutants† | ||
| WT laGFP | 502 | 511 |
| a | 505 | 514 |
| a + b | 505 | 514 |
| a + b + c | n/d | n/d |
| a + b + c + d (laGFP-Red) | 512/518 | 523/591 |
a, Thr142Asp + Asp154Gly; b, Glu211Gln + Glu35Gln; c, Gln56Asn + Val87Met ; d, Gly101Ser + Tyr103His.
3.4.1. LaRFP mutagenesis
Deletion of Ser83 in wild-type laRFP created the laRFP-ΔS83 variant, which had the same spectral characteristics but greatly improved folding and a greatly improved chromophore-maturation rate; the latter decreased from several days (laRFP) to only several hours. This deletion apparently influences the slowest phase of protein folding, which is responsible for the rate of the whole process. It should be pointed out that the residue at position 83 is absent in copFPs (Fig. 4 ▶).
Replacement with Phe, Gln or Ser of the most critical residue, Tyr62, which is covalently bridged to the laRFP chromophore, shifted the fluorescence to the green-yellow range, with emission at ∼525 nm (Table 2 ▶). Elimination of a specific hydrogen bond between Gln211 and Gln35 by the introduction of the single mutations Gln211Glu or Gln35Met also yielded green-emitting variants. Surprisingly, the replacement of Cys140, which presumably favors a fluorescent cis chromophore, with the more potent hydrogen donor Ser produced a green-emitting variant with a considerably dimmer fluorescence. To tackle the role of an unusual cis configuration of the peptide bond preceding Pro55, we generated two mutants, Pro55Ala or Pro55His, favoring a trans conformation of this peptide bond. These two variants were green-emitting with only weak fluorescence.
3.4.2. LaGFP mutagenesis
Based on a comparison of the structures of laGFP and laRFP, which differ at a total of 50 positions, we made an attempt to convert the green-emitting laGFP into a red-emitting variant using the lowest possible number of replacements by stepwise site-directed mutagenesis. We chose four positions in the immediate chromophore environment that differ in laGFP and laRFP: Glu211Gln, Glu35Gln, Gln56Asn and Val87Met. Two other positions, chosen for the importance of their hydrogen bonding to Tyr62, were Thr142Asp and Asp154Gly, located ∼4.5–6 Å from the chromophore. The mutagenesis was carried out in four steps, with two replacements introduced at each step in the variant obtained in the preceding step. The variants produced at each step were characterized by their excitation/emission spectra (Table 2 ▶). The first step (Thr142Asp + Asp154Gly) resulted in a negligible red shift and some decrease in fluorescence brightness. The second step [(Thr142Asp + Asp154Gly) + (Glu211Gln + Glu35Gln)] did not change the emission of the protein but resulted in a considerable decrease in the fluorescence brightness. The third step [(Thr142Asp + Asp154Gly) + (Glu211Gln + Glu35Gln) + (Gln56Asn + Val87Met)] resulted in a colorless nonfluorescent variant, probably because of steric hindrance between Met87 and Tyr103. Finally, the fourth step [(Thr142Asp + Asp154Gly) + (Glu211Gln + Glu35Gln) + (Gln56Asn + Val87Met) + (Gly101Ser + Tyr103His)] led to a mutant named laGFP-Red that, similar to laRFP, underwent a gradual (but incomplete) green-to-red conversion, producing an emission spectrum with 523 and 591 nm bands (Fig. 2 ▶ f and Table 2 ▶). Notably, the excitation spectrum for the red emission band of laGFP-Red possessed both the major 517 nm and the minor 364 nm peaks that are also characteristic of laRFP.
4. Discussion
4.1. Structural features
In spite of their considerable diversity, a characteristic structural feature of all naturally occurring red-emitting FPs (RFPs) from the Cnidaria phylum is the presence of a double bond at the Cα atom of the first chromophore-forming residue (Petersen et al., 2003 ▶; Pletnev et al., 2012 ▶; Pletneva et al., 2007 ▶; Tubbs et al., 2005 ▶; Wall et al., 2000 ▶; Wilmann et al., 2005 ▶; Yarbrough et al., 2001 ▶). This bond lies in the chromophore plane and extends the conjugated π-electron system of the chromophore, causing a bathochromic shift of its fluorescence relative to green-emitting FPs. In some cases, the formation of red chromophores leads to a break in the protein backbone just before the chromophore (Hayashi et al., 2007 ▶). In contrast, the red-emitting laRFP possesses neither a double bond nor a backbone break at the position preceding its chromophore. Thus, previously determined red-shifting modifications do not contribute to the conjugated system of the laRFP chromophore.
A unique structural feature of laRFP that has not been observed in FPs reported previously is a covalent bond (∼1.4 Å) between the Cβ atom of the chromophore Tyr59 and the hydroxyl O atom of the proximal Tyr62, (GYG)–Tyr62 (Fig. 5 ▶ a). The resulting emitting unit can be described as a basic GFP-like chromophore core with an oxygen-bridged aromatic substituent, presumably causing red emission of laRFP even in the absence of the N-acylimine double bond Cα=N. There are a number of examples of fluorescent dyes that emit at significantly different wavelengths upon the introduction of appropriate substituents (Christie, 2008 ▶; Hermanson, 2008 ▶; Lakowicz, 2006 ▶). The hydroxyl O atom of Tyr62 forms a direct hydrogen bond to the proximal Arg89, which may influence the (Tyr62) OH electronic distribution prior to the reaction, potentially facilitating proton transfer. It also forms a water-mediated hydrogen bond to Asp142, which is somewhat distant from the chromophore but could act as a potential proton acceptor in the chemical reactions leading to the formation of a covalent bond. It should be noted that this position is occupied by Thr142 in green laGFP. Replacement of Tyr62 with Phe, Gln or Ser prevents the formation of a covalent bond (GYG)–Tyr62 and yields variants with emission maxima at ∼525 nm (Table 2 ▶). In green-emitting laGFP the distance of 3.45 Å between the Cβ atom of Tyr59 of the chromophore and the hydroxyl of Tyr62 unambiguously indicates the absence of their interaction.
When considering the influence of the (GYG)–Tyr62 covalent bond on the photophysical properties of the laRFP chromophore, it should be noted that the appearance of red emission mainly results not from a strong absorption change (it is only ∼13 nm) but from a dramatic increase of the Stokes shift, which approaches 70 nm, a so-called large Stokes shift (LSS). In conventional FPs, an LSS is typically associated with excited-state proton transfer (ESPT; Henderson et al., 2009 ▶; Piatkevich et al., 2010 ▶; Shu et al., 2007 ▶; Stoner-Ma et al., 2005 ▶), which could not be envisioned for laRFP since it carries a chromophore that is already deprotonated (see Fig. 2 ▶). The exact role of the (GYG)–Tyr62 bond in the manifestation of the spectral properties of laRFP is currently unclear. Further studies, such as the chemical synthesis of corresponding model chromophores as well as quantum-chemical calculations, are required to clarify this issue.
Another distinguishing feature in the area of the chromophore is that Pro55 has a carbonyl at a hydrogen-bonding distance from the chromophore Gly58 and Tyr59 N atoms with the preceding peptide bond in a rare cis conformation. Whereas the overwhelming majority of peptide bonds in proteins are trans, nearly ∼6% of X–Pro bonds adopt a cis conformation (Pal & Chakrabarti, 1999 ▶; Stewart et al., 1990 ▶). The cis and trans isomers of the X–Pro bond, where X may be any residue, are nearly equal energetically, which is not true for all other X–Y bonds where Y ≠ Pro. Trans–cis isomerization of an X–Pro peptide bond shortens the corresponding Cα⋯Cα distance by ∼0.8 Å and can strongly affect the direction of the protein chain. Which of the two configurations will be adopted usually depends on which one will provide optimal protein folding and the tendency of the protein structure to minimize local steric tensions. The presence of a cis-peptide bond X–Pro55 in laFPs is most likely to account for the observed ∼1 Å shift of their chromophores along the β-barrel axis relative to copFPs. The replacement of Pro55 in laRFP by His or Ala resulted in green-emitting variants with a considerably dimmer fluorescence, presumably owing to the opposite cis–trans isomerization of the peptide bond between residues 54 and 55.
4.2. Key sites
Among all known wild-type fluorescent proteins, laRFP is the first example of an FP in which Gln211 replace the strictly conserved catalytic Glu211. In laRFP, Gln211 and Gln35 are hydrogen bonded to each other and interact with the chromophore, forming direct and water-mediated hydrogen bonds with the imidazolinone (Tyr59) N atom and the backbone of Phe61, respectively. In green-emitting counterparts hydrogen bonding between residues at positions 211 and 35 is absent because Glu211 and Glu35 in laGFP and Glu211 and Met35 in copFPs (laFP numbering here and in the following; Fig. 4 ▶) are unable to form hydrogen bonds to each other. Disruption of the Gln211⋯Gln35 hydrogen bond in laRFP by the single-point mutations Gln211Glu or Gln35Met caused an ∼70 nm blue shift of the chromophore fluorescence (Table 2 ▶). Such a drastic change suggests that the hydrogen-bonded Gln211–Gln35 pair is one of the key prerequisites in the proton/electron-transport system supporting specific chemistry in the formation of the (GYG)–Tyr62 covalent bond.
The SH group of Cys140, which is located 3.3 Å from the OH group of Tyr59, occupies a position providing a hydrogen bond that stabilizes the fluorescent cis conformation of the chromophore. However, its unfavorable orientation practically disables effective hydrogen bonding to the chromophore. The Cys140Ser mutation in laRFP was expected to enable the creation of a hydrogen bond to Tyr59 and to affect the brightness of red fluorescence, since Ser is known to form stronger hydrogen bonds. A Cys140Ser replacement, however, resulted in a green-yellow-emitting variant, presumably indicating the absence of the (GYG)–Tyr62 covalent bond. The reason for the observed effect is not quite clear. We rationalize that Ser140 affects the chromophore, preventing the formation of the (GYG)–Tyr62 bond essential for red fluorescence. We assume that a small spatial shift of the Tyr59 phenolic ring from the optimal reaction position stops chromophore maturation at the conventional green state.
Among a total of 50 amino-acid residues that differ between the green laGFP and red laRFP sequences (Fig. 4 ▶), only four are located in the immediate environment of the chromophore (∼4 Å): Glu211Gln, Glu35Gln, Gln56Asn and Val87Met (Fig. 3 ▶). Two other important residues that differ in the two proteins, Asp154Gly and Thr142Asp, belong to the second shell of amino-acid residues surrounding the chromophore. In laGFP, Asp154 forms a direct hydrogen bond to the hydroxyl of Tyr62, partially neutralizing the reactivity of Tyr62, whereas Gly154 in laRFP is not capable of hydrogen bonding. In laRFP, Asp142 forms a water-mediated hydrogen bond to the hydroxyl of Tyr62, making it a potential proton acceptor required for formation of the (GYG)–Tyr62 bond. In laGFP, this site is occupied by the less efficient Thr142.
The four consecutive steps of the double point mutations in green laGFP [(Thr142Asp + Asp154Gly) + (Glu211Gln + Glu35Gln) + (Gln56Asn + Val87Met) + (Gly101Ser + Tyr103His)] resulted in the appearance of a red emission peak at 591 nm corresponding to that of the wild-type laRFP (Fig. 2 ▶ f). The red peak, which was present together with the green peak, suggests incomplete maturation of the red chromophore. A complete green-to-red conversion apparently requires the introduction of additional mutations. The results of mutagenesis suggest that a considerable number of amino-acid replacements in the nearest and more remote chromophore environmental shells of laGFP are required to provide an optimal stereochemical scaffold enabling formation of the GYG–Tyr62 covalent bond crucial for red emission of laRFP.
Very recent work has demonstrated that lancelets might provide a source of highly useful FPs for practical applications (Shaner et al., 2013 ▶). On the basis of a computer-modeled three-dimensional structure of a tetrameric green FP from B. lanceolatum, the authors succeeded in the development of an extremely bright monomeric variant of this protein called mNeonGreen. The experimentally determined crystal structure of the closely related (85% identity) laGFP presented here should provide a structural basis for additional improvement of mNeonGreen, as well as for engineering novel lancelet-derived monomeric FPs of different colors.
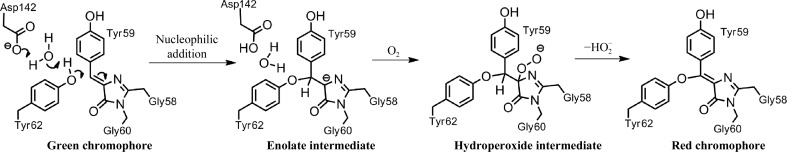
4.3. Proposed mechanism
Based on the data shown above, we are now able to propose a mechanism for the formation of the red chromophore from the intermediate green state (Fig. 6 ▶). The first event in this consecutive series is likely to be nucleophilic addition of Tyr62 to the Tyr59 Cβ=Cα double bond of the green chromophore (Dong et al., 2008 ▶) catalyzed by Asp142. Migration of the Tyr62 phenolic proton to the Asp142 carboxylate acceptor is mediated by a water molecule. As a result, an enolate intermediate is formed, which is attacked by an oxygen molecule to form a peroxide, similarly to the key event in the maturation mechanism of the green chromophore itself (Craggs, 2009 ▶). Finally, β-elimination of hydrogen peroxide results in regeneration of the Tyr59 Cα=Cβ double bond, with the overall oxidation state being increased by two.
Figure 6.
Proposed mechanism for oxidative green-to-red chromophore maturation in laRFP.
The role of Asp142 in the chemistry of the proposed process is essential, but not sufficient. Our mutagenesis results show that a relatively large number of other residues support the process. In particular, the key chromophore-proximal residues in laRFP, Gln211, Gln35 and Asn56 are incorporated in a hydrogen-bond network which is expected to provide proton transport for the specific bonding reaction between Tyr62 and the chromophore. The other significant residues that were identified, namely Met87, His103 and Ser101, are most likely to adjust the scaffold for the functional group primarily involved in the reaction chain.
Supplementary Material
PDB reference: laRFP, 4jf9
PDB reference: laRFP-ΔS83, 4jge
PDB reference: laRFPdam, 4jeo
PDB reference: laGFP, 4hvf
Acknowledgments
Diffraction experiments were carried out on beamline 22-ID of the Southeast Regional Collaborative Access Team (SER-CAT) located at the Advanced Photon Source, Argonne National Laboratory. Use of the APS was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. W-31-109-Eng-38. This project has been supported in part with Federal funds from the National Cancer Institute, National Institutes of Health (NIH) contract No. HHSN261200800001E, by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, by a grant from the Russian Foundation for Basic Research 12-04-31615 and by the Ministry of Education and Science of the Russian Federation (project 14.512.11.0052). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the US Government.
References
- Alieva, N. O., Konzen, K. A., Field, S. F., Meleshkevitch, E. A., Hunt, M. E., Beltran-Ramirez, V., Miller, D. J., Wiedenmann, J., Salih, A. & Matz, M. V. (2008). PLoS One, 3, e2680. [DOI] [PMC free article] [PubMed]
- Barondeau, D. P., Kassmann, C. J., Tainer, J. A. & Getzoff, E. D. (2006). J. Am. Chem. Soc. 128, 4685–4693. [DOI] [PubMed]
- Barondeau, D. P., Putnam, C. D., Kassmann, C. J., Tainer, J. A. & Getzoff, E. D. (2003). Proc. Natl Acad. Sci. USA, 100, 12111–12116. [DOI] [PMC free article] [PubMed]
- Baumann, D., Cook, M., Ma, L., Mushegian, A., Sanders, E., Schwartz, J. & Yu, C. R. (2008). Biol. Direct, 3, 28. [DOI] [PMC free article] [PubMed]
- Bomati, E. K., Manning, G. & Deheyn, D. D. (2009). BMC Evol. Biol. 9, 77. [DOI] [PMC free article] [PubMed]
- Christie, R. M. (2008). Rev. Prog. Coloration Relat. Top. 23, 1–18.
- Chudakov, D. M., Matz, M. V., Lukyanov, S. & Lukyanov, K. A. (2010). Physiol. Rev. 90, 1103–1163. [DOI] [PubMed]
- Craggs, T. D. (2009). Chem. Soc. Rev. 38, 2865–2875. [DOI] [PubMed]
- Deheyn, D. D., Kubokawa, K., McCarthy, J. K., Murakami, A., Porrachia, M., Rouse, G. W. & Holland, N. D. (2007). Biol. Bull. 213, 95–100. [DOI] [PubMed]
- DeLano, W. L. (2002). PyMOL http://www.pymol.org.
- Dong, J., Abulwerdi, F., Baldridge, A., Kowalik, J., Solntsev, K. M. & Tolbert, L. M. (2008). J. Am. Chem. Soc. 130, 14096–14098. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Evdokimov, A. G., Pokross, M. E., Egorov, N. S., Zaraisky, A. G., Yampolsky, I. V., Merzlyak, E. M., Shkoporov, A. N., Sander, I., Lukyanov, K. A. & Chudakov, D. M. (2006). EMBO Rep. 7, 1006–1012. [DOI] [PMC free article] [PubMed]
- Field, S. F. & Matz, M. V. (2010). Mol. Biol. Evol. 27, 225–233. [DOI] [PMC free article] [PubMed]
- Haddock, S. H., Mastroianni, N. & Christianson, L. M. (2010). Proc. Biol. Sci. 277, 1155–1160. [DOI] [PMC free article] [PubMed] [Retracted]
- Hayashi, I., Mizuno, H., Tong, K. I., Furuta, T., Tanaka, F., Yoshimura, M., Miyawaki, A. & Ikura, M. (2007). J. Mol. Biol. 372, 918–926. [DOI] [PubMed]
- Henderson, J. N., Osborn, M. F., Koon, N., Gepshtein, R., Huppert, D. & Remington, S. J. (2009). J. Am. Chem. Soc. 131, 13212–13213. [DOI] [PMC free article] [PubMed]
- Hermanson, G. T. (2008). Bioconjugate Techniques, 2nd ed. Amsterdam: Elsevier.
- Ho, S. N., Hunt, H. D., Horton, R. M., Pullen, J. K. & Pease, L. R. (1989). Gene, 77, 51–59. [DOI] [PubMed]
- Holland, L. Z. et al. (2008). Genome Res. 18, 1100–1111. [DOI] [PMC free article] [PubMed]
- Israelsson, O. (2006). US Patent 20090286314 A1.
- Labas, Y. A., Gurskaya, N. G., Yanushevich, Y. G., Fradkov, A. F., Lukyanov, K. A., Lukyanov, S. A. & Matz, M. V. (2002). Proc. Natl Acad. Sci. USA, 99, 4256–4261. [DOI] [PMC free article] [PubMed]
- Lakowicz, J. R. (2006). Principles of Fluorescence Spectroscopy, pp. 63–94. New York: Springer.
- Lam, A. J., St-Pierre, F., Gong, Y., Marshall, J. D., Cranfill, P. J., Baird, M. A., McKeown, M. R., Wiedenmann, J., Davidson, M. W., Schnitzer, M. J., Tsien, R. Y. & Lin, M. Z. (2012). Nature Methods, 9, 1005–1012. [DOI] [PMC free article] [PubMed]
- Matz, M. V., Fradkov, A. F., Labas, Y. A., Savitsky, A. P., Zaraisky, A. G., Markelov, M. L. & Lukyanov, S. A. (1999). Nature Biotechnol. 17, 969–973. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Pal, D. & Chakrabarti, P. (1999). J. Mol. Biol. 294, 271–288. [DOI] [PubMed]
- Petersen, J., Wilmann, P. G., Beddoe, T., Oakley, A. J., Devenish, R. J., Prescott, M. & Rossjohn, J. (2003). J. Biol. Chem. 278, 44626–44631. [DOI] [PubMed]
- Piatkevich, K. D., Malashkevich, V. N., Almo, S. C. & Verkhusha, V. V. (2010). J. Am. Chem. Soc. 132, 10762–10770. [DOI] [PMC free article] [PubMed]
- Pletneva, N., Pletnev, V., Tikhonova, T., Pakhomov, A. A., Popov, V., Martynov, V. I., Wlodawer, A., Dauter, Z. & Pletnev, S. (2007). Acta Cryst. D63, 1082–1093. [DOI] [PubMed]
- Pletnev, S., Pletneva, N. V., Souslova, E. A., Chudakov, D. M., Lukyanov, S., Wlodawer, A., Dauter, Z. & Pletnev, V. (2012). Acta Cryst. D68, 1088–1097. [DOI] [PMC free article] [PubMed]
- Pouwels, L. J., Zhang, L., Chan, N. H., Dorrestein, P. C. & Wachter, R. M. (2008). Biochemistry, 47, 10111–10122. [DOI] [PMC free article] [PubMed]
- Prasher, D. C., Eckenrode, V. K., Ward, W. W., Prendergast, F. G. & Cormier, M. J. (1992). Gene, 111, 229–233. [DOI] [PubMed]
- Putnam, N. H. et al. (2008). Nature (London), 453, 1064–1071.
- Saiki, R. K., Scharf, S., Faloona, F., Mullis, K. B., Horn, G. T., Erlich, H. A. & Arnheim, N. (1985). Science, 230, 1350–1354. [DOI] [PubMed]
- Shagin, D. A., Barsova, E. V., Yanushevich, Y. G., Fradkov, A. F., Lukyanov, K. A., Labas, Y. A., Semenova, T. N., Ugalde, J. A., Meyers, A., Nunez, J. M., Widder, E. A., Lukyanov, S. A. & Matz, M. V. (2004). Mol. Biol. Evol. 21, 841–850. [DOI] [PubMed]
- Shaner, N. C., Lambert, G. G., Chammas, A., Ni, Y., Cranfill, P. J., Baird, M. A., Sell, B. R., Allen, J. R., Day, R. N., Israelsson, M., Davidson, M. W. & Wang, J. (2013). Nature Methods, 10, 407–409. [DOI] [PMC free article] [PubMed]
- Shu, X., Leiderman, P., Gepshtein, R., Smith, N. R., Kallio, K., Huppert, D. & Remington, S. J. (2007). Protein Sci. 16, 2703–2710. [DOI] [PMC free article] [PubMed]
- Sniegowski, J. A., Lappe, J. W., Patel, H. N., Huffman, H. A. & Wachter, R. M. (2005). J. Biol. Chem. 280, 26248–26255. [DOI] [PubMed]
- Stewart, D. E., Sarkar, A. & Wampler, J. E. (1990). J. Mol. Biol. 214, 253–260. [DOI] [PubMed]
- Stoner-Ma, D., Jaye, A. A., Matousek, P., Towrie, M., Meech, S. R. & Tonge, P. J. (2005). J. Am. Chem. Soc. 127, 2864–2865. [DOI] [PubMed]
- Suto, K., Masuda, H., Takenaka, Y., Tsuji, F. I. & Mizuno, H. (2009). Genes Cells, 14, 727–737. [DOI] [PubMed]
- Tubbs, J. L., Tainer, J. A. & Getzoff, E. D. (2005). Biochemistry, 44, 9833–9840. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Wall, M. A., Socolich, M. & Ranganathan, R. (2000). Nature Struct. Biol. 7, 1133–1138. [DOI] [PubMed]
- Wallace, A. C., Laskowski, R. A. & Thornton, J. M. (1995). Protein Eng. 8, 127–134. [DOI] [PubMed]
- Wilmann, P. G., Battad, J., Petersen, J., Wilce, M. C. J., Dove, S., Devenish, R. J., Prescott, M. & Rossjohn, J. (2006). J. Mol. Biol. 359, 890–900. [DOI] [PubMed]
- Wilmann, P. G., Petersen, J., Pettikiriarachchi, A., Buckle, A. M., Smith, S. C., Olsen, S., Perugini, M. A., Devenish, R. J., Prescott, M. & Rossjohn, J. (2005). J. Mol. Biol. 349, 223–237. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Wood, T. I., Barondeau, D. P., Hitomi, C., Kassmann, C. J., Tainer, J. A. & Getzoff, E. D. (2005). Biochemistry, 44, 16211–16220. [DOI] [PubMed]
- Wu, B., Piatkevich, K. D., Lionnet, T., Singer, R. H. & Verkhusha, V. V. (2011). Curr. Opin. Cell Biol. 23, 310–317. [DOI] [PMC free article] [PubMed]
- Yarbrough, D., Wachter, R. M., Kallio, K., Matz, M. V. & Remington, S. J. (2001). Proc. Natl Acad. Sci. USA, 98, 462–467. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: laRFP, 4jf9
PDB reference: laRFP-ΔS83, 4jge
PDB reference: laRFPdam, 4jeo
PDB reference: laGFP, 4hvf